Abstract
Regeneration of tissues that have been damaged by cell loss requires new growth, often via proliferation of precursor cells followed by differentiation to replace loss of specific cell types. When regeneration occurs after normal differentiation of the tissue is complete, developmental pathways driving differentiation must be re-activated. How proliferation and differentiation are induced and balanced during regeneration is not well understood. To investigate these processes, we utilized a paradigm for tissue damage and regeneration in the developing Drosophila melanogaster eye. Previous studies have demonstrated that tissue damage resulting from extensive cell death stimulates quiescent, undifferentiated cells in the developing larval eye to re-enter the cell cycle and proliferate. Whether these cells are restricted to certain fates or can contribute to all retinal cell types and thus potentially be fully regenerative is not known. Here we found by fate mapping experiments that these cells are competent to differentiate into all accessory cell types in the retina but do not differentiate into photoreceptors, likely because cell cycle re-entry in response to damage occurs after photoreceptor differentiation has completed. We conclude that the ability to re-enter the cell cycle in response to tissue damage in the developing Drosophila eye is not restricted to precursors of a specific cell type and that cell cycle re-entry following damage does not disrupt developmental programs that control differentiation.
Keywords: Drosophila, compensatory proliferation, cell cycle, fate mapping, regeneration
Introduction
Different organisms have varying degrees of regenerative potential after tissue damage. Some organisms (e.g. salamanders) can regenerate entire limbs (Haas and Whited, 2017) and others (e.g. flatworms) can regenerate an entire body plan (Elliott and Sanchez Alvarado, 2013). Even organisms that are incapable of such dramatic regeneration contain tissues that have high regenerative capacity. For example, the mammalian liver regenerates entirely after hepatectomy removes two thirds of the tissue (Miyaoka et al., 2012). In all of these cases, several criteria must be fulfilled at the cellular level to carry out faithful tissue regeneration. First, new growth must occur. This growth often occurs by stimulating proliferation of stem cells or undifferentiated precursor cells. In other instances, such as the Drosophila ovary or intestine, growth after damage occurs not by cell proliferation but by cellular endoreduplication (Tamori and Deng, 2013; Losick et al., 2013). Second, cellular differentiation must occur, particularly in complex tissues that have sustained a sufficient level of damage that many different cell types were lost. In these instances, the cells that proliferate in response to tissue damage likely have some degree of pluripotency in order to repopulate multiple cell types.
Previous work has shown that cell cycle progression can influence differentiation outcomes (Sela et al., 2012; Pauklin and Vallier, 2013; Ruijtenberg and van den Heuvel, 2016; Krentz et al., 2017; Azzarelli et al., 2017; Meserve and Duronio, 2017). The degree to which re-entering the cell cycle in response to tissue loss impacts programs of differentiation of multiple cell types has not been extensively investigated. Here we explore this question using a tissue damage paradigm in the developing eye of Drosophila. When tissue damage is genetically induced in larval eye imaginal discs by expression of the pro-apoptotic protein, Hid, undifferentiated cells that are normally quiescent re-enter the cell cycle and divide to replenish the pool of precursor cells depleted by apoptosis (Fan and Bergmann, 2008; Meserve and Duronio, 2015). This process is called compensatory proliferation (CP) (Huh et al., 2004; Worley et al., 2012). Inhibition of CP exacerbates the developmental defects caused by cell death-induced tissue damage, suggesting cells resulting from CP contribute to the adult eye (Meserve and Duronio, 2015). However, whether cells that undergo CP can differentiate into all retinal cell types or are restricted to a subset of retinal cell types is not known. We addressed this question by using fate mapping to determine the types of differentiated cells resulting from CP. We found that CP provides a pool of cells that differentiate into all accessory cell types of the adult retina except photoreceptor neurons, likely because cell cycle re-entry after tissue damage occurs after photoreceptor differentiation has occurred. We conclude that cells reentering the cell cycle during CP in damaged Drosophila imaginal eye tissue are not highly restricted in terms of cell fate.
Materials and Methods
Drosophila culture conditions and stocks
All experiments were carried out at 25° C. For experiments with pupae, white prepupae were designated (counted as 0hr APF; after puparium formation) and aged for the appropriate number of hours. Stocks used are as follows (Bloomington Stock numbers are listed where applicable): w1118 as wild type, GMR-Gal4 [w*;P{GAL4-ninaE.GMR}12] #1104, UAS-E2fFUCCI/UAS-CycBFUCCI [w1118;P{UASp-GFP.E2f1.1–230}64, P{UASp-mRFP1.NLS.CycB.1–266}5] #55111, UAS-stg [w1118; P{UAS-stg.N}4] #4778, UAS-p35 [w*; P{UAS-p35.H}BH2] #5073, GMR-hid [P{GMR-hid}G1] #5771, tub-Gal80TS [w*; P{tubP-GAL80ts}7/TM6B] #7018. Transgenic genotypes described in all experiments (except GMR-hid alone) are heterozygous.
EdU feeding experiments
For EdU-containing food, 0.6 g of crushed Carolina Formula 4–24 Instant Drosophila Medium was placed in a narrow culture vial and mixed with 3.4 mL of 0.2mM EdU (5-Ethynyl-2′-deoxyuridine, Santa Cruz) and 25 mg/mL amaranth (Sigma) in H2O. Early 3rd instar larvae were placed on EdU-containing media for 10 hours. Larvae that had eaten food (confirmed by presence of amaranth dye in the gut) were transferred to unlabeled food and cultured until they were selected for dissection. Because larvae may not have eaten for the entire 10-hour period, there may be variation in the actual amount of time that EdU was incorporated into DNA among individual animals.
For quantification of EdU-labeling of each cell type, the total number of SMW cell types and EdU+ SMW cell types for WT were displayed based on previously determined numbers of different cell types in a stereotypical ommatidium (Kumar, 2012). Cells shared between ommatidia were accounted for in this calculation (for example, there are six secondary pigment cells surrounding each ommatidium, but each is shared by two ommatidia, so the number per ommatidium is three). We did this because the quantification for GMR-hid, Gal80TS, GMR>p35 retinas is displayed as total cell type percentage from a defined region with many ommatidia, not from numbers of cells per ommatidum. Therefore, we indicate four cone cells, eight photoreceptors, six pigment cells (two primary, three secondary, and one tertiary), and three non-neuronal bristle cells (ELAV-) and one neuronal bristle cell (ELAV+) per ommatidium. For SMW EdU+ cells in wild type, the numbers are the same except that only three photoreceptors will be labeled.
Immunostaining
Immunostaining of larval and pupal retinas was performed using standard protocols (Klein, 2008). Cleaved Caspase 3 (CC3) staining and detection of EdU incorporation were performed as previously described (Meserve and Duronio, 2015). For EdU pulse labeling of larval discs (such as in Fig. 1C), eye discs were incubated in EdU for 1 hour before fixation. Primary antibodies: 1/1000 rabbit α-PH3 (Millipore 06–570), 1/1000 mouse α-GFP (Abcam ab1218), 1/1000 rabbit α-RFP (Clontech 632496), 1/100 rat α-ELAV (DSHB 9F8A9), 1/100 mouse α-Cut (DSHB 2B10), 1/200 guinea pig α-Sens (H. Bellen), 1/100 rabbit α-CC3 (Cell Signaling 9661). Secondary antibodies (all 1/1000): Oregon Green 488-conjugated goat α-mouse (Invitrogen O6380), Cy3-conjugated goat α-mouse (Jackson ImmunoResearch 115–165-003), Cy5-conjugated goat α-mouse (Jackson ImmunoResearch 115–175-146), Alexa 488-conjugated goat anti-rabbit (Jackson ImmunoResearch 111–545-144), Rhodamine Red-conjugated goat α-rabbit (ThermoFisher Scientific R6394), Cy5-conjugated goat α-rabbit (Abcam ab6564), Cy2-conjugated goat α-rat (Jackson ImmunoResearch 112–225-143), Cy3-conjugated goat α-rat (Jackson ImmunoResearch 112–165-143), Cy5-conjugated goat anti-rat (Jackson ImmunoResearch 712–175-153), Cy5-conjugated donkey α-guinea pig (Jackson ImmunoResearch 706–175-148).
Fig. 1: G1-arrested cells re-enter the cell cycle in response to apoptosis in the larval eye.
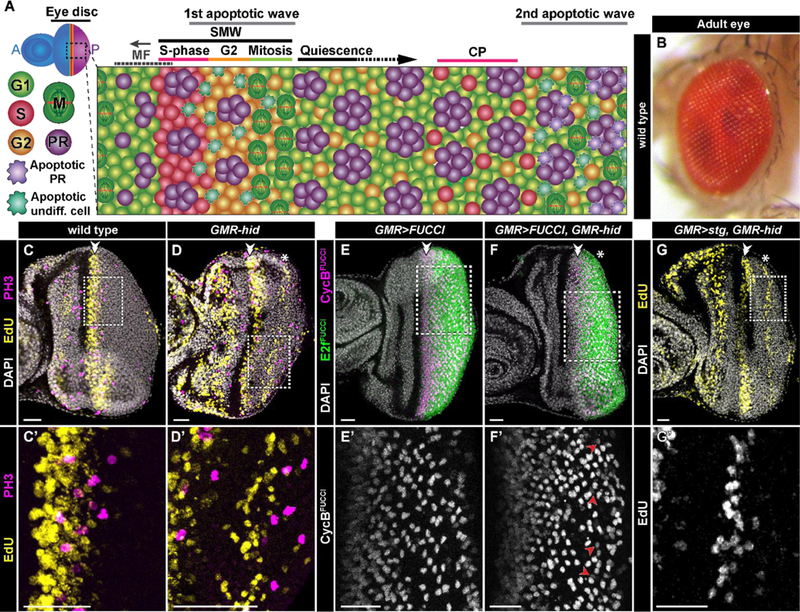
A) Schematic of eye disc development in a GMR-hid third instar larva. Following G1 arrest in the morphogenetic furrow (MF), cells either begin differentiation into photoreceptors (PR) or progress through the cell cycle in the second mitotic wave (SMW). Cells then either divide and arrest in G1, awaiting differentiation cues, or remain arrested in G2. Apoptosis induced by hid expression following the MF results in some cells re-entering the cell cycle and undergoing compensatory proliferation (CP). B) Adult eye of a wild type Drosophila melanogaster. C-C’) In a wild type larval disc, no cells posterior to the SMW (double arrowhead here and in subsequent panels) undergo S-phase (EdU, yellow) or mitosis (PH3, magenta). For this and subsequent panels the dashed box indicates area of magnification in the corresponding panel (e.g. C’). (D-D’) GMR-hid expression results in CP (asterisk here and in subsequent panels), wherein cells enter S phase (EdU, yellow) and undergo division (PH3, magenta). E-F’) GMR-Gal4 drives UAS-CyclinBFUCCI (magenta, E,F; grey, E’,F’) and UAS-E2f1FUCCI (green, E,F) in wild type (E-E’) and GMR-hid (F-F’) larval discs. Cells that express CyclinBFUCCI posterior to the SMW are arrested in G2 in wild type (E’), and are either arrested in G2 (the majority) or undergoing CP in GMR-hid (F’ red arrowheads, which indicate late S-phase cells that accumulate CycBFUCCI and lack E2f1FUCCI). A similar pattern was observed in 6/6 GMR>FUCCI, GMR-hid retinas. (G-G’). Ectopic GMR>stg expression prevents G2 arrest but does not inhibit CP in GMR-hid larval eye discs. Scale bar=20 µM.
Results and Discussion
G1-arrested cells enter into compensatory proliferation
Differentiation of cells within the Drosophila eye begins during third instar larval development in a wave that sweeps across the eye imaginal disc. This wave initiates at the posterior of the disc and progresses anteriorly across the disc and is identifiable by an associated transient apical constriction called the morphogenetic furrow (MF). Cells within the MF arrest in G1 phase of the cell cycle and differentiation commences with the specification of R8 photoreceptors from a subset of these cells. R8 cells are required for the subsequent differentiation of other retinal cell types, including R2, R5, R3, and R4 photoreceptors, which are specified shortly after R8 (Treisman, 2013). G1 arrested cells that haven’t begun differentiation into the first five photoreceptors enter a synchronous S phase just posterior to the MF (Fig. 1A, C-C’). A number of cells that enter S phase arrest in the subsequent G2, while the majority divide and subsequently arrest in G1 (Baker and Yu, 2001) (Fig. 1A). Although not all cells that undergo S phase following the MF divide during larval development, for historical reasons this coordinated entry into S phase is commonly referred to as the “second mitotic wave” (SMW) (Wolff and Ready, 1991; Baker, 2007). The G1 and G2 populations resulting from the SMW are the pool of precursor cells from which the remaining photoreceptors (i.e. R1, R6 and R7) and all accessory cell types (e.g. cone cells, pigment cells, and mechanosensory bristles) are recruited during retinal development. The end result is a fully differentiated compound eye with ~800 ommatidia, the photoreception units of the adult retina (Kumar, 2012) (Fig. 1B).
Our model of regeneration in the Drosophila eye makes use of GMR-hid, a transgene that activates extensive apoptosis posterior to the MF (Fig. 1A and Fig. 3). The tissue damage resulting from this cell death induces normally quiescent cells posterior to the SMW (Fig. 1C) to reenter the cell cycle. The resulting wave of CP can be visualized as EdU-labeled cells well posterior to the MF (Fig. 1D-D’; quantified in Fig. 3F) (Fan and Bergmann, 2008; Meserve and Duronio, 2015).
We considered the possibility that the ability to re-enter the cell cycle might be restricted to a subset of undifferentiated cells with a specific fate. We previously demonstrated that G2-arrested cells posterior to the MF are selected as sensory organ precursors (SOP) that divide during pupal development and differentiate into the interommatidial bristle groups of the adult eye (Meserve and Duronio, 2017). This result suggests that the cell cycle influences subsequent differentiation. Therefore, to begin asking whether CP cells have restricted cell fates, we first determined whether they originate from the G1 or G2 cell populations posterior to the MF. Cell cycle position in whole tissues can be visualized by expressing fluorescent ubiquitination-based cell cycle indicators (FUCCI) ((Zielke et al., 2014), Fig. 1E-E’). These indicators are comprised of fluorescent proteins fused to a degron sequence recognized by an E3 ubiquitin ligase that is active only at a particular phase of the cell cycle, resulting in accumulation of the fluorescent protein only in the cell cycle phase in which the E3 is not active (e.g. S and G2 for CyclinBFUCCI, a substrate of the Anaphase Promoting Complex, which is active only during mitosis and G1; and G1, G2, and M for E2f1FUCCI, a substrate of CRL4Cdt2, which is only active during S phase; Fig 1E, F). We hypothesized that because G2-arrested cells are primed to divide and re-enter S-phase during pupal development, they may be precociously activated to divide and then enter S-phase following tissue damage in larvae. However, in GMR-hid eye discs, the wave of CP S-phase precedes mitosis (Fig. 1D-D’), suggesting that CP cells enter the cell cycle from G1 and not G2. In addition, the pattern of G2-arrested cells is not greatly disrupted by hid expression (Fig. 1F-F), as would be expected if many G2 cells were re-entering the cell cycle. Finally, stg expression, which prevents G2 arrest (Baker and Yu, 2001; Meserve and Duronio, 2017), does not disrupt CP in GMR-hid larval discs (Fig. 1G-G’). These data suggest that cells undergoing CP primarily originate from the G1 population of undifferentiated cells posterior to the MF.
The CP wave induced by GMR-hid occurs after all eight photoreceptors have differentiated (Fan and Bergmann, 2008). Therefore, in order for cells that undergo CP to replace lost photoreceptors, differentiation signals driving photoreceptor recruitment would likely have to be re-activated in the damaged tissue. Interestingly, while some photoreceptors undergo hid-induced apoptosis, R8 is resistant to Hid and most R8 cells survive to at least the end of larval development in GMR-hid discs (Fan and Bergmann, 2014). We hypothesized that perhaps these apoptosis-resistant R8 cells could re-initiate photoreceptor differentiation in response to cell loss. This re-programming and re-specification has been observed during regeneration in wing imaginal discs, where cells temporarily lose cell fate following tissue damage (Smith-Bolton et al., 2009; Repiso et al., 2013; Ahmed-de-Prado et al., 2018;). Therefore, we next determined whether cells resulting from CP are restricted to certain fates or are competent to become any retinal cell type.
EdU labeling for fate mapping of proliferating cells
Our strategy for fate mapping involved feeding third instar larvae EdU in order to label cells in S phase as they undergo CP, followed by detection of EdU-positive cells in pupal retinas at a stage when cell differentiation is complete. First, we developed an EdU-labeling protocol in which third instar wild type larvae are pulse-fed EdU for 10 hours, then allowed to develop and pupate on unlabeled food. We subsequently dissected retinas from these pupae 24hrs after puparium formation (APF) (Fig. 2A-B). Using this protocol for pulse labeling, we found EdU+ cells in the anterior portion of the pupal retina, where cells anterior to the MF were proliferating asynchronously during larval stages, and a stripe of EdU+ cells resulting from the SMW (Fig. 2B), similar to previous studies where EdU was injected in third instar larvae and stained during pupal stages (Wolff and Ready, 1991). All accessory cells (e.g. cone and pigment cells) are derived from the SMW (Wolff and Ready, 1991) and can be identified by expression of specific markers and apical/basal location of their nuclei within the ommatidium (Fig. 2C-F’). Accessory cells were labeled with EdU in a broad stripe along the D/V axis of pupal retinas (Fig. 2C-F’). In contrast, only the photoreceptors derived from the SMW were EdU+ (i.e. R1, R6, and R7), while photoreceptors that began to differentiate within the MF and never entered S phase were EdU-(i.e. R8, R5, R4, R3, and R2) (Fig. 2E-E’’). Importantly, no cells in the posterior of the retina were EdU+ because they were derived from a population of older precursor cells that had completed S phase of the SMW prior to our pulse labeling. These data indicate that we can precisely identify cells that are derived from the SMW and that any cells resulting from S phases that occur posterior to the SMW, such as during CP in GMR-hid larva, will be a distinct population of EdU+ cells in the posterior of the pupal retina.
Fig. 2: Cells that underwent S phase during larval development can be detected in pupal retinas.
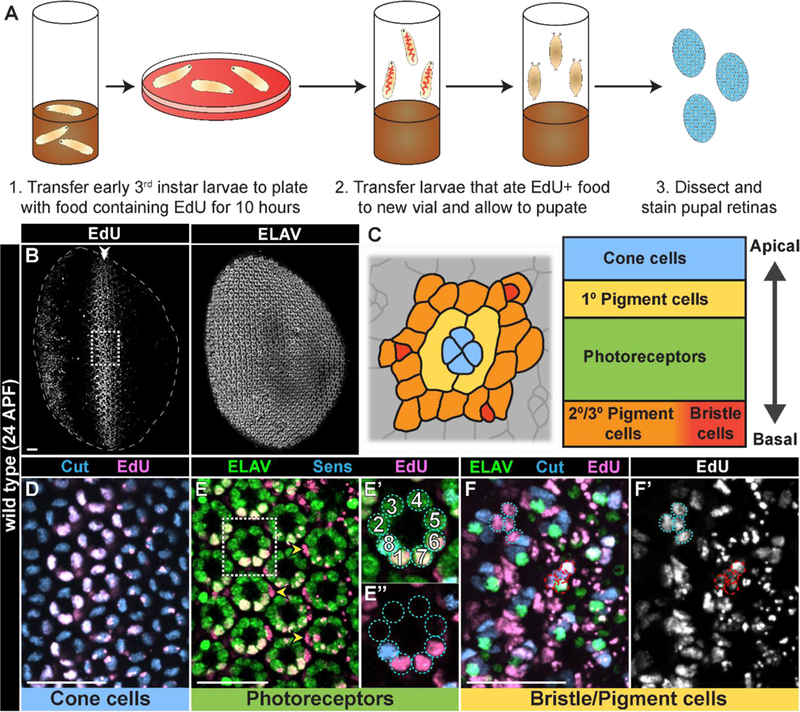
A) Diagram of EdU feeding protocol for detecting pupal retinal cells that underwent S phase during the 3rd larval instar stage. (B-B’) Wild type ~24hrs APF (after puparium formation) retina derived from 3rd instar larvae fed with EdU for 10 hours and stained for EdU incorporation (B) and ELAV (B’). Boxed area in B indicates representative area of magnification in D-F’ (panels are different retinas). Posterior of the retina is oriented to the right, and SMW-derived EdU+ cells are indicated with a double arrowhead. C) Top down schematic of an ommatidium at 24hrs APF (based on membrane staining; indicates cells boundaries) and apical/basal location of cell nuclei at this stage. Staining in D-F are nuclear. D-F’) EdU+ cells within the SMW label (D) Cut-expressing cone cells (blue); (E-E’’) primary pigment cells (yellow arrowheads; based on absence of ELAV and apical/basal location of nuclei) and ELAV (green)-expressing photoreceptors R1, R7, and R6, identified based on orientation to Sens-expressing R8 (blue). (F,F’) Cut-expressing basal bristle groups (red dashed circles; Cut, blue; neurons express ELAV (green)); and secondary/tertiary pigment cells (blue dotted circles; Cut/ELAV-negative). Scale bar=20 µM.
Compensatory proliferating cells differentiate into retinal accessory cells
We applied our EdU-labeling strategy to GMR-hid discs to determine which retinal cell types result from CP. Because GMR-hid adult eyes are drastically reduced in size as a result of continued apoptosis during pupal development (Grether et al., 1995) (Fig. 3A), we attenuated Hid activity by expressing UAS-p35 under control of GMR-Gal4 (GMR>p35). Expression of p35 alone inhibits both apoptosis and CP (Fan and Bergmann, 2008); therefore, we modulated the amount of p35 expression using a temperature sensitive Gal80 (a Gal4 repressor expressed constitutively using the tubulin promoter). At 25° C, apoptosis is present in the posterior eye disc of both GMR-hid, Gal80TS and GMR-hid, Gal80TS, GMR>p35 larvae (Fig. 3C, D), and CP is induced in GMR-hid, Gal80TS, GMR>p35 larvae (Fig. 3E-E’). We observe significantly more cells undergoing CP in GMR-hid, Gal80TS, GMR>p35 discs versus GMR-hid alone (Fig. 3F), likely because a reduced amount of apoptosis allows more undifferentiated cells to re-enter the cell cycle. Importantly, Gal80 does not fully suppress UAS-p35 at 25° C, as evidenced by a rescued adult eye phenotype in GMR-hid, Gal80TS, GMR>p35 flies; their eyes are much larger compared to GMR-hid, Gal80TS adults, although still rough (Fig. 3A, B; compare to Fig. 1B). This genotype allowed us to assay CP fate in largely intact pupal retinas.
Fig. 3: Gal80-attenuated expression of p35 rescues eye development but does not inhibit CP.
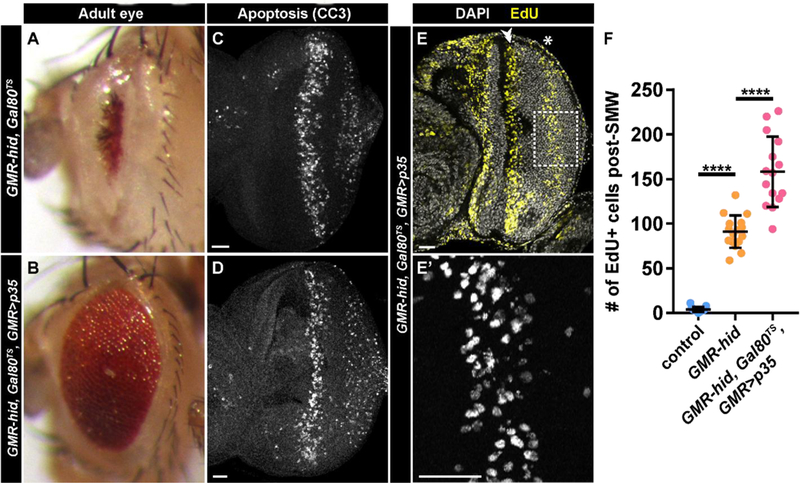
(A,B) Adult eyes from GMR-hid, Gal80TS (A) or GMR-hid, Gal80TS, GMR>p35 (B). (C,D) GMR-hid, Gal80TS (C) or GMR-hid, Gal80TS, GMR>p35 (D) larval eye discs stained with antibodies recognizing cleaved-caspase 3 (CC3). (E) GMR-hid, Gal80TS, GMR>p35 larval eye discs labeled with EdU (yellow) and DAPI (grey). Double arrowhead indicates SMW and asterisk indicates CP. (E’) Higher magnification (area indicated by dotted box in E) of EdU incorporation during CP GMR-hid, Gal80TS, GMR>p35 discs.
F) Quantification of the number of EdU+ (S-phase) cells posterior to the SMW. Control is w1118 or GMR-Gal4 (not significantly different). n=13 discs for control, 16 discs for GMR-hid, and 15 discs for GMR-hid, Gal80TS, GMR>p35. ****p<0.0001. Scale bar=20 µM.
We applied our EdU feeding protocol to GMR-hid, Gal80TS, GMR>p35 larvae, which resulted at 24hrs APF in robust labeling of pupal retinal cells derived from the SMW (Fig. 4A, arrowhead) as well as from CP (Fig. 4A, asterisk). Within the population of cells derived from CP, we observe EdU+ cone cells (Fig. 4B), pigment cells, and interommatidial bristle group cells (Fig. 4C-C’). We quantified the proportion of each cell type that was EdU+ using co-staining of ELAV (photoreceptors and bristle neurons) and Cut (cone cells and bristle groups) (Fig. 4D). We also quantified EdU+ cell types in retinas with only ELAV or only Cut staining to confirm our results (Fig. 4D). We identified the pigment cell population by lack of expression of ELAV or Cut. Consequently, some EdU+ cells may actually be undifferentiated cells, and we acknowledge this possibility in our quantification (Fig. 4D, first graph). Cone cells were labeled relatively infrequently and most often when the CP wave was at the far posterior end of the retina, likely because cone cells differentiate shortly after photoreceptors during larval development (Kumar, 2012). Bristle groups may arise from G2-arrested cells that are selected as SOPs since we observe CycBFUCCI positive cells in a subset of EdU-labeled cells that underwent CP (Fig. 4E-F). Alternatively, or in addition, these SOPs may be recruited from G1-arrested cells, which we previously have shown can occur (Meserve and Duronio, 2017). These data indicate that cells resulting from CP can differentiate into any accessory cell type.
Fig. 4: Compensatory proliferating cells contribute to non-photoreceptor, accessory cells types in the retina.
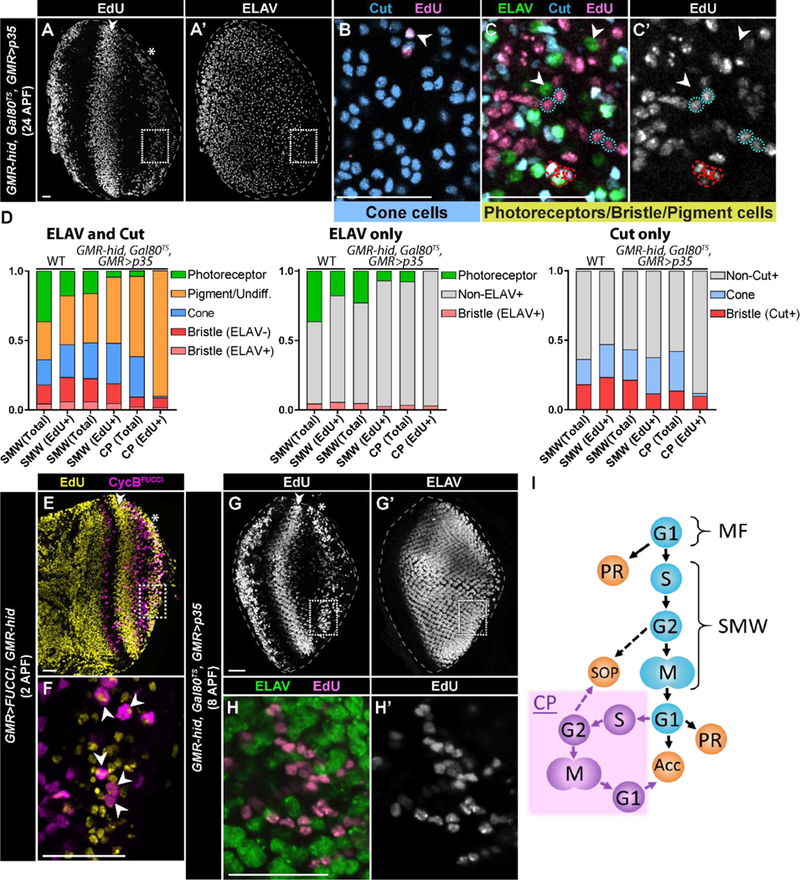
(A-C’) GMR-hid, Gal80TS, GMR>p35 retina at 24hrs APF stained for EdU incorporation (A), ELAV (A’,C), and Cut (B,C) (A, B, and C from different retinas). Retinas were derived from 3rd instar larvae fed with EdU for 10 hours. The SMW (double arrowhead) and CP (asterisk) are visible in A. (B) Magnification (dotted box in A, A’) of an apical section of the CP wave displays Cut expressing cone cells (blue) that are EdU+. (C,C’) Magnification of a basal section of the CP wave displays EdU+ (pink), Cut-expressing bristle groups (red dashed circles; Cut, blue; neurons express ELAV (green)) and pigment cells (blue dotted circles; Cut/ELAV-). ELAV expressing photoreceptors (Cut-) are EdU-(arrowheads). Note that the number of photoreceptors in these retinas is reduced relative to wild type due to the continual expression of hid. (D) Quantification of the types of EdU+ pupal retinal cells after feeding EdU to 3rd instar larvae for 10 hours. Retinas were stained for ELAV and Cut, only ELAV, or only Cut. Data are presented as a percentage of the total (i.e. 1.0). SMW (Total) and SMW (EdU+) for WT were calculated based on the numbers of cells in an ommatidium (see Methods). One retina was counted per graph for SMW in GMR-hid, Gal80TS, GMR>p35; CP analysis was performed on n=5 retinas (ELAV and Cut), n=2 retinas (ELAV only), and n=2 retinas (Cut only). At least 50 EdU+ cells and 250 total cells were counted per retina. Pigment cells were identified by lack of expression of ELAV or Cut; this population may include undifferentiated (Undiff.) cells as well. (E-F) GMR>FUCCI, GMR-hid retina at ~2hrs APF derived from 3rd instar larvae fed with EdU for 10 hours and stained for EdU incorporation. Box in E,E’ indicates area of magnification in F, where a subset of cells that were EdU-labeled in the CP wave express CycBFUCCI (arrowheads). (G,G’) GMR-hid, Gal80TS, GMR>p35 retina at ~8hrs APF derived from 3rd instar larvae fed with EdU for 10 hours and stained for EdU incorporation (G) and ELAV (G’). (H,H’) Magnification (dotted box in G) demonstrates that ELAV expressing photoreceptors (green) are not labeled with EdU (pink in H, gray in H’). Scale bars=20 µM. (I) Schematic of the relationship between cell cycle phase (G1,S,G2,M) and precursor cell fate in damaged GMR-hid larval retinas. SOP denotes sensory organ precursor cells that give rise to the interommatidial bristle group of cells; PR denotes photoreceptor; Acc denotes accessory cell. Purple denotes compensatory proliferation (CP). MF, morphogenetic furrow. SMW, second mitotic wave.
In contrast, photoreceptor cells present at 24hrs APF in GMR-hid, Gal80TS, GMR>p35 pupal retinas (Fig. 4A’) were not labeled with EdU (Fig. 4C-D). One possibility for this result is that CP cells differentiate into photoreceptors that are eliminated before 24hrs APF (compare drastic reduction in numbers of photoreceptors between Fig. 2B’ and Fig. 4A’, 4D). To address this question, we dissected EdU-labeled retinas at ~8hrs APF when the majority of photoreceptors are still present, though a significant proportion have undergone apoptosis (Fig. 4G-G’). At this time point of retinal development we did not detect any EdU+ photoreceptors in cells resulting from CP (Fig. 4H-H’) (n=8 discs, >75 EdU+ cells counted per disc). This result suggests that cells resulting from CP do not differentiate into photoreceptors.
A critical aspect of repairing tissue damage is the replacement of cells lost due to cell death. In damaged tissues, cell cycle re-entry of quiescent precursor cells can promote regeneration (Heber-Katz et al., 2013), as occurs in the developing Drosophila larval eye disc following genetically induced tissue damage using GMR-hid (Meserve and Duronio, 2015). In order to approach full regeneration, cells derived from CP would have to contribute to multiple types of differentiated cells. Indeed, we found here that cells that undergo CP contribute to all accessory cell types in the Drosophila retina. However, CP cells do not differentiate into photoreceptors (Fig. 4I), suggesting R8 photoreceptors are likely not reactivated to drive photoreceptor differentiation following damage. It will be interesting to determine whether death of photoreceptors earlier in development, shortly after they are specified, would result in replacement of those photoreceptors, or if R8 cells can be genetically re-activated at later stages of development to drive recruitment and replacement of other lost photoreceptors. In this GMR-hid paradigm, compensatory proliferating cells contribute to an undifferentiated cell population that can subsequently differentiate into each type of retinal accessory cell. Thus, in addition to signals stimulating cell cycle re-entry, developmental programs driving differentiation may need to be re-activated in a regenerating tissue to ensure that all necessary cell types are replaced. Moreover, in this tissue, reactivation of the cell cycle doesn’t interfere with these differentiation programs.
HIGHLIGHTS.
Quiescent cells re-enter the cell cycle in response to damage in the Drosophila eye.
These cells differentiate into retinal accessory cell types but not photoreceptors.
Multiple different programs of differentiation can still occur after tissue damage.
Compensatory proliferating cells in damaged tissue are not highly lineage restricted.
Acknowledgements
We thank the Bloomington Drosophila Stock Center, supported by P40-OD018537, for stocks used in this study. We also thank Hugo Bellen and Norman Zielke for stocks and reagents used in this study.
Funding
This work was supported by the National Institutes of Health (F31-AG044957 to JHM and R01-GM057859 and R01-GM058921 to RJD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed-de-Prado S, Diaz-Garcia S, Baonza A, 2018. JNK and JAK/STAT signalling are required for inducing loss of cell fate specification during imaginal wing discs regeneration in Drosophila melanogaster. Dev. Biol [DOI] [PubMed]
- Azzarelli R, Hurley C, Sznurkowska MK, Rulands S, Hardwick L, Gamper I, Ali F, McCracken L, Hindley C, McDuff F, Nestorowa S, Kemp R, Jones K, Gottgens B, Huch M, Evan G, Simons BD, Winton D, Philpott A, 2017. Multi-site Neurogenin3 Phosphorylation Controls Pancreatic Endocrine Differentiation. Dev. Cell 41, 274–286 e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M, 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol 66, 57–80. [PubMed] [Google Scholar]
- Baker NE, 2007. Patterning signals and proliferation in Drosophila imaginal discs. Curr. Opin. Genet. Dev 17, 287–293. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY, 2001. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell 104, 699–708. [DOI] [PubMed] [Google Scholar]
- Elliott SA, Sanchez Alvarado A., 2013. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip. Rev. Dev. Biol 2, 301–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A, 2008. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev. Cell 14, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A, 2014. Multiple mechanisms modulate distinct cellular susceptibilities toward apoptosis in the developing Drosophila eye. Dev. Cell 30, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H, 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 9, 1694–1708. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Whited JL,2017. Advances in Decoding Axolotl Limb Regeneration. Trends Genet 33, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber-Katz E, Zhang Y, Bedelbaeva K, Song F, Chen X, Stocum DL, 2013. Cell cycle regulation and regeneration. Curr. Top. Microbiol. Immunol 367, 253–276. [DOI] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA, 2004. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol 14, 1262–1266. [DOI] [PubMed] [Google Scholar]
- Klein T, 2008. Immunolabeling of imaginal discs. Methods Mol. Biol 420, 253–263. [DOI] [PubMed] [Google Scholar]
- Krentz NAJ, van Hoof D, Li Z, Watanabe A, Tang M, Nian C, German MS, Lynn FC, 2017. Phosphorylation of NEUROG3 Links Endocrine Differentiation to the Cell Cycle in Pancreatic Progenitors. Dev. Cell 41, 129–142 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, 2012. Building an ommatidium one cell at a time. Dev. Dyn 241, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Fox DT, Spradling AC, 2013. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr. Biol 23, 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meserve JH, Duronio RJ, 2015. Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage. Development 142, 2740–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meserve JH, Duronio RJ, 2017. A population of G2-arrested cells are selected as sensory organ precursors for the interommatidial bristles of the Drosophila eye. Dev. Biol 430, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A, 2012. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol 22, 1166–1175. [DOI] [PubMed] [Google Scholar]
- Pauklin S, Vallier L, 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repiso A, Bergantiños C, Serras F, 2013. Cell fate respecification and cell division orientation drive intercalary regeneration in Drosophila wing discs. Development 140, 3541–3551. [DOI] [PubMed] [Google Scholar]
- Ruijtenberg S, van den Heuvel S, 2016. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela Y, Molotski N, Golan S, Itskovitz-Eldor J, Soen Y, 2012. Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells 30, 1097–1108. [DOI] [PubMed] [Google Scholar]
- Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK, 2009. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell 16, 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamori Y, Deng WM, 2013. Tissue repair through cell competition and compensatory cellular hypertrophy in postmitotic epithelia. Dev. Cell 25, 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, 2013. Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol 2, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF, 1991. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113, 841–850. [DOI] [PubMed] [Google Scholar]
- Worley MI, Setiawan L, Hariharan IK, 2012. Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet 46, 289–310. [DOI] [PubMed] [Google Scholar]
- Zielke N, Korzelius J, van Straaten M, Bender K, Schuhknecht GF, Dutta D, Xiang J, Edgar BA, 2014. Fly-FUCCI: A versatile tool for studying cell proliferation in complex tissues. Cell Rep 7, 588–598. [DOI] [PubMed] [Google Scholar]


