Abstract
Aims:
To examine Salmonella and Escherichia coli in storm runoff and irrigation ponds used by fresh produce growers, and compare Salmonella serovars with those found in cases of human salmonellosis.
Methods and Results:
We collected water before and after rain events at two irrigation ponds on farms in southern Georgia, USA, and collected storm runoff/storm flow within the contributing watershed of each pond. Salmonella and E. coli concentrations were higher in ponds after rain events by an average of 0.46 (P < 0.01) and 0.61 (P < 0.05) log10 most probable number (MPN) 100 ml−1, respectively. Salmonella concentrations in storm runoff from fields and forests were not significantly higher than in ponds before rain events, but concentrations in storm flow from streams and ditches were higher by an average of 1.22 log10 MPN 100 ml−1 (P < 0.001). Eighteen Salmonella serovars were identified from 155 serotyped isolates, and eight serovars were shared between storm runoff/storm flow and ponds. Seven of the serovars, including five of the shared serovars, were present in cases of human illness in the study region in the same year. However, several serovars most commonly associated with human illness in the study region (e.g. Javiana, Enteritidis, and Montevideo) were not found in any water samples.
Conclusions:
Salmonella and E. coli concentrations in irrigation ponds were higher, on average, after rain events, but concentrations of Salmonella were low, and the ponds met FDA water quality standards based on E. coli. Some similarities and notable differences were found between Salmonella serovars in water samples and in cases of human illness.
Significance and Impact of Study:
This study directly examined storm runoff/storm flow into irrigation ponds and quantified increases in Salmonella and E. coli following rain events, with potential implications for irrigation pond management as well as human health.
Keywords: Salmonella, agriculture, water, environmental, epidemiology
INTRODUCTION
Salmonella enterica causes the largest number of bacterial foodborne illnesses as well as deaths related to any foodborne illness each year in the United States (Scallan et al. 2011). In some cases, outbreaks of Salmonella from fresh fruit and vegetables have been traced back to contaminated surface water sources used for crop irrigation (Greene et al. 2008; Behravesh et al. 2011). Surface water sources, especially ponds that collect storm runoff, are widely used in the southeastern USA for crop irrigation. Recent surveys in southern Georgia found that long-term geometric mean levels of Salmonella in these types of ponds are generally very low, less than 0.055 MPN 100 ml−1 (Luo et al. 2015) but levels of Salmonella in individual samples may occasionally reach levels capable of contributing to illness in consumers of fresh produce (Stine et al. 2005). Above-average rates of Salmonella infections occur in the southeastern USA, especially in parts of southern Georgia, where 83 cases per 100,000 people were reported for 2012 (Georgia Department of Public Health, unpublished data), compared to the US FoodNet average of only 16 cases per 100,000 people (CDC 2014).
Sources of Salmonella in waterways include fecal matter from humans, livestock, wildlife, or pets, which may be deposited directly or transported by surface runoff or subsurface flow during rain events (Haley et al. 2009). Salmonella also survives and may multiply in sediments on land and in water (Fish and Pettibone 1995; Davies and Wray 1996; Winfield and Groisman 2003). A study of the impacts of rain events on Salmonella loading in rivers in southern Georgia found higher levels of Salmonella in storm flow compared to base flow (Martin 2009). Other studies of rivers have found correlations between Salmonella or indicator Escherichia coli levels and recent rainfall (Haley et al. 2009; Walters et al. 2011). However, none of these studies have directly examined the impact of rain events in relation to irrigation ponds used by growers of fresh produce.
This study examined Salmonella and indicator E. coli concentrations in irrigation ponds before and after rain events, in surface runoff from fields and forests during rain events, and in storm flow in streams and ditches in the contributing watershed of each irrigation pond. Additionally, we examined similarities between serovars of Salmonella found in these landscapes and in cases of salmonellosis in the same year among people living in the same rural region of southern Georgia as the study irrigation ponds.
MATERIALS AND METHODS
Study region.
This study was located in southern Georgia in the Little River watershed (USGS HUC-8 03110204). The Little River watershed covers 2,309 km2 and includes dense networks of low-gradient stream channels and wide riparian forest wetlands typical of the larger Coastal Plain region in the southeastern USA (Sullivan et al. 2007). Rainfall occurs throughout the year in this watershed; storms are generally small and frequent during the long, humid summers, and are generally larger and less frequent during the short, mild winters (Bosch et al. 1999). The Hawthorne geologic formation extends across most of the watershed, with primarily loamy sand surface soils underlain by clay (Sullivan et al. 2007) Land cover in the watershed as of 2011 was 48% natural or forested (including planted pine forests), 45% agricultural (including row crops and pastures), and 7% developed (including residential/commercial/industrial areas) (USGS 2014). Animal agriculture in the watershed in 2013 included an estimated 18,000 cattle and the production of 13.5 million broiler chickens per year (only 2% and 1% of Georgia’s cattle population and broiler chicken production, respectively) (USDA National Agricultural Statistics Service 2014). Fruits, vegetables, and nuts—often produced using irrigation water from surface water sources or groundwater from the Floridan aquifer—accounted for a market value of about $55 million in the watershed (USDA National Agricultural Statistics Service 2014).
In this study, we focused on two produce farms (Farm 1 and Farm 2) with irrigation ponds. Like most irrigation ponds in southern Georgia, the ponds at both farms were constructed by excavating and damming small, slow-moving, heavily vegetated streams. Both ponds were part of previous studies of Salmonella, Campylobacter jejuni, and Escherichia coli O157:H7 in fruit and vegetable irrigation water sources (Gu et al. 2013a,b; Luo et al. 2015).
Study overview.
Samples of water were collected from the irrigation ponds at the two farms before and after twelve rain events between January and August 2013. During those rain events, samples were also collected in the contributing watershed of each irrigation pond, including storm runoff from fields and forests and storm flow from streams and ditches. Samples were collected from one farm per rain event, with six rain events per farm. Sampling equipment was set up when weather forecasts predicted rain within the next few hours or overnight. All sampled rain events produced surface runoff and storm flow.
Tipping bucket rain gauges and data loggers (Onset HOBO) set up at each pond recorded rainfall throughout the study period. Rainfall amounts during the rain events sampled for this study ranged from 4–80 mm at Farm 1 and 3–11 mm at Farm 2, and the duration of rain events ranged from a few hours to overnight. The highest rainfall intensity during the most intense five minutes of any rain event was 72 mm h−1, which was not extreme for this region (Frederick et al. 1977). During the study period, the region experienced more rainfall than during the same months in the previous three years; nearby weather stations recorded 101 days with rainfall and a total rainfall of 132 cm from January through August, compared to the same months in 2010–2012 with only 46–94 cm of rainfall (www.georgiaweather.net, accessed 2014-04-25).
Study sites.
The boundaries of the subwatersheds were calculated using BASINS with 1/3 arc-second resolution elevation data and hydrography data (USGS 2014; EPA 2015). Characteristics of the subwatersheds were calculated using QGIS, 30-meter resolution land cover data, 1-meter resolution orthoimagery, and in-person observations (USGS 2014; QGIS Development Team 2015).
The subwatershed of Farm 1 covers 2.8 km2 and includes approximately 40% forested land cover, 35% agricultural land cover, and a small number of developed areas with septic tank systems (Fig. 1). The pond at the base of the subwatershed of Farm 1 has a surface area of 0.08 km2, and was used to irrigate produce and peanuts in nearby fields. The subwatershed of Farm 2 covers 0.7 km2 and includes approximately 55% forested land cover, 45% agricultural land cover, and no developed areas (Fig. 1). The pond at the base of the subwatershed of Farm 2 has a surface area of 0.05 km2, and nearby crop fields included produce, peanuts, cotton, and perennial grasses used as biofuel feedstock. The ponds at both Farm 1 and Farm 2 receive supplemental groundwater from wells.
Figure 1.
The pond watersheds (---), sampling areas (○), and land cover types at Farm 1 and Farm 2. See Table 1 for sampling area map code descriptions.
Samples of pond water from irrigation ponds.
Before and after each rain event, two composite samples of water were collected from the pond edges—one composite from three grab samples at the pond edge near the intake pipe for the irrigation system and the other from grab samples at three separate edges of the pond to characterize the pond as a whole (Fig. 1). Each grab sample was collected in a sterile disposable 2 L Whirl-Pak bag (Nasco) and grab samples from each sampling location were mixed together in the laboratory to create 6 L composite samples.
Samples of storm runoff from fields and forests.
Storm runoff was collected during rain events from multiple sampling locations surrounding the ponds (Fig. 1). The locations were classified as either agricultural fields or forested areas, including one forested residential yard. Downslope of each selected sampling location, four to six Whirl-Pak bags were spread out across a 50–100 m transect, usually near the pond edges. The bags were set up before each rain event, and were fully opened and pinned securely to the ground using autoclaved nails. A built-in wire, with the bottom side secured flush with the ground, allowed the bags to remain open during rain events (Fig. 2). In the laboratory, bags were combined into one composite for each sampling location. Composite volumes totaled 5–7 L per area of land use depending on the size of the rain event.
Figure 2.
A storm runoff sampling bag after a storm.
Samples of storm flow from streams and ditches.
Storm flow was collected during rain events from a major inflow stream, as well as a roadside ditch entering the pond at Farm 1, and from an intermittent stream entering the pond at Farm 2. At the ditch and intermittent stream, composite samples totaling 6 L were collected in Whirl-Pak bags. At the major inflow stream, an automated sampler collected water at regular intervals when the stream rose above base flow, for composite volumes totaling 5–7 L, depending on the duration of the rain event.
Bacterial analyses.
All samples were stored on ice in coolers during transport and then refrigerated in the laboratory. Analysis began within 24 hours of sample collection. In total, 107 composite samples were analysed for Salmonella (48 samples of pond water before or after rain events, 47 samples of storm runoff from fields and forests during rain events, and 12 samples of storm flow from streams and ditches) and 105 of the 107 were also analysed for E. coli.
We used a most-probable-number (MPN) enumeration protocol for Salmonella developed by Luo et al. (2014). Briefly, three replicates of three sample volumes (500 ml, 100 ml, and 10 ml) were analysed for each composite, resulting in a total of nine replicates analysed per composite. For primary enrichment, equal volumes of lactose broth (Difco) were added to each replicate and incubated for 24 h at 37°C. Next, for selective enrichment, 1 ml from each replicate was transferred into 10 ml of tetrathionate broth (Difco) and incubated again. Each replicate was streaked onto XLT4 agar (Remel), and after 24 h at 37°C, presumptive positive colonies were transferred to CHROMagar Salmonella Plus agar (CHROMagar) for 20–24 h at 37°C. All broths and media were prepared from powdered stocks, and negative and positive controls (Salmonella ser. Newport from the Wright strain collection, University of Florida) were used throughout the protocol.
For presumptive positive colonies, at least one isolate per replicate was stored in Luria broth (Difco) for 24 h at ambient temperature, then analysed using polymerase chain reaction (PCR) with primers targeting the invA gene (Chiu and Ou 1996). For each isolate, 1 ml of the inoculated Luria broth was centrifuged at ≥14000 × g for 3 min. The pellet was resuspended in 500 μl of sterile molecular grade water, mixed well, and boiled for 10 min before centrifuging again. A commercially available master mix (Takara Bio Inc. TaKaRa Ex Taq or Promega PCR Master Mix) was added to 5 μl of the supernatant, and thermal cycling was performed at 95°C for 90 s, then 30 cycles of 95°C for 90 s, 50°C for 30 s, and 72°C for 60 s, followed by 72°C for 5 min and held at 4°C. After PCR, each isolate underwent agarose gel electrophoresis on a 100 ml 1% agarose gel containing 2 μl of undiluted ethidium bromide. Salmonella-positive isolates displayed an amplicon of 244 base pairs. All PCR-confirmed isolates were re-cultured and archived at −80°C in a 50/50 mixture of glycerol and Luria broth.
Estimates of Salmonella MPN were calculated based on the presence of PCR-confirmed Salmonella in each of the nine replicates analysed per composited water sample (Jarvis et al. 2010; Andrews et al. 2014). The lower and upper limits of Salmonella detection were 0.0548 and 11 MPN 100 ml−1, respectively. Samples without detectable Salmonella (54 of 107 samples) were assigned half the lower limit of detection (Clarke 1998). Samples exceeding the upper limit of detection (2 of 107 samples) were assigned the upper limit of detection.
Estimates of E. coli MPN for each composite were calculated using 24-hour Colilert with Quanti-Tray/2000 (IDEXX Laboratories). The lower and upper limits of detection in undiluted samples were 1 and 2419 MPN 100 ml−1, respectively. Samples without detectable E. coli (3 of 105 samples) were assigned half the lower limit of detection (Clarke 1998). Samples anticipated to contain E. coli levels above 2419 MPN 100 ml−1 (28 samples) were diluted before analysis, but some (11 samples) still exceeded the upper limit of detection after dilution and were assigned the value of the upper limit based on the dilution used.
Salmonella serotyping.
PCR-confirmed Salmonella isolates (182 in total) were revived in tetrathionate broth and streaked onto CHROMagar Salmonella Plus agar. Revived isolates were first tested with Salmonella O poly antisera (each containing multiple O antigens) (Difco) in the following order until a positive agglutination reaction was observed: B, A, D, G, C, E, F. Next, up to three isolates representing each O poly group found in each composite sample were chosen at random (163 in total), streaked onto tryptic soy broth agar slants, and sent to the National Veterinary Services Laboratory in Ames, Iowa, for complete serovar identification.
Solids.
To measure total suspended solids (TSS) in each composite sample for comparison with microbe concentrations, known volumes of water were filtered through 1.5 μm filters (Hach) and baked, desiccated, and weighed according to standards methods (APHA 2005).
Statistical analyses.
The rates of presence of Salmonella and E. coli in pond water before and after rain events were compared using generalized linear mixed-effects models fit by restricted maximum likelihood with a binomial family (R Core Team 2013; Bates et al. 2014). In each model, sample type (pond water before rain events and pond water after rain events) and sample location (near the pond irrigation intake and near the pond edges) were included as fixed effects, farm was included as a random effect, and the presence of Salmonella or E. coli in each sample was the response variable (Table S1).
Concentrations of Salmonella and E. coli in each sample type were compared using linear mixed-effects models fit by restricted maximum likelihood, with Wald tests of significance (R Core Team 2013; Bates et al. 2014; Kuznetsova et al. 2014). In each model, sample type (pond water before rain events, pond water after rain events, storm runoff from fields, storm runoff from forests, and storm flow from streams and ditches) and sample location were included as fixed effects, farm was included as a random effect, and the log10 concentration of Salmonella or E. coli in each sample was the response variable (Table S1). Residual plots for each model did not show any obvious deviations from normality or homoscedasticity.
Geometric mean concentrations of Salmonella and E. coli were calculated for each sampling location, and geometric mean concentrations and bootstrapped 95% confidence intervals of Salmonella and E. coli were also calculated for each sample type at each farm (Wickham 2009; Canty and Ripley 2014). Correlations between Salmonella concentrations, E. coli concentrations, and TSS were estimated using Spearman’s rs using a Bonferroni correction for multiple comparisons (Harrell, Jr. 2014).
E. coli concentrations at each pond were compared to the FDA rules issued in 2015 as part of the implementation of the Food Safety Modernization Act (FDA 2015). According to these rules, the geometric mean E. coli concentration calculated from the 20 most recent samples of irrigation water must be less than 126 colony-forming units (CFU) 100 ml−1, and the statistical threshold value (STV) approximating the 90th percentile of the lognormal distribution of E. coli concentrations must be less than 410 CFU 100 ml−1.
RESULTS
Microbe presence.
Salmonella was detected in all but one of the sampling locations (a field of peanuts/cotton at Farm 2) during at least one of the 12 rain events (Table 1). Overall, 52 of 107 samples (49%) contained detectable Salmonella, and 103 of 105 samples (98%) contained detectable E. coli. In pond water, Salmonella was present in 33% (N = 24) of samples before rain events and 58% (N = 24) of samples after rain events, although the difference was only marginally significant according to generalized linear mixed-effects model results (P = 0.08). In storm flow from streams and ditches, Salmonella was present in 100% of samples.
Table 1.
Summary of samples collected (N) and percent of samples positive (Pos.) for Salmonella at each sampling area, with geometric means of Salmonella (Salm.), E. coli, and total suspended solids (TSS)
| Farm 1 | Farm 2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample type | Pos. | Salm. | E. coli | TSS | Sample type | Pos. | Salm. | E. coli | TSS | ||||
| Map code/description | N | (%) | (MPN 100 ml−1) | (mg l−1) | Map code/description | N | (%) | (MPN 100 ml−1) | (mg 1−1) | ||||
| Pond before storms | Pond before storms | ||||||||||||
| P1 | Pond at intake | 6 | 33% | 0.045 | 6 | 10 | P1 | Pond at intake | 6 | 17% | 0.032 | 5 | 24 |
| P2 | Pond edges | 6 | 33% | 0.035 | 14 | 15 | P2 | Pond edges* | 6 | 50% | 0.055 | 7 | 17 |
| Pond after storms | Pond after storms | ||||||||||||
| P1 | Pond at intake | 6 | 50% | 0.128 | 64 | 10 | P1 | Pond at intake | 6 | 50% | 0.083 | 11 | 17 |
| P2 | Pond edges | 6 | 67% | 0.162 | 98 | 9 | P2 | Pond edges | 6 | 67% | 0.108 | 11 | 17 |
| Runoff from fields | Runoff from fields | ||||||||||||
| A1 | Peanut field | 6 | 50% | 0.070 | 5426 | 1647 | A1 | Biofuel field | 6 | 33% | 0.036 | 900 | 3483 |
| A2 | Tomato field | 3 | 67% | 0.046 | 4987 | 205 | A2 | Peanuts/cotton | 4 | 0% | 453 | 902 | |
| Runoff from forests | Runoff from forests | ||||||||||||
| F1 | Forested yard | 5 | 40% | 0.056 | 709 | 287 | F1 | Shrubland† | 6 | 17% | 0.031 | 1040 | 1911 |
| F2 | Pine forest | 6 | 67% | 0.088 | 2727 | 305 | F2 | Mixed forest | 6 | 17% | 0.031 | 685 | 205 |
| F3 | Mixed forest* | 5 | 60% | 0.414 | 918 | 935 | |||||||
| Storm flow from streams | Storm flow from streams | ||||||||||||
| S1 | Stream | 6 | 100% | 0.767 | 275 | 14 | S | Stream | 3 | 100% | 0.828 | 6319 | 200 |
| S2 | Road ditch | 3 | 100% | 0.413 | 5517 | 553 | |||||||
One sample was not analysed for E. coli or solids.
One sample was not analysed for solids.
Microbe concentrations.
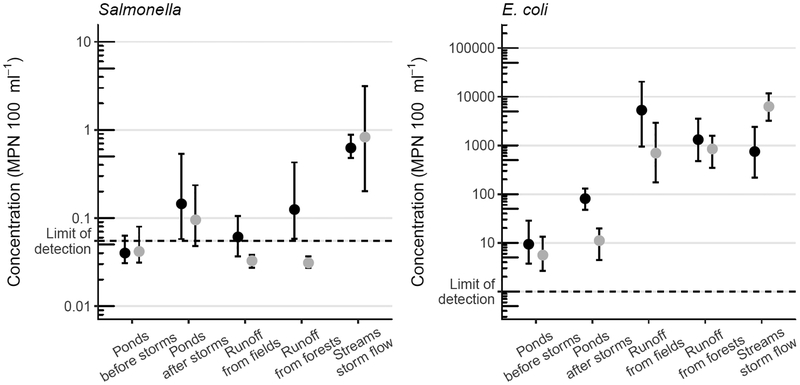
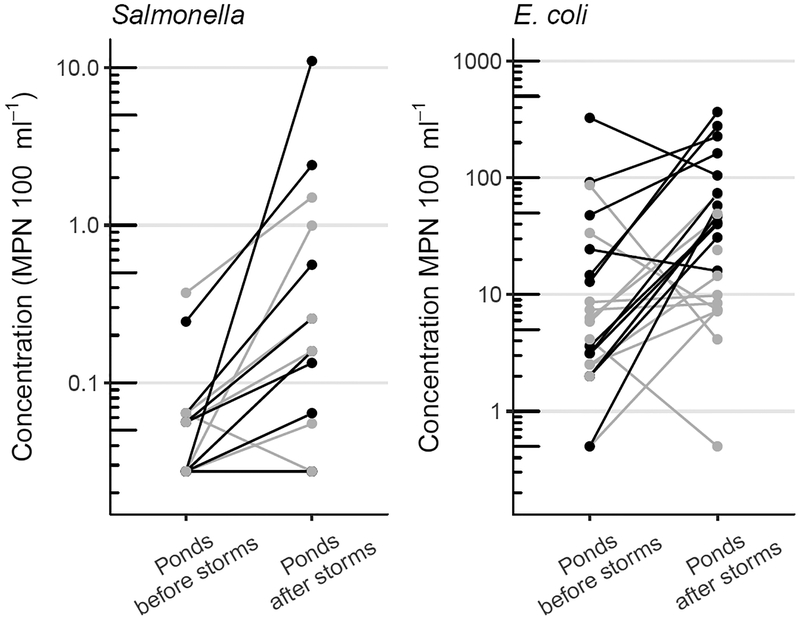
Compared to pond water before rain events, concentrations of Salmonella were 0.46 log10 MPN 100 ml−1 higher (P < 0.01) in pond water after rain events, and 1.22 log10 MPN 100 ml−1 higher (P < 0.001) in storm flow from streams and ditches, based on the linear mixed-effects model. Concentrations in storm runoff from fields were lower by 0.01 log10 MPN 100 ml−1 (P = 0.96), and concentrations in storm runoff from forests were lower by 0.09 log10 MPN 100 ml−1 (P = 0.76), neither of which were statistically significant. Geometric mean Salmonella concentrations and bootstrapped 95% confidence intervals for each sample type at each farm are shown in Fig. 3. Salmonella concentrations after rain events were higher in 58% of paired pond samples, lower in 4%, and showed no change in 38%; these paired comparisons are shown in Fig. 4.
Figure 3.
Geometric means and bootstrapped 95% confidence intervals of Salmonella and E. coli concentrations for each sample type at Farm 1 (●) and Farm 2 ().
Figure 4.
Paired comparisons (before and after rain events) of Salmonella and E. coli concentrations in pond water samples at Farm 1 (●) and Farm 2 ().
Compared to pond water before rain events, concentrations of E. coli were 0.61 log10 MPN 100 ml−1 higher (P < 0.05) in pond water after rain events, 2.0 log10 MPN 100 ml−1 higher (P < 0.001) in storm runoff from fields, 2.1 log10 MPN 100 ml−1 higher (P < 0.001) in storm runoff from forests, and 1.9 log10 MPN 100 ml−1 higher (P < 0.001) in storm flow from streams and ditches, based on the linear mixed-effects model. Geometric mean concentrations and bootstrapped 95% confidence intervals of E. coli for each sample type at each farm are shown in Fig. 3. E. coli concentrations after rain events were higher in 78% of paired pond samples and lower in 22%. Each set of paired pond samples, indicating concentrations in each sampling location before and after each storm, are shown in Fig. 4.
Comparison of E. coli concentrations with FDA rules.
The irrigation ponds at both farms in this study would have met FDA requirements, both before and after rain events. The geometric mean E. coli concentrations in pond water before and after rain events were 9 and 79 CFU 100 ml−1 at Farm 1, and 6 and 11 CFU 100 ml−1 at Farm 2. The STVs of E. coli concentrations in pond water before and after rain events were 102 and 274 CFU 100 ml−1 at Farm 1, and 33 and 61 CFU 100 ml−1 at Farm 2.
Comparison among Salmonella, E. coli, and TSS.
According to Spearman’s correlation tests across all samples, Salmonella concentrations were not correlated with E. coli concentrations or TSS. However, E. coli concentrations were correlated with TSS (rs = 0.6, N = 104, P < 0.001).
Salmonella serotyping.
Eighteen serovars of Salmonella were identified (Table 2). At Farm 1, Bareilly, I 38:k:-, and Saintpaul were found in all sample types. Other common serovars at Farm 1 were Muenchen (found in all sample types except the pond before storms) and Rubislaw (found in all sample types except runoff from fields). At Farm 2, Gaminara and Muenchen were found in samples of pond water before and after storms and in samples of storm flow from streams, but not in runoff from fields or forests.
Table 2.
Summary of Salmonella serovars by sample type at each farm, shown in comparison with the number of cases of human salmonellosis in counties within the Little River watershed in 2013 that were attributed to each serovar

|
Number of samples yielding any Salmonella among the total number of samples collected. Some samples yielded more than one serovar.
Human cases were attributed to at least 20 different serovars, seven of which were also found in the present study.
The 141 cases not shown in the table were attributed to Javiana (52 cases), Typhimurium (21), Enteritidis (16), Montevideo (7), Miami (6), I 13,23:b:- (6), Carrau (1), Heidelberg (1), Infantis (1), Kintambo (1), Mississippi (1), Sentftenberg (1), I 4,[5],12:i:- (1), and untested Salmonella serovars (26 cases).
Eight serovars were found in both storm runoff/storm flow and irrigation pond water samples, and six of these serovars were found in both storm runoff/storm flow and irrigation pond water samples after the same rain events. Salmonella serovars Bareilly, Inverness, I 38:k:-, Rubislaw, Saintpaul, and III 60:r:e,n,x,z15 at Farm 1 and only Bareilly at Farm 2 were found in pond water after rain events as well as in storm runoff/storm flow during the same rain events. Serovars found during at least three of the six rain events sampled per farm included Bareilly, I 38:k:-, Muenchen, Rubislaw, Saintpaul, and III 60:r:e,n,x,z15 at Farm 1, and only Muenchen and Gaminara at Farm 2. More than one Salmonella serovar was often found per water sample.
In 2013, 217 cases of human salmonellosis in counties within the Little River watershed were reported to the Georgia Department of Health (Georgia Department of Public Health, unpublished data). Of the 217 cases, at least 76 cases were caused by serovars also present in the farm landscapes we sampled (Table 2, bottom row). Overall, 37 of the 52 water samples containing Salmonella in our study contained serovars also present in cases of human salmonellosis in counties in the Little River watershed during the same year. However, several of the most common serovars responsible for cases of human salmonellosis were not found at all among our water samples, such as Javiana, Enteritidis, or Montevideo (Table 2, footnote c.).
DISCUSSION
In this study of the impact of rain events on irrigation pond water quality in southern Georgia, we found higher concentrations of both Salmonella and E. coli as well as higher rates of Salmonella presence in pond water after rain events. We found the highest concentrations of Salmonella in storm flow from streams and ditches, compared to pond water before/after rain events or storm runoff from fields/forests. Several of the serovars of Salmonella found in these agricultural landscapes have also been present in human salmonellosis cases in the same region during the same year as the study. We note a limitation of this study is the relatively small number of rain events sampled, with six rain events sampled per farm.
The result showing higher concentrations of Salmonella in pond water after rain events is consistent with several other studies of waterways in Georgia, California, and New York (Haley et al. 2009; Walters et al. 2011; Strawn et al. 2013). Studies in Ontario and Florida, however, found no correlations between Salmonella and recent rainfall (Thomas et al. 2013; McEgan et al. 2013). The impact of rain events on Salmonella and indicator E. coli levels in waterways may depend on the characteristics of the waterway and sources of microbes in the surrounding landscape.
Dose-response relationships for Salmonella are variable across serotypes and across different studies, and outbreaks have occurred even at low estimated doses (Bollaerts et al. 2008). However, the Salmonella concentrations detected in this study are likely below levels of major public health concern. According to Stine et al. (2005), Salmonella concentrations of 2.5 CFU 100 ml−1 in irrigation water would result in a 1:10,000 annual risk of infection. That concentration, based on a worst-case scenario where fresh produce is harvested and consumed the day after the last irrigation, is more than ten times higher than the geometric mean Salmonella concentrations in pond water after storms in this study.
The result showing that storm runoff from forests and agricultural fields tended to have lower levels of Salmonella than ponds after rain events indicates that the increase in Salmonella seen in ponds after rain events did not necessarily come from storm runoff from forests and agricultural fields. This is important to note, as some growers of fresh produce have expressed concern about storm runoff from wildlife habitats like the forested areas in this study (Gennet et al. 2013). While forested areas and vegetated buffers adjacent to irrigation ponds may provide habitat for potentially Salmonella-shedding wildlife, these parts of farm landscapes may also serve to filter and trap contaminants in storm runoff and erosion (Lowrance et al. 2007; Cardoso et al. 2012).
Storm flow from streams and ditches, on the other hand, typically had the highest concentrations of Salmonella of all sample types, suggesting that recharging streams may be a major source of Salmonella in these irrigation ponds. Streams concentrate runoff from the entire watershed, accumulating microbial contamination from a variety of landscapes and sources, and eventually depositing pathogens directly into irrigation waters. The consequences of more sudden storm runoff from paved areas, such as in the roadside ditch in this study, as well as the influences of septic tanks, domestic pets, and even small numbers of livestock (dogs, chickens, guineafowl, horse, cattle) upstream as at Farm 1 in this study should be considered. Future efforts could potentially focus on reducing microbial contamination where discharging streams enter waterbodies used for irrigation, such as the use of a series of ponds.
The Salmonella serovars in water samples in our study did not reflect the most common serovars among the local human population during the same time period. Javiana, Newport, Typhimurium, and Enteritidis were the four most common Salmonella serovars in cases of human salmonellosis in counties in the Little River watershed in 2013, but we found only one of those serovars at either farm (Newport, in one sample at Farm 1). However, the fifth most common serovar in cases of human salmonellosis was Saintpaul, and it was the most commonly isolated serovar at Farm 1 in our study. In addition, the seventh most common serovar in cases of human salmonellosis was Muenchen, and it was found in all sample types at Farm 1 and in samples of pond water and storm flow from streams at Farm 2. Muenchen has consistently been a common serovar found in previous studies of waterways in south Georgia (Haley et al. 2009; Martin 2009; Rajabi et al. 2011).
The diversity of serovars in our study (18 serovars among 52 positive samples) was similar to the diversity-per-sample in other studies involving Salmonella in waterways, but the most common serovars vary widely between geographic regions. A study in California reported 16 serovars among 55 positive samples, most commonly Typhimurium and Give (Gorski et al. 2011); a study in New York reported 7 serovars among 26 positive samples, most commonly Cerro, Newport, and Thompson (Strawn et al. 2013); a study in Ontario reported 38 serovars among 91 positive samples, most commonly Heidelberg and Typhimurium (Thomas et al. 2013); and a study in North Carolina reported 12 serovars among 47 positive samples, most commonly Anatum, Gaminara, and Inverness (Patchanee et al. 2010). The most common serovars in our study in south Georgia were Saintpaul, Muenchen, Rubislaw, Bareilly, I 38:k:-, and III 60:r:e,n,x,z15. The differences between serovars detected in our study and other geographic regions may indicate the importance of local transmission and/or the ability to persist in particular environments or climates. Additionally, half of the serovars we detected appeared in no more than one sampling area during no more than one rain event during the study, possibly indicating turnover in the Salmonella serovars present over time as well as the difficulty of detecting very low concentrations of Salmonella.
High levels of total suspended solids in this study did not appear to be related to higher concentrations of Salmonella. This result is corroborated by a study of ponds, creeks, rivers, and canals in central Florida that found Salmonella in 100% of 202 concentrated 10 L water samples collected over 12 months, all from rural areas away from animal agriculture, all collected without disturbing bottom sediments, and all with low levels of total solids (McEgan et al. 2013). Previous studies in south Georgia and central Florida have noted persistent populations of Salmonella in streams even at base flow (Martin 2009; McEgan et al. 2013).
Salmonella and E. coli concentrations were not significantly correlated in this study, and the irrigation ponds at both farms in this study would have met FDA water quality standards for produce safety based on E. coli, even immediately after storms. Limited research is available on the ability of E. coli to indicate the presence of specific pathogens in natural irrigation waters (Winfield and Groisman 2003; Pachepsky et al. 2011; Wu et al. 2011). The use of E. coli to indicate health risks from other microbes is complicated by its ability to survive and even proliferate in natural waters under some conditions (Byappanahalli and Fujioka 1998; Anderson et al. 2005; Byappanahalli et al. 2006). Future work should evaluate how well FDA standards based on E. coli perform at predicting health risks from surface water sources, and explore potential alternative indicators for use in agricultural landscapes.
Our study highlights the likelihood of higher Salmonella and E. coli levels in irrigation ponds after rain events. Farmers are less likely to irrigate soon after rain events, but those that do should be aware of the potential for increased risk of microbial contamination of produce. Several of the serovars found in the farm landscapes in our study are capable of causing human illness, even if they are not currently among the most common causes of salmonellosis in the study region. Future work should also explore strategies for reducing inputs of Salmonella into waterways and reducing the transport of Salmonella from pond water into farm irrigation systems.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the fruit and vegetable growers who allowed us to collect water samples from their farms. Georgia Department of Public Health provided data on serovars in cases of salmonellosis for 2012–2013. Camilla Borgato, Debbie Coker, Charles Gruver, Jill Johnson, Debbie Lee, and Katy Summers provided field and lab assistance, Berry Brosi provided statistical assistance, and Richard Lowrance, Mia Mattioli, and Catherine Pringle provided design and/or editorial assistance.
This work was supported by grant SCB11063 from the Center for Produce Safety, Davis, California. The Center for Produce Safety had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Any mention of trade names or commercial products does not constitute endorsement or recommendation.
Footnotes
CONFLICT OF INTEREST
No conflict of interest declared.
REFERENCES
- Anderson KL, Whitlock JE and Harwood VJ (2005) Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol 71, 3041–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews WH, Wang H, Jacobson A and Hammack T (2014) Bacteriological Analytical Manual: Salmonella. FDA. [Google Scholar]
- APHA (2005) Standard methods for the examination of wastewater, Method 2540 D. American Public Health Association, Washington, DC. [Google Scholar]
- Bates D, Maechler M, Bolker B and Walker S (2014) lme4: Linear mixed-effects models using Eigen and S4, v1.1-7. Available at: https://cran.r-project.org/package=lme4.
- Behravesh CB, Moday RK, Jungk J, et al. (2011) 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. N Engl J Med 364, 918–927. [DOI] [PubMed] [Google Scholar]
- Bollaerts K, Aerts M, Faes C, Grijspeerdt K, Dewulf J and Mintiens K (2008) Human salmonellosis: estimation of dose-illness from outbreak data. Risk Anal. 28, 427–440. [DOI] [PubMed] [Google Scholar]
- Bosch DD, Sheridan JM and Davis FM (1999) Rainfall characteristics and spatial correlation for the Georgia coastal plain. Trans ASAE 42, 1637–1644. [Google Scholar]
- Byappanahalli MN and Fujioka RS (1998) Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci Technol 38, 171–174. [Google Scholar]
- Byappanahalli MN, Whitman RL, Shively DA, Sadowsky MJ and Ishii S (2006) Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ Microbiol 8. [DOI] [PubMed] [Google Scholar]
- Canty A and Ripley B (2014) boot: Bootstrap R (S-Plus) functions, v1.3-13. Available at: https://cran.r-project.org/package=boot.
- Cardoso F, Shelton D, Sadeghi A, Shirmohammadi A, Pachepsky Y and Dulaney W (2012) Effectiveness of vegetated filter strips in retention of Escherichia coli and Salmonella from swine manure slurry. J Environ Manage 110, 1–7. [DOI] [PubMed] [Google Scholar]
- CDC (2014) FoodNet surveillance report for 2012. US Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- Chiu C-H and Ou JT (1996) Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 34, 2619–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JU (1998) Evaluation of censored data methods to allow statistical comparisons among very small samples with below detection limit observations. Environmental Science & Technology 32, 177–183. [Google Scholar]
- Davies RH and Wray C (1996) Seasonal variations in the isolation of Salmonella typhimurium, Salmonella enteritidis, Bacillus cereus and Clostridium perfringens from environmental samples. J Vet Med B, 119–127. [DOI] [PubMed] [Google Scholar]
- EPA (2015) BASINS 4.1 Modeling Framework. Available at: https://www.epa.gov/exposure-assessment-models/basins.
- FDA (2015) Standards for the growing, harvesting, packing, and holding of produce for human consumption. Fed Regist 80, 74353–74568. [Google Scholar]
- Fish JT and Pettibone GW (1995) Influence of freshwater sediment on the survival of Escherichia coli and Salmonella sp. as measured by three methods of enumeration. Lett Appl Microbiol 20, 277–281. [DOI] [PubMed] [Google Scholar]
- Frederick RH, Myers VA and Auciello EP (1977) Five- to 60-minute precipitation frequency for the eastern and central United States. NOAA NWS, Silver Spring, MD. [Google Scholar]
- Gennet S, Howard J, Langholz J, Andrews K, Reynolds MD and Morrison SA (2013) Farm practices for food safety: an emerging threat to floodplain and riparian ecosystems. Front Ecol Environ 11, 236–242. [Google Scholar]
- Gorski L, Parker CT, Liang A, et al. (2011) Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl Environ Microbiol 77, 2734–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene SK, Daly ER, Talbot EA, et al. (2008) Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect 136, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Luo Z, Cevallos-Cevallos JM, Adams P, Vellidis G, Wright A and van Bruggen AHC (2013a) Factors affecting the occurrence of Escherichia coli O157 contamination in irrigation ponds on produce farms in the Suwannee River Watershed. Can J Microbiol 59, 175–182. [DOI] [PubMed] [Google Scholar]
- Gu G, Luo Z, Cevallos-Cevallos JM, Adams P, Vellidis G, Wright A and van Bruggen AHC (2013b) Occurrence and population density of Campylobacter jejuni in irrigation ponds on produce farms in the Suwannee River Watershed. Can J Microbiol 59, 339–346. [DOI] [PubMed] [Google Scholar]
- Haley BJ, Cole DJ and Lipp EK (2009) Distribution, diversity, and seasonality of waterborne salmonellae in a rural watershed. Appl Environ Microbiol 75, 1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE Jr. (2014) Hmisc: Harrell Miscellaneous Available at: https://cran.r-project.org/package=Hmisc.
- Jarvis B, Wilrich C and Wilrich PT (2010) Reconsideration of the derivation of Most Probable Numbers, their standard deviations, confidence bounds and rarity values. J Appl Microbiol 109, 1660–1667. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff P and Christensen R (2014) lmerTest: Tests in linear mixed effects models, v2.0-6. Available at: https://cran.r-project.org/package=lmerTest.
- Lowrance R, Sheridan JM, Williams RG, et al. (2007) Water quality and hydrology in farm-scale coastal plain watersheds: Effects of agriculture, impoundments, and riparian zones. J Soil Water Conserv 62, 65–76. [Google Scholar]
- Luo Z, Gu G, Ginn A, et al. (2015) Distribution and characterization of Salmonella enterica isolates from irrigation ponds in the southeastern USA. Appl Environ Microbiol 81, 4376–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Gu G, Giurcanu MC, Adams P, Vellidis G, van Bruggen AHC and Wright AC (2014) Development of a novel cross-streaking method for isolation, confirmation, and enumeration of Salmonella from irrigation ponds. J Microbiol Methods 101, 86–92. [DOI] [PubMed] [Google Scholar]
- Martin G (2009) Discrete storm impacts on the loading of Salmonella and campylobacters within a south Georgia rural watershed, PhD thesis University of Georgia. [Google Scholar]
- McEgan R, Mootian G, Goodridge LD, Schaffner DW and Danyluk MD (2013) Predicting Salmonella populations from biological, chemical, and physical indicators in Florida surface waters. Appl Environ Microbiol 79, 4094–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachepsky Y, Shelton DR, McLain JET, Patel J and Mandrell RE (2011) Irrigation waters as a source of pathogenic microorganisms in produce: A Review In: Advances in Agronomy, Vol. 113 pp 73–138. [Google Scholar]
- Patchanee P, Molla B, White N, Line DE and Gebreyes WA (2010) Tracking Salmonella contamination in various watersheds and phenotypic and genotypic diversity. Foodborne Pathog Dis 7, 1113–1120. [DOI] [PubMed] [Google Scholar]
- QGIS Development Team (2015) QGIS Geographic Information System. Available at: www.qgis.com.
- R Core Team (2013) R: A language and environment for statistical computing, v3.1.0. Available at: https://www.r-project.org.
- Rajabi M, Jones M, Hubbard M, Rodrick G and Wright AC (2011) Distribution and genetic diversity of Salmonella enterica in the Upper Suwannee River. Int J Microbiol 2011, 461321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, et al. (2011) Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine SW, Song I, Choi CY and Gerba CP (2005) Application of microbial risk assessment to the development of standards for enteric pathogens in water used to irrigate fresh produce. J Food Prot 68, 913–918. [DOI] [PubMed] [Google Scholar]
- Strawn LK, Fortes ED, Bihn EA, et al. (2013) Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl Environ Microbiol 79, 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DG, Batten HL, Bosch D, Sheridan J and Strickland T (2007) Little River Experimental Watershed, Tifton, Georgia, United States: A geographic database. Water Resour Res 43, W09471. [Google Scholar]
- Thomas JL, Slawson RM and Taylor WD (2013) Salmonella serotype diversity and seasonality in urban and rural streams. J Appl Microbiol 114, 907–922. [DOI] [PubMed] [Google Scholar]
- USDA National Agricultural Statistics Service (2014) 2012 Census of Agriculture. Available at: https://quickstats.nass.usda.gov.
- USGS (2014) Land cover, elevation, hydrography, and imagery datasets. Available at: https://nationalmap.gov.
- Walters SP, Thebo AL and Boehm AB (2011) Impact of urbanization and agriculture on the occurrence of bacterial pathogens and stx genes in coastal waterbodies of central California. Water Res 45, 1752–1762. [DOI] [PubMed] [Google Scholar]
- Wickham H (2009) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Winfield MD and Groisman EA (2003) Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol 69, 3687–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Long SC, Das D and Dorner SM (2011) Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J Water Health 9, 265–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.