Abstract
Aims
At the time of analysis, two widely used, drug-specific, tumour-cell programmed death ligand 1 (PD-L1) assays were approved by the US Food and Drug Administration for anti-PD-1 therapies: the Dako PD-L1 immunohistochemistry (IHC) 28-8 pharmDx assay and the Dako PD-L1 IHC 22C3 pharmDx assay. Given that the majority of current PD-L1 testing in US clinical practice is performed at commercial reference laboratories, we aimed to evaluate the concordance of the 28-8 and 22C3 assays in a real-world setting.
Methods
Matched PD-L1 IHC 28-8 and 22C3 results from routine assessment were obtained from 1930 patients, including 412 confirmed to have lung cancer, submitted from hospitals in over 38 US states/territories. Biopsies were stained, reviewed and scored by trained/certified pathologists at a single cancer reference laboratory between 2015 and 2017. Rate of concordance between assay findings was assessed by Bland-Altman analysis; overall per cent agreement (OPA), positive per cent agreement and negative per cent agreement; and Cohen’s kappa.
Results
PD-L1 IHC 28-8 and 22C3 displayed strong correlation across all samples and in samples with a confirmed lung cancer diagnosis irrespective of biopsy site. The OPA was 97%–98% for all samples, depending on the expression level defining PD-L1 positivity. In the Bland-Altman analysis, the mean difference in percentage of tumour cells positively stained for PD-L1 between the paired assay findings was –0.80% for all samples and –0.93% in samples with a confirmed lung cancer diagnosis.
Conclusions
These data, in conjunction with recent findings, support the analytical concordance of the PD-L1 IHC 28-8 and 22C3 assays for assessing per cent tumour-cell membrane PD-L1 expression.
Keywords: lung cancer, cancer, immunohistochemistry, diagnostics
Introduction
The programmed death-1 (PD-1)/programmed death ligand 1 (PD-L1) signalling axis has a critical role in tumour immune evasion.1 Therapeutically targeting PD-1/PD-L1 has yielded impressive clinical outcomes2–11 and there are currently five US Food and Drug Administration (FDA)-approved agents that target the PD-1 pathway: nivolumab12 and pembrolizumab (anti-PD-1)13; atezolizumab,14 avelumab15 and durvalumab (anti-PD-L1)16; with approvals across a wide variety of tumour types, including non-small-cell lung cancer (NSCLC), melanoma, classical Hodgkin’s lymphoma, squamous cell carcinoma of the head and neck, renal cell carcinoma, urothelial carcinoma, hepatocellular carcinoma, gastric cancer, cervical cancer, primary mediastinal large B-cell lymphoma, Merkel cell carcinoma and microsatellite instability-high or mismatch repair-deficient solid tumours, including colorectal cancer.12–16 Higher levels of PD-L1 expression can be associated with greater therapeutic efficacy from anti-PD-1 agents, although patients with lower PD-L1 expression levels can derive clinical benefit in certain tumour types and histologies.12–16
At the time of the analysis, two drug-specific PD-L1 assays associated with the use of PD-1-targeted agents in different treatment scenarios were widely used. For example, in NSCLC, the Dako PD-L1 immunohistochemistry (IHC) 22C3 pharmDx was approved as a companion diagnostic (testing required) for the use of first-line or greater pembrolizumab17 18; the Dako PD-L1 IHC 28-8 pharmDx was approved as a complementary diagnostic (testing optional) for second-line nivolumab therapy.19 Both assays were initially approved during the same time frame.20
In clinical practice, many physicians order a PD-L1 test without specifying which assay to use. This ambiguity is mainly due to clinicians not having a specific anti-PD-1 or anti-PD-L1 antibody in mind when ordering a particular test, as well as a potential lack of familiarity that multiple assays are approved. Several organisations, including the American Association for Cancer Research and the International Association for the Study of Lung Cancer, have highlighted the impracticality of having various assays for multiple anti-PD-1 agents,21–23 prompting the investigation of the interchangeable use of PD-L1 assays.21 23
Analytical concordance between the two assays has been evaluated through multiple concordance analyses.21 23–27 These studies suggest strong concordance between 22C3 and 28-8 assay results when evaluated by the diagnostic manufacturer or academic laboratories. Furthermore, clinical cut-offs for each assay are meaningful as they were validated from clinical trials.21
The present US-based study was performed to evaluate the real-world concordance between the two assays in a single cancer reference laboratory using a wide array of samples submitted by the community where a PD-L1 test was not specified.
Materials and methods
Samples
The analysis was performed on a data set provided by Symphony Health Solutions matching unique identifiers to associate clinical characteristics with PD-L1 biomarker test results and annotations reported by NeoGenomics Laboratories between 26 October 2015 and 5 July 2017. Samples for PD-L1 expression testing were sent to NeoGenomics Laboratories by individual community oncology practices. Samples were recorded as ‘lung cancer’, ‘melanoma’, ‘other’ or ‘blank’, based on diagnostic information voluntarily provided by the physician requisitioning the test and with no additional information on tumour type for ‘blank’ and ‘other’ samples. All staining was performed with the FDA-cleared PD-L1 IHC 22C3 pharmDx assay or the PD-L1 IHC 28-8 pharmDx assay on the Dako Autostainer Link 48.18 19 At the time of this analysis, the 22C3 assay was approved for determining PD-L1 protein expression in NSCLC only, whereas the 28-8 assay was approved for non-squamous NSCLC and melanoma.
Pathological assessment of PD-L1 staining
Sample testing requisitions from the individual practices either did not order a specific PD-L1 IHC assay or ordered both assays, the 22C3 assay only, or the 28-8 assay only. When no PD-L1 IHC assay was specified or both assays were ordered, PD-L1 expression was assessed using both Dako assays; when only the 22C3 or 28-8 assay was ordered, PD-L1 expression was tested using only the specified assay. Biopsies were reviewed and scored by pathologists at NeoGenomics Laboratories who received training and certification in scoring by Dako. FDA-cleared diagnostic kits and protocols/scoring algorithms were used by pathologists to score biopsies.
With the 22C3 assay, PD-L1 protein expression was determined by using tumour proportion score (TPS), which is the percentage of viable tumour cells showing partial or complete membrane staining. By the end of the study period, the NSCLC sample was considered to have PD-L1 expression if the TPS was ≥1% and high PD-L1 expression if the TPS was ≥50%. Non-squamous NSCLC samples were considered to have PD-L1 expression with the 28-8 assay when complete circumferential and/or partial linear plasma membrane staining of tumour cells was observed at any intensity.18 19 Tumour PD-L1 expression is continuously distributed, and TPS was estimated at every integer between 0% and 10% and in increments of 5% thereafter. Statistical analysis was conducted on these values as continuous.
As is commonly done, pathologists randomly received samples to review and score, and all testing was blinded. Pathologists were not aware of the presence of other testing using a different clone, or that this would be used to compare results. PD-L1 expression results were reported to the ordering physician to inform treatment decision-making.
Statistical analyses
Statistical tests used to evaluate agreement between assays included the Bland-Altman analysis; calculation of the positive per cent agreement (PPA), the negative per cent agreement (NPA) and the overall per cent agreement (OPA); and associated Cohen’s kappa statistics.28 All statistical analyses were performed in R and validated in SAS (version 9.3).
Results
Samples
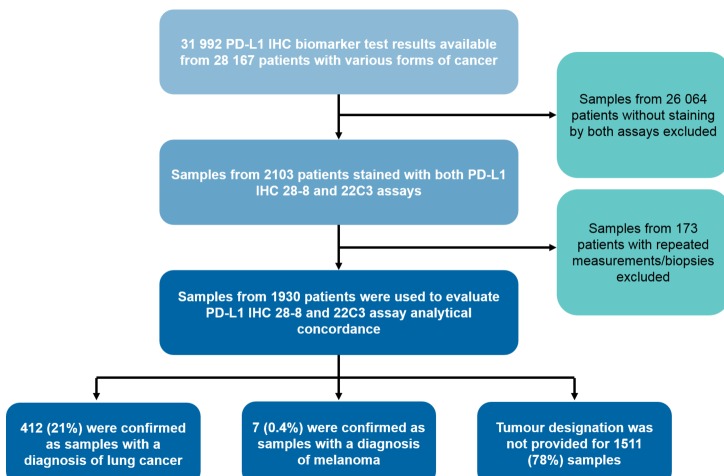
A total of 31 992 PD-L1 biomarker test results were available for 28 167 patients with various forms of cancer (figure 1). Of these patients, 2103 had staining with both 28-8 and 22C3 assays on the same biological sample. The biopsies obtained from 1930 patients with unique 28-8 and 22C3 paired results were used to evaluate assay concordance, following the removal of patients with repeated measurements/biopsies to avoid mixed effects on data analysis. Samples were collected from hospitals in more than 38 US states/territories with more than 250 unique zip codes. Of the biopsies received, 412 of 1930 (21%) were confirmed as samples with a diagnosis of NSCLC, containing a mix of histologies, including squamous and non-squamous, and 7 of 1930 (0.4%) were confirmed as samples with a diagnosis of melanoma. The remaining 1511 of 1930 (78%) samples were either obtained from tumour types not specified by the requisitioning physician or accurate documentation in the Symphony Health Solutions database was not available, based on the lack of diagnostic information provided.
Figure 1.
Consolidated Standards of Reporting Trials (CONSORT) diagram of samples assessed. IHC, immunohistochemistry; PD-L1, programmed death ligand 1.
Tumour-cell membrane staining
Paired biopsy samples stained with both 22C3 and 28-8 displayed high concordance. Across all biopsies, 1825 of 1930 (94.6%) exhibited ≤10% difference in tumour-cell membrane staining when assessed with either PD-L1 IHC 22C3 or 28-8 (table 1). In samples with a confirmed lung cancer diagnosis, the rate of similar scoring (≤10%) was 393 of 412 (95.4%). There was no clear trend of one assay displaying a greater staining frequency than the other. Among all samples, 209 of 1930 (10.8%) reported that the value of PD-L1 IHC 22C3 was greater than PD-L1 IHC 28-8 and 132 of 1930 (6.8%) reported that PD-L1 IHC 28-8 was greater than PD-L1 IHC 22C3. Values were similar in samples with a confirmed lung cancer diagnosis: 45 of 412 (10.9%) reported PD-L1 IHC 22C3>28-8 and 27 of 412 (6.6%) reported PD-L1 IHC 28-8>22C3.
Table 1.
Summary of agreement between PD-L1 IHC 22C3 and 28-8 assays
| All biopsies (n=1930) n (%) |
Biopsies with a confirmed lung cancer diagnosis (n=412) n (%) |
|
| PD-L1 IHC difference 0%–10% | 1825 (94.6) | 393 (95.4) |
| PD-L1 IHC difference 11%–20% | 60 (3.1) | 10 (0.5) |
| PD-L1 IHC difference >20% | 45 (2.3) | 9 (2.2) |
| PD-L1 IHC difference 22C3=28-8 | 1589 (82.3) | 340 (82.5) |
| PD-L1 IHC difference 22C3<28-8 | 132 (6.8) | 27 (6.6) |
| PD-L1 IHC difference 22C3>28-8 | 209 (10.8) | 45 (10.9) |
IHC, immunohistochemistry; PD-L1, programmed death ligand 1.
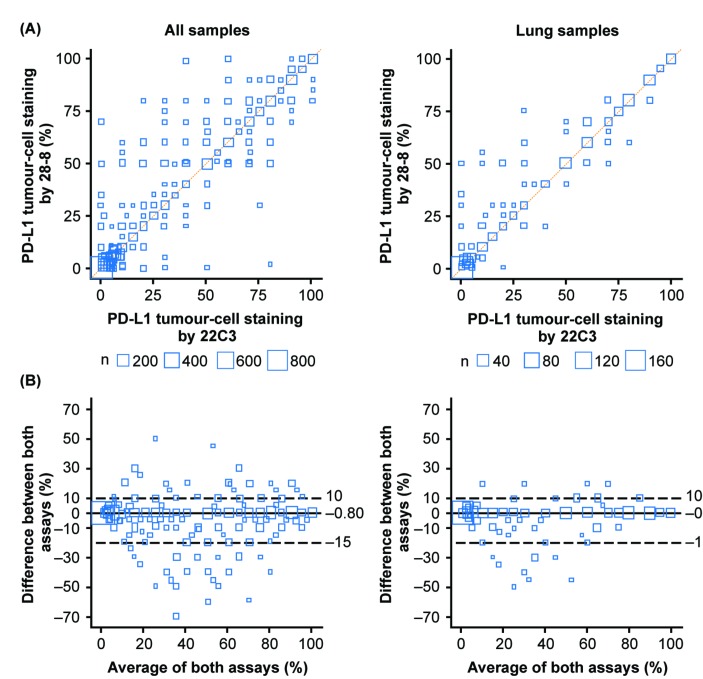
When measured against the identity line, where both PD-L1 IHC 22C3 and 28-8 assays are expected to yield the same staining results, a strong correlation was observed across all samples and samples with a confirmed lung cancer diagnosis (figure 2A). Bland-Altman analysis was performed to visualise the mean difference in the percentage of tumour cells against the average PD-L1 expression of the two assays (figure 2B). Analysis determined a mean difference of <1% in percentage of tumour-cell membrane stained for PD-L1 between the PD-L1 IHC 22C3 and 28-8 assays across all tumour samples (–0.80%; lower 2.5th and upper 97.5th percentiles of the differences were –15% and 10%, respectively) and samples with a confirmed lung cancer diagnosis (–0.93%; lower 2.5th and upper 97.5th percentiles of the differences were –15% and 10%, respectively). Of the 45 cases in which PD-L1 IHC 22C3 and 28-8 values differed by >20%, the PD-L1 IHC 22C3 assay produced the greater tumour-cell membrane staining result in 37 (82.8%) of the samples, whereas the PD-L1 IHC 28-8 assay produced the greater staining result in 8 (17.8%) of the samples. A scatter plot of the assays for these patients did not reveal a pattern because it covered the range of expression.
Figure 2.
Analysis of PD-L1 tumour-cell membrane staining concordance between PD-L1 immunohistochemistry 22C3 and 28-8 assays across all matched tumour biopsies or biopsies with a confirmed lung cancer diagnosis. (A) Bubble plot of matched tumour biopsies stained with PD-L1 immunohistochemistry 28-8 and 22C3 assays against the identity line (orange). Node size is proportional to the number of measurements. (B) Bland-Altman plot of the difference in the percentage of stained tumour-cell membrane. Dashed lines represent the lower 2.5th and upper 97.5th percentiles for the difference between both readings; solid lines represent the mean difference. Bland-Altman analysis visualises the mean difference in percentage of tumour-cell membranes against the average PD-L1 expression of the two assays. PD-L1, programmed death ligand 1.
To understand the clinical impact, the OPA, PPA, NPA and Cohen’s kappa were calculated across PD-L1 expression levels (≥1%, ≥5%, ≥10%, ≥25% and ≥50%). These expression levels represent those used in clinical trials involving both anti-PD-1 and anti-PD-L1 agents. The OPA was between 97% and 98% for all samples and between 95% and 98% for samples with a confirmed lung cancer diagnosis across all expression thresholds (table 2). Cohen’s kappa coefficient, a statistic between 0 and 1 that is used to assess agreement between categorical items, was between 0.92 and 0.95 for all samples and 0.90 and 0.95 in lung cancer.
Table 2.
Analysis of PD-L1 tumour-cell staining concordance between PD-L1 IHC 22C3 and 28-8 assays across PD-L1 expression levels
| PD-L1 expression | All biopsies (n=1930) | Biopsies with a confirmed lung cancer diagnosis (n=412) | ||||||||
| ≥1% | ≥5% | ≥10% | ≥25% | ≥50% | ≥1% | ≥5% | ≥10% | ≥25% | ≥50% | |
| PPA (95% CI) |
97% (96 to 98) |
97% (95 to 98) |
98% (96 to 98) |
98% (97 to 99) |
99% (97 to 99) |
97% (95 to 99) |
97% (93 to 98) |
98% (95 to 99) |
98% (94 to 99) |
99% (96 to 100) |
| NPA (95% CI) |
97% (96 to 98) |
97% (96 to 98) |
98% (96 to 98) |
97% (96 to 98) |
96% (95 to 97) |
96% (92 to 98) |
94% (89 to 96) |
96% (93 to 98) |
96% (93 to 98) |
97% (94 to 99) |
| OPA (95% CI) |
97% (96 to 98) |
97% (96 to 97) |
98% (97 to 98) |
98% (97 to 98) |
97% (96 to 97) |
97% (95 to 98) |
95% (93 to 97) |
97% (95 to 98) |
97% (94 to 98) |
98% (96 to 99) |
| Cohen’s kappa (95% CI) |
0.94 (0.93 to 0.96) |
0.93 (0.92 to 0.95) |
0.95 (0.94 to 0.96) |
0.95 (0.93 to 0.96) |
0.92 (0.90 to 0.94) |
0.94 (0.90 to 0.97) |
0.90 (0.86 to 0.94) |
0.94 (0.91 to 0.97) |
0.93 (0.89 to 0.97) |
0.95 (0.92 to 0.98) |
PPA and NPA calculations use 28-8 as the reference standard.
IHC, immunohistochemistry; NPA, negative per cent agreement; OPA, overall per cent agreement; PD-L1, programmed death ligand 1; PPA, positive per cent agreement.
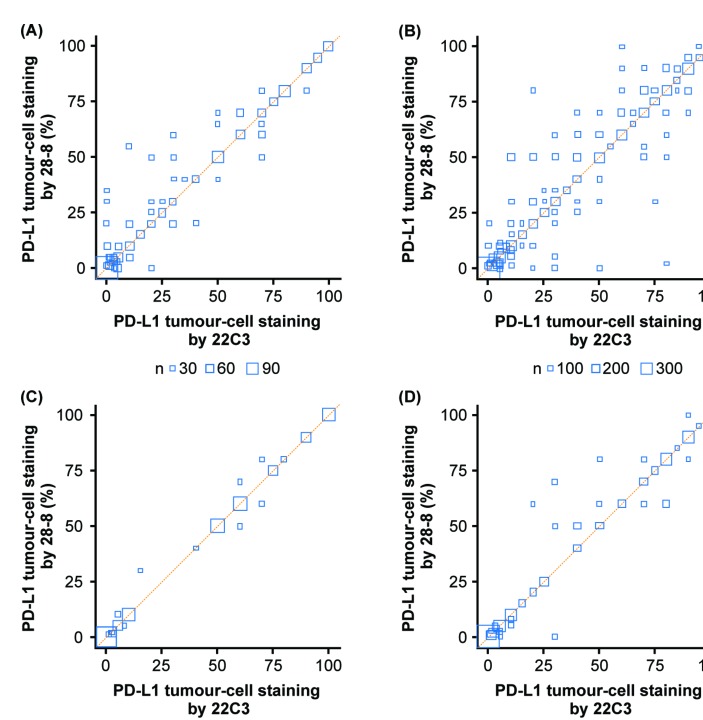
In lieu of missing cancer diagnosis information, assay concordance was compared by most common sites of biopsy. For patients with a diagnosis of lung cancer and those with an unspecified cancer diagnosis, the two most common biopsy sites were lung and lymph nodes (table 3), highlighting the consistency of biopsy sites regardless of whether a diagnosis of lung cancer was made. Irrespective of cancer type and biopsy site, there was strong concordance between the two assays for patients with lung or lymph node biopsy sites (figure 3).
Table 3.
Samples with lung cancer or unspecified cancer diagnosis categorised by site of biopsy
| Biopsy site | Lung cancer diagnosis (n=412) |
Cancer diagnosis unspecified (n=1511) |
||
| Samples | % | Samples | % | |
| Lung | 275 | 66.7 | 804 | 53.2 |
| Lymph node | 44 | 10.7 | 162 | 10.7 |
| Liver | 14 | 3.4 | 75 | 5.0 |
| Pleural fluid | 13 | 3.2 | 55 | 3.6 |
| Bone | 12 | 2.9 | 87 | 5.8 |
| Other | 54 | 13.1 | 328 | 21.7 |
Figure 3.
Pairwise comparison for lung cancer or unspecified cancer diagnosis for most frequent biopsy sites (lung or lymph node). Bubble plot of matched tumour biopsies stained with PD-L1 immunohistochemistry 28-8 and 22C3 assays against the line of agreement (orange). (A) Lung cancer diagnosis, lung biopsy site, n=275. (B) Unspecified cancer diagnosis, lung biopsy site, n=804. (C) Lung cancer diagnosis, lymph node biopsy site, n=44. (D) Unspecified cancer diagnosis, lymph node biopsy site, n=162. Node size is proportional to the number of observations with the same 22C3 and 28-8 expression values. PD-L1, programmed death ligand 1.
Discussion
The present study evaluated real-world concordance between PD-L1 IHC 22C3 and 28-8 pharmDx, as reported by pathologists at NeoGenomics Laboratories, of tumour samples submitted from hospitals in more than 38 US states/territories. Consistent with other reports of assay concordance,21 23 29 30 the rate of concordance between these two assays was high at all levels.
In the present analysis, we did not detect a meaningful difference in the percentage of PD-L1-positive tumour cells between the assays. Despite a negligible decrease in the mean staining frequency of tumour cells (–0.80% across all samples and –0.93% in lung) with the PD-L1 IHC 28-8 test, both tests seem to be identical across all samples and specifically in lung cancer. While there was no clear direction in terms of platforms detecting a greater fraction of PD-L1-positive tumour cells, it was noted that outliers with >20% difference between platforms were associated with increased staining reported by 22C3.
The limitations of this real-world study include unspecified cancer diagnosis by the test requester and treating physician for 1511 of 1930 samples, despite both most likely knowing the diagnosis. Real-world limitations for testing between community prescribers and diagnostic service providers have been documented. Romano et al showed that clinical impression/diagnosis was absent from nearly a quarter of biopsy requisition forms (n=249).31 The lack of information from the requesting physician afforded to the biomarker testing facility in the current study reflects a similar trend. Missing clinical information from the requisition form may impact reference laboratories from performing adequate PD-L1 testing as approvals in newer tumour types now include new scoring algorithms, cell types and cut-offs. Nonetheless, analyses showed that the FDA-cleared PD-L1 IHC 22C3 pharmDx assay and the PD-L1 IHC 28-8 pharmDx assay remained concordant regardless of cancer type and biopsy site. Biopsy sites in this analysis included the most common sites of lung cancer metastases (eg, pleural/pericardial fluid, bone, lungs, brain, adrenal glands, liver, extrathoracic lymph nodes, pleura).32 Furthermore, the first PD-L1 companion and complementary diagnostics were approved in NSCLC and melanoma, and as a result, it would be likely to have more lung and melanoma samples than reported. Test requesters should be encouraged to complete IHC requisition forms with additional patient and disease information. This will allow the pathologist testing the sample to provide a more accurate assessment that will determine treatment strategy.31 Consistent with real-world data and reference lab testing/processing practices, limitations in the available data set preclude us from confirming whether concordance analyses were performed on serial samples from singular blocks. The study may also have been improved by obtaining details regarding the evaluating pathologist and the date of testing at NeoGenomics Laboratories; however, interpathologist variability was addressed through required training, certification in scoring and blinded testing. Although these data do not provide information on clinical validation for the two assays, combined with other efforts these real-world analytical data support the potential analytical interchangeability of the PD-L1 IHC 28-8 pharmDx and 22C3 pharmDx for assessing tumour-cell membrane PD-L1 expression.
Conclusion
Given the real-world nature of the study, patient samples were sent to the laboratory for a generic PD-L1 test with no information as to which anti-PD-1 antibody the clinician intended to treat the patient—a common challenge that faces clinical practice today. Samples were then tested using both the PD-L1 IHC 28-8 pharmDx and 22C3 pharmDx and, independent of cancer diagnosis and validated cut-offs for a particular drug or assay, the findings demonstrated that a patient’s PD-L1 expression report would provide the same result for at least 97% of patients.
Take home messages.
Analytical concordance between the programmed death ligand 1 (PD-L1) immunohistochemistry 28-8 and 22C3 pharmDx assays has been evaluated through multiple concordance analyses.
We evaluated the real-world concordance between the two assays in a single cancer reference laboratory using a wide array of samples from 1930 patients submitted from hospitals in over 38 US states/territories where both PD-L1 tests were ordered or a test was not specified.
Our data suggest that, independent of cancer diagnosis and validated cut-offs for a particular drug or assay, a patient’s PD-L1 expression report would provide the same result for at least 97% of patients.
Acknowledgments
Medical writing support was provided by Jason Hoffman, PharmD, RPh, and editorial support was provided by Jay Rathi, MA, both of Spark Medica Inc. (USA), supported by Bristol-Myers Squibb.
Footnotes
Handling editor: Runjan Chetty.
Correction notice: This article has been made Open Access since it was published Online First.
Contributors: CB, MA, SS, VC and VB designed the study. MA, SS and VC performed all experiments. KZ and CJ performed the biostatistical analysis. All authors reviewed and approved the manuscript.
Funding: This study was supported by Bristol-Myers Squibb.
Competing interests: CB was employed by Bristol-Myers Squibb during the conduct of the study, and is currently an employee of Verily Life Sciences. KZ, CJ and VB are employed by Bristol-Myers Squibb. MA, VC and SS are employed by NeoGenomics Laboratories.
Patient consent: Not required.
Ethics approval: No ethics approval was required for this study because this study consisted of real-world patient samples assessed for PD-L1 expression by NeoGenomics Laboratories as a commercial provider of PD-L1 expression testing.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Bristol-Myers Squibb’s policy on data sharing can be found at https://www.bms.com/researchers-and-partners/independent-research/data-sharing-request-process.html.
Author note: At the time of the study, Dr C Batenchuk was an employee of Bristol-Myers Squibb. Dr C Batenchuk is currently an employee of Verily Life Sciences.
References
- 1.Zitvogel L, Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 2012;1:1223–5. 10.4161/onci.21335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garon EB, Rizvi NA, KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 4.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015;372:320–30. 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- 6.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med 2015;372:311–9. 10.1056/NEJMoa1411087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. 10.1056/NEJMoa1602252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Escudier B, CheckMate 025 Investigators. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–13. 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma P, Callahan MK, Bono P, et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol 2016;17:1590–8. 10.1016/S1470-2045(16)30496-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017;389:2492–502. 10.1016/S0140-6736(17)31046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017;18:1182–91. 10.1016/S1470-2045(17)30422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bristol-Myers Squibb. 2018. OPDIVO [prescribing information]http://packageinserts.bms.com/pi/pi_opdivo.pdf (accessed 28 Jun 2018).
- 13.Merck. 2018. KEYTRUDA [prescribing information]https://www.merck.com/product/usa/pi_circulars/k/keytruda/keytruda_pi.pdf (accessed 28 Jun 2018).
- 14.Genentech. 2018. TECENTRIQ [prescribing information]https://www.gene.com/download/pdf/tecentriq_prescribing.pdf (accessed 28 Jun 2018).
- 15.EMD Serono. 2017. BAVENCIO [prescribing information]https://www.bavencio.com/en_US/document/Prescribing-Information.pdf (accessed 28 Jun 2018).
- 16.AstraZeneca. 2018. IMFINZI [prescribing information]https://www.azpicentral.com/imfinzi/imfinzi.pdf#page=1 (accessed 28 Jun 2018).
- 17.Roach C, Zhang N, Corigliano E, et al. Development of a companion diagnostic PD-L1 immunohistochemistry assay for pembrolizumab therapy in non-small-cell lung cancer. Appl Immunohistochem Mol Morphol 2016;24:392–7. 10.1097/PAI.0000000000000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dako. 2017. PD-L1 IHC 22C3 pharmDxhttps://www.agilent.com/cs/library/packageinsert/public/P03951%20SK006%20NSCLC%20GC%20Rev.%2006.pdf (accessed 28 Jun 2018).
- 19.Dako. 2017. PD-L1 IHC 28-8 pharmDxhttps://www.agilent.com/cs/library/packageinsert/public/P04163_rev_05_2017Sep15.pdf (accessed 28 Jun 2018).
- 20.Jørgensen JT. Companion diagnostic assays for PD-1/PD-L1 checkpoint inhibitors in NSCLC. Expert Rev Mol Diagn 2016;16:131–3. 10.1586/14737159.2016.1117389 [DOI] [PubMed] [Google Scholar]
- 21.Hirsch FR, McElhinny A, Stanforth D. PD-L1 immunohistochemistry assays for lung Cancer: results from phase 1 of the Blueprint PD-L1 IHC assay comparison Project. J Thorac Oncol 2017;12:208–22. 10.1016/j.jtho.2016.11.2228 [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Kwon HJ, Park SY, et al. PD-L1 immunohistochemical assays for assessment of therapeutic strategies involving immune checkpoint inhibitors in non-small cell lung cancer: a comparative study. Oncotarget 2017;8:98524–32. 10.18632/oncotarget.21567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsao MS, Kerr KM, Kockx M, et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of Blueprint phase 2 project. J Thorac Oncol 2018. 10.1016/j.jtho.2018.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rimm DL, Han G, Taube JM, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol 2017;3:1051–8. 10.1001/jamaoncol.2017.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratcliffe MJ, Sharpe A, Midha A, et al. Agreement between programmed cell death ligand-1 diagnostic assays across multiple protein expression cutoffs in non-small cell lung cancer. Clin Cancer Res 2017;23:3585–91. 10.1158/1078-0432.CCR-16-2375 [DOI] [PubMed] [Google Scholar]
- 26.Gaule P, Smithy JW, Toki M, et al. A quantitative comparison of antibodies to programmed cell death 1 ligand 1. JAMA Oncol 2017;3:256–9. 10.1001/jamaoncol.2016.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunnström H, Johansson A, Westbom-Fremer S, et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability. Mod Pathol 2017;30:1411–21. 10.1038/modpathol.2017.59 [DOI] [PubMed] [Google Scholar]
- 28.FDA Statistical guidance on reporting results from studies evaluating diagnostic statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests. 2007. Available from: https://www.fda.gov/RegulatoryInformation/Guidances/ucm071148.htm [Accessed 28 Jun 2018].
- 29.Scheel AH, Baenfer G, Baretton G, et al. Interlaboratory concordance of PD-L1 immunohistochemistry for non-small-cell lung cancer. Histopathology 2018;72:449–59. 10.1111/his.13375 [DOI] [PubMed] [Google Scholar]
- 30.Hendry S, Byrne DJ, Wright GM, et al. Comparison of four PD-L1 immunohistochemical assays in lung cancer. J Thorac Oncol 2018;13:367–76. 10.1016/j.jtho.2017.11.112 [DOI] [PubMed] [Google Scholar]
- 31.Romano RC, Novotny PJ, Sloan JA, et al. Measures of completeness and accuracy of clinical information in skin biopsy requisition forms: an analysis of 249 cases. Am J Clin Pathol 2016;146:727–35. 10.1093/ajcp/aqw186 [DOI] [PubMed] [Google Scholar]
- 32.Tamura T, Kurishima K, Nakazawa K, et al. Specific organ metastases and survival in metastatic non-small-cell lung cancer. Mol Clin Oncol 2015;3:217–21. 10.3892/mco.2014.410 [DOI] [PMC free article] [PubMed] [Google Scholar]