Chronic lung infections with P. aeruginosa are the main cause of morbidity and mortality in patients with cystic fibrosis. This bacterium overproduces a capsular polysaccharide called alginate (also known as mucoidy), which aids in bacterial persistence in the lungs and in resistance to therapeutic regimens and host immune responses. The current study explores a previously unknown link between pyrimidine biosynthesis and mucoidy at the level of transcriptional regulation. Identifying/characterizing this link could provide novel targets for the control of bacterial growth and mucoidy. Inhibiting mucoidy may improve antimicrobial efficacy and facilitate host defenses to clear the noncapsulated P. aeruginosa bacteria, leading to improved prognosis for patients with cystic fibrosis.
KEYWORDS: AlgU (AlgT), alginate, cystic fibrosis (CF), mucoidy, Pseudomonas aeruginosa, RpoN, sigma factor competition, small-colony variant (SCV)
ABSTRACT
Mucoidy due to alginate overproduction by the Gram-negative bacterium Pseudomonas aeruginosa facilitates chronic lung infections in patients with cystic fibrosis (CF). We previously reported that disruption in de novo synthesis of pyrimidines resulted in conversion to a nonmucoid small-colony variant (SCV) in the mucoid P. aeruginosa strain (PAO581), which has a truncated anti-sigma factor, MucA25, that cannot sequester sigma factor AlgU (AlgT). Here, we showed that supplementation with the nitrogenous bases uracil or cytosine in growth medium complemented the SCV to normal growth, and nonmucoidy to mucoidy, in these mucA25 mutants. This conversion was associated with an increase in intracellular levels of UMP and UTP suggesting that nucleotide restoration occurred via a salvage pathway. In addition, supplemented pyrimidines caused an increase in activity of the alginate biosynthesis promoter (PalgD), but had no effect on PalgU, which controls transcription of algU. Cytosolic levels of AlgU were not influenced by uracil supplementation, yet levels of RpoN, a sigma factor that regulates nitrogen metabolism, increased with disruption of pyrimidine synthesis and decreased after supplementation of uracil. This suggested that an elevated level of RpoN in SCV may block alginate biosynthesis. To support this, we observed that overexpressing rpoN resulted in a phenotypic switch to nonmucoidy in PAO581 and in mucoid clinical isolates. Furthermore, transcription of an RpoN-regulated promoter increased in the mutants and decreased after uracil supplementation. These results suggest that the balance of RpoN and AlgU levels may regulate growth from SCV to mucoidy through sigma factor competition for PalgD.
IMPORTANCE Chronic lung infections with P. aeruginosa are the main cause of morbidity and mortality in patients with cystic fibrosis. This bacterium overproduces a capsular polysaccharide called alginate (also known as mucoidy), which aids in bacterial persistence in the lungs and in resistance to therapeutic regimens and host immune responses. The current study explores a previously unknown link between pyrimidine biosynthesis and mucoidy at the level of transcriptional regulation. Identifying/characterizing this link could provide novel targets for the control of bacterial growth and mucoidy. Inhibiting mucoidy may improve antimicrobial efficacy and facilitate host defenses to clear the noncapsulated P. aeruginosa bacteria, leading to improved prognosis for patients with cystic fibrosis.
INTRODUCTION
Pseudomonas aeruginosa, a Gram-negative opportunistic pathogen, is associated with chronic lung infections in patients with cystic fibrosis (CF) (1). There are several bacterial species that can colonize the lungs in children with CF; however, P. aeruginosa eventually emerges as the dominant pathogen as patients grow older. The persistence of this organism is aided by conversion to a mucoid phenotype, which renders the immune response and antimicrobial therapies less effective (2, 3). Alginate production is regulated by regulatory and biosynthetic operons. The major regulatory operon (algU-mucABCD) encodes an alternative sigma factor, AlgU (AlgT) (σ22), along with the cognate anti-sigma factor, MucA (4, 5). The alginate biosynthetic operon (algD) encodes most of the biosynthetic enzymes needed to convert fructose-1-phosphate into the exopolysaccharide alginate (6, 7). MucA sequesters AlgU to the inner membrane, and through proteolysis of MucA, AlgU is released into the cytoplasm to induce transcription at the promoters of both operons (8).
The alternative sigma factor RpoN (σ54) (9) regulates nitrogen metabolism in Pseudomonas (10–12), as well as the production of several virulence factors, such as those related to motility, quorum sensing, biofilm formation, and alginate production (10, 13–19). The two sigma factors AlgU and RpoN both direct transcription of the promoter for the alginate biosynthetic operon (PalgD), with the recognition sequences overlapping (20). This suggests that a dual regulation system is present at PalgD, in which either AlgU or RpoN activates alginate production under certain conditions (20). Damron et al. showed that deletion of RpoN in a kinB PAO1 mucoid mutant resulted in nonmucoidy and a growth-related phenotype known as the small-colony variant (SCV) (13). Interestingly, like mucoidy, the SCV is observed in CF patients with persistent infections and is associated with slow growth and an increase in antibiotics resistance (21–23).
The de novo pyrimidine biosynthesis pathway is an intricate pathway that begins with the conversion of l-glutamate to UMP, which is later used to synthesize UTP and CTP for RNA and DNA production (24). Pyrimidine synthesis is required for the regulation of several cell functions in P. aeruginosa. For example, strains of P. aeruginosa with mutations in the pyrimidine synthesis pathway exhibited deficiencies in production of biofilms and quorum-sensing signaling molecules, which were restored with supplementation of uracil to the medium (25–27). Another study showed that the activity of nucleotide diphosphate kinase (ndk) was increased in mucoid cells compared to that in nonmucoid cells (28). Additionally, the levels of UTP were also significantly higher in mucoid cells than nonmucoid cells (28–30). Yet to date, no work has specifically linked pyrimidine biosynthesis to SCV and alginate production in P. aeruginosa, and it remains unclear how the level of cytosolic nucleotides regulates alginate overproduction. Interestingly, CF sputum has been shown to contain DNA at 20 mg/ml due to the extracellular DNA (eDNA) present in the biofilms, as well as genomic DNA released when host cells are lysed (31).
In this study, we showed that loss of de novo pyrimidine biosynthesis inhibited mucoidy in the mucoid strain, PAO581 (PAO1 mucA25) (32, 33) and resulted in the SCV. Addition of uracil to the growth medium restored the colonies’ mucoid phenotype by restoring transcription at the alginate biosynthetic operon promoter (PalgD) and the normal colony morphology. We show that disruptions in the pyrimidine biosynthetic pathway disrupted nitrogen metabolism in the cell, which resulted in upregulation of rpoN. Consequently, increased levels of RpoN allowed for competition with AlgU for binding at PalgD, which resulted in inhibition of transcription of the alginate biosynthetic operon and thus the observed loss of alginate overproduction.
RESULTS
Supplementation with pyrimidines restores normal growth and mucoidy in a mucA25 mutant with mutations in the de novo pyrimidine biosynthetic pathway.
Previously, we reported that three mutations in de novo pyrimidine biosynthesis genes (carA, carB, and pyrD) in the mucoid strain PAO581 (PAO1 mucA25) each resulted in a SCV and nonmucoid phenotype (33). We thought this might be due to a link between pyrimidine and alginate biosynthesis in P. aeruginosa. To test this hypothesis, PAO581 and the three mutants, PAO581 carA, PAO581 carB, and PAO581 pyrD (all 3 mutants will be referred to collectively as PAO581pmt pyrimidine mutants throughout this paper), were grown with or without supplementation with pyrimidines. When grown on Pseudomonas isolation agar (PIA), the pyrimidine mutants remained nonmucoid and exhibited the SCV phenotype (Fig. 1A and B). However, supplementation with 0.1 mM uracil (Fig. 1A and C) or cytosine (see Fig. S1A in the supplemental material) restored mucoidy and normal colony morphology. We repeated the experiment using synthetic CF medium (SCFM2) (34), which is formulated to more closely represent the lung environment, and observed similar results (Fig. S2A). In addition, SCFM2 and PIA were comparable in inducing mucoidy in PAO581 and a mucoid CF isolate. (Fig. S2B and C). We used the carbazole assay to directly measure the amounts of alginate produced by the wild-type P. aeruginosa strain PAO1, PAO581, and three PAO581pmt mutants when grown on PIA or PIA supplemented with uracil or cytosine (Fig. 1D). Supplementation of the medium with either uracil or cytosine significantly increased alginate production in the pyrimidine mutant strains compared to alginate produced on PIA alone, while supplementation with adenine or guanine had no effect, i.e., the mutants remained SCV and nonmucoid. Clearly, addition of free pyrimidine to the growth medium restored the growth defect and loss of alginate production due to disruption in pyrimidine biosynthesis. These results suggest that pyrimidine biosynthesis may be linked to alginate synthesis.
FIG 1.
De novo pyrimidine biosynthesis is linked to the formation of SCV and mucoidy in P. aeruginosa. (A) PAO581 cells grown at 37°C for 48 h on PIA plates (left) and PIA plates with uracil (right). (B) The effect of disrupting the de novo pyrimidine pathway in a stable alginate-producing strain, P. aeruginosa PAO581. PAO581 carA, PAO581 carB, and PAO581 pyrD were grown on PIA plates at 37°C for 48 h. (C) The effect of adding uracil (0.1 mM) to the medium (PIA) on the growth of three pyrimidine biosynthetic mutants (carA, carB, and pyrD) at 37°C for 48 h. (D) Alginate production for PAO1, PAO581, PAO581 carA, PAO581 carB, and PAO581 pyrD when grown on PIA plates with and without uracil at 37°C for 24 h. Alginate was collected and measured using the standard carbazole assay. Values shown are mean alginate ± standard deviation from triplicate reads. (E) The β-galactosidase activity of PalgD measured using pLP170-PalgD-lacZ reporter constructs in PAO1, PAO581, PAO581 carA, PAO581 carB, and PAO581 pyrD grown on PIA plates containing carbenicillin and uracil at 37°C for 24 h. Relative expression mean values ± standard deviations from triplicate reads are shown. Two-way analysis of variance (ANOVA) was used with statistical significance at a P value of <0.01 (****, P < 0.0001).
Reduced levels of UMP and UTP are associated with SCV and loss of mucoidy in pyrimidine mutants.
The ability to restore the intracellular loss of pyrimidine synthesis with an extracellular source of uracil suggested that uracil was being transported into the cells. To test this, we measured intracellular concentrations of UMP and UTP. P. aeruginosa strains PAO1, PAO581, and the three PAO581pmt mutants were grown on PIA or PIA supplemented with uracil. The cytosolic fraction was collected by centrifugation and the amount of UMP/UTP in this fraction determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS). As expected, intracellular levels of both UMP (Fig. 2A) and UTP (Fig. 2B) increased in the PAO581pmt pyrimidine mutants grown on PIA containing uracil. Yet, no statistically significant changes were observed in our control strains, PAO1 and PAO581. This increase in UMP/UTP was correlated with an increase in alginate production, as determined by the carbazole assay. This suggested that uracil was imported into cells and recycled by the pyrimidine salvage pathway, thereby relieving the inhibitory effect of the mutations on the growth and alginate synthesis.
FIG 2.
Disruption of de novo pyrimidine biosynthesis results in reduction of intracellular UMP and UTP, and supplementation of uracil to the medium restores the levels of UMP and UTP in P. aeruginosa. P. aeruginosa strains were grown on PIA at 37°C for 24 h. The cells of each type in triplicate were harvested, washed, and resuspended in PBS. The cell-free lysates were used for measurement of intracellular concentration of UMP (A) and UTP (B) measured using LC-MS/MS. Two-way ANOVA was used with statistical significance at P < 0.01 (*, P < 0.01; **, P < 0.001; ***, P < 0.0001).
Uracil-mediated alginate activation through modulation of algD promoter activity (PalgD).
To identify how uracil affected alginate production, we used the previously published reporter promoter constructs PalgU_lacZ and PalgD_lacZ in pLP170 to quantify promoter activity at the alginate regulatory operon and the alginate biosynthetic operon, respectively (35). After introduction of the constructs into PAO1, PAO581, and our three PAO581pmt mutants, the strains were grown on either PIA or PIA supplemented with uracil, and we used the β-galactosidase activity assay to quantify the activity of both PalgU (Fig. S1B) and PalgD (Fig. 1E). The promoter activity at PalgU was not affected by the disruption of pyrimidine biosynthesis nor by the addition of uracil to the medium. Conversely, the activity at PalgD in the three PAO581pmt mutants was relatively low when grown on PIA, but promoter activity was increased upon addition of uracil to the medium. Thus, loss of pyrimidine biosynthesis decreased promoter activity at the alginate biosynthesis operon (algD operon) but not at the regulatory operon (algU operon).
Inactivation of the de novo pyrimidine biosynthetic pathway increases RpoN (σ54).
Pyrimidine starvation increases the levels of an alternative sigma factor, RpoN, due to the disruption of nitrogen metabolism (11, 12, 36). We hypothesized that an increase in RpoN might disrupt the balance between sigma factors RpoN and AlgU. Since RpoN and AlgU have overlapping binding sites at PalgD, RpoN could now outcompete AlgU for binding to PalgD, leading to loss of alginate biosynthesis (20). To test whether loss of pyrimidine biosynthesis affected sigma factor levels, we performed Western blotting to analyze the intracellular amounts of RpoN and AlgU (Fig. 3 and 4). In the PAO581 pyrimidine mutant strains, RpoN was present at relatively higher levels than in PAO581, and addition of uracil to the medium decreased RpoN. On the other hand, AlgU levels in the pyrimidine mutants (PAO581pmt) were similar to those in the parent PAO581 strain, and upon the addition of uracil, AlgU remained relatively consistent. Thus, loss of de novo pyrimidine biosynthesis resulted in the upregulation of RpoN, and adding uracil to the medium reduced RpoN levels but did not affect the levels of AlgU.
FIG 3.

The effect of the de novo pyrimidine pathway on intracellular levels of RpoN. RpoN levels from PAO581, PAO581 carA, PAO581 carB, and PAO581 pyrD grown on PIA plates and PIA plates with uracil at 37°C for 24 h from three separate Wes runs using anti-RpoN antibody normalized by total protein levels. Values shown are mean ratios ± standard deviations. Two-way ANOVA was used with statistical significance at P < 0.01 (*, P < 0.01; **, P < 0.001; ****, P < 0.0001).
FIG 4.

Disruption of de novo pyrimidine biosynthetic pathway does not affect the levels of free AlgU in the cytosol. AlgU levels from PAO581, PAO581 carA, PAO581 carB, and PAO581 pyrD grown on PIA plates and PIA plates with uracil at 37°C for 24 h from three separate Wes runs using anti-AlgU antibody normalized by total protein levels. Values shown are mean ratios ± standard deviations. Two-way ANOVA was used with statistical significance at P < 0.01 (*, P < 0.01).
Transcription of PpilA is altered by disruption in pyrimidine biosynthesis due to increased RpoN levels.
Previously, Withers et al. showed that transcription of PpilA is regulated by RpoN (35). To test whether the pyrimidine biosynthesis is linked to RpoN regulation, we measured the promoter activity of PpilA_lacZ in pLP170 (35) in PAO1, PAO581, and our three PAO581pmt mutants (Fig. 5). A significant increase in PpilA promoter activity was observed in the mutants compared to that in PAO581. When the PAO581pmt strains were grown on uracil-supplemented medium the promoter activity decreased and was similar to that in PAO581 strains on PIA with uracil. This supports our above results that cytosolic RpoN levels are increased in the mutants, which in turn leads to inhibition of alginate biosynthesis.
FIG 5.
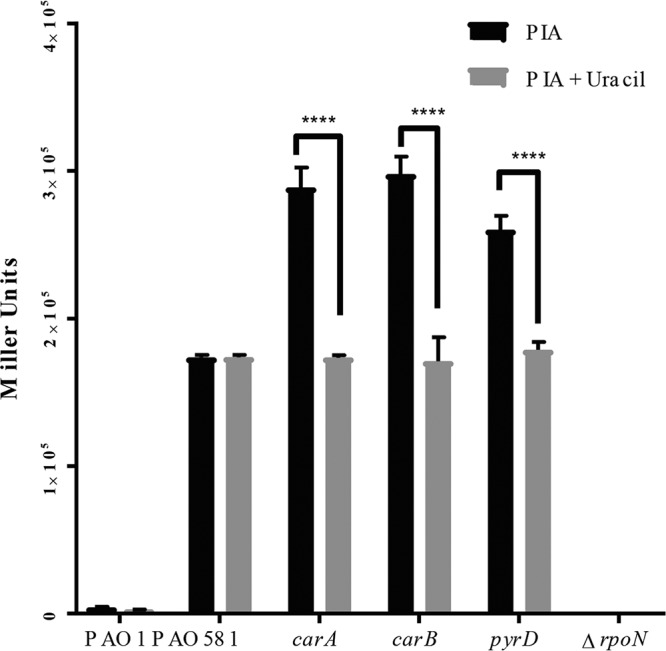
Transcriptional activity of RpoN-dependent type IV pilin promoter PpilA was increased in pyrimidine biosynthesis mutants compared to the parent PAO581. The β-galactosidase activity of PpilA measured using pLP170-PpilA-lacZ reporter constructs in PAO1, PAO581, PAO581 carA, PAO581 carB, and PAO581 pyrD grown on PIA plates containing carbenicillin and uracil at 37°C for 24 h. Relative expression mean values ± standard deviations from triplicate reads are shown. Two-way ANOVA was used with statistical significance at P < 0.01 (****, P < 0.0001).
Overexpression of RpoN in PAO581 reduced transcriptional activity at PalgD but not at PalgU.
Previously, Yin et al. (37) showed that overexpression of rpoN in PAO581 resulted in the loss of mucoidy, suggesting there is sigma factor competition between AlgU and RpoN. Boucher et al. showed that RpoN has a binding sequence at PalgD, and inactivation of RpoN in the strain carrying muc23 resulted in loss of transcription from PalgU (20). To test how RpoN affects alginate production, we overexpressed RpoN and measured promoter activity at the regulatory and biosynthetic alginate operons. To do this, we conjugated the pHERD20T vector carrying rpoN into PAO581 with PalgU-lacZ and PalgD-lacZ inserted at the CTX site on the chromosome (38) (Fig. 6B and C). The activity of both promoters was tested when PAO581 was grown on PIA or PIA supplemented with arabinose at increasing concentrations (0.05%, 0.1%, and 0.5%) to induce rpoN expression. Activity at PalgU was not affected by the overexpression of rpoN. However, the activity at PalgD decreased in a concentration-dependent manner as the RpoN levels increased in the cell. This suggested that increased levels of RpoN may lead to increased RpoN binding to PalgD and loss of the mucoid phenotype in PAO581. To test whether the loss of mucoidy observed with overexpression of RpoN might be a strain-specific effect, we overexpressed RpoN in PAO579 (muc23) and in two stable mucoid CF isolates, CF10 and C7447 (Fig. S3). All three strains exhibited reduction in mucoidy similar to our observations in PAO581.
FIG 6.
Overexpression of RpoN suppresses mucoidy through interference with PalgD but not PalgU in P. aeruginosa PAO581. (A) Image of PAO581 carrying empty vector pHERD20T and pHERD20T-rpoN grown on a PIA plate with carbenicillin and 0.1% arabinose at 37°C for 24 h. (B) The β-galactosidase activity of PalgU and of (C) PalgD using PalgU-lacZ and PalgD-lacZ reporter system shuttled in the chromosome of PAO581 via mini-CTX. These two strains were introduced pHERD20T (control) and pHERD20T-rpoN. Strains were grown on PIA plates with tetracycline, carbenicillin, and various concentrations of arabinose (0.05%, 0.1%, and 0.5%) at 37°C for 24 h. Values shown are mean relative expression ± standard deviation from triplicate reads. One-way ANOVA was used with statistical significance at P < 0.01.
Pyrimidine mutants can become mucoid through overexpression of AlgU.
To investigate if the PAO581pmt mutants can still produce alginate regardless of the mutations in the pyrimidine pathway and overcome the higher levels of RpoN, algU was overexpressed in PAO1, PAO581, and the three PAO581pmt pyrimidine mutants using the pHERD20T-algU plasmid. After growing the PAO581pmt pyrimidine mutants carrying the pHERD20T_algU plasmid on PIA supplemented with 0.1% arabinose, the mucoid phenotype was restored without the need to supplement the medium with pyrimidines (Fig. 7). This shows that by overexpressing algU, mucoidy can be returned regardless of pyrimidine starvation and suggests a restoration in the balance of RpoN and AlgU.
FIG 7.
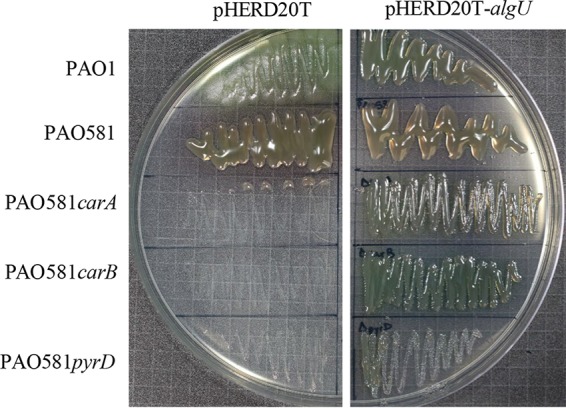
Overexpressing algU leads to mucoidy in the pyrimidine biosynthesis mutants. Shown is the effect of overexpressing algU on the growth of PAO1, PAO581, PAO581 carA, PAO581 carB, and PAO581 pyrD carrying empty vector pHERD20T and pHERD20T-algU and grown on PIA with carbenicillin and 0.1% arabinose at 37°C for 24 h.
Generation of in-frame deletions of pyrD in PAO1 and PAO581.
Our work showed that disruption in de novo pyrimidine synthesis results in the SCV phenotype. To identify if the transposon disruption of the pyrimidine pathway was polar, we in-frame deleted the pyrD gene in PAO1 and PAO581, resulting in the SCV for PAO1 and nonmucoidy and the SCV for PAO581. The addition of free uracil to PAO1 ΔpyrD results in the return of the normal phenotype of PAO1 but does not induce mucoidy (Fig. 8A and B). Similar to PAO581 pyrD, PAO581 ΔpyrD also becomes mucoid when grown on PIA with uracil (Fig. 8C and D). These results show that the phenotype of the transposon mutants was true and was unrelated to the method by which the mutants were generated.
FIG 8.

In-frame deletion of pyrD results in SCV in PAO1, and SCV and nonmucoid phenotype in PAO581. (A and B) PAO1 and PAO1 ΔpyrD grown on PIA plates (A) and PIA plates with uracil (B) at 37°C for 24 h. (C and D) PAO581 and PAO1 ΔpyrD grown on PIA plates (C) and PIA plates with uracil (D) at 37°C for 48 h.
DISCUSSION
The presence of alginate in the lungs of CF patients hinders the function of the patient’s immune system and the efficacy of antimicrobial therapy. A better understanding of the regulation of alginate production would aid in development of pharmaceuticals that target the process and eventually eliminate the sources of chronic lung infections. Our previous work (33) showed that loss of de novo pyrimidine biosynthesis resulted in loss of mucoidy in an otherwise mucoid P. aeruginosa strain, PAO581. There has been little work investigating the relationship between de novo pyrimidine biosynthesis and alginate biosynthesis. Here, we aimed to map the link between these pathways to identify new targets for antialginate therapies.
In this study, we found that the addition of free uracil or cytosine to the medium restored the mucoid phenotype in PAO581 pyrimidine mutants, likely through the pyrimidine salvage pathway. This effect appears to be pyrimidine specific, as the addition of purine bases to the medium had no effect on the phenotype of these pyrimidine mutant strains. To show that the addition of uracil is linked to the return of mucoidy by stimulating the pyrimidine salvage pathway, we used LC-MS/MS to measure the intracellular UMP and UTP concentrations. UMP is the common end product of both de novo and salvage pathways. Intracellular concentrations of UMP in the three pyrimidine mutants was lower than that in PAO581 when grown on PIA; however, when the medium was supplemented with uracil, intracellular UMP and UTP concentrations were restored to PAO581 levels. This indicated that the addition of uracil stimulated the pyrimidine salvage pathway, which in turn restored the mucoid phenotype. This result is consistent with early observations that mucoid strains were associated with higher levels of intracellular nucleotides (28, 29).
Alginate biosynthesis in P. aeruginosa occurs via two main operons, the regulatory algU-mucABCD and the biosynthetic algD operons. We used a promoter reporter fusion plasmid to measure transcriptional activities from both operons to identify the level at which the loss of pyrimidine biosynthesis affected the alginate pathway. The promoter activity of the algU operon was not affected by the loss of pyrimidine synthesis, nor was the activity changed when the strains were grown in the presence of uracil. Therefore, regulation of the algU promoter is not affected by the intracellular pyrimidine concentration. Importantly, the normal algU promoter activity showed that loss of pyrimidine biosynthesis does not cause a universal decrease in transcription, which would be expected to cause a reduction in alginate production. On the other hand, PalgD exhibited a significant decrease in activity in the three PAO581 mutants compared to that in the parent strain. When uracil was added to the medium, promoter activity was restored to levels comparable to the activity observed in PAO581. This increase in promoter activity was concurrent with the return of mucoidy in all three pyrimidine mutant strains. This suggests a direct relationship between intracellular pyrimidine concentration and PalgD activity.
Boucher et al. found that AlgU and RpoN have overlapping binding sequences at PalgD, and that overexpression of rpoN inhibited mucoidy in PAO578 (20). However, the muc23 mutation was later characterized to be pilA108, which produces a truncated type IV pilin with an FTF tail at the C terminus of PilA108 to activate AlgW, which initiates the proteolytic cascade to degrade the full length of MucA (39). Similarly, Yin et al. reported that overexpression of rpoN using pHERD20T resulted in the loss of mucoidy in PAO581 (37). RpoN is a sigma factor that regulates expression of numerous important genes that control virulence, stress response, motility, and metabolism, among other functions (10, 15, 17–19, 40). Our Western blot analysis results showed that the levels of RpoN increased in the PAO581pmt mutants and were restored to PAO581 levels after the addition of uracil. On the contrary, the levels of AlgU remained unchanged before and after uracil induction in the PAO581pmt mutants. This suggests that RpoN binding to the promoter was probably the major inhibitory factor in the loss of mucoidy in the pyrimidine mutants. However, proteolysis of MucA25 is significant in the augmentation of alginate in the parent strain, because the alginate production was increased significantly when uracil was used. It is likely that disruption in pyrimidine de novo synthesis results in disruption in the nitrogen metabolism in the cell, which is regulated by RpoN (11, 12, 36). To resolve this disruption, rpoN is upregulated in the mutants, as verified by our Western blot analyses. This increase in RpoN disrupts the RpoN/AlgU balance, leading to competition with AlgU for binding at PalgD. Moreover, the effect of the increased RpoN levels is evident by the increased promoter activity of PpilA in the pyrimidine mutants, which is restored to levels comparable to those of the wild-type PAO581 strain when uracil is added to the medium (Fig. 5). With the addition of free uracil to the medium, the mucoid phenotype returned, along with a decrease in RpoN levels and an increase in UMP/UTP. So, addition of uracil to the medium restores intracellular pyrimidine levels by supplementing the salvage pathway, and it relieves the stress presented on the nitrogen cycle. Moreover, upon overexpression of algU using pHERD20T, we observed a return of the mucoid phenotype without the need to supplement the medium with uracil. This shows that by artificially increasing the levels of AlgU in the cell, it can outcompete RpoN for the PalgD site, which is shown by the increase of PalgD promoter activity and the consequent return of mucoidy. These results show that there is an important balance between AlgU and RpoN levels in Pseudomonas species, with AlgU at those normal levels being the dominant sigma factor (41) binding and activating PalgD. By deleting the pyrimidine biosynthesis enzymes, RpoN is overexpressed to overcome the lower levels of pyrimidine, thereby becoming the dominant sigma factor due to its higher levels.
In-frame deletion of pyrD in PAO1 resulted in the SCV, and uracil restored the normal colony morphology. In PAO581, a similar deletion resulted in both the SCV and nonmucoidy, and the addition of uracil restored both phenotypes. In the case of PAO1 ΔpyrD, addition of uracil only returns the SCV phenotype to the normal phenotype because mucA is still wild type. These details show that two separate phenotypes are found with these gene deletions. The first is the SCV, due to pyrimidine starvation resulting in overexpression of rpoN. The second phenotype is the loss of mucoidy due to the increased levels of RpoN that competes with AlgU for binding to PalgD.
Antibiotic sensitivities were tested for the PAO581pmt mutants using the standard disc diffusion assay on PIA and PIA with uracil (see Tables S1 and S2 in the supplemental material). Our results showed that, compared to PAO581, the PAO581pmt mutants were statistically more sensitive to all eight antibiotics tested (aztreonam, meropenem, imipenem, ceftazidime, cefepime, levofloxacin, ciprofloxacin, and tobramycin) (Table S1). However, addition of uracil cannot restore the level of antibiotic sensitivity despite the restoration of the normal growth rate in mutants (Table S2). This suggests that the pyrimidine biosynthetic enzymes may have a previously unknown function that mediates resistance to antibiotics. Further studies need to be performed to test this possibility.
Our results showed that free uracil induced mucoidy in PAO581pmt pyrimidine mutants through mechanisms independent of PalgU transcriptional activity. Therefore, the activation of AlgU to induce mucoidy is likely posttranslational. AlgU is regulated posttranslationally through the proteolytic degradation of MucA, and PAO581 has a truncated MucA variant, MucA25 (33), which is rapidly degraded by the intracellular protease ClpXP. While adding uracil to the medium induced a higher level of AlgU in the parent PAO581, which corresponds to more alginate production (Fig. 1D), there was no statistically significant difference in the levels of AlgU between untreated and treated mutants. This result seems to indicate that alteration of proteolytic degradation of MucA25 in the PAO581pmt mutants was less likely. It is more likely that loss of RpoN-mediated inhibition at PalgD is responsible for uracil-mediated alginate induction in the mutants. Another explanation is that RpoN has a secondary effect on mucoidy. Overexpressing RpoN results in loss of PalgD activity and, subsequently, loss of mucoidy; this may be due to an inhibitor regulated by RpoN that is causing the observed effects rather than to a direct inhibition exhibited by RpoN.
Our work demonstrates that there is an indirect link between de novo pyrimidine synthesis and alginate biosynthesis through regulation of sigma factor levels. The disruption of the de novo pyrimidine pathway results in increased levels of RpoN and causes a loss of mucoidy in PAO581. Addition of free uracil to the medium results in a return to normal pyrimidine levels, causing a decrease in the RpoN levels and thereby a return of mucoidy. Similarly, overexpression of algU causes the balance between AlgU and RpoN to shift to favor AlgU and the mucoid phenotype (Fig. 9). Overall, our findings demonstrate that both sigma factors need to exist in a balanced state to maintain the desired phenotype. A possible explanation of the role of the dual regulation system at PalgD is to regulate cellular energy under stress toward essential pathways necessary for cellular survival and away from the energy expensive alginate synthesis.
FIG 9.
Schematic showing the link between de novo pyrimidine synthesis, SCV, and alginate biosynthesis through sigma factors RpoN and AlgU competition for PalgD. (A) Disruption in the pyrimidine de novo biosynthetic pathway results in decreased levels of UMP. This causes an increase in RpoN that competes with AlgU to bind to the algD promoter (PalgD), thereby inhibiting alginate production. (B) The addition of free uracil to the medium causes the levels of UMP to increase through the salvage pathway and thereby decrease the levels of RpoN, which allows AlgU to bind to the algD promoter (PalgD); this results in alginate production. Moreover, overexpression of algU using pHERD20T and arabinose results in increased AlgU in the cell that outcompetes RpoN and results in alginate production.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains used in this study are listed in Table 1. P. aeruginosa strains were all grown on Pseudomonas isolation agar (PIA) plates (Difco, Detroit, MI) or Pseudomonas isolation broth (PIB; Difco, Detroit, MI) at 37°C, prepared by adding 20 ml of glycerol/liter of medium as per manufacturer recommendation. PIA was supplemented with 300 μg/ml of carbenicillin (RPI, Mt. Prospect, PA), 0.1%/1% arabinose (MP Biomedicals, Solon, OH) by weight, 10% sucrose (Fisher Scientific, Waltham, MA) by weight, or 0.1 mM nitrogenous bases (adenine, cytosine, guanine, and uracil; Arcos Organics, Morris, NJ) when necessary. All Escherichia coli strains were grown in Lennox broth (LB) (Difco, Detroit, MI) or on LB agar plates and supplemented with 100 μg/ml of carbenicillin or 50 μg/ml of kanamycin (RPI, Mt. Prospect, PA) when needed. Synthetic CF medium (SCFM2) was prepared according to the protocol cited by Turner et al. (34), with 0.1 mM uracil (11 μg/ml) supplemented when necessary.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype, phenotype, or descriptiond | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Prototroph, NM | P. Phibbsa |
| PAO579 | PAO381 derivative with muc-23 (pilA23 resulting in PilA108), M | J. Govanb |
| PAO581 | PAO1 mucA25, M | J. Govanb |
| PAO581 carA::aacC1 | PAO581DR68/PAO581 carA::Gmr, NM, SCV | 33 |
| PAO581 carB::aacC1 | PAO581DR26/PAO581 carB::Gmr, NM, SCV | 33 |
| PAO581 pyrD::aacC1 | PAO581DR50/PAO581 pyrD::Gmr, NM, SCV | 33 |
| PAO581 algD::aacC1 | PAO581DR1/PAO581 algD::Gmr, NM | 33 |
| PAO1 ΔpyrD | PAO1 with in-frame deletion of pyrD, NM, SCV | This study |
| PAO581 ΔpyrD | PAO581 with in-frame deletion of pyrD, NM, SCV | This study |
| CF001 | CF isolate, M | M. Ansteadc |
| CF10 | CF isolate, M | 42 |
| C7447 | CF isolate, M | 43 |
| E. coli TOP10 | DH5α derivative | Invitrogen |
| Plasmids | ||
| pRK2013 | Kmr Tra Mob ColE1 | 44 |
| pHERD20T | pUCP20T Plac replaced with 1.3-kb AflIII-EcoRI fragment of araC-PBAD cassette | 45 |
| pHERD20T-algU | algU (PA0762) from PAO1 in pHERD20T (pBAD/pUCP20), XbalI/HindIII | 33 |
| pHERD20T-rpoN | rpoN (PA4462) from PAO1 in pHERD20T, XbaI/HindIII | 38 |
| pEX100T-NotI | Suicide vector with NotI restriction site fused into SmaI of pEX100T, sacB oriT Cbr | 38 |
| pEX100T-NotI-ΔpyrD | 1.2-kb fragment flanking pyrD (PA3050) gene ligated into pEX100T-NotI with in-frame deletion of pyrD | GeneScript |
| pLP170 | 8.3 kb, promoterless lacZ, Apr, multiple cloning site | 46 |
| pLP170-PalgU | Complete PalgU promoter (541 bp upstream of ATG) fused with lacZ in pLP170, BamHI/HindIII | 35 |
| pLP170-PalgD | Complete PalgD promoter (989 bp upstream of ATG) fused with lacZ in pLP170, BamHI/HindIII | 35 |
| pLP170-PpilA | Complete PpilA promoter (500 bp upstream of ATG) fused with lacZ in pLP170, BamHI/HindIII | 35 |
| MiniCTX-lacZ | Gene delivery vector for inserting genes at the CTX phage att site on the P. aeruginosa chromosome; Tcr | 47 |
| MiniCTX-PalgU-lacZ | Complete PalgU promoter (541 bp upstream of ATG) EcoRI/HindIII fused with lacZ for integration at the CTX phage att site in P. aeruginosa |
38 |
| MiniCTX-PalgD-lacZ | Complete PalgD promoter (1,525 bp upstream of ATG) HindIII/BamHI fused with lacZ for integration at the CTX phage att site in P. aeruginosa |
38 |
P. Phibbs, East Carolina University, Greenville, NC.
J. Govan, University of Edinburgh, Scotland, United Kingdom.
M. Anstead, University of Kentucky Department of Pediatrics, Lexington, KY.
NM, nonmucoid; M, mucoid; SCV, small-colony variant.
Genomic DNA, plasmid and protein extractions.
Bacterial genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Germantown, MD) from 1 × 109 CFU/ml of cells grown in PIB or LB broth at 37°C for 24 h. Plasmids were extracted using the QIAprep spin miniprep kit (Qiagen, Germantown, MD) from 1 × 108 CFU/ml of cells grown in LB containing the necessary selective antibiotics at 37°C for 24 h. The plasmid DNA was extracted using the provided protocol. DNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Whole-cell proteins were extracted using the NoviPure microbial protein kit (Mo Bio, Carlsbad, CA; Qiagen, Germantown, MD). Cells were grown on PIA plates containing the necessary selection medium supplement. Plates were scraped into 1× phosphate buffered saline (PBS), and 1 × 108 CFU/ml of cells were pelleted and used to extract the whole-cell protein, using the protocol provided with the kit. Proteins were eluted in 100 μl of PE elution buffer. Protein concentration was measured using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA) and the optical density (OD) read on the SpectraMax i3x (Molecular Devices, Downingtown, PA) at 562 nm. All extracted samples were stored at −20°C.
PCR.
All PCRs were run using the Hot Start Taq 2× master mix (New England Biolabs, Ipswich, MA). A total of 25 μl of the master mix was mixed with 1 μl of each primer and 23 μl of DNA plus DNase/RNase-free water. Thermocycling reaction conditions were 95°C for 30 s for initial denaturation, 30 cycles of 95°C for 30 s, 58°C for 30 s, and 68°C for 60 s, and a final extension of 68°C for 5 min, and then samples were held at 4°C.
Conjugation.
Pseudomonas strains that were used to acquire the plasmid were grown from a purified culture in LB broth at 42°C for 24 h. The plasmid-containing E. coli was grown in LB broth at 37°C for 24 h. A total of 300 μl of each of the three strains [donor strain containing the plasmid, E. coli(pRK2013), and recipient strain] was mixed in a 1.7-ml sterile microcentrifuge tube. The tube was vortexed on high for 15 s using a benchtop vortex. The tube was then centrifuged at 17,900 × g for 3 min to obtain a cell pellet. The supernatant was discarded, and the pellet was resuspended in the remaining 50 μl of supernatant and was then blotted on an LB plate that was dried for 30 min at 37°C. The LB plate containing the blot was left face up to dry under a hood; once dry, the LB plate was covered and placed in the incubator face down at 37°C for 6 h. Half of the blot was streaked using an inoculating loop on PIA plates containing selective antibiotics (300 μg/ml carbenicillin with or without 150 μg/ml gentamicin, depending on the strain) and allowed to incubate at 37°C for 24 h. Single colonies were selected from the PIA plate and isolated twice on selective PIA plates to obtain single-colony isolates.
Transformation.
Transformations were performed with TOP10 electrocompetent cells (Invitrogen, Carlsbad, CA). A total of 500 ng of plasmid was mixed with 50 μl of E. coli cells (TOP10) that were thawed on ice and mixed. Samples were electrically shocked and 250 μl of super optimal broth with catabolite repression (SOC) medium was added to the cells and allowed to shake at 37°C for 1 to 2 h in a 1.7-ml elution tube. Aliquots of 10 μl and 50 μl and the pellet were then spread on Luria agar plates containing 100 μg/ml of carbenicillin and grown at 37°C for 24 h.
In-frame gene deletion.
In-frame deletions were conducted using pEX100T-ΔpyrD through a two-step allelic exchange procedure (38). Single-crossover merodiploid strains were selected based on sensitivity to sucrose (sacB) and resistance to carbenicillin. Selected merodiploid strains where then grown in LB broth at 37°C. Double-crossover strains were selected based on sensitivity to carbenicillin and confirmed through PCR amplification of the flanking region of the target gene.
Alginate assay.
The alginate assay is also referred to in this study as the carbazole assay. Strains were grown in PIB at 37°C to a McFarland turbidity of 0.5 (1.5 × 108 CFU/ml), 450 μl of the broth was then spread on 150-mm by 10-mm petri dishes (Fisherbrand, Waltham, MA) containing 50 ml of PIA or 50 ml of PIA containing the respective supplement (0.1 mM nitrogenous bases or specified concentration of 5-FU). Plates were grown at 37°C for 24 h. Using 30 ml of 0.85% NaCl solution, the colonies were scraped and placed in a 50-ml conical tube on ice. Sulfuric acid-borate solution (18 M) was aliquoted (3 ml) into culture tubes and placed on ice. The collected sample (350 μl) was added in triplicates to culture tubes containing the sulfuric acid-borate solution and then briefly vortexed. A total of 100 μl of 0.1% carbazole (Sigma-Aldrich, St. Louis, MO) in ethanol was added to the tubes and then vortexed for 5 s. Tubes were then incubated at 55°C for 30 min. The optical density at 600 nm (OD600) of the initial collected samples and OD530 of the culture tubes were read. Using a standard curve, the total alginate concentration per OD600 was then calculated using the formula (alginate concentration)/OD600.
β-Galactosidase activity assay.
Strains carrying the lacZ fusion were streaked on PIA or PIA supplemented with 0.1 mM uracil at 37°C for 24 h. The colonies were then scraped into 4 ml 1× PBS and then diluted to an OD600 of 0.3 to 0.7. Triplicates of 100 μl of the sample were added to 900 μl of Z-buffer and 20 μl toluene in a 1.5-ml elution tube. After mixing by inverting 4 or 5 times, tubes were placed with tops open in a shaking incubator at 37°C for 40 min. After 200 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg/ml; Thermo Scientific, Waltham, MA) was added, and the time of color change was recorded, the reaction was stopped by adding 500 μl of 1 M Na2CO3 (Fisher Scientific, Waltham, MA) after 20 min. OD420 and OD550 were measured using a SpectraMax i3x plate reader (Molecular Devices, Downingtown, PA). Miller units were calculated using the following formula:
Liquid chromatography-mass spectrometry measuring intracellular levels of UMP/UTP.
All strains were grown on PIA or PIA supplemented with 0.1mM uracil at 37°C for 24 h. Using 1× PBS, the colonies were scraped off the plates and centrifuged at 6,800 × g for 5 min to obtain 100 mg pellets, which were shipped on dry ice to the Proteomics and Mass Spectrometry Facility at the Danforth Plant Science Center (St. Louis, MO). A total of 10 samples in triplicate were analyzed using LC-MS. The samples were extracted in 200 μl of 80% methanol containing 10 μM the internal standards, 15N2, 13C9 UMP, and 15N2, 13C9 UTP, by vortexing for 10 min. Samples were centrifuged and then filtered through 8.0 μm polyethersulfone (PES) spin filters to remove particulates. Samples (1 μl) were loaded onto a 0.5- by 100-mm custom-packed Proteomix-SAX column using 250 mM ammonium carbonate in water (A) and 25% methanol (B). The gradient started at 100% B for 3 min, followed by a ramp to 100% B over 10 minutes, with a hold at 100% B for a minute and then ramp back down to 100% B over 1 min. The column was then reequilibrated for 15 minutes. Data were recorded in negative ion mode using a Q-Exactive mass spectrometer set to a resolution setting of 140,000 (at m/z 200), with a scan range of 200 to 700. The data were integrated using the Quan Browser application of Xcalibur. The concentrations were calculated based on comparison to the internal standards and plotting against a calibration curve made from various concentration of unlabeled standards and a constant concentration of isotope-labeled standards (10 μM).
Western blot analysis.
Whole-cell lysates extracted using a NoviPure microbial protein kit were used to run Western blots on the Wes system (ProteinSimple, San Jose, CA). Samples were diluted to 250 μg/ml using 0.1× sample buffer and mixed with 5× fluorescent master mix at a 4:1 ratio and denatured at 95°C for 5 min. Samples were then loaded (3 μl) into the Wes plate (Wes 12- to 230-kDa prefilled plate), with the first lane containing 5 μl of the biotinylated protein ladder provided along with blocking reagent, primary antibodies, ProteinSimple horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody, luminol peroxide, and washing buffer. The primary antibodies used were anti-AlgU (6C6) (5) and anti-RNA σ54 (RpoN) (BioLegend, San Diego, CA) mouse IgG monoclonal antibodies at dilutions of 1:20 and 1:1,000, respectively. The Wes plate was run using the Wes default settings: 27-min separation time at 375 V, blocking with Antibody Diluent 2 (ProteinSimple) for 5 min (30 min for total protein assay), primary antibody for 30 min, secondary antibody for 30 min, and exposure times ranging from 1 to 120 s. Gel images were generated and results were analyzed on the Compass for Simple Western software, version 3.1.7 (ProteinSimple, San Jose, CA).
Statistical analysis.
The unpaired two-tailed Student t test and one-way and two-way analysis of variance (ANOVA) were run with statistical significance set at a P value of <0.01. All statistical analysis was done using Prism version 7.02 for Windows (GraphPad Software, La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grants P20GM103434 and 1R43GM113545-01A1. H.D.Y. is also the cofounder and chief science officer (CSO) of Progenesis Technologies, LLC.
We thank Richard Niles and Meagan Valentine from Progenesis Technologies and Lexie Blalock from the Marshall University Joan C. Edwards School of Medicine for critical reviews of the manuscript. We thank Michael J. Schurr from the University of Colorado for constructive comments on this work.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00575-18.
REFERENCES
- 1.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoiby N. 2002. Understanding bacterial biofilms in patients with cystic fibrosis: current and innovative approaches to potential therapies. J Cyst Fibros 1:249–254. doi: 10.1016/S1569-1993(02)00104-2. [DOI] [PubMed] [Google Scholar]
- 3.Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 4.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehm BHA. 2009. Alginate production: precursor biosynthesis, polymerization and secretion, p 55–71. In Rehm BHA. (ed), Alginates: biology and applications, Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- 7.Remminghorst U, Rehm BH. 2006. Bacterial alginates: from biosynthesis to applications. Biotechnol Lett 28:1701–1712. doi: 10.1007/s10529-006-9156-x. [DOI] [PubMed] [Google Scholar]
- 8.Damron FH, Goldberg JB. 2012. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol 84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potvin E, Sanschagrin F, Levesque RC. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 10.Köhler T, Harayama S, Ramos JL, Timmis KN. 1989. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J Bacteriol 171:4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kustu S, Santero E, Keener J, Popham D, Weiss D. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiological Rev 53:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiau SP, Schneider BL, Gu W, Reitzer LJ. 1992. Role of nitrogen regulator I (NtrC), the transcriptional activator of glnA in enteric bacteria, in reducing expression of glnA during nitrogen-limited growth. J Bacteriol 174:179–185. doi: 10.1128/jb.174.1.179-185.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, Goldberg JB, Schurr MJ, Yu HD. 2012. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol 194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. 2003. negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J Bacteriol 185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd MG, Lundgren BR, Hall CW, Gagnon LB, Mah TF, Moffat JF, Nomura CT. 2017. Targeting the alternative sigma factor RpoN to combat virulence in Pseudomonas aeruginosa. Sci Rep 7:12615. doi: 10.1038/s41598-017-12667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson LS, Webb JS, Rice SA, Kjelleberg S. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol Lett 220:187–195. doi: 10.1016/S0378-1097(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 17.Totten PA, Lara JC, Lory S. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol 172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. 2016. RpoN modulates carbapenem tolerance in Pseudomonas aeruginosa through Pseudomonas quinolone signal and PqsE. Antimicrob Agents Chemother 60:5752–5764. doi: 10.1128/AAC.00260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. 2017. RpoN promotes Pseudomonas aeruginosa survival in the presence of tobramycin. Front Microbiol 8:839. doi: 10.3389/fmicb.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boucher JC, Schurr MJ, Deretic V. 2000. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol 36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- 21.Evans TJ. 2015. Small colony variants of Pseudomonas aeruginosa in chronic bacterial infection of the lung in cystic fibrosis. Future Microbiol 10:231–239. doi: 10.2217/fmb.14.107. [DOI] [PubMed] [Google Scholar]
- 22.Haussler S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ Microbiol 6:546–551. doi: 10.1111/j.1462-2920.2004.00618.x. [DOI] [PubMed] [Google Scholar]
- 23.Schniederjans M, Koska M, Haussler S. 2017. Transcriptional and mutational profiling of an aminoglycoside-resistant Pseudomonas aeruginosa small-colony variant. Antimicrob Agents Chemother 61. doi: 10.1128/AAC.01178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donovan GA, Neuhard J. 1970. Pyrimidine metabolism in microorganisms. Bacteriological Rev 34:278–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaume M, Köhler T, Fontana T, Tognon M, Renzoni A, van Delden C. 2015. Metabolic pathways of Pseudomonas aeruginosa involved in competition with respiratory bacterial pathogens. Front Microbiol 6:321. doi: 10.3389/fmicb.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralli P, Srivastava AC, O'Donovan G. 2007. Regulation of the pyrimidine biosynthetic pathway in a pyrD knockout mutant of Pseudomonas aeruginosa. J Basic Microbiol 47:165–173. doi: 10.1002/jobm.200610248. [DOI] [PubMed] [Google Scholar]
- 27.Ueda A, Attila C, Whiteley M, Wood TK. 2009. Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb Biotechnol 2:62–74. doi: 10.1111/j.1751-7915.2008.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlictman D, Kubo M, Shankar S, Chakrabarty AM. 1995. Regulation of nucleoside diphosphate kinase and secretable virulence factors in Pseudomonas aeruginosa: roles of algR2 and algH. J Bacteriol 177:2469–2474. doi: 10.1128/jb.177.9.2469-2474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlictman D, Kavanaugh-Black A, Shankar S, Chakrabarty AM. 1994. Energy metabolism and alginate biosynthesis in Pseudomonas aeruginosa: role of the tricarboxylic acid cycle. J Bacteriol 176:6023–6029. doi: 10.1128/jb.176.19.6023-6029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundin GW, Shankar S, Chakrabarty AM. 1996. Mutational analysis of nucleoside diphosphate kinase from Pseudomonas aeruginosa: characterization of critical amino acid residues involved in exopolysaccharide alginate synthesis. J Bacteriol 178:7120–7128. doi: 10.1128/jb.178.24.7120-7128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fyfe JA, Govan JR. 1980. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol 119:443–450. doi: 10.1099/00221287-119-2-443. [DOI] [PubMed] [Google Scholar]
- 33.Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. 2008. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology 154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan Withers T, Heath Damron F, Yin Y, Yu HD. 2013. Truncation of type IV pilin induces mucoidy in Pseudomonas aeruginosa strain PAO579. Microbiologyopen 2:459–470. doi: 10.1002/mbo3.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunt TP, Magasanik B. 1985. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci U S A 82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Y, Withers TR, Wang X, Yu HD. 2013. Evidence for sigma factor competition in the regulation of alginate production by Pseudomonas aeruginosa. PLoS One 8:e72329. doi: 10.1371/annotation/4d6c4127-82e4-408d-af89-5f2e207d523b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol 191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, Withers RT, Dai J, Ruan J, Li W, Dai Y, An W, Yu D, Wei H, Xia M, Tian C, Yu HD, Qiu D. 2016. Truncated type IV pilin PilA(108) activates the intramembrane protease AlgW to cleave MucA and PilA(108) itself in vitro. Arch Microbiol 198:885–892. doi: 10.1007/s00203-016-1248-y. [DOI] [PubMed] [Google Scholar]
- 40.Cai Z, Liu Y, Chen Y, Yam JK, Chew SC, Chua SL, Wang K, Givskov M, Yang L. 2015. RpoN regulates virulence factors of Pseudomonas aeruginosa via modulating the PqsR quorum sensing regulator. Int J Mol Sci 16:28311–28319. doi: 10.3390/ijms161226103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E). Proc Natl Acad Sci U S A 92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Head NE, Yu H. 2004. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect Immun 72:133–144. doi: 10.1128/IAI.72.1.133-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Y, Withers TR, Johnson SL, Yu HD. 2013. Draft genome sequence of a mucoid isolate of Pseudomonas aeruginosa strain C7447m from a patient with cystic fibrosis. Genome Announc 1:e00837-13. doi: 10.1128/genomeA.00837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. P(BAD)-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston MJ, Seed PC, Toder DS, Iglewski BH, Ohman DE, Gustin JK, Goldberg JB, Pier GB. 1997. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect Immun 65:3086–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






