Abstract
Objectives
Detection of genomic alterations in diseases can be achieved with current molecular technologies. However, the molecules extracted from formalin-fixed, paraffin-embedded (FFPE) bio-samples are often limited possibly due to DNA fragmentation and crosslinking caused by the sample fixation and processing. The study objective was to design a droplet digital PCR (ddPCR) assay to assess the quality and quantity of DNA derived from various DNA extraction conditions on FFPE samples.
Methods
We used 10 μm-thick sections from 5 FFPE oral tumoral blocks, each consisting of 10–15 sections. The protocol variables tested included: 1) tissue staining; 2) duration and 3) temperature of post-digestion heat treatment; and 4) DNA extraction method. DNA quantity was assessed using the NanoDrop 2000 (Thermo Fisher Scientific, USA), the Qubit fluorometer (Thermo Fisher Scientific, USA), and a ddPCR-based assay. DNA quality was assessed using a ddPCR assay for the degree of fragmentation and the effectiveness of removing crosslinks with varying guanine-cytosine (GC)-content.
Results
Deparaffinization with xylene helped to increase the DNA yield. Tissue staining (methyl green staining, pH 6) prior to microdissection, comparing to no staining, caused additional DNA fragmentation. Compared to column-based method, DNA extracted with phenol chloroform and ethanol precipitation increased the degree of fragmentation and lowered the yield of amplifiable DNA. The cross-linking derived from GC-contents may not be the only factor impacting on the DNA quality.
Conclusions
Samples undergoing different pre-treatment conditions prior to extraction can impact the yield of amplifiable DNA. Our ddPCR assay can be used to assess for both DNA quantity and quality.
Keywords: Droplet digital PCR, Formalin-fixed paraffin-embedded tissues, DNA extraction, GC-content, DNA fragmentation, Formalin-induced crosslinking
1. Introduction
Advancements in molecular technologies provide opportunities to detect key genomic events during carcinogenesis that can be used as biomarkers for progression and prognosis, and as therapeutic targets [1]. Formalin-fixed, paraffin-embedded (FFPE) bio-samples are used for routine histology diagnosis [2] and are most easily accessible and often used for molecular studies. However, the quality and quantity of the nucleic acids from FFPE samples may vary and can have significant impact on the results [3,4].
Droplet digital PCR (ddPCR) is a relatively new and robust molecular technique to detect copy number alterations (CNAs), one of the key genomic events in carcinogenesis [5,6]. The method uses oil and microfluidics to partition a single PCR mixture into ∼20,000 droplets [7]. Each droplet acts as an individual reaction and the results are counted by scoring signals as either positive or negative. The high sensitivity of ddPCR is suitable for absolute quantification of copy numbers and has potential to be clinically translatable with appropriate assay designs [8].
We recently developed a multiplex ddPCR assay to analyze oral bio-samples, including FFPE, for CNAs on oral lesions [9,10]. It is known that tissue fixation can cause DNA fragmentation [11]; thus, one aspect of our assay targeted the same gene, but used different sets of primers to generate different amplicon sizes. Therefore, by comparing the absolute counts of large- and small-sized amplicons, we were able to assess DNA fragmentation.
Tissue fixation in formalin has known to induce DNA crosslinking [12]. Studies have indicated that in the presence of formaldehyde, nucleotide sequences containing guanine-cytosine (GC) bases can form DNA interstrand crosslinking [13] and crosslink to amino acids [14]. Crosslinking with formaldehyde has also been found to weaken the hydrogen bonds between paired DNA bases and contributes to the denaturation of DNA [15,16]. In addition, genomic regions with an abundance of GC bases can form secondary structures [17] and the percentages of GC-content positively correlate with melting temperatures through base stacking interactions [18]. With the inherent factors of GC bases and the additive effects of crosslinking at GC bases, we hypothesize that inadequate reversal of formalin-induced crosslinking can influence the quantity and quality of DNA extracts especially when targeting genomic regions with higher GC-content.
In this study, we designed ddPCR primer sets that target the same gene while varying the GC-content of the amplicons as an attempt to use the absolute quantitation properties of ddPCR to test our hypothesis. In addition, we also tested different DNA extraction methods and focused on conditions that may reverse the crosslinking induced by formalin. The objectives of the study were to use ddPCR-based approach to quantify the amplifiable DNA, and to investigate the fragmentation and the reversibility of crosslinking at GC regions of FFPE samples in various DNA extraction conditions.
2. Materials and methods
2.1. Tissue preparation
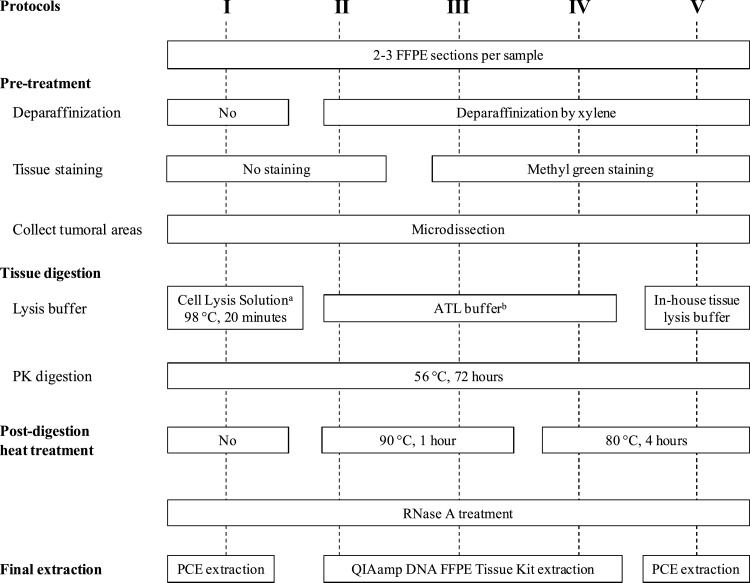
Five surgically obtained FFPE oral tumor samples with minimum 1.0 × 1.0 x 0.3 cm tumor volumes were collected in order to study five extraction protocols. From each tumor FFPE block, we cut 10–15 sections. To make sure the variation of the amount of DNA is not due to the various tumor content of each section, we carefully divided the sections serially among the different protocols, e.g., the tumor dissected from the first, 6th, and 11th sections will be collected for protocol I and the second, 7th, and 12th sections will be collected for protocol II, and so forth. Thus, 2–3 sections from each tumor FFPE block were used to analyze each protocol (see Fig. 1). To ensure a similar volume and to reduce normal contamination, we circled and microdissected areas of the same size on each slide under a direct microscope (Fisher Scientific, USA).
Fig. 1.
Schematic workflow for testing various factors in extracting DNA from formalin-fixed, paraffin-embedded (FFPE) tissue samples.
aCommercially available from QIAGEN; bIncluded in the QIAamp DNA FFPE Tissue Kit.
Abbreviations: PK: Proteinase K; PCE: Phenol-chloroform extraction and ethanol precipitation.
2.2. DNA extraction
The general workflow for FFPE DNA extraction consisted of tissue pre-treatment, tissue dissection, digestion, post-digestion heat treatment, and final extraction (Fig. 1).
2.2.1. Pre-treatment
Four sets (Protocols II, III, IV, V) of tissue sections were baked in the oven (60 °C, 1 h), deparaffinized with xylene (3 times of 10 min), rehydrated with decreasing percentages of ethanol (70–100%), and (only III, IV, V) stained with 0.2% methyl green (MG; pH 6).
2.2.2. Microdissection
We used a fine point needle to manually separate the epithelium from the connective tissue by viewing tissue sections under a light microscope. MG staining helped to visually differentiate the tissue morphology. For Protocols I and II (unstained sections), we referenced the hematoxylin-eosin stained slides to obtain tumoral areas.
2.2.3. Digestion
We used three tissue lysis conditions. Protocol I, without xylene pre-treatment, deparaffinized tissues with heat by incubating in Cell Lysis Solution (QIAGEN, Germany) for 20 min at 98 °C. Protocol V used in-house tissue lysis buffer (10 mM Tris−HCl, 25 mM EDTA, 100 mM NaCl, 0.5% SDS, pH 8–9). Protocols II, III, and IV used ATL buffer (QIAGEN, Germany). All tissues were enzymatically digested with 20–30 μL of Proteinase K (QIAGEN, Germany) per sample by incubating for 72 h in a 56 °C water bath.
2.2.4. Post-digestion heat treatment
After complete digestion of tissues, samples received either without (Protocol I) or with heat treatment using a heat block at either 90 °C for 1 h (Protocol II, III) or at 80 °C for 4 h (Protocol IV, V).
2.2.5. Final extraction
All protocols treated the samples with RNase A (QIAGEN, Germany) prior to extraction. In Protocols I and V, DNA was extracted using phenol-chloroform and ethanol precipitation (PCE). Briefly, samples were cleaned up using a mixture of phenol:chloroform:isoamyl alcohol (25:24:1), chloroform, and 2 mL Heavy Phase Lock Gel tubes (QuantaBio, USA). The DNA pellets were precipitated using 7.5 M ammonium acetate (1/3 of sample volume) and 100% ethanol (2-2.5x of sample volume), then centrifuged for 30 min at 4 °C. After 70% ethanol washes and air drying, the pellets were resuspended in 1x IDTE buffer, pH 8.0 (Integrated DNA Technologies, USA). In Protocols II, III, and IV, DNA samples were extracted using the commercially available, column-based, QIAamp DNA FFPE Tissue Kit (QIAGEN, Germany) following the manufacturer’s protocol. We used 20 μL of buffer to resuspend the DNA pellet from PCE or to elute the DNA from spin-columns.
2.3. Assessment of quality and quantity of DNA
2.3.1. ddPCR workflow
We used genes HFE2 and CPT2 to design our ddPCR assay for the evaluation of DNA yields and quantification. These 2 genes were chosen because our previous study and the Cancer Genome Atlas (TCGA) of the head and neck cancer specimens found that these two genes were copy number neutral, i.e., stabilization across genome of head and neck cancers [5]. We have used these 2 genes as part of the control panel in our previous publication [9].
Reactions were performed in duplicates and the workflow was as previously described [9]. Each replicate had a final volume of 20 μL, consisting of 10 μL of 2x ddPCR Supermix for Probes (no dUTP) (Bio-Rad Inc., USA), primers at final concentrations of 900 nM, FAM and/or HEX-labeled probes at final concentrations of 150–300 nM (Table 1), and 15 ng of Qubit quantified FFPE DNA. We loaded the reaction mixtures and droplet generation oil for probes (Bio-Rad Inc., USA) into the respective wells of a DG8 cartridge (Bio-Rad Inc., USA) to generate droplets using a QX200 droplet generator (Bio-Rad Inc., USA). The emulsions were transferred to a 96-well plate (Bio-Rad Inc., USA) and the plate was heat sealed with pierceable foil using a P X 1 PCR plate sealer (Bio-Rad Inc., USA) prior to loading onto a PTC-200 thermal cycler (Bio-Rad Inc., USA). The thermocycling was initiated at 95 °C for 10 min, then 50 cycles consisting of 94 °C for 30 s, 60 °C for 1 min, and 65 °C for 30 s, and then final extension at 98 °C for 10 min. The ramp rate was set to 2.5 °C/second. After thermocycling, the fluorescent signals of droplets were captured by the FAM and HEX channels of a QX200 droplet reader (Bio-Rad Inc., USA).
Table 1.
Sequences of primers and probes for ddPCR analyses.
| Locus | Amplicon size | Amplicon GC% | Forward primera | Reverse primera | Probea,b |
|---|---|---|---|---|---|
| Fragmentation analysis | |||||
| CPT2c | 106 bp | 54% | CCA GCA GTG AAC CTT GGG | CTG GGT AGG AAG AGA CAT TGC | CCT GTG GTC TCT GAT GGC TTT GGT |
| CPT2 | 125 bp | 54% | TAC GGG CAG ATA AAC CAC AA | CAG TTG TCA TGA ACA GCA TAC C | CCT GTG GTC TCT GAT GGC TTT GGT |
| CPT2 | 151 bp | 54% | CCT GCA TAC GGG CAG ATA AA | AAG AGA CAT TGC AGC CTA TCC | CCT GTG GTC TCT GAT GGC TTT GGT |
| CPT2 | 179 bp | 54% | GAT CAT CTT GCC TGA GCT CTA C | AAG AGA CAT TGC AGC CTA TCC | CCT GTG GTC TCT GAT GGC TTT GGT |
| CPT2 | 199 bp | 55% | TAC GGG CAG ATA AAC CAC AAT | CTA AGG CCT TCT CCA CAC ATT | CCT GTG GTC TCT GAT GGC TTT GGT |
| CPT2 | 252 bp | 56% | TGA CCG ACA CTT GTT TGC TCT G | AAC TCC CGG GCA TTG CG | CCT GTG GTC TCT GAT GGC TTT GGT |
| Analysis of reversing cross-linking | |||||
| RPP30c | 98 bp | 44% | GGG AAG GAA GTA TGA CAG ATG TT | GAA GCC ATC CTT GAG TCC TTA G | AGC AAA GTA CAA CAG GAA GAC ACC TTG G |
| RPP30 | 97 bp | 46% | TGT ACC CTC CCA GCT CTT TA | CAT GTG GCA TAA CTT CAA CGT G | TGG TAT CTG CCT CAA ATC CAC CTC C |
| RPP30 | 97 bp | 55% | GCT ACA GAA TAA GGC TCC TGT G | TGG GTG AGT CTT CTC TAC CTA AT | ACA GTG CCC TCT CAG CTG CA |
| RPP30 | 99 bp | 57% | ACC TGA AGA TAC CTG GGA AGT T | GTA CCA CCG AGG CCA GT | ACT GAT GCA GGA CAT TAC AGC CTG G |
| RPP30 | 97 bp | 58% | TCC AGC TAC TCA CTC TGT CTT T | CTG CAT GTG GCG GTG AG | CCT CAC AGG CGA TAA GAT GCT CCG |
| RPP30 | 97 bp | 61% | GAC GGT CAT GGG ACT TCA G | GCG GCT GTC TCC ACA AG | ATG GCG GTG TTT GCA GAT TTG GAC |
| Quantification of DNA | |||||
| HFE2c | 97 bp | 49% | GGG ATC CAG TTT GTC GAT TCA | AGC TGT CTG CCG AAT GAT TAT AG | AAC TGC TAA CCC TGG GAA CCA TGT |
Sequences written in 5’ to 3’ direction.
RPP30 and HFE2 probes are labeled with 5’ FAM and 5’ HEX, respectively. Two different probes labeled with 5’ FAM and 5’ HEX are used for each CPT2 reaction.
Sequences have been previously published by our group [9].
2.3.2. Evaluation of DNA yield
Samples were quantified using the NanoDrop 2000 (Thermo Fisher Scientific, USA) and Qubit dsDNA high sensitivity assay kit (ThermoFisher Scientific, USA) to determine DNA concentrations for the reaction setup. After ddPCR, samples were further quantified based on the copies of HFE2 as shown by the equations below [9].
| (1) |
| (2) |
| (3) |
2.3.3. Evaluation of DNA quality
We used primers and probes that targeted CPT2 and RPP30 genes to evaluate the degree of fragmentation and effectiveness of crosslinking reversal, respectively [9]. Six pairs of primers and probes were designed to target CPT2 with amplicon sizes between 106–252 bp. Another six pairs of primers and probes targeted RPP30 with amplicons between 44–61% of GC-content. The GC-content was calculated as the ratio of the number of GC bases to the total number of bases for each amplicon. Each reaction also contained a constant control of HFE2 for quantifying amplifiable DNA and for normalization, as described previously [9]. Sequences for the primers and probes can be found in Table 1.
2.4. Statistical analyses
We used Student’s t-test to determine the statistical significance of DNA yield extracted, as quantified with Qubit and ddPCR, between each protocol. Regression analysis was used to observe the effectiveness of reversing formalin induced crosslinks at varying GC-content of RPP30 amplicons.
3. Results and discussion
3.1. Quantity of DNA
We tested five different protocols and the DNA quantity was assessed using NanoDrop, the Qubit fluorometer, and ddPCR quantification of HFE2. One sample using Protocol I yielded ∼150 ng of DNA, as quantified by Qubit, and did not meet the minimum amount of DNA needed for our ddPCR analysis (180 ng for 6 multiplex, duplicate reactions). We omitted the case from further analysis.
Using the Qubit fluorometer to measure the yield of double stranded DNA (dsDNA), Protocol I produced the lowest average DNA concentration (28 ng/μL; range 19–42 ng/μL). This suggested that using xylene, rather than heat, to deparaffinize FFPE tissues may helped to increase the DNA yield, which contrasted a previous report that deparaffinization does not impact the DNA yield [19]. Although Protocols IV and V showed the highest average DNA concentrations, Protocol V showed a wide range of DNA concentration over samples. The only difference between Protocols IV and V was at the final extraction step, using QIAamp DNA FFPE Tissue Kit, not PCE. This suggested that compared to PCE, using spin-columns to extract similar sized tissue sections may be robust and to better obtain a more predictable DNA yield. Comparing Protocols III and IV, post-digestion heat treatment with a lower temperature and longer incubation time gave a better yield of dsDNA (p = 0.02).
Using the NanoDrop to quantify the DNA yield, Protocols I and V had the widest ranges of DNA concentrations (Table 2). When comparing the ratio of NanoDrop to Qubit concentrations, Protocols I and V again showed the highest ratios. These observations were likely due to the varying degrees of phenol contamination since DNA and phenol have similar absorbance wavelengths [20]; hence, nucleic acid quantification using NanoDrop should be interpreted with caution.
Table 2.
Summary of concentrations and quality of DNA extracted from 5 different protocols.
| Extraction protocol | Qubit (ng/μL)a |
NanoDrop (ng/μL)a |
ddPCR (ng/μL)a |
Nanodrop/Qubit ratiob | ddPCR/Qubit ratiob | Fragmentation (252/106 bp)a |
|---|---|---|---|---|---|---|
| I | 28 (19-42) | 165 (81-279) | 27 (15-44) | 5.73 ± 1.24 | 0.92 ± 0.10 | 7% (5-9%) |
| II | 62 (43-83) | 176 (124-259) | 38 (25-60) | 2.80 ± 0.24 | 0.60 ± 0.09 | 17% (15-20%) |
| III | 47 (35-58) | 144 (124-167) | 29 (26-36) | 3.15 ± 0.45 | 0.64 ± 0.10 | 15% (13-17%) |
| IV | 77 (65-99) | 174 (145-236) | 44 (33-65) | 2.25 ± 0.13 | 0.56 ± 0.09 | 14% (11-16%) |
| V | 72 (26-155) | 342 (193-679) | 38 (11-89) | 5.31 ± 1.37 | 0.49 ± 0.05 | 12% (11-13%) |
Please see Fig. 1 for detailed flowchart. Key points of differences among protocols:
I: Heat deparaffinization, tissues unstained, cell lysis buffer, no post-digestion heat treatment, phenol-chloroform DNA extraction.
II: Xylene deparaffinization, tissues unstained, post-digestion heat treatment (90 °C, 1 h), column-based DNA extraction.
III: Xylene deparaffinization, Methyl Green (MG) staining, post-digestion heat treatment (90 °C, 1 h), column-based DNA extraction.
IV: Xylene deparaffinization, MG staining, post-digestion heat treatment (80 °C, 4 h), column-based DNA extraction.
V: Xylene deparaffinization, MG staining, post-digestion heat treatment (80 °C, 4 h), phenol-chloroform DNA extraction.
Values: mean, range in the bracket.
Values: average, standard deviation.
Using the ddPCR assay, we counted the absolute number of droplets with positive signals of targeted genes as the means to quantify amplifiable DNA [9]. We found that there was no significant difference on the average amount of amplifiable DNA for 97 bp amplicons using HFE2 among different protocols. However, the ratio of ddPCR to Qubit ranged from 49% to 92%. This observation suggested that the amount of DNA quantified using Qubit may be overestimating the perceived amount of amplifiable DNA by measuring both amplifiable DNA and fragmented DNA, which may not be amplifiable depending on the amplicon size. This may impact on or explain some PCR-based studies with low and/or negative target gene signals [3,4].
3.2. Quality of DNA
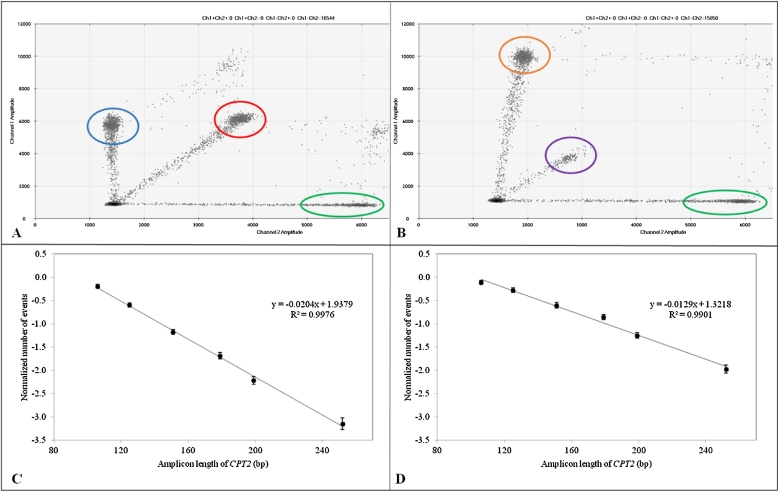
We designed different amplicon sizes of CPT2 (106-252 bp) to further investigate the fragmentation effect of the FFPE samples under various protocols. DNA fragmentation could be visually observed from the ‘rains’ and the number of signals (Fig. 2A and B) [9]. Additionally, smaller amplicons would have relatively higher counts in comparison to those of larger amplicons, especially using the same gene with varying amplicon sizes as shown in Fig. 2C and D. The absolute value of the slope from the best-fit line would therefore allow us to compare the degree of fragmentation, i.e., the larger slope (example in 2C) representing the less amount of yields of larger sized amplicons in the same sample. The steepness of the slopes can act as a proxy for the degree of fragmentation (Fig. 2C and D). By comparing the ratios of the two amplicons with the largest-size discrepancy, i.e. 252/106 bp, Protocols I and II had the smallest and largest average values, respectively (Table 2). Comparing Protocols II and III, the latter, stained with MG, showed relatively more fragmentation, but the difference was not statistically significant. This was consistent with other studies that MG staining had minimal impact on fragmentation [21,22]. The observed fragmentation likely resulted from low pH staining solution that could cause DNA damage [23]. A low pH environment during the extraction process, causing hydrolyzation of DNA from hydrogen binding to phosphodiester bonds and break the bases off, have shown to affect both the yield and amplification of DNA [24]. Further tissue processing with acidic pH reagents can also cause degradation of DNA and decreases the yield of quantifiable nucleic acids [23]. In conjunction with other studies that found MG to have had minimal impact on PCR [21,22], we thus believe that the fragmentation was induced by the acidic pH of the MG staining solution. On the other hand, the purpose of tissue counterstaining prior to microdissection is to visualize the tissue structure and differentiate cell types, to increase the tumor purity, and to avoid normal tissue contamination. MG staining has shown to have less damage to the DNA with the highest pH and acceptable visualization to the details of the tissue allowing us to perform tissue microdissection. Comparing Protocols I and II, both unstained samples, the differences in fragmentation may be associated with other factors, e.g., the removal of formalin-induced crosslinking especially at GC region [17].
Fig. 2.
Using ddPCR assay to assess DNA fragmentation. Upper panels (A and B) showed ddPCR outputs of 2 experiments: A) HFE2 (97 bp, green), CPT2 (106 bp, red), and RPP30 (98 bp, blue) and B) HFE2 (97 bp, green), CPT2 (252 bp, purple), and RPP30 (97 bp, orange). Comparing the droplet numbers of different-size amplicons of the same gene, the number of droplets in the purple circle (CPT2, 252 bp) was comparably less than that in the red circle (CPT2, 106 bp). Lower panels (C and D) showed examples of different extraction protocols (Protocols I and II) of a sample (V8559). Higher degree of fragmentation in Protocol I (absolute slope = 0.020) is observed, compared to Protocol II (absolute slope = 0.013). Error bars represent the 95% confidence intervals of the sample replicates. Please see Fig. 1 for detailed parameters in each protocol. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
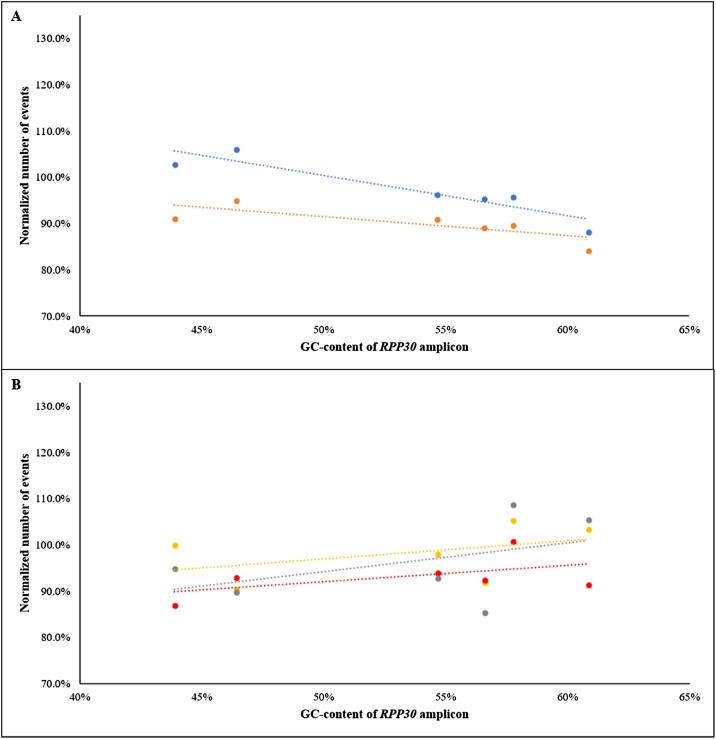
To test this hypothesis, we designed primer sets that targeted different regions within RPP30 to generate varying GC-content, but similar-sized, amplicons (Table 1). We chose RPP30 for it is commonly used as a reference gene [5,7,10]. Our regression analysis suggested that comparing Protocols I and V, i.e., PCE extraction method, post-digestion heat treatment helped to decrease the variation of qualities among samples as the GC-content of RPP30 amplicons increased (Fig. 3A), which supports our hypothesis that DNA quality can be influenced from inadequate reversal of formalin-induced crosslinking. Among samples extracted with column-based methods (Protocols II, III and IV), the results unexpectedly showed the trends that the relative counts of RPP30 amplicons increased with increasing GC-content (Fig. 3B). We observed minimal effects on reversing formalin-induced crosslinking at GC regions using different extraction conditions. Due to the proprietary nature of the commercial kit, we were unable to explain the differences observed between PCE and column-based extraction methods. Additionally, nucleotide sequence with preference cross-linking associated with the Adenine-Thymine dinucleotide has been reported [13]. Other inherited factors prior to FFPE processing and storage may also play some roles.
Fig. 3.
Evaluating the effectiveness of reversing formalin-induced crosslinking at GC regions (44–61% GC) of RPP30 amplicons. The data points represent the average values of the four samples in each extraction protocol with respect to their RPP30 amplicons. The dotted lines represent the best-fit line and the colours correspond to their respective extraction protocols. A) DNA extracted using phenol-chloroform and ethanol precipitation (PCE), Protocols I (blue) and V (orange), showed trend of decreasing averaged number of events with increasing GC-content of RPP30 amplicons. B) DNA extracted using column-based method from a commercial kit, Protocols II (yellow), III (red), and IV (grey), showed trend of slightly increasing averaged number of events with increasing GC-content of RPP30 amplicons. Please refer to Fig. 1 for detailed parameters of each extraction protocol (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.3. Potential applications of quality assessment by ddPCR
In this study, we proposed the use of a ddPCR assay to perform quantity and quality assessment of FFPE-derived DNA samples prior to any PCR-based assays, including NGS, qPCR, SNP, or methylation arrays, etc. Our assay is relatively inexpensive and has fast turnaround time. Using our study as an example, we can generate report in one day for ∼8 samples and we use ∼$100 for consumables (less than $15 per sample). Due to the one-day report, this will not significantly impact on the workflow.
There is an increasing number of downstream PCR-based applications utilizing FFPE DNA due to the ease of storage and availability of FFPE tissues for routine diagnosis and retrospective studies [25]. In addition, the cost per sample for these large scale analyses can be quite substantial and thus, a simple quality assessment test of FFPE DNA prior to downstream applications would be of great value. This is a critical step to avoid negative impacts on result interpretation due to fragmentation and cross-links of FFPE samples. Therefore, for any PCR-based assays that prioritize the quality of FFPE samples, the assessment results using ddPCR may outweigh the cost.
3.4. Limitations
There were some limitations to this study. First, we only had a small set of samples to test five different extraction conditions. However, our results showed that the most suitable method may depend on the overall workflow and downstream applications. For example, a clinical lab may prioritize quick turnover rates and the spin columns (Protocol III) would provide that benefit. On the other hand, a research lab conducting next-generation sequencing with FFPE DNA may incorporate prolong post-digestion heat treatment to reverse formalin-induced crosslinking with phenol-chloroform extraction (Protocol V). Second, we used tumor samples for quality analysis using ddPCR assay. Heterogeneity between tumor cells may have influenced our results. Third, the unknown compositions of reagents from commercial extraction kits limited the comparisons to be only made for the sample pre-treatment steps. Finally, other factors might be related to formalin-induced crosslinking were not investigated.
4. Conclusion
This is the first time to demonstrate the effectiveness using ddPCR assay to assess the quantity and quality (fragmentation and formalin-induced GC crosslinking) of amplifiable DNA from FFPE samples. Although preliminary, we have shown that various extraction variables might impact DNA quantity and quality for PCR-based reactions. Further studies for increasing the understanding of the associations between GC regions and formalin-induced crosslinking are warranted.
Conflict of interest
The authors declare that there are no conflicts of interest.
Handled by Justin O’Grady
References
- 1.Majewski I.J., Bernards R. Taming the dragon: genomic biomarkers to individualize the treatment of cancer. Nat. Med. 2011;17:304–312. doi: 10.1038/nm.2311. [DOI] [PubMed] [Google Scholar]
- 2.Psifidi A., Dovas C.I., Bramis G. Comparison of eleven methods for genomic DNA extraction suitable for large-scale whole-genome genotyping and long-term DNA banking using blood samples. PLoS One. 2015;10 doi: 10.1371/journal.pone.0115960. e0115960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapp J.R., Diss T., Spicer J. Variation in pre-PCR processing of FFPE samples leads to discrepancies in BRAF and EGFR mutation detection: a diagnostic RING trial. J. Clin. Pathol. 2015;68:111–118. doi: 10.1136/jclinpath-2014-202644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simbolo M., Gottardi M., Corbo V. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062692. e62692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zack T.I., Schumacher S.E., Carter S.L. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindson B.J., Ness K.D., Masquelier D.A. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindson C.M., Chevillet J.R., Briggs H.A. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughesman C.B., Lu X.J., Liu K.Y., Zhu Y., Poh C.F., Haynes C. A robust protocol for using multiplexed droplet digital PCR to quantify somatic copy number alterations in clinical tissue specimens. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161274. e0161274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughesman C.B., Lu X.J.D., Liu K.Y.P. Detection of clinically relevant copy number alterations in oral cancer progression using multiplexed droplet digital PCR. Sci. Rep.-UK. 2017;7 doi: 10.1038/s41598-017-11201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong S.Q., Li J., Salemi R. Targeted-capture massively-parallel sequencing enables robust detection of clinically informative mutations from formalin-fixed tumours. Sci. Rep. 2013;3:3494. doi: 10.1038/srep03494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell. 1978;15:945–954. doi: 10.1016/0092-8674(78)90278-7. [DOI] [PubMed] [Google Scholar]
- 13.Huang H., Hopkins P.B. DNA interstrand cross-linking by formaldehyde: nucleotide sequence preference and covalent structure of the predominant cross-link formed in synthetic oligonucleotides. J. Am. Chem. Soc. 1993;115(9402) [Google Scholar]
- 14.Lu K., Ye W., Zhou L. Structural Characterization of Formaldehyde-Induced Cross-Links Between Amino Acids and Deoxynucleosides and Their Oligomers. J. Am. Chem. Soc. 2010;132:3388–3399. doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGhee J.D., Von Hippel P.H. Formaldehyde as a probe of DNA structure. I. Reaction with exocyclic amino groups of DNA bases. Biochemistry. 1975;14:1281–1296. doi: 10.1021/bi00677a029. [DOI] [PubMed] [Google Scholar]
- 16.Do H., Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin. Chem. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 17.Strien J., Sanft J., Mall G. Enhancement of PCR amplification of moderate GC-containing and highly GC-rich DNA sequences. Mol. Biotechnol. 2013;54:1048–1054. doi: 10.1007/s12033-013-9660-x. [DOI] [PubMed] [Google Scholar]
- 18.Yakovchuk P., Protozanova E., Frank-Kamenetskii M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert M.T., Haselkorn T., Bunce M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desjardins P., Conklin D. NanoDrop microvolume quantitation of nucleic acids. J. Vis. Exp. 2010 doi: 10.3791/2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrig T., Abdulkadir S.A., Dintzis S.M., Milbrandt J., Watson M.A. Quantitative amplification of genomic DNA from histological tissue sections after staining with nuclear dyes and laser capture microdissection. J. Mol. Diagn. 2001;3:22–25. doi: 10.1016/S1525-1578(10)60645-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murase T., Inagaki H., Eimoto T. Influence of histochemical and immunohistochemical stains on polymerase chain reaction. Mod. Pathol. 2000;13(147) doi: 10.1038/modpathol.3880028. [DOI] [PubMed] [Google Scholar]
- 23.Williams N.H. DNA hydrolysis: mechanism and reactivity. In: Zenkova M.A., editor. Artificial Nucleases. Springer Berlin Heidelberg; Berlin, Heidelberg: 2004. pp. 3–17. [Google Scholar]
- 24.Shi S.R., Cote R.J., Wu L. DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J. Histochem. Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 25.Kamps R., Brandao R.D., Bosch B.J. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]