ABBREVIATIONS
- BBB
blood–brain barrier
- CNS
central nervous system
- CSF
cerebrospinal fluid
- EGFRvIII
epidermal growth factor receptor variant III
- GBM
glioblastoma
- MHC
major histocompatibility complexes
- MRI
magnetic resonance imaging
- PSSM
position-specific scoring matrix
- TCGA
The Cancer Genome Atlas
- TIL
tumor-infiltrating lymphocytes
- TMG
tandem minigene
Glioblastoma (GBM) remains a disease with a poor prognosis. Unfortunately, over the past decade, no new treatment options have improved survival in patients beyond the current standard-of-care radiation plus temozolomide following maximal surgical resection. The initial enthusiasm that extensive genomic profiling of driver mutations, of which GBM was one of the first to be characterized by The Cancer Genome Atlas (TCGA),1,2 would lead to effective molecularly targeted therapy for central nervous system (CNS) malignancies has yet to come to fruition. The reason for the failure of this “mutation-to-drug” paradigm is likely multifactorial, including the subclonal heterogeneity of GBM3 and the necessity of systemically delivered drugs to penetrate the blood–brain barrier(BBB) at a sufficient concentration to be efficacious. As such, any new treatment approach will need to address these complexities of GBM if it is to be successful.
It is to this end that immunotherapy offers renewed promise. The systemic immune system has the ability to attack multiple targets simultaneously, and has the capacity to penetrate the BBB. As our understanding of CNS immunosurveillance and tumor immunity continues to deepen, novel strategies to prime and augment a potent antitumor immune response will emerge. Recent interest has been focused on the identification of tumor-specific mutations, termed neoantigens, which can serve as immunodominant targets for antitumor immune effector cells to maximize “on-tumor” effect and minimize “off-tumor” toxicities. In this review, we will discuss: (1) the current perspective on CNS immunosurveillance, (2) the process of neoantigen identification focusing on the cancer immunogenomics approach, and (3) how this strategy can be used to target GBM specifically.
EVIDENCE OF ACTIVE IMMUNOSURVEILLANCE IN GBM: CNS IMMUNOBIOLOGY
The potential of immunotherapy in CNS malignancies has long been thought to be futile given the immunoprivileged and immunosuppressive nature of the intracranial environment. However, recent data have demonstrated that the CNS is not wholly a sanctuary site due to immune isolationism. On the contrary, the immune system actively surveys the CNS, and is capable of mounting an effective immunological response when necessary supporting the renewed enthusiasm for immunotherapy in combating CNS disease.
Immunoprivilege in the CNS
The topic of CNS immunosurveillance has been extensively reviewed recently,4-8 and is not within the scope of this article. However, given that the historic viewpoint of an “immunoprivileged CNS” has often been interpreted as an “immunocompromised CNS,” several key concepts must be discussed in order to understand the rationale for pursuing immune therapy in GBM. As summarized eloquently by Engelhardt and colleagues,4 the immunoprivileged phenotype of the CNS was based on the experimental observation that tissues grafted into the brain parenchyma are not rejected due to the lack of an induced cell-mediated immunity.9 Importantly, the simultaneous implantation of skin homografts subcutaneously led to equivalent rejection of both the skin and brain grafts9 implying that the effector arm of the systemic immune system is able to sufficiently locate, penetrate, and remove CNS-based antigens. Similar results were demonstrated following intraparenchymal injection of bacillus Calmette-Guerin that resulted in a demyelinating delayed type hypersensitivity reaction following subsequent systemic immunization despite a minimal local reaction initially.10 These experiments support the notion that a deficient afferent limb of the immune response may be largely responsible for the immunoprivileged phenotype of the brain parenchyma, and that the effector arm is functionally intact.
Importantly, it should be noted that this observation is perhaps most relevant under steady state circumstances. For instance, the intraparenchymal injection of immunostimulatory agents such as lipopolysaccharide,11 TNFα, or IL-1αβ12 leads to robust, albeit delayed, influx of innate immune cells such as neutrophils, monocytes, and macrophages as well as activation of resident microglial cells demonstrating that local inflammation does indeed drive recruitment and infiltration of systemic immune cells. Therefore, under inflammatory conditions, the immunoprivileged nature of the brain parenchyma is subverted.5 Furthermore, it should also be pointed out that these observations are limited to the brain parenchyma as implantation of virus or tissue grafts into the cerebrospinal fluid (CSF) or choroidal plexus results in robust immune responses equivalent to systemic sites.13-15
Mechanisms of CNS Antigen Drainage to Lymph Nodes
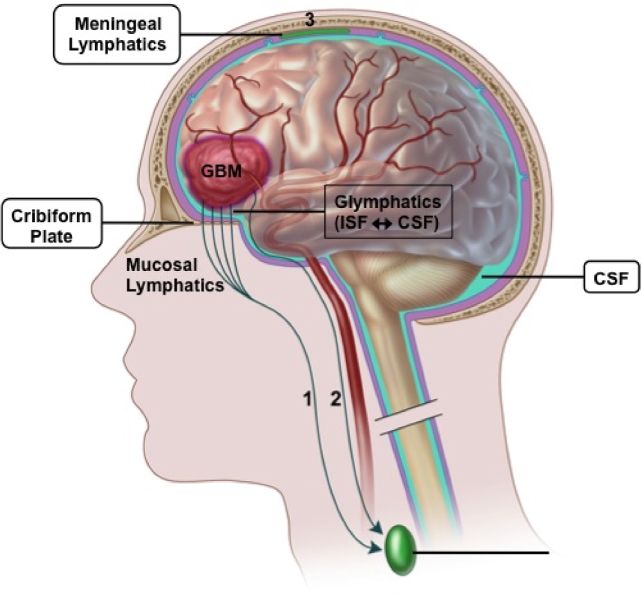
The apparent deficiency of the afferent limb within the brain parenchyma was initially attributed to the presence of the BBB and lack of classic lymphatics. While the CNS is certainly immunologically distinct from other organs anatomically, the immune system is still able to actively survey CNS antigens through various mechanisms. Specifically, there are 3 primary routes by which intracranial antigens, and presumably tumor-derived antigens, are able to drain from the CNS into locoregional and systemic lymphoid tissue (Figure 1). The first is via ventricular and subarachnoid CSF that is able to cross the cribiform plate and enter the lymphatics of the nasal mucosal ultimately draining into the deep cervical lymph nodes.7 Secondly, CSF is able to enter recently described meningeal lymphatics located in the dura that also drain to the deep cervical lymph node chain.16,17 These routes of CNS drainage are amenable both to soluble antigens as well as immune cells such as T cells, monocytes, and dendritic cells. The third route CNS-derived antigens use to reach regional lymph nodes results from parenchymal interstitial fluid trafficking through the basement membrane of the wall of capillaries and arteries of the brain.18 Unlike the CSF, arterial-based drainage is limited to acellular antigen transportation due to size exclusion. Alternatively, intraparenchymal interstitial fluid is also exchanged with CSF in a process termed glymphatics.19 It is interesting to note that drainage of parenchymal antigens is abrogated in mice lacking meningeal lymphatics16 despite only ∼15% of interstitial fluid from the parenchyma draining through the CSF.20 Therefore, it seems reasonable to assume that GBM-derived antigens are able to reach draining lymph nodes via both routes, though the relative contribution in human disease remains unclear and may be largely dependent on geography of the tumor.
FIGURE 1.
Routes of CNS-based tumor antigen drainage to regional lymph nodes. Tumor-derived antigens can reach draining cervical lymph nodes in several ways. Antigen that gains access to the CSF either by direct extension of the tumor, breakdown of the BBB, cellular trafficking by APC, or through glymphatic exchange can enter the lymphatic system by traversing the cribiform plate into the nasal mucosa (1) or through meningeal lymphatics of the dura (3). Alternatively, acellular antigen can enter the wall of intraparenchymal capillaries and arteries to migrate retrograde toward local lymph nodes (2). BBB, blood–brain barrier; CSF, cerebrospinal fluid; GBM, glioblastoma; ISF, interstitial fluid. Adapted from Engelhardtet al.4
Effector Immune Responses to GBM
Regardless of the pathway used by GBM-derived antigens to translocate to local draining lymph nodes, it is clear that such antigens are able to elicit effector immune responses. For example, spontaneously arising autoantibodies to GBM-specific proteins: GLEA1, GLEA2, and PHF3 have been demonstrated in 24%, 48%, and 57% of adult GBM patients, respectively,21 providing support for the generation of a naturally occurring antiglioma humoral response. Likewise, the cellular arm of the immune system also appears to be primed against GBM tumor cells. Barcia et al22 observed activated cytotoxic CD8+ T cells in close proximity to GBM tumor cells in Situ, characterized by CD3/T cell receptor (TCR) clusters, cytoskeletal rearrangement, and granzyme B polarization toward the tumor cells supporting recognition of cognate antigen:MHC complexes on GBM cells by antigen-specific T cells. Additionally, Berghoff and colleagues23 reported that the majority of newly diagnosed patients (72.6%) and recurrent patients (83.3%) had tumor-infiltrating lymphocytes (TILs) present in tumor specimens, indirectly pointing toward an interaction between tumor and the host immune system.
While TILs are largely confined to the perivascular space of postcapillary venules and peripheral zones of tumor invasion,23 numerous studies have demonstrated a positive correlation between the presence of TIL and clinical outcome for patients with GBM.24 For example, Brooks et al25 examined clinical records and biopsy specimens of 149 patients from 1962 to 1976 and noted that perivascular lymphocyte infiltration correlated with a 2 to 4 mo increase in survival over patients without such infiltrate. Obviously, TIL represent a heterogeneous group of immune cells, comprising both effector and suppressive subsets. Thus, as one might expect, the effect on survival largely seems to be dependent upon the ratio of effector T cells (ie, CD4+ or CD8+ subsets) to suppressor T cells (Tregs).26-30
Together, these data support the notion that both the humoral and cellular arms of the immune system are able to be primed against GBM antigens. However, it remains unclear by which mechanism these adaptive immune responses are generated; which antigens they recognize; the functional capacity of such naturally occurring responses; and the role these spontaneous immune responses play in driving immune escape.
THE BBB IN GBM
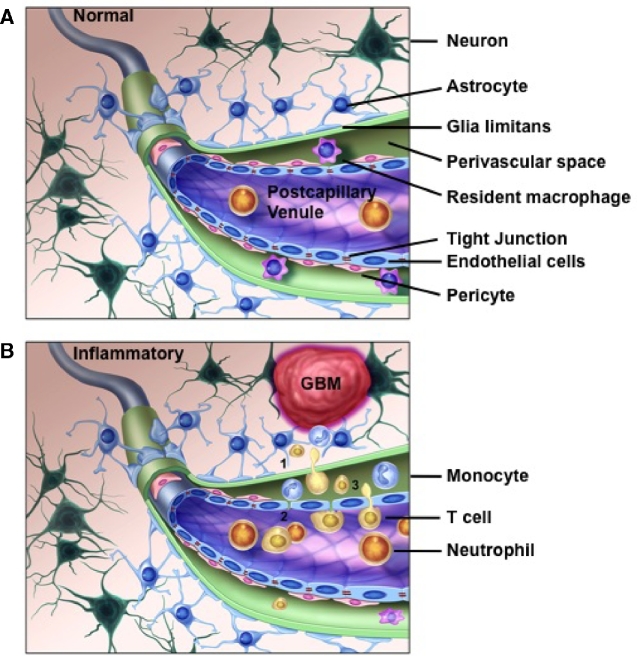
The exact mechanism leading to recruitment of infiltrating lymphocytes into the GBM microenvironment is not understood. One potential explanation is that the BBB is compromised in the setting of GBM, and this “leaky” BBB could serve as a conduit for interactions between lymphocytes and the tumor.31,32 At steady state, the BBB is composed of tight junctions between specialized capillary endothelial cells supported by an extracapillary layer of cells including pericytes and astrocytic end-foot processes, which form the glia limitans (Figure 2). It is this dual layered barrier that restricts transfer of solute and cells into the brain parenchyma. However, in GBM, this barrier is compromised (Figure 2). For example, Nduom and colleagues33 observed by immunohistochemistry that in patients with GBM, regions of magnetic resonance imaging (MRI) enhancement corresponded with breakdown of the normal astrocyte-endothelial cell relationship demonstrated by gaps between GFAP (glial fibrillary acidic proteins; expressed by astrocytic cells) and the aquaporin molecule, AQP4, which is expressed on the luminal side of glial processes. These proteins should normally demarcate a tight boundary on the basolateral side of astrocytes that surround the endothelial cells to secure the BBB as demonstrated in nonenhancing lesions.33 A similar phenomenon has been observed in brain metastases and pediatric high-grade gliomas, but not in low-grade, non-MRI-enhancing tumors.33-35
FIGURE 2.
Proposed model of leukocyte recruitment due to altered BBB integrity in GBM. A, Under normal conditions, the dual layers of the BBB is maintained through tight junctions between capillary endothelial cells and the glia limitans, which is comprised of astrocytic end-foot processes. In the postcapillary venules, these 2 layers separate creating a perivascular (Virchow-Robin) space, which contains resident macrophages. B, In the context of GBM or inflammation, the BBB is disrupted. The glia limitans loses polarity due to altered expression of AQP4 in the astrocytic end-foot processes leading to expansion of the perivascular space and communication with the underlying parenchyma (1). The capillary tight junctions are disrupted due to reduced expression of claudin-3, which permits exchange of solutes, antigens, and chemokines/cytokines (2). It also allows circulating leukocytes, such as neutrophils, monocytes, and T cells, to gain access to the perivascular space where they interact with APC that present tumor antigen from the parenchyma (3). AQP4, aquaporin 4 molecule; BBB, blood–brain barrier; GBM, glioblastoma.
In addition to disruption of the BBB due to altered polarity of the astrocytic end-foot processes, the endothelial layer is also perturbed. Particularly, the interendothelial cell tight junctions, which are essential to maintaining the integrity of the BBB, also becomes dysregulated in GBM. Wolburg et al35,36 observed a loss of the tight junction molecule, claudin-3, in GBM, which greatly contributed to the increased permeability noted in intratumoral capillary vessels. Together, these findings demonstrate a dramatic loss of integrity of the BBB adjacent to GBM that affects both the endothelial layer and glia limitans (Figure 2).
What is interesting to note is that the perivascular space, which forms between the endothelial cell layer and the glia limitans in the postcapillary venules, becomes expanded at the site of BBB disruption in GBM.35,36 The perivascular space is also the site of resident macrophages. Therefore, one potential mechanism by which a local immune reaction is incited against GBM is that the breakdown of the BBB facilitates detection by and activation of resident macrophages to GBM-associated antigens leading to recruitment of circulating immune cells that are then able to recognize cognate antigens on local antigen-presenting cells(APC) (Figure 2). Consistent with this model, Proescholdt et al37 noted that it is not until the BBB is disrupted that an immune infiltrate is detected in a rat brain tumor model.
CANCER IMMUNOGENOMICS AND THE IDENTIFICATION OF NEOANTIGENS IN GBM
Cancer immunogenomics represents a complementary approach to the application of genomics in developing novel treatment strategies for malignancies. Using this approach, putative tumor-specific neoantigens derived from expressed, nonsynonymous missense or frameshift mutations in the exome are prioritized based on predicted processing and binding affinity to a patient's individual HLA (human leukocyte antigen) molecules.38 Thus, rather than stratifying mutational targets based on the “drivers” and “passengers” classification, the predicted immunodominance of a mutational alteration is given precedence, creating a “mutation-to-antigenic target” paradigm. This approach is increasingly being applied to neoantigen identification both preclinically and clinically. The actual process of neoantigen discovery using this approach will be discussed here.
Definition of Neoantigen
We now know that endogenous T cells recognize tumor antigens presented by major histocompatibility complexes (MHC) on the surface of malignant cells. These recognition events are mediated by specific interactions between MHC-bound tumor antigens and T cell receptors.39 To date, 3 classes of MHC-binding tumor antigens have been documented40: (1) shared tumor antigens which are nonmutant, normally expressed proteins that are aberrantly overexpressed in tumor cells, (2) cancer-testis antigens that are normally only found in healthy adult germ cell tissues but exhibit re-expression in some cancers, and (3) tumor-specific mutant antigens, referred to as neoantigens, which represent novel peptide sequences encoded by somatic mutations in the cancer genome. To date, cancer vaccine clinical trials that have used peptide-based vaccines comprising of shared-tumor antigens or cancer-testis antigens have not yielded promising results despite concomitant induction of a high frequency of antigen-specific T cells.41 One theory underlying the lack of success may be attributable to issues of central tolerance whereby high-affinity endogenous T cells specific to these conserved tumor antigens are eliminated due to expression in normal tissue during development. Additional challenges common to both types of antigens include limited expression in tumor cells compared to levels in nonmalignant cells; lack of known binding within less common HLA alleles precluding their broader use in many patients; as well as increased risks of “off-target” immune recognition of nonmalignant cells. Compared to nonmutant tumor-associated antigens, neoantigens circumvent issues of immune tolerance as they consist of peptides derived from somatic, nonsynonymous mutations only present in the tumor genome, and therefore would appear as “foreign” to the host immune system. Likewise, immunodominant neoantigens are tailored to a patient's specific HLA alleles, permitting the broader application of this approach to a larger, more diverse patient population.38
Cancer Immunogenomics: Pipeline for Neoantigen Discovery
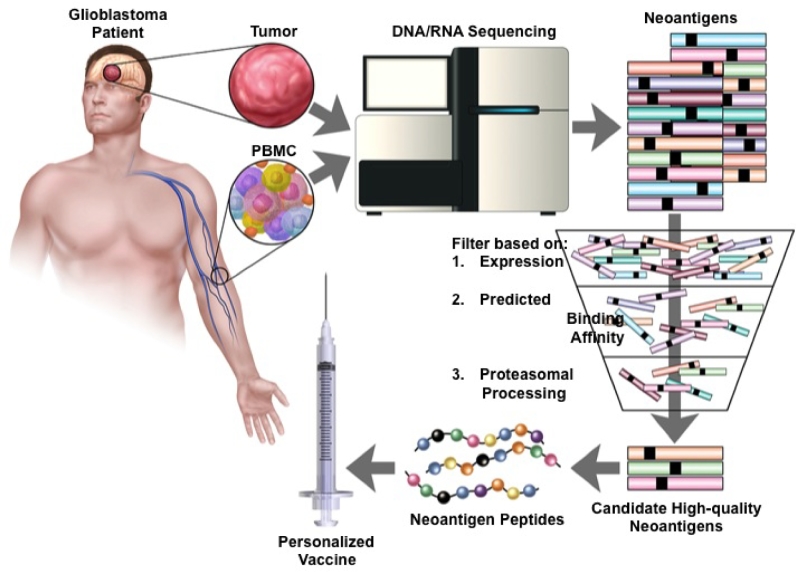
Cancer immunogenomics refers to a concept in which genomic alterations inherent to cancer cells are leveraged as targets for immune-based therapies.42-45 One example of this strategy is the identification and targeting of neoantigens. Until recently, the identification of patient tumor-specific neoantigens required highly labor-intensive laboratory techniques that precluded its use in the clinical setting. However, the development of next-generation sequencing technologies and advances in the downstream computational analyses have revitalized these efforts by facilitating rapid characterization of the tumor mutational landscape. By removing these technological barriers, genomic breakthroughs have paved the way for high-throughput and cost-effective personalized neoantigen identification. The initial step in identifying neoantigens begins with DNA whole exome and RNA sequencing of matched patient normal and tumor tissue (Figure 3). Using one of many currently available variant calling and annotation software programs, the raw exome sequence data are mined for nonsynonymous missense tumor variants and integrated with transcriptome analysis to select for expressed mutations.46 Peptide sequences containing the encoded amino acid mutations are then generated according to predesignated residue length settings to accommodate the different binding grooves of MHC class I or class II molecules. Due to the vast number of candidate neoantigen peptides that can be generated for a given tumor, in silico algorithms are used to aid in the selection of immunogenic neoantigens by predicting the binding affinity of each candidate peptide for patient-specific HLA alleles. Ultimately, candidate peptides with the highest predicted binding affinity are synthesized and used in either personalized vaccines or in a variety of immunological assays to validate the presence/generation of neoantigen-specific T cell responses in an individual (Figure 3).47,48
FIGURE 3.
Schematic representation of cancer immunogenomics workflow for neoantigen discovery. Normal reference tissue (ie, PBMC) and tumor tissue is obtained and undergoes DNA whole exome and RNA sequencing to identify somatic, nonsynonymous mutations. Tumor-specific mutations are then filtered using computational software to prioritize neoantigens based on expression, predicted patient-specific HLA binding affinity, and likelihood of endogenous proteosomal processing. Peptides corresponding to candidate high-quality neoantigens are then manufactured and administered back to the patient as a personalized vaccine. PBMC, peripheral blood mononuclear cell.
Genomic Analysis of Raw Sequence Data
The computational analysis employed in the context of neoantigen discovery can be thought of as occurring in 2 phases, beginning with the initial processing of raw genomic data and subsequently moving into the use of in silico immunogenomics tools to characterize the tumor–immune cell interactions. A number of different software packages are available for the initial processing of cancer sequence data. These programs include tools to identify single nucleotide polymorphism, indels, or gene fusions, as well as annotation algorithms that infer alterations in protein structure and function based on genomic and transcriptomic data. Of note, the functional annotation of a variant depends on the transcripts or isoforms used. For example, the most widely used annotation browsers include ENSEMBL, REFSEQ, and UCSC, which contain sets of transcripts used to determine the functional consequences of a given genetic variant.49,50 Significantly, in an analysis by McCarthy and colleagues,51 the authors found that using different transcript sets results in nonoverlapping variant annotation results. Thus, as variant annotation is not yet fully resolved, this step must be carefully considered in the broader picture of neoantigen identification.
Predicting Peptide:MHC Binding Affinity for MHC Class I Neoantigens
In the second phase of neoantigen discovery, mutant peptide sequences generated from prior genomic analyses are filtered in order to select only for candidates that are likely to elicit T cell responses. To aid in the selection of immunogenic neoantigens from lists that often contain thousands of candidate peptides, in silico algorithms are used to predict peptide binding affinities for patient-specific MHC alleles.52 Though less complex in murine studies, predicting peptide binding affinities in humans is considerably more difficult given the multiple HLA alleles present in a given patient. Located on chromosome 6, the HLA locus is among the most polymorphic regions in the human genome, with current HLA databases comprising 6000 distinct alleles. Inheriting 3 HLA class I (HLA-A, -B, and -C) and 3 HLA class II (HLA-DQ, -DP, -DR) molecules from each parent, each person can possess up to 12 unique HLA complexes, each capable of presenting a distinct set of neoantigens. As such, this immense genetic diversity must be addressed by prediction algorithms in order to prioritize candidates that could ultimately be displayed on a specific patient's MHC molecules.
Currently, the vast majority of computational methods for predicting neoantigen binding affinity use a combination of statistical models and machine learning algorithms. The earliest prediction algorithms, such as SYFPEITHI, Rankpep, and BIMAS, calculate peptide:MHC binding affinities using position-specific scoring matrices (PSSMs), and therefore the accuracy of these predictions depends on the abundance of empirically validated peptide-binding data for specific MHC alleles.50 Entities such as the Immune Epitope Database (IEDB) have attempted to improve prediction accuracy by incorporating multiple different prediction algorithms; however, benefits over the use of single algorithm methods is so far marginal. Currently, IEDB contains over 120 000 curated epitopes and for this reason is frequently used for neoantigen discovery.50,53
PSSM prediction methods are limited by their dependence on available validated data sets, and therefore perform suboptimally when predicting binding affinities for less common HLA alleles, about which little is known regarding peptide:MHC interactions. Therefore, new computational tools, such as NetMHC, have been developed that predict peptide binding affinity for any MHC molecule without the need for empirical data. These tools are built on machine-learning neural networks that are “trained” on available peptide:MHC binding data and then extrapolate to predict novel binding interactions.54 While still limited by the size and quality of training sets, improvements in neural-network prediction methods have been made by expanding training sets to include those from all species (NetMHCpan). Though there is no general consensus on recommendations for specific neoantigen prediction tools, results from a number of recent studies rank machine-learning methods such as NetMHCpan among the highest performing algorithms. For example, Fritsch and colleagues55 performed a retrospective analysis to evaluate the accuracy of conventional prediction algorithms to distinguish immunogenic neoantigens from inert ones. Based on a set of 31 previously identified immunogenic neoantigens, NetMHCpan correctly identified 27 (87%) of the 31 naturally occurring epitopes to be immunogenic based on the conventional binding affinity cutoff of ic50 < 500 nM. However, while considered the most selective event in the antigen presentation pathway, peptide:MHC binding alone cannot account for all of the confirmed immunogenic neoantigens, as a number of low affinity, immunogenic peptides have been documented elsewhere.56
MHC Class II-Restricted Neoantigens
Compared to in silico predictions of MHC class I-binding peptides, there has been considerably less progress in developing computational methods for predicting peptide-binding interactions with MHC class II molecules. Multiple MHC class II-binding prediction algorithms do exist (TEPITOPE, netMHCII, and SMM-align); however, the focus in neoantigen discovery has predominantly been on class I peptides, and therefore, the class II prediction algorithms generally lack much needed empirically validated training sets.57 Improving MHC class II peptide predictions will also likely involve a number of additional considerations to address the fundamental molecular differences between the 2 MHC complexes. MHC class II molecules are only expressed on professional APC such as dendritic cells, macrophages, and B cells, and play an important role in activating helper T cells. Differences also extend to features of the peptides that bind each class. Unlike MHC class I molecules, the MHC class II peptide-binding groove is open at both ends such that significantly longer peptides of up to 30 amino acids can bind.41 This feature generates considerable variation in both the length of compatible peptides and in the location of the peptide-binding core, as the peptide is free to “slide” along the open binding groove. The latter poses a substantial challenge, as peptide:MHC class II binding affinity is determined not only by binding core sequence but also by flanking residues.58 Thus, due to the molecular complexity of peptide:MHC class II interactions and the general lack of available training data, class II peptide prediction algorithms are significantly less accurate than those for class I peptides.
Despite the challenges, new efforts are being directed towards improving shortcomings in class II predictions in light of recent studies demonstrating the substantial contribution of CD4+ T cells in antitumor immunity. A study by Kreiter and colleagues59 that analyzed T cell responses to neoantigen vaccination in tumor-bearing mice found that 95% of the neoantigen-specific T cells were CD4+. Further investigation of this phenomenon in human malignancies resulted in the finding that infusion with CD4+ T cells specific for an ERBB2IP-derived neoantigen elicited significant tumor regression in a patient with metastatic cholangiocarcinoma. Studies in melanoma patients similarly identified a significant proportion of neoantigen-specific intratumoral CD4+ T cells.60 Together, these clinical data support the need to continue to optimize class II-restricted neoantigen prediction alogrithms, particularly as class II antigens expand the pool of potential targets that can be incorporated into personalized vaccines, which may have important implications for patients with tumors like GBM that possess a relatively lower mutational burden.
Alternative Approaches to in silico Neoantigen Predictions
While current computational algorithms predict peptide: MHC class I-binding events with moderately high sensitivity, this methodology ignores other factors in the antigen presentation pathway including peptide processing. These steps preceding peptide:MHC binding, including proteasomal cleavage and transport of peptides via TAP proteins into the endoplasmic reticulum, also significantly impact which putative neoantigens are ultimately presented by MHC complexes.41 Recently, new computational tools have been developed that quantify the probability of peptide cleavage by the proteasome (NetChop) and interactions with TAP (PredTAP, SVMTAP) in order to apply cutoffs that may be useful in identifying naturally processed neoantigens.61 Schumacher and colleagues47 performed a similar retrospective study to simultaneously evaluate peptide:MHC class I binding affinity and NetChop filters (probability > 0.5) to accurately identify neoantigens. From a set of 17 previously identified immunogenic neoantigens, NetMHC predictions (cutoff of ic50 <500 nM) correctly identified 14 neoantigens, while NetChop filters correctly identified 15 out of 17 neoantigens. Furthermore, the authors evaluated the use of a “similarity-to-self” filter and found that 15 of the 17 studied neoantigens contained mutations within the TCR-binding domain of the peptide. These data suggest that current shortcomings in neoantigen predictions will likely be solved by the development of computational tools that harness a deeper understanding of the antigen presentation pathway.
While advancements of current in silico neoantigen prediction methods have greatly improved the feasibility of peptide-based screening approaches, researchers have begun to look towards alternative approaches in order to circumvent inherent challenges with HLA coverage and endogenous peptide processing. Recently, Lu and colleagues62 developed a novel tandem minigene (TMG) approach to rapidly identify neoantigens recognized by autologous T cells in 2 melanoma patients who had experienced durable remission following adoptive TIL therapy. In brief, TMG constructs encoding 6 to 24 mutant tumor peptides were transfected into patient autologous APC. In Vitro validation assays to detect T cell response to TMG-expressing APCs were performed, and subsequent deconstruction of T cell recognition events identified KIF2C and POLA2 neoantigens as TIL targets. These results strongly suggest the utility of a TMG approach for high-throughput neoantigen screening; however, further studies will be needed to directly compare it to conventional peptide-screening methods.
Mass Spectrometry
Recent technological advancements in the field of mass spectrometry (MS) have enabled its application in neoantigen discovery as a method to investigate true in Vivo peptide:MHC interactions on the tumor cell surface. In silico prediction methods are inherently plagued by large numbers of false positives (peptides predicted to be MHC-binders but ultimately are not immunogenic) while limitations in MS peptide-detection sensitivity predispose to high rates of false negatives.63 A number of studies have recently begun to use an approach that combines these 2 complementary techniques for neoantigen predictions. Yadav and colleagues64 demonstrated that vaccination with 2 neoantigens predicted by both in silico binding-affinity algorithms and MS structural analysis resulted in therapeutic T cell responses in tumor-bearing mice. Similarly, Bassani-Sternberg and colleagues65 compared neoantigen predictions generated by NetMHC with those predicted by MS in 5 patients with melanoma. Interestingly, none of the 11 mutant peptides identified by MS were listed among the top 10 predicted candidate peptides as determined by NetMHC, though 2 of the MS neoantigens were found to elicit patient autologous T cell responses.
These studies suggest that MS may provide much needed refinement of in silico prediction algorithms, particularly for the less common HLA alleles. MS data undoubtedly represent tumor heterogeneity more accurately than do studies in tumor cell lines and also provides validation of true in Vivo neoantigen presentation that is absent from conventional peptide-screening methods. Thus, incorporation of MS data into existing neoantigen identification methods holds promise to reduce rates of false-positive predictions and lessen the burden of unnecessary empirical validation. While the small number of mutant peptides identified by MS in comparison to the large quantity predicted by in silico methods may be attributed to issues of instrument sensitivity, inferences also congruent with this observation include that unaccounted for restrictions of proteasome processing and TAP transport may result in over-representation of the true neoantigen pool.65 Currently, more widespread clinical use of this technology is limited by the relatively large tissue sample size required for analysis as well as the considerable training needed to operate such advanced equipment. However, as advancements in MS instrumentation and computational tools give way to more robust immunopeptidomics studies, results from future experiments will likely help inform the next generation of personalized neoantigen discovery.
NEOANTIGENS AS TARGETS IN GBM
The ultimate goal of neoantigen discovery is to target immunogenic neoantigens using various immunotherapeutic strategies. The advent of high-throughput DNA and RNA sequencing has made the possibility of identifying actionable neoantigens on a per patient basis feasible from both a cost and time standpoint. Importantly, the translational implications of such an approach have already been demonstrated clinically. Rosenberg and colleagues have successfully treated 2 patients, 1 with metastatic colorectal carcinoma and the other with metastatic cholangiocarcinoma, using an adoptive cell transfer approach with ex vivo expanded TIL specific to patient-specific neoantigens.60,66 The neoantigens targeted included a MHC class I-restricted KRASG12D mutation and a MHC class II-restricted ERBB2IPE805G mutation, respectively. In both circumstances, the infused CD8+ or CD4+ neoantigen-specific T cells resulted in regression of the metastatic disease. Similarly, a number of centers are attempting to administer polyvalent neoantigen-based personalized vaccines to patients with malignancies from various histologies, including GBM, using multiple vaccine platforms, including peptide, nucleic acid, and dendritic cells.67 Interestingly, several neoantigens have already been targeted in GBM and provide some critical insight to guide future trials.
Neoantigen Target: Epidermal Growth Factor Receptor Variant III
Epidermal growth factor receptor variant III (EGFRvIII) is a mutant form of EGFR that is present in roughly 25% to 30% of GBM.1,68 EGFRvIII forms from an in-frame deletion of 801 base pairs within the extracellular domain of EGFR. The resulting junctional sequence spanning the in-frame deletion represents a bona fide neoantigen and vaccination using a 13-mer peptide incorporating this novel junction was the basis of Celldex's Rindopepimut vaccine. Although Rindopepimut resulted in significant improvement in overall survival for patients with EGFRvIII+ GBM in early phase studies compared to matched historic controls,69 the recent ACT IV randomized phase III trial somewhat surprisingly failed to show a survival benefit despite a comparable effect on survival as seen with the vaccine in earlier trials.70 The difference was due to a better than expected median overall survival in the control group.
Several key issues stem from the EGFRvIII experience however. First, EGFRvIII is known to be heterogeneously expressed in only a subset of tumor cells,71 and emergence of EGFRvIII-negative subclones has been seen in early phase studies,72 suggesting that targeting a single subclonal neoantigen may be insufficient due to selective pressure on escape variants. Alternatively, it may be necessary to target multiple subclonal neoantigens if a high priority clonal neoantigen is not present. Another important caveat is that patients were given Rindopepimut irrespective of HLA haplotype. As such, correlative studies largely demonstrated a predominate humoral response rather than a cell-mediated response.73 Thus, it is possible that a cell-mediated immune response would be more efficacious, in which case, identifying which HLA alleles are capable of presenting EGFRvIII may lead to improved outcomes. Overall, the results from EGFRvIII-based vaccination provide encouraging and insightful results into targeting neoantigens in GBM moving forward.
Neoantigen Target: Mutant IDH1R132H
Recurrent mutations in the isocitrate dehydrogenase 1 (IDH1) gene, of which over 90% possess a R132H mutation, represent an attractive actionable neoantigen.74 In general, IDH1 mutations are thought to be an early transformative event, so unlike EGFRvIII, IDH1 mutations are usually present in the founding clone and thus expressed by all tumor cells. Moreover, this mutation is found in the majority of grade II-III gliomas and a high percentage of “secondary” GBMs.75,76 Furthermore, Schumacher et al77 demonstrated both cellular and humoral immune responses reactive to an IDH1R132H-containing long peptide in patients with IDH1-mutant gliomas suggesting that it is a potential neoantigenic target.77 Consistently, in silico analysis suggested that IDH1R132H-containing peptides are predicted to bind with high affinity to the MHC class II molecule, HLA-DRB, providing additional supportive evidence that IDH1R132H mutations are an immunodominant neoantigenic target of CD4+ T cells. Overall, these preliminary data not only provide support for pursuing the mutant IDH1R132H neoantigen as a vaccine target but also highlight the potential importance of incorporating MHC class II neoantigen predictions in vaccine design to expand the neoantigenic pool of candidate targets.
Neoantigens as Polyvalent Vaccines and as Aggregate Biomarkers
Ultimately, selected neoantigens for vaccine development will focus on clonality as well as polyvalency. For example, in the case of IDH1 wild-type or EGFRvIII-mutated GBM where there are potentially no clonal neoantigens present, likely targets will be pooled from high-quality subclonal mutations. In preclinical models, the feasibility and efficacy of targeting neoantigens with bivalent vaccines was demonstrated by Gubin et al78 in which the administration of a therapeutic vaccine protected mice against a progressively growing methylcholanthrene-derived sarcoma.
As proof-of-principle for this concept in GBM, we recently reported the identification of immunogenic neoantigens in two preclinical mouse models using the cancer immunogenomics approach described in previous sections. This approach initially identified a large pool of high-affinity putative neoantigens in both the carcinogen-induced GL261 and spontaneously derived SMA-560 tumors.79 Using a validation strategy that combined IFN-γ ELISPOT and tetramer-based FACS analysis, we credentialed 2 neoantigens, IMP3D81N (GL261) and ODC1Q129L (SMA-560), as immunogenic and capable of eliciting spontaneous neoantigen-specific CD8 T cell responses detectable both in intracranially implanted tumors and corresponding draining lymph nodes. Importantly, of the 24 combined total candidate neoantigens evaluated between these 2 models, these were the only 2 that were fully confirmed as immunodominant neoantigens. Consistently, this false-positive rate is comparable to other preclinical and clinical studies encompassing multiple tumor types.59,60,62,64,65,78,80-100 As such, this further demonstrates the need to improve upon the current in silico prediction algorithms used to assign binding affinity in an attempt to increase the true-positive rate of neoantigen identification. Furthermore, it provides additional rationale for incorporating a polyvalent vaccine approach, as it will increase the likelihood of immunizing with a truly immunogenic neoantigen.
Clinical trials using the cancer immunogenomics pipeline to design personalized neoantigen-based vaccines for patients with GBM are underway. One of the major objectives in these studies aside from assessing the immunogenicity and efficacy of this approach will be to determine the frequency of identifying immunogenic neoantigens given the relatively low mutational burden in GBM. Rooney et al101 applied a similar neoantigen discovery pipeline to a cohort of GBM tumors in the TCGA and found that on average these tumors harbored approximately 70 nonsynonymous mutations (range 2-258) from which roughly 10 potential neoantigens (range 0-51) were predicted. The 1 caveat from this study to note is that the neoantigen predictions were limited to HLA class I-restricted candidates. Thus, the incorporation of a pipeline to identify HLA class II-restricted neoantigens would expand the pool of potential candidates, which may be particularly important in these lower mutational load malignancies. Moreover, the efficacy of neoantigen-specific CD4+ T cells observed by Tran and colleagues60 also provides credence for focusing on class II neoantigens in future studies. Collectively, these and other studies will generalize the feasibility of personalized polyvalent neoantigen vaccine approaches, but a strong emphasis must be placed on the correlative studies to determine how effective these vaccines are at inducing specific immune responses and, most importantly, controlling disease.
Aside from serving as select targets for polyvalent vaccination strategies, 1 additional clinical implication for characterizing the neoantigen landscape of individual tumors is that a high neoantigen burden can also serve as a biomarker for responsiveness to other forms of immunotherapy, such as checkpoint inhibitor therapy. This concept was initially reported by Le and colleagues102 in colorectal cancer patients, where they demonstrated a near dichotomous response to anti-PD-1 therapy in patients whose tumors were deficient in the DNA mismatch repair pathway compared to those with proficient machinery. This sentinel study established a correlation between high mutational burden, and by extension high neoantigen burden, with response to checkpoint inhibitor therapy. This same association has since been demonstrated in melanoma,94,103,104 nonsmall cell lung cancer,92 and bladder cancer.105
The use of neoantigen load as a biomarker to identify patients that may benefit from checkpoint inhibitor therapy also appears to apply to patients with GBM. In 2 recent studies, both an adult patient with a hypermutated GBM secondary to a germline POLE mutation106 as well as 2 pediatric patients with GBM due to germline biallelic mismatch repair deficiencies107 were shown to have dramatic responses to anti-PD-1 therapy after having progressed on standard of care treatment. While the overall incidence of patients with hypermutated GBM is relatively low at time of diagnosis,108 there is growing appreciation that a subset of recurrent GBM, approximately 20% to 25%, acquire a hypermutated phenotype at time of recurrence due to an acquired deficiency in the mismatch repair pathway.109,110 This population seems to be particularly enriched in those patients with methylated promoters for 06-methylguanine methyltransferase (MGMT) and IDH mutations previously treated with temozolomide.2,111-113 Together, these data would suggest a possible neoantigen-based treatment strategy in GBM whereby those patients with high neoantigen burden would be stratified to receive checkpoint inhibitor therapy and those with low neoantigen burden could receive a polyvalent vaccine (+/– checkpoint inhibitor). With that being said, additional clinical trials are necessary to determine the optimal platform for incorporating immunotherapy and cancer immunogenomics into the treatment algorithm of GBM.
CONCLUDING REMARKS
Immunotherapy represents a unique approach to treating GBM, as well as other malignancies, as it is mechanistically distinct from conventional treatments. By taking advantage of the highly specific nature of the immune system to target tumor-specific neoantigens, immunotherapy offers an ideal strategy to maximize “on-tumor” effects while minimizing “off-target” adverse events. With the recent developments in high-throughput genomic sequencing technologies and computational analyses, the possibility of generating personalized neoantigen-based immunotherapies has become a reality. Moreover, the early clinical success seen using such strategies has fueled both the enthusiasm and effort in further developing these treatment modalities for a greater number of cancer patients. However, a considerable amount of work remains. Ongoing studies will be aimed at (1) attempting to optimize the pipelines through novel filters and machine-learning algorithms to increase the likelihood of identifying high-quality neoantigen targets; (2) understanding mechanisms of resistance or escape; (3) comparing immunization methods in order to induce the most effective immune response to candidate neoantigens; and (4) determining how to incorporate or sequence immunotherapy with current cytotoxic chemotherapy, radiotherapy, and molecular therapies to fully exploit the expanding multimodality cancer treatment options available.
Disclosures
Dr Dunn is a cofounder of Immunovalent Therapeutics. This study was funded by National Institutes of Health grant K08NS092912 (G.P.D.), American Cancer Society-Institutional Research grant (G.P.D), the Physician-Scientist Training Program (T.M.J.), and the Medical Scientist Training Program (J.B.K.) at Washington University School of Medicine. The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
REFERENCES
- 1. Brennan CW, Verhaak RG, McKenna A et al. . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37(6):E11. [DOI] [PubMed] [Google Scholar]
- 4. Engelhardt B, Vajkoczy P, Weller RO. The movers and shapers in immune privilege of the CNS. Nat Immunol. 2017;18(2):123-131. [DOI] [PubMed] [Google Scholar]
- 5. Galea I, Bechmann I, Perry VH. What is immune privilege (not)? Trends Immunol. 2007;28(1):12-18. [DOI] [PubMed] [Google Scholar]
- 6. Ransohoff RM, Engelhardt B. The anatomical and cellular basis of immune surveillance in the central nervous system. Nat Rev Immunol. 2012;12(9):623-635. [DOI] [PubMed] [Google Scholar]
- 7. Cserr HF, Knopf PM. Cervical lymphatics, the blood-brain barrier and the immunoreactivity of the brain: a new view. Immunol Today. 1992;13(12):507-512. [DOI] [PubMed] [Google Scholar]
- 8. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991-998. [DOI] [PubMed] [Google Scholar]
- 9. Medawar PB. Immunity to homologous grafted skin; the fate of skin homografts transplanted to the brain, to subcutaneous tissue, and to the anterior chamber of the eye. Br J Exp Pathol. 1948;29(1):58-69. [PMC free article] [PubMed] [Google Scholar]
- 10. Matyszak MK, Perry VH. Demyelination in the central nervous system following a delayed-type hypersensitivity response to bacillus Calmette-Guerin. Neuroscience. 1995;64(4):967-977. [DOI] [PubMed] [Google Scholar]
- 11. Andersson PB, Perry VH, Gordon S. The acute inflammatory response to lipopolysaccharide in CNS parenchyma differs from that in other body tissues. Neuroscience. 1992;48(1):169-186. [DOI] [PubMed] [Google Scholar]
- 12. Blond D, Campbell SJ, Butchart AG, Perry VH, Anthony DC. Differential induction of interleukin-1beta and tumour necrosis factor-alpha may account for specific patterns of leukocyte recruitment in the brain. Brain Res. 2002;958(1):89-99. [DOI] [PubMed] [Google Scholar]
- 13. Mason DW, Charlton HM, Jones AJ, Lavy CB, Puklavec M, Simmonds SJ. The fate of allogeneic and xenogeneic neuronal tissue transplanted into the third ventricle of rodents. Neuroscience. 1986;19(3):685-694. [DOI] [PubMed] [Google Scholar]
- 14. Murphy JB, Sturm E. Conditions determining the transplantability of tissues in the brain. J Exp Med. 1923;38(2):183-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stevenson PG, Hawke S, Sloan DJ, Bangham CR. The immunogenicity of intracerebral virus infection depends on anatomical site. J Virol. 1997;71(1):145-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aspelund A, Antila S, Proulx ST et al. . A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J Exp Med. 2015;212(7):991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Louveau A, Smirnov I, Keyes TJ et al. . Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carare RO, Bernardes-Silva M, Newman TA et al. . Solutes, but not cells, drain from the brain parenchyma along basement membranes of capillaries and arteries: significance for cerebral amyloid angiopathy and neuroimmunology. Neuropathol Appl Neurobiol. 2008;34(2):131-144. [DOI] [PubMed] [Google Scholar]
- 19. Tarasoff-Conway JM, Carare RO, Osorio RS et al. . Clearance systems in the brain-implications for Alzheimer disease. Nat Rev Neurol. 2015;11(8):457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szentistvanyi I, Patlak CS, Ellis RA, Cserr HF. Drainage of interstitial fluid from different regions of rat brain. Am J Physiol. 1984;246(6 pt 2):F835-, 844. [DOI] [PubMed] [Google Scholar]
- 21. Pallasch CP, Struss AK, Munnia A et al. . Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor-associated autoantibodies correlated with prolonged survival. Int J Cancer. 2005;117(3):456-459. [DOI] [PubMed] [Google Scholar]
- 22. Barcia C Jr, Gomez A, Gallego-Sanchez JM et al. . Infiltrating CTLs in human glioblastoma establish immunological synapses with tumorigenic cells. Am J Pathol. 2009;175(2):786-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berghoff AS, Kiesel B, Widhalm G et al. . Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dunn GP, Dunn IF, Curry WT. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun. 2007;7:12. [PMC free article] [PubMed] [Google Scholar]
- 25. Brooks WH, Markesbery WR, Gupta GD, Roszman TL. Relationship of lymphocyte invasion and survival of brain tumor patients. Ann Neurol. 1978;4(3):219-224. [DOI] [PubMed] [Google Scholar]
- 26. Han S, Zhang C, Li Q et al. . Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br J Cancer. 2014;110(10):2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sayour EJ, McLendon P, McLendon R et al. . Increased proportion of FoxP3+ regulatory T cells in tumor infiltrating lymphocytes is associated with tumor recurrence and reduced survival in patients with glioblastoma. Cancer Immunol Immunother. 2015;64(4):419-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang I, Tihan T, Han SJ et al. . CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17(11):1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yue Q, Zhang X, Ye HX et al. . The prognostic value of Foxp3+ tumor-infiltrating lymphocytes in patients with glioblastoma. J Neurooncol. 2014;116(2):251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lohr J, Ratliff T, Huppertz A et al. . Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-beta. Clin Cancer Res. 2011;17(13):4296-4308. [DOI] [PubMed] [Google Scholar]
- 31. Long DM. Capillary ultrastructure and the blood-brain barrier in human malignant brain tumors. J Neurosurg. 1970;32(2):127-144. [DOI] [PubMed] [Google Scholar]
- 32. Vajkoczy P, Menger MD. Vascular microenvironment in gliomas. J Neurooncol. 2000;50(1-2):99-108. [DOI] [PubMed] [Google Scholar]
- 33. Nduom EK, Yang C, Merrill MJ, Zhuang Z, Lonser RR. Characterization of the blood-brain barrier of metastatic and primary malignant neoplasms. J Neurosurg. 2013;119(2):427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong CS, Ho W, Piazza MG, Ray-Chaudhury A, Zhuang Z, Heiss JD. Characterization of the blood brain barrier in pediatric central nervous system neoplasms. J Interdiscip Histopathol. 2016;4(2):29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolburg H, Noell S, Fallier-Becker P, Mack AF, Wolburg-Buchholz K. The disturbed blood-brain barrier in human glioblastoma. Mol Aspects Med. 2012;33(5-6):579-589. [DOI] [PubMed] [Google Scholar]
- 36. Wolburg H, Wolburg-Buchholz K, Kraus J et al. . Localization of claudin-3 in tight junctions of the blood-brain barrier is selectively lost during experimental autoimmune encephalomyelitis and human glioblastoma multiforme. Acta Neuropathol. 2003;105(6):586-592. [DOI] [PubMed] [Google Scholar]
- 37. Proescholdt MA, Merrill MJ, Ikejiri B et al. . Site-specific immune response to implanted gliomas. J Neurosurg. 2001;95(6):1012-1019. [DOI] [PubMed] [Google Scholar]
- 38. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69-74. [DOI] [PubMed] [Google Scholar]
- 39. Rammensee HG, Singh-Jasuja H. HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev Vaccines. 2013;12(10):1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14(2):135-146. [DOI] [PubMed] [Google Scholar]
- 41. Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsushita H, Vesely MD, Koboldt DC et al. . Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Segal NH, Parsons DW, Peggs KS et al. . Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889-892. [DOI] [PubMed] [Google Scholar]
- 44. DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482(7385):405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boon T, Coulie PG, Van den Eynde BJ, van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:175-208. [DOI] [PubMed] [Google Scholar]
- 46. Hundal J, Carreno BM, Petti AA et al. . pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3(5):e28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gupta SK, Jaitly T, Schmitz U, Schuler G, Wolkenhauer O, Vera J. Personalized cancer immunotherapy using Systems Medicine approaches. Brief Bioinform. 2016;17(3):453-467. [DOI] [PubMed] [Google Scholar]
- 49. Hackl H, Charoentong P, Finotello F, Trajanoski Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nat Rev Genet. 2016;17(8):441-458. [DOI] [PubMed] [Google Scholar]
- 50. Pabinger S, Dander A, Fischer M et al. . A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2014;15(2):256-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCarthy DJ, Humburg P, Kanapin A et al. . Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6(3):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 2008;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang X, Yu X. An introduction to epitope prediction methods and software. Rev Med Virol. 2009;19(2):77-96. [DOI] [PubMed] [Google Scholar]
- 54. Hoof I, Peters B, Sidney J et al. . NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2(6):522-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Duan F, Duitama J, Al Seesi S et al. . Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211(11):2231-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Linnemann C, van Buuren MM, Bies L et al. . High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21(1):81-85. [DOI] [PubMed] [Google Scholar]
- 58. Wang RF, Wang HY. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 2017;27(1):11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kreiter S, Vormehr M, van de Roemer N et al. . Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tran E, Turcotte S, Gros A et al. . Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lu YC, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28(1):22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu YC, Yao X, Crystal JS et al. . Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20(13):3401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Polyakova A, Kuznetsova K, Moshkovskii S. Proteogenomics meets cancer immunology: mass spectrometric discovery and analysis of neoantigens. Expert Rev Proteomics. 2015;12(5):533-541. [DOI] [PubMed] [Google Scholar]
- 64. Yadav M, Jhunjhunwala S, Phung QT et al. . Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572-576. [DOI] [PubMed] [Google Scholar]
- 65. Bassani-Sternberg M, Braunlein E, Klar R et al. . Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun. 2016;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tran E, Robbins PF, Lu YC et al. . T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375(23):2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. www.clinicaltrials.gov. Accessed June 14, 2017. [Google Scholar]
- 68. Dunn GP, Rinne ML, Wykosky J et al. . Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Swartz AM, Li QJ, Sampson JH. Rindopepimut: a promising immunotherapeutic for the treatment of glioblastoma multiforme. Immunotherapy. 2014;6(6):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weller M, Butowski N, Tran D et al. . ACT IV: An international, double-blind, phase 3 trial of rindopepimut in newly diagnosed, EGFRvIII-expressing glioblastoma. Neuro Oncol. 2016;18(suppl 6):vi17-vi18. [DOI] [PubMed] [Google Scholar]
- 71. Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21(2):53-56. [DOI] [PubMed] [Google Scholar]
- 72. Sampson JH, Heimberger AB, Archer GE et al. . Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28(31):4722-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schuster J, Lai RK, Recht LD et al. . A phase II, multicenter trial of rindopepimut (CDX-110) in newly diagnosed glioblastoma: the ACT III study. Neuro Oncol. 2015;17(6):854-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yan H, Parsons DW, Jin G et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cancer Genome Atlas Research Network Brat DJ, Verhaak RG et al. . Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eckel-Passow JE, Lachance DH, Molinaro AM et al. . Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schumacher T, Bunse L, Pusch S et al. . A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324-327. [DOI] [PubMed] [Google Scholar]
- 78. Gubin MM, Zhang X, Schuster H et al. . Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Johanns TM, Ward JP, Miller CA et al. . Endogenous neoantigen-specific CD8 T cells identified in two glioblastoma models using a cancer immunogenomics approach. Cancer Immunol Res. 2016;4(12):1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Anagnostou V, Smith KN, Forde PM et al. . Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7(3):264-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Carreno BM, Magrini V, Becker-Hapak M et al. . Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Castle JC, Kreiter S, Diekmann J et al. . Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081-1091. [DOI] [PubMed] [Google Scholar]
- 83. Cohen CJ, Gartner JJ, Horovitz-Fried M et al. . Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015;125(10):3981-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Creaney J, Ma S, Sneddon SA et al. . Strong spontaneous tumor neoantigen responses induced by a natural human carcinogen. Oncoimmunology. 2015;4(7):e1011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gros A, Parkhurst MR, Tran E et al. . Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22(4):433-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Martin SD, Brown SD, Wick DA et al. . Low mutation burden in ovarian cancer may limit the utility of neoantigen-targeted vaccines. PLoS One. 2016;11(5):e0155189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. McGranahan N, Furness AJ, Rosenthal R et al. . Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parkhurst MR, Gros A, Pasetto A et al. . Isolation of T cell receptors specifically reactive with mutated tumor associated antigens from tumor infiltrating lymphocytes based on CD137 expression. Clin Cancer Res. 2017;23(10):2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Prickett TD, Crystal JS, Cohen CJ et al. . Durable complete response from metastatic melanoma after transfer of autologous T cells recognizing 10 mutated tumor antigens. Cancer Immunol Res. 2016;4(8):669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pritchard AL, Burel JG, Neller MA et al. . Exome sequencing to predict neoantigens in melanoma. Cancer Immunol Res. 2015;3(9):992-998. [DOI] [PubMed] [Google Scholar]
- 91. Rajasagi M, Shukla SA, Fritsch EF et al. . Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rizvi NA, Hellmann MD, Snyder A et al. . Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Robbins PF, Lu YC, El-Gamil M et al. . Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Snyder A, Makarov V, Merghoub T et al. . Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stronen E, Toebes M, Kelderman S et al. . Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352(6291):1337-1341. [DOI] [PubMed] [Google Scholar]
- 96. Tran E, Ahmadzadeh M, Lu YC et al. . Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van Rooij N, van Buuren MM, Philips D et al. . Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31(32):e439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Verdegaal EM, de Miranda NF, Visser M et al. . Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature. 2016;536(7614):91-95. [DOI] [PubMed] [Google Scholar]
- 99. Wick DA, Webb JR, Nielsen JS et al. . Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res. 2014;20(5):1125-1134. [DOI] [PubMed] [Google Scholar]
- 100. Woller N, Gurlevik E, Fleischmann-Mundt B et al. . Viral infection of tumors overcomes resistance to PD-1-immunotherapy by broadening neoantigenome-directed T-cell responses. Mol Ther. 2015;23(10):1630-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Le DT, Uram JN, Wang H et al. . PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Van Allen EM, Miao D, Schilling B et al. . Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Johnson DB, Frampton GM, Rioth MJ et al. . Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4(11):959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rosenberg JE, Hoffman-Censits J, Powles T et al. . Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Johanns TM, Miller CA, Dorward IG et al. . Immunogenomics of hypermutated glioblastoma: A patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6(11):1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bouffet E, Larouche V, Campbell BB et al. . Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206-2211. [DOI] [PubMed] [Google Scholar]
- 108. Erson-Omay EZ, Caglayan AO, Schultz N et al. . Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol. 2015;17(10):1356-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cahill DP, Levine KK, Betensky RA et al. . Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13(7):2038-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hunter C, Smith R, Cahill DP et al. . A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66(8):3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Johnson BE, Mazor T, Hong C et al. . Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kim J, Lee IH, Cho HJ et al. . Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318-328. [DOI] [PubMed] [Google Scholar]
- 113. Wang J, Cazzato E, Ladewig E et al. . Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]