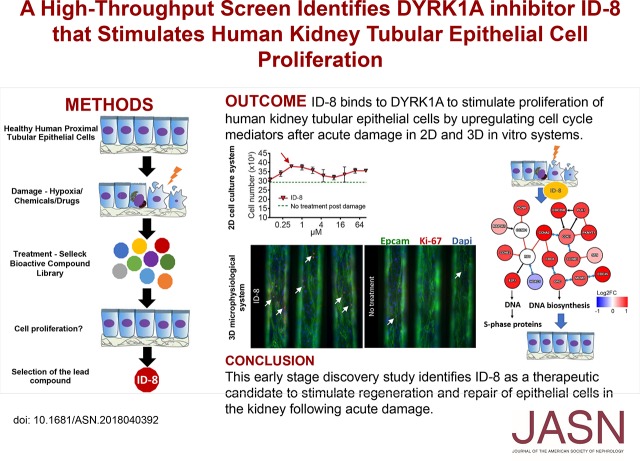
Abstract
Background
The death of epithelial cells in the proximal tubules is thought to be the primary cause of AKI, but epithelial cells that survive kidney injury have a remarkable ability to proliferate. Because proximal tubular epithelial cells play a predominant role in kidney regeneration after damage, a potential approach to treat AKI is to discover regenerative therapeutics capable of stimulating proliferation of these cells.
Methods
We conducted a high-throughput phenotypic screen using 1902 biologically active compounds to identify new molecules that promote proliferation of primary human proximal tubular epithelial cells in vitro.
Results
The primary screen identified 129 compounds that stimulated tubular epithelial cell proliferation. A secondary screen against these compounds over a range of four doses confirmed that eight resulted in a significant increase in cell number and incorporation of the modified thymidine analog EdU (indicating actively proliferating cells), compared with control conditions. These eight compounds also stimulated tubular cell proliferation in vitro after damage induced by hypoxia, cadmium chloride, cyclosporin A, or polymyxin B. ID-8, an inhibitor of dual-specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A), was the top candidate identified as having a robust proproliferative effect in two-dimensional culture models as well as a microphysiologic, three-dimensional cell culture system. Target engagement and genetic knockdown studies and RNA sequencing confirmed binding of ID-8 to DYRK1A and upregulation of cyclins and other cell cycle regulators, leading to epithelial cell proliferation.
Conclusions
We have identified a potential first-in-class compound that stimulates human kidney tubular epithelial cell proliferation after acute damage in vitro.
Keywords: Acute kidney injury, renal tubular epithelial cells, DYRK inhibitor, ID-8, Regenerative medicine, proliferation
Visual Abstract
AKI affects one in five hospitalized patients worldwide and its incidence is currently increasing.1,2 AKI is associated with substantial morbidity and mortality and is recognized as a leading cause of CKD.3,4
The death of epithelial cells in the proximal tubules is thought to be the primary cause of AKI5 as these cells serve as sensors, effectors, and targets of injury.6 However, they also have a remarkable ability to proliferate and repair tubules after damage. During tissue repair after kidney injury, the surviving epithelial cells are responsible for repopulating the tubule through a process of dedifferentiation, proliferation, and redifferentiation.7 Recent evidence also shows that the AKI-to-CKD transition is triggered by the incomplete repair of the renal tubules after injury, which may eventually lead to interstitial renal fibrosis.8 Therefore, we hypothesized that discovery of new therapeutics that promote efficient tubular epithelial cell proliferation may allow regression of kidney injury, thereby preventing AKI and the development of fibrosis, and halting progression to CKD.
In vitro phenotypic high-throughput screens (HTS) have enabled the discovery of mitogenic small-molecule drugs that promote proliferation of pancreatic β cells and hepatocytes as potential therapeutics for diabetes and liver disease.9,10 We therefore conducted HTS to identify compounds that can stimulate kidney tubular epithelial cell proliferation. Primary human proximal tubular epithelial cells (HPTECs) have previously been characterized as a relevant in vitro model for studying kidney cell damage and recovery in both two-dimensional (2D) culture models and a three-dimensional (3D) microphysiologic system (MPS).11 These in vitro systems retain many features of the differentiated kidney proximal tubular epithelium, such as polar architecture; junctional assembly; expression and activity of transporters; the ability to respond to physiologic stimuli, stress, and toxicity; and the ability to perform critical biochemical synthetic activities.11,12 We screened primary HPTECs against the Selleck Bioactive Compound Library, which contains structurally diverse, medicinally active, and cell-permeable FDA-approved compounds, active pharmaceutical and chemotherapeutic agents, and a small number of natural products. Serial rounds of phenotypic HTS identified ID-8 (1-[4-Methoxyphenyl]-2-methyl-3-nitro-1H-indol-6-ol), an inhibitor of the dual-specificity tyrosine-phosphorylation-regulated kinase 1A13 (DYRK1A) that induces epithelial cell proliferation after injury in 2D and 3D culture systems. We propose that this compound may have the potential to be developed into a therapeutic for AKI.
Methods
Cell Culture
Primary HPTECs (Biopredic International, Saint-Grégoire, France) from three different unique donors and NIH/3T3 fibroblasts (American Type Culture Collection no. CRL-1658) were used. Detailed methods are described in Supplemental Material.
Primary Screen
A primary screen of 1902 compounds was performed at the Institute of Chemistry and Cell Biology, Longwood Facility, Harvard Medical School. Primary HPTECs were automatically seeded in 96-well plates (WellMate; Thermo Scientific) in DMEM/Ham-F12 GlutaMAX medium (Thermo Scientific) supplemented with penicillin/streptomycin, hydrocortisone, EGF, insulin-transferrin-selenium, and triiodothyronine (full medium, see Supplemental Material for a detailed description). On day 1, full medium was replaced with DMEM/Ham-F12 GlutaMAX medium containing only penicillin/streptomycin (free medium) to deprive cells of growth signals and increase their sensitivity to proliferative stimuli. On day 3, cells were treated in duplicates with 11 µM dilutions of the Selleck library or with a panel of controls—full medium (positive control), free medium (negative control), or 0.1 µM digoxin (toxic control)—using a Seiko Compound Transfer Robot. The 11 µM concentration was on the basis of previous studies that performed similar assays.9,14,15 After treatment, live-cell imaging was performed using digital phase contrast to generate a baseline cell count at 0 hour. On day 5, cells were fixed and permeabilized, and nuclei were stained and counted at 48 hours (Operetta High-Content Imaging System; PerkinElmer).
Raw images were automatically analyzed for nuclei segmentation, nuclei/cell counting, and cell area (Columbus 2.4.2 Software; PerkinElmer). Proliferation rate or normalized cell count (NCC) was calculated on the basis of nuclei counts at day 5 normalized to (1) live-cell count at day 3 (0 hour), (2) cell area at day 5, and (3) mean of eight free medium–treated control wells on each plate. Cells treated with library compounds or with the panel of controls were assigned as proliferating if NCC>1, nonproliferating if NCC=1, or dying if NCC<1. The assay robustness, reproducibility, and variability were evaluated by determining the Z-values across multiple replicates using the panel of controls.
Secondary Screen
Compounds with an average NCC>1.1 in the primary screen were taken forward to secondary screening. As in the primary screen, primary HPTECs were seeded in full medium. On day 1, full medium was replaced with free medium. On day 3, cells were treated in triplicate with the selected compounds using a D300 drug dispenser (Hewlett Packard) at 1, 3, 10, and 30 µM for 48 hours. After treatment, cells were counted as in the primary screen. On day 5, 4 hours before fixing the cells, the modified thymidine analog EdU (Click-iT EdU Plus; Invitrogen) was added to mark proliferating cells. NCC was calculated as described in the primary screen section. We also evaluated the rate of proliferating cells on the basis of the percentage of EdU-labeled cells in compound-treated wells compared with free medium controls to catch differences in proliferation based not only in the cell number. Detailed EdU assay protocol is described in the Supplemental Material.
In Vitro Damage Models
Induction of proliferation in primary HPTECs after damage was assessed using four different in vitro models of acute cell damage: (1) hypoxia (1% O2, 29% of cell death), (2) 15 µM cadmium chloride (CdCl2; 16% of cell death), (3) 5 µM cyclosporin A (CsA; 12% of cell death), or (4) 75 µM polymyxin (PMB; 11% of cell death) for 24 hours as described in the Supplemental Material. In contrast to the screening phases, all supplements with the exception of growth factor EGF were added back into the cell medium (EGF-free medium). Damaged cultures were treated with two concentrations of the hit compounds from the secondary screen using D300 drug dispenser (Hewlett Packard). Cells were treated for 24, 72, or 96 hours and the proliferation effect was measured by comparison of compound-treated cell counts with the untreated control. After treatment, cells were fixed and nuclei were stained and counted (Operetta High-Content Imaging). Compounds that promoted cell proliferation in at least two different damage models were used to treat cells after damage in a ten-point dose range (2.15-fold serial dilution) from 0.1 to 100 µM for 96 hours, spanning the primary screen concentration.
The compound that demonstrated strongest proliferative potential using NCC was used to treat cells along with inactive and active DYRK inhibitors analogs. Cells damaged for 24 hours with hypoxia, CdCl2 (15 µM), CsA (5 µM), or PMB (75 µM) were treated with 1 µM of the lead compound or its active and inactive analogs for 48 hours. Proliferation was assessed by the percentage of EdU-labeled cells as this method evaluate actively proliferating cells and not just cell number. The compounds that demonstrated strongest proliferative effects were confirmed by a dose response from 0.1 to 1 µM to identify proliferation that could be induced by concentrations lower than 1 µM. To test the specificity of the proliferation effect, the compounds were also tested in NIH/3T3 fibroblasts using the same experimental design. Proliferation was measured on the basis of the percentage of EdU-labeled cells compared with the untreated control (EGF-free medium for HPTECs and 1% bovine calf serum for fibroblasts) in both analyses.
Cell Culture in a 3D MPS
Human kidney tissues were obtained from surgical resection of renal cell carcinoma performed at the University of Washington Medical Center (Seattle, Washington). Primary HPTECs were isolated, seeded, and cultured as previously described.12 Detailed protocols are described in the Supplemental Material.
Cell Proliferation in a 3D MPS Platform via Immunocytochemistry
The ability of hit compounds to induce proliferation of primary HPTECs was tested in a 3D MPS. Cells were maintained for 48 hours in EGF-free medium (control) or damaged with 50 µM of PMB. Subsequently, control cells were kept in EGF-free medium and PMB was substituted by EGF-free medium (untreated) or treated for 48 hours with either 1 µM of ID-8 or harmine. Cell proliferation was assessed by staining for Ki-67 and epcam, using immunocytochemistry.16 EdU-labeling was not used to assess proliferation in the 3D MPS because of extensive background signal from the matrix in the device. Antibodies are listed in Supplemental Table 1.
Kidney Injury Molecule-1 Expression
Effluents of kidney 3D MPS were analyzed for human kidney injury molecule-1 (KIM-1) using the Mesoscale Diagnostics Human KIM-1 Kit (K141JHD-2). Effluents from controls devices were collected at 24, 48, 72, and 96 hours. Effluents from a 3D MPS damaged with 50 µM of PMB were collected at 24 and 48 hours and subsequently collected at 24 and 48 hours after been treated with 1 µM of harmine, 1 µM of ID-8, or EGF-free medium (untreated).
Target Engagement
To test DYRK activation, we performed a cell-free, active site–dependent, competition binding assay commercially known as KINOMEscan (DiscoverX), which quantitatively measured the ability of a compound to compete with an immobilized, active-site directed ligand. We tested ID-8 and harmine in 11-point, three-fold serial dilution starting at 30 µM against DYRK1A and DYRK2. The assay was performed by combining three components: DNA-tagged kinase, immobilized ligand, and ID-8 or harmine in different concentrations. The ability of both compounds to compete with the immobilized ligand was measured via quantitative PCR of the DNA tag.17
Small Interfering RNA Transfection
Small interfering RNA (siRNA) transfections were done in 384-well plates following the same experimental design described in the in vitro damage models section. Transfection complexes were prepared in Opti-MEM medium using Lipofectamine RNAiMax (Thermo Scientific) and human DYRK1A siRNA (10 nM final concentration, #s4401, Silencer Select; Thermo Scientific), following the manufacturer’s protocol. After damage, cells were treated for 24 hours with either DYRK1A siRNA, ID-8 (1 µM), DYRK1A siRNA+ID-8 (1 µM), or siRNA negative control (short hairpin RNA). Transfection efficiency was measured by DYRK1A protein expression, and knockdown effects on proliferation were on the basis of the percentage of EdU-labeled cells across tested groups.
Library Preparation and RNA Sequencing
RNA samples (n=3 per group) were checked for quality (RIN value >8.0) and quantity using Agilent 2200 Bioanalyzer instrument and nanodrop (Thermo Scientific), respectively. Library preparation, quality control, and bioinformatics analysis using bcbio-nextgen18,19 are fully described in the Supplemental Material. The dataset is available with the National Center for Biotechnology Information’s Gene Expression Omnibus database under accession number GSE113039 (reviewer token: utgzgyeiftozbgp).
Immunofluorescence
Immunofluorescence was used to validate upregulated genes found in the transcriptomics study at the protein level (proliferating cell nuclear antigen [PCNA], E2F transcription factor 1 [E2F1], and cyclins B1, E2, and D1) and to analyze the effect of ID-8 and harmine on the cell cycle. Detailed protocols and antibodies are described in the Supplemental Material.
Western Blotting
Protein expression of DYRK1A was confirmed in HPTECs and NIH/3T3 fibroblasts treated with ID-8 and harmine after damage, using Western blotting. Detailed protocol and antibodies are described in the Supplemental Material.
Statistical Analyses
Data are presented as mean±SEM. Statistical difference as calculated by t test. Multiple group comparison was conducted by two-way ANOVA followed by Dunnett multiple comparisons post hoc test. P<0.05 was considered significant and represented by * when compared with corresponding controls or by # when compared with other groups.
Results
Primary Screen Reveals 129 Compounds that Promote HPTEC Proliferation
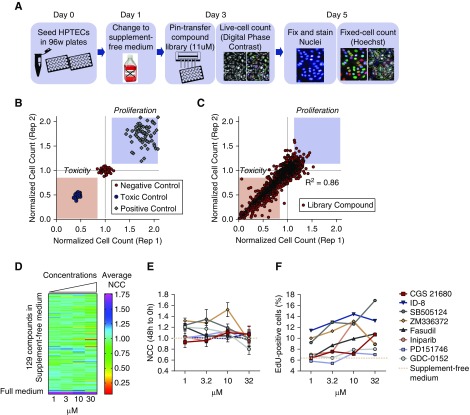
We tested 1902 compounds from the Selleck library for the ability to increase proliferation of HPTECs at 11 µM. Cells cultured in supplement-free medium (free) were treated with individual compounds or maintained in free medium (control). Cell counts were performed immediately after treatment (0 hour) in live cells and 48 hours post-treatment after fixing and staining of the nuclei (Figure 1A). Cell proliferation was also measured in a panel of control conditions, full medium (positive control), free medium (negative control), or 0.1 µM digoxin medium (toxic control), and assayed for cell number at 0 and 48 hours as described above (Figure 1B). NCC was used as a surrogate to the proliferation rate. Proproliferative compounds had an NCC>1, nonproliferative compounds had an NCC of 1, and toxic compounds had an NCC<1. On the basis of the NCC values, the panel of controls showed a clear separation of positive, negative, and toxic controls (NCC=1.85, 1, and 0.58, respectively) (Figure 1C). Assay reproducibility was shown by a coefficient of variation of 7.9, 4.8, and 6.6, respectively, and by a correlation coefficient of 0.86 among duplicates. Assay sensitivity was demonstrated by Z-factor >0.32 among the 22 tested plates (Supplemental Figure 1, A–C). We chose NCC>1.1 as cut-off for proproliferative effects, which lead to the selection of 129 compounds (Figure 1D, Supplemental Table 2) involved in the activation of a variety of pathways (Supplemental Figure 1D).
Figure 1.
Screening selects eight potential proproliferative compounds in primary HPTECs. (A) Schematic of the experimental design for the identification of compounds with proliferative potential in primary HPTECs. (B) Control scatter plot shows NCC of positive control (cells maintained in supplemented full medium shown by red squares), negative control (cells maintained in supplement-free medium shown by green circles), and toxic control (cells treated with 0.1 µM digoxin shown by purple circles). (C) Library scatter plot showing NCC of primary HPTECs maintained in supplement-free medium and treated for 48 hours with each of the 1902 compounds of the Selleck library at 11 µM. Blue circles represent library molecules. Blue boxed region indicates zone of increased proliferation. Pink boxed region indicates zone of decreased proliferation and/or cell death. Correlation coefficient of duplicates =0.86. (D) Heat map depicting the NCC of HPTECs maintained in supplement-free medium followed by 48 hours of treatment with the 129 compounds selected in the primary screen, in four concentrations (1, 3, 10, and 30 µM). (E) Detailed dose response curve after 48 hours of treatment with the eight compounds that produced increase in the NCC. Dose response curves compared with cells maintained in supplement-free medium (control). (F) Increase in the percentage of EdU-positive cells after 48 hours of treatment with the eight compounds. Data are represented by mean±SEM of the FC over cells maintained in supplement-free medium (control). n=3 biologic replicates per group.
A Secondary Screen Reveals Eight Compounds that Promote HPTEC Proliferation
To confirm the activity of the 129 hit compounds identified in the primary screen with an NCC>1.1, HPTECs were rescreened against all 129 hits over four concentrations (1, 3, 10, and 30 µM) because many of the compounds selected are more specific at low concentrations and activate multiple secondary targets at higher concentrations (Figure 1D). Proliferation was measured using the same experimental design as described in the primary screen (Figure 1A) using two different methods: cell number increment, using NCC; and by induction of cells actively cycling, measured by EdU incorporation. The secondary screen identified eight compounds that produced NCC>1.1 in at least two of the four tested concentrations compared with cells maintained in free medium (NCC=1, coefficient of variation <15%; Figure 1E) and that increased the number of actively proliferating cells from 6% (free medium) to 10.6% (CGS21680), 8% (GDC-0152), 16.9% (SB505124), 13.1% (ZM336372), 10.8% (fasudil), 13% (iniparib), 7.3% (PD151746), and 14.4% (ID-8) (Figure 1F).
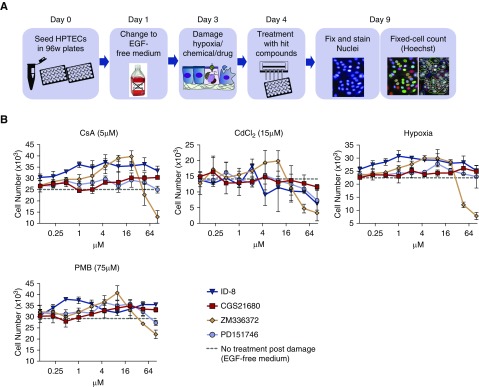
Confirmatory Screen Reveals Four Compounds that Promote HPTEC Proliferation after Damage
We next assayed the eight compounds selected in the secondary screen for the ability to promote cell proliferation after injury. To that end, cells cultured in EGF-free medium were damaged for 24 hours by hypoxia (1% O2), CsA (5 µM), PMB (75 µM), or CdCl2 (15 µM), followed by treatment with each of the eight compounds at each of the two concentrations that produced the strongest proproliferative effect in the secondary screen (Figure 1, E and F): 10 and 30 µM (CGS21680, SB505124, iniparib, and ID-8), 1 and 3 µM (GDC-0152 and fasudil), 3 and 10 µM (ZM336372 and PD151746) (Supplemental Figure 2). Cells that received only EGF-free medium postdamage were used as controls. Cells were treated for 96 hours, fixed, and nuclei were stained, counted, and compared with the control group (Columbus Image Analysis Suite) (Figure 2A).
Figure 2.
Confirmatory screen reveals four compounds promoting proliferation of primary HPTECs after damage. (A) Schematic of the experimental design for the identification of compounds with proliferative potential in primary HPTECs after in vitro drug-/chemical-induced damage and hypoxia-induced damage. (B) A ten-point dose range (2.15-fold serial dilution) shows the change in cell number promoted by 96 hours of treatment with the four selected hit compounds in ten different concentrations (0.1–100 µM) after 24 hours of damage with 5 µM CsA, 75 µM PMB, 15 µM CdCl2, or hypoxia (1% O2). Data are represented as mean±SEM of the FC over the untreated group (EGF-free medium). n=3 per group, three biologic replicates.
Treatment with compounds CGS21680 (30 µM), ZM336372 (3 µM), PD151746 (10 µM), and ID-8 (10 µM) promoted significant increases in cell number after 24 hours of damage in at least two different damage models. Time course studies using ten-point dose range (2.15-fold serial dilution) confirmed the observed increase in proliferation after injury when compared with cells receiving full medium or no treatment (EGF-free medium) (Figure 2B). In the confirmatory assay, only ID-8 produced an increase in cell number after damage across all damage models (P<0.05; fold change [FC]=1.48 after CsA, 1.29 after CdCl2, 1.29 after PMB, and 1.36 after hypoxia) and was therefore selected for further characterization in the next set of experiments.
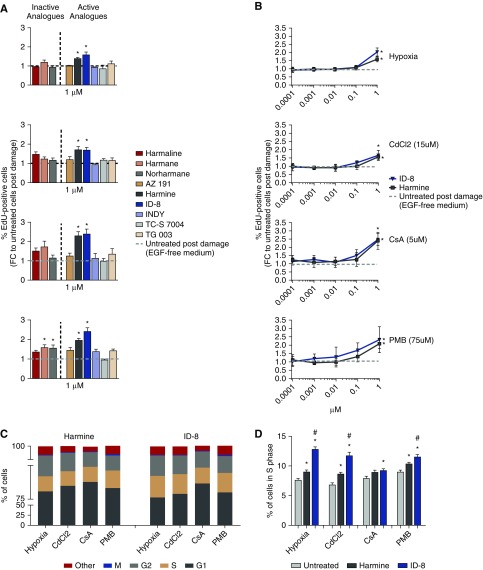
DYRK Inhibitors Induce Tubular Cell Proliferation after Damage
ID-8, the top hit identified in the damage repair assay, is a DYRK1A inhibitor.13 To evaluate the ability of ID-8 and other DYRK inhibitors to promote proliferation after damage, cells were treated with three inactive analogs of DYRK inhibitors (harmaline, harmane, and norharmane) and six active analogs of DYRK inhibitors (AZ191, harmine, ID-8, INDY, TC-S 7004, and TG 003) at 1 µM and proliferation was assessed by the percentage of EdU-labeled cells. Treatment for 48 hours with harmine and ID-8 produced a significant increase in cell proliferation (P<0.05) compared with the untreated group (EGF-free medium) after hypoxia (FC=1.39 and 1.58, respectively), CdCl2 (FC=1.7 and 1.69, respectively), CsA (FC=2.3 and 2.39, respectively), and PMB (FC=1.96 and 2.41, respectively) (Figure 3A). Harmane and norharmane also promoted significant cell proliferation after damage with PMB (P<0.05; FC=1.59 and 1.56, respectively) but this effect was only observed in the PMB group and was lower than the observed effects of ID-8 and harmine, so these compounds were not tested further. To investigate whether proliferation promoted by ID-8 and harmine could be induced at concentrations lower than 1 µM, HPTECs were damaged as previously described and treated for 48 hours with ID-8 and harmine at range of concentrations from 0.0001 to 1 µM. Treatment with 1 µM of harmine or ID-8 promoted significant cell proliferation (P<0.05) compared with the untreated group after hypoxia (FC=1.58 and 2, respectively), CdCl2 (FC=1.56 and 1.66, respectively), CsA (FC=2.4 and 2.39, respectively), and PMB (FC=2.08 and 2.32, respectively). No significant proliferation effect was observed in concentrations <1 µM (Figure 3B) and it was selected as the concentration for the next experiments. The specificity of the proliferation effect of ID-8 and harmine was tested in NIH/3T3 fibroblasts after damage. Harmine (1 µM) increased the number of actively cycling cells (P<0.05) compared with untreated cells (1% bovine calf serum medium) after hypoxia (FC=1.6), CsA (FC=2.1), and PMB (FC=1.9). ID-8 did not induce any proliferation in NIH/3T3 fibroblasts, thereby suggesting ID-8 to have specificity in stimulating proliferation of HPTECs (Supplemental Figure 3).
Figure 3.
DYRK inhibitors ID-8 and harmine promote proliferation of primary HPTECs after damage. (A) Proliferation effect promoted by 48 hours of treatment with 1 µM of inactive (harmaline, harmane, and norharmane) and active (AZ191, harmine, ID-8, INDY, TC-S 7004, and TG 003) DYRK inhibitors analogs after 24 hours of damage with hypoxia (1% O2), 15 µM of CdCl2, 5 µM of CsA, or 75 µM of PMB (n=2 per group, 3–5 biologic replicates). (B) Dose response curve after 48 hours of treatment with ID-8 and harmine in HPTECs after 24 hours of damage with hypoxia (1% O2), 15 µM CdCl2, 5 µM CsA, or 75 µM PMB (n=3 per group, three biologic replicates). EdU-positive cells were normalized to the total cell number per well; Data are presented as mean±SEM of the FC over the untreated group (EGF-free medium). (C) Quantification of cells in different cell cycle phases after damage followed by 24 hours of treatment with ID-8 or harmine. (D) Increment of cells in S phase promoted by the treatment with 1 µM of ID-8 or harmine compared with the untreated group after four types of damage (n=12–24 per group, two biologic replicates). Data are presented as the percentage of the total cell count. *P<0.05 compared with the untreated group; #P<0.05 compared with the harmine group.
We next analyzed the effect of harmine and ID-8 on cell cycle using HPTECs. Regardless of the group cells were mostly in G1 phase (75%–85%), followed by S (9%–11%), G2 (5%–10%), and M phase (0.1%–0.2%; Figure 3C). Treatment with 1 µM ID-8 significantly increased (P<0.05) cells in S phase compared with untreated cells (EGF-free medium) after hypoxia (12.8% versus 7.6%, respectively), CdCl2 (11.7% versus 6.8%, respectively), CsA (9.5% versus 7.9%, respectively), and PMB (11.5% versus 9%, respectively). The fraction of cells in S phase in the ID-8 group was also superior compared with the harmine group, suggesting higher potency of ID-8 compared with harmine in stimulating proliferation (Figure 3D).
ID-8 Enhances HPTEC Proliferation after Damage in a 3D Culture MPS
We next examined whether the proproliferative effects of ID-8 and harmine observed in primary HPTECs in a 2D culture system would be recapitulated in 3D MPS. Under normal conditions, primary HPTECs cultured in MPSs show little or no expression of the kidney damage biomarker KIM-1, indicating the absence of tissue damage. Exposure of HPTECs to PMB (50 µM) significantly increased KIM-1 protein expression after 24 and 48 hours (P<0.05; mean, 226.5 pg/ml versus 36.2 pg/ml and 228.5 pg/ml versus 47.8 pg/ml, respectively), and was not reversed by 24 hours of PMB washout (untreated) or by treatment with 1 µM of either ID-8 or harmine (P<0.05; mean: 143, 145, and 130 pg/ml, respectively) (Supplemental Figure 4A). KIM-1 expression reverted to normal levels after 48 hours of PMB withdrawal, independently of treatment with DYRK inhibitors. Proliferation after HPTEC treatment with 1 µM of ID-8 after PMB-induced damage was not statistically significant but did trend toward increased proliferation (Supplemental Figure 4B), as shown by a higher number of Ki-67–positive nuclei (P=0.07; FC=2.37) compared with the untreated group (EGF-free medium). Harmine treatment did not show the same trend (P=0.9; FC=1.09). This result corroborates our previous 2D experiments showing a stronger induction of cell proliferation by ID-8 as compared with harmine and to cells receiving no treatment after damage.
Target Engagement and Genetic Manipulation Studies Demonstrate Pharmacologic Activity of ID-8 via Binding to DYRK1A
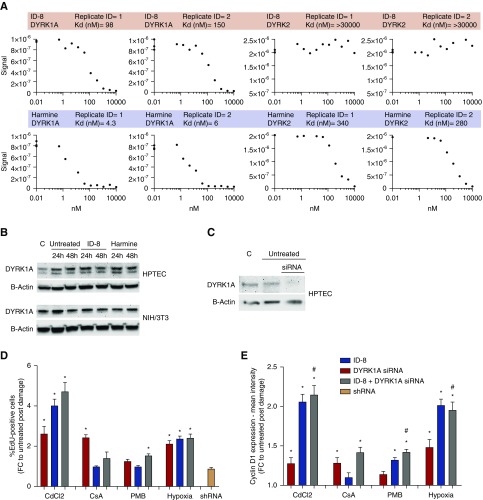
To confirm target engagement of DYRK by ID-8 we used a cell-free, active-site dependent, competition binding assay (KINOMEscan; DiscoverX) and investigated binding of ID-8 and harmine to DYRK1A and DYRK2. We demonstrated that ID-8 targets DYRK1A (Kd=120 nM) but not DYRK2 (Kd>30,000 nM). Harmine, on the other hand, targeted DYRK1A (Kd=5.1 nM) and also DYRK2 (Kd=310 nM; Figure 4A). Expression of DYRK1A was confirmed in HPTECs and NIH/3T3 fibroblasts protein lysates in control conditions and after damage (Figure 4B). Treatment of HPTECs with DYRK1A siRNA for 24 hours resulted in >90% knockdown of DYRK1A (Figure 4C) and promoted increased proliferation (P<0.05) compared with the untreated group after CdCl2 (FC=2.6), CsA (FC=2.4), and hypoxia (FC=2.1) (Figure 4D). Cells treated with siRNA negative control (short hairpin RNA [shRNA]) showed no increase in proliferation compared with the control. Combination of ID-8+DYRK1A siRNA led to higher levels of proliferation (P<0.05) compared with the untreated group after hypoxia (FC=2.4), CdCl2 (FC=4.1), and mildly after PMB (FC=1.53; Figure 4D), demonstrating a potential additive/synergistic effect on proliferation after pharmacologic and genetic inhibition of DYRK1A. Because this was observed only after hypoxia and CdCl2 damage, it is possible that the mechanism to initiate proliferation after DYRK1A inhibition depends on the distinct mechanism of initiation of damage.
Figure 4.
Inhibition of DYRK1A by harmine and ID-8 leads to proliferation of primary HPTECs. (A) Matrix of binding constants (Kd) and curve images for ID-8 and harmine competition binding assay against DYRK1A and DYRK2. The amount of kinase measured by quantitative PCR (Signal; y-axis) is plotted against the corresponding compound concentration in nanomolar in log10 scale (x-axis). (B) Expression of DYRK1A in HPTECs and NIH/3T3 fibroblasts protein lysates in control cells, in damaged untreated cells (EGF-free in HPTECs and 1% bovine calf serum in fibroblasts), or treated with ID-8 or harmine for 24 and 48 hours. (C) Confirmation of DYRK1A knockdown by Western blot. (D) Proliferation induced by DYRK1A knockdown after damage with 15 µM CdCl2, 5 µM CsA, 75 µM PMB. and hypoxia (1% O2). (E) Upregulation of cyclin D1 after inhibition of DYRK1A by siRNA, ID-8, or DYRK1A siRNA+ID-8. Data are presented as mean±SEM of the FC over the untreated group (EGF-free medium). *P<0.05 compared with the untreated group; #P<0.05 compared with DYRK1A siRNA group (n=12–24 technical replicates per group, two biologic replicates). C, control cells.
DYRK1A has been shown to control cell cycle entry via cyclin D1 upregulation in neonatal foreskin fibroblasts.20 We therefore measured expression of cyclin D1 by immunofluorescence in the cells receiving ID-8 and DYRK1A siRNA (Figure 4E) to further clarify the mechanism by which ID-8 could be inducing proliferation. Cells treated with DYRK1A siRNA for 24 hours showed mild increased cyclin D1 expression (P<0.05) compared with the untreated group after CdCl2 (FC=1.2), CsA (FC=1.2), and hypoxia (FC=1.4). However, in the groups treated with the combination of ID-8+DYRK1A siRNA, we observed a significant increase in proliferation compared not only with the untreated control after CdCl2 (FC=2.1), CsA (FC=1.4), PMB (FC=1.4), and hypoxia (FC=1.9), but also compared with DYRK1A siRNA treatment alone after CdCl2 (FC=2.2), PMB (FC=1.4), and hypoxia (FC=1.85), extending the role of cyclin D1 in HPTEC proliferation induced by DYRK1A.
ID-8 Modulates Cell Cycle Responses Upregulating More Proproliferative Genes
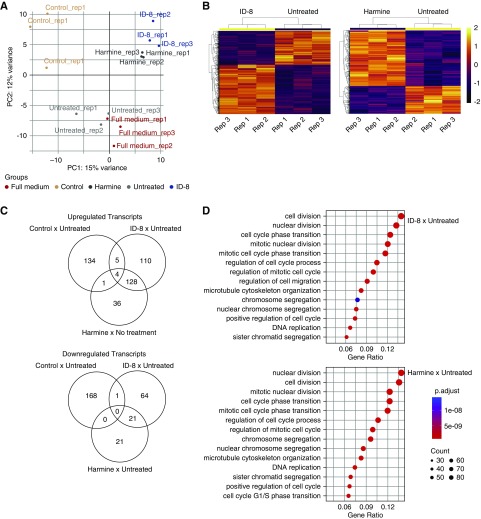
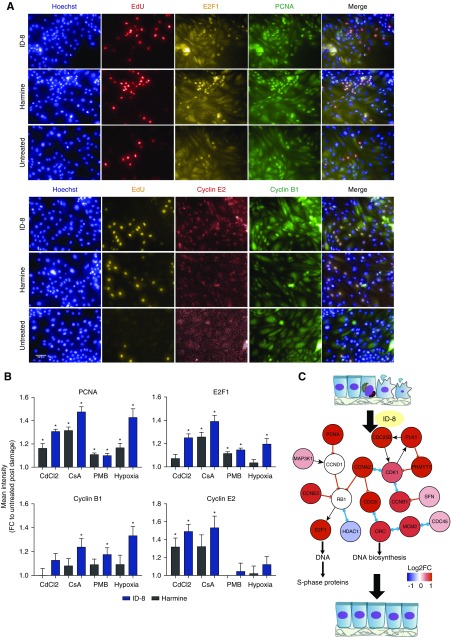
To investigate the mechanisms underlying stimulation of proliferation by ID-8 and harmine after damage, we performed RNA sequencing in undamaged cells (control) and in cells damaged by 24 hours of hypoxia followed by treatment with 1 µM of ID-8, harmine, EGF (full medium), or EGF-free medium (untreated). Quality control data for the sequencing protocol is shown in Supplemental Figure 5, A and B. Principal component analysis showed clear separation of the cells treated with ID-8 or harmine away from the untreated and full medium groups, as well as a separation from the control group (Figure 5A). Hierarchical clustering showed concordance among the biologic replicates and a distinct pattern of differentially expressed genes in the groups treated with ID-8 or harmine and the untreated group (Figure 5B). A higher number of genes (n=764) was significantly modulated by ID-8 compared with the harmine (n=486) and the full medium (n=19) groups, showing that ID-8 promoted higher transcription modulation than the harmine treatment (Figure 5C). Pathway analysis revealed that ID-8 treatment was more potent than harmine on upregulating proproliferative cell cycle genes, notably PCNA, E2F1, MYC, cyclins E2, A2, and B1, and genes belonging to the prereplication complex (cell division cycle 6 [CDC6], Origin recognition complex subunit 1 [ORC1], and Minichromosome maintenance complex component 2–7 [MCM2–7]) as compared with the untreated group (Figure 5D, Supplemental Figure 5C). On the other hand, the most upregulated genes in the untreated group compared with undamaged cells were related to immune response and inflammation: leukocyte antigen complex (Major histocompatibility complex, class I, G [HLA-G] and B [HLA-B.5] and major histocompatibility complex, class II, DQ beta 1 [HLA-DQB1]) and the complement system (complement C1r [C1R] and 3 [C3]). Upregulation of PCNA, E2F1, and cyclins B1 and E2 was confirmed at the protein level across different damage models (Figure 6, A and B, Supplemental Table 3) after ID-8 treatment. As observed in the transcriptomics data, ID-8 produced higher upregulation of cell cycle genes at the protein level compared with harmine, which did not induce upregulation of cyclin B1 and produced lower modulation of PCNA, E2F1, and cyclin E2. Therefore, ID-8 promoted upregulation of genes involved in cell cycle machinery showing the potential mechanism of proliferation after injury (Figure 6C).
Figure 5.
ID-8 (1 µM) modulates cell cycle response, upregulating proproliferative genes. (A) Principal component analysis shows distinct distribution of the cells treated for 48 hours with ID-8 and harmine in comparison with undamaged cells (control), no treatment after damage, or treated with EGF (full medium) after hypoxia-induced damage. (B) Hierarchical clustering of genes differentially expressed between cells treated for 48 hours with ID-8 or harmine and untreated cells. (C) Venn diagrams showing the number of differentially upregulated and downregulated genes in cells treated for 48 hours with ID-8, harmine, and untreated cells; FC>1.5. (D) Gene ontology of the top modulated pathways in cells treated for 48 hours with ID-8 or harmine compared with untreated cells.
Figure 6.
ID-8 upregulates proproliferative proteins across different damage models. (A) Immunostaining images of all nuclei (Hoechst), actively cyclin cells (EdU), PCNA, E2F1, and cyclins B1 and E2. (B) Relative quantitation of PCNA, E2F1, and cyclins B1 and E2 in HPTECs treated with 1 µM of harmine or ID-8 after 24 hours of damage with hypoxia (1% O2), 15 µM CdCl2, 5 µM CsA. or 75 µM PMB. *P<0.05 compared with the untreated group, (n=3 per group, two biologic replicates). Data are presented as mean±SEM of the FC over the untreated group (EGF-free medium). (C) Schematic representation depicting how ID-8 treatment after damage upregulates cell cycle genes, potentially leading to cell proliferation (generated by Integrated Network and Dynamical Reasoning Assembler: msb.embopress.org/content/13/11/954).
Discussion
Collectively, our early-stage discovery study identifies ID-8 as a potential therapeutic candidate to stimulate kidney tubular epithelial cell proliferation. By measuring proliferation of HPTECs in 2D and 3D in vitro models, we demonstrated that inhibition of DYRK1A by ID-8 induces proliferation of HPTECs after multiple forms of tubular damage. Mechanistically, the target engagement studies suggest the specificity of ID-8 to bind to DYRK1A and transcriptomics experiments identified key cell cycle regulators upregulated by ID-8 to mediate cell proliferation.
Although HPTECs are known to promote tubular regeneration after injury,21 the regenerative processes can be inefficient, impaired, and dysregulated, resulting in extensive tissue remodeling and fibrosis.22 One reason for inadequate repair may be that the mechanisms of tissue repair after AKI are complex and involve epithelial, endothelial, stromal, and inflammatory cell types. This cellular complexity makes the task of inducing repair through specific pathways difficult.7 Despite this complexity and heterogeneity, there are a number of potential therapeutic approaches such as α-melanocyte–stimulating hormone, recently licensed as ABT-719 for the prevention of AKI in patients undergoing cardiac surgery; and QPI-1002, a siRNA targeting the p53 gene, currently in phase 1 clinical trials.23 Another promising approach uses mesenchymal stem cells and, although no serious adverse effects have been reported in trials with mesenchymal stem cells, concerns about maldifferentiation, tumorigenesis, and overimmunodepression are still to be rigorously addressed.7
With the advent of large compound libraries and HTS methods, remarkable success was made in identifying potential therapeutic targets and lead candidate compounds stimulating proliferation of pancreatic β cells, hepatocytes, or podocytes.9,10,14,24 In this study we took a similar approach and screened compounds under basal conditions for those that promoted increased cell proliferation. As a second step we focused on investigating stimulation of repair after injury and assessed the compounds with the strongest proproliferative effects after different types of damage and validated the results in a 3D model. As a final step, we looked at the biologic pathways targeted by the hit compounds. Among the four final compounds that promoted proliferation of HPTECs after damage, the adenosine A2A receptor agonist CGS21680 was shown to preserve renal function, reversing fibrosis and reducing macrophage infiltration and inflammatory activation in rat nephrotoxic nephritis.25 The remaining compounds ZM336372, a potent and selective c-Raf inhibitor, and PD151746, a selective, cell-permeable calpain inhibitor have not been extensively studied in kidney regeneration and remain interesting as potential new therapeutic agents.
The lead compound validated in our screen is a DYRK inhibitor that was recently described as targeting DYRK1A.13 Previously, DYRK2 and DYRK4 were described as the putative targets for ID-8,26 but a comprehensive evaluation of ID-8 revealed its high specificity for DYRK1A and confirmed its lack of activity against DYRK2 and DYRK4.13 DYRKs are a conserved family of eukaryotic kinases that are related to the cyclin-dependent kinases (Cdks), mitogen-activated protein kinases, glycogen synthase kinases, and Cdk-like kinases, playing key roles on cell proliferation and apoptosis induction.27 Among its isoforms, DYRK1A and DYRK1B are negative regulators of the cell cycle promoting a switch to quiescent cellular state.28–30 Recently, DYRK1A inhibitors showed stimulation of human pancreatic β cell replication, holding therapeutic promise for human diabetes.9,31 After testing different active and inactive DYRK inhibitors analogs, we identified harmine, a DYRK1A inhibitor as well32 as a possible proproliferative compound along with ID-8. However, harmine’s induction of cell proliferation was not corroborated by the 3D model and it had a smaller effect modulating gene and protein expression compared with ID-8. Besides, harmine also induced proliferation of NIH/3T3 fibroblasts showing less specificity, thereby increasing the potential for off-target adverse effects.
Recent studies have shown the role of cell cycle control in AKI repair postdamage. After the first 24 hours of ischemic injury, tubular cells undergo apoptotic and necrotic cell death. In response, many of the surviving, normally quiescent proximal tubule epithelial cells proliferate and enter the cell cycle,33 sequentially activating Cdks. This cell cycle reentry after injury is viewed as a protective response. Transient expression of Cdk2 or Cdk4/6 inhibitors therefore represents a novel strategy to improve renal repair and could provide protection against early tubular cell death and still allow for normal repopulation of injured tubules via subsequent proliferation.7 In accordance with previous studies on DYRK inhibitors,9,31,34,35 we observed that ID-8 upregulated genes involved in cell cycle machinery like PCNA, E2F1, and cyclins E2 and B1 (Figure 6C), showing the potential mechanism of proliferation after treatment. Cyclin D1 was not modulated in our transcriptomics study; however, it has been described that knockdown of DYRK1A increase cyclin D1 at the protein level splitting cells into two fates, with one subpopulation accelerating the cell cycle and the other entering an arrested state.20 Our results showed increase in cyclin D1 expression after pharmacologic and genetic knockdown of DYRK1A, corroborating previous studies.20
The main caveat of our study is the absence of in vivo data to corroborate the in vitro efficacy findings. However, ID-8 inhibits the same family of kinases that induce proliferation in other in vivo studies,9,31 supporting our hypothesis that this compound may have therapeutic potential. Moreover, ID-8 consistently promoted proliferation across different 2D damage models and were reproduced by the 3D in vitro model, demonstrating efficacy in a complex biologic system.
In summary, we demonstrate that ID-8 is a potential first-in-class compound that stimulates proliferation of primary HPTECs after AKI, inducing the expression of proproliferative genes in proximal tubule cells. This compound may provide a path for new therapies toward kidney tubule repair after damage.
Disclosures
S.R. is an employee of AstraZeneca and V.S.V. is an employee of Pfizer, Inc.
Supplementary Material
Acknowledgments
The authors appreciate the support of Institute of Chemistry and Cell Biology Longwood Facility at Harvard Medical School for their invaluable assistance with the design of the high-throughput screen, the Biopolymers Facility at Harvard Medical School for performing the RNA sequencing, the Harvard Chan Bioinformatics for performing the RNA-sequencing analysis (with the support from Harvard Medical School Tools and Technology Committee and Harvard Catalyst, The Harvard Clinical and Translational Science Center; National Institutes of Health [NIH] award UL1RR025758), and Petar Todorov for the support generating the proliferation diagram using the Integrated Network and Dynamical Reasoning Assembler (msb.embopress.org/content/13/11/954).
V.S.V. and S.R. designed the study; M.B.M., S.R., V.C., and S.A.B. carried out the two-dimensional experiments; E.J.W., K.A.L., and E.J.K. carried out the three-dimensional experiments; M.B.M. and S.R. analyzed the data and prepared the figures; and M.B.M., S.R., and V.S.V. wrote the manuscript. All authors approved the final version of the manuscript.
Work in the Vaidya laboratory is supported by Outstanding New Environmental Sciences award from NIH/National Institute of Environmental Health Sciences (NIEHS) (ES017543) and Innovation in Regulatory Science Award from Burroughs Wellcome Fund (BWF-1012518). M.B.M. was supported by the São Paulo Research Foundation (grant 2016/04935-2). S.R. was supported by the Carl W. Gottschalk Research Scholar of the American Society of Nephrology Foundation for Kidney Research. S.A.B. was supported by the NIH P50 GM107618 and the Harvard Program in Therapeutic Science. E.J.K. is supported by NIH/National Center for Advancing Translational Sciences (UH3TR000504 and UG3TR002158) and NIH/NIEHS (P30ES007033).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018040392/-/DCSupplemental.
References
- 1.Li PK, Burdmann EA, Mehta RL; World Kidney Day Steering Committee 2013 : Acute kidney injury: Global health alert. Kidney Int 83: 372–376, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Susantitaphong P, Cruz DN, Cerda J, Abulfaraj M, Alqahtani F, Koulouridis I, et al.: Acute Kidney Injury Advisory Group of the American Society of Nephrology : World incidence of AKI: A meta-analysis. Clin J Am Soc Nephrol 8: 1482–1493, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin A, Tonelli M, Bonventre J, Coresh J, Donner JA, Fogo AB, et al.: ISN Global Kidney Health Summit participants : Global kidney health 2017 and beyond: A roadmap for closing gaps in care, research, and policy. Lancet 390: 1888–1917, 2017 [DOI] [PubMed] [Google Scholar]
- 4.Coca SG, Singanamala S, Parikh CR: Chronic kidney disease after acute kidney injury: A systematic review and meta-analysis. Kidney Int 81: 442–448, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonventre JV: Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J Am Soc Nephrol 14[Suppl 1]: S55–S61, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Molitoris BA: Therapeutic translation in acute kidney injury: The epithelial/endothelial axis. J Clin Invest 124: 2355–2363, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphreys BD, Cantaluppi V, Portilla D, Singbartl K, Yang L, Rosner MH, et al.: Acute Dialysis Quality Initiative (ADQI) XIII Work Group : Targeting endogenous repair pathways after AKI. J Am Soc Nephrol 27: 990–998, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramann R, Kusaba T, Humphreys BD: Who regenerates the kidney tubule? Nephrol Dial Transplant 30: 903–910, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, Alvarez-Perez JC, Felsenfeld DP, Liu H, Sivendran S, Bender A, et al.: A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication. Nat Med 21: 383–388, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan J, Logan DJ, Root DE, Carpenter AE, Bhatia SN: High-throughput platform for identifying molecular factors involved in phenotypic stabilization of primary human hepatocytes in vitro. J Biomol Screen 21: 897–911, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adler M, Ramm S, Hafner M, Muhlich JL, Gottwald EM, Weber E, et al.: A quantitative approach to screen for nephrotoxic compounds in vitro. J Am Soc Nephrol 27: 1015–1028, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, et al.: Development of a microphysiological model of human kidney proximal tubule function. Kidney Int 90: 627–637, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellmaine SF, Ovchinnikov DA, Manallack DT, Cuddy CE, Elefanty AG, Stanley EG, et al.: Inhibition of DYRK1A disrupts neural lineage specificationin human pluripotent stem cells. eLife 6: e24502, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HW, Khan SQ, Faridi MH, Wei C, Tardi NJ, Altintas MM, et al.: A podocyte-based automated screening assay identifies protective small molecules. J Am Soc Nephrol 26: 2741–2752, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, et al.: Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol 9: 514–520, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Ness KP, Chang SY, Weber EJ, Zumpano D, Eaton DL, Kelly EJ: Microphysiological systems to assess nonclinical toxicity. Curr Protoc Toxicol 73: 14.18.1–14.18.28, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian MA, Biggs WH 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, et al.: A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol 23: 329–336, 2005 [DOI] [PubMed] [Google Scholar]
- 18.bcbio - nextgen. Available at: https://bcbio-nextgen.readthedocs.io/en/latest/. Accessed October 19, 2017
- 19.Steinbaugh MJ, Pantano L, Kirchner RD, Barrera V, Chapman BA, Piper ME, et al. : bcbioRNASeq: R package for bcbio RNA-seq analysis, 2017. Available at: https://f1000research.com/articles/6-1976/v2. Accessed October 19, 2017
- 20.Chen JY, Lin JR, Tsai FC, Meyer T: Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle. Mol Cell 52: 87–100, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphreys BD, Czerniak S, DiRocco DP, Hasnain W, Cheema R, Bonventre JV: Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci U S A 108: 9226–9231, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S: Cellular and molecular pathways of renal repair after acute kidney injury. Kidney Int 93: 27–40, 2018 [DOI] [PubMed] [Google Scholar]
- 23.Demirjian S, Ailawadi G, Polinsky M, Bitran D, Silberman S, Shernan SK, et al.: Safety and tolerability study of an intravenously administered Small Interfering Ribonucleic Acid (siRNA) post on-pump cardiothoracic surgery in patients at risk of acute kidney injury. Kidney Int Rep 2: 836–843, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widmeier E, Tan W, Airik M, Hildebrandt F: A small molecule screening to detect potential therapeutic targets in human podocytes. Am J Physiol Renal Physiol 312: F157–F171, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia GE, Truong LD, Chen JF, Johnson RJ, Feng L: Adenosine A(2A) receptor activation prevents progressive kidney fibrosis in a model of immune-associated chronic inflammation. Kidney Int 80: 378–388, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa K, Yasuda SY, Teo JL, Nguyen C, McMillan M, Hsieh CL, et al.: Wnt signaling orchestration with a small molecule DYRK inhibitor provides long-term xeno-free human pluripotent cell expansion. Stem Cells Transl Med 1: 18–28, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker W, Sippl W: Activation, regulation, and inhibition of DYRK1A. FEBS J 278: 246–256, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA: DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev 25: 801–813, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer SE, Friedman E: Mirk/Dyrk1B: A multifunctional dual-specificity kinase involved in growth arrest, differentiation, and cell survival. Cell Biochem Biophys 45: 303–315, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Thompson BJ, Bhansali R, Diebold L, Cook DE, Stolzenburg L, Casagrande AS, et al.: DYRK1A controls the transition from proliferation to quiescence during lymphoid development by destabilizing Cyclin D3. J Exp Med 212: 953–970, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dirice E, Walpita D, Vetere A, Meier BC, Kahraman S, Hu J, et al.: Inhibition of DYRK1A stimulates human β-cell proliferation. Diabetes 65: 1660–1671, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Göckler N, Jofre G, Papadopoulos C, Soppa U, Tejedor FJ, Becker W: Harmine specifically inhibits protein kinase DYRK1A and interferes with neurite formation. FEBS J 276: 6324–6337, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Price PM, Safirstein RL, Megyesi J: The cell cycle and acute kidney injury. Kidney Int 76: 604–613, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen W, Taylor B, Jin Q, Nguyen-Tran V, Meeusen S, Zhang YQ, et al.: Inhibition of DYRK1A and GSK3B induces human β-cell proliferation. Nat Commun 6: 8372, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida K: Role for DYRK family kinases on regulation of apoptosis. Biochem Pharmacol 76: 1389–1394, 2008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.