Abstract
For the past three decades, the use of genomics to inform drug discovery and development pipelines has generated both excitement and scepticism. Although earlier efforts successfully identified some new drug targets, the overall clinical efficacy of developed drugs has remained unimpressive, owing in large part to the heterogeneous causes of disease. Recent technological and analytical advances in genomics, however, have now made it possible to rapidly identify and interpret the genetic variation underlying a single patient’s disease, thereby providing a window into patient-specific mechanisms that cause or contribute to disease, which could ultimately enable the ‘precise’ targeting of these mechanisms. Here, we first examine and highlight the successes and limitations of the earlier phases of genomics in drug discovery and development. We then review the current major efforts in precision medicine and discuss the potential broader utility of mechanistically guided treatments going forward.
Since the beginning of the human genome project, there has been exceptional enthusiasm for how genetics and eventually genomics would transform drug discovery. With the benefit of hindsight, it is now clear that much of this enthusiasm was, at best, premature. With the recent rapid evolution in genomic technologies, we are entering a new phase in genomics, one in which it is now possible to comprehensively characterize the genomes of both patients and healthy individuals. Importantly, the development of sequencing technologies has been paired with a transition towards integrating genomic data with electronic medical records, ultimately facilitating the generation of a data commons useful for identifying relationships between genomic variation and clinical presentation1.
This new phase of genomics, which is increasingly referred to as precision medicine, has sparked a new chapter in the relationship between genomics and drug development — one we argue will be laborious and lengthy but eventually substantially more productive than any of the earlier phases. Here, we first review the key recent phases of the relationship between genomics and drug development. Next, we describe the core elements of precision medicine and how advances in the field are expected to influence both drug development and drug use. We end with additional considerations and challenges we expect precision medicine to confront. Our key message is that above all, precision medicine is a new window into the biology of disease, and this new understanding of the physiological and molecular basis of disease will transform drug development and clinical use.
Early efforts, ESTs and drug discovery
The mid-20th century began what has since been termed the ‘golden age’ of drug discovery2–4, the start of which was largely characterized by observation-based discoveries from the phenotypic screening of animals, whole organs or tissues using synthetic small molecules5,6. This ‘molecular roulette’ approach gave rise to many of the medicines in current use, including anti biotics and immunosuppressants4,5. However, the overall productivity of this approach began to decline, and by the 1970s, a general push towards understanding the biological functions and structural properties of putative drugs and their targets (that is, rational drug development) arose4,7. For a few decades thereafter, drug discovery was dominated by a ‘function-to-gene’ approach8, which focused on investigating a specific protein as a potential drug target, such as one associated with or causative of a particular disease. This type of approach gave rise to drugs that treat a variety of chronic diseases, such as captopril (an angiotensin-converting enzyme (ACE) inhibitor) and other cardio vascular medicines4. Nevertheless, by the early 1990s, an appreciable decline in pharmaceutical productivity began, partly owing to a lack of appropriately validated new drug targets. This period, however, coincided with major advances in both computation and biotechnology, which together cultivated the field of genomics as an innovator for the pharmaceutical industry.
In the early 1990s, the discovery and use of expressed sequencing tags (ESTs) as a tool to transcriptionally profile populations of cells were the first major technological advances that led to the widespread and systematic integration of genomics into drug discovery pipelines and spearheaded a transition to a ‘gene-to-screen’ approach by industry8–10. The main assumption underlying this approach is that the complex combination of genes expressed in a given cell is a primary contributor to the overall cellular phenotype. Thus, in the context of disease, differentially expressed genes in dysfunctional cell populations or tissues could be causally involved in the disease process and could therefore serve as possible drug targets11,12. EST profiling not only provided the ability to detect these differences in gene expression but also led to the identification of new genes and dysregulated pathways, all of which delivered a plethora of new targets with potential therapeutic implications. The more ideal candidates, such as those with predicted druggability and/or those with gene expression limited to the cell or tissue types of interest, were then chosen for cloning and additional functional analyses, and if still promising, these analyses were followed up with high-throughput compound screening or rational drug design8. Of course, the challenge in such work was to discriminate causal from incidental differences. In fact, this highlights a general challenge in ‘omic’ studies, in which a signal is easy to find, but very low P values can often make it too easy to confuse causation and correlation, and discriminating between these two is crucial in the context of drug development.
By shedding light on disease-associated genes and pathways, EST profiling began to shift the focus in drug discovery and development from alleviating symptoms to attacking the underlying mechanisms of disease. Furthermore, this approach substantially expanded the array of potential targets, in turn providing wide-spread optimism that genomics could help resolve the burgeoning pharmaceutical productivity crisis of the early 1990s6. Accordingly, the late 1990s and early 2000s saw a surge of partnerships between genomics firms that developed proprietary databases of expression data and pharmaceutical companies that had expertise and resources in drug development4,6,13. By 2001, pharmaceutical companies allocated nearly 60–70% of their discovery portfolios to drugs with novel targets14. Nevertheless, from 1991 to 2000, drugs based on novel targets had nearly a 40% lower portfolio retention rate compared with those with clinically validated targets14. Although numerous factors have been demonstrated to be at fault, one potential contributor was that early genomics transformed the bottleneck of the productivity crisis from target discovery to effective prioritization of numerous candidate targets. Given that the genomics leads were both plentiful and often of little direct value in terms of elucidating the biological functions of the targets6, there was no immediately available basis for prioritization. Consequently, from 1978 to 2002, there was a tenfold increase in research and development spending, and this increase, paired with an inflated number of false positives and growing drug attrition rates, led to an overall continued decline in both pharmaceutical productivity and optimism in this approach15. Target validation, through either a robust genetic link between the target and the disease or a greater understanding of the role played by the target in the disease aetiology, improved the value of genetic information. For example, in the AstraZeneca portfolio from 2005 to 2010, targets with stronger validation of the biological role of the target in human disease were less likely to fail in clinical development owing to a lack of efficacy16. Furthermore, a broad analysis identified an increased proportion of targets with direct genetic evidence as the development pipeline progresses through to approval17.
GWAS and drug discovery
The rising challenge of clinical trial failures led drug developers to seek information that better identified targets worth pursuing. This coincided with the publication of the human genome sequence in 2003, followed by the characterization and arrangement of nearly three million single nucleotide polymorphisms (SNPs) into genome-wide haplotype maps as part of the International HapMap project18, both of which provided the tools necessary to begin to understand the relationship between common human genomic variation and complex diseases. This was primarily accomplished through genome-wide association studies (GWAS) — an unbiased and cost effective approach to genotyping and comparing much of the common genomic variation of cohorts with particular diseases or traits to that of control populations. GWAS are grounded on the principle of linkage disequilibrium at a population level between common SNPs or ‘markers’ and disease-causing variants. Markers that associate with the disease more often than expected indicate the presence of causal variants in the genomic vicinity of the associated variants, which, in theory, could be identified through additional genetic analyses, such as fine mapping or re-sequencing.
From a drug discovery standpoint, identifying genetic determinants of common disease through GWAS provided valuable knowledge about the genetic architecture of disease and potential pointers to underlying causative mechanisms and engendered the promise of discovering better candidate targets for common, complex diseases such as diabetes, cancer and autoimmune conditions19,20. From 2007 to 2012, nearly 2,000 significantly associated loci of complex diseases and traits were discovered, which translated to nearly 2,000 new potential pharmacological leads21. Some of these loci were found to be associated with multiple diseases, such as SNPs discovered at the IL23R locus that were associated with several autoimmune conditions including psoriasis, inflammatory bowel disease and ankylosing spondylitis22–25. These pleiotropic genes were of particular interest given their potential as targets for blockbuster drugs treating several diseases19. Monoclonal antibodies targeting interleukin-23 (IL-23) and/or IL-12 (along with the downstream cytokine IL-17) for the treatment of various immune-mediated conditions are already used clinically or are in clinical trials with promising results26–31 (NCT02407223, NCT02204397 and NCT02698475).
Despite these efforts, the vast majority of well-accepted disease-associated loci have yet to be biologically explained, which has hindered the widespread implementation of GWAS findings in drug discovery pipelines. GWAS data often fail to uncover causal variants and, in many cases, even fail to implicate specific genes. GWAS findings often implicate numerous candidate genomic regions, which can include nearly all the genes close to the identified signal: as such, concrete guidance for drug development is limited. A hallmark example involves the FTO locus, in which multiple intronic SNPs within the gene encoding FTO (α-keto-glutarate-dependent dioxygenase) were shown to be strongly enriched in individuals with increased body mass. This finding was highly significant and replicated across different ethnic populations and age groups with an overall estimated 1.2-fold increase in the risk of obesity32–36. Although FTO has been the subject of intense investigation to determine its contribution to obesity, a solid causal link has yet to be established, and the roles of other genes under regulatory control of the FTO locus, such as IRX3 and IRX5, have recently been implicated37,38.
The challenge in making effective use of most GWAS findings in the context of drug development has been formally explained recently by Pritchard39 and colleagues in the form of their omnigenic model. They note that for many human traits and diseases, there are as many as 100,000 contributing SNPs that are distributed throughout the genome. In the early days of GWAS, a similar explanation was proposed for the genetics of human height using a strictly orthogonal approach. Relying on the effect size distribution of height-associated SNPs as a basis for estimating the full distribution of effect size for all height variants, including those that remained undiscovered, it was inferred that as many as 93,000 independent variants would be required to account for the heritability of height40. To explain this observation, Pritchard and colleagues argue in their omnigenic model that these small-effect ‘peripheral’ genes (theoretically any gene expressed in the disease-relevant cell type) are part of such highly interconnected cellular networks that any single weak-effect perturbation on peripheral genes could affect the regulation of strong-effect ‘core’ disease genes (consistent with the ‘small-world’ property of networks). The model suggests that the sum of these numerous small effects on core genes could substantially alter disease expression. In addition, although information on peripheral gene involvement from GWAS could be useful in determining individualized disease risks, drug discovery and development would instead clearly benefit far more from the firm identification and targeting of the strong-effect core genes. To the extent that this model is accurate, it provides a compelling explanation of why the vast majority of GWAS findings have had little relevance to drug discovery.
PCSK9: ‘nature’s gift’ to drug discovery
We can identify one final phase before the transition to precision medicine, which relies upon the use of genomics to identify gene variants that point directly to new targets for treating common conditions. The overarching idea was that more successful preclinical models could be developed using these ‘experiments of nature’ for the initial target validation41. Recent technological advances have increased the efficacy and speed and reduced the cost of high-throughput sequencing, which has improved our ability to characterize and interpret human genomic variation — particularly rare variation. By understanding the phenotypic effects of a spectrum of rare mutations ranging from loss-of-function to gain-of-function mutations within a single gene, genetics provides information on the putative efficacy and/or toxic effects resulting from the modulation of that particular gene product in humans. This knowledge thereby builds confidence in the rationale for targeting that gene product for the treatment of a more common human disease, rather than relying on information gained from less predictive animal or cellular models. Although this approach still relies on genomics to directly inform drug development, it is distinct from precision medicine in that the choice of treatment does not take into account the patient’s underlying mechanism of disease.
The poster child for this approach is proprotein convertase subtilisin/kexin type 9 (PCSK9), a target for the treatment of high cholesterol. In 2003, gain-of-function mutations in PCSK9 were identified in French families with autosomal dominant hypercholesterolaemia, a condition associated with an increase in the risk of early-onset cardiovascular disease42. Loss-of-function mutations were also subsequently identified in association with both low plasma levels of low-density lipoprotein cholesterol (LDL-C) in African American and white cohorts and an overall decreased risk of coronary events with no notable health problems43–46. Functional analyses demonstrated that PCSK9 is a hepatic secretory protein that can enter the circulation and bind to LDL receptors (the primary source for clearance of circulating cholesterol), ultimately mediating LDL receptor endocytosis and subsequent degradation47,48. The combination of these genetic, phenotypic and molecular findings served as the basis for pursuing PCSK9 as a drug target with the rationale that PCSK9 inhibition would result in a surplus of available LDL receptors, thereby reducing plasma LDL-C levels and lowering the risk of adverse cardiac events49. A series of clinical trials were conducted using humanized monoclonal antibodies against PCSK9 in treating populations of patients stratified based on cholesterol levels, cardiovascular disease risks, use of other lipid-modifying enzymes and duration of treatment, and the US Food and Drug Administration (FDA) has since approved two monoclonal antibodies (alirocumab and evolocumab) for the treatment of high cholesterol not adequately controlled by statins or diet49 (see Further information). These approvals were based largely on evidence demonstrating a significant reduction (36–60%) in LDL-C following treatment. Additional trials evaluating the efficacy in preventing adverse cardiovascular events (such as cardiovascular death, myocardial infarction and stroke) are ongoing (NCT01624142, NCT01764633 and NCT01663402). Thus far, results from the FOURIER trial support a lower incidence of adverse cardiovascular events in patients with atherosclerotic cardiovascular disease treated with evolocumab while on a statin therapy, with no apparent increase in other adverse outcomes such as new-onset diabetes or neurocognitive side effects50.
The promise of this approach in identifying and validating drug targets with overall higher chances for successful clinical trial outcomes and lower drug attrition rates has seemingly revived the excitement for genetics in drug discovery41,51,52. In the past few years, new partnerships and initiatives among the pharmaceutical industry have been established, at least partly in an effort to carry out larger, systematic hunts for these ‘experiments of nature’ through genome sequencing and extreme phenotyping of large patient populations. In 2012, Amgen acquired the Iceland-based genomics company deCODE and have since published a set of nearly 7,000 rare loss-of-function mutations in a large Icelandic population for use in their drug discovery pipeline53. Regeneron Pharmaceuticals has partnered with Geisinger Health System, together forming the DiscovEHR collaboration, with the aim of performing genomic analyses on 250,000 patients and correlating these data with patients’ clinical records. This initiative has already revealed a role for ANGPTL4 in coronary artery disease, as loss-of-function mutations are associated with good lipid profiles and a lower risk of coronary artery disease54. These findings mirror the effects of monoclonal antibodies in animal models54, further supporting ANGPTL4 as a candidate drug target. Although this overall approach to drug discovery is certainly compelling and has led to enthusiastic reviews, it remains unclear how many of these pointers human genomic variation will ultimately provide. With the notable exceptions of drugs targeting sclerostin (encoded by SOST) and the α-subunit of the voltage-gated sodium channel protein type 9 Nav1.7 (encoded by SCN9A) (BOX 1), early attempts to utilize this approach have rarely resulted in new drugs being discovered and developed. Hence, it seems likely that such pointers to generally useful medicines, regardless of underlying causes of disease in individual patients, may be fairly rare.
Box 1 |. Gene to drug: SOST and SCN9A.
In the late 1990s, before the advent of next-generation sequencing, Brunkow and colleagues149 examined 22 families from an Afrikaner population in South Africa who had sclerosteosis, a rare, severe sclerosing skeletal dysplasia that results in massive bone overgrowth. Through genetic mapping and positional cloning, they identified mutations in a novel gene, SOST149. The product of SOST, sclerostin, is an inhibitor of the activity of osteoblasts, key bone-resorbing cells, and is produced in the bone by osteocytes150. Sclerostin was identified as a drug target for a potential anabolic treatment to restore lost bone. The monoclonal antibody romosozumab, which binds to sclerostin, was developed with the therapeutic intention of increasing bone formation151. Romosozumab increases bone formation and decreases bone resorption152, and phase III data in postmenopausal women demonstrated a 73% lower risk of spine fracture over 12 months compared with placebo153. If approved, romosozumab would be the first in a new class of anabolic therapies for osteoporosis and would build confidence in the use of genomics to identify new drug targets and thus the promise of precision medicine beyond oncology. Following a complete response letter for the biologics licence application in July 2017, two further phase III romosozumab studies have been included in a resubmission to the FDA (see Further information).
Another promising example of a human-validated target is the discovery of the role of SCN9A in pain. In 2004, a group of investigators published results from a linkage analysis performed on a Chinese family with a rare autosomal dominant form of erythromelalgia154. This analysis led to the identification of a missense mutation in SCN9A, which encodes the α-subunit of the voltage-gated sodium channel Nav1.7. This study also identified a second missense mutation in SCN9A in a sporadic case of erythromelalgia. Subsequently, linkage analysis of a cohort of 11 families and 2 sporadic cases of paroxysmal extreme pain disorder reported 8 missense mutations in SCN9A155. Functional analyses of three of these mutations demonstrated a persistent sodium current, suggestive of gain-of-function effects. Interestingly, a separate study found three different homozygous null mutations in SCN9A in three consanguineous families with congenital indifference to pain, a condition characterized by the absence of nociception156. Single nucleotide polymorphisms in SCN9A have also been suggested to alter pain perception in the general population157,158. Following these discoveries, there has been a push to develop inhibitors and modulators of Nav1.7 for the treatment of pain. Drugs including raxatrigine and funapide are currently in phase II/III clinical trials (NCT02935608 and NCT02365636).
Precision medicine and drug development
At its core, precision medicine embodies an effort to understand the underlying cause of disease in individual patients. By definition, this opportunity in drug development is distinct from any of the earlier described phases. Once the underlying cause of a patient’s disease is identified, this information can then be used to gain new insights into the underlying basic biology and disease pathogenesis, which will ultimately foster the discovery of treatments targeting the precise cause of disease. Development of the medicine will then be done for those patient populations most likely to benefit. Inevitably, this will result in a transition away from the production of ‘one-size-fits-all’ treatments towards targeted treatments that are more efficacious in small groups of patients. Driving the transition from a stochastic clinical practice model, precision medicine therapies will also require the co-development of diagnostic tools to identify the optimal treatment for individual patients. In the rest of this section, we discuss developments in precision medicine along with the opportunities they present for drug development. We draw examples in particular from those therapeutic areas that are expected to advance most quickly towards a precision medicine framework, including genetic disorders with a strong genetic component (particularly neurological diseases such as epilepsy) and cancer.
Cancer
The field of oncology has been a clear pioneer of precision medicine. The move towards personalized therapies was likely in part due to the general cytotoxicity and severe side effects of existing ‘one-size-fits-all’ cancer drugs along with the identification of associated tumour-specific vulnerabilities as potential drug targets. Classic chemotherapy agents comprise a broad spectrum of drugs with variable efficacies for different tumour types that often affect both cancerous and vulnerable healthy cells. Therapies can now be designed to more precisely target cancer cells through two primary methods: selectively disrupting pathways necessary for cancer cell survival or growth (pathway-based targeted therapy), and artificially modulating patients’ immune systems to generate a response against cancer cells (immunotherapy) (FIG. 1).
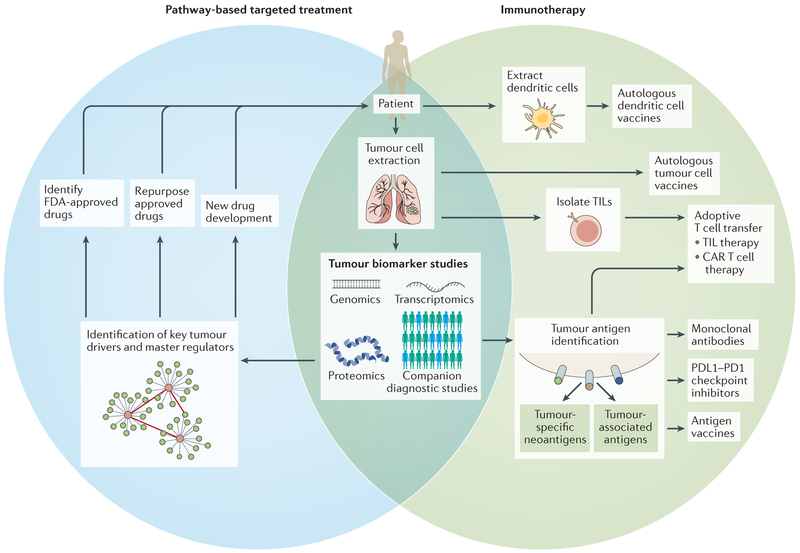
Figure 1 |. Precision therapy approaches in oncology.
Precision therapies in cancer generally use two primary approaches: pathway-based targeted therapies and immunotherapies. For both approaches, access to tumour cells (through resection or biopsy of solid tumours, or blood sample for haematological cancers or circulating tumour cells) enables an investigation into tumour biomarkers using various tools, including companion diagnostics, next-generation sequencing, gene expression profiling and proteomics. For pathway-based targeted treatments, these biomarker studies are used for the discovery of key drivers and master regulators of networks and pathways that promote tumour proliferation and survival. US Food and Drug Administration (FDA)-approved drugs that target these particular pathways can then be identified, or opportunities for drug repurposing or development may be explored for new targets. Alternatively, precision immunotherapy approaches include cell-based therapies, vaccines and biologics. Autologous (patient-derived) tumour cell and dendritic cell vaccines are generated from extracted tumours and dendritic cells, respectively. Extracted tumours may also be used to isolate tumour-infiltrating lymphocytes (TILs). This, in combination with tumour antigen data obtained from biomarker studies, has given rise to TIL-based adoptive cell therapies and chimeric antigen receptor (CAR) T cell therapies. The identification of tumour antigens, such as tumour-specific neoantigens or tumour-associated antigens, is also important for other personalized therapies, including antigen vaccines, programmed cell death 1 ligand 1 (PDL1)–PD1 checkpoint inhibitors and other monoclonal antibodies aimed at targeting tumour-promoting antigens.
Targeted therapies.
Pathway-based targeted therapies rely on pre-existing knowledge of the underlying biology of the specific tumour, with the overall objective of modulating an aberrant protein or pathway essential for cancer subsistence. Not only does this approach help spare healthy cells, it also promotes the substratification of tumour types, allowing treatments to be tailored correspondingly. The small-molecule kinase inhibitor imatinib (Gleevec; Novartis), which is used to target the constitutively active protein product of the BCR–ABL fusion gene in chronic myelogenous leukaemia (CML) and gain-of-function mutations in the genes encoding mast/stem cell growth factor receptor KIT (also known as SCFR) or platelet-derived growth factor receptor-α (PDGFRα) in gastrointestinal stromal tumours (GISTs), served as one of the first clinical success stories for targeted cancer treatment. Whereas previous CML treatments, such as interferon-α and allogeneic bone marrow transplantation, had low efficacy with substantial risks of morbidity and mortality55, Gleevec demonstrated complete responses in >90% of patients with interferonresistant, chronic-phase CML and in 75–90% of those with advanced GISTs56–58.
Drugs targeting other oncogenic kinases, including receptor tyrosine-protein kinase erbB-2 (HER2), BRAF or epidermal growth factor receptor (EGFR), have also demonstrated clinical responses in specific groups of patients56. Overexpression of HER2 is associated with a particularly aggressive form of breast cancer with overall poor prognosis. Treatment with trastuzumab (Herceptin; Genentech/Roche), a monoclonal antibody that targets HER2, has shown a significant survival benefit, with a 20% reduction in the risk of death at 30 months59. Activating mutations in BRAF, which are present in nearly half of all melanomas, are now targeted by the mutation-selective inhibitors vemu-rafenib and dabrafenib. Both inhibitors have shown substantially increased objective response rates compared with standard chemotherapy60,61. Small-molecule and monoclonal antibody inhibitors that target EGFR have since become first-line therapy for EGFR-mutant non-small-cell lung cancer (NSCLC) with overall improved objective response rates, better quality of life and longer progression-free survival62–67. Importantly, this move to a precision medicine era in oncology has necessitated growth in the development of companion diagnostics to enable the right patient to be matched with these targeted medicines, as illustrated by the increase in FDA approvals of such diagnostics (TABLE 1).
Table 1 |.
Companion diagnostics currently approved by the FDA
| Drug (brand name) | Indication | Biomarker | Technology (year PMA first approved) |
|---|---|---|---|
| Afatinib(Cilotrif) | NSCLC | ECFR exon 19 deletions or exon 21 (L858R) substitution mutations | RT-PCR(2013) |
| Cetuximab (Erbitux) and panitumumab (Vectibix) | Colorectal cancer | KRAS mutation negative or KRAS and NRAS* mutation negative | IHC (2004); RT-PCR (2012); NGS* (2017) |
| Crizotinib (Xalkori) | NSCLC | ALK overexpression or gene fusion | FISH (2011); IHC (2015) |
| Crizotinib(Xalkori) | NSCLC | ROS1 fusions | NGS(2017) |
| Dabrafenib (Tafinlar) and trametinib (Mekinist) | Melanoma | BRAFV600E or BRAFV600K mutations | RT-PCR(2013);NGS (2017) |
| Dabrafenib (Tafinlar) and trametinib (Mekinist) | NSCLC | BRAFV600E or BRAFV600K mutations | NGS(2017) |
| Deferasirox (Exjade) | Non-transfusion-dependent thalassaemia | Liver iron concentration | MRI(2013) |
| Enasidenib(ldhifa) | AML | IDH2 mutation positive | PCR(2017) |
| Erlotinib(Tarceva) | NSCLC | EGFR exon 19 deletions or exon 21 (L858R) substitution mutations | RT-PCR(2013) |
| Cefitinib(lressa) | NSCLC | EGFR exon 19 deletions or exon 21 (L858R) substitution mutations | RT-PCR(2O15);NGS (2017) |
| Imatinib(Cleevec) | Gastrointestinal stromal tumours | KIT expression | IHC (2005) |
| Midostaurin (Rydapt) | AML | FLT3 mutation-positive | PCR(2017) |
| Olaparib (Lynparza) | Ovarian cancer | BRCA1 or BRCA2 mutation | PCR and Sanger sequencing (2014) |
| Osimertinib (Tagrisso) | NSCLC | EGFRT790M mutation | RT-PCR(2016) |
| Pembrolizumab (Keytruda) | NSCLC | PDL1 expression | IHC (2016) |
| Rucaparib (Rubraca) | Ovarian cancer | BRCA1 or BRCA2 mutation | NGS(2016) |
| Trastuzumab (Herceptin) | Breast cancer | HER2 expression and/or ERBB2 amplification | ISH (2011); CISH (2011); IHC (2012) |
| Trastuzumab (Herceptin); pertuzumab (Perjeta) and ado-trastuzumab emtansine (Kadcyla) | Breast cancer, gastric cancer | ERBB2 amplification | FISH (2005); ICC (1998) |
| Vemurafenib (Zelboraf) | Melanoma | BRAfV600E mutations | RT-PCR(2011) |
| Venetoctax (Venclexta) | B cell chronic lymphocytic leukaemia | Deletion of 17p (which contains TP53) | FISH (2016) |
All FDA-cleared or FDA-approved companion diagnostic devices on 14 August 2017, not including humanitarian device exemptions. The FDA describes a companion diagnostic as an “in vitro diagnostic device that provides information that is essential for the safe and efficacious use of the corresponding therapeutic product” (see the FDA website). Thus, the number of approvals of companion diagnostics is a measure of progress in the approval of new precision medicines. Notably, all but two companion diagnostics were approved after 2005, and all but one were developed for oncology indications. AML, acute myelogenous leukaemia; CISH, chromogenic in situ hybridization; EGFR, epidermal growth factor receptor; FISH, fluorescence in situ hybridization; HER2, receptor tyrosine-protein kinase erbB-2; IHC, immunohistochemistry; ISH, in situ hybridization; KIT, mast/stem cell growth factor receptor KIT; MRI, magnetic resonance imaging; NSCLC, non-small-cell lung cancer; NGS, next-generation sequencing; PDL1, programmed cell death 1 ligand 1; PMA, premarket approval application; RT-PCR, real-time PCR.
Panitumumab only.
Precision medicine extends beyond kinase inhibitors: treatment with poly(ADP-ribose) polymerase (PARP) inhibitors results in the synthetic lethality of tumour cells containing mutations in BRCA1 or BRCA2, which has provided a new therapy option for ovarian cancer68. Specifically, the defect in homologous recombination repair (HRR) caused by loss-of-function mutations in BRCA1 or BRCA2 renders the tumour cells sensitive to PARP inhibition, ultimately resulting in tumour cell death69. Interestingly, PARP inhibitors are efficacious for BRCA-mutated ovarian tumours70–72, but they may also prove beneficial for other subgroups of patients without identifiable germline BRCA mutations but with similar defects in HRR (also known as ‘BRCAness’). Clinical trials evaluating the efficacy of PARP inhibitors for the treatment of other tumour types containing HRR mutations are now under way (NCT01682772 and NCT01585805), and the PARP inhibitor olaparib has been granted breakthrough therapy designation by the FDA for the treatment of metastatic castration-resistant prostate cancer with mutations in BRCA1, BRCA2 or ATM (see Further information).
Although a pathway-based approach was broadly thought to become the primary approach to treating cancer, this concept has been challenged by the emergence of acquired drug resistance in patients — a phenomenon primarily resulting from ancillary mutations in the targeted protein or compensatory modifications to other components of the same or parallel pathways. Advances in understanding tumour resistance and the key mutations responsible have led to the development of the next generation of kinase inhibitors and a combinatorial approach to targeted therapies. The power of precision medicine to drive drug development is demonstrated by osimertinib, a third-generation EGFR tyrosine kinase inhibitor designed to target the T790M mutation that provides resistance to other EGFR inhibitors, which was approved by the FDA less than 2 years and 9 months after the first dose was given to a patient in a clinical study73.
Immunotherapies.
Decades of progress in our understanding of tumour immunology have recently propelled immunotherapy to the forefront of cancer treatments. Despite the capacity of the immune system to detect and eliminate cancer cells, intrinsic tumour cell and tumour microenvironmental changes can augment immune evasion, which enables tumour progression. Immunotherapies aim to harness and reinforce the inherent power of the cancer–immunity cycle (BOX 2) through a multitude of innovative methods, including the stimulation of immune cells to boost and strengthen the host response — through treatments such as cancer vaccines and adoptive T cell therapy (ACT) — or by counteracting immunosuppression, for example, through immune checkpoint blockades74,75 (FIG. 1).
Box 2 |. The cancer–immunity cycle.
As reviewed by Chen and Mellman89, a spontaneous anticancer immune response — coined the cancer–immunity cycle — is initially generated from the uptake of cancer cell antigens (such as debris from cancer cell death) by antigen-presenting cells (APCs, predominantly dendritic cells). These antigens may subsequently bind to the major histocompatibility complex, which presents antigens on the surface of APCs and activates T cells by binding to cognate T cell receptors. These activated killer T cells are then capable of infiltrating tumour beds (thus becoming tumour-infiltrating lymphocytes), where they target and destroy tumour cells. Tumour cell death results in more antigenic debris available for uptake by APCs, thus further propagating the immune response. Despite the potential potency of this anticancer cycle, the adaptability and evolution of tumour cells, along with natural host immune checkpoint mechanisms, can lead to an impaired and dampened immune response, causing tumour progression and metastasis.
Cancer vaccines deliver cancer cell antigens for subsequent uptake by antigen-presenting cells and presentation to future cytotoxic T cells. Work in this field initially focused on developing vaccines to prime the immune system to recognize cancer germline antigens or antigens differentially overexpressed in tumour cells. Although discouraging clinical data have historically plagued this approach76, growing evidence supporting the targeting of tumour-specific neoantigens by T cells has revitalized interest in therapeutic vaccines76–80. These neoantigens, which are the product of mutations that accumulate throughout oncogenesis, can serve to refine the capability of the immune system to distinguish self from non-self81, thereby fostering the induction of a potent anticancer immune response while sparing normal, healthy cells. Various clinical trials are now under way to evaluate personalized vaccines for the treatment of cancers including melanoma, pancreatic cancer and glioblastoma (NCT01970358, NCT03122106 and NCT02510950). Currently, the selection of neoantigens that are the most immunogenic and hence best to include in the vaccine remains a key challenge.
ACT is yet another new and encouraging immunotherapeutic approach that takes advantage of the tumour-targeting nature of T cells. For ACT of tumour-infiltrating lymphocytes (TILs), activated T cells are extracted from tumour samples, expanded ex vivo and subsequently infused back into the patient. This approach, coupled with prior lymphodepletion, led to a breakthrough in the treatment of melanoma, with a clinical response rate of approximately 50–70% in patients with advanced melanoma82–84. Clinical trials are currently in progress for TIL-based treatment of melanoma and other metastatic solid tumours, including ovarian, renal cell, nasopharyngeal and hepatocellular carcinomas (NCT02360579, NCT02482090, NCT02926053 and NCT01174121). Notably, ACT of TILs predominantly targeting tumour-specific neoantigens has also been associated with long-lasting tumour regression in cases of melanoma and has been described in a case of metastatic cholangiocarcinoma85,86.
Another burgeoning ACT approach is the genetic modification of patients’ own T cells to express receptors that not only recognize tumour-associated antigens but also foster T cell activation, expansion and persistence. This form of treatment, also referred to as chimeric antigen receptor (CAR) T cell therapy, has shown the most benefit thus far in patients with B cell malignancies through the targeting of the tumour-associated B lymphocyte antigen CD19 (REF. 87). Additional phase I and II clinical trials are currently evaluating CAR T cells that target various tumour-associated antigens (such as epithelial cell adhesion molecule (EPCAM), HER2, glypican 3 (GPC3), GD2 and mucin 1 (MUC1)) for the treatment of numerous other tumour types, such as breast, ovarian, gastric, brain, colorectal, nasopharyngeal, oesophageal, pancreatic, prostate, lung and hepatic cancers (NCT02723942, NCT03013712, NCT02713984 and NCT02617134).
Immunosuppression within the tumour microenvironment, resulting from the activation of immune checkpoints, is another obstacle in cancer treatment. Under normal circumstances, these checkpoint mechanisms function to prevent an excessive immune response; however, for tumour cells, exploitation of checkpoints promotes immune evasion, such as through the T cell negative regulators programmed cell death protein 1 (PD1)–programmed cell death 1 ligand 1 (PDL1) and cytotoxic T lymphocyte antigen 4 (CTLA4)88,89. Inhibition of the PD1–PDL1 interaction or the CTLA4 pathway using monoclonal antibodies has shown significant and long-lasting clinical responses in addition to an acceptable safety profile and improved tolerability versus chemotherapy, and these antibodies are now a dominant class of therapeutic90–95. Recent impressive long-term aggregate data for advanced NSCLC demonstrated 2-year survival and 5-year survival of up to 37% and 16%, respectively, compared with 14.5% and 4.9% based on standard treatment96 (see Further information). Across many indications, including NSCLC, head and neck squamous cell carcinoma and urothelial bladder cancer, tumour expression of PDL1 identifies those patients more likely to respond to monotherapy94,96–100; however, the complexity of tumour biology will ultimately determine the future utility of immune therapy in combination strategies with other therapeutic modalities.
As discussed by Vanneman and Dranoff101, the benefits and limitations of pathway-based targeted treatments and immunotherapy imply potential synergistic roles in cancer therapy. Although targeted therapies have been associated with the development of acquired tumour resistance, they are effective at inducing rapid tumour regression in defined subsets of patients. Immunotherapy has also demonstrated successful clinical responses with the added benefit of long-term immune memory of tumour cells, yet these responses typically occur in a fraction of patients — for example, approximately 40–60% of individuals receiving both therapies targeting CTLA4 and those targeting PD1 or PDL1 for metastatic melanoma and renal cell carcinoma respond102,103 — likely owing in part to untargeted tumour-mediated immunosuppressive mechanisms. Evidence suggests, however, that targeted therapies can help to alleviate these immunosuppressive effects104, and numerous clinical studies are now under way evaluating the efficacy of combined targeted therapies and immunotherapies. For example, clinical trials of immune checkpoint inhibitors in combination with mitogen-activated protein kinase (MAPK) and vascular endothelial growth factor (VEGF) pathway inhibitors for treatment of metastatic melanoma, NSCLC and renal cell carcinoma are planned or in progress, with the prospect that MAPK and VEGF pathway inhibition will not only directly affect tumour cell growth and angiogenesis but also affect tumour antigenicity and lymphocyte infiltration, thereby complementing immune checkpoint therapy105 (NCT02027961, NCT02130466 and NCT02724878).
Clinical trial design.
The transition to precision medicine approaches in cancer has also sparked a much-needed shift in the design and implementation of clinical trials. In order for targeted cancer therapies to be adequately assessed for their efficacy, they need to be tested in the appropriate group of patients: those who are predicted to respond. Basket trials, for instance, evaluate the effectiveness of a drug based on its underlying mode of action rather than strictly on the specific form of cancer it was intended to treat106. For example, the ongoing CREATE trial evaluates the use of crizotinib in treating patients with a variety of tumour types, all with targetable variants in the hepatocyte growth factor (HGF) receptor (also known as MET) and/or AKT pathways (NCT01524926). Alternatively, in umbrella trials, genomically guided targeted treatments are provided to groups of patients with the same cancer type, and outcomes are compared to controls receiving only standard therapy. The ALCHEMIST trial, for instance, tests the effectiveness of a variety of therapies that target either EGFR-driven or ALK tyrosine kinase receptor-driven early-stage NSCLC following completion of standard therapy (NCT02194738).
An even more personalized approach to clinical trials are the n-of-1 trials, which aim to treat patients individually, yet in a controlled and consistent manner, in an effort to determine whether the patient is a responder or non-responder to targeted therapy, with the long-term goal of aggregating these results in ways that could inform how to treat other subsets of patients107. An n-of-1 clinical trial that is currently in process at Columbia University Medical Center aims to recruit 260 patients with various tumour types to assess the effectiveness of targeting the master regulators that drive cancer formation and progression in single patients108. These master regulators are at ‘bottlenecks’: points at which tumour-driving cellular networks converge on one or a few proteins that could serve as an Achilles heel for tumour cells109. Data from whole-genome sequencing and RNA transcriptome analysis will be evaluated to find master regulators that are known targets of drugs that are either FDA-approved or at advanced stages of clinical testing. Candidate compounds will then be functionally tested using the patient’s tumour sample to determine the effects on tumour growth and survival.
Highly genetic conditions
Large-scale exome and genome sequencing efforts and improvements in variant interpretation are resulting in the fast-paced discovery of disease-associated genes and pathogenic mutations. Thus, precision medicine may soon become a reality for many highly genetic conditions. A clear illustration of this paradigm is the targeted treatment of genetically identified causes of disease. Disease-causing and disease-contributing genetic variants provide a window into underlying pathological mechanisms and can thus serve as a starting point for identifying treatments that act upon these mechanisms. In fact, precision medicine-based therapies, such as those that replace deficient proteins, directly target underlying molecular defects and disease-associated pathways or interfere with the expression of disease-relevant genes, have already received FDA approval and are currently in clinical use — we highlight a few of these therapies below. We also draw upon recent examples of precision medicine efforts in genetically explained epilepsies as an illustration of our expectations for the direction of the field.
Molecular replacement and pathway modification.
Unsurprisingly, initial precision medicine efforts have focused largely on common Mendelian diseases, likely owing in part to the increasing knowledge of implicated genes, variant classification and the functional effects of pathogenic mutations. Classic early examples include diet modifications for certain inborn errors of metabolism, such as phenylketonuria (PKU), a dis order caused by the toxic build-up of phenylalanine in the brain due to a deficiency in the enzyme phenylalanine hydroxylase. Low phenylalanine diets instituted from birth, which have been the standard of care for PKU for 50 years, are imperative for normal brain development and function. Another success story has been enzyme replacement therapy for various lysosomal storage disorders, a group of nearly 50 disorders caused primarily by a deficiency in lysosomal enzymes. The discovery that lysosomal enzymes are targeted to lysosomes by the mannose-6-phosphate receptor pathway110 set the stage for the use of exogenously supplied, functional lysosomal enzymes as a form of treatment111. Enzyme replacement therapy is now used clinically for the treatment of several lysosomal storage disorders, such as Gaucher disease, Fabry disease, Hunter syndrome and Pompe disease.
More recently, drugs targeting mutant proteins directly have engendered interest in conditions with historically limited treatment options. For example, in 2006, Vertex Pharmaceuticals entered a collaboration with Cystic Fibrosis Foundation Therapeutics for the accelerated development of ivacaftor, a drug that has since been approved by the FDA for the treatment of cystic fibrosis in patients with mutations in CFTR that result in a glycine to aspartate substitution at residue 551 in the protein product, cystic fibrosis transmembrane conductance regulator (CFTR) (see Further information). Ivacaftor is a CFTR channel potentiator that functions by stabilizing the open state of the channel, thus targeting the underlying CFTR gating defect associated with the G551D mutation112.
Molecular chaperones have been used to directly target the underlying molecular defect in Fabry disease, a multisystemic X-linked lysosomal storage disorder caused by a functional deficiency in the lysosomal enzyme α-galactosidase A (αGALA, encoded by GLA). Two recent phase III studies (the FACETS and ATTRACT studies, NCT00925301 and NCT01218659, respectively) evaluating the safety and efficacy of the chaperone drug migalastat, which targets certain GLA mutations that affect enzyme conformation, have demonstrated favourable clinical outcomes for patients carrying these mutations compared with placebo or enzyme replacement therapy113,114. Migalastat has since been approved for the treatment of Fabry disease by the European Medicines Agency, and a new drug application will be submitted to the FDA by the end of this year (see Further information).
Targeting disease-associated pathways has also proved successful, such as targeting the mechanistic target of rapamycin (mTOR) pathway for the treatment of tuberous sclerosis complex (TSC). Everolimus, an mTOR complex 1 (mTORC1) inhibitor, received FDA approval in 2012 for the treatment of patients with TSC-related subependymal giant cell astrocytomas and renal angiomyolipomas. Results of a recent phase III trial (the EXIST-3 trial) evaluating the use of everolimus as adjunctive treatment for TSC-related intractable epilepsy also revealed a significant decrease in seizure frequency compared with the placebo group115. As such, everolimus may be considered for the treatment of other genetic epilepsies that have mTORC1 activation, such as DEPDC5-related epilepsy.
Most targeted drugs affect either the mutant protein product or associated pathway, but some drugs have been designed to interfere at the level of gene expression. For example, the antisense oligonucleotide (ASO) drug nusinersen (Spinraza; Biogen) was granted accelerated approval by the FDA in December 2016 for the treatment of spinal muscular atrophy (SMA) (see Further information). Nusinersen aims to rescue the disease-causing impaired function of SMN1 by promoting the production of a stable form of survival motor neuron protein (SMN) from the nearly identical SMN2. A specific C to T change in SMN2 relative to SMN1 causes exon 7 to be spliced out in SMN2, which leads to an unstable protein product. This process is targeted and blocked by nusinersen, leading to production of a full-length, functional SMN protein from SMN2116. Interim analysis of a phase III clinical trial of infantile-onset SMA demonstrated improved motor milestones in 40% of patients receiving treatment, whereas no improvement was observed in sham-treated patients (see Further information). This capacity of ASOs to modulate gene expression, even in the central nervous system, has generated enthusiasm for this approach in the treatment of other diseases of the central nervous system117. Phase I and II clinical trials evaluating the safety and tolerability of ASOs for treatment of adults with early-manifest Huntington disease are also ongoing (NCT02519036).
Epilepsy.
With rapid gene discovery and good in vitro and animal models, epilepsy is arguably uniquely positioned to serve as a model for precision medicine for genetic conditions (FIG. 2). Epilepsy is a common neurological disorder characterized by recurrent, spontaneous seizures, ranging in frequency and severity, stemming from neuronal hyperexcitability and neuronal network hypersynchrony. Epilepsy is extremely heterogeneous in both clinical presentation and aetiology, thereby complicating seizure prevention and treatment. Even with more than 24 anti-epileptic drugs available on the market, seizures cannot be controlled in nearly one-third of patients118,119. Recent, large-scale sequencing studies of the rare and more severe group of epilepsy disorders, the epileptic encephalopathies, have made major progress in uncovering new causative genes120,121. These include, but are not limited to, genes involved in ion channel activity, synaptic transmission, cell signalling and growth. In addition, various model systems and electrophysiology assays are available that can assess the functional consequences of these disease-associated mutations and, importantly, can also serve as platforms for drug screening122,123. Armed with these tools, we can now take the causative epilepsy mutations of individual patients, rapidly assess their functional effects using the plethora of model systems and assays available and screen for drugs that alleviate these effects (FIG. 2).
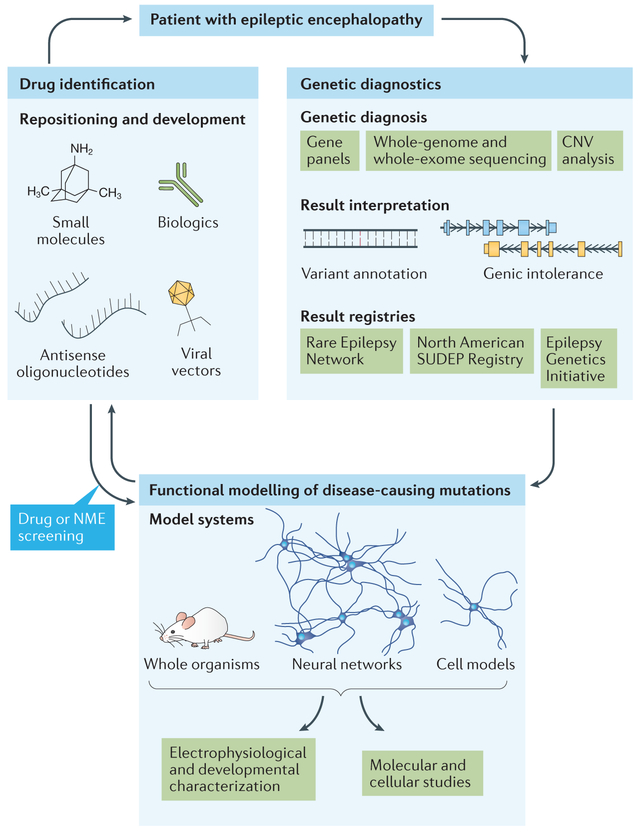
Figure 2 |. Precision medicine for highly genetic diseases — epileptic encephalopathy as a model.
A patient with epileptic encephalopathy can undergo genetic testing, including screening of an epilepsy gene panel or whole-exome sequencing for detection of single nucleotide variants, or microarray analysis for identification of copy number variants (CNVs) (right panel). Novel variants are interpreted using existing variant annotation tools and gene-level intolerance scores to determine likely pathogenicity. Patient registries have been established to house the data on disease-causing mutations and their associated phenotypes for future diagnostic efforts. Advances in gene-editing technologies have revolutionized the ability to generate functional models of pathogenic variants (bottom panel). In vivo modelling of whole organisms and in vitro modelling of neural networks along with individual neurons (derived from mouse or human induced pluripotent stem cells) and heterologous cell models can be thoroughly evaluated for pro-epileptic states using a variety of electrophysiological platforms, including electroencephalography, electroconvulsive threshold studies, multielectrode arrays and patch–clamp studies. Additional molecular and cellular studies, such as those assessing protein–protein interactions, protein localization or gene expression, can also be performed to further dissect disease pathogenesis and identify potential drug targets. These drug targets can be used as the basis for drug repositioning or drug development efforts (left panel). The efficacy of candidate compounds can then be tested using the previously established electrophysiological screening platforms. Compounds that are already US Food and Drug Administration (FDA)-approved and demonstrate amelioration of the disease phenotype in functional models may be considered for use in the patient under the care and surveillance of their physician. Efforts in epilepsy precision medicine have been thoroughly reviewed by the EpiPM Consortium122. NME, new molecular entity; SUDEP, Sudden Unexplained Death in Epilepsy.
Recent research evaluating gain-of-function mutations in KCNT1, which encodes potassium channel subfamily T member 1, in association with epilepsy of infancy with migrating focal seizures and autosomal dominant nocturnal frontal lobe epilepsy has revealed that quinidine, a drug approved for the treatment of cardiac arrhythmias, could reversibly block KCNT1 channels in in vitro systems124. Subsequent functional analyses of these epilepsy-associated KCNT1 mutations using patch clamp analysis of a heterologous expression system (Xenopus laevis oocytes) confirmed a gain-of-function effect on current amplitude that was mitigated by quinidine125. Three patients with KCNT1 mutations have since been clinically treated with quinidine and reported in the literature, two of whom showed partial responses with a reduction in seizure frequency, and one who showed no response126,127. Although this particular targeted therapy does not seem to work well and is limited by its therapeutic index, this example still serves to illustrate the paradigm. This experience with KCNT1 also highlights the clear necessity for careful and objective clinical evaluation of candidate targeted treatments.
Another epilepsy example has been the use of memantine for treatment of GRIN2A-related epileptic encephalopathy. Analysis of a single child’s GRIN2A mutation via patch clamp in a heterologous expression system (X. laevis oocytes) identified a gain-of-function effect with increased N-methyl-D-aspartic acid receptor (NMDAR) activity128. Subsequent screening of an NMDAR antagonist library using this system revealed substantial inhibition of NMDAR by memantine, a drug previously demonstrated to have anticonvulsive effects in animal models of epilepsy but traditionally used in the treatment of Alzheimer disease. The use of memantine as an adjuvant therapy for this patient was reported to result in a reduction in seizure burden and a subsequent decrease in the number of seizure medications needed. Memantine has also recently demonstrated therapeutic effects in two children with gain-of-function GRIN2D mutations, with an overall mild to moderate improvement in seizures noted129.
Thus far, it has proven easier to target gain-of-function mutations through the use of inhibitors, but there is some promise for targeting loss-of-function or dominant-negative mutations. For instance, KCNQ2-related epileptic encephalopathy has recently been targeted with retigabine (also known as ezogabine), a potassium channel activator. KCNQ2, which encodes a subunit of the voltage-gated potassium channel KV7.2, functions to modulate neuronal excitability130. Investigation of epileptic encephalopathy-associated mutations via patch clamp analysis of the X. laevis oocyte heterologous expression system revealed loss-of-function and dominant-negative effects on KV7.2 channel activity131. Additional in vitro and mouse in vivo studies found that treatment with retigabine ameliorates the effects of these dominant-negative mutations and can attenuate seizure activity in mouse models132,133. A small retrospective study of 11 children with KCNQ2-related epileptic encephalopathy treated with retigabine found that 3 of 4 patients treated before 6 months of age had seizure improvement, whereas clinical response was less impressive for patients treated after 6 months, with only 2 out of 7 showing improvement134.
Although a diagnosis of epilepsy conveys no information about the underlying mechanism of disease, identifying and uncovering the functional effects of a causal mutation suggests potential therapies. Precision medicine for epilepsy remains in its infancy, but these examples serve to illustrate how the paradigm is evolving. As this work progresses, we envision that the targeted treatments will become increasingly more effective. For instance, it is clear that the effectiveness of quinidine for the treatment of KCNT1-related epilepsy is restricted by dose-limiting cardiotoxicity. Yet, it is also clear that, using this same paradigm, it could be possible to identify more potent inhibitors as targeted treatments for this disease.
Outlook
It remains unclear how generalizable this precision medicine approach will be for other diseases — especially those in which a single responsible and strongly contributing genetic mutation is found in only a small number of individuals. Three factors, however, suggest that precision medicine will eventually reach multiple areas of medicine. First, for relatively common diseases and traits that are traditionally considered to be complex, there are now many examples of cases influenced by rare, strongly acting mutations. For instance, genes initially identified to be associated with the more severe and devastating epilepsies are also clearly implicated in more common epilepsies, such as genetic generalized epilepsy or the non-lesional focal epilepsies135. This has also been demonstrated for other complex diseases, including autism, congenital heart disease, idiopathic pulmonary fibrosis and immunodeficiencies, as well as quantitative traits, including height, lipid levels and blood pressure136–146. Second, there may be scope for precision medicine approaches in other conditions in which there is genetic heterogeneity, but pathway homogeneity. For example, there are multiple genes altered in patients with amyotrophic lateral sclerosis, including TBK1, OPTN and SQSTM1, that seem to act at similar stages in the autophagy pathway147. We therefore find it plausible that targeted treatments developed for specific genetically defined conditions, such as for patients with TBK1 mutations, may be useful in cases with different causes that act through the same pathway, such as patients with OPTN mutations. Third, we postulate that genetically complex causes of disease may often affect similar pathways and patients may share commonalities in optimal treatment with individuals with strongly acting mutations. We thus imagine that some fraction of treatments used for defined genetic conditions may yet work for individuals without those genetic causes but who share similar disease mechanisms.
The role of precision medicine in drug development for cancer is already clear, as outlined above. Outside of cancer, the precision medicine paradigm is contributing to the drug development process by focusing on targets responsible for disease in individual patients and the stratification of clinical trials based on the underlying mechanistic causes of disease. There will be a growing number of trials that are targeted to the precise genetic and mechanistic cause of disease, as the examples of GRIN2A and KCNT1 illustrate. Although this may seem a slow and cumbersome approach to identify a medicine for wide use, it is worth emphasizing that some of the treatments targeted to specific underlying causes of disease may have wider application beyond individuals carrying those precise genetic causes. Beyond such examples of explicitly targeted treatments, current clinical trials routinely include patients who have diseases with a broad range of underlying causes. It is reasonable to assume that treatments, even when not explicitly targeted, will work better for diseases driven by particular underlying mechanisms. Currently, this is a source of variation in treatment response that is often ignored in most clinical trials. As two simple examples, we now know that clinical trials in refractory epilepsy and in chronic kidney disease include patients who have disease caused by very different underlying mechanisms, which can be revealed through genetic evaluation. Even if drugs are studied in such all-comer populations, dividing patients into sub-groups that are mechanistically distinct may identify populations in whom these drugs are more effective, as demonstrated by the clinical benefit of olaparib in patients with recurrent platinum-sensitive serous ovarian cancer and BRCA mutations148. Evaluation of a candidate medicine without recognition of this underlying diversity in the patient population is likely to be a key contributor to drug development failures due to insufficient overall efficacy14. Utilizing genetic and genomic approaches to stratify clinical populations into mechanistic subgroups is very likely to permit a molecular classification of disease that results in a higher success rate within those molecularly defined subpopulations, thereby benefiting patients, physicians, drug developers, regulators and payers.
FURTHER INFORMATION
Amgen news release for romosozumab: http://wwwext.amgen.com/en-gb/media/news-releases/2017/07/amgenand-ucb-provide-update-on-regulatory-status-of-evenityromosozumab-in-the-us/
Amicus Therapeutics press release for migalastat (2016): http://ir.amicusrx.com/releasedetail.cfm?releaseid=973355
Amicus Therapeutics press release for migalastat (2017): http://ir.amicusrx.com/releasedetail.cfm?releaseid=1032662
AstraZeneca press release for olaparib: https://www.astrazeneca.com/media-centre/press-releases/2016/Lynparza-Olaparib-granted-Breakthrough-Therapy-Designation-by-USFDA-for-treatment-of-BRCA1-2-or-ATM-gene-mutated-metastatic-Castration-Resistant-Prostate-Cancer-28012016.html
Bristol-Myers Squibb press release for nivolumab:
deCODE: http://investors.amgen.com/phoenix.zhtml?c=61656&p=irol-newsArticle&ID=1765710
DiscoverEHR collaboration: http://investor.regeneron.com/releasedetail.cfm?releaseid=818844
US Food and Drug Administration (FDA) label for evolocumab: https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/125522s000lbl.pdf
FDA label for alirocumab: https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/125559Orig1s000lbledt.pdf
FDA label for ivacaftor: https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/203188lbl.pdf
FDA label for nusinersen: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/209531lbl.pdf
FDA list of companion diagnostic devices: https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Acknowledgements
The authors thank J. Carulli, M. Lalioti and C. Bostick for their valuable feedback on this manuscript.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease (National Academies Press, 2011). [PubMed] [Google Scholar]
- 2.Raviña Rubira E The Evolution of Drug Discovery: From Traditional Medicines to Modern Drugs. (Wiley-VCH, 2011). [Google Scholar]
- 3.Zanders ED The Science and Business of Drug Discovery. (Springer, 2011). [Google Scholar]
- 4.Hopkins MM, Martin PA, Nightingale P, Kraft A & Mahdi S The myth of the biotech revolution: an assessment of technological, clinical and organisational change. Res. Policy 36, 566–589 (2007). [Google Scholar]
- 5.Nightingale P & Madhi S in Knowledge Accumulation and Industry Evolution: The Case of Pharma-Biotech (eds Mazzucatu M & Dosi G) 73–111 (Cambridge Univ. Press, 2006). [Google Scholar]
- 6.Martin P, Hopkins M, Nightingale P & Kraft A in The Handbook of Genetics & Society: Mapping the New Genomic Era (eds Atkinson P, Glasner P & Lock M 145–162 (Routledge, 2009). [Google Scholar]
- 7.Nightingale P Economies of scale in experimentation: knowledge and technology in pharmaceutical R&D. Ind. Corp. Change 9, 315–359 (2000). [Google Scholar]
- 8.Debouck C & Metcalf B The impact of genomics on drug discovery. Annu. Rev. Pharmacol. Toxicol 40, 193–207 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Adams MD et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252, 1651–1656 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Adams MD et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377, 3–174 (1995). [PubMed] [Google Scholar]
- 11.Haseltine WA Discovering genes for new medicines. Sci. Am 276, 92–97 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Haseltine WA Genomics and drug discovery. J. Am. Acad. Dermatol 45, 473–475 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Gershon D Smithkline backs sequencing company. Nature 363, 387 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Ma P & Zemmel R Value of novelty? Nat. Rev. Drug Discov 1, 571–572 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Hopkins M, Kraft A, Martin P, Nightingale P & Madhi S in Comprehensive Medicinal Chemistry II (eds Taylor JB & Triggle DJ) 591–613 (Elsevier, 2007). [Google Scholar]
- 16.Cook D et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov 13, 419–431 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat. Genet 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Manolio TA, Brooks LD & Collins FSA HapMap harvest of insights into the genetics of common disease. J. Clin. Invest 118, 1590–1605 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsmore SF, Lindquist IE, Mudge J, Gessler DD & Beavis WD Genome-wide association studies: progress and potential for drug discovery and development. Nat. Rev. Drug Discov 7, 221–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopes MC, Zeggini E & Panoutsopoulou K Do genome-wide association scans have potential for translation? Clin. Chem. Lab. Med 50, 255–260 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Visscher PM, Brown MA, McCarthy MI & Yang J Five years of GWAS discovery. Am. J. Hum. Genet 90, 7–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duerr RH et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4, e1000041 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Australo-Anglo-American Spondyloarthritis Consortium (TASC) et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet 42, 123–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaffen SL, Jain R, Garg AV & Cua DJ The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol 14, 585–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feagan BG et al. Ustekinumab as induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med 375, 1946–1960 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Krueger JG et al. Anti–IL-23A mAb BI 655066 for treatment of moderate-to-severe psoriasis: safety, efficacy, pharmacokinetics, and biomarker results of a single-rising-dose, randomized, double-blind, placebo-controlled trial. J. Allergy Clin. Immunol 136, 116–124.e7 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Feagan BG et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 389, 1699–1709 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Kopp T et al. Clinical improvement in psoriasis with specific targeting of interleukin-23. Nature 521, 222–226 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Poddubnyy D, Hermann K-GA, Callhoff J, Listing J & Sieper J Ustekinumab for the treatment of patients with active ankylosing spondylitis: results of a 28-week, prospective, open-label, proof-of-concept study (TOPAS). Ann. Rheum. Dis 73, 817–823 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Fragoulis GE, Siebert S & McInnes IB Therapeutic targeting of IL-17 and IL-23 cytokines in immune-mediated diseases. Annu. Rev. Med 67, 337–353 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Frayling TM et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dina C et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet 39, 724–726 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Scuteri A et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tung YCL, Yeo GSH, O’Rahilly S & Coll AP Obesity and FTO: changing focus at a complex locus. Cell Metab. 20, 710–718 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Loos RJF & Yeo GSH The bigger picture of FTO: the first GWAS-identified obesity gene. Nat. Rev. Endocrinol 10, 51–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claussnitzer M et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med 373, 895–907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smemo S et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle EA, Li YI & Pritchard JK An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).This paper proposes an ‘omnigenic’ model of complex disease and trait inheritance, arguing in favour of a few ‘core’ disease variants, with the bulk of heritability caused by an immense amount of ‘peripheral’ variants expressed in disease-relevant cell types.
- 40.Goldstein DB Common genetic variation and human traits. N. Engl. J. Med 360, 1696–1698 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Plenge RM, Scolnick EM & Altshuler D Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov 12, 581–594 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Abifadel M et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet 34, 154–156 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Fasano T et al. A novel loss of function mutation of PCSK9 gene in white subjects with low-plasma low-density lipoprotein cholesterol. Arterioscler. Thromb. Vasc. Biol 27, 677–681 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Cohen J et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet 37, 161–165 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Cohen JC, Boerwinkle E, Mosley TH & Hobbs HH Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med 354, 1264–1272 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Kotowski IK et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet 78, 410–422 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang D-W et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem 282, 18602–18612 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Poirier S et al. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J. Biol. Chem 283, 2363–2372 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Dadu RT & Ballantyne CM Lipid lowering with PCSK9 inhibitors. Nat. Rev. Cardiol 11, 563–575 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Sabatine MS et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med 376, 1713–1722 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Kamb A, Harper S & Stefansson K Human genetics as a foundation for innovative drug development. Nat. Biotechnol 31, 975–978 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Eisenstein M Amgen and Regeneron push for a genetic renaissance in drug discovery. Nat. Biotechnol 32, 208–209 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Sulem P et al. Identification of a large set of rare complete human knockouts. Nat. Genet 47, 448–452 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Dewey FE et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N. Engl. J. Med 374, 1123–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silver RT et al. An evidence-based analysis of the effect of busulfan, hydroxyurea, interferon, and allogeneic bone marrow transplantation in treating the chronic phase of chronic myeloid leukemia: developed for the American Society of Hematology: presented in part at the Education Session of the American Society of Hematology, December 5, 1998, Miami Beach, FL: Blood; 94, 1517–1536 (1999). [PubMed] [Google Scholar]
- 56.Stegmeier F, Warmuth M, Sellers WR & Dorsch M Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin. Pharmacol. Ther 87, 543–552 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Druker BJ Translation of the Philadelphia chromosome into therapy for CML. Blood 112, 4808–4817 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Demetri GD et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med 347, 472–480 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Slamon DJ et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med 344, 783–792 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Hauschild A et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380, 358–365 (2012). [DOI] [PubMed] [Google Scholar]
- 61.McArthur GA et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 15, 323–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maemondo M et al. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N. Engl. J. Med 362, 2380–2388 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Zhou C et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12, 735–742 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Mitsudomi T et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 11, 121–128 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Rosell R et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 13, 239–246 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Sequist LV et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol 31, 3327–3334 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Wu Y-L et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 15, 213–222 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Tewari KS, Eskander RN & Monk BJ Development of olaparib for BRCA-deficient recurrent epithelial ovarian cancer. Clin. Cancer Res 21, 3829–3835 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Lord CJ & Ashworth A PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pujade-Lauraine E et al. Treatment with olaparib monotherapy in the maintenance setting significantly improves progression-free survival in patients with platinum-sensitive relapsed ovarian cancer: results from the phase III SOLO2 study. Gynecol. Oncol 145, 219–220 (2017).28431684 [Google Scholar]
- 71.Kaufman B et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol 33, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mirza MR et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med 375, 2154–2164 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Yver A Osimertinib (AZD9291) — a science-driven, collaborative approach to rapid drug design and development. Ann. Oncol 27, 1165–1170 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Yang Y Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Invest 125, 3335–3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farkona S, Diamandis EP & Blasutig IM Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 14, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mullard A The cancer vaccine resurgence. Nat. Rev. Drug Discov 15, 663–665 (2016). [DOI] [PubMed] [Google Scholar]
- 77.van Rooij N et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol 31, e439–e442 (2013).This is the first report of T cell reactivity to tumour-specific neoantigens in human patients.
- 78.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Linnemann C et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med 21, 81–85 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Gubin MM et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Rosenberg SA et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dudley ME et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J. Clin. Oncol 23, 2346–2357 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dudley ME et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 298, 850–854 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu Y-C et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J. Immunol 190, 6034–6042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tran E et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sadelain M CAR therapy: the CD19 paradigm. J. Clin. Invest 125, 3392–3400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma P & Allison JP Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen DS & Mellman I Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Brahmer JR et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med 366, 2455–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Powles T et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515, 558–562 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ansell SM et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med 372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seiwert TY et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 17, 956–965 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Schadendorf D et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J. Clin. Oncol 33, 1889–1894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herbst RS et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Borghaei H et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med 373, 1627–1639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rebelatto MC et al. Development of a programmed cell death ligand-1 immunohistochemical assay validated for analysis of non-small cell lung cancer and head and neck squamous cell carcinoma. Diagn. Pathol 11, 95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Massard C et al. Safety and efficacy of durvalumab (MEDI4736), an anti-programmed cell death ligand-1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J. Clin. Oncol 34, 3119–3125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rosenberg JE et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vanneman M & Dranoff G Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 12, 237–251 (2012).This is an insightful review that highlights the opportunities for and potential clinical benefits of combined targeted therapies and immunotherapies in oncology precision medicine.
- 102.Weber JS et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Hammers HJ et al. Phase I study of nivolumab in combination with ipilimumab in metastatic renal cell carcinoma (mRCC). J. Clin. Oncol 32, 4504–4504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rakhra K et al. CD4+ T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18, 485–498 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hughes PE, Caenepeel S & Wu LC Targeted therapy and checkpoint immunotherapy combinations for the treatment of cancer. Trends Immunol. 37, 462–476 (2016). [DOI] [PubMed] [Google Scholar]
- 106.Renfro LA & Sargent DJ Statistical controversies in clinical research: basket trials, umbrella trials, and other master protocols: a review and examples. Ann. Oncol 28, 34–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schork NJ Personalized medicine: time for one-person trials. Nature 520, 609–611 (2015). [DOI] [PubMed] [Google Scholar]
- 108.Rubin RA Precision medicine approach to clinical trials. JAMA 316, 1953–1955 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Alvarez MJ et al. Functional characterization of som a tic mutations in cancer using network-based inference of protein activity. Nat. Genet 48, 838–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kornfeld S Lysosomal enzyme targeting. Biochem. Soc. Trans 18, 367–374 (1990). [DOI] [PubMed] [Google Scholar]
- 111.Desnick RJ & Schuchman EH Enzyme replacement and enhancement therapies: lessons from lysosomal disorders. Nat. Rev. Genet 3, 954–966 (2002). [DOI] [PubMed] [Google Scholar]
- 112.Van Goor F et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl Acad. Sci. USA 106, 18825–18830 (2009).This study reports the in vitro mechanism of the now-approved cystic fibrosis drug ivacaftor (VX-770) that selectively targets the molecular defect underlying the G551D mutation in CFTR.
- 113.Germain DP et al. Treatment of Fabry’s disease with the pharmacologic chaperone migalastat. N. Engl. J. Med 375, 545–555 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Hughes DA et al. Oral pharmacological chaperone migalastat compared with enzyme replacement therapy in Fabry disease: 18-month results from the randomised phase III ATTRACT study. J. Med. Genet 54, 288–296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.French JA et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet 388, 2153–2163 (2016).This study reports clinical trial data for the use of the mTOR inhibitor everolimus in the treatment of TSC-related seizures, thus serving as an example of pathway-based treatment.
- 116.Hua Y, Vickers TA, Okunola HL, Bennett CF & Krainer AR Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet 82, 834–848 (2008).This study describes the use of an ASO tiling method to interfere with the splicing silencers heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) and HNRNPA2 as a therapeutic mechanism for SMA and provides an example of how drugs may be designed to exert effects at the level of gene expression.
- 117.Corey DR Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci 20, 497–499 (2017). [DOI] [PubMed] [Google Scholar]
- 118.Brodie MJ, Barry SJE, Bamagous GA, Norrie JD & Kwan P Patterns of treatment response in newly diagnosed epilepsy. Neurology 78, 1548–1554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sankaraneni R & Lachhwani D Antiepileptic drugs — a review. Pediatr. Ann 44, e36–e42 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Epi4K Consortium et al. De novo mutations in epileptic encephalopathies. Nature 501, 217–221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Epilepsy Phenome/Genome Project, Epi4K Consortium & EuroEPINOMICS-RES Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet 100, 179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Consortium EpiPM. A roadmap for precision medicine in the epilepsies. Lancet Neurol. 14, 1219–1228 (2015).This article provides a conceptual framework for developing and implementing targeted therapies for epilepsy.
- 123.Dhindsa RS & Goldstein DB Genetic discoveries drive molecular analyses and targeted therapeutic options in the epilepsies. Curr. Neurol. Neurosci. Rep 15, 70 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Yang B et al. Pharmacological activation and inhibition of Slack (Slo2.2) channels. Neuropharmacology 51, 896–906 (2006). [DOI] [PubMed] [Google Scholar]
- 125.Milligan CJ et al. KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Ann. Neurol 75, 581–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bearden D et al. Targeted treatment of migrating partial seizures of infancy with quinidine. Ann. Neurol 76, 457–461 (2014). [DOI] [PubMed] [Google Scholar]
- 127.Mikati MA et al. Quinidine in the treatment of KCNT1-positive epilepsies. Ann. Neurol 78, 995–999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pierson TM et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann. Clin. Transl Neurol 1, 190–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li D et al. GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers. Am. J. Hum. Genet 99, 802–816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang HS et al. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 (1998). [DOI] [PubMed] [Google Scholar]
- 131.Orhan G et al. Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy. Ann. Neurol 75, 382–394 (2014). [DOI] [PubMed] [Google Scholar]
- 132.Ihara Y et al. Retigabine, a Kv7.2/Kv7.3-channel opener, attenuates drug-induced seizures in knock-in mice harboring Kcnq2 mutations. PLoS ONE 11, e0150095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wuttke TV et al. Peripheral nerve hyperexcitability due to dominant-negative KCNQ2 mutations. Neurology 69, 2045–2053 (2007). [DOI] [PubMed] [Google Scholar]
- 134.Millichap JJ et al. KCNQ2 encephalopathy: features, mutational hot spots, and ezogabine treatment of 11 patients. Neurol. Genet 2, e96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.International League Against Epilepsy Consortium on Complex Epilepsies. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 13, 893–903 (2014).This meta-analysis implicates mutations in genes initially known to cause the severe, monogenic epilepsies in the more common generalized and focal epilepsies, thus serving as an example of the allelic and corresponding phenotypic heterogeneity that may render certain targeted treatments applicable to patients carrying a range of mutations in the same gene.
- 136.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zaidi S et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Al Turki S et al. Rare variants in NR2F2 cause congenital heart defects in humans. Am. J. Hum. Genet 94, 574–585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Petrovski S et al. An exome sequencing study to assess the role of rare genetic variation in pulmonary fibrosis. Am. J. Respir. Crit. Care Med 196, 82–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Asgari S et al. Exome sequencing reveals primary immunodeficiencies in children with community-acquired Pseudomonas aeruginosa sepsis. Front. Immunol 7, 357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maffucci P et al. Genetic diagnosis using whole exome sequencing in common variable immuno-deficiency. Front. Immunol 7, 220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dickinson RE et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marouli E et al. Rare and low-frequency coding variants alter human adult height. Nature 542, 186–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Surakka I et al. The impact of low-frequency and rare variants on lipid levels. Nat. Genet 47, 589–597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Surendran P et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet 48, 1151–1161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Weishaupt JH, Hyman T & Dikic I Common molecular pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Trends Mol. Med 22, 769–783 (2016).This review summarizes findings from amyotrophic lateral sclerosis genetic studies and the convergence of implicated genes into functional pathways, thus underscoring the potential utility of these disease-associated pathways as therapeutic targets.
- 148.Oza AM et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 16, 87–97 (2015). [DOI] [PubMed] [Google Scholar]
- 149.Brunkow ME et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet 68, 577–589 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Winkler DG et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 22, 6267–6276 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Padhi D, Jang G, Stouch B, Fang L & Posvar E Single-dose, placebo-controlled, randomized study of AMG 785, a sclerostin monoclonal antibody. J. Bone Miner. Res 26, 19–26 (2011). [DOI] [PubMed] [Google Scholar]
- 152.McClung MR et al. Romosozumab in postmenopausal women with low bone mineral density. N. Engl. J. Med 370, 412–420 (2014). [DOI] [PubMed] [Google Scholar]
- 153.Cosman F et al. Romosozumab treatment in postmenopausal women with osteoporosis. N. Engl. J. Med 375, 1532–1543 (2016). [DOI] [PubMed] [Google Scholar]
- 154.Yang Y et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J. Med. Genet 41, 171–174 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fertleman CR et al. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 52, 767–774 (2006). [DOI] [PubMed] [Google Scholar]
- 156.Cox JJ et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reimann F et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc. Natl Acad. Sci. USA 107, 5148–5153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duan G et al. A single-nucleotide polymorphism in SCN9A may decrease postoperative pain sensitivity in the general population. Anesthesiology 118, 436–442 (2013). [DOI] [PubMed] [Google Scholar]