This work presents a comprehensive quantification of the parameters of transport by the Sec general secretory system in the three in vitro systems. The standard reconstitution used by most investigators can be enhanced to yield six times as many active translocons simply by adding SecA to SecYEG during reconstitution. This robust system faithfully reflects the properties of translocation in native membrane vesicles. We have expanded the number of precursors studied to nine. This has allowed us to conclude that the rate constant for translocation varies with precursor species.
KEYWORDS: protein translocation, protein export, secretion, E. coli, SecA, SecYEG, membrane protein, proteoliposomes, translocon
ABSTRACT
In all cells, a highly conserved channel transports proteins across membranes. In Escherichia coli, that channel is SecYEG. Many investigations of this protein complex have used purified SecYEG reconstituted into proteoliposomes. How faithfully do activities of reconstituted systems reflect the properties of SecYEG in the native membrane environment? We investigated by comparing three in vitro systems: the native membrane environment of inner membrane vesicles and two methods of reconstitution. One method was the widely used reconstitution of SecYEG alone into lipid bilayers. The other was our method of coassembly of SecYEG with SecA, the ATPase of the translocase. For nine different precursor species we assessed parameters that characterize translocation: maximal amplitude of competent precursor translocated, coupling of energy to transfer, and apparent rate constant. In addition, we investigated translocation in the presence and absence of chaperone SecB. For all nine precursors, SecYEG coassembled with SecA was as active as SecYEG in native membrane for each of the parameters studied. Effects of SecB on transport of precursors faithfully mimicked observations made in vivo. From investigation of the nine different precursors, we conclude that the apparent rate constant, which reflects the step that limits the rate of translocation, is dependent on interactions with the translocon of portions of the precursors other than the leader. In addition, in some cases the rate-limiting step is altered by the presence of SecB. Candidates for the rate-limiting step that are consistent with our data are discussed.
IMPORTANCE This work presents a comprehensive quantification of the parameters of transport by the Sec general secretory system in the three in vitro systems. The standard reconstitution used by most investigators can be enhanced to yield six times as many active translocons simply by adding SecA to SecYEG during reconstitution. This robust system faithfully reflects the properties of translocation in native membrane vesicles. We have expanded the number of precursors studied to nine. This has allowed us to conclude that the rate constant for translocation varies with precursor species.
INTRODUCTION
In all three domains of life, a specific subset of newly synthesized polypeptides must pass through membranes to reach their final destination. The pathway across the membrane is provided by a highly conserved protein in the translocation apparatus. In eubacteria and archaea, the channel is SecY, and in eukaryotes it is Sec61α (1, 2). In Escherichia coli, the general secretory (Sec) system is comprised of two heterotrimeric integral membrane protein complexes: the translocon SecY, SecE, and SecG (SecYEG) (2) and a second trimer, SecD, SecF, and YajC (SecDFYajC). In addition, there is an essential peripheral component of the apparatus, SecA, an ATPase. SecA is found both associated with the membrane and in the cytoplasm. Free in solution, SecA exhibits an equilibrium between monomer and dimer (3, 4); however, it remains unclear what the oligomeric state of functional SecA is at the various steps of export (2). The Sec system transports polypeptides that carry an amino-terminal leader sequence and can only translocate polypeptides that have not yet acquired stable, tertiary structure. In vivo, some precursors are captured directly by SecA, which recognizes the leader peptide (5, 6). A subset of precursors is maintained in a soluble state devoid of stable tertiary structure with the help of the small cytosolic chaperone, SecB, a homotetramer (7). SecB does not directly recognize the leader peptide but binds to unstructured regions of the mature domain (8–15). The ATPase activity of SecA with a precursor bound is maximally activated when it engages SecYEG (16). The energy provided by hydrolysis of ATP is required for translocation. SecDF functions late in the cycle. There is evidence that SecDF couples proton motive force to the release of the translocated polypeptides from the secretory apparatus (17–20). It is not clear whether leader peptidase I, which has its active site on the periplasmic side of the inner membrane, removes the leader sequence before or after the release.
Most in vitro systems are based on vesicles from cells that overproduce only SecYEG or on proteoliposomes reconstituted from purified proteins and lipids and do not include SecDF. These systems transport precursors, indicating that SecDF is not absolutely essential for transport across the membrane. A comparative study from the laboratory of Ian Collinson (21) showed that whereas SecYEG translocated 12% of the precursor added to the system, the holotranslocon that included SecDFYajC and YidC translocated only 4%.
The reconstitution of purified SecYEG into liposomes, originally established in the early 1990s, provided a powerful tool for biochemical studies of translocation (22, 23). In the standard reconstitution, currently used by most research groups, purified SecYEG is assembled with lipids to form proteoliposomes. SecA is added subsequently along with the precursor to be translocated (PLYEG+SecA). We developed a robust reconstitution in which SecYEG is coassembled with SecA into the liposomes. We refer to those proteoliposomes as PLYEG·SecA. In these proteoliposomes the percentage of translocons that are active compared to those in standard reconstitution is increased from 10% to 55%. This brings the percentage of active translocons to the same level as that observed in the isolated inner membrane vesicles (24). When we developed PLYEG·SecA, we did not determine whether the translocons that showed activity were fully functional and quantitatively similar to those in the native state. Here, we address that question with a comprehensive study of the parameters that characterize the translocation of nine different precursors using native membrane vesicles and the two types of reconstituted proteoliposomes.
RESULTS
The system.
Inner membrane vesicles (IMV) were isolated from E. coli and are taken to represent SecYEG in its native membrane environment for comparison with high-activity coassembled proteoliposomes (PLYEG·SecA) and low-activity standard proteoliposomes (PLYEG) to which SecA is added after assembly (PLYEG+SecA). In an attempt to determine whether the characteristics of precursors influence the parameters of translocation in the process of export, we chose nine precursors (see Materials and Methods and Table 1) that varied in final location, i.e., the periplasm or outer membrane, in secondary structure, in β-strands versus α-helices, and in molar mass. Three are precursors of outer membrane proteins, outer membrane protein A (proOmpA, 37.2 kDa), maltoporin (pLamB, 49.9 kDa), and phosphoporin E (pPhoE, 38.9 kDa); these proteins are β-barrels in the outer membrane. Four are precursors of soluble periplasmic proteins, alkaline phosphatase (pPhoA, 49.4 kDa), galactose-binding protein (pGBP, 35.7 kDa), maltose-binding protein (pMBP, 43.4 kDa), and ribose-binding protein (pRBP, 31.0 kDa); the periplasmic proteins are α-β structures. The precursors of MBP and RBP fold too rapidly in vitro to be efficiently captured by SecA or SecB and thus cannot be translocated; therefore, polypeptides carrying mutations that slow folding, pMBPY283D and pRBPA248T, were used (25, 26). To determine whether the leader peptide and/or the mature region plays a role in the rate-limiting step of transport, we took advantage of the fact that the monomer of OmpA comprises two distinct domains, each having similar molar masses: an N-terminal eight-stranded β-barrel that is embedded in the outer membrane and a C-terminal α-β structure that extends into the periplasmic space. We created two separate precursor polypeptides, each carrying the same native OmpA leader sequence followed by one of the two structurally distinct domains. These polypeptides are referred to as proOmpAN176 (21.2 kDa) and properiOmpA (18.5 kDa).
TABLE 1.
Plasmids and host strains
| Host straina | Plasmid | Protein | Mol wt (kDa) | Mutations |
|---|---|---|---|---|
| BL21.19 | pAL800 | properiOmpA | 18.5 |
ompA C290S and G244C residues, including signal sequence followed by residue 172 to the end, residue 325 |
| pAL942 | pRBPA248T | 31.0 | rbsB A248T, A188C, E246C | |
| pAL928 | pPhoA | 49.4 | phoA C169S, C179S, C287S, C337S, A369C, A427C | |
| pAL950 | pPhoE | 38.9 | phoE A249C, Q307C | |
| pAL831 | pLamB | 49.9 | lamB C22S, C38S, D329C, N386C | |
| pAL832 | proOmpAN176 | 21.2 | ompA T95C, I153C, truncated after residue 176 | |
| MM52 | pAL612 | proOmpA | 37.2 | ompA C290S, G244C |
| pAL725 | pGBP | 35.7 | mglB L244C, D287C | |
| pAL829 | pMBPY283D | 43.4 | malE Y283D, G289C, V347C |
BL21.19 (61) is a derivative of BL21(DE3) E. coli B F− dcm ompT hsdS(rB− mB−) gal λ(DE3) with secA13(Am), supF(Ts), trp(Am), zch::Tn10, clpA::KAN, and recA::CAT. MM52 is a derivative of MC4100 (56): F− (araD139)B/r secA51(ts) Δ(argF-lac)169 λ− flb-5301 Δ(fruK-yeiR)725(fruA25) relA1 rpsL150(strR) rbsR22 Δ(fimB-fimE)632(::IS1) deoC1.
Translocation assays of the nine radiolabeled precursors were carried out. Transfer of the precursors through lipid bilayers was assessed by protection of the polypeptides from protease added to the assay mixture in the presence of denaturants to render all untranslocated precursors sensitive to proteolysis. The denaturing conditions varied among the precursors. Studies showed that inclusion of 1 M urea during proteinase K digestion was sufficient for five of the precursors, three requiring 2 M urea and one requiring 1 N guanidine hydrochloride (for specific details see Materials and Methods and Table 3). Translocation is ATP dependent; thus, to ensure that protection of precursors reflects genuine translocation by SecYEG and not simply resistance to proteolysis, a translocation assay for each precursor was done in the absence of ATP.
TABLE 3.
Conditions for proteinase K digestion
| Denaturant | Precursor(s) |
|---|---|
| 1 M urea | proOmpA, pGBP, pMBP(Y283D), pPhoA, properiOmpA |
| 2 M urea | pPhoE, pLamB, proOmpAN176 |
| 1 N GnHCl | pRBP(A248T) |
Time courses of translocation and the associated ATPase were determined for each precursor in the presence of SecB to maximize the amount of competent precursors and, thus, the level of translocation observed (Fig. 1, for example). Four precursors, proOmpA, pGBP, pPhoA, and properiOmpA, were assessed in all three in vitro systems. The remaining five precursors, when assayed with the standard reconstitution system, showed levels of translocation that were too low to obtain reliable results. In those cases, translocation was compared between the robust proteoliposomes, PLYEG·SecA, and vesicles of native membrane, IMV. The following parameters were determined by fitting the data to a single exponential rise to maximum (see Materials and Methods): the maximal amplitude (A), the apparent rate constant (k), which is the rate constant of the slowest step in the cycle, and the coupling of energy of ATP hydrolysis to the work of translocation.
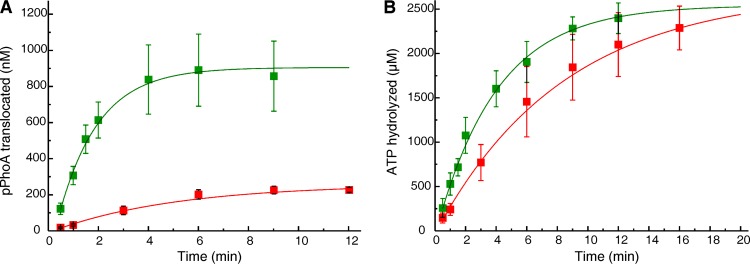
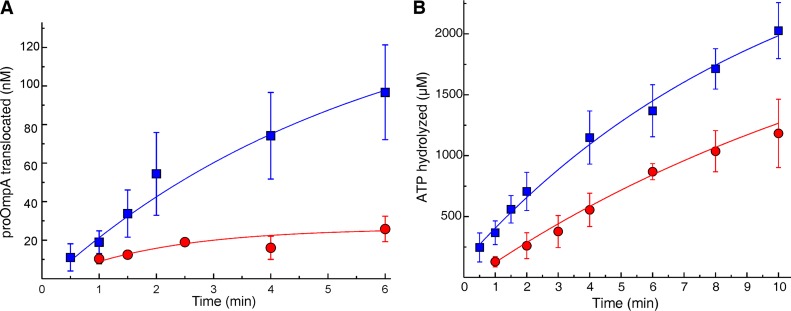
FIG 1.
In vitro translocation and associated ATPase of 14C-labeled pPhoA. (A and B) The extent of translocation (A) and SecA ATPase activity (B) during in vitro translocation of 14C-labeled pPhoA into PLYEG·SecA (green) and PLYEG+SecA (red). The average values for each time point, with standard deviations, are shown (green, n = 9; red, n = 3). When error bars are not visible, they fall within the symbols. These data and the data in all subsequent figures were fitted to a single exponential rise to maximum (see Materials and Methods). The fits are weighted with 1/σ2, where sigma is the standard deviation.
Maximal amplitude.
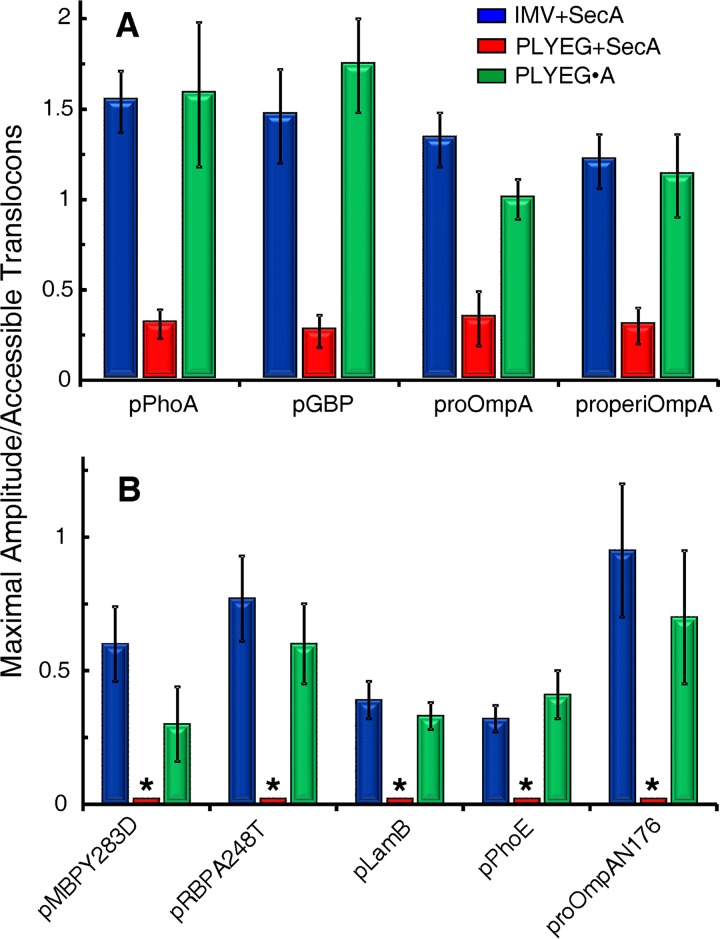
To compare the final extent of translocation among the three in vitro systems, the maximal amplitudes were normalized to the number of translocons that are accessible to proteins added externally in each assay mixture; 55% for proteoliposomes (24) and 95% for IMV (27, 28). The averages of the normalized amplitudes from multiple experiments are shown in Fig. 2. Native IMV and PLYEG·SecA display approximately the same maximal level of translocation. The precursors that could be assayed for translocation in PLYEG, having SecA added subsequently (PLYEG+SecA), show approximately 15% to 25% of the level of translocation seen with IMV or PLYEG·SecA. Since SecYEG is limiting in these assays (see Materials and Methods), the difference in amplitude can be explained readily by the fact that only 10% of the accessible translocons are active in the standard reconstitution compared to approximately half in both IMV and PLYEG·SecA (24).
FIG 2.
Extent of precursor translocation in three in vitro systems: IMV+SecA, PLYEG+SecA, and PLYEG·SecA. The values represent averages of normalized amplitudes with their standard deviations. (A) Numbers of replicates for translocation of each species were the following for blue, red, and green, respectively: pPhoA, 7, 5, and 9; pGBP, 6, 5, and 6; proOmpA, 7, 5, and 10; and properiOmpA, 3, 4, and 3. (B) Translocation assays were performed at least 3 times. The bars marked by an asterisk indicate that assays were done but translocation levels were too low to obtain reliable fits.
Apparent rate constant.
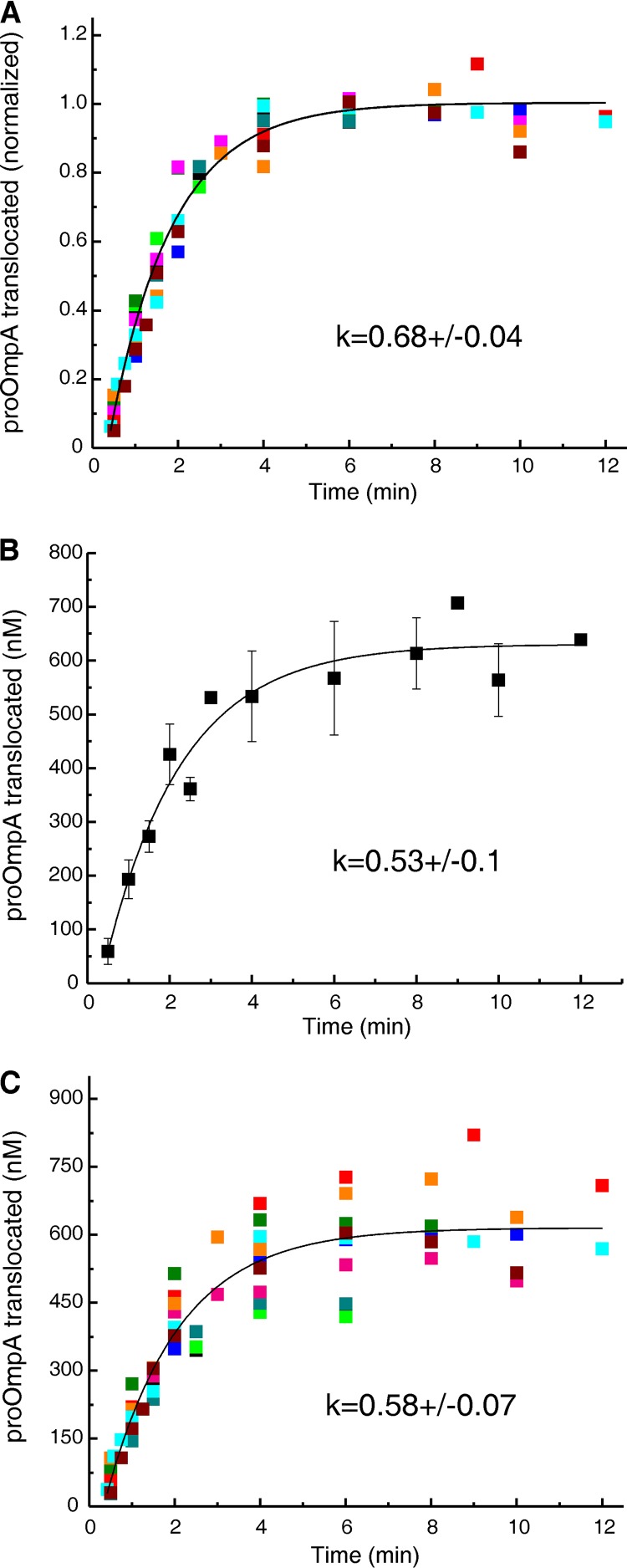
To determine whether those translocons that are active in the reconstitution systems are completely undamaged or show a diminished activity, we turned to the rate constant (k). The rate constant is a characteristic parameter of the secretory apparatus that expresses the probability of a translocation event per unit of time. Since k is a constant, it is not affected by the variation in the maximal amplitude that is observed among the experiments (see error bars in Fig. 2). Therefore, instead of averaging values from the fits of multiple experiments, as was done for Fig. 2, the data were globally fit after normalizing each data point in a time course to the maximal amplitude obtained from the fit for that individual experiment. The global fit of the normalized data (Fig. 3A) displayed an error of 6%. This is a dramatic, 3-fold improvement over the error of 19% seen both when averaging the data from multiple experiments followed by fitting (Fig. 3B) and also when fitting the individual experiments followed by averaging the obtained rate constants (data not shown). In the case of globally fitting all data from multiple experiments but without the normalization, the error was 12% (Fig. 3C).
FIG 3.
Fits of multiple data sets and associated errors. Translocation of proOmpA into PLYEG·SecA was performed multiple times (n = 10) under the condition of limiting SecYEG. Each color represents an individual experiment. The ratio of precursor to accessible SecY in the assays ranged from 1.5 to 3. (A) Global fit of the normalized data. (B) Fit of averaged data. (C) Global fit of data from ten translocation experiments without normalization. See the text for a description of normalization and fitting.
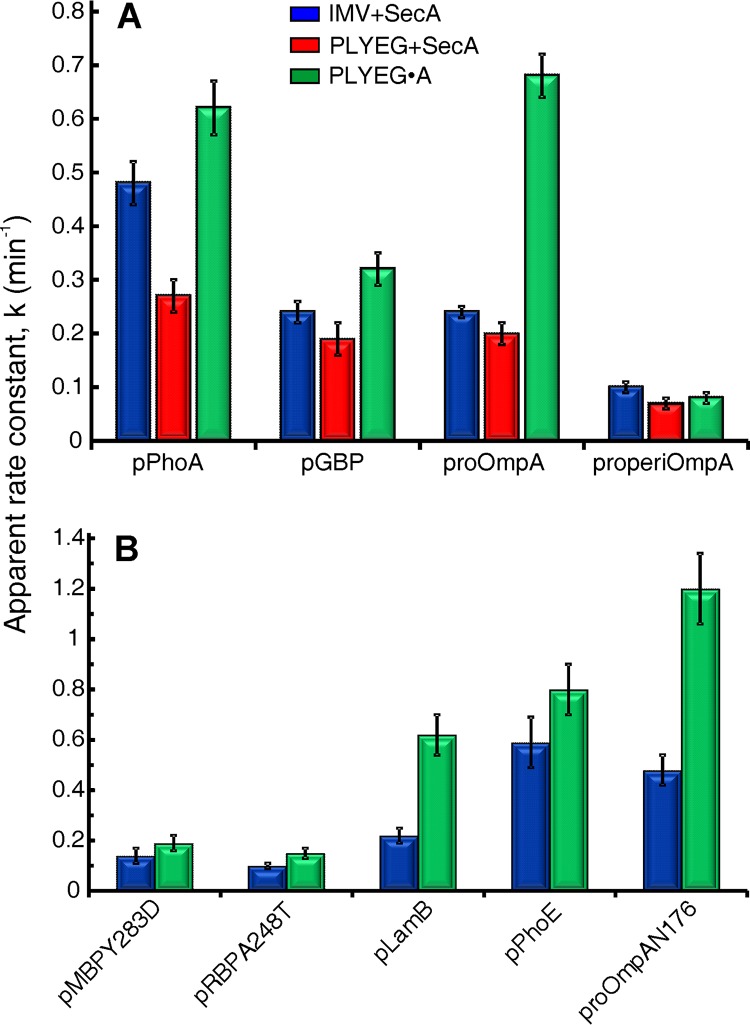
The apparent rate constants between the translocons that are active in the standard reconstitution and those in IMV (Fig. 4A, compare red and blue) are the same within error, with the exception of pPhoA, which has a 1.7-fold-higher rate constant in IMV. We conclude that despite the lower translocation activity in the standard reconstitution system, the rate constants of the active translocons are essentially the same as those observed with IMV. The few translocons that are active after standard reconstitution most likely escaped damage during membrane preparation and solubilization and therefore are fully functional. Those translocons that were inactivated by solubilization can be restored to the active state by coassembly with SecA but do not spontaneously recover during standard reconstitution.
FIG 4.
Apparent rate constants of translocation. The apparent rate constants were determined for each precursor using IMV+SecA (blue), PLYEG+SecA (red), and PLYEG·SecA (green). See the text for a description of normalization and global fitting. The error bars represent the errors in the global fit. The ratio of precursor to accessible SecYEG varied over the ranges given here for precursors. (A) pPhoA, 1.3 to 3.5; pGBP, 1.5 to 3; proOmpA, 1.3 to 3.0; properiOmpA, 2 to 4; (B) pMBPY283D, 2 to 4; pRBP(A248T), 1.5 to 3; pLamB, 4 to 5.5; pPhoE, 2.6 to 4; proOmpAN176, 3 to 6.
For translocation of precursors using PLYEG·SecA (Fig. 4), the apparent rate constants of all nine precursors were restored to values at least as high as those observed in IMV. Six precursors showed rate constants that were either within error or 1.3-fold higher, whereas three are approximately 3-fold higher. We have no explanation for the rate constants that are unexpectedly high. Here, we briefly discuss differences among the systems that might contribute to the observed results. First, proteoliposomes have a greater surface area of exposed lipids than do IMV. The in vivo ratio of lipid to protein is reported to be approximately 400:1 (29). However, since optimization of our system showed maximal translocation at a lipid-to-protein ratio of 1,000:1 (data not shown), the proteoliposomes in our study were assembled at that ratio. The membranes of IMV and proteoliposomes also differ in protein content. The membrane of IMV contains a wide variety of proteins, many with soluble domains on both sides of the membrane, whereas the proteoliposomes contain only integral SecYEG or SecYEG with SecA bound. Thus, more surface area is devoid of protein cover in proteoliposomes than in IMV. The three proteins that show higher k values in PLYEG·SecA, proOmpA, proOmpAN176, and pLamB are outer membrane β-barrels. Beta-barrels have been shown to assemble spontaneously in vitro when in contact with lipid bilayers (30); first the polypeptide adsorbs to the lipid surface, and the extended strands then come together into antiparallel β-sheets (31). On the cytoplasmic face of the proteoliposome the precursors would be prevented from association with lipid because they would be bound to SecB and SecA. It is feasible that as precursors emerge inside the liposomes they begin to fold on the surface, providing energy to pull the polypeptide through the channel. However, if the greater surface of exposed lipid explained the phenomenon, then one would expect both types of proteoliposomes, PLYEG·SecA and PLYEG, to exhibit higher rate constants than those observed with IMV. Unfortunately, only one of the β-barrel proteins, proOmpA, had a level of translocation sufficiently high with PLYEG+SecA to obtain a reliable rate constant. The rate constant observed was essentially the same as that with IMV. One clear difference between PLYEG·SecA and PLYEG+SecA is the presence of SecA both inside and outside the liposomes in PLYEG·SecA. Since SecA has affinity for precursors, it might bind proOmpA as the polypeptide emerges inside the proteoliposome and thereby provide energy to aid in pulling the precursor through the translocon. In this scenario, we cannot explain why only β-barrel precursors exhibit unexpectedly high rate constants. Further work will be required to discover the basis for the differences. In any case, the enhancement of rate is not likely to be physiologically important.
Coupling of ATP hydrolysis to translocation.
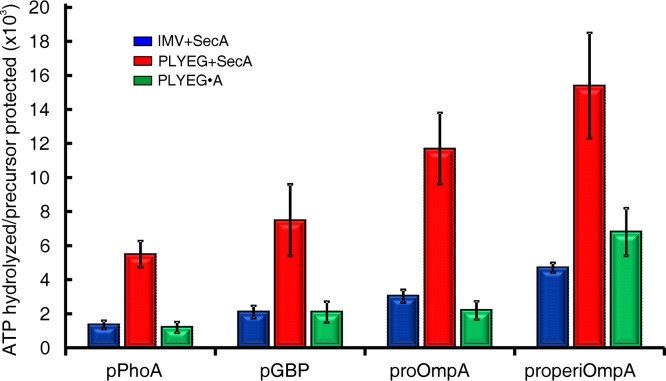
The hydrolysis of ATP by SecA is required for translocation of precursors across the membrane (32). We assessed ATP consumption during translocation by monitoring [γ-32P]phosphate release from ATP (see Materials and Methods). Both translocation data and the associated ATPase data were obtained from the same in vitro mixture, and the two data sets were each fitted to a single exponential rise to maximum (Fig. 1). Using the parameters (k and A) obtained from the fits, we calculated the ratio of moles of ATP hydrolyzed to the moles of precursor translocated in the first 15 s after the onset of translocation. This ratio is an expression of the coupling of the energy to the work of translocation. The less ATP was consumed, the more efficient the coupling was.
Comparison among the three systems showed that both PLYEG·SecA and the native membrane were equally efficient in coupling the energy of ATP hydrolysis to translocation (Fig. 5, green [PLYEG·SecA] and blue [native membrane]). Therefore, coassembly with SecA results in a state that is near native. The efficiency of energy coupling varied among the precursor species over a range of 1,000 to 7,000 mol of ATP hydrolyzed per mol of precursor protected. These quantities of ATP hydrolyzed are similar to those observed previously for proOmpA and pGBP translocated into IMV (32–34). In the case of PLYEG (Fig. 5, red), the ATP consumption was much higher than that observed in the other two systems. Thus, one might conclude that when SecA is added after PLYEG is assembled, ATP hydrolysis by SecA is much more loosely coupled to translocation. However, an alternative interpretation is that the few translocons in PLYEG, which are fully functional in terms of amplitude and rate constant of translocation, are equally efficient in coupling the energy of ATP, as are those in PLYEG·SecA. The ATP hydrolysis we observe is that of the entire ensemble; thus, the high level of hydrolysis could be due to both the active and the defective translocons. To test whether SecYEG species that are defective for translocation could activate SecA ATPase, we used a mutated form of SecY, SecY(F328R), which has been shown to be defective in translocation (35). As expected, proteoliposomes containing translocation-defective SecY(F328R)EG, to which SecA was added during the assay, displayed less than 20% of the translocation seen with wild-type SecYEG (Fig. 6A). However, the translocation-defective SecY(F328R)EG stimulated the ATPase activity of SecA to reach 95% of the maximal level of ATP hydrolyzed with a rate constant that is 56% of the wild-type value seen when SecA is activated by wild-type SecYEG (Fig. 6B). We do not know what the specific defect is in the ∼90% of the SecYEG that is inactive in the standard reconstitution. It is possible that, like SecY(F328R), those translocons can activate the SecA ATPase. This would result in two populations of complexes between SecYEG and SecA, those with the coupling characteristics of actively translocating SecYEG complexes and a population that is completely uncoupled, simply hydrolyzing ATP without translocation. Thus, the ensemble would display an artificially high ratio of coupling.
FIG 5.
Coupling of ATP hydrolysis to translocation of precursors. Translocation ATPase in three in vitro systems: IMV+SecA, PLYEG+SecA, and PLYEG·SecA. The values are an average of the coupling energy with the standard deviations. Higher values represent lower efficiency of coupling the energy to the translocation. The numbers of replicates were 5 to 8 for pPhoA, 5 for pGBP, 4 to 7 for proOmpA, and 3 or 4 for properiOmpA.
FIG 6.
Comparison of activity of SecYEG wild type and SecY(F328R)EG. Translocation and ATPase assays of 14C-labeled proOmpA were done using proteoliposomes reconstituted with wild-type SecYEG (squares, n = 3) and with SecY(F328R)EG (circles, n = 3). SecA was added to both translocation mixtures. (A and B) Precursor translocated (A) and SecA ATPase (B) activity. The values are averages with standard deviations. When error bars do not show, they lie within the data points.
Effect of SecB on translocation.
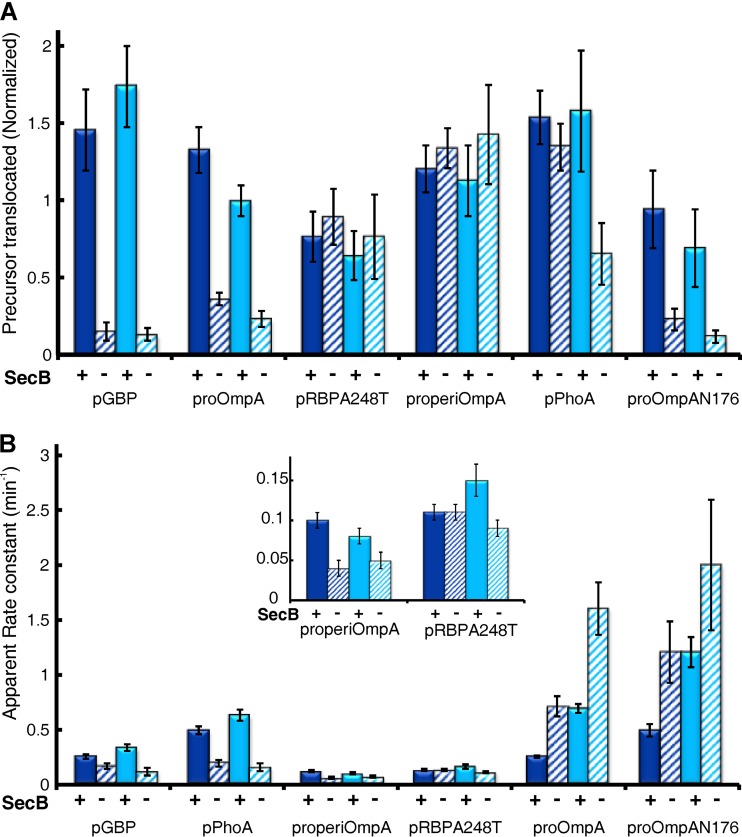
All data discussed in the previous sections were obtained from assays in the presence of SecB. In E. coli, SecB functions to bind a subset of precursors and keep them in a state compatible with translocation (7). It is known that in vivo SecB dependence varies with the precursor species. SecB-null strains display defects in secretion of several proteins, including MBP, LamB, and GBP, proteins which we have studied here (36–38). Our in vitro results agree well with these in vivo observations. Although pPhoE has not been studied in vivo, the level of translocation of pPhoE as well as pMBPY283D and pLamB, in the absence of SecB, was too low to generate reliable results; all three were translocated in the presence of SecB (Fig. 2B). In the case of pGBP, the presence of SecB increased the level of translocation approximately 10-fold (Fig. 7A). In vivo, proOmpA can be translocated without SecB (39, 40); however, translocation is enhanced in its presence. The presence of SecB increased translocation of proOmpA and proOmpAN176 approximately 4- to 5-fold in our in vitro systems (Fig. 7).
FIG 7.
Effect of SecB on translocation of precursors. Precursors were assessed for translocation using IMV (dark blue) and PLYEG·SecA (light blue) in the presence (+) and absence (−) of SecB. The normalized amplitude and the apparent rate constant were obtained as described previously. All experiments were performed at least 3 times. The inset in panel B is an expansion of the y axis to make values visible. The error bars in panel A represent standard deviations, and those in panel B represent the error in the global fit.
Studies in vivo using an SecB-null strain indicate that neither RBP nor PhoA requires SecB (37). Consistent with the in vivo studies, the presence of SecB had no significant effect on the level of translocation of either pRBP or pPhoA when assayed using IMV (Fig. 7A, dark blue). We also did not observe an effect of SecB on the transport of properiOmpA, which has not been studied in vivo. In contrast to RBP, the translocation of pPhoA in the presence of SecB appears to be enhanced when it was translocated in PLYEG·SecA, as judged by the ratio of the amplitude in the presence of SecB to the amplitude in its absence (Fig. 7A, PhoA, light blue, solid and hatched; ratio, 2.4). However, this effect results not from a true stimulation by addition of SecB: the amplitudes of translocated pPhoA are the same in IMV and in PLYEG·SecA when SecB is present (Fig. 7A, compare IMV with SecB, 1.54, solid dark blue, and PLYEG·SecA with SecB, 1.58, solid light blue). Rather, the apparent stimulation results from a decrease in translocation in the absence of SecB in PLYEG·SecA (Fig. 7A, compare IMV, no SecB, 1.35, hatched dark blue, and PLYEG·SecA, no SecB, 0.66, hatched light blue). This decrease probably arises from the need for pPhoA to bind SecB in order to avoid nonspecific interaction with the large lipid surface area exposed in proteoliposomes, which would not be present in IMV or cells. Thus, our results are not in conflict with the studies in cells.
The rate constants of transport were also affected by SecB. In the case of pGBP, pPhoA, and properiOmpA the rate constants were increased by the presence of SecB, whereas for both proOmpA and proOmpAN176 the rate constants were decreased (Fig. 7B). SecB had no significant effect on the rate constant for pRBPA248T using IMV but showed a slight but significant stimulation with PLYEG·SecA.
DISCUSSION
When investigating the mechanism of protein export in vitro, it is vital to know that reconstituted model systems assembled from purified proteins and lipids reflect the function in the native membrane environment. Solubilization of SecYEG from isolated membrane followed by standard reconstitution results in only 10% of the translocons in the active state, whereas coassembly with SecA brings the level of active translocons back to that observed in IMV (approximately half). The translocons that are inactive at the time of isolation of the IMV are not restored to activity by reconstitution in the presence of SecA. We have previously suggested (24) that SecA could function to stabilize an active conformation of SecYEG during assembly in vivo. In cells grown for preparation of IMV, SecYEG is expressed at approximately 50-fold over the chromosomal level. This abnormally large quantity of SecYEG might result in a situation in which the amount of SecA is insufficient to assist in the correct assembly of SecYEG, causing irreversible damage.
We have determined the parameters that are characteristics of transport using three in vitro systems: the native membrane and the two reconstituted systems. We begin discussion of our results with the amplitude and the effects of SecB on the amplitude. The amplitude, expressed in nanomolar concentrations, varies with the precursor species; however, for a given species it is similar between IMV and PLYEG·SecA, indicating full restoration of activity by coassembly of SecYEG with SecA. A subset of proteins interacts with SecB in vivo to remain competent for translocation. The effect of SecB on the maximal amplitude in vitro faithfully mimics the degree of SecB dependence in vivo. Protein proOmpA does not absolutely require SecB to be transported either in vivo or in vitro, but the level of translocation is increased by SecB (34, 41). This is in contrast to LamB and PhoE, which are not translocated without SecB. The tendency to aggregate and lose competence might result from structural differences among the proteins, even though all three are β-structures. Whereas LamB and PhoE are embedded in the membrane as β-barrels, in addition to the embedded β-barrel domain, OmpA has a soluble domain that comprises 50% of its mass. This soluble domain has been shown to act as an endogenous chaperone to keep the β-barrel from aggregating (42), explaining why proOmpA is not completely dependent on SecB.
In the late 1980s, work in the laboratory of Wickner et al. (32) showed that thousands of moles of ATP were hydrolyzed to translocate one mole of precursor, indicating that translocation is only loosely coupled to ATP hydrolysis. Later, Schiebel and colleagues (43) proposed that the loose coupling was the result of precursor polypeptides sliding backwards during translocation. In the case of PLYEG to which SecA was added, the extremely high level of ATP hydrolyzed per mole of precursor translocated is not likely to result only from backsliding in the 10% of translocons that are active. It more probably reflects activation of the SecA ATPase by the great majority of translocons that are inactive for translocation. This conclusion is supported by our demonstration that a translocation-defective SecY stimulates the ATPase activity of SecA. Using ensemble approaches such as the assays described here, it will be difficult to demonstrate tight coupling. Unless 100% of the translocons in the system are active, the true amount of ATP required for translocation will be smaller than that observed.
The rate constant for transport that is observed is that of the slowest step in the cycle, that is, the step that limits the rate. A change in the rate constant indicates that there has been a change either in the step limiting the rate or in the features of that step. Therefore, identification of factors that change the rate constants will help us learn what is involved in the slowest step of the cycle. As a chaperone, SecB increases the amplitude of precursor translocation by increasing the quantity of translocation-competent precursor available. This will result in an increase in the rate of translocation but not in the rate constant, since the rate (measured as molar concentration per minute) of a reaction is the product of the concentration of substrates (precursor; measured as molar concentration) and the rate constant of the reaction (k; units are per minute). The rate constant is an inherent property of the reaction, and it expresses the probability of the reaction occurring. Nonetheless, the presence of SecB does change the rate constant for translocation of five of the precursors studied. The k is increased for pGBP, properiOmpA, and pPhoA but decreased for proOmpA and proOmpAN176. The increase in k might be the result of delivering the precursors from SecB to SecA in an unfolded state ready to be translocated. Transfer of the precursor from SecB to SecA occurs within a SecB:SecA complex, not through solution, where it might acquire structural elements (44). The effect of SecB on the rate constant is supported by a single-molecule fluorescence study that concludes that the slow step that precedes rapid transit through the channel is the delivery of the precursor from SecB to SecA (41). A possible explanation for the decrease in k of the two related β-barrels is a high affinity for SecB that would decrease the probability of transfer to SecA.
With the nine precursors studied here, we did not observe a correlation of rate constant with molar mass or final location (i.e., soluble in the periplasm or embedded in the outer membrane). The three species carrying the same leader sequence have significantly different rate constants: proOmpA, k = 0.24 ± 0.01; proOmpAN176, k = 0.48 ± 0.06; properiOmpA, k = 0.10 ± 0.01. Therefore, the leader peptide itself is not the determining feature; rather, the rate constant is likely to be determined by the nature of the amino acyl residues in the mature portion of the protein.
Previous studies of intact proOmpA have indicated that the rate of transfer is dependent on the primary sequence of the polypeptide. Mizushima and his colleagues (45) reported discontinuous transfer of proOmpA in 1989. In a subsequent study (46) they concluded this was caused by a retardation of transfer that occurs when hydrophobic stretches of 4 aminoacyl residues interact with the translocon. Work from the laboratory of Driessen et al. (47) demonstrated a decrease in rate of translocation by insertion of stretches of either positively or negatively charged aminoacyl residues into the periplasmic domain of proOmpA. Rapoport and his coworkers (48) showed that amino acyl residues vary in strength of interaction, with the two-helix finger of SecA resulting in periods of active and passive translocation through the SecYEG channel. Thus, the rate of transfer per residue is not constant. A conclusion which is in conflict with the reports discussed above (46–48) is based on work from Tomkiewicz et al. (33) as well as Fessl et al. (41). Their work indicates that SecA drives translocation at a constant rate that is independent of the nature of the aminoacyl residues. Our data reported here contradict the conclusion that the rate of transfer is constant per aminoacyl residue. This is most simply seen by the 5-fold difference in the rate constants of the two domains of proOmpA, proOmpAN176, and properiOmpA, which are approximately the same length, 176 aminoacyl residues for OmpAN176 and 154 residues for periOmpA.
The difference in rate constants observed among the precursors is not likely to reflect different rate-limiting steps but rather differences in the details of interactions during that step. There are at least two steps that would be consistent with our results: movement through the channel (48) and delivery of the precursor to the channel in a state optimal for passage. It is easy to envision different interactions that would be specific for each precursor (46) occurring in either step. However, it is possible that some other step in the cycle is rate limiting but still dependent on the aminoacyl residues of the precursor, for example, release of precursor from the secretory apparatus in order to reset it for the next polypeptide. SecDF in conjunction with proton motive force has been proposed to function in release of translocated precursors (18, 19, 49). If this were the case, our in vitro systems would be missing the release function. The IMV can generate proton motive force, but SecYEG has been overproduced and SecDF has not; thus, they are not present in the correct ratio. The proteoliposomes cannot generate proton motive force, and they do not contain SecDF. Work is in progress to develop a system in which the role of SecDF can be assessed.
MATERIALS AND METHODS
Determination of lipid and protein concentration and specific activity of radiolabeled precursors.
The concentrations of lipids in membrane vesicles, liposomes, and proteoliposomes were determined as described on the Avanti Polar Lipids website (www.avantilipids.com/tech-support/analytical-procedures/determination-of-total-phosphorus). A total phosphorus assay was carried out in which the phosphorus standard solution (0.65 mM; Sigma-Aldrich) and the unknown lipid samples went through an acidic digestion. The released inorganic phosphate was reacted with ammonium molybdate and ascorbic acid to generate a colored solution. The absorbance of the samples was measured spectrophotometrically at 820 nm. A calibration curve was generated using the standard solutions, and the amount of phospholipids in the unknown samples was calculated.
The concentrations of pure proteins were determined spectrophotometrically at 280 nm using molar extinction coefficients: SecB tetramer, 47,600 M−1 cm−1; SecA dimer, 78,900 M−1 cm−1; and SecYEG, 45,590 M−1 cm−1. The concentration of SecY in the cytoplasmic inner membrane vesicles and in proteoliposomes was determined using a quantitative Western blot and a quantitative Coomassie brilliant blue R-250-stained gel, respectively. A sample with unknown SecY concentration was loaded in increasing quantities along with a range of purified SecY at 5 or 6 known concentrations on the same gel to generate a standard curve. Blots were incubated with primary antibody, a rabbit polyclonal antiserum raised to pure SecY, followed by a goat antibody raised to a rabbit immunoglobulin and conjugated with horseradish peroxidase. A 4-chloro-1-naphthol-hydrogen peroxide solution was used to stain the protein bands. By comparing the intensity of the protein bands with those of standards, the concentrations of SecY in inner membrane vesicles and proteoliposomes were determined. The intensity of protein bands for stained gels and for Western blots was quantified using TotalLab software (version 13.0; TotalLab Ltd., UK). Radiolabeled precursors (greater than 95% pure) were subjected to analyses to determine the amino acid composition from which the concentration of the protein was determined (in milligrams per milliliter). The analyses of amino acid composition were performed by the AAA Service Laboratory, Inc. (Damascus, OR). The radioactivity in the pure protein preparation (counts per minute per microliter) was determined by scintillation counting. Five microliters was removed in duplicate into 100 μl water held in scintillation vials to which 3 ml of 30% Scintisafe (Fisher Scientific) was added. The specific activity of radiolabeled precursors (counts per minute per microgram) was calculated from the concentration and radioactivity.
Purification of SecYEG, SecA, and SecB.
The translocons SecYEG and SecY(F328R)SecESecG were purified as described previously (24) from strain C43(DE3) (50) and transformed with a plasmid (pEXP2) that expresses secYC329S, secYC385S, and secE with a His tag at the N terminus and secG under the control of an arabinose-inducible promoter (51). The ATPase, SecA, was purified as described previously (52). The chaperone, SecB, was purified as described previously (53). SecYEG was stored in 20 mM Tris-Cl, pH 8, 0.3 M NaCl, 10% (wt/vol) glucose, 0.6 mM dodecyl-beta-maltoside (DBM), and 2 mM dithiothreitol (DTT). SecB and SecA were stored in 10 mM HEPES, pH 7.6, 0.3 M potassium acetate (KAc), 2 mM DTT, at −80°C.
Preparation of radiolabeled precursors.
All precursors were purified from a temperature-sensitive SecA strain of E. coli, harboring a plasmid that carried the precursor gene of interest. Each plasmid carried a gene altered by site-directed mutagenesis to generate a polypeptide with only two cysteines (Table 1). These cysteines were introduced from a previous study (24). The genes for ribose-binding protein (RBP) and maltose-binding protein (MBP) also carried mutations that have been shown to slow folding of the precursors (25, 26). For precursors of the integral domain (proOmpAN176), the plasmid carried a gene that encodes the leader and the aminoacyl residues through 176. For properiOmpA, the leader sequence of proOmpA was attached directly to the first aminoacyl residue of the periplasmic domain, i.e., residue 172.
The [14C]leucine (Perkin-Elmer)-radiolabeled precursors were purified from a strain, either BL21.19 or MM52, each carrying a SecA temperature-sensitive mutation (Table 1). The bacterial cells were grown in M9 minimal media in the presence of 19 amino acids (no leucine), MgSO4 (1 mM), CaCl2 (0.1 mM), and vitamin B1 (0.0004%, wt/vol) (54). The culture was supplemented with glycerol (0.4%, wt/vol) as the carbon source and the antibiotic ampicillin (100 μg/ml). For precursor OmpA (proOmpA), the cells were also supplemented with glucose (34). After growth at 30°C to an optical density at 560 nm (OD560) of 0.25 to 0.3, the cells were pelleted and suspended in medium prewarmed to 39°C to apply a heat shock and inactivate SecA (55, 56). Further growth was continued at 39°C to an optical density at 560 nm of 0.8, and isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added to induce expression of the precursor protein. Thirty minutes after induction, [14C]leucine (328 mCi/nmol; final concentration of 4 µM) was added and growth continued for another 2.5 h (39°C). The cells were harvested by centrifugation (16,000 × g, 10 min at 4°C; GSA rotor; Sorvall) and washed with and then suspended in 20 mM Tris-HCl at pH 8. The cell suspension was frozen by dripping into liquid nitrogen and stored at −80°C. After thawing, lysozyme (0.2 mg/ml) and EDTA (4 mM) were added and the suspension was subjected to three cycles of freezing and thawing to lyse the cells. The lysate was incubated with micrococcal nuclease (314 U/ml) and calcium chloride (5 mM) for 1 h on ice to degrade nucleic acids. EDTA (10 mM) then was added to chelate calcium. Inclusion bodies containing the precursor were collected by centrifugation (4,000 × g, 10 min at 4°C; SS-34 rotor; Sorvall) and washed once with 20 mM Tris-Cl, pH 7.6, 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF). The washed pellets were exposed to differential solubilization procedures (see below and Table 2 for steps in solubilization). Each pellet suspension was incubated on ice for 2 to 3 h, followed by centrifugation (543,000 × g, 15 min at 4°C; TL100.4 rotor; Beckman). The supernatant and pellet fractions were examined by SDS gel electrophoresis to determine the solubility and purity of the precursor of interest. The precursors of RBP and GBP were purified by one step of solubilization. The precursors of OmpA, LamB, PhoE, and PhoA and the integral OmpAN176 were pure after two steps of solubilization. The precursors of MBP and periplasmic OmpA were solubilized, followed by column chromatography, using QAE-825 in 4 M urea to achieve purity. Precursor MBP was eluted using a gradient of NaCl from 0 to 150 mM, and the precursor of the periplasmic domain of OmpA eluted in the flowthrough. All precursors were stored at −80°C with tris(2carboxyethyl)phosphine (TCEP) to maintain the sulfhydryls in reduced forms.
TABLE 2.
Purification scheme for precursors
| Precursor | Solubilization/purification method | Storage buffer |
|---|---|---|
| Maltose-binding protein (pMBPY283D, 43.4 kDa) | 1.5 N GnHCl/QAE column | 1.5 N GnHCl, 10 mM HEPES (pH 7.6), 0.15 M KAc, 2 mM TCEP |
| Periplasmic domain of OmpA (properiOmpA, 18.5 kDa) | 2.5 N GnHCl/QAE column | 4 M urea,10 mM HEPES (pH 7.6), 0.1 M KAc, 5 mM TCEP |
| Ribose-binding protein (pRBPA248T, 31.0 kDa) | 1 N GnHCl | 1N GnHCl, 10 mM HEPES (pH 7.6), 0.15 M KAc, 2 mM TCEP |
| Galactose-binding protein (pGBP, 35.7 kDa) | 2 N GnHCl | 1 N GnHCl, 10 mM HEPES (pH 7.6), 0.3 M KAc, 1 mM EGTA, 2 mM TCEP |
| Alkaline phosphatase (pPhoA, 49.4 kDa) | a, 6 M urea; b, 0.8 N GnHCl | 6 M urea, 10 mM HEPES (pH 7.6), |
| Phosphoporin E (pPhoE, 38.9 kDa) | a, 1.5 N GnHCl; b, 2.5 N GnHCl | 4 N GnHCl, 10 mM HEPES (pH 7.6), 0.15 M KAc, 2 mM TCEP |
| Maltoporin (pLamB, 49.9 kDa) | a, 2 N GnHCl; b, 3 N GnHCl | 8 M urea, 10 mM HEPES (pH 7.6), 0.15 mM KAc, 2 mM TCEP |
| Outer membrane protein A (proOmpA, 37.2 kDa) | a, 1 N GnHCl; b, 2 N GnHCl | 4 M urea, 10 mM HEPES (pH 7.6), 0.1 M KAc, 2 mM TCEP |
| Integral membrane domain of OmpA (proOmpAN176, 21.2 kDa) | a, 1.5 N GnHCl; b, 2.5 N GnHCl | 5 M urea, 10 mM HEPES (pH 7.6), 2 mM TCEP |
Preparation of cytoplasmic inner membrane vesicles.
IMV were prepared from E. coli strain HB4105, which carries a ΔuncBC mutation to eliminate the proton-translocating ATPase, which would interfere with the translocation assay by hydrolyzing ATP. The strain harbors the same plasmid (pEXP2) as that used for purification of SecYEG. Bacterial cells were grown in a rich medium (LB) with glucose (0.5% wt/vol) and ampicillin (100 µg/ml) at 35°C. When the preculture reached an OD560 of ∼0.7, glucose was removed from the culture by centrifugation (14,000 × g for 10 min at 4°C; SLC-6000; Sorvall), followed by suspension in cold LB medium with ampicillin (100 µg/ml), and stored at 7°C overnight. The culture was diluted to an OD560 of 0.1, and growth was continued the next day. To induce the expression of the translocon SecYEG, arabinose (0.5%, wt/vol) was added when the culture reached an OD560 of 0.6. The culture was further grown to an OD560 of 1.5 and harvested by centrifugation. Cells were disrupted using a chilled French pressure cell at 8,000 lb/in2. The cytoplasmic membranes were isolated by equilibrium density gradient centrifugation (57). The fractions containing sucrose with densities of 1.14 to 1.18 g·ml−1, which corresponds to the density of cytoplasmic inner membrane vesicles, were collected, brought to 4 M urea, and incubated for 30 min on ice, followed by centrifugation to remove bound SecA. The time of centrifugation was corrected for the viscosity and density of urea. The pellet was washed once in 10 mM HEPES, 0.3 M KAc, and 5 mM magnesium acetate [Mg(Ac)2], pH 7.6, and suspended in the same buffer. The suspensions were stored in aliquots at −80°C. The level of production of SecYEG over the chromosomal level was determined to be ∼50-fold by Western blotting of the uninduced and induced cultures.
Preparation of proteoliposomes.
E. coli polar lipid extracts (Avanti) in chloroform were taken to dryness using a stream of dry nitrogen. The lipid films were held in a vacuum chamber overnight and then hydrated by suspension in 10 mM HEPES, pH 7.6, 30 mM KAc, and 1 mM Mg(Ac)2. The suspension was forced through a polycarbonate membrane (100-nm pore diameter; Liposofast; Avestin) and passed back and forth between two gas-tight glass syringes to get unilamellar liposomes. Proteoliposomes were formed as previously described (24). Liposomes in the presence of the detergent dodecyl-β-maltoside at a ratio of 4.65 mM detergent to 5 mM lipids, a level sufficient to swell liposomes but not disrupt them (58), were incubated at room temperature for 3 h in a rotary mixer. The proteins to be incorporated were then added. The following concentrations were used for the standard reconstitution (PLYEG): SecYEG, 5.2 µM; for the coassembled proteoliposomes (PLYEG·SecA), SecYEG, 5.2 µM; SecA dimer, 5 µM (24). After another hour of incubation, detergent was removed by addition of BioBeads SM-2 (Bio-Rad). The proteoliposome suspension was centrifuged at 436,000 × g for 20 min at 4°C (TL100.1 rotor; Beckman Coulter), and the pellet was washed twice with the same buffer as that used for lipid suspension to give a concentration of 10 mM lipids and 10 µM SecY. The suspensions were stored in aliquots at −80°C.
In vitro protein translocation and ATPase assays.
Both translocation and ATPase assays were carried out in the same reaction mixture, made up in glass tubes (12 by 75 mm) on ice. The mixture contained 20 mM HEPES, 230 to 250 mM KAc, 5 mM Mg(Ac)2, 1 mM DTT, 1 mM EGTA, 3.3 mM ATP, and an ATP-regenerating system consisting of 7.5 mM phosphocreatine and 37 U/ml creatine phosphokinase. NADH (1.7 mM) was added when translocation was carried out with IMV. SecA was added if the translocation mixtures included IMV or proteoliposomes containing SecYEG only (PLYEG). No SecA was added to coassembled proteoliposomes (PLYEG·SecA), since it was already present. SecB and either proteoliposomes or inner membrane vesicles were added to the mixture, followed by addition of the [14C]leucine radiolabeled precursor. The level of radioactivity (counts per minute per microliter) in each mixture was determined by moving 5 μl of duplicate into 100 μl water held in scintillation vials to which 3 ml of 30% Scintisafe (Fisher Scientific) was added. The concentration of precursor (in micrograms per milliliter) in each mix was determined from the specific activity (counts per minute per microgram) of purified proteins. To assess ATP hydrolysis, [γ-32P]ATP (specific activity, 9 µCi/µmol; final concentration, 3 to 5 nM; Perkin Elmer) was added to the remaining translocation mix. It is important to remove samples for determination of the level of 14C radioactivity before adding [γ-32P]ATP.
The reaction was initiated by transferring the glass tube to a water bath at 30°C. Samples were taken on ice (t = 0) and at given times after transfer to 30°C. At each time, to stop ATP hydrolysis and translocation, 8 μl of the reaction mixture was added to tubes held on ice containing 6 μl of 42 mM EDTA to stop ATP hydrolysis, 25 mM DTT, and denaturant (Table 3 lists specific denaturant required for each precursor). After an incubation of 10 min, 2 µl was removed from the sample tubes for the ATPase assay. The remainder of each sample was subjected to proteolysis by addition of proteinase K (4.5 U/ml) and incubation for 15 min on ice, except for proOmpAN176, which was incubated at 40°C. The protease activity was terminated by addition of trichloroacetic acid (final concentration, 11%) to precipitate the proteins. The washed precipitate was dissolved in sample buffer containing DTT (10 mM), boiled for 4 to 5 min, and subjected to SDS–12% PAGE.
To ensure that SecYEG was limiting during the assay, precursor was added in excess (1.2 to 3 µM) over the active translocons (0.2 to 0.4 µM) with SecA at 1.2 µM dimer. In both coassembled proteoliposomes (PLYEG·SecA) and vesicles, the active translocons represent 55% of the total accessible translocons (95% accessible in vesicles and 55% accessible in proteoliposomes) (24, 27, 28). The same ratio of precursor to active translocons was used when comparing activity of proteoliposomes with activity of vesicles. SecB4 was added at a 1:1 molar ratio to precursor to keep the precursor unfolded. Precursors were stored in solutions containing either urea or guanidine hydrochloride (GnHCl) (Table 2) and diluted into the translocation assay. A final denaturant concentration of up to 200 mM urea or 46 mM GnHCl had no effect on translocation.
Analysis of translocation data.
All samples from a given translocation experiment were loaded on a gel in equivalent volumes. The gels were dried, and the radioactivity of proteins in the gels was measured using a phosphorimager (Fujifilm FLA 3000) in the linear range of its response. The intensities of the radioactive bands were quantified (Image Gauge 4.0), and the percentage of full-length polypeptides that were protected at each time point, were estimated by comparing the band intensities to those of the input, i.e., the sample taken from the same reaction mix on ice (t = 0) in duplicate but not subjected to proteinase K treatment. In addition to the full-length species, for some precursors there are protected intermediates, which were partially translocated at the time the assay was stopped. In such scenarios, molarities of both full-length and intermediate-length species were included in the calculation of molarity translocated. No intermediate-length species were observed for the three smallest precursors, pRBPA248T, properiOmpA, and proOmpAN176. The amount of precursor protected was plotted as a function of time. Origin software was used to fit data to a single exponential rise to maximum with the equation y = y0 + A(1 – e-k(t – t0)), where A is the maximal amplitude of the reaction, k is the apparent rate constant, and t – t0 corrects for the initial time lag. The time for the reaction mixture to come to 30°C was measured using a thermocouple, and it was found to be between 15 and 30 s depending on the volume.
Determination of number of active translocons.
To determine the number of active translocons in the in vitro systems studied here, assays were carried out as described above, except the 14C-labeled proOmpA was oxidized to form a disulfide-bonded loop that could not pass through the channel and stops translocation. The number of translocons that were active but stalled was determined by quantification of the radiolabeled intermediate (24). For the preparations used in this study, the active fractions varied between 50% and 55% for PLYEG · SecA and 10% for PLYEG+SecA.
Analysis of SecA ATPase activity by thin-layer chromatography.
One-microliter samples for each time point that had been subjected to EDTA treatment (18 mM) as described above were analyzed by thin-layer chromatography using a polyethyleneimine (PEI)-cellulose F plate (20 by 20 cm; Merck). The plate (stationary phase) was developed in a closed container with 125 mM KH2PO4 as the mobile phase (59). The plate was dried and radioactivity was determined using a phosphorimager (Fujifilm FLA 3000). ATP remained at the origin, and the inorganic phosphate migrated with an Rf of ∼0.4. The ATP hydrolyzed (in micromolar concentration) was plotted as a function of time and fitted to a single exponential rise to maximum as described above. The spontaneous hydrolysis of [γ-32P]ATP was determined to be less than 1% by dilution of [γ-32P]ATP into a translocation mixture without SecA.
Calculation of the coupling of energy of ATP hydrolysis to translocation.
The coupling of ATP hydrolysis to translocation of precursors is expressed as the moles of ATP hydrolyzed per mole of precursor protected (34). Data from both translocation of precursors and the associated ATPase activities were fitted individually using a single exponential rise to maximum. From the fits, the moles of precursor protected and the moles of ATP hydrolyzed during the first 15 s were calculated. Only early time points were considered, because the affinity of SecA for ADP is five times higher than that for ATP (60) and accumulation of ADP at later time points would suppress the activity. Also, the generation of 32Pi continues even though translocation has stopped, because translocation ATPase activity is replaced by membrane ATPase activity, which occurs when SecA binds SecYEG in the absence of a precursor.
Accession number(s).
All plasmids used in this study that carry precursor protein genes have been deposited in Addgene and are available under the following accession numbers: pAL800, 119306; pAL942, 119721; pAL928, 119853; pAL950, 119748; pAL831, 119301; pAL832, 119305; pAL612, 119860; pAL725, 119862; pAL829, 119697.
ACKNOWLEDGMENTS
We are grateful to Chunfeng Mao for many helpful discussions of the work and for critically reading the manuscript. We thank Angela Lilly and Yuying Suo for determination of the level of production of SecYEG, Angela Lilly for construction of all plasmids used in this work, and Yuying Suo for purification of the precursors.
This work was supported by an endowment from the Hugo Wurdack Trust at the University of Missouri and National Institutes of Health grant GM29798 (to L.L.R.).
We have no conflicts of interest regarding this report.
REFERENCES
- 1.Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 2.Crane J, Randall L. 2017. The Sec system: protein export in Escherichia coli. EcoSal Plus 2017. doi: 10.1128/ecosalplus.ESP-0002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabelli RJ, Dolan KM, Qian LP, Oliver DB. 1991. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J Biol Chem 266:24420–24427. [PubMed] [Google Scholar]
- 4.Matsuyama S-I, Mizushima S. 1995. Biochemical analyses of components comprising the protein translocation machinery of Escherichia coli. Adv Cell Mol Biol Membr Organelles 4:61–84. doi: 10.1016/S1874-5172(06)80007-6. [DOI] [Google Scholar]
- 5.Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. 2007. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. 2008. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol 382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randall LL, Hardy SJ. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci 59:1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topping TB, Randall LL. 1994. Determination of the binding frame within a physiological ligand for the chaperone SecB. Protein Sci 3:730–736. doi: 10.1002/pro.5560030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khisty VJ, Munske GR, Randall LL. 1995. Mapping of the binding frame for the chaperone SecB within a natural ligand, galactose-binding protein. J Biol Chem 270:25920–25927. doi: 10.1074/jbc.270.43.25920. [DOI] [PubMed] [Google Scholar]
- 10.Smith VF, Hardy SJ, Randall LL. 1997. Determination of the binding frame of the chaperone SecB within the physiological ligand oligopeptide-binding protein. Protein Sci 6:1746–1755. doi: 10.1002/pro.5560060815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randall LL, Topping TB, Suciu D, Hardy SJ. 1998. Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci 7:1195–1200. doi: 10.1002/pro.5560070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall LL, Hardy SJ, Topping TB, Smith VF, Bruce JE, Smith RD. 1998. The interaction between the chaperone SecB and its ligands: evidence for multiple subsites for binding. Protein Sci 7:2384–2390. doi: 10.1002/pro.5560071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crane JM, Suo Y, Lilly AA, Mao C, Hubbell WL, Randall LL. 2006. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J Mol Biol 363:63–74. doi: 10.1016/j.jmb.2006.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilly AA, Crane JM, Randall LL. 2009. Export chaperone SecB uses one surface of interaction for diverse unfolded polypeptide ligands. Protein Sci 18:1860–1868. doi: 10.1002/pro.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C, Rossi P, Saio T, Kalodimos CG. 2016. Structural basis for the antifolding activity of a molecular chaperone. Nature 537:202–206. doi: 10.1038/nature18965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lill R, Dowhan W, Wickner W. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60:271–280. doi: 10.1016/0092-8674(90)90742-W. [DOI] [PubMed] [Google Scholar]
- 17.Arkowitz RA, Wickner W. 1994. SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J 13:954–963. doi: 10.1002/j.1460-2075.1994.tb06340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. 2011. Structure and function of a membrane component SecDF that enhances protein export. Nature 474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukazaki T. 2018. Structure-based working model of SecDF, a proton-driven bacterial protein translocation factor. FEMS Microbiol Lett 365:fny112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuyama S, Fujita Y, Mizushima S. 1993. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J 12:265–270. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze RJ, Komar J, Botte M, Allen WJ, Whitehouse S, Gold VAM, Lycklama A Nijeholt JA, Huard K, Berger I, Schaffitzel C, Collinson I. 2014. Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG–SecDF–YajC–YidC. Proc Natl Acad Sci U S A 111:4844–4849. doi: 10.1073/pnas.1315901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. 1990. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62:649–657. doi: 10.1016/0092-8674(90)90111-Q. [DOI] [PubMed] [Google Scholar]
- 23.Nishiyama K, Mizushima S, Tokuda H. 1993. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J 12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao C, Cheadle CE, Hardy SJS, Lilly AA, Suo Y, Gari RRS, King GM, Randall LL. 2013. Stoichiometry of SecYEG in the active translocase of Escherichia coli varies with precursor species. Proc Natl Acad Sci U S A 110:11815–11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu GP, Topping TB, Cover WH, Randall LL. 1988. Retardation of folding as a possible means of suppression of a mutation in the leader sequence of an exported protein. J Biol Chem 263:14790–14793. [PubMed] [Google Scholar]
- 26.Kim J, Lee Y, Kim C, Park C. 1992. Involvement of SecB, a chaperone, in the export of ribose-binding protein. J Bacteriol 174:5219–5227. doi: 10.1128/jb.174.16.5219-5227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futai M. 1974. Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J Membr Biol 15:15–28. doi: 10.1007/BF01870079. [DOI] [PubMed] [Google Scholar]
- 28.Seckler R, Wright JK. 1984. Sidedness of native membrane vesicles of Escherichia coli and orientation of the reconstituted lactose: H+ carrier. Eur J Biochem 142:269–279. doi: 10.1111/j.1432-1033.1984.tb08281.x. [DOI] [PubMed] [Google Scholar]
- 29.Gennis RB. 1989. Biomembranes–molecular structure and function. Springer-Verlag, New York, NY. [Google Scholar]
- 30.Tamm LK, Hong H, Liang B. 2004. Folding and assembly of β-barrel membrane proteins. Biochim Biophys Acta 1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Kleinschmidt JH. 2015. Folding of beta-barrel membrane proteins in lipid bilayers–unassisted and assisted folding and insertion. Biochim Biophys Acta 1848:1927–1943. doi: 10.1016/j.bbamem.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Lill R, Cunningham K, Brundage LA, Ito K, Oliver D, Wickner W. 1989. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J 8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomkiewicz D, Nouwen N, van Leeuwen R, Tans S, Driessen AJM. 2006. SecA supports a constant rate of preprotein translocation. J Biol Chem 281:15709–15713. doi: 10.1074/jbc.M600205200. [DOI] [PubMed] [Google Scholar]
- 34.Mao C, Hardy SJS, Randall LL. 2009. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J Bacteriol 191:978–984. doi: 10.1128/JB.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. 2008. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers EL, Randall LL. 1995. Export of periplasmic galactose-binding protein in Escherichia coli depends on the chaperone SecB. J Bacteriol 177:1906–1907. doi: 10.1128/jb.177.7.1906-1907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumamoto CA, Beckwith J. 1985. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol 163:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collier DN, Bankaitis VA, Weiss JB, Bassford PJ Jr.. 1988. The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell 53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 39.Kumamoto CA. 1990. SecB protein: a cytosolic export factor that associates with nascent exported proteins. J Bioenerg Biomembr 22:337–351. doi: 10.1007/BF00763171. [DOI] [PubMed] [Google Scholar]
- 40.Kumamoto CA. 1989. Escherichia coli SecB protein associates with exported protein precursors in vivo. Proc Natl Acad Sci U S A 86:5320–5324. doi: 10.1073/pnas.86.14.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fessl T, Watkins D, Oatley P, Allen WJ, Corey RA, Horne J, Baldwin SA, Radford SE, Collinson I, Tuma R. 2018. Dynamic action of the Sec machinery during initiation, protein translocation and termination. Elife 7:e35112. doi: 10.7554/eLife.35112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danoff EJ, Fleming KG. 2011. The soluble, periplasmic domain of OmpA folds as an independent unit and displays chaperone activity by reducing the self-association propensity of the unfolded OmpA transmembrane β-barrel. Biophys Chem 159:194–204. doi: 10.1016/j.bpc.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiebel E, Driessen AJ, Hartl FU, Wickner W. 1991. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64:927–939. doi: 10.1016/0092-8674(91)90317-R. [DOI] [PubMed] [Google Scholar]
- 44.Suo Y, Hardy SJS, Randall LL. 2015. The basis of asymmetry in the SecA:SecB complex. J Mol Biol 427:887–900. doi: 10.1016/j.jmb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tani K, Shiozuka K, Tokuda H, Mizushima S. 1989. In vitro analysis of the process of translocation of OmpA across the Escherichia coli cytoplasmic membrane. A translocation intermediate accumulates transiently in the absence of the proton motive force. J Biol Chem 264:18582–18588. [PubMed] [Google Scholar]
- 46.Sato K, Mori H, Yoshida M, Tagaya M, Mizushima S. 1997. Short hydrophobic segments in the mature domain of ProOmpA determine its stepwise movement during translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem 272:5880–5886. doi: 10.1074/jbc.272.9.5880. [DOI] [PubMed] [Google Scholar]
- 47.Nouwen N, Berrelkamp G, Driessen AJ. 2009. Charged amino acids in a preprotein inhibit SecA-dependent protein translocation. J Mol Biol 386:1000–1010. doi: 10.1016/j.jmb.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 48.Bauer Benedikt W, Shemesh T, Chen Y, Rapoport TA. 2014. A “push and slide” mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell 157:1416–1429. doi: 10.1016/j.cell.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa A, Nakayama S, Yoshikaie K, Tanaka Y, Tsukazaki T. 2018. Remote coupled drastic beta-barrel to beta-sheet transition of the protein translocation motor. Structure 26:485–489. doi: 10.1016/j.str.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 50.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 51.Cannon KS, Or E, Clemons WM, Shibata Y, Rapoport TA. 2005. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Physiol 169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJ. 2005. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol 348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 53.Randall LL, Topping TB, Smith VF, Diamond DL, Hardy SJS, 1998. SeeB: a chaperone from Escherichia coli. Methods Enzymol 290:444–459. doi: 10.1016/S0076-6879(98)90037-4. [DOI] [PubMed] [Google Scholar]
- 54.Low KB. 1974. Experiments in molecular genetics. Jeffrey H. Miller. Q Rev Biol 49:151. doi: 10.1086/408025. [DOI] [Google Scholar]
- 55.Mitchell C, Oliver D. 1993. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol 10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 56.Danese PN, Murphy CK, Silhavy TJ. 1995. Multicopy suppression of cold-sensitive sec mutations in Escherichia coli. J Bacteriol 177:4969–4973. doi: 10.1128/jb.177.17.4969-4973.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thom JR, Randall LL. 1988. Role of the leader peptide of maltose-binding protein in two steps of the export process. J Bacteriol 170:5654–5661. doi: 10.1128/jb.170.12.5654-5661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lambert O, Levy D, Ranck J-L, Leblanc G, Rigaud J-L. 1998. A new “gel-like” phase in dodecyl maltoside–lipid mixtures: implications in solubilization and reconstitution studies. Biophys J 74:918–930. doi: 10.1016/S0006-3495(98)74015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levit MN, Grebe TW, Stock JB. 2002. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J Biol Chem 277:36748–36754. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 60.Fak JJ, Itkin A, Ciobanu DD, Lin EC, Song XJ, Chou YT, Gierasch LM, Hunt JF. 2004. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry 43:7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 61.Das S, Grady LM, Michtavy J, Zhou Y, Cohan FM, Hingorani MM, Oliver DB. 2012. The variable subdomain of Escherichia coli SecA functions to regulate SecA ATPase activity and ADP release. J Bacteriol 194:2205–2213. doi: 10.1128/JB.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]