ABSTRACT
Celiac disease (CD) is an immune-mediated enteropathy involving genetic and environmental factors, whose interaction influences disease risk. The intestinal microbiota, including viruses and bacteria, could play a role in the pathological process leading to gluten intolerance. In this study, we investigated the prevalence of pathogens in the intestinal microbiota of infants at familial risk of developing CD. We included 127 full-term newborns with at least one first-degree relative with CD. Infants were classified according to milk-feeding practice (breastfeeding or formula feeding) and HLA-DQ genotype (low, intermediate or high genetic risk). The prevalence of pathogenic bacteria and viruses was assessed in the faeces of the infants at 7 days, 1 month and 4 months of age. The prevalence of Clostridium perfringens was higher in formula-fed infants than in breast-fed over the study period, and that of C. difficile at 4 months. Among breastfed infants, a higher prevalence of enterotoxigenic E. coli (ETEC) was found in infants with the highest genetic risk compared either to those with a low or intermediate risk. Among formula-fed infants, a higher prevalence of ETEC was also found in infants with a high genetic risk compared to those of intermediate risk. Our results show that specific factors, such as formula feeding and the HLA-DQ2 genotype, previously linked to a higher risk of developing CD, influence the presence of pathogenic bacteria differently in the intestinal microbiota in early life. Further studies are warranted to establish whether these associations are related to CD onset later in life.
KEYWORDS: celiac disease, gut microbiota, HLA genotype, pathogenic bacteria, PCR
Introduction
Celiac disease (CD) is an autoimmune enteropathy triggered by dietary gluten in genetically predisposed individuals. Disease development is mediated by the recognition of gluten peptides associated with the HLA class II DQA1/DQB1 heterodimers on antigen presenting cells (APCs) by T cells which trigger an aberrant immune response against self-intestinal structures.1 Gluten-containing foods are common in the Western diet and, on average, the daily gluten intake is estimated to be 5–20 g/day.2 In spite of this generalized exposure to dietary gluten of the population, not all the individuals carrying the HLA-DQ risk genotype finally develop CD, which means that additional genetic and environmental factors are needed to trigger disease onset.3,4
In recent years, alterations in the gut microbiota are being investigated as part of the modifiable factors possibly involved in the CD puzzle. CD patients have a different faecal and duodenal microbiota structure, displaying increases in pathobionts (Clostridium spp. and enterobacteria) and decreases in potential protective bacteria (Bifidobacterium spp. and Lactobacillus group).5,6 Associations have also been established between the HLA-DQ2/DQ8 genotype and the gut microbiota composition, suggesting that the microbiota could also act as a predisposing factor for CD.7,8 Early viral and bacterial infections have been associated with the subsequent development of CD.9,10 A population-based cohort study estimated that children who suffered from more than ten episodes of respiratory or gastrointestinal infections presented a higher risk of developing CD compared to children with less than four infection events.11 Likewise, similar associations have been found by other epidemiological studies enrolling large cohorts and recording only the gastrointestinal infections.12,13 These observations were based on parent or hospital reports where the causal pathogen was unidentified and subclinical infections were not considered.11-13
Some studies have aimed to identify the etiological agents responsible for the association between CD and intestinal infections. A prospective study in children genetically predisposed to CD showed that the high frequency of rotavirus infections increased the risk of developing the disease.14 Also a role was recently established for reovirus in the loss of gluten tolerance.15 In the case of bacterial pathogens causing gastrointestinal infections, the development of CD was linked to previous Campylobacter spp. infection by one case study16; and with a higher incidence of Clostridium difficile infection17; whereas animal models have proven the role of specific enteric bacteria (E. coli ENT CAI:5) in the gluten-induced immunopathology.18 Interestingly, some of these pathogenic bacteria previosly associated with the disease can produce toxins that disrupt the tight junction proteins and increase the intestinal permeability,19-22 a condition linked to the break-down of gluten tolerance.1 Although some potential candidates have been proposed, we lack strong evidence for pathogenic bacteria as trigger factors for CD development in humans.
In the present study, we analysed the prevalence of pathogenic bacteria and virus in the gut microbiota of infants at familial risk of CD development. The broader goal of our research is to gain a greater understanding of how early postnatal environmental factors, and their interaction with host factors, could influence the risk of developing CD.
Materials and methods
Ethical considerations
This study was conducted in accordance with the ethical rules of the Helsinki Declaration (Hong Kong revision, September 1989), following the EEC Good Clinical Practice guidelines (document 111/3976/88 of July 1990) and current Spanish law regulating clinical research in humans (Royal Decree 561/1993 regarding clinical trials). The study was approved by the local Ethic Committees of the CSIC and of the hospitals involved. Written informed consent was obtained from the parents of infants included in the study.
Subjects and sampling
This study included a subset of 127 full-term newborns with at least one first-degree relative suffering from CD, selected from an ongoing larger prospective observational 5-year study described elsewhere.7
The DNA typing for CD HLA-DQA1 and HLA-DQB1 genes was elucidated using sequence-specific primers (Polymerase Chain Reaction-Sequence Specific Primers (PCR-SSP)).23
Infants were classified into three groups by HLA-DQ genotyping. The high risk (HR) included those individuals carrying the DQ2 haplotype in both cis (DQA1*05:01-DQB1*02:01 in homozygosis) and trans conformations (DQA1*02:01-DQB1*02:02 with DQA1*05:05-DQB1*03:01 in heterozygosis), associated with the highest probability (20%) of developing CD. The intermediate risk (IR) included those infants carrying the DQ2 haplotype in cis conformation along with any other haplotype, as well as infants carrying the DQ8 haplotype (DQA1*03:01 DQB1*03:02) in homozygosis. This genotype is associated with a 7% probability of developing CD. The low risk (LR) included the infants with other common genotypes not associated with CD.24,25
Faecal samples were collected at home by the parents at 7 days, 1 month and 4 months of age. Samples were immediately frozen at −20°C and delivered to the hospital for centralization, and stored at −80°C.
DNA extraction and PCR conditions
Faecal samples were prepared as previously described in Palma et al.7 PCR amplification reactions were carried out in a 50 μl volume containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 1 μM of each primer, 0.5 μM of each deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Ecotaq, Ecogen, Spain), and 50 ng of DNA. The amplification products were subjected to gel electrophoresis in 1% agarose gels stained with ethidium bromide. The pathogenic reference strains were from the CECT (Spanish Type Culture Collection) or from bacteria isolated in our laboratory identified by 16S rRNA gene sequencing as described elsewhere.26 Specifically, the strains were: Clostridium perfringens CECT 376, Clostridium difficile CECT 531, Shigella ENT CBD8, enterotoxigenic E. coli (ETEC) CECT 685, enteropathogenic E. coli (EPEC) CECT 729, and Campylobacter jejuni CECT 7572. The primers used were: C. perfringens (16S rDNA) F: ATG CAA GTC GAG CGA (G/T)G, R: TAT GCG GTA TTA ATC T(C/T)C CTT T27; C. difficile (16S rDNA) F: TTG AGC GAT TTA CTT CGG TAA AGA, R: CCA TCC TGT ACT GGC TCA CCT27; Shigella group (ipaH gene coding for invasion plasmid antigen H) F: GTT CCT TGA CCG CCT TTC CGA TAC, R: CAT TTC CTT CAC GGC AGT GGA28; ETEC (elt gene coding for thermolabile toxin) F: GCG ACA AAT TAT ACC GTG CT, R: CCG AAT TCT GTT ATA TAT ATG T29; EPEC (eae gene coding for adhesin) F: AAA CAG GTG AAA CTG TTG CC, R: CTC TGC AGA TTA ACC CTC TGC30; C. jejuni (hip gene coding for the hippuricase enzyme) F: GAA GAG GGT TTG GGT GGT G, R: AGC TAG CTT CGC ATA ATA ACT TG.31
Virus detection
The presence of rotavirus and enteric adenovirus were detected in faecal suspensions (1:10 w/v in PBS) by immunochromatographic technique with commercially available kits (CORIS BioConcept, Gembloux, Belgium) following the manufacturer's instructions.
Statistical analyses
Differences in the prevalence (positive or negative) of the pathogenic bacteria were analysed with the Chi-square test with the Yates' correction to prevent overestimation of statistical significance for small data (count smaller than 5 in one of the cells in the table). Analyses were carried out with GraphPad Prism 7. Classification and Regression Tree (CRT) analysis was also performed to predict the influence of the type of feeding, genetic risk, and age in the prevalence of pathogenic bacteria and virulence factors with SPSS (version V24). Recursive partitioning with the binary cut of entered variables was used for decision tree development and the best separator explaining the presence of each pathogen was chosen for tree root. Statistically significant differences were established at p<0.05.
Results
Subjects included in the study
This study included 127 full-term newborns with at least one first-degree relative suffering from CD. Gestation lasted an average of 38.34 ± 3.80 weeks. A total of 92 cases were born by vaginal delivery, and 35 cases by caesarean section. The mean size of the infants at birth was 49.88 ± 2.47 cm, and the mean weight was 3361 ± 603 grams. Infants were grouped according to feeding practices at 7 days, 1 month and 4 months of age, into formula-fed infants and breast-fed infants. Infants were also classified into three groups according to their genetic risk of developing CD: low risk group (n = 43), intermediate risk group (n = 59), and high risk group (n = 25).
Influence of milk-feeding practices on the prevalence of pathogens
The prevalence of pathogens according to feeding practices (breast-fed versus formula) is shown in Table 1. For all three ages analysed (7 days, 1 month and 4 months) infants fed with formula milk presented a higher prevalence of C. perfringens than breast-fed infants (p = 0.004, p = 0.016 and, p = 0.042, respectively). Besides, at 4 months old, the prevalence of C. difficile was higher in infants fed with formula (p = 0.016).
Table 1.
Prevalence of pathogenic bacteria according to the type of milk feeding at 7 days, 1 month and 4 months of infant's age.
| 7 days |
1 month |
4 months |
|||||||
| Pathogen |
Breast feeding |
Formula feeding |
p value |
Breast feeding |
Formula feeding |
p value |
Breast feeding |
Formula feeding |
p value |
| C. perfringens | 24/73 (32.9) | 23/36 (63.9) | 0.004 | 22/78 (28.2) | 25/49 (51.0) | 0.016 | 15/46 (32.6) | 38/71 (53.5) | 0.042 |
| C. difficile | 21/73 (28.8) | 12/36 (33.3) | 0.790 | 21/78 (26.9) | 17/49 (34.7) | 0.464 | 11/46 (23.9) | 34/71 (47.9) | 0.016 |
| Shigella group | 5/73 (6.8) | 3/36 (8.3) | 1.000 | 8/78 (10.3) | 5/49 (10.2) | 1.000 | 3/46 (6.5) | 9/71 (12.7) | 0.447 |
| Enterotoxigenic E. coli | 4/73 (5.5) | 5/36 (13.9) | 0.258 | 13/78 (16.7) | 4/49 (8.2) | 0.270 | 4/46 (8.7) | 12/71 (16.9) | 0.324 |
| Enteropathogenic E. coli | 9/73 (12.3) | 1/36 (2.8) | 0.203 | 5/78 (6.4) | 3/49 (6.1) | 1.000 | 1/46 (2.2) | 6/71 (8.5) | 0.318 |
| Campylobacter jejuni | 5/73 (6.8) | 0/36 (0.0) | 0.262 | 3/78 (3.8) | 3/49 (6.1) | 0.874 | 5/46 (10.9) | 3/71 (4.2) | 0.310 |
Data are expressed as prevalence (positive/total). The percentage of positive samples is shown in parentheses.
Differences were calculated with the Chi-square test with the Yates' correction. Significant differences were established at p<0.050.
Concerning the detection of virus, of the total number of 353 faecal samples analysed, only two were positive for rotavirus and one for enteric adenovirus. These three positive samples were collected at 4 months of age and corresponded to formula-fed infants.
Influence of the genotype on the prevalence of pathogens
At 4 months of age, infants with a high genetic risk presented a higher prevalence of ETEC, assessed by detecting the elf virulence gene (coding for thermolabile toxin), than the infants with an intermediate genetic risk in both breast-fed (p<0.001) and formula fed infants (p = 0.018) (Table 2). The same trend was observed when comparing the prevalence of this pathogen in low risk versus high risk infants, but the differences were only significant in the sub-group of breast-fed infants (p = 0.019). At 4 months, also in this sub-group of infants, the prevalence of C. difficile was higher in the infants with an intermediate genetic risk compared to a high genetic risk (p = 0.043) and the same trend was observed comparing infants with a low and high genetic risk (p = 0.124).
Table 2.
Prevalence of pathogenic bacteria in breast-fed and formula fed infants with different HLA-DQ genotype at 7 days, 1 month and 4 months of age.
|
p value, |
|||||||||||||
| Breast feeding |
Formula feeding |
Breast feeding |
Formula feeding |
||||||||||
| Pathogen |
Age |
LR |
IR |
HR |
LR |
IR |
HR |
p A |
p B |
p C |
p A |
p B |
p C |
| 7 D | 9/24 (37.5) | 9/36 (25.0) | 6/13 (46.2) | 8/12 (66.7) | 9/15 (60.0) | 6/9 (66.7) | 0.455 | 0.872 | 0.285 | 1.000 | 1.000 | 1.000 | |
| C. perfringens | 1 M | 9/27 (33.3) | 9/38 (23.7) | 4/13 (30.8) | 11/16 (68.8) | 10/21 (47.6) | 4/12 (33.3) | 0.565 | 1.000 | 0.891 | 0.342 | 0.140 | 0.665 |
| 4 M | 5/15 (33.3) | 7/25 (28.0) | 3/6 (50.0) | 15/27 (55.6) | 15/27 (55.6) | 8/17 (47.1) | 1.000 | 0.831 | 0.583 | 1.000 | 0.811 | 0.811 | |
| 7 D | 5/24 (20.8) | 10/36 (27.8) | 6/13 (46.2) | 5/12 (41.7) | 4/15 (26.7) | 3/9 (33.3) | 0.761 | 0.218 | 0.387 | 0.681 | 1.000 | 1.000 | |
| C. difficile | 1 M | 6/27(22.2) | 9/38 (23.7) | 6/13 (46.2) | 5/16 (31.3) | 8/21 (38.1) | 4/12 (33.3) | 1.000 | 0.239 | 0.237 | 0.933 | 1.000 | 1.000 |
| 4 M | 3/15 (20.0) | 4/25 (16.0) | 4/6 (66.7) | 13/27 (48.1) | 15/27 (55.6) | 6/17 (35.3) | 1.000 | 0.124 | 0.043 | 0.785 | 0.600 | 0.317 | |
| 7 D | 2/24 (8.3) | 3/36 (8.3) | 0/13 (0.0) | 1/12 (8.3) | 1/15 (6.7) | 1/9 (11.1) | 1.000 | 0.758 | 0.690 | 1.000 | 1.000 | 1.000 | |
| Shigella group | 1 M | 2/27 (7.4) | 5/38 (13.2) | 1/13 (7.7) | 0/16 (0.0) | 4/21 (19.0) | 1/12 (8.3) | 0.741 | 1.000 | 0.977 | 0.189 | 0.883 | 0.748 |
| 4 M | 0/15 (0.0) | 3/25 (12.0) | 0/6 (0.0) | 2/27 (7.4) | 4/27 (14.8) | 3/17 (17.6) | 0.438 | — | 0.901 | 0.665 | 0.579 | 1.000 | |
| Enterotoxigenic E.coli (ETEC) | 7 D | 1/24 (4.2) | 2/36 (5.6) | 1/13 (7.7) | 0/12 (0.0) | 3/15 (20.0) | 2/9 (22.2) | 1.000 | 1.000 | 1.000 | 0.304 | 0.334 | 1.000 |
| 1 M | 5/27 (18.5) | 6/38 (15.8) | 1/13 (7.7) | 1/16 (6.3) | 0/21 (0.0) | 3/12 (25.0) | 1.000 | 0.671 | 0.791 | 0.890 | 0.391 | 0.974 | |
| 4 M | 1/15 (6.7) | 0/25 (0.0) | 4/6 (66.7) | 4/27 (14.8) | 1/27 (3.7) | 6/17 (35.3) | 0.794 | 0.019 | <0.001 | 0.348 | 0.227 | 0.018 | |
| Enterotoxigenic E.coli (ETEC) | 7 D | 2/24 (8.3) | 4/36 (11.1) | 3/13 (23.1) | 1/12 (8.3) | 0/15 (0.0) | 0/9 (0.0) | 1.000 | 0.454 | 0.552 | 0.909 | 1.000 | — |
| 1 M | 1/27 (3.7) | 2/38 (5.3) | 2/13 (15.4) | 3/16 (18.8) | 0/21 (0.0) | 0/12 (0.0) | 1.000 | 0.501 | 0.566 | 0.144 | 0.332 | — | |
| 4 M | 0/15 (0.0) | 1/25(4.0) | 0/6 (0.0) | 1/27 (3.7) | 4/27 (14.8) | 1/17 (5.9) | 1.000 | — | 1.000 | 0.348 | 1.000 | 0.674 | |
| C. jejuni | 7 D | 2/24 (8.3) | 3/36 (8.3) | 0/13 (0.0) | 0/12 (0.0) | 0/15 (0.0) | 0/9 (0.0) | 1.000 | 0.758 | 0.690 | — | — | — |
| 1 M | 2/27 (7.4) | 0/38 (0.0) | 1/13 (7.7) | 1/16 (6.3) | 2/21 (9.5) | 0/12 (0.0) | 0.330 | 1.000 | 0.570 | 1.000 | 1.000 | 0.730 | |
| 4 M | 0/15 (0.0) | 4/25 (16.0) | 1/6 (16.7) | 3/27 (11.1) | 0/27 (0.0) | 0/17 (0.0) | 0.276 | 0.627 | 1.000 | 0.235 | 0.418 | — | |
Data are expressed as prevalence (positive/total). The percentage of positive samples is shown in parentheses.
Genetic risk of developing CD was established according to the HLA-DQ genotype. Low risk, LR; Intermediate risk, IR; High risk, HR.
Differences were calculated with the Chi-square test with the Yates' correction. p A comparison between LR vs IR; p B comparison between LR and HR; p C comparison between IR vs HR. Significant differences were established at p<0.05. Low risk, LR; Intermediate risk, IR; High risk, HR. 7 days, 7 D; 1 month, 1 M; 4 months, 4.
Classification and regression tree (CRT) analysis.
We use decision tree methodology as a tool for predicting the factors (type of feeding, genetic risk or age) or the combination thereof that predominately influence the presence of pathogenic bacteria in the infants' microbiota.32
In agremeent with the results based of the Chi-square test, the CRT analyses showed that formula was the milk feeding type that better predicted the presence of C. difficile (Fig 1a). According to the CRT analysis, formula feeding was also a better predictor of the presence of C. perfringens (Fig 1b) in infants with an intermediate and high genetic risk. A higher genetic risk (intermediate and high genetic risk groups) also predicted a higher prevalence of C. perfringens (Fig 1b) compared to the low genetic risk. However, the relationship between the high genetic risk and the higher prevalence of ETEC and C. difficile detected by the Chi-square test was not confirmed by the CRT analysis (Fig 1c and 1d). Finally, at the age of 1 month and 4 months the number of positive samples for ETEC was higher than at 7 days (Fig 1e) according to the CRT analysis.
Figure 1.
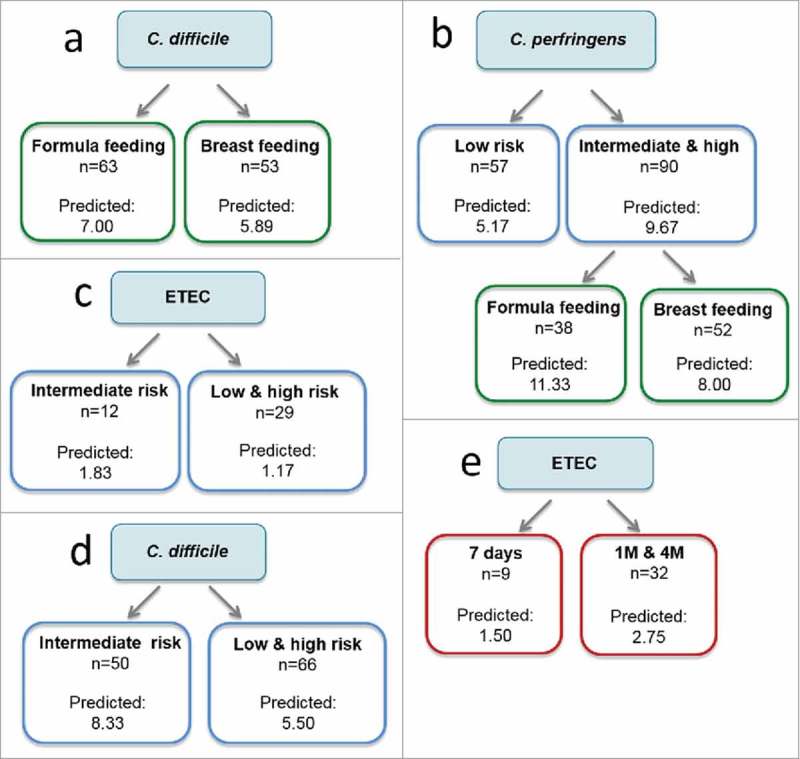
Classification and Regression Trees (CRT) to predict the influence of the type of feeding (green), genetic risk (blue), and age (7 days, 1 month and 4 months, in red) on the prevalence of pathogenic bacteria. CRT splits the data into segments that are as homogeneous as possible with respect to the dependent variable.
Discussion
The prevalence of CD has increased in the last decades, a trend that is not fully explained by an increase in disease awareness and diagnosis efficiency.33,34 This suggests that changes in the interaction between environmental and genetic factors are contributing to a real disease rate increase. Some epidemiological studies have proposed that milk feeding practices and intestinal infections could play a role as predisposing factors, but to date, a causal relationship remains unconfirmed.3,35,36 In the present study, we have assessed the prevalence of six potential pathogenic bacteria and two viruses causing gastrointestinal infections in healthy infants at familial risk of developing CD. Overall, we found that some pathogenic bacteria are specifically associated with the formula feeding pattern (C. perfringens and C. difficile), while others (enterotoxigenic E. coli) seem to be related to the HLA-DQ2 genotype. The performance of the Chi-square test and the CRT algorithms was similar regarding the associations between Clostridium spp. and formula feeding, while some discrepancies were found regarding associations with the genotype.
A previous study incluing of most of the cases enrolled in the PROFICEL project showed that breast-feeding promotes the colonization of Clostridium leptum group and Bifidobacterium species; whereas formula-feeding prometed that of C. coccoides-E. rectale group by quantitative PCR using genus- and group-specific primers.7 Here, using species-specific primers, we have analysed the prevalence of two potentially pathogenic species of the Clostridium genus. The increased prevalence of C. perfringens found at 7 days, 1 month and 4 months of age in the formula-fed infants of our cohort is in agreement with previous studies reporting that formula feeding promoted the presence of C. perfringens.37 This bacterial species has also been considered as a causal agent of necrotizing enterocolitis in infants.38 Similarly, a higher prevalence of C. perfringens has been proposed to possibly increase the risk of suffering other chronic intestinal disorders. Altogether, this evidence could support the protective role attributed to breast-feeding in reducing the rate of infections and, in turn, has been related to a reduced risk of developing CD in some studies.39
Our previous studies with this cohort of infants also led to the conclusion that the HLA genotype influences the early intestinal microbiota composition.7,8 In particular, we described that breast-fed infants with a high genetic risk (HLA-DQ2) of developing CD had reductions in Bifidobacterium spp., a feature that theoretically could be related to the development of the disease in the later life.7,8 Besides, we observed that infants with a high genetic risk (HLA-DQ2) had a higher relative abundance of Proteobacteria, and of unclassified Enterobacteriaceae.8 However, all the preceding studies were taxonomy-based, and the pathogenic potential of the increased abundance of the family Enterobacteriaceae remained unexplored. Here, we describe that, at 4 months of age, the infants with the highest genetic risk of CD development showed a higher prevalence of ETEC, irrespective of milk feeding practices, although this result was only detected by applying a Chi-square test and was not corroborated by the CRT analysis. A potentially pathogenic role of E. coli in the pathogenesis and development of CD has already been proposed.18,26 In humans, the E. coli clones isolated from active and non-active celiac patients carry more virulent genes than the species isolated from healthy controls.26 Moreover, in NOD-DQ8 mice the colonization with E. coli ENT CAI:5 isolated from one CD patient increased the sensitivity to gluten-induced immunopathology and suggested that this could be a contributing factor to the disease.18 The ETEC type is a major cause of diarrhoea,40,41 but it has also been detected in asymptomatic subjects acting as carriers,42-44 who would presumably have an increased risk of developing gastrointestinal disorders.
The influence of the presence of potentially pathogenic bacteria in the overall bacterial ecosystem is uncertain. Correlation analysis of our previous data shows negative associations between Bifidobacterium and Clostridium, and between Bifidobacterium and Escherichia/Shigella.8 Then, it can be hypothesized that the higher prevalence of C. perfringens and C. difficile, and of enterotoxigenic E. coli might be a consequence of the reductions in Bifidobacterium numbers as previously observed in formula-fed infants and the carriers of the HLA-DQ2 genotype.7,8 Unravelling which is the cause or the consequence, as well as the mechanisms that favor the colonization of certain bacterial groups and the co-exclusion of others should be investigated in future studies.
In conclusion, our results show that specific environmental factors, such as formula feeding and the HLA-DQ2 genotype, already linked with a higher risk of developing CD in epidemiological and intervention studies, are also associated with an increased prevalence of pathogenic bacteria in the gut microbiota of infants at a very early age. Theoretically this could predispose subjects to a pro-inflammatory gut ecosystem that could, in turn, favour mucosal permeability alterations and ultimately trigger CD development. Further studies are, however, necessary to demonstrate a causal link between these risk factors and CD development.
Funding Statement
This work was supported by grants AGL2011-25169, AGL2014-52101-P and AGL2007-66126-C03-03/ALI (YS and FP) from the Spanish Ministry of Economy and Competitiveness (MINECO). The scholarship to MO from CSIC (JAEpre) and the contract to ABP from the European Union's Seventh Framework Program under the grant agreement no 613979 (MyNewGut) are also fully acknowledged.
Abbreviations
- APC
Antigen presenting cells
- CD
Celiac disease
- CECT
Spanish Type Culture Collection
- CRT
Classification and Regression Trees
- EPEC
Enteropathogenic E. coli
- ETEC
Enterotoxigenic coli
- PCR-SSP
Polymerase Chain Reaction-Sequence Specific Primers
Contributors
YS conceived the study design, MO and AB-P performed the experimental work, MO and AB-P analysed the data, GDP created the data base, EN, GC, VV, AM, JAG, IP, ED, CR-K, CC and LO recruited and followed-up the infants, AC and FP genotyped the infants, MO and YS drafted the manuscript and all authors read and approved its final version.
Footnotes
AC currently works in the Center for regenerative medicine, Boston university school of medicine, Boston (United States). JAG currently works in the Laboratorio de Genética, Servicio de Análisis Clínicos, Hospital Universitario Rio Hortega y Grupo de Inmunidad de las Mucosas, Facultad de Medicina-IBGM, Universidad de Valladolid (Spain). FP currently works in the Institut de Recerca Sant Joan de Déu and CIBERER, Hospital Sant Joan de Déu, Barcelona (Spain).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Laura Barrios working at the Departmet of Statistics from the Spanish National Research Council (CSIC) for her help with statistical assistance.
References
- 1.Ciccocioppo R, Di Sabatino A, Corazza GR. The immune recognition of gluten in coeliac disease. Clin Exp Immunol. 2005;140(3):408–16. doi: 10.1111/j.1365-2249.2005.02783.x. PMID:15932501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesiekierski JR. What is gluten?. J Gastroenterol Hepatol. 2017;32(Suppl 1):78–81. doi: 10.1111/jgh.13703. PMID:28244676. [DOI] [PubMed] [Google Scholar]
- 3.Vriezinga SL, Auricchio R, Bravi E, Castillejo G, Chmielewska A, Crespo Escobar P, et al.. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. 2014;371(14):1304–15. doi: 10.1056/NEJMoa1404172. PMID:25271603. [DOI] [PubMed] [Google Scholar]
- 4.Dieli-Crimi R, Cenit MC, Nunez C. The genetics of celiac disease: A comprehensive review of clinical implications. J Autoimmun. 2015;64:26–41. doi: 10.1016/j.jaut.2015.07.003. PMID:26194613. [DOI] [PubMed] [Google Scholar]
- 5.Cenit MC, Olivares M, Codoner-Franch P, Sanz Y. Intestinal Microbiota and Celiac Disease: Cause, Consequence or Co-Evolution? Nutrients. 2015;7(8):6900–23. PMID:26287240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marasco G, Di Biase AR, Schiumerini R, Eusebi LH, Iughetti L, Ravaioli F, et al.. Gut Microbiota and Celiac Disease. Dig Dis Sci. 2016;61(6):1461–72. doi: 10.1007/s10620-015-4020-2. PMID:26725064. [DOI] [PubMed] [Google Scholar]
- 7.Palma GD, Capilla A, Nova E, Castillejo G, Varea V, Pozo T, et al.. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS One. 2012;7(2):e30791. doi: 10.1371/journal.pone.0030791. PMID:22319588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivares M, Neef A, Castillejo G, Palma GD, Varea V, Capilla A, et al.. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2015;64(3):406–17. doi: 10.1136/gutjnl-2014-306931. PMID:24939571. [DOI] [PubMed] [Google Scholar]
- 9.Fasano A, Shea-Donohue T. Mechanisms of disease: the role of intestinal barrier function in the pathogenesis of gastrointestinal autoimmune diseases. Nat Clin Pract Gastroenterol Hepatol. 2005;2(9):416–22. doi: 10.1038/ncpgasthep0259. PMID:16265432. [DOI] [PubMed] [Google Scholar]
- 10.Plot L, Amital H. Infectious associations of Celiac disease. Autoimmun Rev. 2009;8(4):316–9. doi: 10.1016/j.autrev.2008.10.001. PMID:18973831. [DOI] [PubMed] [Google Scholar]
- 11.Canova C, Zabeo V, Pitter G, Romor P, Baldovin T, Zanotti R, et al.. Association of maternal education, early infections, and antibiotic use with celiac disease: a population-based birth cohort study in northeastern Italy. Am J Epidemiol. 2014;180(1):76–85. doi: 10.1093/aje/kwu101. PMID:24853109. [DOI] [PubMed] [Google Scholar]
- 12.Beyerlein A, Donnachie E, Ziegler AG. Infections in early Life and development of celiac disease. Am J Epidemiol. 2017;186(11):1277–80. doi: 10.1093/aje/kwx190. PMID:28637333. [DOI] [PubMed] [Google Scholar]
- 13.Marild K, Kahrs CR, Tapia G, Stene LC, Stordal K. Infections and risk of celiac disease in childhood: a prospective nationwide cohort study. Am J Gastroenterol. 2015;110(10):1475–84. doi: 10.1038/ajg.2015.287. PMID:26346866. [DOI] [PubMed] [Google Scholar]
- 14.Stene LC, Honeyman MC, Hoffenberg EJ, Haas JE, Sokol RJ, Emery L, et al.. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am J Gastroenterol. 2006;101(10):2333–40. doi: 10.1111/j.1572-0241.2006.00741.x. PMID:17032199. [DOI] [PubMed] [Google Scholar]
- 15.Bouziat R, Hinterleitner R, Brown JJ, Stencel-Baerenwald JE, Ikizler M, Mayassi T, et al.. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science. 2017;356(6333):44–50. doi: 10.1126/science.aah5298. PMID:28386004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdu EF, Mauro M, Bourgeois J, Armstrong D. Clinical onset of celiac disease after an episode of Campylobacter jejuni enteritis. Can J Gastroenterol. 2007;21(7):453–5. doi: 10.1155/2007/169591. PMID:17637949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebwohl B, Nobel YR, Green PHR, Blaser MJ, Ludvigsson JF. Risk of Clostridium difficile Infection in Patients With Celiac Disease: A Population-Based Study. Am J Gastroenterol. 2017;112:1878–1884. doi: 10.1038/ajg.2017.400. PMID:29087398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galipeau HJ, McCarville JL, Huebener S, Litwin O, Meisel M, Jabri B, et al.. Intestinal microbiota modulates gluten-induced immunopathology in humanized mice. Am J Pathol. 2015;185(11):2969–82. doi: 10.1016/j.ajpath.2015.07.018. PMID:26456581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glotfelty LG, Zahs A, Hodges K, Shan K, Alto NM, Hecht GA. Enteropathogenic E. coli effectors EspG1/G2 disrupt microtubules, contribute to tight junction perturbation and inhibit restoration. Cell Microbiol. 2014;16(12):1767–83. doi: 10.1111/cmi.12323. PMID:24948117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubreuil JD. Enterotoxigenic Escherichia coli targeting intestinal epithelial tight junctions: An effective way to alter the barrier integrity. Microb Pathog. 2017;113:129–34. doi: 10.1016/j.micpath.2017.10.037. PMID:29079214. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein J, Morris WE, Loidl CF, Tironi-Farinati C, McClane BA, Uzal FA, et al.. Clostridium perfringens epsilon toxin increases the small intestinal permeability in mice and rats. PLoS One. 2009;4(9):e7065. doi: 10.1371/journal.pone.0007065. PMID:19763257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johal SS, Solomon K, Dodson S, Borriello SP, Mahida YR. Differential effects of varying concentrations of clostridium difficile toxin A on epithelial barrier function and expression of cytokines. J Infect Dis. 2004;189(11):2110–9. doi: 10.1086/386287. PMID:15143480. [DOI] [PubMed] [Google Scholar]
- 23.Olerup O, Aldener A, Fogdell A. HLA-DQB1 and -DQA1 typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Tissue Antigens. 1993;41(3):119–34. doi: 10.1111/j.1399-0039.1993.tb01991.x. PMID:8316943. [DOI] [PubMed] [Google Scholar]
- 24.Donat E, Planelles D, Capilla-Villanueva A, Montoro JA, Palau F, Ribes-Koninckx C. Allelic distribution and the effect of haplotype combination for HLA type II loci in the celiac disease population of the Valencian community (Spain). Tissue Antigens. 2009;73(3):255–61. doi: 10.1111/j.1399-0039.2008.01191.x. PMID:19254257. [DOI] [PubMed] [Google Scholar]
- 25.Bourgey M, Calcagno G, Tinto N, Gennarelli D, Margaritte-Jeannin P, Greco L, et al.. HLA related genetic risk for coeliac disease. Gut. 2007;56(8):1054–9. doi: 10.1136/gut.2006.108530. PMID:17344279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez E, Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Reduced diversity and increased virulence-gene carriage in intestinal enterobacteria of coeliac children. BMC Gastroenterol. 2008;8:50. doi: 10.1186/1471-230X-8-50. PMID:18983674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–77. doi: 10.1111/j.1365-2672.2004.02409.x. PMID:15546407. [DOI] [PubMed] [Google Scholar]
- 28.Islam MS, Hossain MS, Hasan MK, Rahman MM, Fuchs G, Mahalanabis D, et al.. Detection of Shigellae from stools of dysentery patients by culture and polymerase chain reaction techniques. J Diarrhoeal Dis Res. 1998;16(4):248–51. PMID:10453122 [PubMed] [Google Scholar]
- 29.Tornieporth NG, John J, Salgado K, de Jesus P, Latham E, Melo MC, et al.. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J Clin Microbiol. 1995;33(5):1371–4. PMID:7615758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, Yamasaki S, et al.. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J Clin Microbiol. 2002;40(6):2009–15. doi: 10.1128/JCM.40.6.2009-2015.2002. PMID:12037056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linton D, Lawson AJ, Owen RJ, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35(10):2568–72. PMID:9316909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song YY, Lu Y. Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry. 2015;27(2):130–5. PMID:26120265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubio-Tapia A, Ludvigsson JF, Brantner TL, Murray JA, Everhart JE. The prevalence of celiac disease in the United States. Am J Gastroenterol. 2012;107(10):1538–44; quiz 7, 45. doi: 10.1038/ajg.2012.219. PMID:22850429. [DOI] [PubMed] [Google Scholar]
- 34.Murray JA, Van Dyke C, Plevak MF, Dierkhising RA, Zinsmeister AR, Melton LJ 3rd. Trends in the identification and clinical features of celiac disease in a North American community, 1950–2001. Clin Gastroenterol Hepatol. 2003;1(1):19–27. doi: 10.1053/jcgh.2003.50004. PMID:15017513. [DOI] [PubMed] [Google Scholar]
- 35.Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, et al.. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med. 2014;371(14):1295–303. doi: 10.1056/NEJMoa1400697. PMID:25271602. [DOI] [PubMed] [Google Scholar]
- 36.Silano M, Agostoni C, Sanz Y, Guandalini S. Infant feeding and risk of developing celiac disease: a systematic review. BMJ Open. 2016;6(1):e009163. doi: 10.1136/bmjopen-2015-009163. PMID:26810996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagpal R, Tsuji H, Takahashi T, Nomoto K, Kawashima K, Nagata S, et al.. Gut dysbiosis following C-section instigates higher colonisation of toxigenic Clostridium perfringens in infants. Benef Microbes. 2017;8(3):353–65. doi: 10.3920/BM2016.0216. PMID:28504574. [DOI] [PubMed] [Google Scholar]
- 38.Dittmar E, Beyer P, Fischer D, Schafer V, Schoepe H, Bauer K, et al.. Necrotizing enterocolitis of the neonate with Clostridium perfringens: diagnosis, clinical course, and role of alpha toxin. Eur J Pediatr. 2008;167(8):891–5. doi: 10.1007/s00431-007-0614-9. PMID:17952466. [DOI] [PubMed] [Google Scholar]
- 39.Szajewska H, Chmielewska A, Piescik-Lech M, Ivarsson A, Kolacek S, Koletzko S, et al.. Systematic review: early infant feeding and the prevention of coeliac disease. Aliment Pharmacol Ther. 2012;36(7):607–18. doi: 10.1111/apt.12023. PMID:22905651. [DOI] [PubMed] [Google Scholar]
- 40.Mansour A, Shaheen HI, Amine M, Hassan K, Sanders JW, Riddle MS, et al.. Diarrhea burden due to natural infection with enterotoxigenic Escherichia coli in a birth cohort in a rural Egyptian community. J Clin Microbiol. 2014;52(7):2595–603. doi: 10.1128/JCM.00215-14. PMID:24829232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18(3):465–83. doi: 10.1128/CMR.18.3.465-483.2005. PMID:16020685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahl JW, Sistrunk JR, Baby NI, Begum Y, Luo Q, Sheikh A, et al.. Insights into enterotoxigenic Escherichia coli diversity in Bangladesh utilizing genomic epidemiology. Sci Rep. 2017;7(1):3402. doi: 10.1038/s41598-017-03631-x. PMID:28611468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabal A, Garcia-Castillo M, Canton R, Gortazar C, Dominguez L, Alvarez J. Prevalence of Escherichia coli Virulence Genes in Patients with Diarrhea and a Subpopulation of Healthy Volunteers in Madrid, Spain. Front Microbiol. 2016;7:641. doi: 10.3389/fmicb.2016.00641. PMID:27199966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins C, Chart H, Willshaw GA, Cheasty T, Tompkins DS. Association of putative pathogenicity genes with adherence characteristics and fimbrial genotypes in typical enteroaggregative Escherichia coli from patients with and without diarrhoea in the United Kingdom. Eur J Clin Microbiol Infect Dis. 2007;26(12):901–6. doi: 10.1007/s10096-007-0388-z. PMID:17899229. [DOI] [PubMed] [Google Scholar]


