Abstract
The UNC-49 receptor is a unique nematode γ-aminobutyric acid (GABA)-gated chloride channel that may prove to be a novel target for the development of nematocides. Here we have characterized various charged amino acid residues in and near the agonist binding site of the UNC-49 receptor from the parasitic nematode Haemonchus contorts. Utilizing the Caenorhabditis elegans GluCl crystal structure as a template, a model was generated and various charged residues [D83 (loop D), E131 (loop A), H137 (pre-loop E), R159 (Loop E), E185 (Loop B) and R241 (Loop C)] were investigated based on their location and conservation. These residues may contribute to structure, function, and molecular interactions with agonists. It was found that all residues chosen were important for receptor function to varying degrees. Results of the mutational analysis and molecular simulations suggest that R159 may be interacting with D83 by an ionic interaction that may be crucial for general GABA receptor function. We have used the results from this study as well as knowledge of residues involved in GABA receptor binding to identify sequence patterns that may assist in understanding the function of lesser known GABA receptor subunits from parasitic nematodes.
Keywords: Haemonchus contortus, UNC-49 GABA receptor, Mutagenesis, Molecular dynamics, Agonist recognition
Graphical abstract
Highlights
-
•
Key functional residues were investigated in the Hco-UNC-49 receptor.
-
•
A glutamic acid in the binding pocket (loop B) is essential for agonist recognition.
-
•
Interaction between residues in different binding loops are important for function.
1. Introduction
The ligand-gated ion channel family or more specifically cys-loop receptors are an important family of proteins which are targets for current insecticides and anthelmintics. Several of these receptors are targets for anthelmintics that are used to treat a variety of parasitic nematodes that infect animals including humans (Wolstenholme, 2011). The two most widely studied cys-loop receptors are the glutamate-gated chloride channels (GluCls) which are targets for macrocyclic lactone anthelmintics such as ivermectin and the nicotinic acetylcholine receptors which are targets for cholinergic anthelmintics such as levamisole (Holden-dye et al., 2014). However, the cys-loop receptor family of nematodes is quite large with several subtypes having no mammalian orthologues (Jones and Sattelle, 2008). This highlights their potential as targets for novel anthelmintics.
Cys-loop GABA receptors are well-studied targets for insecticides and also play an important role in nematode biology. These receptors are also targets for the anthelmintic piperizine (Accardi et al., 2012). The nematode GABA receptor most studied is the UNC-49 receptor which plays a role in muscle contraction essential for locomotion (Bamber et al., 1999). The importance of this receptor to the biology of nematode parasites is evident from the fact that genes encoding the various subunits of the UNC-49 receptor family are found in many parasitic nematode genomes (Accardi et al., 2012). Moreover, the UNC-49 receptor is not analogous to GABA receptors found in mammals and exhibits a unique pharmacology suggesting structural differences in the binding pocket (Bamber et al., 2003; Kaji et al., 2015). In several cases, differences in binding site residues between the UNC-49 receptor and mammalian GABA receptors have partially explained differences in function (Kaji et al., 2015; Kwaka et al., 2018). However, there is much to be learned about how various residues in and around the binding pocket contribute to the overall function of the nematode UNC-49 receptor as well as other nematode GABA receptors and GABA receptors in general. Such knowledge is important for our understanding of the evolution and function of GABA neurotransmission and the potential of these receptors as targets for future nematocides.
The crystal structure of the Caenorhabditis elegans GluCl (Hibbs and Gouaux, 2011) has provided the means to begin to examine the structure of the UNC-49 receptor from H. contortus which has been key to understanding the residue requirements for receptor function and pharmacology (Kaji et al., 2015). There are many residue types that play key roles in overall receptor function. However, charged residues are particularly important in that they play essential roles in the structure and function of the various binding loops and they have been shown to interact directly with GABA (Ashby et al., 2012; Newell et al., 2004). While many of these residues have been studied in other GABA receptors there is little information on their role in GABA receptors from nematodes.
Molecular dynamic (MD) simulations have been extremely useful in visualizing and quantifying inter- and intramolecular interactions between residues (Ashby et al., 2012; Kwaka et al., 2018). In addition, when coupled with mutational analysis this approach can provide a deeper understanding of receptor function, particularly receptors from parasitic nematodes where there has been limited study. Here we have investigated the function of various charged residues in the UNC-49 receptor from the parasitic nematode H. contortus using mutational analysis, homology modelling and molecular dynamic simulations. We show that this approach can provide novel insight into the function of cys-loop receptors from parasitic organisms which may aid research that focuses on the development of novel anthelmintics. In addition, using the results from this study along with knowledge of human GABAA receptors, we have examined other nematode GABA receptor subunits for the presence or absence of key residues that can be used to explain or predict function.
2. Materials and methods
2.1. Homology modelling
The C. elegans GluCl crystal structure (PDB 3RIF) was used as a template in MODELLER 9.14 (Sali and Blundell, 1993) for the generation of a Hco-UNC-49B extracellular domain homodimer (GenBank # EU939734.1). The most energetically favorable models were determined based on their DOPE and molpdf scores as well as a PROCHECK Ramachandran plot analysis as described in Kaji et al. (2015). The final model chosen was visually inspected to ensure the binding loops were in the proper positions.
2.2. Computational agonist docking
The energetically reduced zwitterion form of GABA was obtained from the Zinc database, http://zinc.docking.org/ (Irwin et al., 2012). The GABA molecule was prepared for docking using AutoDock Tools and was docked using AutoDock Vina (Trott and Olson, 2010). The center of the 30 × 30 × 30 Å search box located in the aromatic box of the agonist binding site was used for agonist docking. A maximum of 50 binding models all within a range of 5 kcal mol−1 from the best scoring pose was generated (Kaji et al., 2015).
2.3. Site-directed mutagenesis and cRNA production
The coding sequence of hco-unc-49b, subcloned into the pT7TS vector was used as template for mutagenesis. Fourteen mutants were generated using Stratagene's web-based QuikChange® Primer Design program (www.stratagene.com/sdmdesigner/decault.aspx) and the QuikChange® site-directed mutagenesis kit. Each mutation was verified by DNA sequencing (Genome Quebec). cRNA was produced by in vitro transcription using T7 RNA polymerase in the mMessage Machine Kit (Ambion) and precipitated using lithium chloride and resuspended in H2O at a concentration of 0.5 ng/L.
2.4. Expression of unc49b mutants in Xenopus oocytes
Expression of unc-49 receptor cRNA was according to methods outlined in Abdelmassih et al. (2018) and adhered to the guidelines of the Canadian Council of Animal Care. Xenopus laevis were anesthetized using 0.15% 3-aminobenzoic acid ethyl ester, methane sulphate salt buffered to pH 7 with sodium bicarbonate. Oocytes were surgically removed and cut into small clumps of approximately 20 eggs per clump. These were incubated in OR-2 (82 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, pH 7.5) with type-II collagenase (Sigma Aldrich, Canada) for 2 h while shaking. These were then placed in ND96 supplemented with pyruvate and gentamycin. Oocytes were injected with 50 nL of either wild-type (hco-unc-49b/c) or mutant hco-unc-49b/ wildtype hco-unc-49c cRNA, using the Drummond Nanoject microinjector. These were stored at 20ᵒC for 48 h, replacing the supplemented ND96 twice before electrophysiology.
2.5. Two-electrode voltage clamp electrophysiology
Approximately 48 h after injection, electrophysiology was performed using the Axoclamp 900A voltage clamp. Responses were recorded using varying concentrations of GABA dissolved in non-supplemented ND96. Microelectrodes were made using Ag|AgCl wires and filled with 3M KCl. Injected X. laevis oocytes were clamped and held at −60 mV and left for 1–2 min to stabilize before GABA was washed over the oocytes at a flow rate of 8 mL/min. For dose-response analysis, oocytes were washed with ND96 for 1–2 min between agonist applications. Data was recorded and analyzed using Clampex 10.3 software and graphs were produced using Graphpad Prism Software 5.0. Data was collected from at least five individual oocytes. To assure consistency, two different batches of oocytes were used.
2.6. Statistical analysis
Dose-response curves were generated using Prism 5.0 (Graphpad Software, San Diego, CA, USA) using the log (agonist) vs. normalized response-variable slope equation:
Where Imax is the maximal response, [D] is concentration of agonist, EC50 is the concentration of agonist required to produce a half maximal response and h is the Hill coefficient. Responses that generated dose-response curves were normalized as a percentage of the maximal current produced by individual oocyte's maximal response to GABA. Statistical analysis comparing EC50 and hill values between wildtype and mutants was performed using Students t-test with the Bonferonni correction. P-values ≤ 0.01 was considered significant.
2.7. MD simulations
MD simulations of the Hco-UNC-49B homodimer with docked GABA were conducted as described in Kwaka et al. (2018). The GROMACS 2016.4 software package (Abraham et al., 2017), employing the CHARMM36 force field (Best et al., 2012), was used to run the simulations. The CHARMM General Force Field (CGenFF) program was used to acquire the CHARMM force field parameters for the GABA molecule (Vanommeslaeghe et al., 2010; Yu et al., 2012; Vanommeslaeghe and MacKerell, 2012; Vanommeslaeghe et al., 2012; CGenFF interface https://cgenff.paramchem.org). The simulation box was solvated with ∼21,000 tip3p water molecules and Cl− counter ions to neutralize the total charge of the system. After a brief energy minimization, a 2 ns equilibration MD simulation was performed under the NVT conditions at 300 K and a time step of 2 fs. The positions of the protein and the ligand were restrained to prevent any structural changes during the equilibration process. In the production run, under the NPT conditions, the system was kept at 300 K and 1 atm with a time step of 2 fs for 500 ns. The Parrinello-Rahman isotropic pressure coupling (with τp = 5 ps and compressibility 4.5 × 10− 5 bar −1) and the Nose-Hoover thermostat (with a time constant of 0.1 ps) were employed for these production runs. Long-range electrostatics were calculated with the particle mesh Ewald method. The trajectory was printed every 10 ps.
The gromacs utilities, such as gmx_mpi mindist, gmx_mpi gyrate and gmx_mpi rms (Humphrey et al., 1996; http://www.ks.uiuc.edu/Research/vmd/) were used to calculate the results. Ionic bonds between oppositely charged residues were determined at d < 3.0 Å.
2.8. Sequence analysis
Amino acid sequences of various GABA receptor subunits were retrieved from GenBank. Sequences were aligned using the Clustal Omega: Multiple Sequence Alignment Tool via the European Bioinformatics Institute portal (www.ebi.ac.uk).
3. Results
3.1. Modelling results
A model of the binding site of Hco-UNC-49B homodimer was generated using the C. elegans GluCl (PDB 3RIF) as a template. This model was used for determining which residues may have functional significance and/or their proximity to residues of opposite charge or the GABA molecule. Based on our model D83, E131, H137, R159, E185 and R241 were chosen for analysis (Fig. 1).
Fig. 1.
(A) Model of Hco-UNC-49B homodimer with docked GABA molecule with residues that were examined in this study. Representative distances are shown in angstroms (B) Sequence alignment of the location of the residues within the major binding loops. The various binding loops corresponding to each residue are indicated by color coding. Residues examined in this study are indicated by * (C) Representative electrophysiological tracings of Hco-UNC-49BC with alanine mutated UNC-49B subunits D83A, E131A, E185A, H137A, R159A, R241A (D) Dose-response curves of all functional Hco-UNC-49B alanine mutants showing differences in GABA sensitivity, with normalized currents. Each point represents a mean ± SE (n ≥ 5). Residue numbering refers to the first methionine in the Hco-UNC-49B subunit (EU939734.1). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Electrophysiology of mutants
Each residue was first examined by alanine mutagenesis. Each mutated receptor was exposed to GABA in increasing concentrations to determine the EC50. Upon recording, it was found that most receptors with mutated subunits responded to GABA, and clean tracings were established for comparisons and determination of the EC50 (Fig. 1; Table 1). Of the six residues that were mutated to alanine, D83A and E185A both had complete lack of function and were unresponsive to GABA concentrations up to 20 mM. The other mutants were functional and EC50 values were determined. E131A exhibited an EC50 of 219 ± 21 μM or a 3.2 fold decrease in sensitivity and had a significant decrease in hill slope (n = 5). H137A was found to have an EC50 of 2174 ± 173 μM for GABA, or a 32 fold decrease in sensitivity (n = 5). R159A was found to have an EC50 of 5469 ± 396 μM, or an 80 fold decrease in sensitivity (n = 7), and R241A was found to have an EC50 of 5174 ± 472.5 μM, or a 76 fold decrease in sensitivity (n = 6). The results are summarized in Fig. 1D and Table 1. From the results of the alanine mutagenesis H137, R159, R241, E185 and D83 were chosen for further analysis since they all appeared to have a major impact on GABA sensitivity.
Table 1.
Summary of EC50 values and hill slopes of each mutation introduced into Hco-UNC-49B. Each EC50 represents the mean ± standard error using n ≥ 5 independent oocytes.
| Mutation | EC50 ± SE (μM) | nH | Mut/WT | N |
|---|---|---|---|---|
| Wild-Type | 68 ± 17 | 2.056 ± 0.2 | 1 | 6 |
| D83A | n.r. <20,000 | – | – | 10 |
| D83E | 276 ± 37a | 2.064 ± 0.2 | 4.1 | 5 |
| D83C | 454 ± 47a | 1.239 ± 0.1a | 6.7 | 5 |
| D83N | 342 ± 30a | 2.285 ± 0.2 | 5.0 | 5 |
| D83T | 520 ± 34a | 1.549 ± 0.1 | 7.6 | 5 |
| E131A | 219 ± 21a | 0.884 ± 0.1a | 3.2 | 5 |
| H137A | 2174 ± 173a | 1.123 ± 0.1a | 32.0 | 5 |
| H137K | 4166 ± 745a | 1.045 ± 0.1a | 61.2 | 5 |
| R159A | 5469 ± 396a | 1.931 ± 0.2 | 80.4 | 7 |
| R159K | n.r. <20,000 | – | – | 11 |
| E185A | n.r. <20,000 | – | – | 13 |
| E185D | n.r. <20,000 | – | – | 5 |
| R241A | 5174 ± 473a | 2.168 ± 0.2 | 76.1 | 6 |
| R241K | 3793 ± 435a | 1.279 ± 0.1a | 55.8 | 6 |
Indicates statistically different from WT (P ≤ 0.005).
3.3. Mutational analysis of residues by conserving charge
As a further analysis of charged residues in and around the binding pocket, we mutated each residue to a different amino acid that was similar in charge. In this case there were two mutants, R159K and E185D, which were non-functional even though the replacement amino acid was the same charge. On the other hand, H137K and R241K produced functional channels but had a 61 and 55-fold reduction in GABA sensitivity, respectively. These results suggested that the original residue is crucial for receptor function and each may have complex roles in the nematode UNC-49 receptor. On the other hand, D83E exhibited only a slight reduction in GABA sensitivity (4-fold reduction) (Table 1). Because D83 appeared to be the most tolerant to mutagenesis this position was further investigated using a series of amino acid changes.
3.4. Examination of D83
While removal of the functional group of D83 through the D83A mutation resulted in non-functional receptors, the introduction of other residues particularity those with a polar group maintained some receptor function. Aside from the D83E mutation, the introduction of similarly sized but polar asparagine (D83N) resulted in only a 5 fold reduction in GABA sensitivity. Similarly, D83T and D83C resulted in an 8 and 6 fold decrease in sensitivity, respectively (Fig. 2; Table 1).
Fig. 2.
Dose-response curves of D83 mutations of Hco-UNC-49B in comparison to WT showing differences in GABA sensitivity, with normalized currents. Each point represents a mean ± SE (n ≥ 5).
3.5. MD simulations
MD simulations were conducted to provide a deeper examination of the above residues and their interactions. During the entire simulation GABA remained with the binding pocket and interacted with nearby residues. First, our simulations revealed that E185 forms an ionic bond with GABA 94% of the time over the course of the simulation (Kwaka et al., 2018). It is not surprising therefore that both E185A and E185D produced non-functional receptors. Simulations also revealed an ionic bond between E185 and R241 which occurred 91% of the time over the course of the simulation (Fig. 3a). These values are similar to a MD study of the Drosophila RDL receptor (Ashby et al., 2012). D83 also interacts with R159 via an ionic bond 45% of the time over the course of the simulation. The simulation shows a tight coupling of D83 and R159 between 80 and 150ns then again from 350 to 500ns (Fig. 3b). However, the simulation did reveal electrostatic interaction between D83 and R159 (ie remained <1 nm distance from each other) during the entire 500ns simulation (Fig. 3b).
Fig. 3.
Molecular simulations of residue interactions in the Hco-UNC-49 receptor. (A) Interactions between E185 and R241 over the course of the simulation. (B) Interactions between D83 and R159 over the course of the simulation. Insets: snapshots of the residues during the simulation. Dashed lines represent distances ≤3 Å. The bonds with the shortest distance are used to calculate bond percentages.
3.6. Identification of key residues in other nematode GABA receptors important for function
The mutational results presented here along with knowledge of agonist recognition residues identified in mammalian GABA binding sites and other studies on the UNC-49 receptor has provided tools to help characterize key features in the nematode GABA receptor family. Fig. 4 provides an alignment of various GABA receptor binding loops from other nematodes along with mammalian αβγ receptors. First, results from the D83 (loop D) mutational analysis revealed that changes from aspartic acid to other polar residues were fairly well tolerated in Hco-UNC-49. Alignments shown in Fig. 4 also suggests variability in amino acids at this position. It appears that the most common residue is D while other nematode GABA receptor subunits exhibit either T, E or N. Interestingly, Ace-LGC-36 from Ancylostoma ceylanicum and Cel-LGC-36 are the only subunits exhibiting an asparagine at this position which we also found to be tolerated in Hco-UNC-49. H137 which was highly sensitive to mutagenizes is highly conserved among GABA receptors with the exception of the EXP-1 receptors which either have an R (Cel-EXP-1) or a K (Bma-EXP-1a). We found that H137K resulted in a large reduction in GABA sensitivity in Hco-UNC-49 so it remains to be seen if Bma-EXP1a is a functional receptor. R159 is highly conserved which is consistent with our results that show that a change to a similarly charged residue (R159K) produced nonfunctional receptors. E185 (Loop B) is a residue that is essential as it interacts with the positive amine group of GABA. All mutations to this position in Hco-UNC-49 produced nonfunctional receptors. LGC-36 and 37 both exhibit a glycine at this position which would explain why Hco-LGC-37 required co-expression with Cel-GAB-1 to produce a functional GABA receptor (Feng et al., 2002). Likewise, R241 (Loop C) was highly sensitive to mutagenesis and is highly conserved among nematode GABA receptors except LGC-36 and 37. Table 2 highlights several of the known key agonist recognition residues that can explain and possibly predict receptor function in parasitic nematodes. First, Hco-UNC-49B, Hco-LGC-38, EXP-1 and Dme-RDL all contain the residues ERRS and all readily form functional homomeric channels in oocytes (Beg and Jorgensen, 2003; Siddiqui et al., 2010; Ashby et al., 2012) (Fig. 5). Cel-GAB-1 (or Hco-GAB-1) and Hco-LGC-37 have been shown to be obligate heteromeric receptor subunits (Feng et al., 2002) and contain ERHG and GVRS respectively. If GAB-1 functions as a beta-subunit (principle subunit) and LGC-37 as an alpha-like subunit (complementary subunit) then the GABA binding site would contain the residues ERRS essential for GABA receptor binding and function. Therefore, we would predict that like LGC-37, Ace-LGC-36 (and other nematode LGC-36 subunits) would require a GAB-1 like subunit to be functionally expressed. Based on the criteria outlined above, Table 2 provides a summary of how these four functionally important resides were used to classify various nematode GABA receptor subunits.
Fig. 4.
Sequence alignment of the major binding loop of Hco-UNC-49B with other Cys-loop GABA receptors. The amino acid residues mutated in this study is indicated by (●) and are numbered above. Residues discussed that are important for agonist recognition described in Table 2 are indicated by (★).
Table 2.
Residues important for agonist recognition and receptor function and used to classify the various cys-loop GABA receptor subunits found in parasitic nematodes.
|
Principle Subunit |
Complementary Subunit |
Comment | |||
|---|---|---|---|---|---|
| Loop B | Loop C | Loop D | Loop E | ||
| Hco-UNC-49B | E | R | R | S | UNC-49 subunit/RDL-like |
| Hco-GAB-1 | E | R | H | G | Beta-like subunit |
| Ace-LGC-36 | G | A | R | S | Alpha-like subunit |
| Hco-LGC-37 | G | V | R | S | Alpha-like subunit |
| Hco-LGC-38 | E | R | R | S | RDL-like subunit |
| Bma-EXP-1 | E | R | R | S | EXP-1 (GABA cation channel) |
| Dme-RDL | E | R | R | S | RDL |
| Hsa-β2 | E | R | Q | G | Beta-subunit |
| Hsa-α1 | G | V | R | T | Alpha-subunit |
| Hsa-γ1 | S | I | A | T | Gamma-subunit |
| Hsa-γ2 | S | V | A | T | Gamma-subunit |
Explanation of residues:
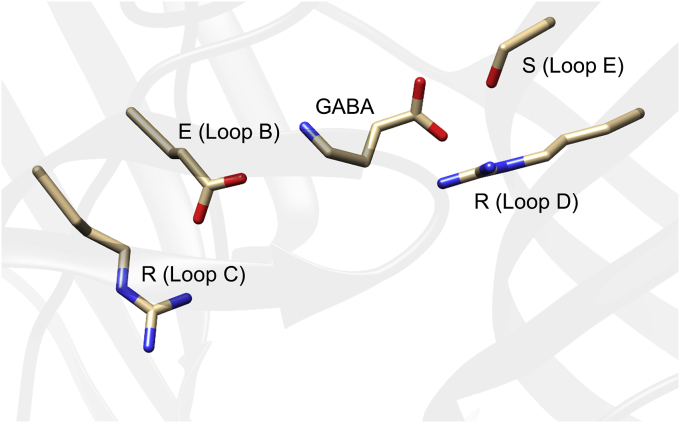
Glutamic acid (E) (Loop B): ionic bond with amine group of GABA (current study; Newell et al., 2004).
Arginine (R) (Loop C): ionic bond the loop B glutamic acid (current study; Ashby et al., 2012).
Arginine (R) (Loop D): ionic bond with carboxyl group of GABA (Accardi and Forrester, 2011; Kwaka et al., 2018).
Serine (S) (Loop E): hydrogen bond with carboxyl group of GABA (Ashby et al., 2012; Kwaka et al., 2018).
NOTE: naming of subunits uses the system outlined in Beech et al. (2010) (Hco- H. contortus).
Fig. 5.
Molecular model of Hco-UNC-49 homodimer with GABA docked. The residues ERRS and their positions are indicated.
4. Discussion
This study has confirmed that a glutamic acid residue in loop B (E185) is absolutely essential for GABA receptor activation in nematode UNC-49 GABA receptor. The analogous residues of E185 are E155 in the mammalian GABAA receptor and E204 in the Drosophila RDL. In both case these residues have been shown to be crucial channel function (Newell et al., 2004; Ashby et al., 2012; Miller et al., 2014). In the Drosophila RDL receptor, like the UNC-49 receptor, all mutations to E204 resulted in channels that were unresponsive. It has been suggested that this loop B glutamic acid acts as a control element where it interacts with the positive amine group of GABA coupling binding to the channel activation (Newell et al., 2004). Not only does E185 form a critical interaction with GABA but it also interacts with an arginine in loop C (R241). Mutations of R241 showed drastically impaired function of the channel, with R241A showing an approximately 80 fold decrease in sensitivity and R241K showing an approximately 59 fold decrease in sensitivity. The crucial role of R241 is also supported by our MD simulations which revealed an ionic bond between R241 and E185. This interaction between analogous residues has also been observed in the human GABAA receptor (Bergmann et al., 2013; Miller et al., 2014) and it may function in the closure of the channel (Miller et al., 2014). It appears therefore that a glutamic acid in loop B and a loop C arginine and their potential interaction are crucial for GABA receptor function across phyla.
D83 appears to be important for receptor function as a mutation to an alanine produced a non-functional channel. Interestingly, maintaining the charge as in D83E produced only a small reduction in GABA sensitivity. Other mutations at this position which included residues capable of hydrogen bonding such as D83N, D83T and D83C also produced functional channels. It had been shown in both MD simulations of the RDL receptor and a homology model of the human GABAA receptor that there is a salt bridge between residues analogous to D83 and R159 which likely play a key role in the stability of binding loops D and E (Ashby et al., 2012; Bergmann et al., 2013). R159 seems to be particular sensitive to mutations to either alanine (current study) or cytosine (Kwaka et al., 2018) which caused severely impaired receptors. Surprisingly, R159K produced non-functional receptors while the same mutation (R178K) in the Drosophila RDL receptor produced a channel that was similar in sensitivity to WT (Ashby et al., 2012). This suggests that in Hco-UNC-49, R159 may have other functions other than interacting with D83. This is also supported by the results of our MD simulations which found that while D83 and R159 interact by an ionic bond, the strength of this interaction and potential frequency was not as high as the equivalent residues in the RDL receptor.
The genomes of parasitic nematodes appear to contain at least five distinct types of genes that encode various subunits of cys-loop GABA-gated chloride channel receptors (Accardi et al., 2012). Of these subunits only two, UNC-49B and LGC-38 have been shown to form functional homomeric channels in Xenopus oocytes which is similar to the Drosophila RDL receptor (Siddiqui et al., 2010, 2012). On the other hand, subunits such as GAB-1 and LGC-37 have been shown to be unable to form functional homomeric channels but can combine to form a GABA sensitive heteromeric channel (Feng et al., 2002). However, the somewhat weak sequence similarity with classical mammalian αβγ GABA receptors makes it difficult to classify with certainty nematode subunits using mammalian nomenclature. Thus, the naming system utilized in Jones and Sattelle (2008) and Beech et al., 2010 is the most practical and useful method for classifying nematode GABA receptors. However, the knowledge that we now have gained though mutational studies, modelling and comparisons to mammalian αβγ GABA receptors can help better describe these subunits and the key functional motifs they possess. Thus, using criteria based on the presence or absence of key functional residues we suggest that GAB-1 is the only true β-like subunit in the nematode genome that provides essential GABA binding residues from loops B and C but lacks the key binding residues in the complementary loops D and E. The loop D and E residues would be provided by LGC-37 which when co-expressed with GAB-1 produces a functional heteromeric receptor (Feng et al., 2002). The LGC-37 residues indicated in Table 1 resulted in our description of LGC-37 as α-like. While both GAB-1 and LGC-37 are not direct orthologues of mammalian β or α subunits, they both appear to share several of the key residues that are essential for the function of each of these subunits. A similar rational can be used to describe the currently uncharacterized subunit LGC-36 as also α-like. This is what makes the UNC-49 receptors different compared to GABAA receptors, in that one subunit, (in this case UNC-49B) possessed all the essential requirements for GABA binding and receptor activation. However, it's important to note that the residues described here do not predict for certainty the function of uncharacterized subunits such as LGC-36 nor provide the complete story about the other nematode subunits as there are several other residues that are essential for agonist recognition and receptor function (Lynagh and Pless, 2014).
In conclusion, this study has uncovered several residues that are important for Hco-UNC-49 receptor structure and agonist recognition. It appears that residues such as E185 and R241 in the nematode UNC-49 receptor play a similar role as GABA receptors from other phyla. On the other hand while R159 appears to be interacting with D83, it may play other roles in the function of the Hco-UNC-49 receptor.
Declarations of interest
None.
Acknowledgements
Research was funded by the Natural Sciences and Engineering Council of Canada (NSERC) (Grant #210290) SGF. The funding body played no role in the design or execution of the study. The authors declare no conflict of interest.
References
- Abdelmassih S.A., Cochrane E., Forrester S.G. Evaluating the longevity of surgically extracted Xenopus laevis oocytes for the study of nematode ligand-gated ion channels. Invertebr. Neurosci. 2018;18:1. doi: 10.1007/s10158-017-0205-z. [DOI] [PubMed] [Google Scholar]
- Abraham M.J., van der Spoel D., Lindahl E., Hess B., the GROMACS development team . 2017. GROMACS User Manual Version 2016.4.www.gromacs.org [Google Scholar]
- Accardi M.V., Forrester S.G. The Haemonchus contortus UNC-49B subunit possesses the residues required for GABA sensitivity in homomeric and heteromeric channels. Mol. Biochem. Parasitol. 2011;178(1–2):15–22. doi: 10.1016/j.molbiopara.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Accardi M.V., Beech R.N., Forrester S.G. Nematode cys-loop GABA receptors: biological function, pharmacology and sites of action for anthelmintics. Invertebr. Neurosci. 2012;12(1):3–12. doi: 10.1007/s10158-012-0129-6. [DOI] [PubMed] [Google Scholar]
- Ashby J.A., McGonigle I.V., Price K.L., Cohen N., Comitani F., Dougherty D.A., Molteni C., Lummis S.C. GABA Binding to an insect GABA receptor: a molecular dynamics and mutagenesis study. Biophys. J. 2012;103(10):2071–2081. doi: 10.1016/j.bpj.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamber B., Beg A., Twyman R., Jorgensen E. The Caenorhabditis elegans unc-49 locus encodes multiple subunits of a heteromultimeric GABA receptor. J. Neurosci. 1999;19(13):5348–5359. doi: 10.1523/JNEUROSCI.19-13-05348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamber B.A., Twyman R.E., Jorgensen E.M. Pharmacological characterization of the homomeric and heteromeric UNC-49 GABA receptors in C. elegans. Br. J. Pharmacol. 2003;138:883–893. doi: 10.1038/sj.bjp.0705119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech R.N., Wolstenholme A.J., Neveu C., Dent J.A. Nematode parasite genes: what's in a name? Trends Parasitol. 2010;26(7):334–340. doi: 10.1016/j.pt.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Beg A.A., Jorgensen E.M. EXP-1 is an excitatory GABA-gated cation channel. Nat. Neurosci. 2003;6(11):1145–1152. doi: 10.1038/nn1136. [DOI] [PubMed] [Google Scholar]
- Bergmann R., Kongsbak K., Sorensen P., Sander T., Balle T. A unified model of the GABAA receptor comprising agonist and benzodiazepine binding sites. PloS One. 2013;8(1) doi: 10.1371/journal.pone.0052323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R.B., Zhu X., Shim J., Lopes P.E.M., Mittal J., Feig M., MacKerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi1 and chi2 dihedral angles. J. Chem. Theor. Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.P., Hayashi J., Beech R.N., Prichard R.K. Study of the nematode putative GABA type-A receptor subunits: evidence for modulation by ivermectin. J. Neurochem. 2002;83(4):870–878. doi: 10.1046/j.1471-4159.2002.01199.x. [DOI] [PubMed] [Google Scholar]
- Hibbs R.E., Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474(7349):54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden-Dye L., Walker R. 2014. Anthelmintic Drugs and Nematicides: Studies in Caenorhabditis elegans. WormBook.http://www.ncbi.nlm.nih.gov/books/NBK116072/ retrieved from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. http://www.ks.uiuc.edu/Research/vmd/ [DOI] [PubMed] [Google Scholar]
- Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.K., Sattelle D.B. The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans. Invertebr. Neurosci. 2008;8(1):41–47. doi: 10.1007/s10158-008-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji M., Kwaka A., Callanan M., Nusrat H., Desaulniers J.P., Forrester S. A molecular characterization of the agonist binding site of a nematode cys-loop GABA receptor. Br. J. Pharmacol. 2015;172:3737–3747. doi: 10.1111/bph.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaka A., Khatami M.H., Foster J., Cochrane E., Habibi S.A., de Hann H.W., Forrester S.G. Molecular characterization of binding loop E in the nematode cys-loop GABA receptor. Mol. Pharm. November 2018;94(5):1289–1297. doi: 10.1124/mol.118.112821. [DOI] [PubMed] [Google Scholar]
- Lynagh T., Pless S.A. Principles of agonist recognition in Cys-loop receptors. Front. Physiol. 2014;5:160. doi: 10.3389/fphys.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Aricescu A. Crystal structure of a human GABA a receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell J.G., McDevitt R.A., Czajkowsi C. Mutation of Glutamate 155 of the GABAA Receptor Β 2 subunit produces a spontaneously open channel: a trigger for channel activity. J. Neurosci. 2004;24(50):11226–11235. doi: 10.1523/JNEUROSCI.3746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Siddiqui S., Brown D., Vijayaraghava T., Forrester S. An UNC-49 GABA receptor subunit from the parasitic nematode Haemonchus contortus is associated with enhanced GABA sensitivity in nematode heteromeric channels. J. Neurochem. 2010;113:1113–1122. doi: 10.1111/j.1471-4159.2010.06651.x. [DOI] [PubMed] [Google Scholar]
- Siddiqui S.Z., Brown D.D., Accardi M.V., Forrester S.G. Hco-LGC-38 is novel nematode cys-loop GABA receptor subunit. Mol. Biochem. Parasitol. 2012;185(2):137–144. doi: 10.1016/j.molbiopara.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., MacKerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force field. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K., MacKerell A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Raman E.P., MacKerell A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme A.J. Ion channels and receptor as targets for the control of parasitic nematodes. Int. J. Parasitol. Drugs Drug Resist. 2011;14(1):2–13. doi: 10.1016/j.ijpddr.2011.09.003. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., He X., Vanommeslaeghe K., MacKerell A.D., Jr. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]