Abstract
Background
MicroRNAs influence almost every genetic pathway and are involved in colorectal cancer (CRC). However, the biological role of miR486-3p in CRC remains to be elucidated.
Methods
In this study, miR486-3p expression in CRC cell lines and normal colonic epithelial cells was determined. After miR486-3p mimic, inhibitor, and BIK siRNA transfection, cell proliferation, apoptosis, and migration were examined. Furthermore, the target of miR486-3p was identified by luciferase-reporter assay and underlying molecular mechanisms studied.
Results
The results revealed that miR486-3p was significantly upregulated in CRC compared with normal colonic epithelial cells, whereas BIK expression was remarkably downregulated in CRC cells. MTT assays demonstrated that suppression of miR486-3p expression reduced CRC cell proliferation, whereas elevated miR486-3p or BIK silencing induced cell proliferation. Wound-healing assays and transwell experiments revealed that both upregulation of miR486-3p and down-regulation of BIK increased CRC cell migration and invasion ability. Moreover, bioinformatic target prediction identified BIK as a putative target of miR486-3p. Knockdown of miR486-3p was shown to upregulate BIK expression, whereas overexpression of miR486-3p suppressed the expression of BIK. Luciferase reporter assay results further confirmed this deduction.
Conclusion
In conclusion, these findings suggest that miR486-3p is an oncogene in CRC. Gene therapy using miR486-3p inhibition may provide a new clue for CRC therapy.
Keywords: colorectal cancer, miR486-3p, BIK, apoptosis, invasion
Introduction
In the last few decades, extensive research has produced remarkable advances in our understanding of biological and genetic cancer mechanisms, among which realization of tumor cell apoptosis and migration is the most important. Apoptosis is a genetically encoded cell death program that directly regulates tumorigenesis.1 Aberrations in the regulation of a restricted number of key pathways that control cell proliferation and cell survival are mandatory for establishment of all tumors. Deregulated cell proliferation with suppressed apoptosis constitute the minimal common platform upon which all neoplastic evolution occurs.2 Also, cancer metastasis, which requires cancer cells to survive and proliferate outside their tissue of origin, is the most life-threatening aspect of cancer.3 It is a complex and highly coordinated cell migration process that is regulated by such genes as the EGFR family, PAK family, cMET, and PTEN.4 As such, therapies that target cancer cell apoptosis and metastasis might have great impact on the survival of cancer patients.
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers in the world and has high risk of affecting the entire length of the large intestine and rectum.5 It remains the third-most prevalent cause of death among the different types of cancer worldwide and the second-leading cause of cancer death in Western society.6,7 Approximately 50%–70% patients have developed distant metastases when diagnosed. Overall survival for patients with unresectable metastatic CRC is poor, with median survival of ~24–27 months and 5-year survival of 10%–15%.8 CRC patients with distant metastasis are not suitable candidates for conventional surgery, chemotherapy, or radiotherapy.9,10 Therefore, better understanding of the underlying molecular mechanism of CRC cell apoptosis and invasion is essential for developing novel diagnostic and therapeutic strategies.
The KRAS protein spans 2 kDa, encodes Ras p21, and is involved in the G-protein signal transduction pathway, influencing cellular proliferation and differentiation.11 KRAS mutations are routinely used as a predictive biomarker for EGFR-targeted therapy.12 BRAF, an RAF gene family serine/threonine kinase, is a downstream effector of KRAS in the RAS–RAF–MAPK signaling pathway. Basically, the BRAFV600E mutation is present in about 8%–10% of CRC cases.13 BRAF mutation has been used to differentiate between familial and sporadic CRC. Currently, there are no prognostic biomarkers available that can provide clear clues about the likelihood recurrence, metastasis, or CRC cases that are difficult to treat with chemotherapy. Therefore, finding new biomarkers is essential.14
Recent studies show that some miRNAs regulate cell proliferation and apoptosis processes that are important in cancer formation. By using multiple molecular techniques, which include Northern blot analysis, real-time PCR, miRNA microarray, up- or down-expression of specific miRNAs, it was found that several miRNAs were directly involved in human cancers, including lung, breast, brain, liver, colon cancer, and leukemia. Deregulation in miRNA expression correlates with various cancers by acting as tumor suppressors and oncogenes. miRNA is an important biomarker for CRC, and a series of studies have indicated the involvement of miRNAs in CRC. The first study on the diagnostic usefulness of serum-based miRNA biomarkers in CRC reported that a subset of 69 miRNAs was highly expressed in CRC patients compared to healthy controls. Additionally, 14 of these serum-based miRNAs from CRC patients were not detectable in those with lung cancer, highlighting the CRC specificity of these miRNAs.15 Other studies have indicated that miR21 is a biomarker for the early detection of CRC, and the direct target of miR21 was confirmed to be Pdcd4.16–18 In addition, studies have revealed that miR598 can inhibit metastasis in CRC by suppressing the JAG1–Notch2 pathway, stimulating epithelial–mesenchymal transition, and that miR363-3p can restrain epithelial–mesenchymal transition and suppress metastasis in CRC by targeting Sox4. A series of studies have indicated the involvement of miRNAs in CRC. Some research has indicated that miR486-3p can be a biomarker for the detection of tongue squamous cell carcinoma and retinoblastoma.19,20 Also, research has shown that KRAS mutation was associated with upregulation of miR486-3p in primary tumors of 60 patients with metastatic CRC.21 However, the biological roles of miR486-3p in CRC are still poorly understood.
BIK is the founding member of the BH3-only family of proapoptotic proteins.22 BIK is predominantly localized in the endoplasmic reticulum (ER) and induces apoptosis through the mitochondrial pathway by mobilizing calcium from the ER to the mitochondria.23,24 BIK interacts strongly with several antiapoptotic proteins, such as BCL2, E1B19K and BCLxL, and exhibits potent cell death activity when expressed autonomously. This activity is suppressed by coexpression of BCL2, BCLxL, and E1B19K, suggesting that BIK might be an important target for cellular and viral antiapoptosis proteins. BIK-mediated apoptosis is mediated by selective activation of BAX, whereas BIK also induces nonapoptotic cell death in certain cells by unknown mechanisms. Several anticancer drugs activate BIK gene expression through transcriptional pathways dependent on such factors as E2F and p53 or by removal of epigenetic marks on chromatin. At the same time, BIK has also been used as a therapeutic molecule to treat difficult cancers in gene therapy.
In this study, miR486-3p expression was examined in CRC cell lines and normal colonic epithelial cells. Roles of miR486-3p in CRC cells were investigated, which included cell proliferation, apoptosis, and metastasis. It was also verified that miR486-3p acted as an oncogene in CRC by targeting BIK. The specific aim of the study was to investigate and verify the role of miR486-3p in CRC. In-depth study on the molecular mechanisms will provide new strategies and methods for the treatment of CRC.
Methods
Cell lines and cell culture
The human CRC cell lines SW620 and HT29 and the normal colon cell line NCM460 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS, 100 mg/mL streptomycin, and 100 U/mL penicillin. Cultures were maintained in an incubator at 37°C in 5% CO2.
Cell transfection
For transfection, Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) was used. miR486-3p mimics, inhibitors, and negative control were purchased from GenePharma (Shanghai, China). BIK siRNA was synthesized and chemically modified by Santa Cruz Biotechnology (Dallas, TX, USA). All transfections were performed according to manufacturers’ recommended protocols. Cells were then cultured for 24 hours for further analysis.
MTT cell viability assay
Cell viability was measured with an MTT assay kit (Beyotime, Haimen, China) 24 hours after transfection according to the manufacturer’s protocol. OD450 was measured using a Synergy 2 multifunction microplate reader (BioTek, Winooski, VT, USA). Each experiment was independently repeated three times.
Cell apoptosis
After transfection with miRNAs or siRNAs, cells were seeded in six-well plates at 3×104 per well and cultured for 48 hours. Cells were subsequently harvested and double-stained with fluorescein isothiocyanate-conjugated annexin V and propidium iodide (BD Biosciences, San Diego, CA, USA). Then, apoptosis rates were analyzed with flow cytometry (FACSCalibur, BD Biosciences).
Wound-healing assays
Cell migration was evaluated by wound-healing assays performed 24 hours after transfection. Briefly, confluent monolayers of CRC cells cultured in the six-well plates were mechanically wounded by dragging a 10 µL sterile plastic tip at the center of the well. Cells were washed with PBS to remove cellular debris and allowed to migrate for 12 hours. Cells migrating into wound areas were visualized and photographed. All experiments were repeated at least three times.
Transwell invasion assay
The migratory and invasive ability of CRC cells were examined basing on their ability to cross the 8 mm pores of migration chambers. Cells were plated in the upper transwell chambers, whereas DMEM with 10% FBS was added to the lower chamber as a chemoattractant. After incubation for 24 hours, nonmigrated cells in the upper chamber were removed using a cotton swab, and migrated cells on the bottom of the transwell were fixed in 4% paraformaldehyde and stained with 0.2% crystal violet (Sigma-Aldrich, St Louis, MO, USA). Migrated cells were counted in five randomly selected microscopic fields and photographed (magnification 100×).
RNA isolation and quantitative reverse transcription (RT)-PCR
Total RNA was isolated from SW620, HT29, or NCM460 cells using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. RT was performed using a PrimeScript RT-reagent kit (Takara Bio, Kusatsu, Japan). For quantitative RT-PCR of miRNA, total RNA was reverse transcribed using a stem-loop primer (RiboBio, Guangzhou, China). Oligo(dT)12–18 or miRNA-specific primers were added to initiate cDNA synthesis. PCR was carried out using a Step One Plus real-time PCR system (Thermo Fisher Scientific) and SYBR premix Ex Taq II (Takara Bio). U6 was used as an endogenous control. For mRNA evaluation, synthesis of cDNA was performed using an RNA PCR kit (Takara, Japan), and quantitative real-time PCR was carried out using the SYBR premix Ex Taq II kit according to the manufacturer’s instructions. GAPDH was used as an endogenous control. All reactions were performed in triplicate. The 2−ΔΔCt method was adopted and applied to calculate the relative quantities of each gene.
Western blot
Primary antibodies for BIK, Apaf1, pERK1/2, and ERK1/2 were obtained from Abcam (Cambridge, UK). Primary antibodies for cytochrome c, caspase 9, and caspase 3 were from Beyotime. Secondary antibodies were from Santa Cruz Biotechnology. Total protein lysates extracted from cells were quantified using a BCA protein assay kit (Beyotime). Equal amounts of total protein lysates were separated with 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Membranes were blocked with 5% nonfat milk, followed by incubation overnight at 4°C with primary antibodies. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies and visualized using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific). GAPDH was used as an internal control.
Luciferase reporter assay
The full-length of the BIK 3′UTR sequence was amplified from human cDNA. Wild-type and mutated 3′UTR regions of BIK were cloned into the XbaI site of pGL3 (Promega, Fitchburg, WI, USA). Site-directed mutagenesis of the miR486-3p target site in the BIK 3′UTR was carried out using a QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. For reporter assays, cells were seeded in 24-well plates and cultured for 14–16 hours before transfection. 3′UTR plasmids were cotransfected in SW620 cells using Lipofectamine 2000. Reporter assays were performed 24 hours posttransfection using a dual luciferase assay system (Promega). Relative Renilla luciferase activities were normalized to firefly luciferase activities and used as an internal control for transfection efficiency.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). Values presented in this study are means±SD with Student’s t-test or one-way ANOVA. Differences were considered significant at P<0.05.
Results
Upregulation of miR486-3p and downregulation of BIK in malignant colorectal cancer cell lines
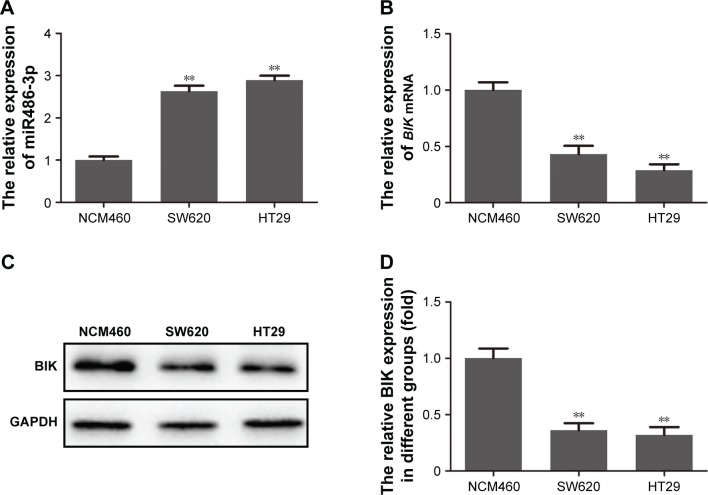
miR486-3p-expression levels in the two malignant CRC cell lines and the normal colorectal cell line were initially determined by RT-PCR. As shown in Figure 1A, compared with the normal colorectal cell line, a notable increase in miR486-3p was observed in both HT29 and SW620 CRC cells. To evaluate the expression of BIK in malignant CRC cell lines, we utilized RT-PCR and Western blot to analyze mRNA and protein levels of BIK, respectively. As shown in Figure 1B–D, both mRNA and protein levels of BIK were obviously decreased in both HT29 and SW620 CRC cells. The results suggested that upregulation of miR486-3p may have some correlation with downregulation of BIK in malignant CRC.
Figure 1.
Upregulation of miR486-3p and downregulation of BIK in malignant colorectal cancer cell lines.
Notes: (A) Expression level of miR486-3p determined by reverse-transcription PCR; (B) mRNA level of BIK determined by reverse-transcription PCR; (C) protein level of BIK determined by Western blot; (D) densitometry analysis of BIK protein level. **P<0.01.
BIK 3′UTR a direct target for miR486-3p
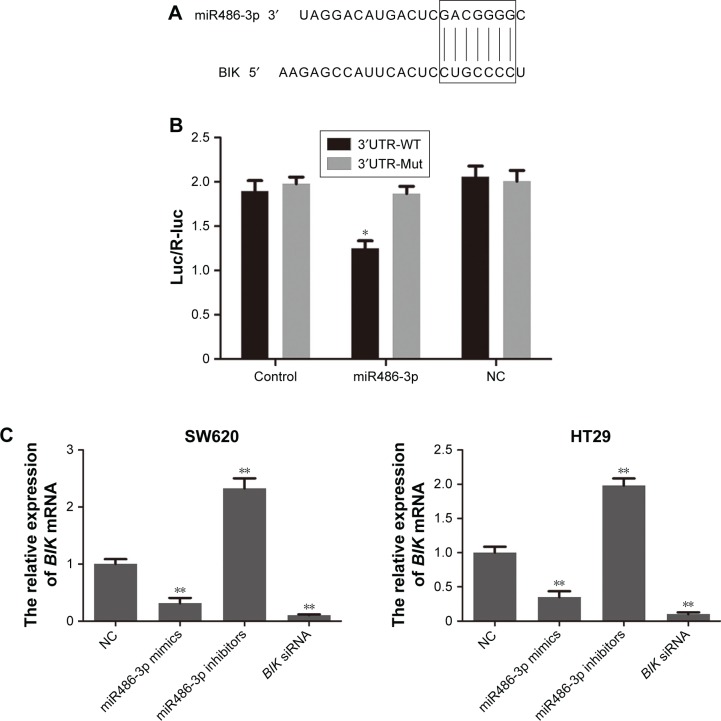
Base pairing between the 3′UTR of mRNA and “seed sequence” located in the 5′-end of miRNA are indispensable for estimating whether targeting miRNA leads to mRNA degradation or translation inhibition.25 To explore the potential target of miR486-3p, we performed bioinformatic analysis on the TargetScan and miRWalk websites and discovered that the 3′UTR of BIK contained regions matching the seed sequences of miR486-3p (Figure 2A). To test whether miR486-3p directly targeted the 3′UTR of BIK, we performed a dual luciferase reporter assay in our study. As shown in Figure 2B, miR486-3p significantly decreased the luciferase activity of the BIK 3′UTR in CRC cells (P<0.05), but not the mutant sequence of the 3′UTR of BIK. These results indicated that the BIK 3′UTR is a direct target for miR486-3p.
Figure 2.
The BIK 3′UTR is a direct target for miR486-3p.
Notes: (A) Base pairing of BIK and miR486-3p; (B) dual luciferase reporter assay; (C) mRNA-expression level of BIK after transfection; (D) protein level of BIK after transfection. *P<0.05; **P<0.01.
Abbreviations: NC, negative control; WT, wild type.
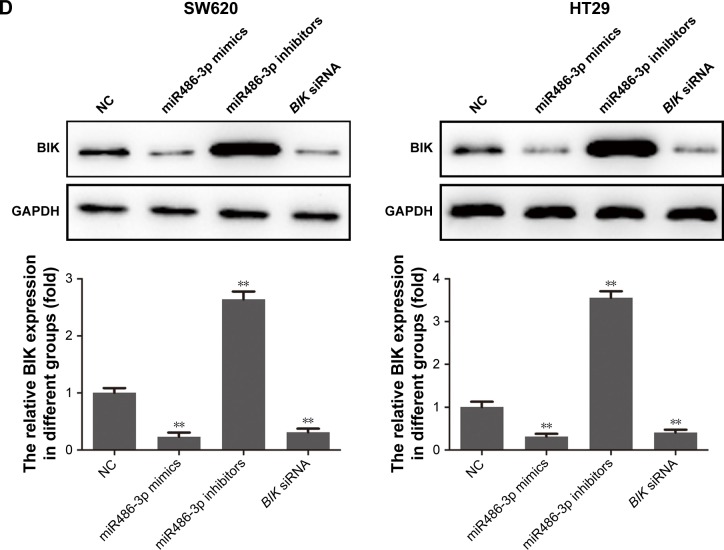
To confirm the targeting effect of miR486-3p on BIK expression, we examined mRNA- and protein-expression levels of BIK by RT-PCR and Western blot, respectively, after transfection of miR486-3p mimics or inhibitors. As demonstrated in Figure 2C, transfection with miR486-3p inhibitors led to a remarkable increase in BIK mRNA level, while transfection with miR486-3p mimics led to significant downregulation of BIK, the trend of which was consistent with BIK siRNA transfection. The results of Western blot assay (Figure 2D) further verified that miR486-3p negatively regulated the expression of BIK.
Downregulation of miR486-3p suppresses cell proliferation, migration, and invasion in vitro
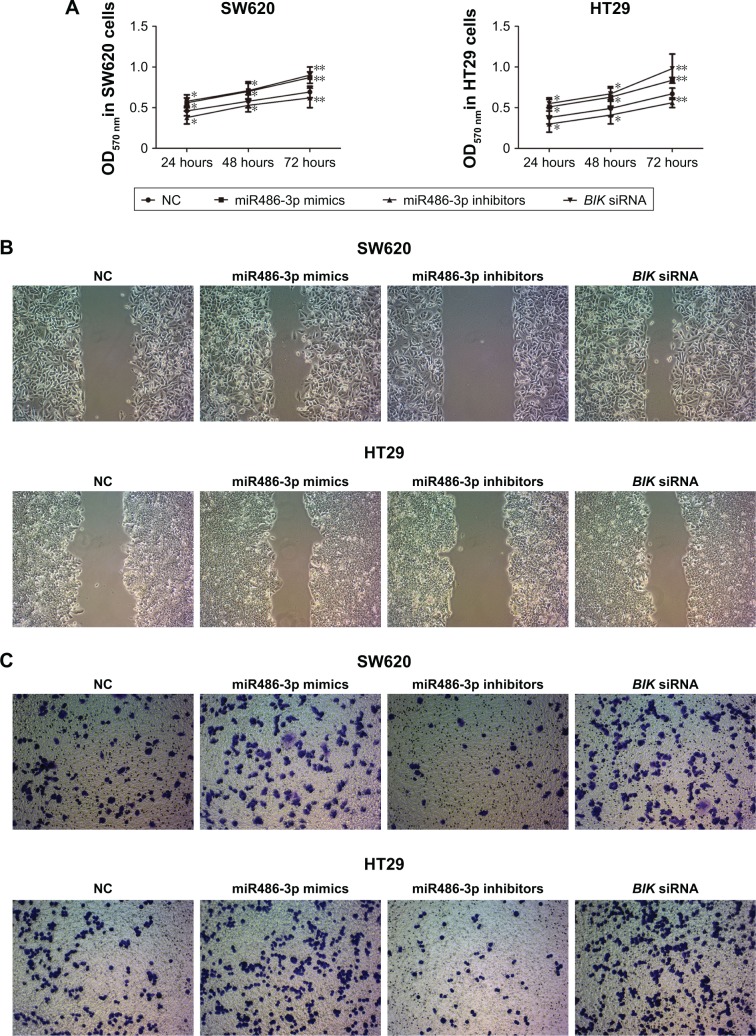
To elucidate further the biological role of miR486-3p in CRC, we investigated the effects of miR486-3p on CRC cell proliferation, migration, and invasion. MTT assay data in Figure 3A shows that BIK silencing enhanced CRC cell proliferation, which was consistent with the group transfected with miR486-3p mimics. However, miR486-3p knockdown significantly inhibited CRC cell proliferation (P<0.01). The results suggested that miR486-3p may act as an oncogene in CRC. Then, wound-healing and transwell assays were conducted to investigate the roles of BIK and miR486-3p in the regulation of CRC cell migration and invasion. As demonstrated in Figure 3B and C, transfection with BIK siRNA or miR486-3p mimics notably promoted CRC cell migration and invasion, whereas transfection with miR486-3p inhibitors played a contrary role in the regulation of migration and invasion of CRC cells in vitro. These results indicated that miR486-3p may play a positive role in CRC metastasis.
Figure 3.
Downregulation of miR486-3p suppresses cell proliferation, migration, and invasion in vitro.
Notes: (A) MTT assay of colorectal cancer (CRC) cells after transfection; (B) microscopy of wound-healing assays in CRC cells after transfection; (C) microscopy of transwell invasion experiments in CRC cells after transfection. *P<0.05; **P<0.01.
Abbreviation: NC, negative control.
Downregulation of miR486-3p promotes CRC cell apoptosis
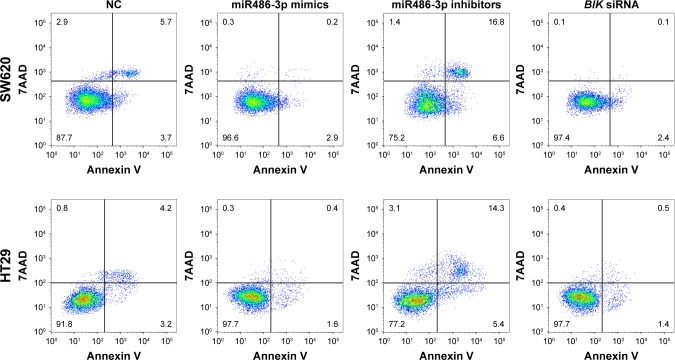
We performed annexin V–fluorescein isothiocyanate double-staining flow cytometry analysis to evaluate the effect of miR486-3p on apoptosis in CRC cells. As shown in Figure 4, compared with the negative control group, cells transfected with miR486-3p inhibitors significantly promoted apoptosis, whereas cells transfected with BIK siRNA or miR486-3p resulted in a small degree of apoptosis. SW620 and HT29 cell lines presented similar apoptosis trends. The data demonstrated that miR486-3p inhibits apoptosis of CRC cells.
Figure 4.
Downregulation of miR486-3p promotes colorectal cancer-cell apoptosis.
Abbreviation: NC, negative control.
Upregulation of miR486-3p inhibits mitochondrial apoptosis pathway
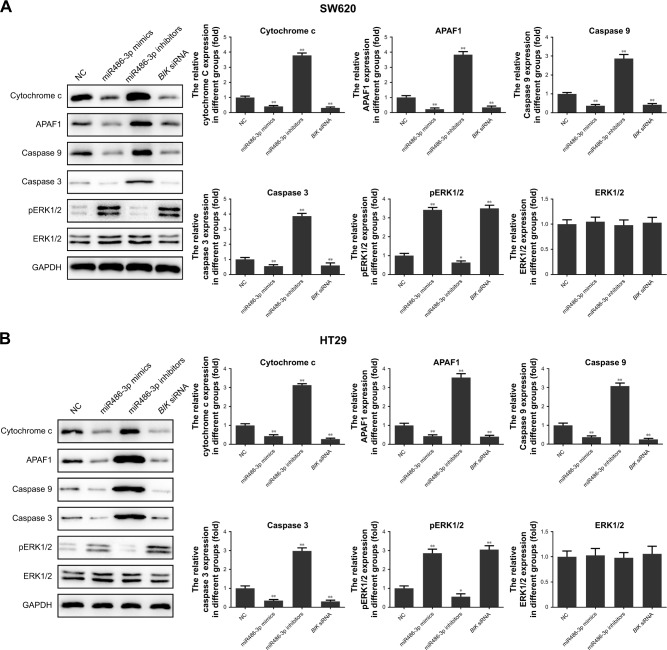
BIK is a proapoptotic tumor suppressor in several human tissue types, and BIK-induced apoptosis may be closely related to the mitochondrial apoptosis pathway.26 In the aforementioned experiments, we demonstrated that miR486-3p directly targeted BIK in CRC tumorigenesis. To reveal the underlying molecular mechanism of this targeting effect, we investigated the alteration in key signals of the mitochondrial apoptosis pathway in CRC cells after transfection. As shown in Figure 5, upregulation of miR486-3p significantly suppressed the expression of cytochrome c, APAF1, caspase 9, and caspase 3, but increased the expression of pERK1/2, while downregulation of miR486-3p presented a contrary effect on the expression of these signals. Also, compared with the miR486-3p-mimic group, cells transfected with BIK siRNA had a parallel effect on the expression level of proteins. The results proved that miR486-3p may work as a carcinogenic gene in the occurrence and development of CRC through modulating the mitochondrial apoptosis pathway.
Figure 5.
Upregulation of miR486-3p inhibits mitochondrial apoptosis pathway.
Notes: (A) Key protein-expression levels of mitochondrial apoptosis pathway in SW620; (B) key protein-expression levels of mitochondrial apoptosis pathway in HT29. *P<0.05; **P<0.01.
Abbreviation: NC, negative control.
Discussion
Behind cardiovascular and infectious diseases, cancer-related mortality is the third-most common cause of death worldwide.27 Uncontrolled proliferation of damaged cells as a result of deregulation of genes involved in cell cycle machinery and apoptosis leads to tumor formation.28 Conventional treatments for cancer depend on radiotherapy, chemotherapy, interventional therapy, and surgery. However, serious side effects and high surgical risk and recurrence rate force us to explore new ways for cancer diagnosis and therapy. Over the years, miRNAs have been detected in many types of tissue and body fluid, and control and regulate several vital cellular processes, such as proliferation, differentiation, cell cycle, and apoptosis. Seen from the molecular level, on the other hand, miRNAs can silence target genes efficiently and regulate a broad set of genes of interest simultaneously, which benefits treatment of cancer as a heterogenic disease.29 Many studies have evaluated the feasibility of employing miRNAs as diagnostic and prognostic markers for CRC. For example, the miR126 class has been used as a prognostic marker for CRC to determine sensitivity toward a particular drug. miR31 acts as an oncomir and is known to modulate response to chemotherapeutic drug and inhibit cell proliferation.
miR48plays a critical role in regulating human erythroid differentiation and has been demonstrated to be abnormally expressed in several tumors. Ye et al30 found that miR486-3p is a tumor suppressor that directly targets ECM1 to repress cell proliferation and metastasis in cervical cancer, and there is a report showing that miR486-3p levels are increased in papillary thyroid carcinoma.31 However, no literature has reported the role of miR486-3p in CRC. In this study, we observed a notable increase in miR486-3p in both HT29 and SW620 CRC cells. By using prediction software, we found that BIK was a target gene with high miR486-3p score in CRC. The targeting relationship between miR486-3p and BIK was verified by dual luciferase reporter assay. BIK is the founding member of the BH3-only family proapoptotic proteins.26 It is a critical effector in apoptosis. Shore et al showed that ER-localized BIK induced apoptosis through the mitochondrial pathway that resulted in cytochrome c release.32–34 Our study demonstrated that BIK mRNA and protein levels were downregulated in malignant CRC cell lines. Downregulation of miR486-3p led to a remarkable increase in BIK mRNA levels and further suppressed cell proliferation, migration, and invasion, but promoted CRC cell apoptosis.
Researchers have discovered that dysregulation of apoptosis occurs in cancer cells. Some of the ways in which aberrant regulation of cell death genes contributes to aggressive behaviors of malignant cells have been highlighted. The mitochondrial apoptosis pathway is one of the most important and well-researched apoptosis pathways of cells. Mitochondria can be induced to release cytochrome c in response to death stimuli. Release of cytochrome c into the cytosol results in APAF activation and subsequent activation of caspases that induce apoptosis.35 In our study, we found that upregulation of miR486-3p significantly suppressed the expression of cytochrome c, APAF1, caspase 9, and caspase 3, which revealed the underlying molecular mechanism of CRC cells’ aggressive behavior. In addition, we discovered that upregulation of miR486-3p significantly increased pERK1/2 expression, which may account for the metastatic behavior of CRC cells.
In conclusion, in this study, we found that miR486-3p was a significantly upregulated miRNA in CRC. Further studies confirmed that miR486-3p regulated CRC cell proliferation, apoptosis, and metastasis by directly targeting BIK. It was also verified that miR486-3p was negatively related to BIK expression. These findings suggest that miR486-3p is an oncogene in CRC. Gene therapy using miR486-3p inhibition may provide a new avenue for CRC therapy.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Fisher DE. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78(4):539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 2.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005;17(5):559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15(1):87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 5.dos Reis SA, da Conceição LL, Siqueira NP, Rosa DD, da Silva Letícia L, do Carmo G Peluzio Maria. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr Res. 2017;37:1–19. doi: 10.1016/j.nutres.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization WHO global health observatory: cancer mortality and morbidity. [Internet] 2014. Available from: http://www.who.int/gho/ncd/mortality_morbidity/cancer_text/en.
- 8.American Cancer Society, Colorectal Cancer Facts and Figures 2014–2016 Am Cancer Soc. 2014 [Google Scholar]
- 9.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 10.Wanebo HJ, Legolvan M, Paty PB, et al. Meeting the biologic challenge of colorectal metastases. Clin Exp Metastasis. 2012;29(7):821–839. doi: 10.1007/s10585-012-9517-x. [DOI] [PubMed] [Google Scholar]
- 11.Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP5genetic mutation analysis for resectable and unresectable colorectal liver metastases: a systematic review of the current evidence. Surg Oncol. 2018;27(2):280–288. doi: 10.1016/j.suronc.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Waring P, Tie J, Maru D, Karapetis CS. RAS Mutations as predictive biomarkers in clinical management of metastatic colorectal cancer. Clin Colorectal Cancer. 2016;15(2):95–103. doi: 10.1016/j.clcc.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418(6901):934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Liang L, Fang JY, Xu J. Somatic gene copy number alterations in colorectal cancer: new quest for cancer drivers and biomarkers. Oncogene. 2016;35(16):2011–2019. doi: 10.1038/onc.2015.304. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 16.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 17.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-2as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-2(miR-21) post-transcriptionally downregulates tumor suppressor Pdcdand stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 19.Venkatesan N, Deepa PR, Khetan V, Krishnakumar S. Computational and in vitro investigation of miRNA-gene regulations in retinoblastoma pathogenesis: miRNA mimics strategy. Bioinform Biol Insights. 2015;9(9):89–101. doi: 10.4137/BBI.S21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Yu T, Cabay RJ, et al. miR-486-3p, miR-139-5p, and miR-2as biomarkers for the detection of oral tongue squamous cell carcinoma. Biomark Cancer. 2017;9:1–8. doi: 10.4137/BIC.S40981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosakhani N, Sarhadi VK, Borze I, et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51(1):1–9. doi: 10.1002/gcc.20925. [DOI] [PubMed] [Google Scholar]
- 22.Boyd JM, Gallo GJ, Elangovan B, et al. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene. 1995;11(9):1921–1928. [PubMed] [Google Scholar]
- 23.Germain M, Mathai JP, Shore GC. BH-3-only BIK functions at the endoplasmic reticulum to stimulate cytochrome c release from mitochondria. J Biol Chem. 2002;277(20):18053–18060. doi: 10.1074/jbc.M201235200. [DOI] [PubMed] [Google Scholar]
- 24.Germain M, Mathai JP, Mcbride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. Embo J. 2005;24(8):1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Chinnadurai G, Vijayalingam S, Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27(Suppl 1):S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittmann J, Jäck HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806(2):200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Vecchione A, Croce CM. Apoptomirs: small molecules have gained the license to kill. Endocr Relat Cancer. 2010;17(1):F37–F50. doi: 10.1677/ERC-09-0163. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye H, Yu X, Xia J, Tang X, Tang L, Chen F. MiR-486-3p targeting ECMrepresses cell proliferation and metastasis in cervical cancer. Biomed Pharmacother. 2016;80:109–114. doi: 10.1016/j.biopha.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Swierniak M, Wojcicka A, Czetwertynska M, et al. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98(8):E1401–E1409. doi: 10.1210/jc.2013-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain M, Mathai JP, Mcbride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. Embo J. 2005;24(8):1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain M, Mathai JP, Shore GC. BH-3-only BIK functions at the endoplasmic reticulum to stimulate cytochrome c release from mitochondria. J Biol Chem. 2002;277(20):18053–18060. doi: 10.1074/jbc.M201235200. [DOI] [PubMed] [Google Scholar]
- 34.Mathai JP, Germain M, Shore GC. BH3-only BIK regulates BAX, BAK-dependent release of Ca2+ from endoplasmic reticulum stores and mitochondrial apoptosis during stress-induced cell death. J Biol Chem. 2005;280(25):23829–23836. doi: 10.1074/jbc.M500800200. [DOI] [PubMed] [Google Scholar]
- 35.Reed JC. Dysregulation of apoptosis in cancer. J Clin Oncol. 1999;17(9):2941–2953. doi: 10.1200/JCO.1999.17.9.2941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.