Abstract
The discovery of human induced pluripotent stem cells (hiPSCs) has provided an unprecedented opportunity to study tissue morphogenesis and organ development as “organogenesis-in-a-dish”. Current approaches of cardiac organoid engineering rely on either direct cardiac differentiation from embryoid bodies or generation of aligned cardiac tissues from pre-differentiated cardiomyocytes from monolayer hiPSCs. To experimentally model early cardiac organogenesis in vitro, our protocol combines biomaterials-based cell patterning with stem cell organoid engineering. Three-dimensional cardiac microchambers are created from two-dimensional hiPSC colonies, which approximates an early developing heart with distinct spatial organization and self-assembly. With proper training on photolithography microfabrication, maintenance of human pluripotent stem cells, and cardiac differentiation, a graduate student with guidance will likely be able to carry out this experimental protocol, which requires approximately three weeks. We envisage that this in vitro model of human early heart development could serve as an embryotoxicity screening assay in drug discovery, regulation, and prescription for healthy fetal development. Currently, this protocol has yet to be implemented to assess the cardiac microchamber formation from the hiPSC lines with inherited disease, which can potentially be used to study the disease mechanisms of cardiac malformations at early stage of embryogenesis.
Keywords: Stem Cell Organoids, Cell Patterning, Early Cardiac Organogenesis, Human Pluripotent Stem Cells, Biomaterials Fabrication
Editorial summary:
This protocol describes the generation early developing cardiac organoids from human pluripotent stem cells. Geometric confinement of the hiPSCs drives spatial organization of the cells from a 2D layer into 3D cardiac microchambers.
INTRODUCTION
Organogenesis is a dynamic and complex process that not only involves differentiation of stem cells to their final fate, but also organizes the differentiating cells into spatial defined tissue patterns. Traditional model organisms for studying organogenesis processes suffer from both species differences and inability to control in utero morphogenetic events for quantitative evaluation of the relationship between stem cell fate choice and three-dimensional (3D) tissue development1. Alternatively, biomaterials-based stem cell organoid technologies have demonstrated promising progress towards overcoming these challenges, especially in combination with rapid development of human induced pluripotent stem cells (hiPSCs)2–6. In this protocol, we describe a robust approach to model the characteristics of early human cardiac organogenesis using a poly (ethylene glycol) (PEG)-based surface patterning method7, which creates the necessary geometric confinement to drive the spatial organization of hiPSCs undergoing cardiac differentiation. We have demonstrated that our method can direct 2D hiPSC colonies to 3D cardiac microchambers, which can approximate an early developing heart with defined spatial distribution of different cell phenotypes within the individual patterns7. Thus, this protocol advances stem cell organoid technology by engineering long-term confinement conditions that allow precise control and manipulation of the biomechanics to induce changes in downstream signal transduction processes involved in organogenesis.
Development of the protocol
Organ development is often thought to be directed by surrounding biochemical signals and biophysical cues in utero. Conventional cardiac differentiation from hiPSCs and human embryonic stem cells (hESCs) were primarily conducted under 2D cell culture systems (6-well plates) without proper biophysical cues, thus morphogenetic events were not induced for modeling of early cardiac organogenesis. We initially studied the cardiac organization of embryoid bodies (EBs) derived from hiPSCs, and found that EBs would spontaneously organize, but with no clear structured cardiac microchamber7,8. The lack of biophysical control of these preliminary models did not permit reproducibility or consistency required for experimental analysis. We sought to take advantage of cell micropatterning because of its’ proven ability to regulate stem cell fate and direct embryonic spatial patterning of pluripotent stem cells to model embryogenesis9,10. The current protocol of a robust long-term cell patterning method supplies biophysical confinement throughout the duration of the cardiac differentiation procedure to generate beating 3D cardiac microchambers to model the early developing human heart.
Comparison with other methods, novelty and advantages
The 3D organoid technology is built upon fundamental concepts of developmental biology, which relies on dynamic multicellular self-organization to form functional biological tissues11. hiPSC or hESC derived organoids generated by simply supplying biochemical agents have been established for numerous organs, including the optic cup12, gut13,14, kidney15, brain16, which, to a degree, exhibit the typical tissue structure of the particular organ17. However, such organoids have manifested variability and heterogeneity in cell type specification, as well as the positioning of notable regions of the tissue. Brain organoids, for example, demonstrate that different regions of the brain can develop independently16. These regions are not reliably organized correctly in relation to one another, presenting challenges with respect to not only the accuracy, but also the consistency and reproducibility of the organoid models.
Particularly, stem cell-based cardiac organoid engineering approaches primarily rely on either direct cardiac differentiation from EBs18–20, or generation of aligned 3D cardiac tissues from pre-differentiated cardiomyocytes from monolayer hiPSCs21–25. EB-mediated cardiac differentiation suffers from variability of EB size, organoid heterogeneity, and poor spatial organization26. For example, in the biowire platform27, microfluidic-based devices28, and cantilever systems29, cardiomyocytes were first differentiated from hiPSCs/hESCs in conventional tissue culture plates and then seeded as single cells or tissue clusters into the fabricated devices to form adult-like aligned cardiac muscle23,30. The advantages of these engineered cardiac tissue models are not only the structural mimicry of adult heart tissue, but also their capability of electromechanical measurements and stimulations31,32. However, these approaches do not allow us to observe cell lineage specification and spatial self-organization during the cardiac development process, since these cells are seeded and reconstructed after differentiation.
Cell micropatterning methods have enabled precise control over the cellular microenvironment (e.g., shape and size of cell colonies) that regulates cellular morphology and functions33–35. Commercially micropatterned substrates (e.g. CYTOO chips etc.) are also available9, but don’t offer the same flexibility of designing and fabricating diverse arrays of geometrical features. The novelty of our protocol is that it combines organoid concepts with cell micropatterning approaches to (1) promote the self-organization of 3D tissue structures to mimic early heart and organ formation; (2) supply biophysical cues during cardiac differentiation of hiPSCs to facilitate cellular polarity and spatial organization of cardiac tissue patterns; and, (3) reduce tissue heterogeneity to create a systematic and experimentally reproducible in vitro cardiac organoid model.
Applications
The heart is one of the first organs to form in mammals, making it highly vulnerable to both intrinsic and extrinsic disease factors36,37. Congenital heart malformation is a prominent hereditary birth defect, which presents greater risks for heart diseases in adulthood7. Cardiac tissue morphogenesis during development has been difficult to model and investigate, because most cardiac tissue engineering techniques utilized pre-differentiated cardiomyocytes. Our engineered human cardiac microchambers represent a model system for studying human-specific heart formation at early developmental stages through the assessment of cardiac microchamber characteristics (height, volume, uniformity, etc). Additionally, we can characterize contractile abnormalities of cardiac microchambers, as well as profiling the distinct disease-specific gene expression of the cardiac diseases associated with early embryonic developmental defects. Using this concept and workflow, we envision this tissue model as a useful platform to study the progression of a diverse array of congenital cardiac defects, such as left ventricle non-compaction cardiomyopathy (LVNC), hypoplastic left heart syndrome (HLHS), Ogden Syndrome etc., with patient-specific hiPSCs and their genetically-corrected counterparts. Although some diseases we have listed are not lethal at these early stages, they can still cause abnormalities in cardiac physiology and formation at these early stages, and we propose that our model can predict based on the organoid formation and contractility analysis. With respect to translational potential, we anticipate that our model will enable study of adverse drug reactions at early cardiac developmental stages and that these drug reactions can be characterized similarly by assessing the microchamber formation, contractility and gene expression. Therefore, this model can be extended as a screening platform for drug candidates that may introduce cardiac related birth defects if administrated during pregnancy.
Limitations
As discussed, this protocol generates beating 3D representations of cardiac microchambers to offer greater significance in replicating in vivo cardiac organogenesis. There are, however, limitations with this approach, since the model is restricted to early stages of cardiac development7 that reflect cellular differentiation and spatial segregation of pluripotent cells to cardiac lineages. We suggest our cardiac microchambers resemble the early stages of heart development after fusion of the cardiac mesoderm into the primitive heart tube, during the onset of heart tube bulges, and before the occurrence looping events. Later events of cardiac development, such as ventricular and atrial differentiation, valve formation and vascularization cannot be replicated with current model. Additionally, other tissue layers, such as pericardium and endocardium, will not form in our cardiac microchambers, which will limit the further tissue maturation and development resulted from the crosstalk among different tissue types38,39. Furthermore, the geometrical constraints of our system are primarily 2D, since we can only fabricate our patterned substrate on the bottom of cell culture 6-well plate. Thus, after starting formation of 3D tissues, no constraint is provided to the microchambers above the substrate (~ 300 μm of total height of cardiac microchambers), resulting in only a partial 3D model. Our current system utilizes motion-tracking analysis to quantify the contraction velocity of the cardiac microchambers, which cannot be substituted for the contractile force measurement because it takes advantage of video-based analysis, as opposed to direct measurement of the force via mechanical sensors.
Experimental Design
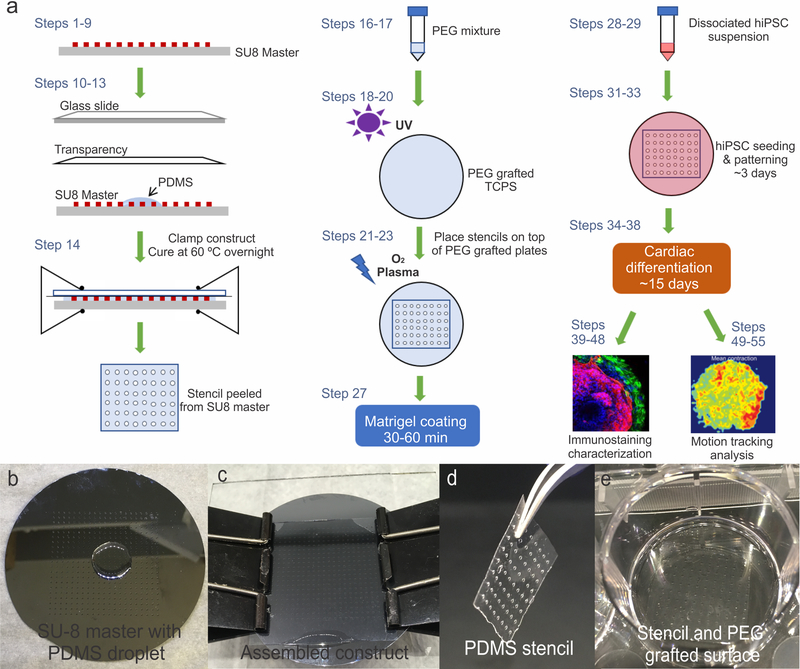
Micropatterned substrates are fabricated using standard lithography techniques and surface grafting of PEG. SU8 masters are first fabricated by spinning a thin layer of SU8 photoresist onto a silicon wafer. The photoresist is exposed to UV for crosslinking under a photomask, and unexposed regions can be dissolved in the developer solution, leaving behind specific patterns to imprint in polydimethylsiloxane (PDMS) and create a stencil. The PDMS stencils are used to expose specific patterns on a layer of PEG that are etched through oxygen plasma. hiPSC suspensions are then seeded into fabricated micropatterns and maintained in pluripotent stem cell culture media, such as mTeSR1 or Essential 8. When the cells are near confluent within their respective micropatterns, the cells are differentiated following a cardiac differentiation protocol that modulates the WNT/β-catenin pathway via CHIR9902 and IWP-4 treatment on specific days40. The formation of beating cardiac microchambers is observed around 15-day post CHIR treatment. The cardiac microchambers can be characterized through immunocytochemistry with cardiomyocyte markers (cardiac troponin T, sarcomeric α-actinin, and myosin heavy chain). The physiological contraction of the beating chambers can be further quantitatively characterized using open-source in-house developed software41. A schematic outlining the entire procedure is illustrated in Fig. 1a. The following procedure-specific information should be regarded prior to adapting the protocol depending on the specific experimental situation.
Figure 1. Schematic of procedure and process of PDMS stencil fabrication.
(a) Schematic outlining key steps of the entire procedure. (b) SU8 master with a small amount of PDMS prepolymer. (c) Completed assembly of the patterned SU8 master-PDMS-transparency-glass slide construct. (d) Thin film of PDMS removed from the assembly after curing and then placed on top of the optically clear PEG grafted surface (e).
Cell type (Step 27)
The expected results shown in this protocol were obtained using a WTC hiPSC line (available at Coriell Institute NIGMS Human Genetic Cell Repository) donated from Bruce R. Conklin at the Gladstone Institute. H9 hESCs (available at WiCell Research Institute, WA09) have also been successfully patterned and differentiated into cardiac microchambers using this procedure. However, our previous experiments using a particular line of H9 cells resulted in smaller microchambers that were approximately half the height of the microchambers derived from WTC hiPSCs. Thus, different cell lines can exhibit variations, but this protocol is potentially acceptable for the self-organization and cardiac morphology of multiple robust hPSC lines.
Culture conditions (Step 27)
This protocol originally made use of hiPSCs cultured in mTeSR1 media. However, high quality and appropriately conditioned media, such as Essential 8, has also been tested for supporting the formation of cardiac microchambers, so long as the growth and expansion of hiPSCs are favorable. After thawing from cryopreservation, hiPSCs should be grown and expanded for at least three passages using standard pluripotent stem cell culture procedures before beginning the protocol in order to establish a stable culture. The choice of coating matrix also must be considered. We have had success with hESC qualified Matrigel™, Geltrex™ basement membrane matrix, and Synthemax™ surface coating solutions. We found that some coatings, such as vitronectin and fibronectin, support the expansion of hiPSCs on the micropatterns, but have limitations in the 3D formation of the microchambers.
Seeding density (Steps 31–33)
It’s imperative the cells are grown to confluence before performing cardiac differentiation. The hiPSC seeding densities described in this protocol were optimized to seed 400 μm and 600 μm circle patterns, as well as triangle and square patterns with the same seeding area as the 400 μm circles. The densities were optimized by determining the seeding densities that could reach confluency in 3 days. This is to ensure that the initial CHIR treatment is applied on uniform cell/surface conditions. Unsuitable seeding densities can result in poor cardiomyocyte differentiation efficiency, as well as irregular differentiation patterns that don’t resemble cardiac microchambers. Depending on the desired results and the sizes of the fabricated patterns, optimizing the seeding density is typically required.
Fabrication of micropatterned PEG substrate (Steps 1–23)
This protocol uses predesigned photomasks to fabricate SU8 patterns on silicon wafers. The photomasks are routinely designed using AutoCAD software and printed on transparencies by CAD/Art Services, Inc. The photolithography process for SU8 master microfabrication is completed in a cleanroom facility and it’s advised to consult with a photolithography expert. If there’s limited access to a photolithography facility, arrange for a microfabrication technician to prepare a mask from appropriate files containing the designs. The predesigned photomasks used for this protocol consist of 400 μm and 600 μm diameter circles, as well as triangles and squares with the same geometric area as the 400 μm circles. The PDMS stencils with clear-through holes are casted from SU8 master. The PEG non-fouling thin polymer film was grafted onto standard tissue culture polystyrene via UV photo-polymerization, masked with PDMS stencils, and patterned via oxygen plasma etching42. The PEG substrate is sterilized with 70% alcohol and coated with Matrigel™ hESC qualified matrix for hiPSCs attachment. With this method, colonies of hiPSCs can be organized into arrays of patterns with different shapes and sizes.
Cardiac differentiation (Steps 34–38)
The cardiomyocyte differentiation protocol described here uses small molecule induction to modulate the WNT/β-catenin pathway40. Our preliminary experiments showed that cardiac differentiation only succeeded on 400- and 600- μm circle patterns, and on triangle patterns with the same surface areas. Hence, successful cardiac tissue formation is dependent on the pattern size. We don’t exclude the possibilities that other differentiation protocols may be also suitable for generation of cardiac microchambers; however, the differentiation protocol based on growth factors (Activin A and BMP4)43 has been tested, but failed to consistently generate uniform cardiac microchambers. Differentiation protocols with chemically-defined medium44 (the basal medium RPMI 1640, L-ascorbic acid 2-phosphate and rice-derived recombinant human albumin) or albumin-free medium45 has not been tested on our patterned substrate for generation of cardiac microchambers.
Microchamber characterization (Steps 39–47)
The cardiac markers to stain for in this protocol include cardiac troponin, sarcomeric α-actinin, myosin heavy chain. Connective cell type is characterized by myofibroblast/smooth muscle cell markers include SM22, calponin, and smooth muscle actin. The options of cardiomyocyte and/or myofibroblast markers are not limited to the ones listed here, and various other markers can be tagged to characterize the microchambers.
Motion tracking analysis (Steps 48–50)
We measure and quantify the beating exhibited by hiPSC derived cardiac tissue to assess the overall physiology and phenotype of the microchambers. Motion tracking of cardiac contraction used in this protocol is based on an open-source in-house developed software capable of generating contraction waveforms, motion heat maps, and various data regarding contraction rate and velocities41. The software used in this protocol is noninvasive, such that it only requires video recordings of the tissue within the culture environment using video microscopy. Thus, no external stimulation or measurement tools are necessary and the tissue remains undisturbed in the process. Other commercially available or open source programs46,47 capable of measuring cardiac contraction and outputting the waveform would also be suitable for this application.
MATERIALS
Reagents
Photomasks (CAD/Art Services Inc. Bandon, OR, Website: http://www.outputcity.com/)
Photoresist (Microchem, SU8–50) CAUTION! Handling of SU8 photoresist should be carried out with adequate ventilation in a laminar flow chemical hood and cleanroom. SU8 photoresist is toxic and can cause skin and eye irritation.
SU8 developer solution (Microchem, contact supplier for purchasing information) CAUTION! Handling of SU8 developer should be carried out with adequate ventilation in a laminar flow chemical hood and cleanroom. SU8 developer solution is volatile and can cause respiratory irritation.
Isopropyl alcohol (IPA) (Sigma Aldrich, cat. no. W292907)
Poly(ethylene glycol) (PEG) 1000 (Polysciences, cat. no. 16666)
PEG (400) diacrylate (Polysciences, cat. no. 01871)
2-Hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959) (Sigma Aldrich, cat. no. 410896) Protect from light
(heptadecafluoro-1,1,2,2-tetrahydrodecyl)trimethoxysilane (Gelest, cat. no. SIH5841.5)
Silicon wafers (University Wafers, cat. no. 447: 3” P(100) 0–100 ohm-cm SSP 406–480um Test Grade)
Color laserjet transparency (e.g. HP, 8.5”X11”)
Poly(dimethylsiloxane) (PDMS) (Dow Corning, Sylgard 184 silicone elastomer kit)
WTC hiPSCs (from Bruce R. Conklins’ lab, Gladstone Institute of Cardiovascular Disease, also available at Coriell Institute NIGMS Human Genetic Cell Repository, GM25256) CRITICAL hiPSC handling requires minimum Biosafety level 2 safety precautions. CAUTION! The cell lines used in the research should be regularly checked to ensure that they are authentic and not infected with mycoplasma. All work using human induced pluripotent stem cell lines must be performed following institutional regulatory board guidelines enforced by the users’ institution. Our experiments were approved to conform to the requirements established by the OSHA Bloodborne Pathogen Standard and Syracuse University Microbiological Safety Program.
Essential 8™ (E8) medium (Life Technologies, cat. no. A1517001)
mTeSR™1 (StemCell Technologies, cat. no. 05850)
StemPro Accutase (Life Technologies, cat. no. A1110501)
Y-27632 (EZSolution) (BioVision, cat. no. 1784–5)
Dulbeccos phosphate buffered saline (DPBS) (Corning, cat.no. 21–031-CV)
Matrigel™ hESC qualified matrix (Corning, cat. no. 354277)
Knockout™ DMEM (Life Technologies, cat. no. 10829018)
RPMI 1640 media (Corning, cat. no. 10–040-CV)
B27 Supplement minus insulin (50X) (Life Technologies, cat. no. A1895601)
B27 Supplement (50X) (Life Technologies, cat. no. 17504044)
CHIR99021 GSK-3 inhibitor (Stemgent, cat. no. 04–0004)
IWP4 Wnt inhibitor (Stemgent, cat. no. 04–0036)
Trypan blue 0.4% (Bio-Rad, cat. no. 1450021)
Triton-X 100 (Sigma Aldrich, cat. no. T8787)
Bovine serum albumin (BSA) (Sigma Aldrich, cat. no. A8022)
Goat serum (Thermo Fisher Scientific, cat. no. 50062Z)
16% Formaldehyde solution (Thermo Fisher Scientific, cat. no. 28908) CAUTION! Formaldehyde is combustible and harmful if swallowed. Causes skin and eye irritation. Handle under laminar flow hood.
DAPI (Invitrogen, cat. no. R37606)
Dimethyl sulfoxide (sterile) (Sigma Aldrich, cat. no. D2650–100ML)
Antibodies for immunofluorescence staining. Refer to Table 1
Table 1:
Antibodies used in this protocol
| Antibodies | Vendor | Cat. No. | Dilution |
|---|---|---|---|
| Primary | |||
| Cardiac troponin T (mouse) | Thermo Fisher Scientific | MS295P | 1:200 |
| Sarcomeric α-actinin (mouse) | Sigma Aldrich | A7811 | 1:300 |
| Myosin heavy chain (mouse) | Abcam | ab97715 | 1:200 |
| SM22 (rabbit) | Abcam | ab14106 | 1:300 |
| Calponin (rabbit) | Abcam | ab46794 | 1:200 |
| Smooth muscle actin (rabbit) | Abcam | ab5694 | 1:100 |
| Secondary | |||
| Alexa Fluor 488 Goat anti-mouse IgG | Thermo Fisher Scientific | A-11029 | 1:200 |
| Alexa Fluor 546 Goat anti-rabbit IgG | Thermo Fisher Scientific | A-11010 | 1:200 |
Equipment
Oxygen plasma treater (e.g. Plasma-Preen II 973 Plasmatic Systems Inc).
Dymax UV Illuminator (2000EC Modular with light shield and manual shutter, Dymax, cat. no. 39723)
Spin caster (Laurell Technologies, cat. no. WS-650Mz-23NPPB)
Laboratory oven (e.g., Fisher Scientific Isotemp oven, Fisher Scientific, cat. no. 13246516GAQ)
Oxygen cylinder (Airgas, UN1072)
Hot plate with temperature display (e.g. Fisher Scientific Isotemp hotplates)
Nitrogen spray gun (NCI Innotech; cat. no. TA-N2–1000)
Vacuum desiccator (e.g. Bel-Art, Fisher Scientific, cat. no. 08 594 15A)
Cell culture biological safety cabinet (e.g. Thermo Fisher Scientific 1300 Series Class II A2, cat. no. 1323TS)
Cell culture CO2 incubator (e.g. VWR Air Jacketed CO2 incubators, VWR, cat. no. 10810–902)
Cell culture centrifuge (e.g. Sorvall Legend X1R, Thermo Fisher Scientific cat. no. 75004261)
Inverted fluorescence microscope with motorized stage (e.g. Nikon Eclipse TS100F)
EMD Millipore Steriflip filter units (Fisher Scientific, cat. no. SCGP00525)
Corning tissue culture 6 well plates (Fisher Scientific, cat. no. 07 200 83)
Hemacytometer (Hausser phase contrast) (Fisher Scientific, cat. no. 02 671 54)
Fisherbrand pipet controller (Fisher Scientific, cat. no. 14 955 202)
Pipetters (Fisherbrand elite pipette kit) (Fisher Scientific, cat. no. 14 388 100)
5 mL serological pipettes (Fisher Scientific, cat. no. 14 955 233)
10 mL serological pipettes (Fisher Scientific, cat. no. 14 955 234)
25 mL serological pipettes (Fisher Scientific, cat. no. 14 955 235)
20 μL barrier pipette tips (Fisher Scientific, cat. no. 02 707 432)
200 μL barrier pipette tips (Fisher Scientific, cat. no. 02 707 430)
1000 μL barrier pipette tips (Fisher Scientific, cat. no. 02 707 404)
Microcentrifuge tubes, 1.5 mL (Fisher Scientific, cat. no. 05 408 130)
Large binder clips, 2” with 1” capacity (Staples, OfficeMax etc.)
Fine tipped tweezers (Fisher Scientific, cat. no. 12 000 131)
ImageJ (https://imagej.nih.gov/ij/)
Matlab version R2016a (or latest) (MathWorks)
Motion tracking software (https://gladstone.org/46749d811)
Reagent Setup
Essential 8 (E8): Prepare the biosafety cabinet by wiping down the working area with 70% EtOH. Lightly spray materials and reagents with 70% EtOH and transfer into the biosafety cabinet (BSC). Use sterile cell culture materials, including pipets, tips, and cultureware to minimize risks of mycoplasma and bacterial contamination. These procedures will be further referred to as “aseptic”. Using a sterile pipet, add the entire contents of E8 Supplement into the 500 mL bottle of E8 basal medium. The medium can be stored for up to 2 weeks in 4 °C, or aliquoted into 40mL working volumes and frozen at −20 °C for up to 6 months.
mTeSR1: Using a sterile pipet in the BSC, add the entire contents of mTeSR1 Supplement into the 500 mL bottle of mTeSR1 basal medium. The medium can be stored for up to 2 weeks in 4 °C, or aliquoted into 40 mL working volumes and frozen at −20 °C for up to 6 months.
Diluted Matrigel™ coating: Thaw the bottle of Matrigel™ on ice and position the bottle to prevent the mouth from coming in contact with ice. Pre-chill 15 mL conical tubes and 200 μL pipette tips at −20 °C. Place items in the BSC. Take a sterile tip and aliquot working volumes of 150 μL of thawed Matrigel™ into each tube. Store aliquots at −20 °C until expiration date stated on the bottle. When needed (Step 27), thaw on ice and add 12mL cold Knockout DMEM and mix by pipetting up and down to make the coating solution.
RPMI/B27-I medium: In the BSC, take a sterile pipet and add the contents of B27 Supplement (10 mL) minus insulin into a 500 mL bottle of RPMI 1640. Aliquot into 40 mL working volumes and store at −20 °C for up to 6 months or at 4 ˚C for up to 4 weeks.
RPMI/B27+C medium: In the BSC, take a sterile pipet and add the contents of B27 Supplement (10 mL) into a 500 mL bottle of RPMI 1640. Aliquot into 40 mL working volumes and store at −20 °C for up to 6 months or at 4 ˚C for up to 4 weeks.
10μM GSK3 inhibitor (CHIR99021): In the BSC, dilute the contents of the bottle in sterile DMSO. Calculate and aliquot working volumes of the small molecules and store at −20 ˚C for up to 6 months. The working volumes of small molecules are dependent on the volume of RPMI/B27-I aliquots to dilute the CHIR concentration to 10μM. Add the CHIR aliquot to the working volume of RMPI/B27-I. CRITICAL The volume of DMSO to add is specified by the supplier to make a 10mM concentration.
5μM Wnt inhibitor (IWP4): In the BSC, dilute the contents of the bottle in sterile DMSO.. Calculate the aliquot volumes accordingly depending on media volumes to dilute the IWP4 to a final concentration of 5 μM. Aliquots can be stored in −20 ˚C for up to 6 months. CRITICAL The volume of DMSO to add is specified by the supplier to make a 2.5 mM concentration.
4% PFA (vol/vol): Break the ampule and dilute 10 mL of formaldehyde solution per 30 mL DPBS. Store at 4 ˚C for up to 3 months. CAUTION! PFA is a carcinogen and causes eye, skin, and respiratory irritation and should be handled under adequate ventilation.
Blocking solution: Prepare 40 mL the blocking solution in DPBS composed with 2% (wt/vol) bovine serum albumin (BSA), and 4% (vol/vol) goat serum in DPBS. Store at 4 ˚C for up to 6 months.
Primary antibody solution: Refer to Table 1 and dilute primary antibodies accordingly in blocking solution. Prepare antibody solutions fresh as needed.
Secondary antibody solution: Refer to Table 1 and dilute secondary antibodies accordingly in blocking solution. Prepare antibody solutions fresh when needed.
DAPI: Dilute DAPI stock in DPBS to a final concentration of 300 nM. DAPI stocks can be stored at 4 ˚C for 12 months.
Equipment Setup
Set tissue/cell culture incubators to 37 °C, 5% CO2 and ensure humidity pans are filled to the vendor recommended levels.
Ensure that biosafety cabinets are functional and up to date on quality checks
Laboratory ovens should be pre-set to 60 °C
Set the gas regulators for the oxygen plasma treater at 10–20 psi.
PROCEDURE
SU8 master fabrication TIMING – 1 day
-
1
Turn on the vacuum for the spin caster. Place the wafer on the spinner and make sure that the wafer is secured by the vacuum and stays in place during the spin casting.
-
2
Slowly pour approximately 2 mL of SU8 50 onto the center of the wafer and begin spinning at 500 rpm at a 100 rpm/second acceleration. Hold at this speed for 10 seconds until the SU8 covers the entire wafer. Once the wafer is covered, increase the speed to 2000 rpm at an acceleration of 300 rpm/second and hold for 30 seconds to achieve a layer of photoresist of ~50 μm in height.
-
3
Cover a hot plate with aluminum foil and smoothen the surface for even heating. Heat the plate to 65 °C and place the wafer on top of the foil. Prebake the wafer for 10 min at 65 °C, ramp the hot plate to 95 °C with wafer on top, and then soft-bake for 25 min at 95 °C. Turn off the hot plate and leave the wafer on the hot plate till cool to room temperature (22 °C) before transferring to UV exposure.
-
4
Turn on the Dymax UV illuminator (100 mW/cm2) with the shutter closed and allow it to warm up for about 5 min. Place the photomask on the wafer, on top of the SU8, with the ink side of the design touching the SU8. Place a UV penetrable glass on the photomask and place everything in the Dymax 32 cm below the lamp source. Close the door to the Dymax and open the shutter to expose the SU8 for 3 seconds.
-
5
After UV exposure, remove the glass and the photomask and post-exposure bake the wafer again on the 65 °C hot plate for 1 min. Then ramp the hot plate to 95 °C with wafer on top, and then bake at 95 °C for 8 min. Turn off the hot plate and leave the wafer on the hot plate until cooled to room temperature (estimated 30–60 minutes).
-
6
Place the cooled wafer into a petri dish and transfer to a sterile biosafety cabinet. In a 1L beaker, pour a sufficient amount of developer to cover the wafer and place the wafer in the developer. Cover the beaker with several layers of parafilm and allow the developer to strip the uncured SU8 for 10 min.
-
7
Remove the wafer from the developer and rinse with IPA (10 mL). Then dry completely with a nitrogen gun. Hard bake the wafers at 150 °C overnight in a convection oven.
PAUSE POINT Fully cured and developed wafers can be stored away in wafer boxes at room temperature indefinitely until use. Note that repeated use of the wafers for stencil fabrication can lead to wear of the SU8 patterns. To extend the usage of wafers, be gentle and careful, and not directly touch the pattern features during the PDMS stencil fabrication process.
-
8
Prior to casting PDMS (Steps 10 – 14), treat the SU8 master with oxygen plasma (200W) for 1 min to increase the hydrophilicity of SU8 master.
CAUTION! After plasma treatment, be certain the vacuum chamber is at atmospheric pressure before attempting to open the door of the plasma-preen unit and lift the glass vacuum chamber.
-
9
Prepare 1 mL (heptadecafluoro-1,1,2,2-tetrahydrodecyl)trimethoxysilane in a 2.0 mL microcentrifuge tube. Place tube and SU8 master in the desiccator. Turn on the vacuum and vacuum deposit the silane overnight to coat the surface of SU8 master. This step ensures easy removal of the PDMS stencils from the SU8 master.
PDMS stencil fabrication TIMING – 2 hours to overnight
-
10
To make the stencils, weigh out PDMS pre-polymer and curing agent at a 10:1 ratio in a weighing boat and mix vigorously in the boat. 2.5g pre-polymer with 0.25g curing agent is enough to make at least 4 stencils. Enclose the mixture inside a vacuum desiccator and open the vacuum valve to degas the mixture for at least 30 min until all bubbles have disappeared.
-
11
Using a 1mL pipette, drop a small amount (about 0.5–1 mL) of the degassed PDMS mixture onto the pretreated SU8 wafer and allow the polymer mixture to spread (Fig. 1b). CRITICAL STEP The amount of PDMS to add can vary, but be sure to not add too much such that the PDMS overfills the patterns.
-
12
Place small square pieces of transparency film on top of the wafer and covering the patterns. The transparency squares should be big enough to cover the patterns.
-
13
Place two glass slides (25mm × 75mm) side by side on top of the transparency and apply pressure so that the PDMS spreads around the patterns, not above, to retain the clear-through holes after curing. CRITICAL STEP Do not apply pressure directly to the transparency (without the glass). It will cause bubbles to form as some parts of the transparency lose and regain contact with the PDMS. CRITICAL STEP Ensure that no polymer mixture is present on top of the SU8 patterns. Otherwise, the holes required for the plasma treatment step will not form.
-
14
Take two large binder clips and clamp down the entire construct consisting of SU8 master-PDMS-transparency-glass (Fig. 1c) to secure all the components in place. Place the construct in a 60 °C oven overnight to cure the PDMS. Afterwards, carefully disassemble the construct and use thin-tipped tweezers to gently remove the stencil (Fig. 1d), as it will be very fragile. Cut the stencils to the size of the wells (6-wells plate) so that they fit appropriately.
PAUSE POINT The stencils can be stored at room temperature in a 50mL tube containing 70% (vol/vol) ethanol (EtOH) until use. They can be stored in this manner indefinitely. Note that PDMS stencils can wear and degrade after repeated exposure to oxygen plasma treatment.
? TROUBLESHOOTING
PEG surface grafting TIMING ~1 hour
-
15
Pretreat two 6-well plates with oxygen plasma (200W) by placing the open plates into a plasma treater and begin pulling the vacuum. Treat the plates for 5 minutes. CAUTION! After plasma treatment, be certain the vacuum chamber is at atmospheric pressure before attempting to open the door of the plasma-preen unit and lift the glass vacuum chamber.
? TROUBLESHOOTING
-
16
Prepare the PEG polymer mixture in a 50 mL tube by combining 0.15g of PEG 1000, 1.8 mL PEG diacrylate (PEGda), 14.55 mL, isopropyl alcohol (IPA), and 0.45 mL MilliQ water. Vortex the mixture for at least 3 minutes to mix evenly and make sure there are no PEG clumps. This will make 16.8 mL of PEG mixture, which is enough to graft up to two 6-well plates.
-
17
Weigh out 40 mg of Irgacure 2959 in the dark and add it to the 50mL conical tube containing the polymer mixture. Cover the tube with foil to protect from light and vortex the mixture for about 3 minutes until thoroughly mixed.
-
18
Take the 1000 μL pipette and transfer 760 μL of the polymer mixture containing Irgacure into each well (up to 12 wells). Make sure to pipet from the center of the wells, not the sides. After filling the wells, cover and wrap the plates in foil and set aside for 10 minutes at RT.
-
19
While waiting for the PEG to settle within the wells, turn on the Dymax UV Illuminator (shutter closed) and allow it to warm-up for about 10 minutes. CRITICAL STEP If the Dymax is turned on too early, the box can get too hot and result in variability. Leave the door open during warm-up and between exposures to prevent the inside temperature from getting too hot.
-
20
Place the PEG coated 6-wells plates, with the lid on, in the center of the platform under the UV (shutter open) for 45 seconds. When the plates finish curing, wash the plates intensively by repeatedly submerging in, or flushing with, MilliQ water to remove any residual polymer. Wash until all chunks have detached and the wells appear optically clear.
Etching using oxygen plasma TIMING ~2 hours
-
21
Using tweezers, gently remove the stencils (from Step 14) from the 70% EtOH. Lightly spray the PEG-grafted wells and dip the stencils into 70% EtOH (or IPA) and gently lay the stencils on top of the PEG layer (Fig. 1e, Fig 2a). The thin covering of alcohol serves as a lubricator to help stencil evenly distribute on top of the PEG layer. CRITICAL STEP Do not simply drop the stencils because this will create bubbles underneath when the alcohol is evaporated.
-
22
Place the plate in a 60 °C oven for two hours to fully evaporate the IPA or EtOH. CRITICAL STEP Make sure that all moisture is evaporated and ensure that the samples are completely dry before plasma treatment. Any residual moisture will inhibit plasma treatment and the PEG will not be etched.
-
23
Treat the plates with oxygen plasma (200 W) for two cycles of 3-min treatment with a 3-min break in between. The plasma treatment will etch away the PEG layer at specific areas, where the PDMS masks have open features. CAUTION! After plasma treatment, be certain the vacuum chamber is at atmospheric pressure before attempting to open the door of the plasma-preen unit and lift the glass vacuum chamber. CAUTION! The glass vacuum chamber can become HOT when operating for long duty cycle. Take proper precautions when handling the hot glass, especially under vacuum.
? TROUBLESHOOTING
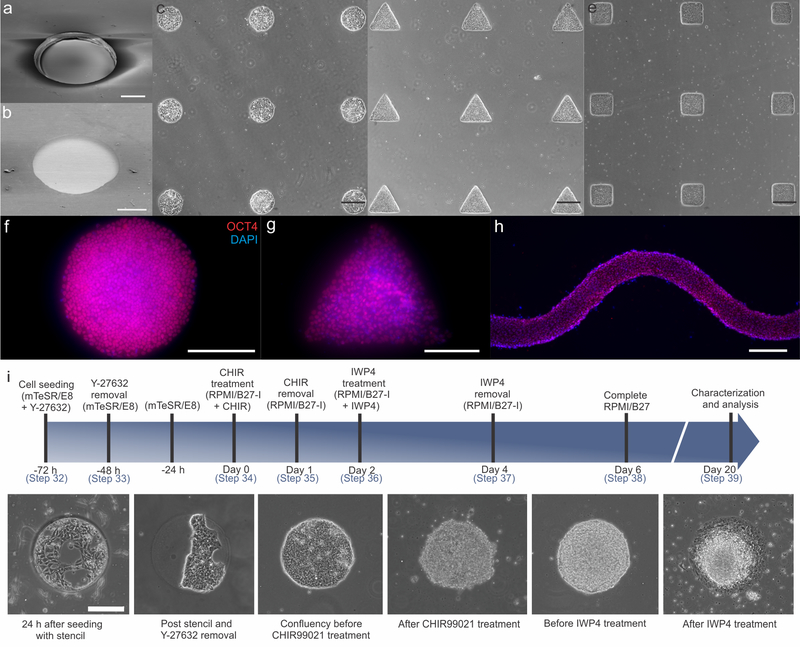
Figure 2. Resulting patterns and seeded cells after oxygen plasma etching.
SEM images of (a) PDMS stencil with clear-through holes (Steps 8–12) and (b) etched PEG surface (Steps 13–20) with circular patterns. (c-e) hiPSCs patterned into an array of circles, triangles and squares, respectively. (f-h) Patterned hiPSCs still maintain pluripotency as indicated by OCT4 expression. (i) Timeline of the differentiation procedure and cell morphologies at key stages of differentiation. Scale bars: 200 μm (a, b, f-i); 400 μm (c-e).
Patterning of hiPSCs TIMING ~2–3 days
-
24
Transfer the newly etched PEG-grafted (Fig. 2b) plates into a tissue culture biosafety cabinet, and add > 3 mL of 70% ethanol to each well for 30 min to sterilize the patterns for cell seeding.
-
25
Aspirate the ethanol and wash the patterned wells by adding and removing 2 mL DBPS three times.
-
26
Grow and expand the hiPSCs for at least three passages in multiple wells within a standard 6-well culturing plate using standard aseptic technique and pluripotent stem cell culture procedure. For seeding, use cells that have expanded to 70–80% confluency (see Experimental Design for details on cells).
-
27
Using tips that have been pre-chilled at −20 °C, add 1 mL of diluted Matrigel coating solution (Corning hESC-qualified) to each patterned well and place the plates in the cell culture incubator (37 °C, 5% CO2 and humidity) for 30 min prior the hiPSC seeding, to allow the Matrigel to form a gel.
-
28
Dissociate confluent hiPSC wells with 1mL Accutase (Invitrogen) per well for 5 min at 37 °C. Quench the Accutase with 1 mL of Essential 8 (E8) or mTeSR1 medium and transfer the cell suspensions into a 15 mL conical tube. Centrifuge the cell suspension at 54 g for two minutes at 22 °C. Be sure to balance the centrifuge.
-
29
Aspirate the supernatant and resuspend the cells in 1 mL of media. Cells can remain in suspension at RT while counting. Take 10 μL of the cell suspension and add to a 1.5 mL microcentrifuge tube. To the tube, add 10 μL Trypan Blue dye and mix well by pipetting up and down 3–5 times (1:1 dilution).
-
30
Load 10μL of the cell/Trypan Blue mixture into a hemocytometer and count the cells following the manufacturer’s instructions. For 400 μm and 600 μm circle patterns, the seeding density has been optimized to 0.8E6 cells/mL and 1.2E6 cells/mL, respectively. CRITICAL STEP If the patterns are different sizes, be sure to optimize the seeding density prior, such that the cells take three days to reach confluence in the different sized patterns.
-
31
Dilute the suspension in E8/mTeSR1 to bring the concentration of cells to the optimized seeding densities. To this, add enough Y-27632 to bring its’ final concentration 10 μM within the entire volume of cell suspension.
-
32
Aspirate excess Matrigel coating solution from the plates in Step 28. Seed the coated PEG-grafted wells by adding 1 mL of the final cell suspensions in E8/mTeSR1 medium containing Y-27632 (from Step 32) to each well.
-
33
Twenty-four hours after seeding, using sterilized sharp tweezers, carefully remove the PDMS masks from the PEG-patterned substrate. Aseptically aspirate the E8/mTeSR1 containing Y-27632 to remove unattached cells and add fresh E8/mTeSR1 without Y-27632. Aspirate and change the E8/mTeSR1 (without Y-27632) daily until the patterns reach confluency (about 2 additional days) to begin cardiac differentiation. Confluent patterns are depicted in Figure 2i.
? TROUBLESHOOTING
Cardiac differentiation TIMING ~15–20 days
CRITICAL Figure 2i contains a timeline and graphical representation Steps 35 – 39
-
34
Once the hiPSCs have reached confluency (Day 0), change the media to RPMI 1640 medium containing B27 supplement minus insulin (RPMI/B27 -I) and 10 μM CHIR and incubate at 37 °C and 5% CO2 for 24h. Washing plates between this and the following media changes is not required
-
35
On Day 1, change the media to RPMI/B27-I (no small molecule) and incubate (37 °C and 5% CO2) for 24 h.
-
36
On Day 2, change the media to RPMI/B27-I containing 5 μM of IWP4 and incubate at 37 °C and 5% CO2 for 48h.
-
37
On Day 4, change the media back to RPMI/B27-I (no small molecule) for 2 additional days.
-
38
On Day 6, change the media to RPMI 1640 containing B27 complete supplement (RPMI/B27+C). Repeat the media change with RPMI/B27+C the following day (Day 7). From here on, change the media every two days until Day 20. The formation of 3D structures should start from Day 1 after CHIR treatment, and beating cardiac microchambers should be visible around Day 10.
? TROUBLESHOOTING
Immunofluorescence staining and imaging of cardiac microchambers TIMING 1 day
-
39
On Day 20, aspirate the media and wash the patterns once with 2 mL of DPBS.
-
40
Fix the cells by adding 2 mL of 4% (vol/vol) PFA per well for 20 min at RT. Discard the PFA as chemical waste and wash the patterns 3X with 2 mL DPBS.
-
41
Permeabilize the cells with 0.1% TritonX-100 (vol/vol) dissolved in PBS for 5 min, then remove TritonX-100 and wash 3 times with 2mL DPBS. Add 2 mL of blocking solution and set aside for 30 min at RT.
-
42
Discard the blocking solution and add 1 mL of primary antibody solution to each pattern (see Table 1 for details on antibody solutions). Incubate for at least 2 h at room temperature.
PAUSE POINT The primary antibody incubation can be left overnight at 4 °C.
-
43
Remove the primary antibody solution and wash the patterns with 1 mL DPBS three times.
-
44
Add 1 mL of the secondary antibody solution to each pattern and incubate for 2–3 h at room temperature in the dark by covering with foil or placing in a drawer
-
45
Remove the secondary antibody solution and wash the patterns with 1 mL DPBS three times.
-
46
Stain nuclei with the addition 1mL of 300 nM DAPI solution and incubate at room temperature for 5 min. Remove the DAPI solution and wash 3X with DPBS.
-
47
Image the patterns using fluorescence microscopy. Use confocal microscopy (Zeiss LSM710 upright confocal microscope with a 40X/NA1.0 water-dipping Plan-Apochromat Objective) to image the microchamber in 3D (XY Resolution 0.346 um/pixel, Z Resolution voxel depth: 6 um) The Z-stack images of cardiac microchamber are reconstructed into 3D in ImageJ to analyze the height and width of the microchambers (ImageJ → Image → Stacks → Orthogonal View/Z projection).
-
48
Using ImageJ, measure the maximum height of the microchamber from the Z orthogonal view. At half of the maximum height, measure the right angle distance to the edge of the microchamber. This distance is the half-width at the half-maximum (HWHM) height of the microchamber. (Refer to Figure 3a for illustration of HWHM measurement).
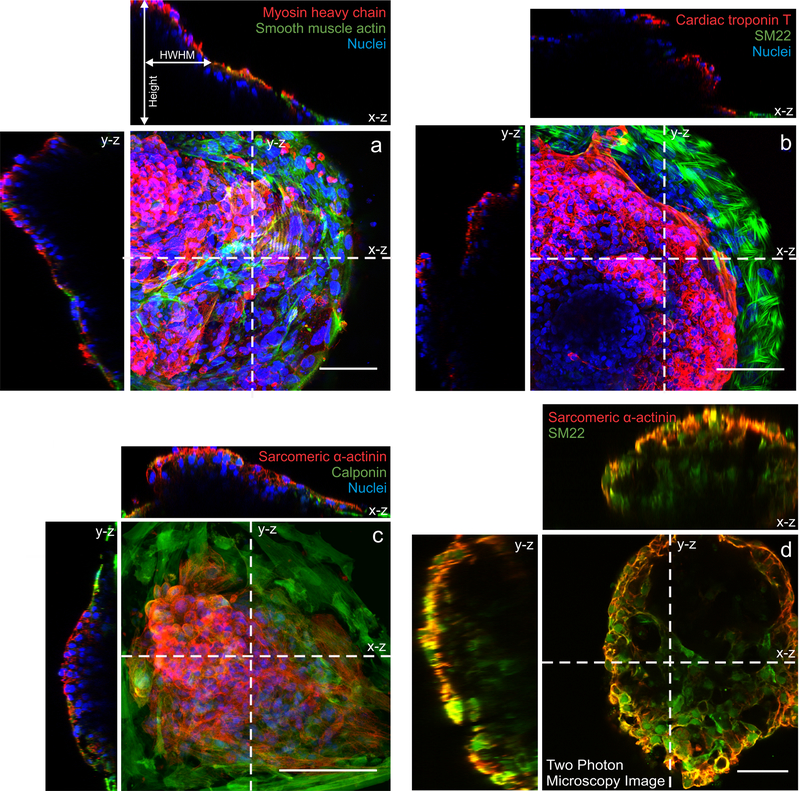
Figure 3. 3D reconstructed images of cardiac microchambers.
Confocal microscopy images showing contracting cardiomyocytes at the center (red) and myofibroblasts (green) along the perimeter characterized by (a) myosin heavy chain and smooth muscle actin, (b) cardiac troponin T and SM22, and (c) sarcomeric α-actinin and calponin. (d) Two-photon microscopy image taken at a single plane of the cardiac microchamber showing a void chamber surrounded by cells. All scale bars 100 μm.
Contractile motion tracking analysis TIMING 1 hour
CRITICAL Motion tracking analysis must be performed using live cells still in culture.
-
49
Using phase contrast microscopy at 10X magnification for video recording, focus on a microchamber and record the cardiomyocytes beating at a minimum of 30 frames per second for the desired number of frames (300–600 frames for a 10–20 second video).
-
50
Export the video as single-frame image sequences.
-
51
Download the Motion Tracking zip folder containing the code required to run the software. Using MATLAB, open the MotionGUI Matlab Code file and load the folder containing the image sequence.
-
52
Following the prompts displayed on the software user-interface, set experiment specific parameters, such the frame rate that was used to acquire the video.
-
53
Once the experiment parameters are set, click on “Get Motion Vectors” to begin the analysis
-
54
When analysis has finished, click “Save” to save the .MAT data file containing information from the analysis. PAUSE POINT Analysis can be directly continued, or this file can be loaded using the LOAD prompt on the user interface and analyzed at a later time.
-
55
Click on “Get Contraction Data” and then click on “Evaluate Data.” A pop-up screen will appear containing the contraction motion waveform and options to further export or analyze the data. Additional parameters, such as beat rate, average/max velocities, x/y velocities etc., can also be analyzed through the software.
? TROUBLESHOOTING
TROUBLESHOOTING
Refer to Table 2 for troubleshooting:
Table 2:
Troubleshooting guidelines
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 14 | No clear-through hole in stencil; incomplete holes | SU8 layer may be uneven; Uneven pressure applied to the wafer-PDMS-transparency-glass construct | Ensure that the wafer is level on the spin caster. Apply even pressure on the transparency/glass to uniformly spread the PDMS |
| 15, 23 | Tissue culture plates melt | Power setting on plasma treater is too high | Lower the power setting and optimize the power level that maintains etching capabilities as well as prevents melting of the plates |
| 33 | hiPSC attachment is poor | PEG layer wasn’t completely etched. Possibly due to too much time under UV (over-polymerized); or not etched with oxygen plasma long enough | Reduce curing time of PEG under UV; treat with oxygen plasma longer |
| 33 | Contamination of cell culture after seeding | Culture plates were not sterilized well | Use ethanol prepared with sterilized DI water; Add more ethanol to each well and swirl to ensure that the entire culture well comes into contact with ethanol; Sterilize for longer times, such as 1–2 h; |
| 33, 34 | Cells proliferating outside patterns after stencil removal | PEG either isn’t present OR it is over etched | Verify that PEG grafting is successful by seeding cells on PEG grafted plates that have NOT been etched. Cells should not attach if PEG is present. If this is a result of over-etching, decrease the power of the plasma or etch for shorter cycles. |
| 38 | Differentiation isn’t efficient (less than 50% microchambers generated from the patterns) | Seeding densities aren’t optimal | Modify and test multiple seeding densities. Make sure that the seeding densities take 3 days to reach confluency. Refer to Figure 2i for ideal confluency to begin differentiation. |
| 49 | Software isn’t detecting motion and can’t compute motion vectors | Contraction motion exhibited by the tissue is too small to be detected with the default time delay. | Increase the time delay incrementally until it’s suitable for the software to calculate motion vectors. |
TIMING
Steps 1–9, SU8 master fabrication: 1 d
Steps 10–14, PDMS stencil fabrication: 2 h to overnight
Steps 15–20, PEG surface grafting: 1 h
Steps 21–23, Oxygen plasma etching: 2 h
Steps 24–33, Patterning of hiPSCs: 3 d
Steps 34–38, Cardiac differentiation of patterned hiPSCs: 15–20 d
Steps 39–48, Immunofluorescence staining and imaging: 1 d
Steps 49–55, Motion tracking analysis: 1 h
ANTICIPATED RESULTS
Upon removal of the PDMS from the SU8 master post curing, the stencils should have physically clear-through holes (Fig. 2a). The clear-through holes are essential, since they expose patterned regions of PDMS to the PEG layer, allowing the oxygen plasma access to the PEG surface. The PEG layer at these exposed regions is etched away to create a small pattern for cell attachment and colonization (Fig. 2b). Because of the flexibility of this approach, a diverse array of patterns can be fabricated to grow confluent hiPSC colonies with different shapes and sizes (Fig. 2c-e). Upon seeding, the cells will only be able to attach and proliferate inside the Matrigel coated patterns, since the PEG layer repels protein adsorption required for cell adhesion7. Prior to cardiomyocyte differentiation via small molecule induction, the hiPSCs patterned into different shapes were maintained in mTeSR1 media and uniformly expressed the pluripotency marker, OCT4 (Fig. 2f-h). The patterned cell morphologies begin changing after CHIR99021 treatment and form 3D tissue structures throughout the entire differentiation process (Fig. 2i)
Spatial differentiation to model cardiac organogenesis is evident with beating cardiomyocytes localized at the center of the microchambers, while connective cell types are localized to the perimeter of the patterns. Cardiomyocyte phenotype is confirmed by the staining of cardiac troponin, sarcomeric α-actinin, and myosin heavy chains, while connective cell types are characterized with myofibroblast/smooth muscle cell markers: SM22, calponin, and smooth muscle actin (Fig. 3). Our previous work showed that spatial cardiac differentiation was successful with WTC hiPSCs as well as H9 hESCs. We also previously showed that cell patterns facilitated the 3D formation of cardiac microchambers that ranged from 100–400 μm in height7.
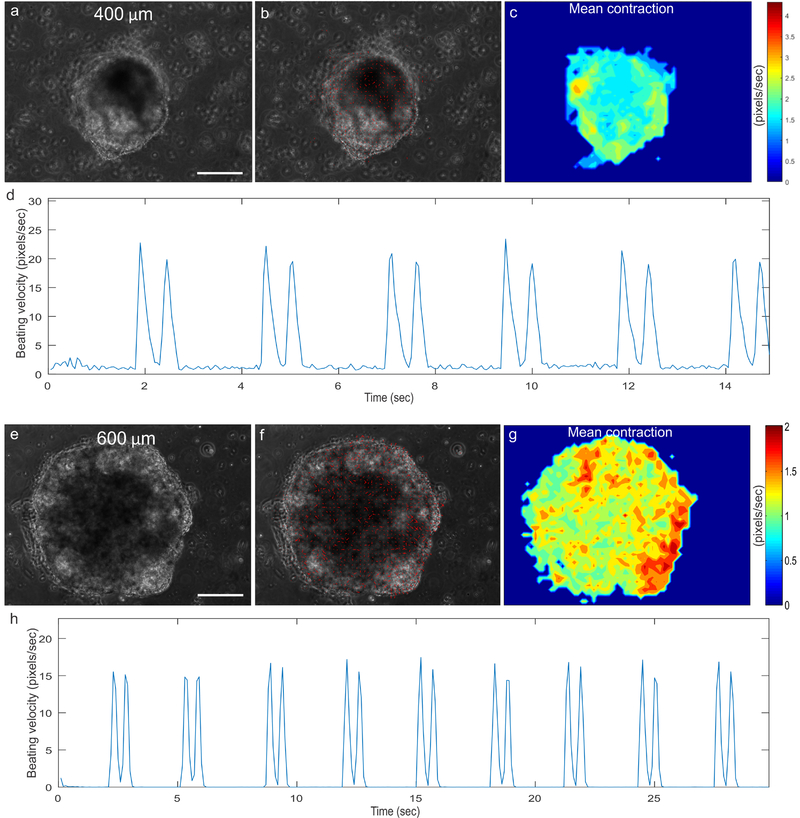
The cardiac microchambers grown from the 400 μm (Fig. 4a-d) and 600 μm (Fig. 4e-h, adapted from Ma, Z. et al., Nature Communications (2015))7, S1 circle patterns should exhibit beating motion that is detectable by the motion tracking software. The software calculates contraction motion by tracking the movement of pixels from one video frame (Fig. 4a, e) to the next, generating motion vectors to quantify the contraction (Fig. 4b, f; Supplementary Video 1, Supplementary Video 2S1). The contraction heat maps can also be output to visualize differences in contraction velocity between different regions of the microchambers (Fig. 4c, g). The mean contraction will be prominent towards the center of the patterns, indicative of actively contracting cardiomyocytes. In contrast, the contraction velocity towards the perimeter of the microchambers is less, which is consistent with myofibroblast lineage. The software can process the contraction data further, by computing motion waveforms from the overall average motion vector velocities (Fig. 4d, h). From the waveforms, we can quantify the cardiac physiology by measuring the beat rate, average contraction, average relaxation etc.
Figure 4. Contractile motion tracking of beating cardiac microchambers grown from 400 μm and 600 μm7 circle patterns.
(a, e) Videos of beating cardiac microchambers are analyzed to calculate motion vectors. (b, f) Motion vectors are plotted as contraction directions. (c, g) Motion vectors are used to compute localized mean contraction velocities as heatmaps. (d, h) Contraction motion waveforms are then generated to characterize beating physiology of the cardiac microchambers. Scale bar: 200 μm.
The efficiency of cardiac differentiation can be determined by counting the percentage of beating cardiac organoids relative to the total number of patterned colonies within each well. Efficiencies of about 80% are expected. Uncoordinated beating associated with arrhythmias can also be identified through the beating waveform. Additionally, the organoid height and size (HWHM: half width at half maximum) can be calculated using confocal microscopy. Lastly, the uniformity of contractile tissue is determined by examining the location of cardiomyocyte region (cTnT+) within each pattern and calculating the area ratio of cardiomyocyte region over an entire single pattern. Area ratios of approximately 0.4–0.5 are expected. We previously assessed the potential applications of this model for embryotoxicity screening by exposing the cardiac microchambers to thalidomide. The drug exposure resulted in reduced differentiation efficiency, defective microchambers, and impaired contractility7.
In summary, we have described here a unique method that takes advantage of cell patterning to engineer stem cell organoids for facilitating tissue morphogenesis of cardiac microchambers. This protocol demonstrates the capabilities of combining biochemical inducing factors with biophysical confinement to direct spatial organization of 3D tissue constructs derived from hiPSCs. We envisage that these early developing cardiac microchambers can potentially be used as embryotoxicity screening assay for drug discovery, and more importantly, to study the disease mechanisms of structural malformations at early stage of cardiac organogenesis with patient-specific hiPSCs.
Supplementary Material
Supplementary Video 1. 400 μm cardiac microchamber. Movie showing the beating cardiac microchamber engineered from 400 μm circle patterns with motion vectors calculated from optical flow analysis of the motion tracking software.
Supplementary Video 2. 600 μm cardiac microchamber7, S1. Movie showing the beating cardiac microchamber engineered from 600 μm circle patterns with motion vectors calculated from optical flow analysis of the motion tracking software7, S1.
ACKNOWLEDGEMENTS
This work was supported by the Nappi Family Foundation Research Scholar Project, NIH-NIBIB R21EB021003, and in part by NIH-NCATS UH3TR000487. Z.M. acknowledges support from American Heart Association (AHA) postdoctoral fellowship 16POST27750031. P.H. acknowledges support from the National Science Foundation Integrative Graduate Education and Research Traineeship (NSF IGERT), DMR-DGE-1068780.
Footnotes
Tweet: Modeling early cardiac organogenesis with spatial-patterned cardiac organoids using human pluripotent stem cells
All the authors declare no competing financial interests.
REFERENCES:
- 1.Lancaster MA & Knoblich JA Organogenesis in a dish: Modeling development and disease using organoid technologies. Science (80-. ). 345, 1247125–1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Yin X et al. Engineering Stem Cell Organoids. Cell Stem Cell 18, 25–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willyard C The boom in mini stomachs, brains, breasts, kidneys and more. Nature 523, 520–522 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Simian M & Bissell MJ Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol 216, 31–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutta D, Heo I & Clevers H Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med 23, 393–410 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Ranga A et al. Neural tube morphogenesis in synthetic 3D microenvironments. Proc. Natl. Acad. Sci 114, E3163–E3163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Z et al. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun 6, 7413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr JC et al. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials 31, 1885–1893 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warmflash A, Sorre B, Etoc F, Siggia ED & Brivanlou AH A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deglincerti A et al. Self-organization of human embryonic stem cells on micropatterns. Nat. Protoc 11, 2223–2232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatehullah A, Tan SH & Barker N Organoids as an in vitro model of human development and disease. Nat. Cell Biol 18, 246–54 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Eiraku M et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Sato T & Clevers H Growing Self-Organizing Mini-Guts from a Single Intestinal Stem Cell: Mechanism and Applications. Science (80-. ). 340, 1190–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Gjorevski N et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 536, 238–238 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Xu C & Ma T In vitro organogenesis from pluripotent stem cells. Organogenesis 10, 159–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breckwoldt K et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc 12, 1177–1197 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Guo XM et al. Creation of Engineered Cardiac Tissue In Vitro From Mouse Embryonic Stem Cells. Circulation 113, 2229–2237 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Shkumatov A, Baek K & Kong H Matrix rigidity-modulated cardiovascular organoid formation from embryoid bodies. PLoS One 9, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun X & Nunes SS Biowire platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Methods 101, 21–6 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Wang G et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20, 616–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathur A et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep 5, 8883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z et al. Three-Dimensional Filamentous Human Cardiac Tissue Model. Biomaterials 35, 1367–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinson JT et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science (80-. ). 349, 982–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettinato G, Wen X & Zhang N Formation of well-defined embryoid bodies from dissociated human induced pluripotent stem cells using microfabricated cell-repellent microwell arrays. Sci. Rep 4, 7402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunes SS et al. Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes. Nat. Methods 10, 781–787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergström G, Christoffersson J, Schwanke K, Zweigerdt R & Mandenius C-F Stem cell derived in vivo-like human cardiac bodies in a microfluidic device for toxicity testing by beating frequency imaging. Lab Chip 15, 3242–3249 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Matsudaira K, Nguyen T & Shoji KH MEMS piezoresistive cantilever for the direct measurement of cardiomyocyte contractile force. (2017). [Google Scholar]
- 30.Huebsch N et al. Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Sci. Rep 6, 24726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavesi A et al. Controlled electromechanical cell stimulation on-a-chip. Sci. Rep 5, 11800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmons CS, Petzold BC & Pruitt BL Microsystems for biomimetic stimulation of cardiac cells. Lab Chip 12, 3235 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Caiazzo M et al. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater 15, 344–352 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Jeon O & Alsberg E Regulation of stem cell fate in a three-dimensional micropatterned dual-crosslinked hydrogel system. Adv. Funct. Mater 23, 4765–4775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBeath R, Pirone DM, Nelson CM, Bhadriraju K & Chen CS Cell shape, cytoskeletal tension, and RhoA regulate stemm cell lineage commitment. Dev. Cell 6, 483–495 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Musunuru K, Domian IJ & Chien KR Stem Cell Models of Cardiac Development and Disease. Annu. Rev. Cell Dev. Biol 26, 667–687 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruneau BG The developmental genetics of congenital heart disease. Nature 451, 943–948 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Bressan M et al. Reciprocal myocardial-endocardial interactions pattern the delay in atrioventricular junction conduction. Development 141, 4149–4157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luxán G et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. 19, 193 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Lian X et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci 109, E1848–E1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huebsch N et al. Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales Tissue Eng. Part C Methods 21, 467–479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tourovskaia A et al. Micropatterns of chemisorbed cell adhesion-repellent films using oxygen plasma etching and elastomeric masks. Langmuir 19, 4754–4764 (2003). [Google Scholar]
- 43.Kattman SJ et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Burridge PW et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei F et al. Chemical-defined and albumin-free generation of human atrial and ventricular myocytes from human pluripotent stem cells. Stem Cell Res. 19, 94–103 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Maddah M et al. A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing. Stem Cell Reports 4, 621–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee EK, Kurokawa YK, Tu R, George SC & Khine M Machine learning plus optical flow: a simple and sensitive method to detect cardioactive drugs. Sci. Rep 5, 11817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- S1.Ma Z et al. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun 6, 7413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. 400 μm cardiac microchamber. Movie showing the beating cardiac microchamber engineered from 400 μm circle patterns with motion vectors calculated from optical flow analysis of the motion tracking software.
Supplementary Video 2. 600 μm cardiac microchamber7, S1. Movie showing the beating cardiac microchamber engineered from 600 μm circle patterns with motion vectors calculated from optical flow analysis of the motion tracking software7, S1.