Abstract
Peptidoglycan is an essential component of the cell wall that protects bacteria from environmental stress. A carefully coordinated biosynthesis of peptidoglycan during cell elongation and division is required for cell viability. This biosynthesis involves sophisticated enzyme machineries that dynamically synthesize, remodel, and degrade peptidoglycan. However, when and where bacteria build peptidoglycan, and how this is coordinated with cell growth, have been long-standing questions in the field. The improvement of microscopy techniques has provided powerful approaches to study peptidoglycan biosynthesis with high spatiotemporal resolution. Recent development of molecular probes further accelerated the growth of the field, which has advanced our knowledge of peptidoglycan biosynthesis dynamics and mechanisms. Here, we review the technologies for imaging the bacterial cell wall and its biosynthesis activity. We focus on the applications of fluorescent d-amino acids, a newly developed type of probe, to visualize and study peptidoglycan synthesis and dynamics, and we provide direction for prospective research.
Keywords: bacterial cell wall, peptidoglycan, fluorescent probes, microscopy, d-amino acids, bacterial morphogenesis
1. INTRODUCTION
The structure and function of the cell wall have intrigued many scientists who are trying to understand bacterial physiology while providing concrete information for the development of antibiotics. Much of our current antimicrobial arsenal targets peptidoglycan (PG) biosynthesis, largely because PG is essential to bacterial growth and owing to the lack of parallel biosynthetic pathways in humans. With antibiotic resistance on the rise, the Centers for Disease Control and Prevention have highlighted the need for new antibiotics (1). It stands to reason that a deeper understanding of PG biosynthesis will lead to the identification of essential targets followed by the development of novel antibiotics.
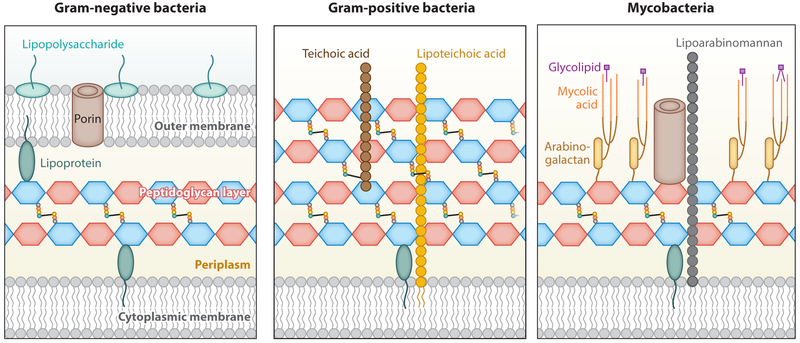
Despite being a highly complex macropolymer, the structure of PG is relatively well conserved across most bacterial species (2, 3). Generally, PG comprises glycan strands of variable length. These strands are cross-linked via oligopeptide bridges, providing mechanical strength as well as resistance to environmental stress. PG can be decorated with a variety of lipoproteins, polysaccharides, and glycolipids, endowing the bacteria cell with very different chemical and physical properties. Bacteria have adopted two fundamentally different PG organization strategies. Gramnegative bacteria contain a thinner PG layer (approximately 5 nm) sandwiched between the cytoplasmic membrane and the outer membrane, whereas Gram-positive bacteria have a thicker PG layer (20–50 nm), on the periplasmic side of the cytoplasmic membrane, as their outermost protective barrier. Although most bacteria can be classified into one of these two classes, a notable exception are the mycobacteria whose cell walls have a unique organization, mainly due to the presence of arabinogalactan and mycolic acids (Figure 1).
Figure 1.
Structure of the bacterial cell wall in different types of bacteria. Left: Gram-negative bacteria cell wall contains a thinner peptidoglycan (PG) layer (~5 nm) sandwiched between the cytoplasmic membrane and the outer membrane. Middle: Gram-positive bacteria cell wall contains a thicker PG layer (20–50 nm) and the cytoplasmic membrane. Right: Mycobacteria cell wall contains a PG layer, the cytoplasmic membrane, and a waxy surface layer made of lipids, mycolic acids, and arabinogalactan.
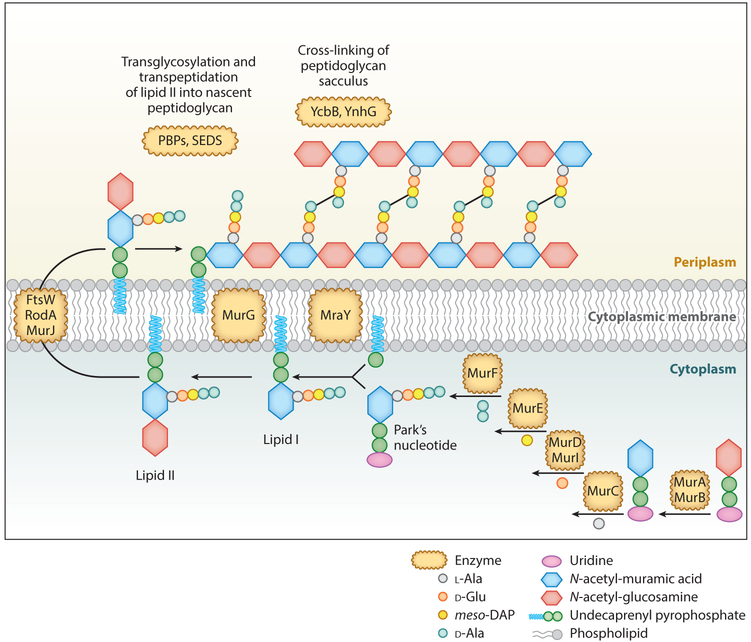
Significant effort has been directed at elucidating the PG biosynthetic pathway (3). This process occurs in three stages. First, Park’s nucleotide is synthesized in the cytoplasm. Next, lipid I and lipid II, both of which are membrane-associated PG precursors, are produced. Finally, after translocation of lipid II across the plasma membrane, PG polymerization and cross-linking occur in the periplasm (or outside the cells in the case of Gram-positives). Bacteria contain sophisticated machinery to synthesize, modify, and degrade PG (Figure 2). Significant past efforts have contributed to our knowledge of PG glycosyltransferase, transpeptidase, amidase, peptidase, and hydrolase enzymes as some of the major participants in PG synthesis and modification. Perhaps even more importantly, we have begun to appreciate how PG biosynthesis is orchestrated by the cytoskeletal system and how it relates to physiological processes such as cell elongation, division, and sporulation (4, 5). A combination of advanced high-resolution microscopy, nanofabrication technology (producing nanocages for entrapment and orientation of individual bacterial cells), and fluorescent labeling techniques [such as fluorescent d-amino acids (FDAAs)] has significantly advanced our understanding of PG synthesis and dynamics and provided insights into how bacteria maintain their shape (6).
Figure 2.
Biosynthesi pathway of PG in Escherichia coli. Some cell wall components are not shown in the figure (e.g., the outer membrane) for simplification. The biosynthesis pathway starts with the formation of Park’s nucleotide in cytoplasm, followed by binding to a lipid component to produce lipid II. Finally, lipid II is translocated across the cytoplasmic membrane and then inserted into the existing PG through transglycosylation and transpeptidation reactions. Abbreviations: PBPs, penicillin-binding proteins; PG, peptidoglycan; SEDS, shape, elongation, division, and sporulation enzyme family.
Considering the complexity of the PG layer, any labeling approach for this macromolecule requires creativity and innovation to uncover finer details about its biosynthesis and structure. For the purpose of this review, we first provide a general introduction to PG imaging techniques and then focus on PG labeling through the lens of fluorescent microscopy and FDAAs. For additional information regarding cell wall labeling, the reader is referred to the thorough review by Siegrist et al. (7) as well as to the in-depth study of cell surface display and biotechnological applications by Gautam et al. (8). Although a number of labeling methods are currently available, FDAAs have profoundly impacted studies on the bacterial cell wall (see Table 1). We discuss some of the most recent discoveries facilitated by FDAAs as well as future directions for their improvement and application.
Table 1.
Examples of fluorescent d-amino acid (FDAA) applications in recent studies. The subheadings classify the use of FDAAs in the experimental designs of these studies
| Article Title | Article Citation |
|---|---|
| Developing d-amino acid–based probes for peptidoglycan study | |
| In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. | Kuru et al. (15) |
| d-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. | Siegrist et al. (97) |
| Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. | Lebar et al. (98) |
| Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. | Kuru et al. (54) |
| d-Amino acid probes for penicillin binding protein-based bacterial surface labeling. | Fura et al. (99) |
| Metabolic profiling of bacteria by unnatural C-terminated d-amino acids. | Pidgeon et al. (100) |
| Metabolic remodeling of bacterial surfaces via tetrazine ligations. | Pidgeon & Pires (101) |
| Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. | Hsu et al. (62) |
| Identifying the formation and/or structure of peptidoglycan | |
| Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. | Pilhofer et al. (102) |
| De novo morphogenesis in L-forms via geometric control of cell growth. | Billings et al. (103) |
| A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. | Liechti et al. (73) |
| Anammox planctomycetes have a peptidoglycan cell wall. | van Teeseling et al. (104) |
| Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. | Liechti et al. (105) |
| Studying bacterial morphogenesis and peptidoglycan growth pattern | |
| Peptidoglycan transformations during Bacillus subtilis sporulation. | Tocheva et al. (106) |
| Site-directed fluorescence labeling reveals a revised N-terminal membrane topology and functional periplasmic residues in the Escherichia coli cell division protein FtsK. | Berezuk et al. (107) |
| Salinity-dependent impacts of ProQ, Prc, and Spr deficiencies on E. coli cell structure. | Kerr et al. (108) |
| Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators. | Möll et al. (109) |
| Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. | Ursell et al. (52) |
| Cell shape dynamics during the staphylococcal cell cycle. | Monteiro et al. (110) |
| Molecular modeling, simulation, and virtual screening of MurD ligase protein from salmonella typhimurium LT2. | Samal et al. (111) |
| Short-stalked Prosthecomicrobium hirschii cells have a Caulobacter-like cell cycle. | Williams et al. (112) |
| Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. | Bisson-Filho et al. (5) |
| Dynamics of the peptidoglycan biosynthetic machinery in the stalked budding bacterium Hyphomonas neptunium. | Cserti et al. (63) |
| Hydrolysis of peptidoglycan is modulated by amidation of meso-diaminopimelic acid and Mg2+ in B. subtilis. | Dajkovic et al. (113) |
| Staphylococcus aureus requires at least one FtsK/SpoIIIE protein for correct chromosome segregation. | Veiga & Pinho (114) |
| GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. | Yang et al. (66) |
| Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. | Yao et al. (115) |
| Determining peptidoglycan synthesis activity and sites | |
| Divin: a small molecule inhibitor of bacterial divisome assembly. | Eun et al. (116) |
| Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. | Cameron et al. (117) |
| Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. | Fleurie et al. (118) |
| MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. | Fleurie et al. (119) |
| Sequential evolution of bacterial morphology by co-option of a developmental regulator. | Jiang et al. (81) |
| Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of S. pneumoniae D39. | Tsui et al. (120) |
| Endopeptidase-mediated β-lactam tolerance. | Dörr et al. (121) |
| Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in E. coli. | Liu et al. (122) |
| A d,d-carboxypeptidase is required for Vibrio cholerae halotolerance. | Möll et al. (123) |
| The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. | Sundararajan et al. (124) |
| Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. | Schirner et al. (125) |
| MreC and MreD proteins are not required for growth of S. aureus. | Tavares et al. (126) |
| The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance and viability. | Yakhnina et al. (127) |
| Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. | Jutras et al. (128) |
| Structure–function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ. | Manuse et al. (129) |
| Roles of the essential protein FtsA in cell growth and division in S. pneumoniae. | Mura et al. (130) |
| FtsZ-dependent elongation of a coccoid bacterium. | Pereira et al. (131) |
| Colanic acid intermediates prevent de novo shape recovery of E. coli spheroplasts, calling into question biological roles previously attributed to colanic acid. | Ranjit & Young (132) |
| Subcompartmentalization by cross-membranes during early growth of Streptomyces hyphae. | Yagüe et al. (133) |
| Pentapeptide-rich peptidoglycan at the B. subtilis cell-division site. | Angeles et al. (134) |
| Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress. | Botella et al. (135) |
| B. subtilis swarmer cells lead the swarm, multiply, and generate a trail of quiescent descendants. | Hamouche et al. (136) |
| Deciphering the mode of action of cell wall-inhibiting antibiotics using metabolic labeling of growing peptidoglycan in Streptococcus pyogenes. | Sugimoto et al. (137) |
2. STRATEGIES FOR PEPTIDOGLYCAN IMAGING
Scientists have always been interested in the fine structure of PG. Methods that enable visualization of fine structural details at high resolution are enhancing our understanding of bacterial growth and division.
2.1. Electron Microscopy
Electron microscopy (EM) is commonly used to observe subcellular structures. After high-energy electron beams have been applied to a specimen, EM instruments collect signals from transmitted electrons (transmission EM) (9), reflected electrons (reflection EM) (10), or secondary electrons (scanning EM) (11). The resolution of EM is determined by the wavelength of the incident electron beam. Recently, high-voltage instruments have been developed, leading to a possible resolution of ∼2 Å (12). Signal intensity is another essential parameter for successful EM application. Generally, specimens with a high electron density produce a strong signal in EM, whereas samples with a low electron density may not be visualized. In the case of bacterial PG, which does not contain electron-dense elements, staining with heavy atoms is necessary for proper imaging. In early studies, mercury-containing chemicals were used. The staining probes selectively react with free amines present on the cell surface, decorating the PG layer with a heavy element. This approach was adopted to study the architecture of PG in Spirillum serpens, a Gram-negative spiral-shaped bacterium, as well as in many other species (13). Results from these studies have shown that isolated PG, also known as sacculus, maintains the shape of actual cell morphology.
After the initial success of observing the bacterial cell wall, the goal for scientists was to link its structure and function via EM application. Two of the most central processes in bacterial biology are elongation and division. To understand bacterial cell growth, fundamental staining methods were developed to differentiate between the synthesis and insertion of nascent PG and the already existing old PG layer. New PG synthesis is required not only for lateral wall elongation but also for transverse wall (septum) construction. The combination of immunostaining and EM resulted in a dynamic understanding of when and where new PG is synthesized and inserted during the bacterial life cycle. The staining method relied on the promiscuous cell wall synthesis machinery and tolerated the incorporation of d-Cys in place of d-Ala, a d-amino acid naturally present in the peptide stem of PG (Figure 2). The side chain thiol reacts with a modified antibody to introduce gold atoms, an electron-dense element, for EM. In their 1997 study, de Pedro et al. (14) applied the above approach to visualize the specific synthesis and insertion of PG in a growth-dependent manner. Furthermore, pulse-and-chase experiments, in the presence and absence of d-Cys, were employed to understand the localization of PG insertion. EM and immunostaining led to the observation that PG insertion is less prominent at the cell poles in Escherichia coli; however, a new cell wall was detected at midcell, suggestive of septum synthesis in preparation for cell division. The basis of this approach has been used for the development of other cell wall–specific and growth-dependent molecular probes, resulting in the ability to observe bacterial growth and division using less energy-intensive fluorescence microscopy (FM) (15).
Despite the many advantages of traditional EM, it has been suggested that sample preparation (fixation and staining) may alter the natural specimen state, leading to the presence of false morphological characteristics (2, 16). Using cryo–transmission EM (cryo-TEM), samples are preserved in a frozen–hydrated state following rapid exposure to liquid nitrogen. This relatively mild sample preparation method preserves the native PG structure (16, 17). For example, the E. coli cell wall structure was observed after complete sample vitrification (18, 19). The outer membrane, PG layer, and inner membrane were clearly distinguishable using cryo-TEM. Importantly, a large fraction of the cells was viable after thawing, suggesting the preservation of their native cell wall structure. Despite some problems with sectioning compression of the specimen, a 6.35-nm thickness of the E. coli PG layer was calculated on the basis of the cryo-TEM images. To improve upon the existing cryo-TEM technology, recent advances have led to the development of a new application, cryo–electron tomography (cryo-ET) (20–22). The usefulness of cryo-ET lies in its ability to provide information about a specimen in three dimensions. To achieve this, samples are tilted in relation to the incident electron beam. Sequential tilting allows the capture of multiple two-dimensional (2D) electromicrographs that are later reconstructed into a three-dimensional (3D) structure. The thickness of specimen is an important determinant of cryo-ET performance. In general, thin specimens result in better cryo-ET images (23). Information from cryo-ET has allowed a further refinement of the bacterial cell wall model. Two models had been suggested in the past, the layered (24, 25) and scaffold (26) models. On the basis of cryo-ET observations using purified sacculi from Gram-negative bacteria (E. coli and Caulobacter crescentus), it was demonstrated that PG exists as a single layer and the glycan strands are perpendicular to the long cell axis. The data are in support of the layered cell wall model (27, 28).
The introduction of EM and its various applications has been tremendously useful for understanding bacterial cell wall architecture. Isolated PG structures were first visualized via immunostaining; the introduction of cryogenic methods enabled visualization of the unaltered cell wall. Subnanoscale improvements in resolution were achieved with high-voltage EM, allowing the reconstruction of molecular-level detail. The development of cryo-ET has further established EM as the tool of choice for 3D PG analysis.
2.2. Atomic Force Microscopy
Technological developments have allowed the application of atomic force microscopy (AFM) to studies of the bacterial cell wall. The technique is based on a cantilever, which is used to monitor the surface of a specimen. The mechanical signal, derived from the contact between the tip and the surface, is transformed into optical signal and further analyzed to produce a corresponding representation of the investigated relief (29, 30). The usefulness of AFM lies in its ability to detect mechanical forces, van der Waals forces, chemical bonds, electrostatic forces, and magnetic forces. Depending on the application, AFM may be the method of choice (31). For example, AFM can provide Z-axis information about a sample on the basis of thickness (or height) measurements. The sample preparation is mild, which allows for imaging in buffer or water, thus preserving the natural state of the specimen’s structure. The cantilever can be adjusted with fine precision, resulting in accurate measurements of surface rigidity and elasticity. These are important physical properties that are usually impossible to obtain via any other technique.
A seminal study relied on AFM to elucidate several physical characteristics of PG shape and structure (32). Using Bacillus subtilis as a model organism, it was determined that the length of a single PG glycan strand can vary between 0.2 to 5 μm, with an average of 1.3 μm. These results provided the first evidence that a glycan strand could be longer than a single bacterial cell, implying that strands are somehow compacted to prevent protrusion from the cell surface. Furthermore, by studying the inner PG relief, cable-like shapes were detected with an average width of 50 nm. Importantly, the cables appeared to be parallel to the short axis of the cell as observed via high-resolution AFM. To account for the presence of these structures in the B. subtilis cell wall, it was proposed that multiple PG strands are combined in a helical manner to produce a fiber. These fibers can subsequently be cross-linked, akin to stem peptide cross-linking, to impart additional strength onto the bacterial cell wall, further protecting the cell against lysis.
2.3. Fluorescence Microscopy
Wide-field epifluorescence and laser-scanning confocal microscopy (LSCM) are the traditional modes of FM. The difference between the two is in the mode of sample illumination. Wide-field epifluorescence leads to excitation of the entire sample on the optical path of the incident light and detection of emitted light from the entire sample, whereas point illumination is used in LSCM. Confocal microscopy allows for focusing of a laser beam onto a much smaller focal volume, followed by a further enhancement of the in-focus signal provided by a pinhole in front of the signal detector, which results in higher signal-to-noise ratio compared with wide-field epifluorescence. In addition to the above FM modes, a near-field technique termed total internal reflection fluorescence microscopy (TIRFM) has also been applied to bacterial cells (4, 33–39). TIRFM is especially useful for visualization of fluorescent molecules localized at the cell surface, within a couple of hundred nanometers from the coverslip. Some of the advantages of this technique are a high signal-to-noise ratio and an increased temporal resolution, resulting in less photobleaching. Due to the inherent spatial resolution limit of conventional FM, imposed by the Abbe diffraction limit (~250 nm in the lateral dimension and ~500 nm in the axial dimension), super-resolution FM methods that address this challenge have been developed for subcellular imaging. Although these advances were first used on larger eukaryotic cells (40), the methods have been successfully applied to bacterial cells (41). Some of the newer super-resolution applications are structured illumination microscopy (SIM) and stimulated emission depletion (STED) microscopy, as well as photoactivation localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM). Their applications are discussed later. Most recently, single-molecule tracking (SMT) has also been applied to study the molecular biology of model bacteria, namely C. crescentus, E. coli, and B. subtilis (5, 42–45). The SMT technique has enormous potential and is expected to provide unprecedented detail of single molecules, similar to the impact of single-cell studies. For a more in-depth description of these techniques and their applications, the reader is directed to a review by Yao and Carballido-López (46).
In addition to the imaging instruments, fluorescent probes are also required to monitor PG structure and synthesis in FM. Such small-molecule probes for site-specific PG labeling have been developed and have become essential tools for improving our understanding of the bacterial cell wall. Some of the first fluorescent probes for PG labeling were antibiotics modified with a fluorescent reporter. This approach utilizes antibiotic molecules that possess a high binding affinity toward PG structural components. For example, vancomycin—a glycopeptide antibiotic that has served as a last-line defense against infections caused by Gram-positive bacteria—binds specifically to peptide stems terminating in d-Ala-d-Ala (47). As a result of drug binding, PG strands cannot be cross-linked via transpeptidation, leading to a much weaker bacterial cell wall. Since the d-Ala-d-Ala subunit is modified as the bacterial cell matures, fluorescein-coupled vancomycin (Van-FL) should label only nascent PG in which the d-Ala-d-Ala target is found, enabling the visualization of PG synthesis/insertion activities (48). After labeling of B. subtilis with Van-FL, a diffuse striped pattern spanning the entire cell was observed, suggesting a circumferential insertion of new PG. The strongest signal was observed at midcell, due to an ongoing cell division that necessitates PG synthesis, while there was a lack of signal at the poles. These findings concurred with previous observations, establishing Van-FL as a tool to monitor PG synthesis (14, 49). Additionally, this fluorescent antibiotic has been used to label Streptococcus pneumoniae and Streptomyces coelicolor to provide visual confirmation of their respective modes of growth (48). Other fluorophore-conjugated antibiotics, including ramoplanin, have been introduced and applied to study PG synthesis. Ramoplanin marks only lipid II insertion sites and has been used to visualize PG synthesis in B. subtilis, confirming the data obtained with Van-FL (50).
An alternative approach of PG labeling is the use of fluorescently labeled wheat germ agglutinin (FWGA), an abundant protein found in wheat kernels, highly specific to N-acetyl-glucosamine (GlcNAc). This molecular probe allows PG labeling in Gram-positive organisms and was initially used as an alternative to Gram staining (51). FWGA binds to PG efficiently (labeling time ~5 min), and the labeling can be carried out with low FWGA concentration (~1 ng/mL), which gives minimal background noise. FWGA has been applied to the study of PG dynamics in E. coli, using complex pulse-chase experiments in conjunction with time-lapse FM (52). These experiments revealed that nascent PG insertion is heterogeneous but correlated with localization of the MreB cytoskeletal protein, suggesting that MreB may have a prominent role in coordinating new PG synthesis. For additional information on different types of PG probes, the reader is referred to a review by Kocaoglu & Carlson (53).
Although fluorescent antibiotics and FWGA have provided a deep understanding of PG synthesis, they have a limitation: Fluorescent antibiotics have growth inhibitory effects on bacteria. Consequently, major optimization efforts are required to prevent aberrant bacterial growth/behavior. Conversely, FWGA has limited use in Gram-negative bacteria because entry is blocked by the outer membrane. FDAAs, developed by the VanNieuwenhze and Brun laboratories, address these limitations and allow PG labeling across many different bacterial species (Gram-positive and Gram-negative) with minimal toxic effect (15, 54). Bacteria produce a variety of d-amino acids and incorporate these—but not the corresponding l-enantiomers—into their PG layer (55, 56). On the basis of these observations, FDAAs were designed in an attempt to metabolically label the bacterial cell wall at sites of active PG growth and/or remodeling. Although FDAAs contain substantial structural modifications, relative to their parent d-amino acid (e.g., d-Ala), they are successfully incorporated into the murein sacculus. On the basis of current data, the mechanism for FDAA incorporation into PG has been proposed to involve either D,D-transpeptidases [penicillin-binding proteins (PBPs)] (57) or the combined action of D,D-carboxypeptidases and L,D-transpeptidases (LDT enzymes) (58–61). FDAA incorporation presumably occurs during PBP-mediated nascent PG cross-linking reactions. They can also be incorporated in a similar fashion during cell wall remodeling, a reaction catalyzed by LDT enzymes. After isolation of sacculi and high-performance liquid chromatography (HPLC)–tandem mass spectrometry (MS/MS) analysis, FDAAs were exclusively found at the fourth position of tetrapeptide stems in E. coli and Agrobacterium tumefaciens. In contrast, Gram-positive organisms such as B. subtilis incorporated FDAAs into the fifth position of pentapeptide stems (15).
Not only are FDAAs specific to PG and do they reflect the activity of PG synthase enzymes, their preparation and application are convenient and streamlined. The synthetic routes are modular and utilize commercially available d-amino acids and fluorophores. A thorough synthesis and labeling protocol has been published (54). Our goal here is to provide an updated summary of FDAA applications, the major advances in cell wall biology facilitated by FDAAs, and some future research directions in which FDAAs will have an essential role.
3. DEVELOPMENT OF FLUORESCENT d-amino ACIDS AND THEIR APPLICATIONS
3.1. Development of Fluorescent d-amino Acids
FDAAs have a wide variety of applications that are discussed further in this section. The first study using FDAAs established that these probes could label PG across a range of phylogenetically diverse bacteria (15). Incubating three model organisms (E. coli, B. subtilis, and A. tumefaciens) with FDAAs revealed labeling signal throughout the entire cell population at both septal and peripheral sites. HPLC–MS/MS analysis of isolated sacculi showed that the incorporation of FDAAs occurs at the 4th or 5th position of the PG peptide chain, suggesting that the labeling is conducted by transpeptidase enzymes, such as the PBPs (D,D-transpeptidases) or the LDTs. To confirm enzyme specificity for the d-enantiomer of the FDAA, experiments utilizing the l-enantiomer revealed no fluorescence incorporation.
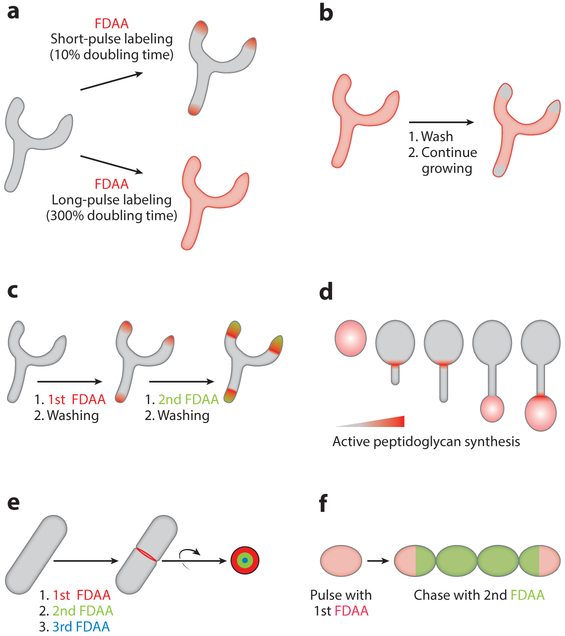
FDAA labeling can be implemented in various forms depending upon the specific application. Long-pulse labeling (>2 generation times) stains all the existing PG throughout the cells (Figure 3a), whereas short-pulse treatment labels only sites of active PG synthesis. For example, a short-pulse labeling led to fluorescence signal only at the poles of Streptomyces venezuelae, a polarly growing bacterium (Figure 3a). This application illustrated a new approach for direct detection of PG synthesis. Another commonly used approach to study new PG synthesis is the pulse-and-chase method, in which cells are labeled with FDAAs with a long pulse, washed, and then grown in fresh culture medium. The newly made PG thus has no signal, but old PG remains fluorescent (Figure 3b).
Figure 3.
Applications of fluorescent d-amino acid (FDAA) labeling. (a) Comparison between short-and long-pulse labeling in Streptomyces venezuelae. (b) Pulse-and-chase labeling; scheme of labeling in S. venezuelae. (c) Virtual time-lapse labeling; scheme of labeling in S. venezuelae. (d) Peptidoglycan growth dynamics in Hyphomonas neptunium; example of FDAA short-pulse labeling. (e) Growth pattern at septal peptidoglycan; example of virtual time-lapse labeling in Bacillus subtilis division septum. (f) Long-pulse-chase-new-labeling; example using two FDAAs in Streptococcus pneumoniae for peptidoglycan turnover rate measurement.
FDAAs also enable successive monitoring of PG growth. This can be achieved by applying sequential short-pulse labeling of different color FDAAs (Figure 3c). Specifically, after performing the first short-pulse labeling, the cells are washed and used for the second short-pulse labeling. This method, referred to as virtual time-lapse microscopy, enables the study of PG growth through identifying the color and location of PG labeling and provides a chronological time stamp of PG growth. With FDAAs that have emission wavelengths spanning the entire visible spectrum, one can perform the virtual time-lapse labeling with up to five unique colors (62).
The highly conserved PG synthesis machineries across bacterial species and their high tolerance of noncanonical d-amino acids have made FDAAs a standard tool for PG labeling (see Table 1) (55). Through the use of FDAAs, the PG growth patterns have been observed in Burkholderia phytofirmans, Brachybacterium conglomeratum, and Verrucomicrobium spinosum, which have proven difficult to achieve using other labeling methods (15). Furthermore, these probes allow the detection of bacteria in environmental samples (e.g., saliva and fresh water) (15). The benefits of such detection are enormous and can improve society immensely (e.g., by identifying if water is contaminated or not).
3.2. Studying the Dynamics of Peptidoglycan Synthesis
There were many challenges to studying PG growth before the FDAAs were discovered. For example, the use of FWGA led to the labeling of all existing PG, making the identification of active PG synthesis sites difficult. FDAAs are powerful tools for studying PG synthesis dynamics because their labeling reports directly on the activity of enzymes involved in PG synthesis/remodeling. Their high biocompatibility also minimizes toxic effects to cells during the labeling process.
In a recent study by Cserti et al. (63), FDAAs were used to unravel the mechanism of PG synthesis during the division of Hyphomonas neptunium. H. neptunium replicates by stalk budding, yet the mechanistic details and enzymes involved were poorly understood. Cryo-ET showed that H. neptunium has a cell envelope typical of a Gram-negative bacterium. Upon initiation of stalk development, the cells grow in length and become more rounded in shape. A stalk forms at the pole of the cell that is both uniform and continuous from the mother cell. Once the stalk reached a maximum length the budding phase begins. As the bud becomes larger in size the length of the stalk decreases. These results indicate a constant remodeling of the stalk to produce the new daughter cell. The use of FDAAs allowed the visualization of distinct growth regions during the different phases of cell division. A fluorescent signal was seen throughout the body of the mother cell during the swimmer-to-stalk phase, in agreement with the proposed growth mode of the cell. Upon stalk initiation, the FDAA signal was concentrated and remained at the site where stalk formation originated (Figure 3d) until the stalk reached maximum length. Then, FDAA signal was transferred from the base of the stalk to its end, where the budding cell initiates, and subsequently was found in the whole daughter cell. FDAAs allowed for the visualization of a distinct PG biosynthesis mechanism in H. neptunium that required several initiation zones of newly synthesized PG, shaping the overall morphology of the cell.
3.3. Studying Peptidoglycan–Enzyme Interactions
Division is essential for cell growth and the sustainment of a bacterial species. A core component of bacterial cell division is PG synthesis and modification and, as such, the process is an extremely valuable target for antibiotics. Yet, detailed information is lacking on the exact mechanism(s) utilized for PG synthesis and modification as well as on the proteins involved in these pathways. Recent studies have utilized FDAAs to correlate new PG synthesis with the requisite enzymatic machinery. A recent study from Bisson-Filho et al. (5) used FDAAs to better understand how B. subtilis creates a septum in preparation for cell division. The septum is the site where the cell is cleaved into two daughter cells. Septal PG synthesis is regulated by two cytoskeletal proteins, FtsZ and FtsA. Together, they determine the precise location of the division site (64, 65) and form the Z ring that provides the scaffold for recruitment of proteins required for septal PG synthesis. Until recently, how the dynamics of synthetic enzymes (i.e., FtsZ/A, PBPs) and septal PG formation activity are coordinated with each other was unknown. Using FDAAs with different fluorescent emission wavelengths, cells were labeled at different time points and visualized by super-resolution microscopy (virtual time-lapse labeling). This revealed a bull’s-eye FDAA pattern that corresponded to the time of labeling at the division site: The first FDAA appeared on the outer ring and the last FDAA at the center (Figure 3e), indicating that newly synthesized PG constricted at the division site over time. Interestingly, using extremely short pulses instead produced arc-like structures at distinct spots around the periphery. These arcs progressively developed into full ring structures as labeling times were increased. Together, these observations led to the hypothesis that PG synthetic enzymes have synchronized movement around the division site for septum synthesis. Using TIRFM, fluorescently labeled FtsZ and FtsA were seen to move around the assembled Z ring directionally either counterclockwise or clockwise. Moreover, a fluorescent fusion of PBP2B, a transpeptidase linked with cell division, showed similar circumferential movement analogous to FtsZ. All of these proteins moved with similar velocities around the Z ring, suggesting coordination between them. More importantly, using quantitative analysis, Bisson-Filho et al. (5) found that the velocity of FtsZ movement regulates the rate of PG synthesis revealed by FDAA labeling. A decreased rate of septal PG formation was observed when FtsZ velocity was attenuated by genetic mutation. Conversely, increasing FtsZ velocity also accelerated septal PG synthesis. These findings provided new insights into how bacteria enzymatically control PG synthesis dynamics to precisely control cell division.
A study by Yang et al. (66) yielded similar results in E. coli. In agreement with Bisson-Filho et al. (5), the GTPase activity of FtsZ was responsible for its movement around the Z ring. However, FtsZ mutants with diminishing GTPase activity did not affect the intensity of FDAA labeling at the septum. Regardless of the reduced GTPase activity, the septal closure was effectively the same. Together, these data indicate that FtsZ is responsible for the recruitment of PG synthases; however, in contrast to the results in B. subtilis from Bisson-Filho et al. (5), FtsZ movement does not affect the rate of PG synthesis in E. coli.
3.4. Studying Peptidoglycan Metabolism
FDAAs can also be used to study PG metabolism. For example, Boersma et al. (67) investigated S. pneumoniae cell division dynamics for a variety of mutants growing both planktonically and in host-relevant biofilms. They utilized a PG analysis adapted from a B. subtilis PG turnover assay in which radiolabeled [14C]GlcNAc is fed to the cell for several generations, followed by washing and then detecting the amount of [14C]GlcNAc released into the growth medium (68, 69). With these data, one can determine the turnover rate of PG. Using an unencapsulated mutant of S. pneumoniae D39, they found a relatively low [14C]GlcNAc turnover rate (5.6 ± 1.6%) in this species (70). However, this low turnover rate could have two possible explanations: (a) S. pneumoniae does in fact have low PG turnover, or (b) PG turnover is followed by rapid recycling of PG degradation products (71). To determine which interpretation is correct, a modified method using FDAAs, termed long-pulse-chase-new-labeling, was tested (Figure 3f). The use of [14C]GlcNAc is replaced by long-pulse labeling with one color FDAA (labeling old PG) followed by second labeling using another color FDAA (labeling new PG), which are then visualized with epifluorescence microscopy at different time points. Using this method, old PG remained present at the initial poles of the first cell, whereas new PG appeared throughout the remainder of the cells over the entire duration. No overlap was observed between the old and new PG; each FDAA color was observed only in distinct regions. The authors concluded that PG has minimal turnover in S. pneumoniae, as was seen previously.
3.5. Identifying the Existence of Peptidoglycan Using Fluorescent d-amino Acid Derivatives
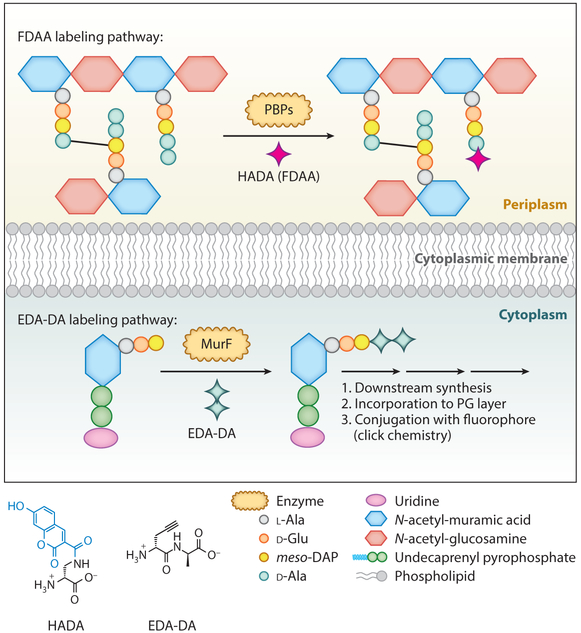
FDAAs have provided solutions to long-standing problems that other methods have been unable to address. For example, the chlamydial anomaly, a problem dating back over 50 years (72), was finally solved through experiments utilizing FDAAs (73). Although Chlamydia sp. were hypothesized to contain PG on the basis of antibiotic susceptibility and genetic screening, the existence of PG in Chlamydia had never been demonstrated (74, 75). Chlamydia encodes the d-Ala-d-Ala ligase (Ddl) and MurF enzymes (76, 77): The Ddl carries out the synthesis of the d-Ala-d-Ala dipeptide; MurF then couples this dipeptide to uridine diphosphate (UDP)-MurNAc-l-Ala-d-Glu-meso-diaminopimelic acid (DAP) to make Park’s nucleotide (Figure 2). These enzymes are responsible for the final steps in the biosynthesis of pentapeptide stems. Because of their presence in Chlamydia sp., it was hypothesized that these species could salvage modified DAAs for PG synthesis, if they possess PG. However, FDAA labeling was unsuccessful at labeling Chlamydia, mainly because the cells possess high carboxypeptidase activity that cleaved the probes from the 5th position of the PG peptide chain after incorporation by the PBP(s) (73). To address this problem, Liechti et al. (73) designed a probe that would enable incorporation into the 4th position of a pentapeptide stem; thus, the idea of d-Ala-d-Ala dipeptide probes was born.
To achieve this goal, the authors synthesized d-Ala-d-Ala derivatives that have a click tag coupled to the N-terminal residue. Once the probe, termed EDA-DA (ethynyl-d-Alanyl-d-Alanine), is incorporated into PG precursor by MurF (Figure 4), the click tag is present at the 4th position of the PG peptide chain. PG formation can thus be visualized by coupling fluorophores to the tag using click chemistry (78). This type of labeling has high resistance to carboxypeptidase activity and was first confirmed in E. coli and B. subtilis. Additionally, EDA-DA treatment could rescue growth in mutant cells lacking natural d-Ala-d-Ala ligases, indicating that the probes are salvaged by the enzymes for PG synthesis in E. coli and B. subtilis.
Figure 4.
Proposed incorporation pathway of FDAA (top) and EDA-DA (bottom). FDAA incorporation occurs in periplasm through transpeptidases activity (e.g., PBP activity). EDA-DA incorporation occurs in cytoplasm through MurF activity. Abbreviations: EDA-DA, ethynyl-d-alanyl-d-alanine; FDAA, fluorescent d-amino acid; HADA, hydroxycoumarin 3-amino-d-alanine; PBP, penicillin-binding protein; PG, peptidoglycan.
To determine if, in fact, Chlamydia contain PG, L2 mouse fibroblast cells were infected with Chlamydia trachomatis with 1 mM EDA-DA. After fixation and permeabilization of the cell membranes, click chemistry was used to attach fluorophores to the probes. Using an antibody for the chlamydial major outer membrane protein (MOMP) for colabeling, click-labeled EDA-DA was observed successfully as rings or lines that bisected the MOMP-labeled cells. The ring structures detected resemble cell divisional rings. The labeling could also be removed enzymatically as seen in B. subtilis, suggesting that the signal came from PG. Moreover, EDA-DA could also rescue growth when C. trachomatis was subjected to d-cycloserine, a known inhibitor of Ddl (79). These labeling results provide strong evidence that C. trachomatis possesses bacterial PG that is essential for cell viability. For the first time, PG was unequivocally demonstrated and visualized in this species. The riddle posed by the chlamydial anomaly was solved.
4. DISCUSSION AND FUTURE DIRECTIONS IN VISUALIZATION OF PEPTIDOGLYCAN BIOSYNTHESIS
PG structures are highly dynamic and can respond quickly to environmental change. For example, C. crescentus controls the synthesis of bacterial stalks—PG-containing antennas extending from the cell surface—to facilitate phosphate uptake from the surroundings (80). This process requires a close crosstalk between existing PG structures, synthetic proteins, and the environment. That is, bacteria can sense the change of environment and precisely adjust their PG structures. However, our knowledge of how bacteria coordinate the activities and dynamics of synthetic proteins for PG synthesis remains limited. To address this issue, scientists have been motivated to investigate PG–protein interactions, and this field of study has been the focus of increasing activity (5, 27, 66, 81). Most of these studies have relied on qualitative and quantitative analyses of bacterial images with tagged targets of interest, using fluorescence, EM, or any other types of microscopy. These studies point to a universal need for the development of new tools and technologies to visualize PG structure, synthetic proteins, and their interactions in high spatiotemporal resolution. In this section, we discuss recent efforts to advance PG studies through the development of new techniques and probes. We talk about current limitations in this field and prospective solutions.
4.1. Advanced Imaging Technologies for Studying Peptidoglycan and Its Synthetic Machineries
Microscope systems have evolved rapidly in recent decades, owing to the improvement of mechanical components, such as cameras, and computational power. The design and functions of microscopes could vary widely but, in general, developers are pursuing high resolution and signal-to-noise ratio with minimal destructive effects to the specimens. The improvement of microscope systems is well appreciated given that the Nobel Prizes in 2014 and 2017 were awarded to scientists developing super-resolution microscopy and EM, respectively.
4.1.1. Super-resolution microscopy.
Optical microscopy offers biocompatible approaches to visualize biological samples, allowing live cell imaging and time-lapse monitoring. With the use of fluorescent probes and/or fluorescent protein fusions, scientists can visualize targets of interest specifically during cell growth for dynamics and mechanism studies. As mentioned in the previous section, the spatial resolution of FM is restricted by the diffraction limit of incident light to ~250 nm, which renders detailed investigation of PG structures and protein behaviors very difficult. To address this problem, super-resolution microscopy has been developed and applied to this field (82). Light microscope systems that generate images with a higher resolution than the diffraction limit are known as super-resolution microscopy. Super-resolution microscopy can be achieved through the mechanical design of the system, the use of specialized molecular probes, or data-processing algorithms. For example, SIM utilizes grid-patterned incident light to image samples. The interaction between the incident light and the structure of the sample generates Moiré interference patterns. By algorithmically decoding the superimposed Moiré patterns in different phases, SIM can reconstruct a molecular structure at a resolution of up to ~100 nm. This technique has been applied in many studies investigating PG biosynthesis and synthetic protein dynamics—for example, FtsZ and PBPs and PG formation (5, 66, 83).
Another commonly used method to achieve super-resolution imaging is point localization-based imaging techniques, such as STORM and PALM (84, 85). Specimens for STORM/PALM experiments are labeled with photo-switchable probes whose fluorescence is kept in an “off “ state. During imaging, low-energy excitation light is cast onto the specimens to turn on a few probes, or even only a single probe molecule, at a time. The accurate localization of the probe-tagged molecules then can be calculated by fitting a Gaussian function to the fluorescence signal. By superimposing thousands of images containing localization information of single molecules, the systems can reconstruct the specimen images with ~10-nm resolution. This method has been employed to study the cytoskeletal proteins, MreB and FtsZ, responsible for PG biosynthesis (86, 87).
The introduction of super-resolution microscopy has greatly advanced the study of PG biosynthesis at the molecular level. Although effort is still required to make these techniques more accessible, super-resolution microscopy could plausibly be widely applied to study PG biosynthesis and dynamics and will significantly impact this field. In addition, the vigorous growth of other super-resolution imaging techniques, such as STED microscopy, will provide additional approaches to investigate PG, and its synthesis machineries, in detail.
4.1.2. Joint microscopy systems.
In comparison with FM, EM possesses ultrahigh spatial resolution for imaging (down to a few Angstroms) but much lower biocompatibility for specimens. The requirement of specimen staining also restricts its utility for studying the function and interaction between biomolecules. To overcome these restrictions, scientists have been developing a joint electron/fluorescent microscope for use in a method known as correlative light and electron microscopy (CLEM) (88). CLEM inherits FM’s ability to visualize fluorescently tagged targets in cells, which provides functional information about the targets. The follow-up EM imaging at the same field of the specimen is then used to collect structural and spatial information about the targets in ultrahigh resolution. This combination of techniques greatly advanced studies of structure–function relationships between biomolecules, for example, neuron–synaptic interactions (89, 90). Since PG biosynthesis involves subtle control of PG structure, which is precisely regulated by synthetic protein localization and activity, CLEM can serve as a powerful technique to visualize protein–protein and protein–PG interactions. For instance, how PBPs are spatially and temporally coordinated with cytoskeletal protein assembly/disassembly during PG synthesis is unclear. With the use of CLEM, scientists might visualize targeted PBP fusions while precisely locating their position on cytoskeletal protein filaments and/or on PG structures. This information will advance our knowledge of how PBP activity and dynamics are regulated. As CLEM has become increasingly accessible, it has the potential of being used to resolve questions regarding protein–PG interactions.
4.2. Advanced Molecular Probes for Studying Peptidoglycan and the Synthetic Machineries
In addition to imaging techniques, the molecular probe is another key player in visualizing macro-molecules in biological systems. As discussed above, a wide variety of probes have been developed for PG and synthetic protein staining. The general design of biomolecular fluorescent probes comprises (a) a targeting moiety that specifically binds to the target of interest, (b) a reporter that generates signal upon excitation, and (c) a linker that joins the two together (91). Each moiety can be customized to enhance specificities and/or function to the probes. The introduction of novel probes could profoundly affect the field by enabling acquisition of information that was not previously accessible. For example, d-amino acids serve as versatile carriers of probe molecules for bacterial cell wall studies because they have high specificity for PG as well as minimized cytotoxicity. In the remaining sections, we discuss possible designs of novel biomolecular probes that are relevant, but not limited, to FDAA application.
4.2.1. Probes having high specificity to peptidoglycan subunits or synthetic enzymes.
Having the ability to visualize individual PG subunits is critical for understanding the dynamics and mechanism of PG biosynthesis. Fluorescent probes have been designed to target these subunits, including the glycan strands (FWGA, 52; fluorescent ramoplanin, 50) and the stem peptide (fluorescent vancomycin, 92; FDAAs, 15). Specific labeling of different PG subunits can further enable studies in this field. For example, researchers have used clickable dipeptides to tag PG precursors (vide supra). The dipeptide probe (EDA-DA) enters the cytoplasm and is incorporated into Park’s nucleotide via the intermediacy of MurF (Figure 4). After additional processing (via lipid I and lipid II) the tagged precursor is flipped to the periplasmic face of the plasma membrane, where it can be conjugated with fluorophores using click chemistry and visualized using FM. This probe serves as a reporter for new PG insertion (via the lipid II pathway) because its incorporation into PG precursors occurs in the cytoplasm. Labeling at the 4th position also helps EDA-DA circumvent trimming by d,d-transpeptidases (and d,d-carboxypeptidases). One notable finding achieved using EDA-DA is identifying the existence of PG in C. trachomatis (73), as mentioned in a previous section. Another notable example of PG subunit-specific probing is the use of a modified glycan unit, N-acetyl-muramic acid (MurNAc), to label the PG backbone. Specifically, Liang et al. (93) synthesized MurNAc mimics containing a biorthogonal handle for click chemistry. The MurNAc mimics can be salvaged by bacteria and used to construct new PG, which results in a specific labeling of PG backbone. Since the labeling process is mediated by PG synthesis and recycling enzymes, it provides a new approach to track PG synthesis activity. The labeled MurNAc mimics can be visualized by conjugating with fluorophores through click chemistry, and this method has been applied to visualize bacterial invasion in mammalian cells (93).
Indeed, probes targeting more PG subunits are in great demand. Scientists are still looking for probes targeting glycine cross-bridges (94), the membrane anchor of lipid II, or even the probes that can distinguish 3–3 and 3–4 cross-linking in PG. The search for novel PG probes will undoubtedly continue to be a very active area of research.
4.2.2. Probes enabling time-lapse monitoring of peptidoglycan formation.
Another challenge in this field is the inability to monitor PG growth continuously. Although fluorescent protein fusions have allowed time-lapse monitoring of protein dynamics, no equivalent technique exists for studying PG formation. Currently, PG is typically visualized using end-point assay(s)/imaging, in which the cells are fixed after PG labeling and then imaged. By synchronizing the cell culture or sorting cells on the basis of morphology, scientists could track PG synthesis and dynamics in different cell growth stages. However, the temporal resolution of this method, so-called pseudo-time-lapse microscopy, is limited. Another approach to time-lapse monitoring of PG is using pulse-and-chase methods, in which the entire PG in live cells is first labeled and then the cells are allowed to grow in the absence of probes (52). In this case, the old PG is fluorescently labeled, whereas newly synthesized PG is not. Researchers can thus monitor the reduction of the fluorescence signal resulting from new PG formation. This method, however, suffers from the issue of low signal-to-background ratio in microscopy imaging because it is relatively difficult to observe signal reduction in the presence of strong background. To address these issues, probes enabling time-lapse monitoring of PG formation are required.
One major obstacle preventing the current probes from time-lapse experiments is the need to wash away excess probe to reduce background fluorescence. The time required for the washing steps might disrupt cell growth and morphology, which makes continuous monitoring problematic. A possible solution to this is using switchable probes, whose fluorescence can be turned “on” and “off.” As mentioned earlier, STORM utilizes the photo-switchable property of fluorophores to achieve single-molecule localization for constructing super-resolution imaging. It provides a proof-of-concept for using switchable probes in FM. Because there are fluorescent molecules sensitive to pH, steric hindrance, or presence of specific functional groups, developing them for sensing of PG incorporation is possible (95, 96). The underlying premise for these probes is to have fluorescence occur only after probe incorporation into PG, whereas the unincorporated probes are nonfluorescent. In this case, washing steps are no longer required. Thus, the formation and/or dynamics of PG could be observed continuously during cell growth. With the combination of fluorescent protein fusions, enzyme–PG interactions could be studied in great detail.
4.2.3. Probes revealing synthetic enzyme activities.
Because the activity and localization of PG synthetic proteins are highly dynamic, knowing both when and where they are functioning is challenging. Being able to visualize their activity during cell growth can advance our understanding of interactions between PG and synthetic proteins. FDAAs represent an example of probes that reveal enzymatic activities. FDAAs are known to incorporate into PG through the transpeptidase activities. Short-pulsed FDAA labeling points out newly made PG in the cells, whereas treatment of transpeptidase inhibitors disrupts FDAA incorporation. Thus, FDAAs serve as reporters of PG–protein interactions, providing information about when and where the proteins are functioning.
Another possible approach to probe PG–enzyme interactions is through use of fluorescence resonance energy transfer (FRET). This technique utilizes energy transfer between two fluorophores (FRET donor and acceptor) embedded on different biomolecules. When the biomolecules interact with each other, the distance between the two fluorophores may be sufficient to enable excitation of the donor fluorophore and observation of the emission signal from the acceptor fluorophore. Since PG serves as the substrate of cell wall synthesis enzymes, dual labeling of PG, and a biosynthetic enzyme incorporating a fluorescent fusion, possibly could be used to report on PG–enzyme interactions.
5. CONCLUSION
PG has been studied for over 100 years. However, our understanding of PG biosynthesis and dynamics remains limited. The introduction of microscopy techniques and molecular probes enabled visualization of PG growth, which has significantly advanced our knowledge in this field. Here, we have reviewed the methods commonly used for imaging PG biosynthesis. We have focused on the application of recently developed tools (FDAAs) and provided a forward-looking analysis of PG imaging strategies. It is our hope that the information contained in this article provides a valuable resource for investigators who study PG synthesis and dynamics and serves as a catalyst for further developments in this rapidly evolving field.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.CDC (Cent. Dis. Control Prev.). 2013. Antibiotic resistance threats in the United States, 2013 US Dept. Health Hum. Serv., Washington, DC [Google Scholar]
- 2.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev 32(2):149–67 [DOI] [PubMed] [Google Scholar]
- 3.Typas A, Banzhaf M, Gross C, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol 10(2):123–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333(6039):222–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, et al. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355(6326):739–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kysela DT, Randich AM, Caccamo PD, Brun YV. 2016. Diversity takes shape: understanding the mechanistic and adaptive basis of bacterial morphology. PLOS Biol. 14(10):e1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegrist MS, Swarts BM, Fox DM, Lim SA, Bertozzi CR. 2015. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol. Rev 39(2):184–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautam S, Gniadek TJ, Kim T, Spiegel DA. 2013. Exterior design: strategies for redecorating the bacterial surface with small molecules. Trends Biotechnol. 31(4):258–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams DB, Carter CB. 1996. The transmission electron microscope In Transmission Electron Microscopy: A Textbook for Materials Science. Boston: Springer [Google Scholar]
- 10.Yagi K 1987. Reflection electron microscopy. J. Appl. Cryst 20:147–60 [Google Scholar]
- 11.Vernon-Parry KD. 2000. Scanning electron microscopy: an introduction. III-Vs Rev. 13(4):40–44 [Google Scholar]
- 12.Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, et al. 2015. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science 348(6239):1147–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Formanek H, Formanek S. 1970. Specific staining for electron microscopy of murein sacculi of bacterial cell walls. Eur. J. Biochem 17:78–84 [DOI] [PubMed] [Google Scholar]
- 14.de Pedro MA, Quintela JC, Höltje J-V, Schwarz H. 1997. Murein segregation in Escherichia coli. J. Bacteriol 179(9):2823–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, et al. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. Engl 51(50):12519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao Y, Zhang T. 2011. Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl. Microbiol. Biotechnol 92(2):381–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühlbrandt W. 2014. Microscopy: cryo-EM enters a new era. eLife 3:e03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matias VRF, Al-Amoudi A, Dubochet J, Beveridge TJ. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol 185(20):6112–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matias VRF, Beveridge TJ. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol 56(1):240–51 [DOI] [PubMed] [Google Scholar]
- 20.Luთić V, Rigort A, Baumeister W. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol 202(3):407–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh R, Nicastro D, Mastronarde D. 2005. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol 15(1):43–51 [DOI] [PubMed] [Google Scholar]
- 22.Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442(7106):1062–64 [DOI] [PubMed] [Google Scholar]
- 23.Diebolder CA, Koster AJ, Koning RI. 2012. Pushing the resolution limits in cryo electron tomography of biological structures. J. Microsc 248(1):1–5 [DOI] [PubMed] [Google Scholar]
- 24.de Petris S 1967. Ultrastructure of the cell wall of Escherichia coli and chemical nature of its constituent layers. J. Ultrastruct. Res 19(1):45–83 [DOI] [PubMed] [Google Scholar]
- 25.Verwer RWH, Nanninga N, Keck W, Schwarz U. 1978. Arrangement of glycan chains in the sacculus of Escherichia coli. J. Bacteriol 136(2):723–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dmitriev BA, Toukach FV, Schaper K, Holst O, Rietschel ET, Ehlers S. 2003. Tertiary structure of bacterial murein: the scaffold model. J. Bacteriol 185(11):3458–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan L, Chen S, Jensen GJ. 2008. Molecular organization of Gram-negative peptidoglycan. PNAS 105(48):18953–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu Y-P, Meng X, VanNieuwenhze MS. 2016. Methods for visualization of peptidoglycan biosynthesis. Methods Microbiol 43:3–48 [Google Scholar]
- 29.Allison DP, Mortensen NP, Sullivan CJ, Doktycz MJ. 2010. Atomic force microscopy of biological samples. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2(6):618–34 [DOI] [PubMed] [Google Scholar]
- 30.Ikai A 2010. A review on: atomic force microscopy applied to nano-mechanics of the cell In Nano/Micro Biotechnology, ed. Endo I, Nagamune T, pp. 47–61. Berlin: Springer; [DOI] [PubMed] [Google Scholar]
- 31.Yao X, Jericho M, Pink D, Beveridge T. 1999. Thickness and elasticity of Gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol 181(22):6865–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. 2008. Cell wall peptidoglycan architecture in Bacillus subtilis. PNAS 105(38):14603–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dempwolff F, Moeller HM, Graumann PL. 2012. Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins. J. Bacteriol 194(17):4652–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dempwolff F, Wischhusen HM, Specht M, Graumann PL. 2012. The deletion of bacterial dynamin and flotillin genes results in pleiotrophic effects on cell division, cell growth and in cell shape maintenance. BMC Microbiol. 12:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soeldner R, Carballido-López R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333(6039):225–28 [DOI] [PubMed] [Google Scholar]
- 36.Gregory JA, Becker EC, Pogliano K. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22(24):3475–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jovanovic G, Mehta P, McDonald C, Davidson AC, Uzdavinys P, et al. 2014. The N-terminal amphi-pathic helices determine regulatory and effector functions of phage shock protein A (PspA) in Escherichia coli. J. Mol. Biol 426(7):1498–1511 [DOI] [PubMed] [Google Scholar]
- 38.Lenn T, Leake MC, Mullineaux CW. 2008. Clustering and dynamics of cytochrome bd-I complexes in the Escherichia coli plasma membrane in vivo. Mol. Microbiol 70(6):1397–1407 [DOI] [PubMed] [Google Scholar]
- 39.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, et al. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol. Microbiol 83(1):41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hell SW. 2007. Far-field optical nanoscopy. Science 316(5828):1153–58 [DOI] [PubMed] [Google Scholar]
- 41.Huang B, Babcock H, Zhuang X. 2010. Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143(7):1047–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. 2011. Single-molecule investiga-tions of the stringent response machinery in living bacterial cells. PNAS 108(31):E365–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. PNAS 103(29):10929–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu L, Yu J. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys. J 95(4):2009–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uphoff S, Reyes-Lamothe R, de Leon FG, Sherratt DJ, Kapanidis AN. 2013. Single-molecule DNA repair in live bacteria. PNAS 110(20):8063–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao Z, Carballido-Lo´ pez R. 2014. Fluorescence imaging for bacterial cell biology: from localization to dynamics, from ensembles to single molecules. Annu. Rev. Microbiol 68:459–76 [DOI] [PubMed] [Google Scholar]
- 47.Hammes WP, Neuhaus FC. 1974. On the mechanism of action of vancomycin: inhibition of peptidoglycan synthesis in Gaffkya homari. Antimicrob. Agents Chemother 6(6):722–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria. Cell 113(6):767–76 [DOI] [PubMed] [Google Scholar]
- 49.Clarke-Sturman AJ, Archibald AR, Hancock IC, Harwood CR, Merad T, Hobot JA. 1989. Cell wall assembly in Bacillus subtilis: partial conservation of polar wall material and the effect of growth conditions on the pattern of incorporation of new material at the polar caps. Microbiology 135(3):657–65 [DOI] [PubMed] [Google Scholar]
- 50.Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. 2006. Imaging peptidoglycan biosyn-thesis in Bacillus subtilis with fluorescent antibiotics. PNAS 103(29):11033–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sizemore RK, Caldwell JJ, Kendrick AS. 1990. Alternate Gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol 56(7):2245–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, et al. 2014. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. PNAS 111(11):E1025–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kocaoglu O, Carlson EE. 2016. Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol 12(7):472–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuru E, Tekkam S, Hall E, Brun YV, VanNieuwenhze MS. 2015. Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc 10(1):33–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam H, Oh D-C, Cava F, Takacs CN, Clardy J, et al. 2009. d-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325(5947):1552–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lupoli TJ, Tsukamoto H, Doud EH, Wang T-SA, Walker S, Kahne D. 2011. Transpeptidase-mediated incorporation of d-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc 133:10748–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev 32(2):234–58 [DOI] [PubMed] [Google Scholar]
- 58.De S, McIntosh LP. 2012. Putting a stop to l,d-transpeptidases. Structure 20(5):753–54 [DOI] [PubMed] [Google Scholar]
- 59.Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. 2008. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol 190(13):4782–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders AN, Pavelka MS. 2013. Phenotypic analysis of Escherichia coli mutants lacking l,d-transpeptidases. Microbiology 159(9):1842–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Heijenoort J 2011. Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev 75(4):636–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsu Y-P, Rittichier J, Kuru E, Yablonowski J, Pasciak E, et al. 2017. Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem. Sci 290:30540–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cserti E, Rosskopf S, Chang Y-W, Eisheuer S, Selter L, et al. 2017. Dynamics of the peptidogly-can biosynthetic machinery in the stalked budding bacterium Hyphomonas neptunium. Mol. Microbiol 103(5):875–95 [DOI] [PubMed] [Google Scholar]
- 64.Chen Y, Erickson HP. 2005. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem 280(23):22549–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3:e04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2017. GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355(6326):744–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boersma MJ, Kuru E, Rittichier JT, VanNieuwenhze MS, Brun YV, Winkler ME. 2015. Minimal peptidoglycan (PG) turnover in wild-type and PG hydrolase and cell division mutants of Streptococcus pneumoniae D39 growing planktonically and in host-relevant biofilms. J. Bacteriol 197(21):3472–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, et al. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol 65(1):180–200 [DOI] [PubMed] [Google Scholar]
- 69.Doyle RJ, Chaloupka J, Vinter V. 1988. Turnover of cell walls in microorganisms. Microbiol. Rev 52(4):554–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomasz A, McDonnell M, Westphal M, Zanati E. 1975. Coordinated incorporation of nascent peptidoglycan and teichoic acid into pneumococcal cell walls and conservation of peptidoglycan during growth. J. Biol. Chem 250(1):337–41 [PubMed] [Google Scholar]
- 71.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann. NY Acad. Sci 1277(1):54–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moulder JW. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect. Agents Dis 2(2):87–99 [PubMed] [Google Scholar]
- 73.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, et al. 2013. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506(7489):507–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moulder JW, Novosel DL, Officer JE. 1963. Inhibition of the growth of agents of the psittacosis group by d-cycloserine and its specific reversal by d-Alanine. J. Bacteriol 85:707–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282(5389):754–59 [DOI] [PubMed] [Google Scholar]
- 76.McCoy AJ, Maurelli AT. 2005. Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting d-Alanyl-d-Alanine ligase activity involved in peptidoglycan synthesis and d-cycloserine sensitivity. Mol. Microbiol 57(1):41–52 [DOI] [PubMed] [Google Scholar]
- 77.Patin D, Bostock J, Chopra I, Mengin-Lecreulx D, Blanot D. 2012. Biochemical characterisation of the chlamydial MurF ligase, and possible sequence of the chlamydial peptidoglycan pentapeptide stem. Arch. Microbiol 194(6):505–12 [DOI] [PubMed] [Google Scholar]
- 78.Kolb HC, Finn MG, Sharpless KB. 2001. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl 40(11):2004–21 [DOI] [PubMed] [Google Scholar]
- 79.Neuhaus FC, Hammes WP. 1981. Inhibition of cell wall biosynthesis by analogues of alanine. Pharmacol. Ther 14(3):265–319 [DOI] [PubMed] [Google Scholar]
- 80.Klein EA, Schlimpert S, Hughes V, Brun YV, Thanbichler M, Gitai Z. 2013. Physiological role of stalk lengthening in Caulobacter crescentus. Commun. Integr. Biol 6(4):e24561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang C, Brown PJB, Ducret A, Brun YV. 2014. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506(7489):489–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coltharp C, Xiao J. 2012. Superresolution microscopy for microbiology. Cell Microbiol 14(12):1808–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jennings PC, Cox GC, Monahan LG, Harry EJ. 2011. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron 42(4):336–41 [DOI] [PubMed] [Google Scholar]
- 84.Rust MJ, Bates M, Zhuang X. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3(10):793–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, et al. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313(5793):1642–45 [DOI] [PubMed] [Google Scholar]
- 86.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. 2008. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat. Methods 5(11):947–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLOS ONE 5(9):e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Begemann I, Galic M. 2016. Correlative light electron microscopy: connecting synaptic structure and function. Front. Synaptic Neurosci 8(28):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bock DD, Lee W-CA, Kerlin AM, Andermann ML, Hood G, et al. 2011. Network anatomy and in vivo physiology of visual cortical neurons. Nature 471(7337):177–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee WA, Bonin V, Reed M, Graham BJ, Hood G, et al. 2016. Anatomy and function of an excitatory network in the visual cortex. Nature 532:370–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou J, Ma H. 2016. Design principles of spectroscopic probes for biological applications. Chem. Sci 7(10):6309–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113(6):767–76 [DOI] [PubMed] [Google Scholar]
- 93.Liang H, DeMeester KE, Hou C-W, Parent MA, Caplan JL, Grimes CL. 2017. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat. Commun 8:15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gautam S, Kim T, Shoda T, Sen S, Deep D, et al. 2015. An activity-based probe for studying crosslinking in live bacteria. Angew. Chem. Int. Ed. Engl 54(36):10492–96 [DOI] [PubMed] [Google Scholar]
- 95.Briggs MS, Burns DD, Cooper ME, Gregory SJ, Ata´z EM, et al. 2000. A pH sensitive fluorescent cyanine dye for biological applications. Chem. Commun 20(23):2323–24 [Google Scholar]
- 96.Haidekker MA, Theodorakis EA. 2010. Environment-sensitive behavior of fluorescent molecular rotors. J. Biol. Eng 4(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. 2013. d-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol 8:500–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lebar MD, May JM, Meeske AJ, Leiman SA, Lupoli TJ, et al. 2014. Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J. Am. Chem. Soc 136:10874–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fura JM, Kearns D, Pires MM. 2015. d-amino acid probes for penicillin binding protein-based bacterial surface labeling. J. Biol. Chem 290:30540–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pidgeon SE, Fura JM, Leon W, Birabaharan M, Vezenov D, Pires MM. 2015. Metabolic profiling of bacteria by unnatural C-terminated d-amino acids. Angew. Chem. Int. Ed. Engl 54:6158–62 [DOI] [PubMed] [Google Scholar]
- 101.Pidgeon SE, Pires MM. 2015. Metabolic remodeling of bacterial surfaces via tetrazine ligations. Chem. Commun 51:10330–33 [DOI] [PubMed] [Google Scholar]
- 102.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, et al. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun 4:2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Billings G, Ouzounov N, Ursell T, Desmarais SM, Shaevitz J, et al. 2014. De novo morphogenesis in L-forms via geometric control of cell growth. Mol. Microbiol 93:883–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Teeseling MCF, Mesman RJ, Kuru E, Espaillat A, Cava F, et al. 2015. Anammox planctomycetes have a peptidoglycan cell wall. Nat. Commun 6:6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liechti G, Kuru E, Packiam M, Hsu Y-P, Tekkam S, et al. 2016. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLOS Pathog. 12:e1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, VanNieuwenhze MS, et al. 2013. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol 88:673–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berezuk AM, Goodyear M, Khursigara CM. 2014. Site-directed fluorescence labeling reveals a revised N-terminal membrane topology and functional periplasmic residues in the Escherichia coli cell division protein FtsK. J. Biol. Chem 289(34):23287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kerr CH, Culham DE, Marom D, Wood JM. 2014. Salinity-dependent impacts of ProQ, Prc, and Spr deficiencies on Escherichia coli cell structure. J. Bacteriol 196(6):1286–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Möll A, Dörr T, Alvarez L, Chao MC, Davis BM, et al. 2014. Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators. J. Bacteriol 196(22):3937–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Ravares AC, et al. 2015. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun 6(1):8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Samal HB, Das JK, Mahapatra RK, Suar M. 2015. Molecular modeling, simulation and virtual screening of MurD ligase protein from Salmonella typhimurium LT2. J. Pharmacol. Toxicol. Methods 73:34–41 [DOI] [PubMed] [Google Scholar]
- 112.Williams M, Hoffman MD, Daniel JJ, Madren SM, Dhroso A, et al. 2016. Short-stalked Prosthecomicrobium hirschii cells have a Caulobacter-like cell cycle. J. Bacteriol 198(7):1149–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dajkovic A, Tesson B, Chauhan S, Courtin P, Keary R, et al. 2017. Hydrolysis of peptidoglycan is modulated by amidation of meso-diaminopimelic acid and Mg2+ in Bacillus subtilis. Mol. Microbiol 104(6):972–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Veiga H, Pinho MG. 2016. Staphylococcus aureus requires at least one FtsK/SpoIIIE protein for correct chromosome segregation. Mol. Microbiol 103(3):504–17 [DOI] [PubMed] [Google Scholar]
- 115.Yao Q, Jewett AI, Chang YW, Oikonomou CM, Beeby M, et al. 2017. Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J 36(11):1577–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eun Y-J, Zhou M, Kiekebusch D, Schlimpert S, Trivedi RR, et al. 2013. Divin: a small molecule inhibitor of bacterial divisome assembly. J. Am. Chem. Soc 135(26):9768–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, Zambryski PC. 2014. Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio. 5(3):e01219–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, et al. 2014. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLOS Genet. 10(4):e1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, et al. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516(7530):259–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsui H-CT, Boersma MJ, Vella SA, Kocaoglu O, Kuru E, et al. 2014. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol. Microbiol 94(1):21–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dörr T, Davis BM, Waldor MK. 2015. Endopeptidase-mediated beta lactam tolerance. PLOS Pathog. 11(4):e1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu B, Persons L, Lee L, de Boer PAJ. 2015. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol 95(6):945–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Möll A, Dörr T, Alvarez L, Davis BM, Cava F, Waldor MK. 2015. A d,d-carboxypeptidase is required for Vibrio cholerae halotolerance. Environ. Microbiol 17(2):527–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Huang KC, Goley ED. 2015. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun 6(1):7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schirner K, Eun Y-J, Dion M, Luo Y, Helmann JD, et al. 2015. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat. Chem. Biol 11(1):38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tavares AC, Fernandes PB, Carballido-Ló pez R, Pinho MG. 2015. MreC and MreD proteins are not required for growth of Staphylococcus aureus. PLOS ONE 10(10):e0140523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yakhnina AA, McManus HR, Bernhardt TG. 2015. The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance and viability. Mol. Microbiol 97(5):957–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jutras BL, Scott M, Parry B, Biboy J, Gray J, et al. 2016. Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. PNAS 113(33):9162–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Manuse S, Jean NL, Guinot M, Lavergne J-P, Laguri C, et al. 2016. Structure–function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ. Nat. Commun 7:12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mura A, Fadda D, Perez AJ, Danforth ML, Musu D, et al. 2016. Roles of the essential protein FtsA in cell growth and division in Streptococcus pneumoniae. J. Bacteriol 199(3):e00608–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pereira AR, Hsin J, Król E, Tavares AC, Flores P, et al. 2016. FtsZ-dependent elongation of a coccoid bacterium. mBio. 7(5):e00908–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ranjit DK, Young KD. 2016. Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid. J. Bacteriol 198(8):1230–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yagüe P, Willemse J, Koning RI, Rioseras B, Ló pez-García MT, et al. 2016. Subcompartmentalization by cross-membranes during early growth of Streptomyces hyphae. Nat. Commun 7:12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Angeles DM, Liu Y, Hartman AM, Borisova M, de Sousa Borges A, et al. 2017. Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol. Microbiol 104(2):319–33 [DOI] [PubMed] [Google Scholar]
- 135.Botella H, Vaubourgeix J, Lee MH, Song N, Xu W, et al. 2017. Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress. EMBO J. 36(4):536–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hamouche L, Laalami S, Daerr A, Song S, Holland IB, et al. 2017. Bacillus subtilis swarmer cells lead the swarm, multiply, and generate a trail of quiescent descendants. mBio. 8(1):e02102–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sugimoto A, Maeda A, Itto K, Arimoto H. 2017. Deciphering the mode of action of cell wall-inhibiting antibiotics using metabolic labeling of growing peptidoglycan in Streptococcus pyogenes. Sci. Rep 7(1):1129. [DOI] [PMC free article] [PubMed] [Google Scholar]