ABSTRACT
Meningioma is the most common brain tumor in adults. Surgical resection remains the primary treatment. No chemotherapy exists. However, gene mutations now could explain ~ 80% of meningioma and targeted therapies based on these are being investigated. Furthermore, with the recent discovery of PD-L1 in malignant meningioma, clinical trials using immunotherapy have commenced. Here, we report for the first time the expression profiles of immune checkpoint proteins PD-L2, B7-H3 and CTLA-4 in meningioma and their association to common gene mutations. PD-L2 and B7-H3 expression was significantly greater than all immune checkpoint proteins studied, and particularly elevated in patients with gene mutations affecting the PI3K/AKT/mTOR pathway. CTLA-4 expressing CD3+ lymphocytes were observed in atypical and malignant meningioma and tumors harboring a PIK3CA or SMO mutation. These results identify novel targets for immunotherapy irrespective of grade and distinguish potential patient populations based on genetic classification for stratification into checkpoint inhibitor clinical trials.
KEYWORDS: Meningioma, immune checkpoint protein, PD-L1, PD-L2, B7-H3, CTLA-4, PI3K/AKT/mTOR, immunotherapy, brain tumor
Introduction
Meningioma, a most common primary brain tumor in adults has a prevalence of ~ 98/100,000 people.1,2 Based on World Health Organization (WHO) criteria that relies on histology alone, ~ 80% of meningioma is classified as grade I or benign, 10–18% grade II or atypical and ~ 2–4% grade III or malignant.3 Grade II and III meningioma have a 5-year recurrence rate of ~ 50% and ~ 100% respectively and ~ 30% of grade I meningioma will recur in a patient’s lifetime, suggesting that a portion of grade I meningioma are biologically more aggressive.4 Meningioma, both grade I and II, may progress to grade III.5 However, it is not clear which of the tumors classified as grade I or II will undergo malignant transformation. This suggests that the current WHO classification may not be a reliable predictor of tumor behavior. Surgical resection remains the primary treatment. Residual tumor, particularly those involving the skull base and/or those invading the draining venous sinuses or of higher grade6,7 are often managed with either radiosurgery or radiotherapy. No chemotherapy exists.
Recent genetic studies have identified mutations that strongly correlate to meningioma subtype, location and growth rate, suggesting molecular characterization as a more reliable biological classifier of meningioma than WHO grading.8–14 Approximately 40–60% of all meningioma harbor NF2 deletions and/or point mutations. NF2 and TERT promotor mutations may be found in each grade but are more frequently observed in grade II and III meningioma and linked to poor prognosis. Several mutations present in the genes AKT1, PIK3CA, SMO, TRAF7, KLF4 and POLR2A are associated with grade I meningioma and are primarily linked to tumors located at the skull base, a particularly difficult location to achieve complete surgical resection.8–14 These mutations are normally found exclusive of NF2 deletions, and collectively referred to as the non-NF2 group which represent ~ 25% of meningiomas. This leaves only ~ 15–20% with an unknown genetic contribution. Importantly, many meningioma-associated mutations are shared with other systemic cancers for which targeted therapies are available. One clinical trial using a smo inhibitor, vismodegib, for patients with SMO L412F and W535L mutations and focal adhesion kinase inhibitor, GSK2256098 for NF2 mutations is currently underway.15
Immunotherapy has also emerged as a treatment for meningioma. Meningioma is located outside the blood brain barrier providing access to peripheral immune cell infiltration. Indeed, studies have shown virtually all meningioma harbor immune cell infiltrates. Macrophages are the most abundant immune cell in the meningioma microenvironment. Macrophage numbers are higher in grades II and III as opposed to grade I.16 Mast cells are also present in large quantities, particularly when edema is present. CD8+ cytotoxic and CD4+ helper T cell lymphocytes are the next most common infiltrating immune cell, followed by regulatory T cells (Tregs) and natural killer (NK) cells.16–20 A few B cells are also infrequently observed in meningioma tissues18,19. Tregs are found to be present in all grades of meningioma, with a tendency to be more frequent in higher grades.16–19 Tregs, together with CD8+ and CD4 + T cells in meningioma microenvironment are antigen experienced and often display an exhaustive phenotype, expressing immune checkpoint proteins, PD-1 and T cell immunoglobulin and mucin-domain containing-3 (TIM-3).17–19 Tregs modulate immune responses and can release cytokines to reduce proliferation of effector T cells. The immunosuppressive activity of PD-1+ Tregs on T cell proliferation is evidenced by a significant drop in CD8+ and CD4 + T cells in grade II and III meningio.17–19 As PD-1 + T cells decrease according to WHO grade, its binding partner on tumor cells, PD-L1, increases. This further confirmed that for malignant meningioma, the tumors appear to establish an immune evading milieu. Recently, three trials have begun recruiting patients with anaplastic meningioma and/or radiation-refractory meningioma for treatment with immune checkpoint protein inhibitors. These trials use an anti-PD-1 antibody ((Nivolumab (NCT02648997),21 Pembrolizomab (NCT03016091)22 and Avelumab (NCT03267836) 23) to block PD-L1-PD-1 binding and thus should block the resulting immunosuppressive signaling. While these trials are promising advances in the treatment of malignant meningioma, in their current design they only encompass a very small subset of patients. Many additional patients irrespective of grade could respond to immunotherapy. In previous studies measuring PD-L1 and PD-1 expressions in meningioma, authors mention high inter-patient variability in expression levels of proteins in each WHO grade. While not as distinct as in malignant meningioma, immunosuppressive microenvironments were still frequently observed in grade I and II patients.16 This led us to propose genetic changes that more accurately reflect meningioma biology than WHO grade also correlate more closely with immune checkpoint protein expression.
In this study, we screened 22 genetically characterized meningioma tissue samples for a panel of immune checkpoint proteins, and have identified for the first time the expression of 3 immune checkpoint proteins PD-L2, B7-H3 and CTLA-4 in addition to PD-L1 and PD-1. Immune checkpoint protein expression was observed across meningiomas of all grades but variation in level of expression was observed between genetic groups of meningioma. Most interestingly, we found that PD-L2 and B7-H3 were expressed at significantly higher quantities than any other immune checkpoint protein analyzed including previously reported PD-L1. This indicates that these two proteins serve important roles in meningioma immune responses. More importantly, PD-L2 and B7-H3 levels were particularly elevated in patients with gene mutations affecting the PI3K/AKT/mTOR pathway. CTLA-4 expression was also measured in meningioma tissues but was restricted to atypical and anaplastic meningioma and tumors harboring a PIK3CA or SMO mutation.
Fluorescent immunohistochemistry revealed B7-H3 and PD-L2 were expressed on cell membranes of tumor cells whilst CTLA-4 was restricted to infiltrating CD3+ immune cells. Finally, this study also evaluated the effect of edema on immune checkpoint protein expression with no correlation observed. These results identify additional checkpoint protein targets for immunotherapy irrespective of grade and warrant further investigation for stratifying patient populations based on genetic classification with checkpoint inhibitor trials.
Methods
This study was approved by the Conjoint Health Research Ethics Board – University of Calgary. Informed written consent was obtained from each patient. Tumor tissue specimens were acquired from patients undergoing surgical resection at the Foothills Medical Center, Calgary. Tumor samples were obtained prior to electrocautery. A portion of the sample was sent to Neuropathology for routine histopathology and WHO grading. The other portion was immediately snap frozen in the operating room and stored at -80°C for molecular characterization. For each case, a 9 ml matched-blood sample was collected prior to surgery and immediately processed to obtain buffycoat containing white blood cells (WBCs). DNA from WBCs was used as a control to establish if a mutation detected in the tumor tissue was somatic or germline. Tissue gDNA was extracted using an in-house phenol-chloroform extraction technique optimized for meningioma.24 Tissue homogenates for immune checkpoint protein multiplex ELISA arrays were processed as described below. All protein and DNA samples were stored at -80°C. For each case, WHO histology classifiers and final WHO grading was obtained from patient record. Patient information (age, sex) and detailed medical history (past resections, cancer, treatments, radiotherapy), baseline cognitive data and information specific to the tumor (size, location, surgical procedure, pre- and post-surgery MR imaging, extent of resection and severity of edema) was also acquired.
NF2, 1p34 deletion screening
The NF2 and 1p34 deletion status of meningioma was performed using multiplex ligation dependent probe amplification (MLPA) on 50 ng samples of gDNA with SALSA MLPA P044 NF2 probemix version B2 together with the MLPA FAM-labeled PCR primer kit (MRC-Holland, Amsterdam, Netherlands). Methods are detailed in Proctor et al 2017. 24 For a baseline comparison to generate deletion and amplification calls in tumor DNA, 50 ng of gDNA extracted from saliva of 4 healthy subjects was included. Additional chromosomal regions (3p22, 4q27, 5p15, 5q31, 6q22, 8q24, 14q31, 15q21, 16q24, 17p13) are also included in the MLPA assay and used as a marker of chromosomal stability.
Next-generation sequencing (NGS)
Targeted NGS was performed on DNA from the tumor tissue using a panel of targets spanning 38 genes with reported clinical association to meningioma. Target regions were enriched with a custom set of small length (80 bp) capture probes (MYbaits®). A tiling density of ~ 66% overlap between baits and stringency filters were applied to the bait design to maximize sequence coverage but reduce non-specific off-target probe binding. Amplified products were quantitated on a Kapa qPCR plate and loaded onto NextSeq150 mid-output 20 GB cartridge and sequenced on Illumina NextSeq 500 sequencer. BAM sequencing read files were analyzed with GATK best practices pipeline and point mutation variants and small indels and amplifications were determined. A 10% stringency was used in calling variants and identified changes were validated by either mutation specific ARMS-PCR or sanger sequencing. Sanger sequencing of matched DNA from WBCs was used to determine whether mutations were somatic or germline. Coverage of target regions (up to 5000X) and off-target sequencing was validated using Geneious software.
Immune checkpoint protein expression
The expression of 10 common immune checkpoint proteins (PD-1, PD-L1, PD-L2, CTLA-4, B7-H2, B7-H3, CD28, CD80, CD86 and ICOS) were determined using a Human Immune Checkpoint Molecule Quantitative Multiplex ELISA Array 1 according to manufacturer instructions (Raybiotech). Tumor tissues were homogenized in RIPA buffer containing proteinase inhibitor cocktail. Lysates were treated with DNAse and protein concentration determined by BCA assay. Microarray glass slides were incubated with 250 µg of tissue lysate protein overnight. Microarrays were sent to Raybiotech for scanning and extracted data was analyzed with custom quantibody analysis tool software (Raybiotech) to determine absolute quantities of each immune checkpoint protein for each sample. Quantities of each protein of interest are determined by an average fluorescent intensity of four spots and compared to a set of protein standard spots included on each glass slide. A representative example of a standard and meningioma tissue scanned microarray are shown in Figure 1C.
Figure 1.
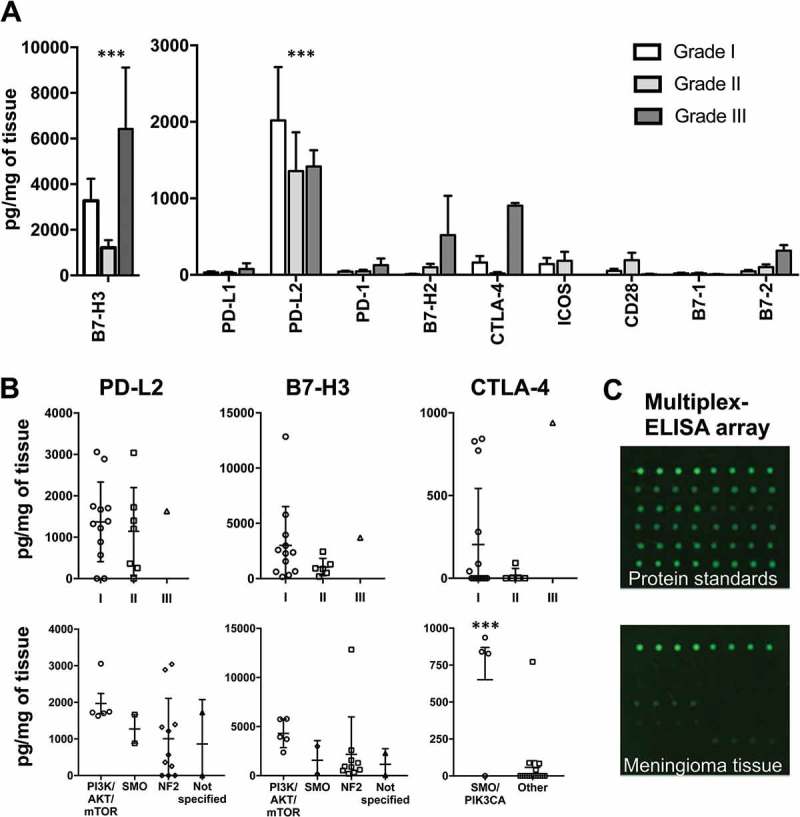
Immune checkpoint protein expression in meningioma tissues according to WHO grade and genetic subtype. (A) Quantities of 10 common immune checkpoint proteins in meningioma tissues according to WHO grade (n = 22). Amounts of B7-H3 and PD-L2 proteins are significantly greater than all other proteins analyzed. (B) Expression of PD-L2, B7-H3 and CTLA-4 for each case (n = 20) is quantified according to WHO grade and gene mutation subtype. (C) Representative scans of a multiplex ELISA array for a single protein standard used in the generation of a protein quantification standard curve and also a meningioma tissue protein sample. *** P-value < 0.001. Bars represent mean protein quantity in pg/mg of meningioma tissue. Error bars represent SEM.
Fluorescent immunohistochemistry
Tissue sections were cut in series from formalin-fixed paraffin embedded (FFPE) tissue blocks for each meningioma case and both control samples (lateral temporal cortex removed during exposure of mesial temporal epilepsy surgery). Standard H&E staining was performed for each tumor and histological features of each section were reviewed by a neuropathologist. For fluorescent IHC, FFPE tissue sections were deparaffinized and rehydrated by washing twice in xylene and 100% EtOH, once with 90 and 70% EtOH and twice in dH2O for 5 min each. Antigen retrieval was achieved by microwaving sections for 15 minutes in 10 mM citrate buffer, ph 6.0. Sections were stained overnight with anti-CD3 antibodies (1:100, CD3 (PC3/188A), alexa-488 Santa Cruz Biotechnology) together with either anti-PD-L2 (1:100, PD-L2 (D7U8CTM) XP® CST), anti-CTLA-4 (1:100, CTLA-4 (F-8), alexa-555 Santa Cruz Biotechnology) or anti-B7-H3 (1;100, B7-H3 (D9M2L) XP® CST). For PD-L2 and B7-H3 a secondary goat anti-rabbit alexa-555 antibody was applied. Sections were then washed in PBST and incubated with 0.05% Sudan black in 70% EtOH for 20 minutes at RT to reduce autofluorescence. Sections were then coverslipped using hard set mounting media containing DAPI (Vectashield). Tissue sections were imaged with a fluorescence microscope (Olympus BX51) with developer’s software and analyzed using ImageJ. Positive staining results were characterized semi-quantitatively for B7-H3 and PD-L2 as percentage of cells expressing membranous immune checkpoint proteins. Samples with no expression was reported as nil (-), 1–10% expression was reported as low (+), 10–50% was reported as moderate (++), and 50–100% was reported as high (+++). For CTLA-4, representative images of each sample at 40x magnification were analyzed for number of cells expressing the checkpoint protein. Representative images with 0–10 cells expressing CTLA-4 was recorded as low (+), 10–40 cells was recorded as moderate (++), and more than 40 cells was recorded as high (+++). Infiltrating T-cells were identified with the maker CD3. Samples were recorded as (+) if CD3 positive cells were present and (-) if staining was absent.
Statistical analysis
Differences in mean immune checkpoint protein levels between groups were tested using an unpaired Student’s t-test. Correlation analyses were performed using the Pearson coefficient analysis. Graphpad Prism 7.0a (GraphPad Software Inc.) was used for all statistical comparisons and correlation analyses.
Results
Comprehensive genetic characterization was performed on 20 meningioma (13 grade I, 6 grade II and 1 grade III) using a custom targeted NGS gene panel to screen for somatic mutations across 38 genes associated with the tumor. MLPA was used to screen for NF2 and 1p32 deletions and evaluate chromosomal instability. Gene mutation data for each patient is summarized in Table 1. NF2, 1p32 co-deletions were the most frequent gene change observed. NF2 and 1p32 mutations were present in both grade I and II tumor specimens. NF2 point mutations were frequently detected in the case set with NF2R57* mutation present in three patients. NF2 gene alterations were present in 50% of cases tested, which is in alignment with previously published studies.9,12 Point mutations associated with meningioma in SMO, TRAF7, PIK3CA, and AKT1 were also identified in tissues. The AKT1 E17K mutation was observed in three patients. Similar to previous reports,9,12 these non-NF2 mutations were found in grade I tumors except for PIK3CA H1047R and TRAF7 R641H that was present in the grade III specimen. Several additional mutations in SMO, IDH1, NEGR1, SUFU, PIK3CA and JARID genes not previously reported in meningioma were also identified in samples.
Table 1.
Patient characteristics and genetics.
| Case ID | Age | Sex | Location | Size – cm | Edema | WHO grade | Gene deletions (MLPA) |
Gene mutations (NGS) |
|---|---|---|---|---|---|---|---|---|
| M2 | 63 | F | Right mid parafalx | 1.8 | Moderate | I | Multiple other | Not specified |
| M3 | 62 | F | Left cerebellopontine angle | 3.5 | Nil | I | NF2, 1p34.1, other | Not specified |
| M4 | 33 | F | Right frontoparietal | 4.4 | Moderate | II | NF2 | Not specified |
| M5 | 71 | F | Left frontotemporal | 2.2 | Moderate | III | None | TRAF R641H, PIK3CA H1047R, IDH1 V178I |
| M6 | 54 | M | Left cerebellopontine angle | 0.8 | Nil | I | None | SMO W535L |
| M7 | 70 | M | Left frontotemporal | 4 | Moderate | I | NF2, 1p34.1 | IDH1 V178I |
| M8 | 52 | M | Right temporal | 6.7 | Mild | II | NF2, 1p34.1 | NF2 R57 STOP |
| M9 | 58 | F | Left frontal parietal parafalx | 6.6 | Nil | II | NF2 | IDH1 V178I, NEGR1 Y347S |
| M11 | 79 | F | Left cerebellopontine angle | 4.8 | Mild | I | None | SMO D25G, FANCD2 G901V |
| M12 | 68 | M | Left frontal medial sphenoid wing | 5 | Mild | I | None | AKT1 E17K |
| M13 | 57 | F | Left frontal | 2.6 | Mild | I | None | AKT1 E17K |
| M14 | 58 | F | Tuberculum sellae | 1.8 | Nil | I | None | NF2 E108 Stop |
| M15 | 75 | F | Left mid posterior parafalx | 7.4 | Moderate | II | NF2, 1p34.1 | IDH1 V178I |
| M17 | 46 | F | Left parietal parafalx | 1.6 | Nil | I | None | NF2 R57 STOP |
| M18 | 28 | F | Midline parafalx right> left | 7.5 | Moderate | II | None | Not specified |
| M19 | 28 | F | Left frontotemporal | 3 | Moderate | I | None | AKT1 E17K, SUFU A340S |
| M20 | 51 | F | Left cerebellopontine angle | 3 | Nil | I | None | PIK3CA I391M, NF2 insertion (frameshift) |
| M21 | 48 | F | Right post frontal parafalx | 4.7 | Nil | I | None | NF2 frameshift (p.L54Lfs*29) |
| M22 | 46 | M | Posterior parafalx parasagittal right > left | 6 | Mild | II | None | NF2 Y66N |
| M23 | 59 | F | Posterior-frontal parafalx | 2.9 | Nil | I | None | NF2 E379 STOP, JARID2 R717Q |
| M45 | 36 | F | Right parasagittal middle third | 7.5 | Nil | II | Not tested | Not tested |
| M65 | 69 | F | Left frontal | 7.8 | Moderate | III | Not tested | Not tested |
| C1 | 40 | F | Right lateral temporal cortex | |||||
| C2 | 28 | M | Left lateral temporal cortex |
We next quantified the expression of immune checkpoint proteins, PD-L2, CTLA-4, B7-H2, B7-H3, CD28, CD80, CD86 and ICOS as well as previously identified PD-1 and PD-L1 in each meningioma tissue using multiplex ELISA arrays. An additional grade II and grade III tumor specimen were included in addition to those that were genetically characterized. All immune checkpoint proteins were identified but amounts differed significantly for each immune checkpoint protein analyzed (Figure 1A). Not all tumor samples evenly expressed each immune checkpoint protein. B7-H3 was the most highly expressed (3070 ± 729 pg/mg of tissue) immune checkpoint protein, and also the only one present in each sample tested. PD-L2 was the next most abundant protein analysed (1793 ± 467 pg/mg of tissue). Levels of B7-H3 and PD-L2 in meningioma tissues were significantly higher than all other proteins analysed (p < 0.001). The most highly expressed immune checkpoint protein of immune cell origin was CTLA-4 (199 ± 78 pg/mg of tissue), which was identified in 8/20 meningiomas. PD-1 was found in most of the tumor specimens but at lower levels than CTLA-4 (51 ± 13 pg/mg of tissue). Expression of several immune checkpoint proteins were elevated in the grade III group (Figure 1A). High inter-patient variance in checkpoint protein expression was present for cases of the same WHO grade which has been mentioned in previous reports for PD-1 and PD-L1 (Figure 1B).
Gene changes may more accurately predict meningioma behavior than WHO grade, particularly for grade I and II tumors. We therefore evaluated checkpoint protein expression according to gene mutation group. As predicted, expression of checkpoint proteins was generally more consistent for meningiomas according to gene subtype than WHO grade (Figure 1B). Most notably, the expressions of PD-L2 and B7-H3 proteins were elevated in a subset of meningioma patients harboring oncogenic mutations disrupting the PI3K/AKT/mTOR pathway. Similarly, immune checkpoint protein CTLA-4 was primarily restricted to meningioma with a PIK3CA or SMO mutation. While these genetic subtypes of meningioma were associated with consistently elevated immune checkpoint proteins, the NF2 mutation group displayed quite variable expression data, particularly for PD-L2 (Figure 1B).
To confirm results obtained by multiplex ELISA and to evaluate the distribution and cellular location of abundant immune checkpoint proteins, fluorescent immunohistochemistry was performed for B7-H3, PD-L2 and CTLA-4 on tissue sections for each meningioma patient as well as two addition control lateral temporal cortex tissues. All 16/21 meningioma specimens showed high expression of B7-H3 with strong membrane staining observed in almost 100% of tumor cells for most cases. The 5 remaining meningioma tissues expressed moderate approaching high expression of B7-H3. Representative fluorescent images of tissues are shown in Figure 2A and semi-quantified in 2B. Importantly, no B7-H3 staining was observed in either lateral temporal cortex tissues. Staining of PD-L2 in tissues was high in 2/21, moderate in 5/21, low in 7/21 and absent in 7 of 21 meningioma tissues and control brain tissues. PD-L2 expression was primarily observed on cell membranes but cytosolic expression was also observed (Figures 2A and 3). PD-L2 staining was not uniform across sections and was present in multiple cell clusters of heterogeneous intensities throughout the tissue section (Figure 2A). Both B7-H3 and PD-L2 expression was present in each tumor grade. CTLA-4 was found in 3/21 meningioma tissues. CTLA-4 staining was only observed in atypical and malignant tumor specimens. CTLA-4 staining occurred in a heterogenous distribution across tumor sections in both singular cells and clusters of aggregated cells (Figure 2A). No CTLA-4 staining was present in lateral temporal cortex tissues. In general, immunofluorescent staining correlated to multiplex ELISA results although differences were observed for a few cases for PD-L2 and CTLA-4 which could be explained by the heterogeneous expression of these two checkpoint proteins.
Figure 2.
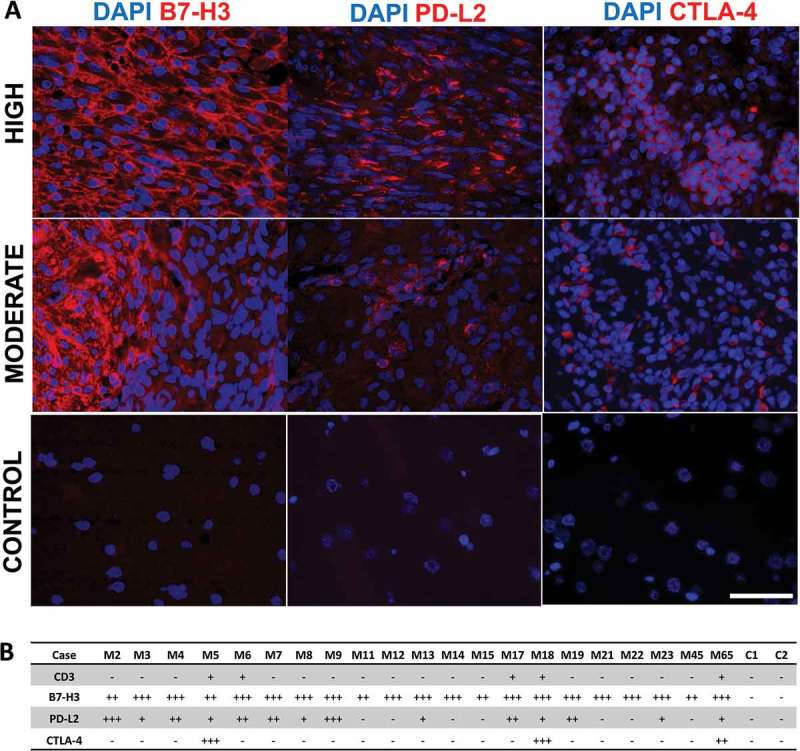
Fluorescence immunohistochemistry of B7-H3, PD-L2, and CTLA-4 in meningioma and lateral temporal cortex tissues. (A) Representative images of B7-H3, PD-L2, and CTLA-4 expression (red) in meningioma specimens and lateral temporal cortex control. Staining severity was determined semi-quantitatively as outlined in methods. Blue represents nuclear DAPI stain. All images were taken with Olympus BX51 fluorescence microscope at 40x magnification with scale bar measuring 50 µm. (B) Summary of CD3, B7-H3, PD-L2, and CTLA-4 expression in 21 meningioma patients (M#) and 2 lateral temporal cortex controls (C#). Staining severity (low, moderate and high expression) follows semi-quantitative characterization as outlined in methods.
Figure 3.
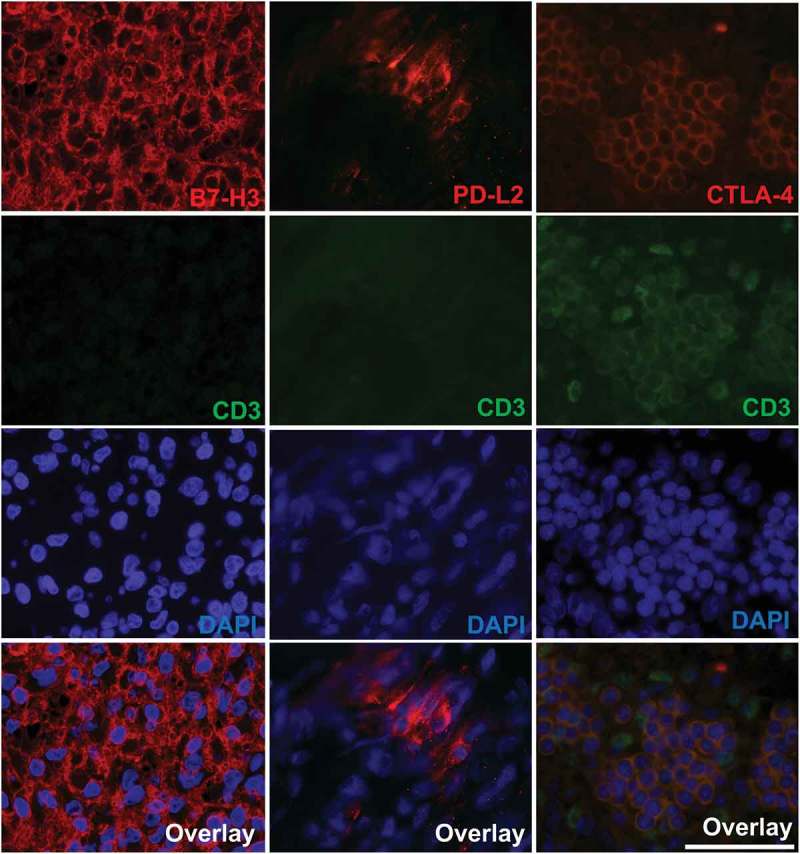
Fluorescence immunohistochemistry co-staining of CD3 together with B7-H3, PD-L2, and CTLA-4 in meningioma tissues. Representative images of DAPI (blue), CD3 (green) and B7-H3, PD-L2 and CTLA-4 (red) stains at 40x magnification (Olympus BX51). Bottom panel shows overlay image of DAPI, CD3 and checkpoint protein. Scale bar measures 50 µm. Mild expression is not shown in the figure as B7-H3 and CTLA-4 showed no mild staining.
Next, to determine whether immune checkpoint proteins were expressed in infiltrating immune cells or in tumor cells, tissues were co-stained for checkpoint proteins together with CD3, a marker of T-cells. Both B7-H3 and PD-L2 were present only on tumor cells Figure 3. All cells expressing CTLA-4 were also positive for CD3 membrane staining, confirming CTLA-4 is expressed on infiltrating T-cells (Figure 3).
Peritumoral brain edema occurs in 38%–67% patients with meningioma25 and accompanied with mast cell infiltration.20 Given that mast cells can modulate immune responses including immune cell recruitment, we next evaluated the effect of edema severity on checkpoint protein expression. Interestingly, no correlation between edema severity and tumor size was observed (r = 0.1142, P value = 0.595; Figure 4A). However, a positive correlation between edema severity and tumor grade was noted (r = 0.5043, P value = 0.012; Figure 4B). In contrast, checkpoint protein expression in meningioma tissue was very similar for patients with no edema and those with either mild or moderate edema (Figure 4C). Similarly, the severity of edema was highly variable in meningioma of the same genetic subtype (Figure 4D). Representative MR images of patients with and without edema and various tumor sizes with associated immune checkpoint protein expression are shown in Figure 4E.
Figure 4.
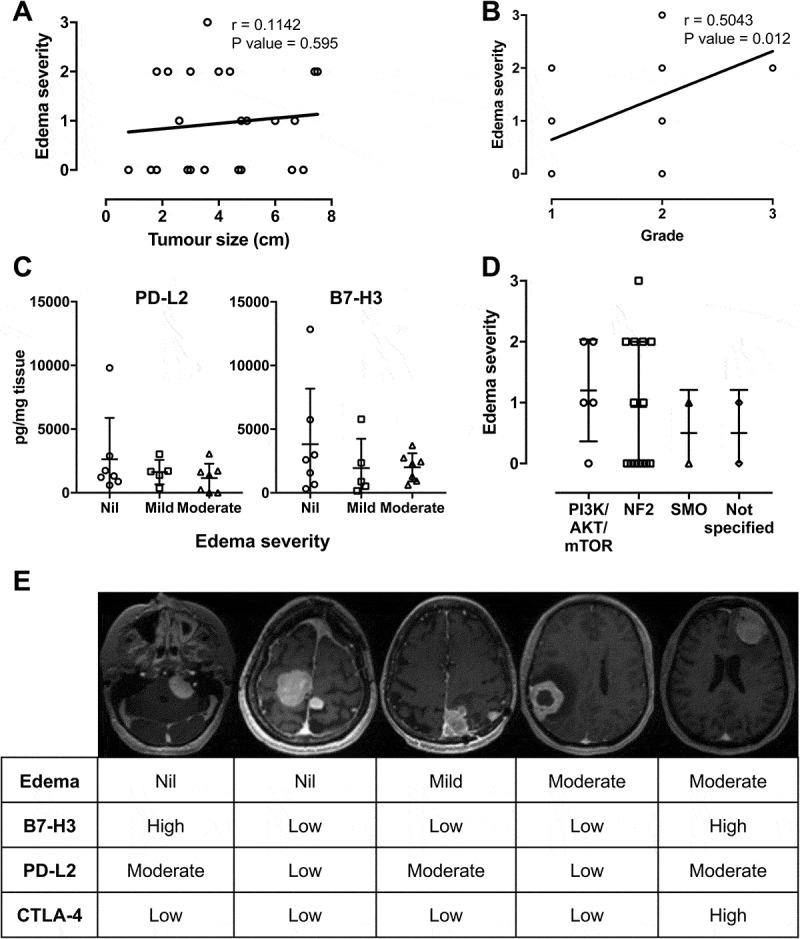
Effects of edema on immune checkpoint expression and correlation with tumor size, grade and genetic subtype. Correlation analysis of edema severity with tumor size (A) and tumor WHO grade (B). (C) Expression of PD-L2 and B7-H3 is quantified according to severity of edema. (D) Edema severity of each patient according to genetic mutation subtype. (Edema severity in A and B; 0 = nil, 1 = mild, 2 = moderate, 3 = severe). (E) Representative axial T1 post – Gd MR images acquired at 1.5T and associated levels of immune checkpoint proteins from 5 patients.
Discussion
In this study, we identified several novel inhibitory immune checkpoint proteins present in meningioma. More intriguing was the discovery that profiles of immune checkpoint protein expression are associated with common gene mutations recently described for meningioma. Immune checkpoint proteins B7-H3 and PD-L2 were consistently elevated in meningioma that also express oncogenic mutations that disrupt the PI3K/AKT/mTOR pathway. Similarly, we found an abundance of immune cell checkpoint protein, CTLA-4, in high-grade meningioma and those harboring a PIK3CA or SMO mutation. This work builds on recent reports that identified checkpoint proteins PD-L1 and its binding partner PD-1 in meningioma tissue, and the interesting observation that expression of PD-L1 in tumor cells increases relative to WHO grade.17,18 Subsequently, three clinical trials were established for treating malignant meningioma with checkpoint inhibitors (an anti-PD-1 antibody).21–23 Unfortunately, only ~ 2–4% of meningioma patients would be eligible for these trials. However, our data demonstrates that molecular classification might be a powerful tool for stratifying additional patient populations eligible for anti-PD-1 immunotherapy. Moreover, having discovered additional checkpoint proteins in meningioma, we have uncovered further molecular targets and patient groups to investigate for possible immunotherapy treatment (e.g. treating PIK3CA, SMO patients and high-grade tumors with the approved anti-CTLA-4 antibody; Ipilimumab).
A unique discovery of this study is that PD-L2 and B7-H3 proteins were much more abundant in meningioma tissues than previously reported tumor cell immune checkpoint protein PD-L1. Indeed, PD-L1 expression was quite low for each tumor specimen examined except for one grade III sample. Limited PD-L1 protein level was expected in Grade I meningioma and high levels are associated with grade III tumors, it was surprising that low levels of PD-L1 were also observed for each grade II specimen. As low expression of PD-L1 has been reported in ~ 40% of grade II meningioma,17,18 this could in part explain our result. Of note, our data indicate that PD-L2 and B7-H3 may play more important roles in regulating immune responses in meningioma, particularly for grade I and II tumors that represent > 96% of all cases. This would represent a significant advancement both in the understanding of immunosuppressive mechanisms present in meningioma and that many PD-L1 negative meningioma could also respond to PD-1 checkpoint blockade.
PD-L2 shares very similar homology to PD-L1 and is the only other known binding partner of PD-1.26 Much less is known about PD-L2, compared to the more extensively studied PD-L1. However, recent findings suggest that PD-L2 is a very important immunological molecule in several cancers.27,28 The PD-L2 ligand, when interacting with its receptor, PD-1, is one of many inhibitory pathways of the B7-CD28 family that inhibits effector T cell proliferation and assist tumor immune evasion. The signal transducer and activator of transcription (STAT) 6 pathway, activated by the ligation of IL-4 and IL13 receptor with their ligands IL-4 or IL-13, in addition to the cytokines TSLP and IL-27, plays an important role in regulating Th2 immune responses and may also induce PD-L2 expression.29
In 2017, Yearley and colleagues screened 400 tumor samples from seven types of cancer for both PD-L1 and PD-L2. They found that PD-L2 was not only present in each cancer and correlated with PD-L1 in both amount and distribution, but also was detected in tumor samples where PD-L1 was absent. In a number of cancer types tested, most notably, gastric and head and neck squamous cell carcinoma, PD-L2 expression was considerably greater than PD-L1.27 Importantly, patients in the KEYNOTE-012 trial with PD-L2+ tumors had more than two times overall response rate to the PD-1 checkpoint blockade drug pembrolizumab than those with PD-L1 only.27 PD-L2 expression in tumors could explain why some patients negative for PD-L1 still respond to anti-PD1 immunotherapies and why a portion of PD-L1 positive tumors does not.30–32 The finding that PD-L2 is abundant in meningioma irrespective of grade is a unique finding suggesting novel treatment strategies for these patients and worth pursuing in additional studies.
B7-H3 was the most prevalent and abundant inhibitory immune checkpoint protein quantified in meningioma. Astonishingly, B7-H3 was expressed in close to 100% of tumor cells in the majority of meningioma specimens tested. B7-H3 is known to be present in both tumor and infiltrating immune cells. However, it was not observed in infiltrating immune cells in the meningiomas examined in this study. B7-H3 is a member of the B7 and CD28 families. While it has been shown to be present in many solid tumors33 B7-H3’s roles in tumor immunity are not well defined. B7-H3’s binding partner is not known. B7-H3 has been described to function as both a stimulatory and inhibitory regulator of the immune response.34 However, many recent publications now focus on its potent inhibitory activity on tumor immune response.35–38 B7-H3 expression has been correlated to poor outcome in patients with non-small cell lung cancer and prostate cancer.36,39,40 Blocking B7-H3 function has been shown to reduce growth rates of cancer and an anti-B7-H3 mAb, enoblituzumab, was developed and is currently in phase I and II clinical trials for treatment of solid tumors.41–43 The fact that we observed moderate to high expression of B7-H3 in each tissue specimen tested, suggests B7-H3 is likely a key molecule in meningioma biology. Importantly, B7-H3 was recently implicated in upstream activation of the PI3K/AKT/mTOR and NF-ĸB pathways, that play a significant role in meningioma biology.41 Knockdown of B7-H3 in bladder cancer cells reduce AKT signaling and the migration and invasion of these cells.44 Furthermore, decreasing expression of B7-H3 in breast cancer cells sensitizes these cells in culture and tumor xenografts to PI3K/AKT/mTOR inhibitors.45 In non-small cell lung cancer, treatment of cells in culture with AKT and mTOR inhibitors lead to decreased expression of B7-H3 suggesting a positive feedback mechanism may exist between PI3K/AKT/mTOR signaling and B7-H3. Although not tested in the current study, this mechanism may help explain why patients with AKT1 and PIK3CA mutations displayed higher than average B7-H3 levels. Application of soluble B7-H3 promotes the malignancy of pancreatic carcinoma cells via NF-ĸB. 46 Furthermore, in a mouse model of pneumococcal meningitis application of recombinant B7-H3 led to production of cytokines also via NF-ĸB signaling.47 As more details are confirmed on the role of this immune checkpoint protein in tumor biology it will be interesting to see how new findings can be incorporated into our understanding of meningioma pathogenesis and whether meningioma patients can benefit from treatment with drugs like enoblituzumab.
Five of our meningioma samples were positive for immune checkpoint protein CTLA-4. CTLA-4 is associated with inhibiting helper T cell activity and enhancing regulatory T cell immunosuppressive activity. Immune responses are supressed by CTLA-4 outcompeting CD28, a T cell co-stimulatory receptor which activates T cells by amplifying T cell receptor signalling, binding to its ligands CD80 and CD86.48 Furthermore, recent studies suggest CTLA-4 also blocks T cell activation by activating the protein phosphatases SHP2 and PP2A, which reduce kinase signalling by T cell receptors.49 CTLA-4 is predominately associated with immune cells but is also expressed in tumor cells.50 In our meningioma samples CTLA-4 was restricted to CD3+ infiltrating T-cells. The discovery that some meningioma contain infiltrating immune cells expressing this protein is exciting, given the success of treating other types of cancer with the anti-CTLA mAb checkpoint blocking drug, Ipilimumab.51 CTLA-4 was not as ubiquitously expressed as B7-H3 or PD-L2, and was primarily found in higher tumor grades. An important observation was that 3 patients with CTLA-4 also carried a mutation in either SMO or PIK3CA. The protein smoothened encoded by SMO, is a class frizzled G protein coupled receptor that is part of the hedgehog signaling pathway. Crosstalk between hedgehog and PI3K/AKT/mTOR signaling pathways is implicated in other cancers.52,53 So it is not surprising that CTLA-4 expression could be similar in patients with mutations in either of these pathways. Combination of the smo inhibitors vismogedib and sonidegib with inhibitors of PI3K/AKT/mTOR is currently being investigated in pre-clinical animal and cell models of various cancers.37 However, we did not detect elevated CTLA-4 in AKT1 mutated meningioma. Smoothened can activate PI3K via the G protein Gi, which is upstream of AKT activity, suggesting CTLA-4 expression may be regulated at the level of PI3K activation (see Figure 5). This will be important to establish in subsequent studies in order to identify patients groups most likely to respond to CTLA-4 blockade. Interestingly, application of PI3Kβ inhibitors was shown to improve the efficacy of anti-CTLA-4 antibodies in treating a mouse model of melanoma,54 suggesting combination of targeted therapy and CTLA-4 blockade could be a strategy to apply to meningioma patients. Finally, we should note that we also observed expression of CTLA-4 in an additional grade III meningioma that did not undergo genetic testing, suggesting CTLA-4 could be important in this meningioma group and it will be worthwhile investigating the use of CTLA-4 checkpoint blockade in addition to anti-PD1 therapy for malignant meningioma.
Figure 5.
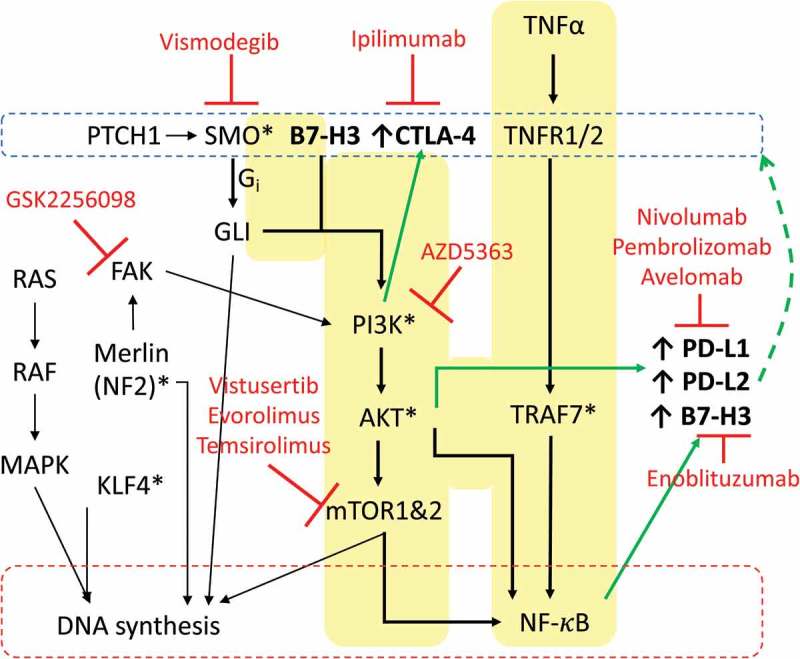
Oncogenic pathways and drug targets important to meningioma and immune checkpoint protein regulation. Key pathways involved in regulating checkpoint proteins in meningioma are highlighted in yellow. Drug therapies in clinical trial or FDA approved towards targets are in red. * Represents mutations in encoding gene is common in meningioma.
Meningioma positive for gene mutations effecting the PI3K/AKT/mTOR signaling also had elevated levels of immune checkpoint proteins PD-L2 and B7-H3. However, unlike CTLA-4 expression profiles, PD-L2 and B7-H3 immune checkpoint proteins were also elevated in patients with the AKT1 E17K mutation. There is evidence for this pathway regulating both gene expression of PD-L1 and PD-L2 via NF-κB and perhaps by enhancing PD-L1 stabilization at the tumor cell membrane via reducing GSK3β phosphorylation (refer to Figure 5).55–57 Importantly, gene expression of PD-L1 can be reduced with application of inhibitors of PI3K/AKT/mTOR signaling.58 This suggests future combination treatment with targeted and immunotherapy could provide synergistic effects in meningioma patients with altered PI3K/AKT/mTOR signaling. This strategy has been proposed for other cancers.59 Disruption of the PI3K/AKT/mTOR pathway is a factor in up to ~ 30% of all cancers making it the most prevalent cellular mechanism contributing to cancer. While many targeted therapies are being developed towards these molecules in this pathway, therapeutic response in patients have been underwhelming. Results from this study showing immune responses may be influenced by gene mutations in PIK3CA and AKT1 offer important insight into how PI3K/AKT/mTOR alterations could contribute to tumor biology and how immunotherapy in combination could improve patient responses to PI3K/AKT/mTOR inhibitors.
Peritumoral brain edema is common in patients with meningioma.25 The cause of edema is not known but is correlated to tumor grade. However, mast cell infiltration is observed in the microenvironment adjacent to edema,20 and we were interested to examine whether this effected immune checkpoint protein expression. No relationship between severity of edema and levels of any immune checkpoint protein was observed. While we cannot rule out a role of edema in regulating immune marker expression, as no comprehensive IHC was included in this study, these results are similar to the previous report that showed macrophage infiltration in meningioma was correlated to the deletion of NF2 gene but was not effected by edema.60
In summary, we show that meningioma invokes a diverse range of immune responses, which in part may be contributed to by common genetic changes associated with this tumor. Immune checkpoint proteins PD-L2 and B7-H3 were the most abundant proteins tested and present in all grades of meningioma, indicating that these proteins may be of more importance to immune responses than previously described immune checkpoint proteins in meningioma. Furthermore, we also describe here the presence of CTLA-4 in high grade meningiomas. It will be interesting to validate these findings in a larger study covering all morphological and genetic subtypes of meningioma and evaluate the effects of these immune markers on clinical outcome.
Funding Statement
This study was funded in part from Advancing iMRI, Western Economic Diversification and Calgary Health Trust.
Acknowledgments
We thank the Division of Neurosurgery – Department of Clinical Neurosciences at Foothills Medical Centre, Calgary Alberta, particularly Dr. Yves Starreveld for providing samples.
Disclosure statement
The authors declare no conflicts of interest.
References
- 1.Wiemels J, Wrensch M, Claus EB.. Epidemiology and etiology of meningioma. J Neurooncol. 2010;99(3):307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowosielski M, Galldiks N, Iglseder S, Kickingereder P, von Deimling A, Bendszus M, Wick W, Sahm F. Diagnostic challenges in meningioma. Neuro Oncol. 2017;19(12):1588–1598. doi: 10.1093/neuonc/nox101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Adegbite AB, Khan MI, Paine KW, Tan LK. The recurrence of intracranial meningiomas after surgical treatment. J Neurosurg. 1983;58(1):51–56. doi: 10.3171/jns.1983.58.1.0051. [DOI] [PubMed] [Google Scholar]
- 5.Rohringer M, Sutherland GR, Louw DF, Sima AA. Incidence and clinicopathological features of meningioma. J Neurosurg. 1989;71(5 Pt 1):665–672. doi: 10.3171/jns.1989.71.5.0665. [DOI] [PubMed] [Google Scholar]
- 6.Minniti G, Amichetti M, Enrici RM. Radiotherapy and radiosurgery for benign skull base meningiomas. Radiat Oncol. 2009;4:42. doi: 10.1186/1748-717X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vera E, Iorgulescu JB, Raper DM, Madhavan K, Lally BE, Morcos J, Elhammady S, Sherman J, Komotar RJ. A review of stereotactic radiosurgery practice in the management of skull base meningiomas. J Neurol Surg B Skull Base. 2014;75(3):152–158. doi: 10.1055/s-0033-1354747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S, Okonechnikov K, Koelsche C, Reuss DE, Capper D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 9.Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, Avşar T, Li J, Murray PB, Henegariu O, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science (New York, NY). 2013;339(6123):1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuzawa S, Nishihara H, Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016;33(4):237–247. doi: 10.1007/s10014-016-0271-7. [DOI] [PubMed] [Google Scholar]
- 11.Abedalthagafi MS, Bi WL, Merrill PH, Gibson WJ, Rose MF, Du Z, Francis JM, Du R, Dunn IF, Ligon AH, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015;208(6):345–350. doi: 10.1016/j.cancergen.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuss DE, Piro RM, Jones DT, Simon M, Ketter R, Kool M, Becker A, Sahm F, Pusch S, Meyer J, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125(3):351–358. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 14.Bujko M, Kober P, Tysarowski A, Matyja E, Mandat T, Bonicki W, Siedlecki JA. EGFR, PIK3CA, KRAS and BRAF mutations in meningiomas. Oncol Lett. 2014;7(6):2019–2022. doi: 10.3892/ol.2014.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vismodegib and FAK inhibitor GSK2256098 in treating patients with progressive meningiomas. https://ClinicalTrials.gov/show/NCT02523014; 2015.
- 16.Domingues PH, Teodósio C, Ortiz J, Sousa P, Otero A, Maillo A, Bárcena P, GarcÌa-Macias MC, Lopes MC, de Oliveira C, et al. Immunophenotypic identification and characterization of tumor cells and infiltrating cell populations in meningiomas. Am J Pathol. 2012;181(5):1749–1761. doi: 10.1016/j.ajpath.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 17.Du Z, Abedalthagafi M, Aizer AA, McHenry AR, Sun HH, Bray MA, Viramontes O, Machaidze R, Brastianos PK, Reardon DA, et al. Increased expression of the immune modulatory molecule PD-L1 (CD274) in anaplastic meningioma. Oncotarget. 2015;6(7):4704–4716. doi: 10.18632/oncotarget.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han SJ, Reis G, Kohanbash G, Shrivastav S, Magill ST, Molinaro AM, McDermott MW, Theodosopoulos PV, Aghi MK, Berger MS, et al. Expression and prognostic impact of immune modulatory molecule PD-L1 in meningioma. J Neurooncol. 2016;130(3):543–552. doi: 10.1007/s11060-016-2256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang L, Lowther DE, Meizlish ML, Anderson RC, Bruce JN, Devine L, Huttner AJ, Kleinstein SH, Lee JY, Stern JN, et al. The immune cell infiltrate populating meningiomas is composed of mature, antigen-experienced T and B cells. Neuro Oncol. 2013;15(11):1479–1490. doi: 10.1093/neuonc/not110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polyzoidis S, Koletsa T, Panagiotidou S, Ashkan K, Theoharides TC. Mast cells in meningiomas and brain inflammation. J Neuroinflammation. 2015;12:170. doi: 10.1186/s12974-015-0388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A study of nivolumab in adult participants with recurrent high-grade meningioma. https://ClinicalTrials.gov/show/NCT02648997; 2016.
- 22.Phase II trial of pembrolizumab in recurrent or residual high grade meningioma. https://ClinicalTrials.gov/show/NCT03279692; 2017.
- 23.Neoadjuvant avelumab and hypofractionated proton radiation therapy followed by surgery for recurrent radiation-refractory meningioma. https://ClinicalTrials.gov/show/NCT03267836; 2017.
- 24.Proctor DT, Yoo EH, Vujadinovic Z, Lama S, van Marle G, Sutherland GR. Optimizing gDNA extraction from fresh frozen meningioma tissue for downstream genetic analysis. Clin Biochem. 2017;50(4–5):194–205. doi: 10.1016/j.clinbiochem.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Go KG, Wilmink JT, Molenaar WM. Peritumoral brain edema associated with meningiomas. Neurosurgery. 1988;23(2):175–179. [DOI] [PubMed] [Google Scholar]
- 26.Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 27.Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, Lunceford J, Cheng J, Chow LQM, Seiwert TY, et al. PD-L2 expression in human tumors: relevance to anti-PD-1 therapy in cancer. Clin Cancer Res. 2017;23(12):3158–3167. doi: 10.1158/1078-0432.CCR-16-1761. [DOI] [PubMed] [Google Scholar]
- 28.Masugi Y, Nishihara R, Hamada T, Song M, da Silva A, Kosumi K, Gu M, Shi Y, Li W, Liu L, et al. Tumor PDCD1LG2 (PD-L2) expression and the lymphocytic reaction to colorectal cancer. Cancer Immunol Res. 2017;5(11):1046–1055. doi: 10.1158/2326-6066.CIR-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozali EN, Hato SV, Robinson BW, Lake RA, Lesterhuis WJ. Programmed death ligand 2 in cancer-induced immune suppression. Clin Dev Immunol. 2012;2012:656340. doi: 10.1155/2012/656340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 32.Schachter J, Ribas A, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet. 2017;390(10105):1853–1862. doi: 10.1016/S0140-6736(17)31601-X. [DOI] [PubMed] [Google Scholar]
- 33.Picarda E, Ohaegbulam KC, Zang X. Molecular pathways: targeting B7-H3 (CD276) for human cancer immunotherapy. Clin Cancer Res. 2016;22(14):3425–3431. doi: 10.1158/1078-0432.CCR-15-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitner J, Klauser C, Pickl WF, Stöckl J, Majdic O, Bardet AF, Kreil DP, Dong C, Yamazaki T, Zlabinger G. B7-H3 is a potent inhibitor of human T-cell activation: no evidence for B7-H3 and TREML2 interaction. Eur J Immunol. 2009;39(7):1754–1764. doi: 10.1002/eji.200839028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Chong KK, Nakamura Y, Nguyen L, Huang SK, Kuo C, Zhang W, Yu H, Morton DL, Hoon DS. B7-H3 associated with tumor progression and epigenetic regulatory activity in cutaneous melanoma. J Invest Dermatol. 2013;133(8):2050–2058. doi: 10.1038/jid.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang X, Thompson RH, Al-Ahmadie HA, Serio AM, Reuter VE, Eastham JA, Scardino PT, Sharma P, Allison JP. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci U S A. 2007;104(49):19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zang X, Sullivan PS, Soslow RA, Waitz R, Reuter VE, Wilton A, Thaler HT, Arul M, Slovin SF, Wei J, et al. Tumor associated endothelial expression of B7-H3 predicts survival in ovarian carcinomas. Mod Pathol. 2010;23(8):1104–1112. doi: 10.1038/modpathol.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingebrigtsen VA, Boye K, Nesland JM, Nesbakken A, Flatmark K, Fodstad Ø. B7-H3 expression in colorectal cancer: associations with clinicopathological parameters and patient outcome. BMC Cancer. 2014;14:602. doi: 10.1186/1471-2407-14-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Wang Y, Zhao J, Gu M, Giscombe R, Lefvert AK, Wang X. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer. 2006;53(2):143–151. doi: 10.1016/j.lungcan.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 40.Benzon B, Zhao SG, Haffner MC, Takhar M, Erho N, Yousefi K, Hurley P, Bishop JL, Tosoian J, Ghabili K, et al. Correlation of B7-H3 with androgen receptor, immune pathways and poor outcome in prostate cancer: an expression-based analysis. Prostate Cancer Prostatic Dis. 2017;20(1):28–35. doi: 10.1038/pcan.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flem-Karlsen K, Fodstad Ø, Tan M, Nunes-Xavier CE. B7-H3 in cancer - beyond immune regulation. Trends Cancer. 2018;4(6):401–404. doi: 10.1016/j.trecan.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Enoblituzumab (MGA271) in children with B7-H3-expressing solid tumors. https://ClinicalTrials.gov/show/NCT02982941; 2016.
- 43.Safety study of enoblituzumab (MGA271) in combination with pembrolizumab in refractory cancer. https://ClinicalTrials.gov/show/NCT02475213; 2015.
- 44.Li Y, Guo G, Song J, Cai Z, Yang J, Chen Z, Wang Y, Huang Y, Gao Q. B7-H3 promotes the migration and invasion of human bladder cancer cells via the PI3K/Akt/STAT3 signaling pathway. J Cancer. 2017;8(5):816–824. doi: 10.7150/jca.17759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nunes-Xavier CE, Karlsen KF, Tekle C, Pedersen C, Øyjord T, Hongisto V, Nesland JM, Tan M, Sahlberg KK, Fodstad Ø. Decreased expression of B7-H3 reduces the glycolytic capacity and sensitizes breast cancer cells to AKT/mTOR inhibitors. Oncotarget. 2016;7(6):6891–6901. doi: 10.18632/oncotarget.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie C, Liu D, Chen Q, Yang C, Wang B, Wu H. Soluble B7-H3 promotes the invasion and metastasis of pancreatic carcinoma cells through the TLR4/NF-κB pathway. Sci Rep. 2016;6:27528. doi: 10.1038/srep27528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Li Y, Blankson S, Liu M, Huang D, Redmond HP, Huang J, Wang JH, Wang J. B7-H3 augments inflammatory responses and exacerbates brain damage via amplifying NF-κB p65 and MAPK p38 activation during experimental pneumococcal meningitis. PLoS One. 2017;12(1):e0171146. doi: 10.1371/journal.pone.0171146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudd CE, Taylor A, Schneider H. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol Rev. 2009;229(1):12–26. doi: 10.1111/j.1600-065X.2009.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Contardi E, Palmisano GL, Tazzari PL, Martelli AM, Falà F, Fabbi M, Kato T, Lucarelli E, Donati D, Polito L, et al. CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int J Cancer. 2005;117(4):538–550. doi: 10.1002/ijc.21155. [DOI] [PubMed] [Google Scholar]
- 51.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brechbiel J, Miller-Moslin K, Adjei AA. Crosstalk between hedgehog and other signaling pathways as a basis for combination therapies in cancer. Cancer Treat Rev. 2014;40(6):750–759. doi: 10.1016/j.ctrv.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Wei L, Xu Z. Cross-signaling among phosphinositide-3 kinase, mitogen-activated protein kinase and sonic hedgehog pathways exists in esophageal cancer. Int J Cancer. 2011;129(2):275–284. doi: 10.1002/ijc.25673. [DOI] [PubMed] [Google Scholar]
- 54.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409–416. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 56.Gowrishankar K, Gunatilake D, Gallagher SJ, Tiffen J, Rizos H, Hersey P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-κB. PLoS One. 2015;10(4):e0123410. doi: 10.1371/journal.pone.0123410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lastwika KJ, Wilson W, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 59.O’Donnell JS, Massi D, Teng MWL, Mandala M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin Cancer Biol. 2018;48:91–103. doi: 10.1016/j.semcancer.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 60.Domingues PH, Teodósio C, Otero Á, Sousa P, Ortiz J, Macias Mdel C, Gonçalves JM, Nieto AB, Lopes MC, de Oliveira C, et al. Association between inflammatory infiltrates and isolated monosomy 22/del(22q) in meningiomas. PLoS One. 2013;8(10):e74798. doi: 10.1371/journal.pone.0074798. [DOI] [PMC free article] [PubMed] [Google Scholar]


