Abstract
TMPRSS2-ERG fusions are common genetic events in prostate cancer. Until now, this genetic alteration was modelled by ERG overexpression. In this short communication, we report the creation of mouse prostate organoids that have undergone gene fusion through a CRISPR/Cas9-based strategy. The genetic fusion of TMPRSS2 and ERG results in ERG overexpression. This effect is androgen receptor-mediated, as expression of the fusion transcript can be restored to wildtype ERG levels by treatment with the androgen receptor antagonist Nilutamide.
Keywords: CRISPR/Cas9, TMPRSS2-ERG, Organoids, Genetic engineering
INTRODUCTION
Gene fusions are genetic events occurring during tumorigenesis of both solid and blood-borne cancers [1]. The best known fusion events are those occurring in leukemia, the first disease in which these genetic events were discovered [2]. In prostate cancer (PC), 40–80% of tumors contain a gene fusion between the androgen receptor (AR) responsive gene Transmembrane protease serine 2 (TMPRSS2) and an ETS family transcription factor, most often the oncogene ETS-related gene (ERG) [3–5]. TMPRSS2 and ERG are located on chromosome 21. A deletion of a region of approximately 3 million basepairs results in the fusion of these two genes, thereby driving AR-induced overexpression of the ERG oncogene. A diverse range of TMPRSS2-ERG hybrids has been described [6,7]. In all cases, the open reading frame (ORF) of exon 1 of TMPRSS2 is included, thereby retaining the androgen response element (ARE) present in the ‘5 UTR of this gene [8]. For ERG, the fusion most often results in an N-terminal truncation of the transcript that retains the functional domains described for this transcription factor [7].
Since the discovery of this molecular alteration in 2005 [4], research studying this fusion has relied on mouse models or cell lines overexpressing ERG [9–11] and human PC cell lines carrying this genetic alteration [9]. Although these mouse models mimic the ERG overexpression observed in TMPRSS2-ERG tumors, they lack the correct genetic setting of TMRPSS2-driven expression, such as the loss of the genes that lie in between TMPRSS2 and ERG [12]. Furthermore, these models do not retain the transcription control of the fusion gene as it occurs in PC. PC cell lines that carry TMPRSS2-ERG fusions contain additional genetic alterations, often of unknown relevance, thereby obscuring the molecular effects of the gene fusion.
In 2013, the technique of CRISPR/Cas9-mediated genome engineering became available as a versatile tool [13,14]. Shortly thereafter, we showed that organoids, 3D structures grown from stem cells that resemble their respective tissue of origin, could be established from both murine and human prostate epithelial cells [15]. As we and others previously showed that CRISPR/ Cas9 can be applied in organoids [16–21], we set out to create a TMPRSS2-ERG gene fusion in mouse prostate organoids. These organoids recapitulate the overexpression of the ERG oncogene that is observed in tumors carrying this genetic alteration. The resulting ERG overexpression is AR-driven as described previously [4,22,23], and can be inhibited by blocking androgen signaling. To our knowledge, we are the first to report the use of CRISPR/Cas9 to create gene fusions in organoids. The TMPRSS2ERG organoids described here, can serve as an in vitro model to study the short and long term molecular consequences of this gene fusion in an otherwise wildtype background.
MATERIALS AND METHODS
Targeting construct cloning
Homology arms containing synonymous mutations to prevent cleavage by Cas9were ordered as g blocks from IDT. eGFP-puro sequence was amplified using from an in-house construct created in the lab. PCR from genomic DNA was used to create the second homology arm for ERG. Short overhangs complimentary to the homology arms were added to these fragments by integrating them into the used primers. Individual fragments plus backbone (created by restriction digestion of cloning vector) were ligated using HiFi DNA Assembly Cloning kit (NEB).
Organoid culture and transfection
Mouse prostate organoids were established and cultured as described previously [15,24]. To obtain mutant organoids, we used the psCas9 vector described by Wright et al. (Ran et al., 2013), that encoded both our sgRNA and the Cas9 protein of S. pyogenes. Transfections were performed using Lipofectamin (Invitrogen). 500 ng DNA was used per construct and resuspended in 50 µLoptimum prior to transfection. 50 µl of a 8% Lipofectamine in optimum solution was mixed with the DNA and incubated at room temperature for 20 minutes. 450 µL of cell suspension (in complete medium - antibiotics) was added to the DNA/Lipofectamine mix in a 48-well cell culture plate. Cells were centrifuged at 600x g for 1 hour to increase transfection efficiency. Subsequently, cells were incubated for 4–6 hours at 37°C 5% CO2. After that, cells were washed, centrifuged, resuspended in BME and plated as normal. After three days of recovery, selection on 1µg/ml puromycin-containing medium was started andTMPRSS2-ERG organoids were maintained on this medium. Five days prior to RNA isolation for RT-qPCR experiments, puromycin was removed from the medium, and - if applicable - Nilutamide (10 µM)was added.
RNA isolation and reverse transcription
RNA was extracted from the organoids using the QIAGEN RNA easy kit. Organoids were pelleted by centrifugation and dissolved in 300µl RLT, 300 µl 70% ethanol was added. The mixture was put on the RNA columns, which were subsequently centrifuged. Columns were washed once with RW1 buffer and twice with RPE buffer. To assure removal of all buffer, columns were spin down once more. RNA was eluted using 30 µl RNAase-free water. RNA was kept on ice at all times. 500 ng RNA was taken as starting material for the reverse transcription reaction. To 500 ng of RNA in 10 µl water, 1,5 µl of a oligo (dT)15primer (0,5 µg per reaction) was added on ice. The mixture was incubated for 5 min at 70°C and cooled on ice. 8,5 µl of GoScript buffer with 0,5 mM dNTPs, 2,5 mM MgCL, 1 µl of GoScript reverse transcriptase and 20 units of RNase-inhibitor (all Promega) was added and the mixture was incubated for 5 min at 25°C, 15 min at 42°C and 15 min at 70°C. cDNA was stored at −20°C until use.
qPCR reaction
25 ng of cDNA was used per reaction. Used primers are depicted in supplementary (Table 1). 25 µl of iQ SYBR Green supermix (Biorad) with a primer concentration of 500 nM was used for each reaction. Actin was used as housekeeping gene. PCR reaction was as follows: 2 minutes 95°C, followed by a repeat of the following incubations: 30 seconds at 95°C, 55°C, 72°C.
Statistical analysis
For expression levels, ΔΔCT values were calculated by making al data relative to actin, and subsequently to wildtype values. For statistical analysis of the RT-qPCR experiments, we used Graphpad software and performed a two-way ANOVA. Subsequent bonferetti posttests revealed a statistically significant deviation of ERG expression in all three TMPRSS2-ERG clones.
RESULTS AND DISCUSSION
Mouse prostate organoids can be genetically modified to carry TMPRSS2-ERG gene fusions
To create organoids carrying a TMPRSS2-ERG gene fusion at the endogenous genomic location, we used a targeting construct in which the genomic region encoding intron 2–3 of TMPRSS2 was connected to the region encodingintron 2–3 of ERG (Figure 1A). As such, fusion would result in a transcript containing exon 1 and 2 of TMPRSS2 and exons3–11 of ERG. This fusion is comparable to the human transcript hybrid most commonly observed in PC [4,25,26], as it fuses the first coding exon of TMPRSS2 to the second coding exon of ERG. In addition, the construct also contained a puromycin selection cassette. This eGFP-puromycin cassette was flanked by two arms homologous to the genomic region downstream of exon 3 of ERG (Figure 1A). As such, the puromycin cassette would be integrated in a non-coding region of the gene. (See supplementary data for the complete sequence of the targeting construct.)
Figure 1.
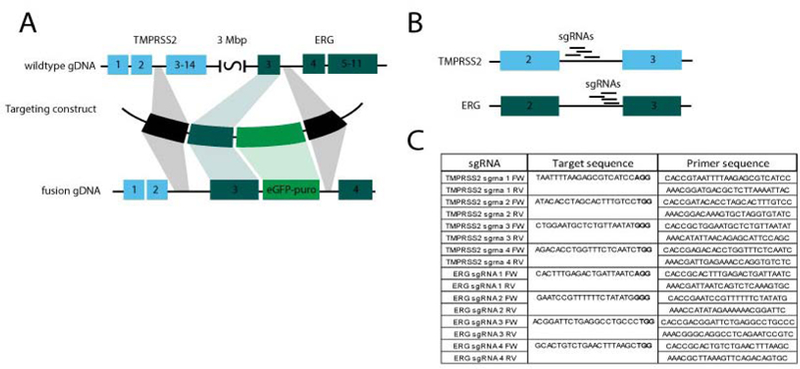
TMPRSS2-ERG fusion targeting construct. A. The developed targeting construct fuses intron 2–3 of TMPRSS2 to intron 2–3 of ERG, which are approximately 3 Mbp apart. In addition, a eGFP-puro cassette is integrated in intron 3–4 of ERG. B. Used sgRNAs target intronic regions of both TMPRSS2 and ERG. C. Sequences targeted by used sgRNAs and the primers used to construct the sgRNA constructs following the protocol of Ran et al [27].
In addition, we developed constructs encoding Cas9 and individual sgRNAs targeting the region of both TMPRSS2 and ERG in the vicinity of the fusion site (Figure 1B). These constructs were created as previously described [27]. sgRNAs targeting intronic regions of TMRPSS2 and ERG were designed using the online CRISPR design tool at http://crispr.mit.edu(for used sequences see Figure 1C). To prevent Cas9-mediated cleavage, sgRNA target region were synonymously mutated in our targeting construct.
Subsequently, we transfected mouse prostate organoids with our targeting construct, one TMPRSS2-targeting sgRNA and one ERG-targeting sgRNA. After approximately one week on selection medium, single organoids growing in selection medium could be observed. We handpicked these organoids, dissociated them into single cells and expanded them as clonal lines.TMRPSS2ERG organoids appeared folded, dense and morphologically very different from their wildtype counterparts (Figure 2A). However, after approximately six to eight passages, the organoids regained the cystic appearance of wildtype organoids (Figure 2B). Ki67staining on paraffin-embedded sections revealed an increase in proliferation in the TMPRSS2-ERG organoids (Figure 2C).
Figure 2.
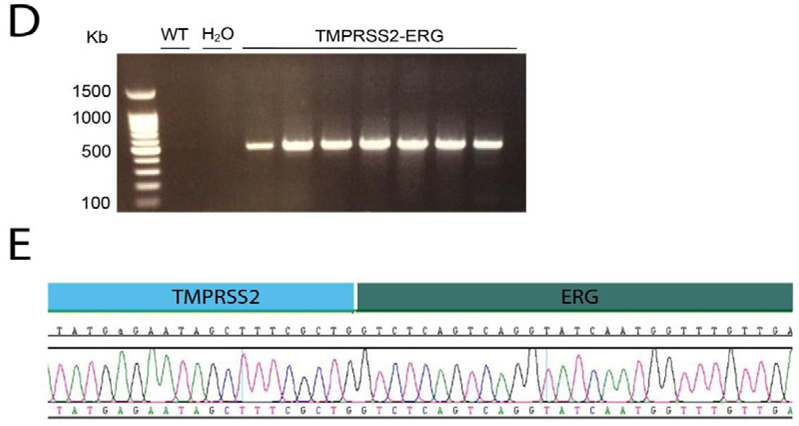
Transfection of mouse prostate organoids with the produced TMPRSS2-ERG targeting construct. A. Morphology of TMPRSS2-ERG organoids compared to WT organoids after clonal expansion of handpicked single organoids. B. Morphology of TMPRSS2-ERG organoid 10 passages after transfection. C. Ki67-staining on paraffin embedded organoids reveals increased proliferation in the fusion organoids compared to their wildtype counterparts. D. Conformation of TMRPSS2-ERG gene fusion on the DNA level. E. Sanger sequencing of the fusion-specific PCR on gDNA shown in D, reveals ligation of TMRPSS2 to ERG.
Genomic DNA was isolated for each line and we performed a PCR reaction with a forward primer in the TMPRSS2 gene and a reverse primer in the ERG gene (for primer sequences, see Supplementary Table (1)). For+/− 85% of the clones we observed a band that upon Sanger sequencing (Figure 2D), revealed the successful fusion of the TMRPSS2 and ERG gene at the genomic level (Figure 2E). We also sequenced the sgRNA-targeted regions of the remaining wildtype alleles of both TMPRSS2 and ERG. We detected small insertions and deletions, caused by incorrect repair of the sgRNA-induced cuts (Supplemental Figure 1). However, as these are located in the intronic regions of the genes, they should not affect the transcript.
By creating the TMPRSS2-ERG fusion at the endogenous location, this model mimics the in vivo situation more specifically than ERG overexpression models that, for example, are driven by a probasin promoter. In our system, the genomic region between TMPRSS2 and ERG is deleted in one of the alleles. This is critical, as this region contains genes such as HMGN1 that is reported to increase N-cadherin expression [28] and alter the G2/M checkpoint [29] upon its loss. Increase of N-cadherin expression is observed in progressing PC [30]. This concomitant hemizygosity of a number of genes cannot be modeled by ERG overexpression.
In addition, this model can be used to better understand the molecular effects of this gene fusion, and more specifically, how it contributes to PC, without the confounding effect of other genetic alterations (which is an issue when using PC cell lines). This might aid the development of new therapies for tumors carrying this genetic alteration. In addition, TMPRSS2-ERG organoids might serve to aid the implementation of already existing targeted therapies for TMPRSS2-ERG tumors in the clinic. It was previously shown that patient-derived organoids can predict tumor response in vivo [31]. Although the organoids described here are of murine origin, technically the described technique should be applicable to human prostate organoids.
To our knowledge, this is the first study showing that CRISPR/Cas9 technology can be applied in organoids to create endogenous gene fusions.
TMRPSS2-ERG fusion organoids are functional and responsive to androgen inhibiting treatment
Next, we investigated if the TMRPSS2-ERG organoids are functional. It was previously shown that tumor lines carrying this fusion over express ERG and, furthermore that this expression is androgen-driven [4]. Therefore, we performed gene expression analysis by RT-qPCR of both TMRPSS2 and ERG relative to β-actin. We observed an increase in ERG expression in TMRPSS2ERG when compared to wildtype. TMRPSS2 expression was not altered significantly (Figure 3A). Of note, the extent of increase in ERG expression varied between the different TMPRSS2-ERG organoid lines.
Figure 3.
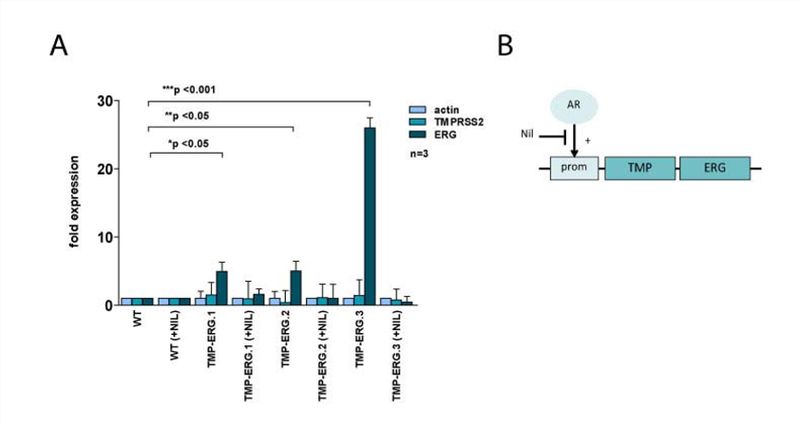
ERG expression in TMPRSS2-ERG organoids is driven by AR signaling. A. Expression analysis of wildtype and three clonal TMPRSS2-ERG organoid lines. Expression of TMPRSS2 and ERG is calculated relative to β-actin. All expression values are calculated relative to wildtype organoids. B. Schematic overview of the effect of Nilutamide on ERG expression in TMRPSS2-ERG organoids. Due to the presence of an ARE in its protomer region, TMRPSS2 gene expression is driven by AR. As such, in TMPRSS2-ERG organoids, AR drives ERG overexpression. However, when Nilutamide is added to the medium, TMPRSS2 expression is prevented and ERG expression levels are restored to wildtype levels.
As our culture medium contains dihydrotestosterone (DHT), the active form of testosterone, we hypothesized that inhibition of AR signaling would result in decreased expression of ERG in the TMPRSS2-ERG organoid lines. To investigate this, we treated organoids for 24 hours with Nilutamide, a competitive antagonist of AR (Figure 3B) and subsequently collected RNA for expression analysis. As expected, we observed a restoration of ERG expression levels in the presence of Nilutamide, which was consistent for all three tested TMRPSS2-ERG organoid lines (Figure 3A). These data support the findings of others that ERG expression in PC carrying this gene fusion is AR-driven and can be inhibited by using AR-inhibiting agents. Our findings imply that AR-blocking treatments directly affect the molecular consequences of TMRPSS2-ERG fusion in PC.
CONCLUSION
Here we demonstrated that CRISPR/Cas9 can be applied in organoids to create endogenous gene fusion in vitro through the creation of a very large intrachromosomal deletion. We generated mouse prostate organoids carrying a variant of TMRPSS2ERG gene fusion that is commonly found in PC. Our approach creates the unique opportunity to study the effect of this genetic alteration in an otherwise wildtype background.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kai Kretzschmar for critically reading this manuscript. E.D. is supported by a ZonMW grant (40-41405-98-208). H.C. is named inventor on several patents related to Lgr5 stem-cell-based organoid technology.
ABBREVIATIONS
- AR
Androgen Receptor
- ARE
Androgen Response Element
- DHT
Dihydrotestosterone
- ERG
ETS-Related Gene
- PC
Prostate Cancer
- TMPRSS2
Transmembrane Protease Serine 2
REFERENCES
- 1.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer 2015; 15: 371–381. [DOI] [PubMed] [Google Scholar]
- 2.Nowell P, Hungerford D. A minute chromosome in human chronic 9 granulocytic leukemia. Science (80-) [Internet] 1960; 132: 1488–1501. [Google Scholar]
- 3.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, et al. TMPRSS2: ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene 2007; 26: 4596–4599. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins S a Rhodes DR, Perner S Dhanasekaran SM, Mehra R Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310: 644–648. [DOI] [PubMed] [Google Scholar]
- 5.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res 2006; 66: 10658–10663. [DOI] [PubMed] [Google Scholar]
- 6.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes, chromosomes & cancer 2006; 45: 717–719. [DOI] [PubMed] [Google Scholar]
- 7.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene 2007; 26: 2667–2673. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 2007; 27: 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE , et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008; 10: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res 2008; 68: 8516–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD , et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A 2008; 105: 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia 2006; 8: 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife 2013; 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014; 159: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015; 521: 43–47. [DOI] [PubMed] [Google Scholar]
- 17.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013; 13: 653–658. [DOI] [PubMed] [Google Scholar]
- 18.Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 2015; 6: 8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, Jansen M, et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med 2016; 8: 745–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, Bali AS, Randell SH, Hogan BLM. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol 2015; 211: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Christin JR, Wang C, Ge K, Oktay MH, Guo W. Mammary Stem Cell Based Somatic Mouse Models Reveal Breast Cancer Drivers Causing Cell Fate Dysregulation. Cell Rep 2016; 16: 3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casey OM, Fang L, Hynes PG, Abou-Kheir WG, Martin PL, Tillman HS, et al. TMPRSS2- driven ERG expression in vivo increases self-renewal and maintains expression in a castration resistant subpopulation. PLoS One 2012; 7: e41668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc 2016; 11: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapointe J, Kim YH, Miller MA, Li C, Kaygusuz G, van de Rijn M, et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol 2007; 20: 467–473. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res 2006; 66: 8347–8351. [DOI] [PubMed] [Google Scholar]
- 27.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinstein YR, Furusawa T, Lim JH, Postnikov YV, West KL, Birger Y, et al. Chromosomal protein HMGN1 modulates the expression of N-cadherin. FEBS J 2005; 272: 5853–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birger Y, Catez F, Furusawa T, Lim JH, Prymakowska-Bosak M, West KL, et al. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res 2005; 65: 6711–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaggi M, Nazemi T, Abrahams NA, Baker JJ, Galich A, Smith LM, et al. N-cadherin switching occurs in high Gleason grade prostate cancer. Prostate 2006; 66: 193–199. [DOI] [PubMed] [Google Scholar]
- 31.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015; 161: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


