Abstract
Alzheimer’s disease (AD) is one of the most serious progressive neurodegenerative diseases among the elderly, therefore the identification of conversion to AD at the earlier stage has become a crucial issue. In this study, we applied multimodal support vector machine to identify the conversion from normal elderly cognition to mild cognitive impairment (MCI) or AD based on magnetic resonance imaging and positron emission tomography data. The participants included two independent cohorts (Training set: 121 AD patients and 120 normal controls (NC); Testing set: 20 NC converters and 20 NC non-converters) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The multimodal results showed that the accuracy, sensitivity, and specificity of the classification between NC converters and NC non-converters were 67.5%, 73.33%, and 64%, respectively. Furthermore, the classification results with feature selection increased to 70% accuracy, 75% sensitivity, and 66.67% specificity. The classification results using multimodal data are markedly superior to that using a single modality when we identified the conversion from NC to MCI or AD. The model built in this study of identifying the risk of normal elderly converting to MCI or AD will be helpful in clinical diagnosis and pathological research.
Keywords: Alzheimer’s disease, classification, magnetic resonance imaging, normal elderly, positron emission tomography, support vector machine
INTRODUCTION
Alzheimer’s disease (AD) is the most serious neurodegenerative disorder in the world. Until 2010, 36.5 million people worldwide lived with AD and other dementias [1]. With the aging of world’s population, more people will suffer from AD. There are different stages in the progress of AD, accompanied by both structural and functional changes in the brain. It is possible to characterize various neuroimaging alterations in AD with the advancement of computational image analysis methods and neuroimaging technologies such as magnetic resonance imaging (MRI) [2, 3] and positron emission tomography (PET) [4, 5]. Researchers have detected or predicted AD with different classification methods based on single-modal or multimodal neuroimaging data [6–9]. As there are no effective treatments for AD, identifying the conversion to AD at the earlier stage has become a crucial issue.
Mild cognitive impairment (MCI), as a transitional stage between normal elderly and AD, has attracted increased attention in this research field. MCI patients exhibit demonstrable memory deficits and neurological changes in the brain, such as gray matter atrophy, hypometabolism, and amyloid-β (Aβ) deposition, which can be examined and detected by MRI [9], 18F-fluorodeoxyglucose PET (FDG-PET) [10] and 18F-florbetapir PET (AV45-PET), respectively. Previous studies classified MCI converters (MCI-c) from MCI non-converters (MCI-nc) using neuroimaging data based on different classification methods [11–14]. For example, Cui et al. used data from MRI, cerebrospinal fluid, and neuropsychological measures to classify MCI-c from MCI-nc based on support vector machine (SVM) [11]. Others have applied the method of Spatial Pattern of Abnormalities for Recognition of Early AD (SPARE-AD) using MRI and cerebrospinal fluid data to predict whether MCI will convert to AD [13, 14]. Contrary to the large number of prediction or classification studies in MCI, similar studies are rare in the normal aging population.
Compared with healthy young people, healthy elderly show various alterations in brain function and structure during normal aging [15, 16]. Previous studies using PET technology showed that Aβ deposition is the major neuropathological feature of AD [17, 18]. In recent years, many studies suggest that approximately 20–30% of cognitively normal elderly also had Aβ deposition in the brain, mainly in the precuneus, temporal lobe, and cingulate cortex accompanied by glucose metabolism reduction and gray matter atrophy [19–22]. The existing documents suggest that a percentage of normal elderly who have Aβ deposition in the brain may convert to AD. The studies of normal aging may improve our understanding of the early pathological alterations of AD. In addition, an accurate classification and identification of subjects in this stage is very important to clinical applications.
Different imaging modalities provide complementary information, such as MRI for gray matter volume, FDG-PET for hypometabolism, and AV45-PET for Aβ deposition [4]. In recent years, SVM has been introduced to study multimodal neuroimaging data [23]. The multimodal SVM, as an improved SVM method, determines the weights of the kernels of each modality; the classification takes advantage of the complementary information within multiple types of the imaging data [24]. Using data from MRI, FDG-PET, and cerebrospinal fluid, Zhang et al. applied multimodal SVM to the classification of AD and MCI [24].
The purpose of our study is to apply the multimodal SVM method to identify the conversion from normal elderly cognition to MCI or AD within 24 months using imaging data from three modalities: MRI, AV45-PET, and FDG-PET. The present study included two datasets: one was from AD subjects and the normal controls (NC) used as the training set to build the classifier; the other dataset was from the independent normal elderly, which included NC-c (NC converted to MCI or AD within 24 months) and NC-nc (NC did not convert) used as the testing set.
MATERIALS AND METHODS
ADNI
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2, and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see http://www.adni-info.org.
Ethics
The ADNI study was approved by Institutional Review Board (IRB) of each participating site, including the Banner Alzheimer’s Institute, and was conducted in accordance with Federal Regulations, the Internal Conference on Harmonization (ICH), and Good Clinical Practices (GCP). Study subjects gave written informed consent at the time of enrollment and completed the questionnaires, according to the IRB rules and local laws.
Participants
According to the ADNI protocols, the diagnosis of probable AD met criteria set by the National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) [25].The severity of cognitive impairment was assessed using the Mini-Mental State Examination (MMSE) [26] and Clinical Dementia Rating (CDR) scores [27]. We selected subjects who had data consisting of three modalities: MRI, FDG-PET and AV45-PET. NC had a CDR of 0 and MMSE scores between 26 and 30. AD patients had a CDR between 0.5 and 2.0 and MMSE scores less than 26. There were a total of 281 participants in the present study. The training set included 121 AD patients and 120 NC. The testing set included 40 normal subjects: 20 NC-nc and 20 NC-c (nineteen subjects converted to MCI and one subject converted to AD). The follow-up period for converters and non-converters is 24 months. The mean conversion time of the converters is 15.36 ± 6.96 months. Normal elderly in the testing set are independent of those in the training set. Table 1 shows the detailed demographic information for all subjects. The AD group did not significantly differ from the NC group with respect to gender ratio or age (t(239) = 0.40, p = 0.69); in addition, the NC-c group did not significantly differ from the NC-nc group with respect to gender ratio or age (t(38) = 0.21, p = 0.83).
Table 1.
Demographic information of the participants
| Training data | Testing data | |||
|---|---|---|---|---|
| AD(n=121) | NC(n=120) | NC-c(n = 20) | NC-nc(n = 20) | |
| Age (years) | 74.9±8.1 | 75.26 ±6.5 | 80.3±4.2 | 79.9±5.9 |
| Gender (Male/Female) | 70/51 | 58/62 | 12/8 | 13/7 |
| MMSE score | 21.30±4.29 | 29.29 ±0.83 | 28.36 ±1.55 | 29.6± 0.68 |
| CDR score (range) | 0.5–2.0 | 0 | 0 | 0 |
| Education (years) | 15.74 ±2.75 | 16.43±2.73 | 17.15±2.76 | 15.32 ±2.67 |
| ADAS-cog | 21.52±7.96 | 5.76±3.02 | 8.65±3.18 | 5.40 ±2.87 |
AD, Alzheimer’s disease; NC, normal control; NC-c, NC converter; NC-nc, NC non-converter; MMSE, Mini-Mental State Examination; CDR, Clinical Dementia Rating; ADAS-Cog, Alzheimer’s Disease Assessment Scale - Cognitive Subscale.
Neuroimaging data acquisition
MRI data
For each participant, a T1-weighted image was obtained from 1.5 T or 3 T scanners. MRI images were obtained from different sites and platforms with different acquisition parameters. Corrections of image intensity were performed for gradient nonlinearity and intensity non-uniformity on all MRI images. Details regarding the protocols of the correction can be found at http://adni.loni.usc.edu/.
PET data
Both FDG-PET and AV45-PET data of the same subject were obtained on the same scanner within one month. The interval between MRI and PET scan was within 3 months. Before the imaging session, the subjects were asked to abstain from eating food and drinking fluids (except water) for at least 4 h. For FDGPET scanning, subjects were injected with 185 MBq of 18F-FDG as a bolus. The subjects were then allowed to rest comfortably in the room for approximately 20 min. Following this, a dynamic 3D scan was acquired, which consisted of six 5-min frames. For AV45-PET scanning, 370 MBq of AV-45 was intravenously injected as a bolus. After a 20-min incorporation period, a PET scan was obtained, consisting of four 5-min frames.
For both types of PET images, all temporal frames were co-registered to decrease the effects of head motion. All image sets were then reoriented to a common spatial orientation and interpolated to a voxel size of 2 mm. The details of the PET protocols can be found at http://adni.loni.usc.edu/.
Image preprocessing
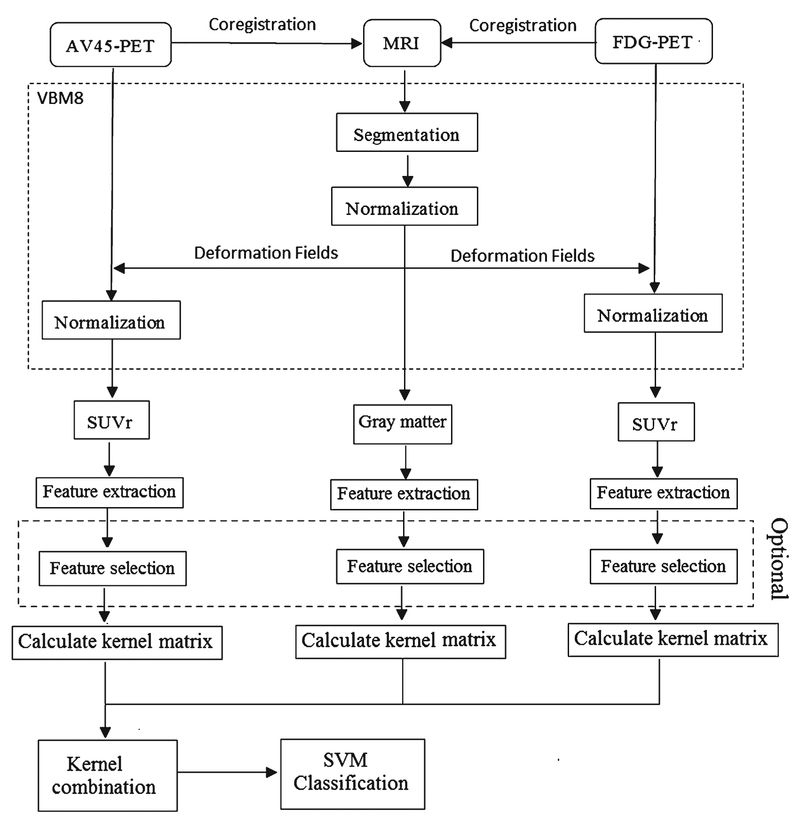
The spatial preprocessing of all MRI and PET images was implemented in SPM8 software (http://www.fil.ion.ucl.ac.uk/spm/) in MATLAB (Mathworks Inc., Sherborn, MA). Figure 1 shows the flowchart of multimodal data processing and classification.
Fig. 1.
The flowchart of multimodal data processing and classification.
The segmentation and normalization of the MRI images were implemented using a VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm8) of SPM8, based on the voxel-based morphometry (VBM) method. First, images were segmented into three parts: gray matter, white matter, and cerebrospinal fluid, using adaptive maximum posterior and partial volume estimation [28, 29]. To further improve segmentation, spatially adaptive non-local means, consisting of a denoising filter [30] and a classical Markov Random Field approach, were used. The gray matter images were normalized using a protocol named as Diffeomorphic Anatomical Registration using Exponential Lie Algebra (DARTEL) [31]. During normalization, a template was created and images were registered iteratively. Finally, gray matter tissue maps were transformed to the Montreal Neurological Institute (MNI) space, which were modulated to preserve the total volume in original space.
For FDG-PET images, each FDG-PET image was first coregistered to its corresponding MRI image for each subject. Each FDG-PET image was then normalized to MNI space using the deformation fields from normalization of the corresponding MRI image. Finally, we computed standardized uptake value ratios (SUVr) for each image by normalizing to the mean uptake of the reference region. AV45-PET and FDGPET images were separately spatially preprocessed using the same protocol; the cerebellum was the reference region for both AV45-PET data and FDG-PET data (Fig. 1).
Feature extraction
Ninety regions of interest (ROIs) on the whole brain, except the cerebellum, were included in our study, which were defined on the Automatic Anatomical Labeling (AAL) atlas [32]. We used the gray matter image to create a mask for each subject, including only voxels that exceeds 0.15 on the gray matter image. These masks were applied to each image, including gray matter, AV45-PET, and FDG-PET images. We then extracted features based on AAL ROIs. For each ROI, mean gray matter volume of the MRI images and mean intensity of the PET images were separately computed as a feature. Therefore, we obtained a total of 270 features for each subject.
Classification
SVM is a supervised classification algorithm for pattern classification; it is based on the statistical learning theory. It constructs a linear boundary to separate samples in high dimensional space through mapping non-linearly separated samples in low dimensional space to high or infinite dimensional space [33]. This method is regarded as one of the most powerful classification algorithms.
The multimodal SVM fuses modalities by weighted coefficients, which uses all of the information of each type of data. After adding weighted coefficients, the kernel function in this method changes into
| (1) |
where is the kernel function for training samples of m-th modality. βm is the weight of the m-th modality.
If x = {x(1), x(2), … , x(M)} is the test sample, we can obtain the decision function
| (2) |
We used a grid search to obtain weights of each modality based on the accuracy of classification between AD and NC, and the size of the grid was 0.1. To reduce the overfitting and ensure the generalizability of our results, we performed 10-fold cross-validation using all training data to gain the weight combination of the three modalities for classifying AD from NC. Firstly, all training data was randomly divided into 10 groups. Then, we used 9 groups to train SVM model and 1 group to test. The accuracy of each grid was entirely obtained by averaging cross-validation 10 results. After obtaining the best combination of weights, a mixed kernel was derived to train a classifier with the training set, which was tested on NC-c and NC-nc group to identify the conversion from normal elderly to AD/MCI. We notice that the training set and the testing set were totally independent in this process. All classification process was performed on the LIBSVM toolbox (http://www.csie.ntu.edu.tw/~cjlin/libsvm/).
Feature selection
A two sample t-test was implemented for each ROI to examine the difference between AD and NC groups for each modality, respectively. And each modality’s ROIs were ranked based on the tests’ significances. We added one ROI a time based on these rankings to distinguish AD and NC subjects in order to obtain the best combination of weights using the same number of ROIs for the three modalities. The optimal number of features was obtained based on the results of classification in the training data.
RESULTS
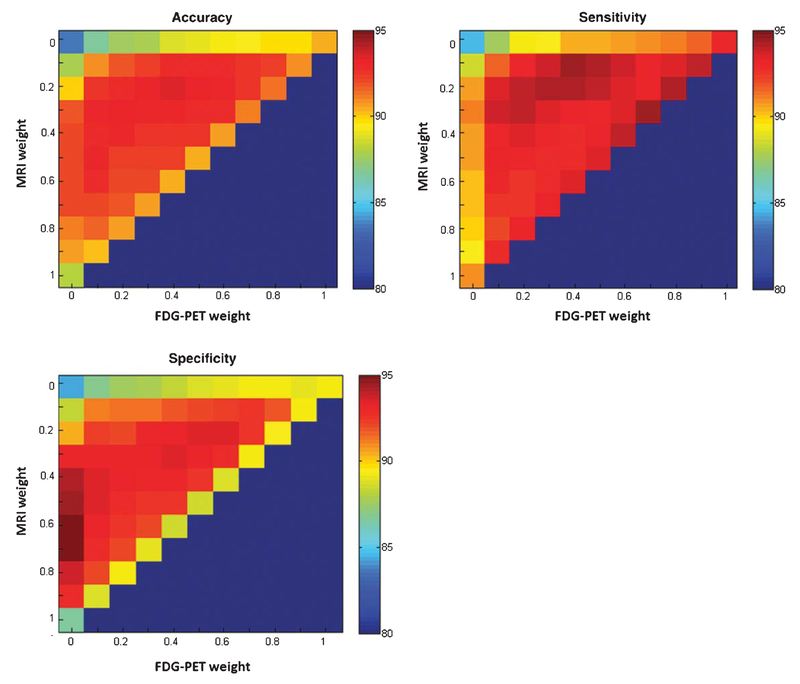
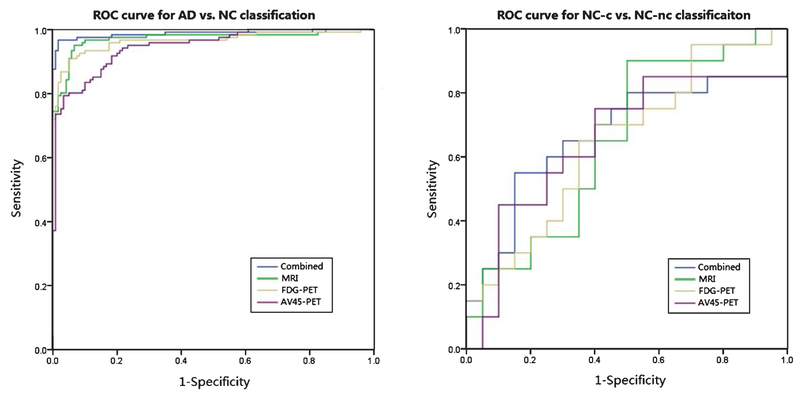
The classification accuracy with different combining weights of MRI, AV45-PET, and FDG-PET data are shown in Fig. 2. According to the results of the grid search, based on the constraint of βMRI + βAV45 + βFDG = 1, the best combination of weights are βMRI = 0.2, βAV45 = 0.4, and βFDG = 0.4. The best accuracy of classification between AD and NC is shown in Table 2. The accuracy, sensitivity, and specificity of the classification are 93.40%, 94.10%, and 93.29%, respectively. Table 2 also shows the confidence intervals of accuracy, sensitivity, and specificity. In addition to Fig. 3, we plotted the ROC curves of the four methods regarding the classification between AD and NC. The area under the ROC curve (AUC) is 0.987 for the multimodal method. The AUCs using only a single modality: MRI, FDGPET, and AV45-PET, are 0.968, 0.963, and 0.951, respectively.
Fig. 2.
The classification accuracy of AD and NC with different combining weights of MRI, FDG-PET, and AV45-PET.
Table 2.
Classification results for AD versus NC without feature selection. The numbers in each bracket denote confidence interval of each index
| Accuracy (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|
| MRI | 88.01 (87.42–88.61)a | 90.98 (90.30–91.66)a | 86.73 (86.04–87.43)a |
| AV45-PET | 83.72 (83.40–84.03)a | 84.62 (84.00–85.24)a | 84.34 (83.87–84.81)a |
| FDG-PET | 90.45 (89.99–90.91)a | 93.04 (92.42–93.66)a | 89.16 (88.68–89.64)a |
| Combined | 93.4 (92.83–93.97) | 94.1 (93.50–94.69) | 93.29(92.47–94.10) |
These results have significant differences between single modal and multimodal (p < 0.05).
Fig. 3.
ROC curves for the classification of AD/NC and NC-c/NC-nc based on the multimodal method and three single-modal methods.
Table 3 shows the classification results of NC-c and NC-nc with both multimodal and single-modal methods using the mixed kernel with the best combination of weights. The accuracy, sensitivity, and specificity, using multimodal SVM method, are 67.5%, 73.33%, and 64%, respectively. The ROC curves also are showed in Fig. 3. The best accuracy, based on the single-modal method, is 62.5%.
Table 3.
Classification results for NC-c versus NC-nc, without and with feature selection
| Without feature selection | With feature selection | |||||
|---|---|---|---|---|---|---|
| Accuracy (%) | Sensitivity (%) | Specificity (%) | Accuracy (%) | Sensitivity (%) | Specificity (%) | |
| MRI | 55 | 55.56 | 54.55 | 55 | 55.56 | 54.55 |
| AV45-PET | 62.5 | 69.23 | 59.26 | 62.5 | 69.23 | 59.26 |
| FDG-PET | 62.5 | 63.16 | 61.91 | 62.5 | 63.16 | 61.91 |
| Combined | 67.5 | 73.33 | 64 | 70 | 75 | 66.67 |
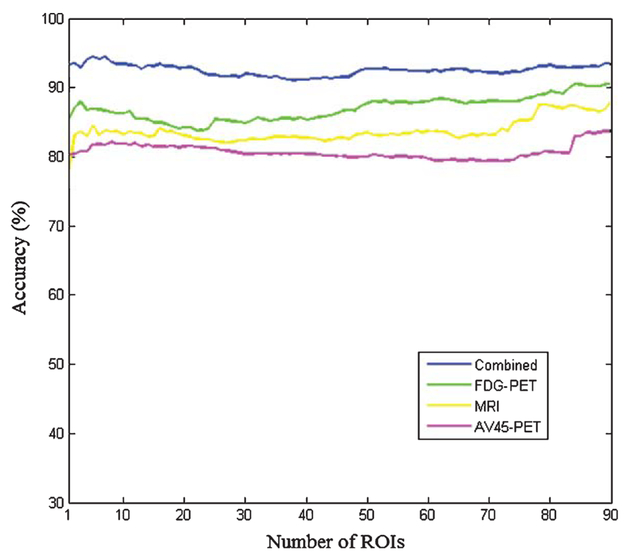
Figure 4 shows the accuracy of four methods with different number of features between AD and NC. The number of ROIs increased with the ranked p values, and the multimodal method achieves better results compared with other three single-modal methods each alone. With the selected 7 features of each modality, the classification accuracy between NC-c and NC-nc is 70%, and the sensitivity and specificity are 75% and 66.67%, respectively (Table 3). Table 4 shows the classification results between AD and NC with feature selection.
Fig. 4.
The classification accuracy of AD and NC based on multi-modal and three single-modal methods, with different numbers of ROIs.
Table 4.
Classification results for AD versus NC with feature selection. The numbers in each bracket denote confidence interval of each index
| Accuracy (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|
| MRI | 83.83 (83.59–84.06) | 84.91 (84.43–85.40) | 84.40 (83.97–84.82) |
| AV45-PET | 81.73(81.71–81.75) | 82.95 (82.51–83.40) | 81.76(81.43–82.10) |
| FDG-PET | 86.72 (86.53–86.92) | 87.80 (87.57–88.02) | 86.72 (86.37–87.08) |
| Combined | 94.44 (94.23–94.64) | 95.69 (95.43–95.95) | 93.85 (93.61–94.09) |
DISCUSSION
The present study identified the conversion from normal elderly cognition to MCI or AD using pattern classification methods. We constructed a multimodal SVM classifier on AD and NC groups. Then we classified NC-c and NC-nc based on data from the three modalities: MRI, FDG-PET, and AV45-PET. The accuracy of the multimodal classification with selected features is 94.44% between AD and NC; the identification accuracy of the conversion from NC to MCI or AD with and without feature selection are 67.5% and 70%, respectively.
MRI and PET techniques have been showed to be sensitive to AD. Thus, a number of studies have used MRI and PET images to detect AD [34–36]. Gray et al. used baseline and 12-month FDG-PET and MRI data to obtain a classification accuracy of 88.4% between AD and NC based on the SVM method [37]. Hinrichs et al. also used FDG-PET and MRI data for AD classification through the multiple kernel learning method and obtained an accuracy of 87% [38]. In our study, we used data from MRI, FDG-PET, and AV45-PET and obtained 94.44% accuracy, which illustrates that our method is useful in classifying AD.
MCI is a transitional stage between AD and NC. Some studies have predicted whether MCI will convert to AD by classifying MCI-c from MCI-nc [9, 14, 39]. For example, based on the regression analysis method, Ewers et al. obtained an accuracy of 72.2% using data from cerebrospinal fluid, MRI, and score of the Rey Auditory Verbal Learning test (RAVLT) [40]. Misra et al. used a high-dimension pattern classification method to obtain an 81.5% accuracy with data from MRI images [39]. These studies indicate that AD can be predicted in the early stages. Meanwhile, the structural and functional changes of the brain related to AD have appeared when the elderly are still cognitively normal. Therefore, our study predicted the conversion by distinguishing between NC-c and NC-nc. We obtained an accuracy, sensitivity, and specificity of 70%, 75%, and 66.67%, respectively. Because brain changes in the healthy elderly are relatively more stable than those in AD or MCI patients, classification between NC-c and NC-nc is more difficult. Thus, it is reasonable that our accuracy is little lower than that of the classification of MCI.
MRI studies have demonstrated the dramatic functional and structural alterations in the brain during normal aging. A few studies related to AD paid attention to the old individuals with normal cognition. De Leon et al. predicted the conversion from normal cognition to MCI in a 3-year longitudinal study of 48 healthy normal elderly, through detecting glucose metabolic alterations of the entorhinal cortex, hippocampus, and temporal neocortex using FDG-PET images. This study suggested that an entorhinal cortex stage of brain involvement can be detected in normal elderly subjects to predict future cognitive and brain metabolism reductions [41]. Pattern classification method has been used in the identification of the conversion. Cui et al. predicted the transition from normal cognition to MCI by classifying NC-c and NC-nc based on SVM method, using a combination of neuropsychological measure scores and multiple morphological measures with the results of 78.51% accuracy, 73.33% sensitivity, and 79.75% specificity [42], which are better than our results. In contrast to Cui et al.’s study, we used data of different modalities and measurement indices based on the different methods. Our results should not be directly compared with theirs due to this aspect. The effect of neuropsychological scores and morphological measures such as cortical thickness and surface area need to be further evaluated in the classification study.
It has been reported that information from different modalities is complementary for classification of AD [43–45]. Dukart et al. obtained an 100% accuracy using FDG-PET and MRI data based on the SVM method; by using only FDG-PET or MRI data, they obtained accuracies of 94.1% and 82.4%, respectively [34]. We can see from Tables 2–4 that when using multimodal data, the accuracy is superior to that when using a single modality, regardless of the classification between AD and NC or between NC-c and NC-nc. In the classification of AD without feature selection, the accuracy of 93.4% based on three modalities is better than the best accuracy of 90.45% among all of the single methods. This is demonstrated by the ROC curves in Fig. 3 and the AUC values. Similarly, the multimodal method obtained the best accuracy of the four methods when we identified the conversion from NC to MCI or AD. Our results are consistent with those in previous studies.
In this study, we conducted feature selection to increase accuracy of classification, since not all brain regions are related to AD. Twenty one features were selected from three modalities in our method. When classifying AD from NC, the accuracy significantly increased from 93.40% to 94.44% using selected features (p < 0.05). In addition, we obtained better results (70% accuracy, 75% sensitivity, and 66.67% specificity) than using all features together for classifying NC-c and NC-nc. The improved classification results illustrated that feature selection can remove redundant information to increase classification accuracy.
SVM, as one of the most popular multivariate methods, has been widely and successfully utilized in the detection of AD with multimodal neuroimaging data [6]. However, the standard SVM only processed concatenated data of different modalities. To take full advantage of the information in each modality, Zhang et al. proposed an improved SVM method, which is referred to as multimodal SVM. This method determines the weights of the kernels of each modality according to the effect on each classification, in an effort to take full advantage of the information within the images [24]. Some articles used the multimodal pattern classification methods to classify AD from NC. Gray et al. used the random forest method and obtained 89% accuracy [36], while Datatzikos et al. obtained 87.6% accuracy based on Multi-Kernel Learning and SVM method [14]. Also, Dukart et al. obtained an 100% accuracy based on the SVM method [34]. Among these pieces of research, Zhang et al. still obtained a high accuracy of 93.2% [24] and we got an accuracy of 94.44%, which was based on the multimodal SVM. As data and subjects were different, the results were reasonable and indicated that the multimodal SVM was an effective method in the classification study of AD.
The aim of the present study is to identify whether the normal elderly converts to MCI or AD. Although we used accuracy as the criterion to identify the optimal classifier, sensitivity and specificity are also important metrics in routine clinical considerations. Higher sensitivity can increase the likelihood of correctly diagnosing AD/MCI patients. However, since the rate of conversion to MCI/AD is much less than equal odds, it may be preferred to emphasize specificity more. In our study, we obtained not only high accuracy 94.44%, but high sensitivity 95.69% in the classification between AD and NC. The accuracy is 70%, but the specificity of 66.67% is lower than the sensitivity of 75% between NC-c and NC-nc. More balanced combination of various indices should be considered in the future studies.
In 2011, the International Working Group (IWG) and the US National Institute on Aging-Alzheimer’s Association developed recommendations towards defining the preclinical stages of AD [46]. The newest diagnostic guidelines (the IWG-2 criteria) [47] indicated that amyloid plaques and neurofibrillary tangles are the core pathophysiological markers on the diagnosis of AD. Meanwhile, FDG-PET and volumetric MRI measures were proposed to track disease progression. Although our results indicated that FDG-PET and MRI data offered more information for AD/NC and NC-c/NC-nc detection, they should not be used as part of the criteria for diagnosis and prognosis of the disease.
One limitation of our study is the small sample size limited by the requirement that each subject must have concurrent MRI, AV45-PET, and FDG-PET data. Additionally, we lack the ability to use the most updated diagnosis criteria since all the ADNI diagnoses were clinical without the use of fluid or imaging based biomarkers (Many ADNI subjects did not have these biomarkers available at the time of their diagnosis).
Present studies predicted MCI or AD in normal elderly using a multimodal SVM method. The prediction was implemented by classifying NC-c and NC-nc subjects using three modalities: MRI, FDGPET, and AV45-PET, after building a classifier based on the AD and NC groups. We fused three complementary modalities with different weights. This multimodal classifier can predict MCI or AD in the stage of normal cognition. In summary, the model we built identified at the risk of converting to MCI or AD in normal elderly; the identification of this population will be helpful in clinical diagnosis and pathological research.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation (NNSF), China (81000603), the Funds for International Cooperation and Exchange of NNSF, China (61210001), the Fundamental Research Funds for the Central Universities, China, the National Institute of Mental Health, US (RO1 MH57899), the National Institute on Aging, US (9R01AG031581–10, P30 AG19610), and the State of Arizona.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bio-engineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; Bio-Clinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Authors’ disclosures available online (http://jalz.com/manuscript-disclosures/14-2820r2).
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
REFERENCES
- [1].Wimo A, Prince MJ (2010) World Alzheimer Report 2010: The global economic impact of dementia, Alzheimer’s Disease International. [Google Scholar]
- [2].Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, Shen D, Alzheimer’s Disease Neuroimaging Initiative (2012) Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging 33, 427. e415–427. e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shi F, Liu B, Zhou Y, Yu C, Jiang T (2009) Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer’s disease: Meta-analyses of MRI studies. Hippocampus 19, 1055–1064. [DOI] [PubMed] [Google Scholar]
- [4].Nordberg A, Rinne JO, Kadir A, Långström B (2010) The use of PET in Alzheimer disease. Nat Rev Neurol 6, 78–87. [DOI] [PubMed] [Google Scholar]
- [5].Herholz K, Ebmeier K (2011) Clinical amyloid imaging in Alzheimer’s disease. Lancet Neurol 10, 667–670. [DOI] [PubMed] [Google Scholar]
- [6].Magnin B, Mesrob L, Kinkingnéhun S, Pélégrini-Issac M, Colliot O, Sarazin M, Dubois B, Lehéricy S, Benali H (2009) Support vector machine-based classification of Alzheimer’s disease from whole-brain anatomical MRI. Neuroradiology 51, 73–83. [DOI] [PubMed] [Google Scholar]
- [7].Adeli H, Ghosh-Dastidar S, Dadmehr N (2005) Alzheimer’s disease and models of computation: Imaging, classification, and neural models. J Alzheimers Dis 7, 187–199. [DOI] [PubMed] [Google Scholar]
- [8].Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM (2008) Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiol Aging 29, 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert M-O, Chupin M, Benali H, Colliot O, Alzheimer’s Disease Neuroimaging Initiative (2011) Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage 56, 766–781. [DOI] [PubMed] [Google Scholar]
- [10].Li Y, Rinne JO, Mosconi L, Pirraglia E, Rusinek H, DeSanti S, Kemppainen N, Någren K, Kim B-C, Tsui W (2008) Regional analysis of FDG and PIB-PET images in normal aging, mild cognitive impairment, and Alzheimer’s disease. Eur J Nucl Med Mol Imaging 35, 2169–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, Zhu W, Park M, Jiang T, Jin JS (2011) Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PloS One 6, e21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang D, Shen D, Initiative AsDN (2012) Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PloS One 7, e33182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davatzikos C, Xu F, An Y, Fan Y, Resnick SM (2009) Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: The SPARE-AD index. Brain 132, 2026–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ (2011) Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging 32, 2322. e2319–2322. e2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ferreira LK, Busatto GF (2013) Resting-state functional connectivity in normal brain aging. Neurosci Biobehav Rev 37, 384–400. [DOI] [PubMed] [Google Scholar]
- [16].Zhu W, Wen W, He Y, Xia A, Anstey KJ, Sachdev P (2012) Changing topological patterns in normal aging using large-scale structural networks. Neurobiology Aging 33, 899–913. [DOI] [PubMed] [Google Scholar]
- [17].Bayer TA, Wirths O, Majtényi K, Hartmann T, Multhaup G, Beyreuther K, Czech C (2001) Key factors in Alzheimer’s disease: β-amyloid precursor protein processing, metabolism and intraneuronal transport. Brain Pathol 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oddo S, Caccamo A, Kitazawa M, Tseng BP, LaFerla FM (2003) Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer’s disease. Neurobiol Aging 24, 1063–1070. [DOI] [PubMed] [Google Scholar]
- [19].Chételat G, Villemagne VL, Pike KE, Baron J-C, Bourgeat P, Jones G, Faux NG, Ellis KA, Salvado O, Szoeke C (2010) Larger temporal volume in elderly with high versus low beta-amyloid deposition. Brain 133, 3349–3358. [DOI] [PubMed] [Google Scholar]
- [20].Mormino EC, Brandel MG, Madison CM, Rabinovici GD, Marks S, Baker SL, Jagust WJ (2012) Not quite PIB-positive, not quite PIB-negative: Slight PIB elevations in elderly normal control subjects are biologically relevant. Neuroimage 59, 1152–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sperling RA, LaViolette PS, O’Keefe K, O’Brien J, Rentz DM, Pihlajamaki M, Marshall G, Hyman BT, Selkoe DJ, Hedden T (2009) Amyloid deposition is associated with impaired default network function in older persons without dementia. Neuron 63, 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rodrigue K, Kennedy K, Devous M, Rieck J, Hebrank A, Diaz-Arrastia R, Mathews D, Park D (2012) β-Amyloid burden in healthy aging Regional distribution and cognitive consequences. Neurology 78, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang GF, Estrada S (2004) Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Annals Neurol 55, 306–319. [DOI] [PubMed] [Google Scholar]
- [24].Zhang D, Wang Y, Zhou L, Yuan H, Shen D (2011) Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage 55, 856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34, 939. [DOI] [PubMed] [Google Scholar]
- [26].Folstein MF, Folstein SE, McHugh PR (1975) Mini-mental state: A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12, 189–198. [DOI] [PubMed] [Google Scholar]
- [27].Morris JC (1993) The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology 43, 2412–2414. [DOI] [PubMed] [Google Scholar]
- [28].Rajapakse JC, Giedd JN, Rapoport JL (1997) Statistical approach to segmentation of single-channel cerebral MR images. IEEE Trans Med Imaging 16, 176–186. [DOI] [PubMed] [Google Scholar]
- [29].Tohka J, Zijdenbos A, Evans A (2004) Fast and robust parameter estimation for statistical partial volume models in brain MRI. Neuroimage 23, 84–97. [DOI] [PubMed] [Google Scholar]
- [30].Manjón JV, Coupé P, Martí-Bonmatí L, Collins DL, Robles M (2010) Adaptive non-local means denoising of MR images with spatially varying noise levels. J Magn Reson Imaging 31, 192–203. [DOI] [PubMed] [Google Scholar]
- [31].Ashburner J (2007) A fast diffeomorphic image registration algorithm. Neuroimage 38, 95–113. [DOI] [PubMed] [Google Scholar]
- [32].Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289. [DOI] [PubMed] [Google Scholar]
- [33].Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20, 273–297. [Google Scholar]
- [34].Dukart J, Mueller K, Horstmann A, Barthel H, Möller HE, Villringer A, Sabri O, Schroeter ML (2011) Combined evaluation of FDG-PET and MRI improves detection and differentiation of dementia. PLoS One 6, e18111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang D, Shen D (2012) Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage 59, 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D (2013) Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. Neuroimage 65, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gray KR, Wolz R, Heckemann RA, Aljabar P, Hammers A, Rueckert D (2012) Multi-region analysis of longitudinal FDG-PET for the classification of Alzheimer’s disease. Neuroimage 60, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hinrichs C, Singh V, Xu G, Johnson SC (2011) Predictive markers for AD in a multi-modality framework: An analysis of MCI progression in the ADNI population. Neuroimage 55, 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Misra C, Fan Y, Davatzikos C (2009) Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: Results from ADNI. Neuroimage 44, 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ewers M, Walsh C, Trojanowski JQ, Shaw LM, Petersen RC, Jack CR Jr, Feldman HH, Bokde AL, Alexander GE, Scheltens P (2012) Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging 33, 1203–1214. e1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].De Leon M, Convit A, Wolf O, Tarshish C, DeSanti S, Rusinek H, Tsui W, Kandil E, Scherer A, Roche A (2001) Prediction of cognitive decline in normal elderly subjects with 2-[18F] fluoro-2-deoxy-D-glucose/positron-emission tomography (FDG/PET). Proc Natl Acad Sci U S A 98, 10966–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cui Y, Sachdev PS, Lipnicki DM, Jin JS, Luo S, Zhu W, Kochan NA, Reppermund S, Liu T, Trollor JN (2012) Predicting the development of mild cognitive impairment: A new use of pattern recognition. Neuroimage 60, 894–901. [DOI] [PubMed] [Google Scholar]
- [43].Walhovd K, Fjell A, Brewer J, McEvoy L, Fennema-Notestine C, Hagler D, Jennings R, Karow D, Dale A (2010) Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. Am J Neuroradiol 31, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer JB, Dale AM (2010) CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer’s disease. J Neurosci 30, 2088–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Landau S, Harvey D, Madison C, Reiman E, Foster N, Aisen P, Petersen R, Shaw L, Trojanowski J, Jack C (2010) Comparing predictors of conversion and decline in mild cognitive impairment. Neurology 75, 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Kaye J, Montine TJ (2011) Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement 7, 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K, DeKosky ST, Gauthier S, Selkoe D, Bateman R (2014) Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol 13, 614–629. [DOI] [PubMed] [Google Scholar]