Abstract
Glioma is the most common malignant tumor of the central nervous system with poor survival. Temozolomide (TMZ) is the first-line chemotherapy drug for initial and recurrent glioma treatment with a relatively good efficacy, which exerts its antitumor effects mainly through cell death induced by DNA double-strand breaks in the G1 and S phases. However, endogenous or acquired resistance to TMZ limits glioma patients’ clinical outcome and is also an important cause of glioma replase. MicroRNA-195 (miR-195) plays an important role in the regulation of G1-phase/S-phase transition, DNA damage repair, and apoptosis of tumor cells. We found that miR-195 expression was significantly decreased in TMZ-resistant glioma cells induced with TMZ and correlated to the resistance index negatively. Also, the exogenous expression of miR-195 reversed TMZ resistance and induced the apoptosis of TMZ-resistant glioblastoma cells. Further bioinformatics analysis showed cyclin E1 (CCNE1) was a potential target gene of miR-195. Knockdown of CCNE1 partially reversed the effect of decreased miR-195 on TMZ resistance. The data from The Cancer Genome Atlas – Cancer Genome further suggested that hsa-miR-195 could negatively regulate the expression of CCNE1 in glioma. In conclusion, miR-195 reverses the resistance to TMZ by targeting CCNE1 in glioma cells and it could act as a potential target for treatment in glioma with TMZ resistance.
Keywords: chemoresistance, cyclin E1, glioblastoma, microRNA-195, temozolomide
Introduction
Glioma is the most common malignant tumor of the central nervous system, with a high recurrence rate, a high disability rate, and a high mortality rate. In high-grade glioma [glioblastoma (GBM)], the median survival is only 12–15 months and the 5-year survival rate is less than 3% 1,2. Chemotherapy is the most common method for the treatment of glioma and it can kill residual tumor cells, prevent tumor recurrence, and prolong the survival time of patients with glioma. Oral alkylating agent temozolomide (TMZ) is the best first-line chemotherapy drug for the treatment of primary and recurrent glioma 3, which plays an antitumor role depending on its ability to alkylate/methylate DNA, through acting on the cell cycle G1/S and interference DNA replication, to induce apoptosis and death of tumor cells. Like other chemotherapy drugs, some glioma patients are resistant to TMZ already at diagnosis (primary resistance), whereas others may develop TMZ resistance during treatment (acquired resistance). TMZ resistance leads to increased drug doses and adverse reactions, prolonging the chemotherapy cycle, and also adversely affects the survival and the quality of life of patients with gliomas. Therefore, TMZ resistance is a major obstacle toward achievement of better survival in the treatment of this disease. It is very important to elucidate the underlying molecular mechanism of TMZ resistance to improve the clinical benefit and the survival of glioma patients.
MicroRNAs (miRNAs) are endogenous noncoding regulatory RNAs with 17–22 nucleotides, which bind to complementary sequences in 3′-untraslated region (3′-UTR) of the target mRNAs and regulate the expression of target genes at the post-transcriptional level. Increasingly more evidence suggests that miRNAs contribute to various human malignancies, including glioma 4–6. Emerging studies showed that abnormal and specific expressions of miRNAs were closely related to tumor cell proliferation and apoptosis, invasion, and migration and drug resistance. It is known that microRNA-195 (miR-195) is downregulated widely in a variety of malignant tumors, and may be the ‘key node’ of tumor cells’ abnormal proliferation and apoptosis 7. Our present study aims to explore whether miR-195 was a specific marker for intrinsic or acquired resistance of glioma cells to TMZ and to elucidate its underlying molecular mechanism of resistance to TMZ.
Materials and methods
Cell lines and treatment
The human glioma cell line U251MG was purchased from Cobioer (Nanjing, China). Cells were cultured in high-glucose DMEM medium (Gibco, Carlsbad, California, USA) supplemented with 10% FBS (Sigma, St. Louis, Missouri, USA), 100 μg/ml streptomycin, and 100 U/ml penicillin in a humid atmosphere containing 5% CO2 at 37°C. Cells were digested and passaged when the confluency reached 70–80%. The concentration gradient method was used to establish the resistant cell lines. The concentration of TMZ was 1, 5, 10, 15, and 20 μg/ml. The cell line with certain resistance to TMZ was named U251R. When the cell line was established, it was cultured with 20 μg/ml of TMZ to stabilize its drug resistance for one week per month.
Transfection
The cells were seeded in six-well plates (3×105/well) and cultured for 24 h. When the cell density reached 70–80%, the old culture medium was replaced with 2 ml of fresh medium. The transfection mix containing the 3 μl HiPerFect Transfection Reagent (Qiagen, Hilden, Germany) and 50 nmol/l miR-195-5p mimics or negative control (NC) [or 100 nmol/l miR-195-5p inhibitor or 50 nmol/l siRNA for cyclin E1 (CCNE1)] was added to the medium and the cells were incubated at 37°C. After 48 h of transfection, the medium was replaced and the cells were collected for further analyses.
Cell proliferation and drug sensitivity
Glioma cells in the logarithmic phase were collected and plated in 96-well plates (4000 cells/well) and incubated at 37°C for 24, 48, 72, 96, and 120 h. 20 μl of methyl thiazolyl tetrazolium (MTT, 5 mg/ml; Sigma, Shanghai, China) was added to each well and the cells were incubated for another 4 h of incubation. After removing the medium, 200 μl dimethylsulfoxide was added and the plate was shaken for 10 min to terminate the reaction. A SpectraMax i3x microplate reader (Molecular Devices, Silicon Valley, California, USA) was used to detect the absorbance at 490 nm. Each experiment was conducted in triplicate and included three replicates.
IC50 determination
The cell viability was detected by MTT at different concentrations of TMZ (20, 50, 100, 200, 300, 400, 500, 600, 800, 1000, and 1200 μg/ml). The inhibition percent of each test compound was calculated by comparing with the vehicle control. The IC50 value was determined by a nonlinear regression analysis of the concentration–response curve using the Hill equation and the resistance index=IC50 (resistant cells)/IC50 (parent cells).
Cell apoptosis analysis
Cell apoptosis was first measured using the Hoechst 33342/propidium iodide (PI) Staining Kit (Solarbio, Beijing, China). Then, the cell apoptosis rate was measured using the Annexin V : FITC Apoptosis Detection Kit (BD Biosciences, San Jose, California, USA) according to the manufacturer’s instructions. Briefly, the cells were collected at 72 h after transfection and adjusted to the concentration of 1×106/ml. Cell suspension of 200 μl was assigned for each sample, washed twice with cold PBS, and resuspended in 100 μl binding buffer. Then, 5 μl Annexin-V–FITC (20 μg/ml) and 5 μl PI (50 μg/ ml) were added and the cells were incubated on ice in the dark for 15 min. Then, 400 μl binding buffer was added and the detection was performed using BD FACSCanto II (BD Biosciences).
Real-time quantitative PCR
Cells were collected and lysed in RNAiso for Small RNA (Takara, Kusatsu, Japan) for miRNA extraction. The One-Step PrimeScript miRNA cDNA Synthesis Kit (Takara) was used for reverse transcription. The expression level of miR-195-5p was detected using the One-Step qRT-PCR method. The miR-195-5p primer, U6 primer, and EzOmics SYBR qPCR kit were purchased from Biomics Biotechnologies (Guangzhou, China). Briefly, for amplification of miR-195-5p, 100 ng RNA was used in a 25-μl reaction system containing 12.5 μl 2× Master Mix, 0.5 μl 50× SYBR-Green, 0.5 μl reverse transcription primer (10 μmol/l), 0.5 μl sense, and 0.5 μl antisense primers (10 μmol/l). One-Step PCR parameters for miRNA quantification were as follows: 37°C for 60 min for reverse transcription, 10 min at 95°C, and then 40 cycles of 20 s at 95°C, 30 s at 62°C, and 30 s at 72°C. Each sample was tested in triplicate.
For RNA analysis, total RNA was isolated from the cultured cells using Trizol reagent (Invitrogen, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Reverse transcription was performed using the PrimeScript RT-PCR kit (Takara). GAPDH mRNA levels were used for normalization. The primer sequences were as follows: CCNE1, left primer: 5′-GGA AGA GGA AGG CAA ACG TG-3′ and right primer: 5′-GCA ATA ATC CGA GGC TTG CA-3′; GAPDH, left primer: 5′-TCG TGG AAG GAC TCA TGA CC-3′, and right primer: 5′-ATG ATG TTC TGG AGA GCC CC-3′. The real-time PCR parameters for relative quantification were as follows: 5 min at 94°C, followed by 30 cycles of 30 s at 94°C, 45 s at 57°C, and 45 s at 72°C. Each sample was tested in triplicate, and the fold change in mRNA expression was calculated using the 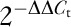 method.
method.
Western blot analysis
Cell protein was extracted using RIPA lysis buffer. The supernatant was quantified using a bicinchoninic acid assay (Pierce, Rockford, Illinois, USA). Next, 25 μg of protein was denatured with 5× SDS loading buffer at 95°C for 5 min. Subsequently, whole-protein samples were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto 0.45-μm nitrocellulose membranes. Following 1 h of blocking in 5% fat-free milk, the membranes were incubated with the CCNE1 antibody (1 : 1000) and the β-actin antibody (1 : 1000) (Sigma, USA) overnight at 4°C. Blots were then washed and incubated for 1 h with the secondary antibodies. After washing with PBST, chemiluminescence was detected using the Odyssey scanning system (LI-COR, Lincoln, Nebraska, USA).
Dual-luciferase reporter assay
The potent target genes of miR-195 were predicted using the online tool TargetScan. CCNE1 3′-UTR (wild type) and its mutant were cloned into the psiCHECK-2 reporter vector, and co-transfected with miR-195-5p mimics (50 nmol/l) or NC when HEK-293 cells reached 80–90% confluence. 72 h after transfection, luciferase activity was measured using the dual-luciferase reporter assay kit (Promega, Madison, Wisconsin, USA). Briefly, the cells were washed twice with PBS and then lysed by incubation at room temperature for 15 min with passive lysis buffer. The supernatants were collected and 20 μl of the aliquots were added to 96-well plates. The firefly luciferase reporter was measured immediately after adding Luciferase Assay Reagent II. Next, 100 μl of Stop & Glo reagent (Promega, Madison, Wisconsin, USA) was added to each well to initiate the renilla luciferase. The psiCHECK-2 vector, which provides constitutive expression of firefly luciferase, was co-transfected as an internal control. All experiments were conducted three times.
Statistical analysis
All experiments were conducted in triplicate and results were presented as the mean±SD or mean±SE. Statistical analysis was carried out using one-way analysis of variance in GraphPad Prism 5 (GraphPad Software, La Jolla, California, USA). t-Test was used for comparison between groups. A P value less than 0.05 was considered statistically significant.
Result
MiR-195 was downregulated in TMZ-resistant glioblastoma cells
First, we established the TMZ-resistant cell line (U251R), which was generated by stepwise (6 months) exposure of parental cells to TMZ. Cell viability was detected using the MTT method and compared at 24, 48, 72, 96, and 120 h after seeding. The viability of U251R had progressed compared with U251 cells, and significant differences were found at 72 h after seeding. The IC50 of U251 cell line was 82.35 μg/ml (Fig. 1a) and the IC50 of the U251R cell line was 289.83 μg/ml (Fig. 1a). The resistance index was 3.52. At the same time, we found that the U251R had an increased proliferation rate (Fig. 1b).
Fig. 1.
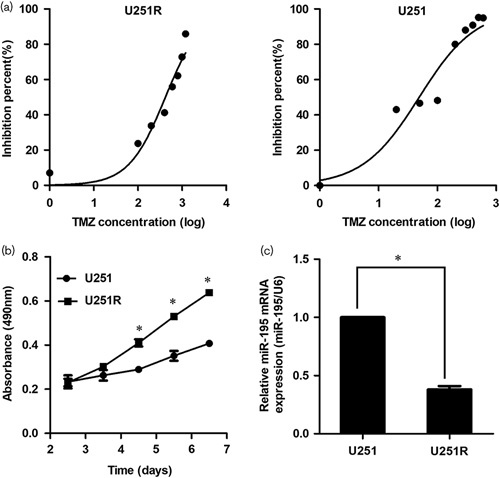
MiR-195 is downregulated in temozolomide (TMZ)-resistant glioblastoma cells. (a) The IC50 of U251 (right) and U251R (left) cell lines. (b) The growth curves of U251 and U251R cell lines. (c) MiR-195 is downregulated in U251R cells. The data are the result of three independent experiments and are presented as mean±SD. *P<0.05.
As mentioned above, miR-195 was abundant in multiple types of cancer. In this study, the downregulated miR-195 was verified by qPCR in U251R cells (Fig. 1c). Results showed a significant reduction in miR-195 expression in U251R cells than in the parent U251 cells (P<0.05), which was consistent with previous studies, implying that miR-195 might be related to the progression and resistance of glioma. Therefore, we furtherly analyzed its roles and mechanism in the parent U251 and U251R cells in the following experiments.
Inhibition of miR-195 promoted cell resistance and suppressed cell apoptosis and cell cycle of glioma cells
The miR-195 mimics and inhibitor were used to overexpress and inhibit miR-195, respectively. Then, chemoresistance was detected using the MTT method in the U251 or U251R cells transfected with the miR-195 inhibitor or miR-195 mimics. These results indicated that decreasing miR-195 leads to the chemoresistance of U251 cell viability (P<0.05, Fig. 2a, left) and overexpressing miR-195 suppressed cell chemoresistance in U251R cells (P<0.05, Fig. 2a, right).
Fig. 2.
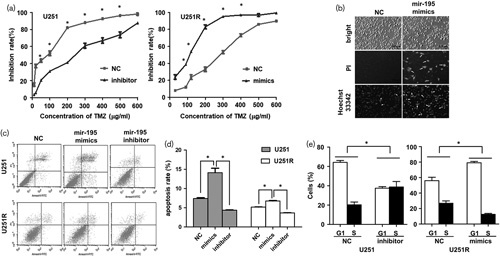
MiR-195 reverses temozolomide (TMZ) resistance in glioma cells. (a) Inhibition of miR-195 promotes TMZ resistance in U251 cells and overexpression of miR-195 reverses TMZ resistance in U251R cells. (b) Overexpression of miR-195 induces apoptosis in U251R cells. Cell apoptosis was measured using the Hoechst 33342/PI Staining Kit. Scale bars=100 μm. (c, d) Inhibition of miR-195 suppresses cell apoptosis, whereas overexpression of miR-195 induces apoptosis in U251 and U251R cells. (e) Ectopic expression of miR-195 blocks G1/S transition, whereas inhibition of miR-195 promotes G1/S transition. The data are the result of three independent experiments and are presented as mean±SD. *P<0.05.
Next, we examined cell apoptosis using the Hoechst 33342/PI staining method. The results showed an obvious increase in the cell apoptosis rate when U251R cells were transfected with miR-195 mimics compared with the NC group (Fig. 2b). Then, the cell apoptosis rate was detected by flow cytometry using an Annexin-V–FITC/PI staining method. Results also showed an obvious increase in cell apoptosis when U251 and U251R cells were transfected with miR-195 mimics, which was inhibited by the miR-195 inhibitor (Fig. 2c and d). Apoptosis was promoted by miR-195 upregulation (P<0.05) and inhibition of miR-195 significantly suppressed cell apoptosis (P<0.05). Meanwhile, ectopic expression of miR-195 blocked G1/S transition in U251R cells (Fig. 2e). Taken together, this evidence showed that miR-195 can inhibit cell resistance and promote cell apoptosis and cell cycle in GBM cells.
CCNE1 was a target gene of miR-195
To explore the potent mechanism of miR-195 in cell resistance and apoptosis, the potential target genes of miR-195 were predicted using the online tool TargetScan (http://www.targetscan.org/). The software showed that CCNE1 was a potential target gene of miR-195 (Fig. 3a). RT-qPCR showed that hsa-miR-195 mimics exerted an obvious downregulation effect on CCNE1 mRNA, whereas the hsa-miR-195 inhibitor promoted the expression of CCNE1 mRNA in U251R and parent U251 cell lines (Fig. 3b).
Fig. 3.
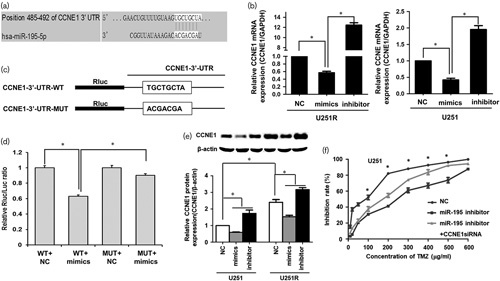
MiR-195 reverses temozolomide (TMZ) resistance by targeting CCNE1 in glioma cells. (a) Targetscan shows that CCNE1 is a potential target gene of miR-195. (b) RT-qPCR shows that hsa-miR-195 mimics exert an obvious downregulation effect on CCNE1 mRNA, whereas the hsa-miR-195 inhibitor promotes the expression of CCNE1 mRNA in U251R and parent U251 cell lines. (c) Luciferase reporter with wild-type (WT) CCNE1 3′-UTR and mutant (MUT) CCNE1 3′-untranslated region (3′-UTR). (d) Hsa-miR-195 mimics decreases the fluorescent value of the reporter with a wild-type CCNE1 3′-UTR. When the binding site is mutated, the fluorescence reporter could not be inhibited by miR-195 mimics. (e) Ectopic expression of miR-195 induces a decrease in CCNE1 protein, whereas inhibition of mi5-195 leads to an increase in CCNE1 in U251R and parent U251 cell lines. (f) Knockdown of CCNE1 partially reverses TMZ resistance induced by decreased miR-195. The data are the result of three independent experiments and are presented as mean±SD. *P<0.05.
To confirm the regulation of miR-195 on CCNE1, the dual-luciferase reporter assay was performed. The result showed that hsa-miR-195 mimics decreased the fluorescent value of the reporter with a wild-type CCNE1 3′-UTR. When the binding site was mutated, the fluorescence reporter could not be inhibited by miR-195 mimics (Fig. 3c and d). Also, western blot verified that the downregulation effect of miR-195 on CCNE1 mRNA led to a significant decrease in CCNE1 at the protein level in U251 and U251R cells (Fig. 3e). The results showed that hsa-miR-195 could suppress the expression of CCNE1 by binding the target site in the 3′-UTR of CCNE1 directly.
Then, the effect of CCNE1 on chemoresistance induced by decreased miR-195 was measured in U251 cells. The result showed that decreasing miR-195 leads to significant TMZ resistance in U251 cell; however, when CCNE1 was knocked down, the TMZ resistance suppressed. This indicated that knockdown of CCNE1 partially reversed the effect of decreased miR-195 on TMZ resistance (Fig. 3f).
MiR-195 was correlated negatively with CCNE1 in glioma samples
To confirm the correlation between miR-195 and CCNE1, we analyzed the data of glioma samples from The Cancer Genome Atlas – Cancer Genome. We selected the cohort of The Cancer Genome Atlas – Cancer Genome Lower Grade Glioma, which consisted of 530 clinical samples, and obtained the complete expression data of mRNA and miRNA. We analyzed the correlation of miR-195 and CCNE1 using the Xena browser and the result showed that there was a probable correlation between the expression of miR-195 and CCNE1 (Fig. 4a). Further nonparametric correlation (Spearman) analysis showed there was a negative correlation between the expression of miR-195 and CCNE1 (r=−0.2160, P<0.0001, Fig. 4b). The results of clinical samples further suggested that hsa-miR-195 could negatively regulate the expression of CCNE1 in glioma.
Fig. 4.
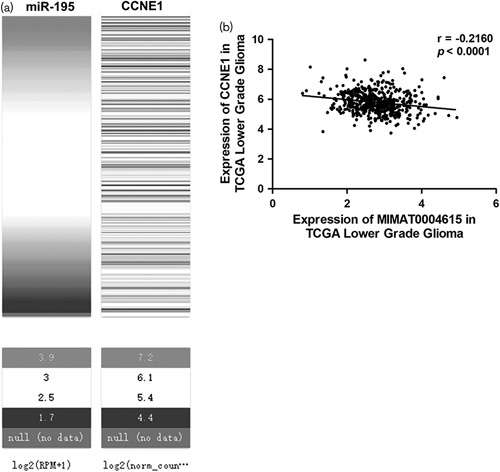
MiR-195 is correlated negatively with CCNE1 in glioma samples. (a) Heatmap of the expression of miR-195 and CCNE1 mRNA in glioma samples. (b) Spearman analysis shows that there is a negative correlation between the expression of miR-195 and CCNE1 in glioma samples from The Cancer Genome Atlas – Cancer Genome (TCGA).
Discussion
In our present study, miR-195 was found to be downregulated in TMZ-resistant glioma cell line – U251R. The function experiment indicated that miR-195 can reverse the resistance to TMZ and inhibit cell proliferation and promote apoptosis of glioma cells. Also, we found that CCNE1 was one of the targets of miR-195 and its expression was suppressed by upregulated miR-195, implying the possible regulatory mechanism of miR-195 in glioma resistance to TMZ chemotherapy.
As a mature miRNA, miR-195 was reported to be involved in multiple tumors. It plays a tumor-suppressor role in multiple tumors, including colorectal cancer 8, osteosarcoma 9, breast cancer 10–12, prostate cancer 13,14, in gliomas 15, cervical cancer 16, bladder tumor 17, and hepatocellular carcinoma 18,19 by targeting different target genes. Some studies have reported that miR-195 participates in the chemotherapy resistance of cancer cells. In non-small-cell lung cancer, miR-195 was associated with gefitinib sensitivity 20. In breast cancer, it has been shown to increase drug responsiveness to doxorubicin 21 and adriamycin 22 by targeting Raf-1. In gastric cancer, miR-195 was involved in chemotherapy/radiotherapy response by targeting CHK1 23. However, its role and mechanism in the TMZ resistance in glioma remains unclear.
Our study showed that miR-195 is downregulated in the TMZ-resistant glioma cells and CCNE1 is its direct target gene. Cyclins were the key regulators of the cell cycle, playing important roles in tumorigenesis 24,25. As a potent oncogene, CCNE1 functions as a regulatory subunit of CDK2 and regulates the transition from the G1 phase to the S phase of the cell cycle. CCNE1 protein accumulates at the G1–S phase boundary and is degraded as cells progress through the S phase. Overexpression of this gene has been observed in many tumors and its deregulation is considered to play a fundamental role in tumorigenesis 25. CCNE1 has also been reported to contribute toward the progression of many types of cancer 26,27. Its gene amplification or overexpression has been observed in esophageal adenocarcinoma 28, gastric carcinoma patient 26, endometrioid endometrial carcinomas 29, ovarian cancer 30, endometrial intraepithelial carcinoma, and uterine serous carcinoma 31. Its amplification was considered an early genetic event during tumor progression 31. Thus, CCNE1 may present as a target for precision cancer therapy 32.
CCNE1 has been also reported to be associated with chemoresistance. In ovarian carcinomas, high CCNE1 copy number and protein expression were associated with primary chemoresistance 33. In multiple myeloma, overexpression of CCNE1 reduced the sensitivity to seliciclib 34. In bladder cancer, upregulation of CCNE1 was associated with cisplatin resistance 35. Our result showed that CCNE1 is upregulated in TMZ-resistant glioma cells and its knockdown suppressed cell viability and reduced the resistance to TMZ. Thus, CCNE1 may be a key gene associated with chemoresistance. Recently, CCNE1 has been reported to play a role in mediating asymmetric cell division, specifying cell fate and, importantly, in driving stem cell self-renewal, independent of its activity in cellular proliferation 36,37. Thus, its effect on cancer stem cells may be its underlying mechanism of chemoresistance. Thus, as one of the targets of miR-195, CCNE1 expression is suppressed by upregulated miR-195, implying the possible regulatory mechanism of miR-195 in glioma resistance to TMZ chemotherapy.
Conclusion
MiR-195 was decreased in TMZ-resistant glioma cells and its overexpression can inverse the resistance to TMZ and CCNE1 is a direct target gene of miR-195. As a promising candidate for the treatment of TMZ-resistant glioma, further research is needed to clarify the function, mechanism, and the potential clinic value of miR-195 in detail in the treatment of glioma.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 81470115 for Hongqin Wang and grant no. 81502135 for Pengzhou Kong).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Cho DY, Lin SZ, Yang WK, Hsu DM, Lin HL, Lee HC, et al. The role of cancer stem cells (CD133(+)) in malignant gliomas. Cell Transplant 2011; 20:121–125. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol 2005; 109:93–108. [DOI] [PubMed] [Google Scholar]
- 3.Graham CA, Cloughesy TF. Brain tumor treatment: chemotherapy and other new developments. Semin Oncol Nurs 2004; 20:260–272. [PubMed] [Google Scholar]
- 4.Besse A, Sana J, Fadrus P, Slaby O. MicroRNAs involved in chemo- and radioresistance of high-grade gliomas. Tumour Biol 2013; 34:1969–1978. [DOI] [PubMed] [Google Scholar]
- 5.Rolle K. miRNA Multiplayers in glioma. From bench to bedside. Acta Biochim Pol 2015; 62:353–365. [DOI] [PubMed] [Google Scholar]
- 6.Costa PM, Cardoso AL, Mano M, de Lima MC. MicroRNAs in glioblastoma: role in pathogenesis and opportunities for targeted therapies. CNS Neurol Disord Drug Targets 2015; 14:222–238. [DOI] [PubMed] [Google Scholar]
- 7.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015; 5:1122–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M, Wu W, Gao M, Zhang J, Ding X, Zhu R, et al. Coactivator-associated arginine methyltransferase 1 promotes cell growth and is targeted by microRNA-195-5p in human colorectal cancer. Tumour Biol 2017; 39:1010428317694305. [DOI] [PubMed] [Google Scholar]
- 9.Qu Q, Chu X, Wang P. MicroRNA-195-5p suppresses osteosarcoma cell proliferation and invasion by suppressing naked cuticle homolog 1. Cell Biol Int 2017; 41:287–295. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Zhang X, Zou C, Kung HF, Lin MC, Dress A, et al. miR-195 inhibits tumor growth and angiogenesis through modulating IRS1 in breast cancer. Biomed Pharmacother 2016; 80:95–101. [DOI] [PubMed] [Google Scholar]
- 11.Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep 2015; 5:17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res 2011; 17:1722–1730. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Tao T, Liu C, Guan H, Huang Y, Xu B, et al. Downregulation of miR-195 promotes prostate cancer progression by targeting HMGA1. Oncol Rep 2016; 36:376–382. [DOI] [PubMed] [Google Scholar]
- 14.Cai C, Chen QB, Han ZD, Zhang YQ, He HC, Chen JH, et al. miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin Cancer Res 2015; 21:4922–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilaz Susluer S, Biray Avci C, Dodurga Y, Ozlem Dogan Sigva Z, Oktar N, Gunduz C. Downregulation of miR-195 via cyclosporin A in human glioblastoma cells. J BUON 2015; 20:1337–1340. [PubMed] [Google Scholar]
- 16.Li Z, Wang H, Wang Z, Cai H. MiR-195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1. Tumour Biol 2016; 37:6457–6463. [DOI] [PubMed] [Google Scholar]
- 17.Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS ONE 2014; 9:e84311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan JJ, Chang Y, Zhang YN, Lin JS, He XX, Huang HJ. miR-195 inhibits cell proliferation via targeting AEG-1 in hepatocellular carcinoma. Oncol Lett 2017; 13:3118–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Zhang J, Tong L, Ma X, Qiu X. MiR-195 is a key negative regulator of hepatocellular carcinoma metastasis by targeting FGF2 and VEGFA. Int J Clin Exp Pathol 2015; 8:14110–14120. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Q, Cao J, Wu YC, Liu X, Han J, Huang XC, et al. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am J Cancer Res 2015; 5:1692–1705. [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzo S, Cangemi A, Galvano A, Fanale D, Buscemi S, Ciaccio M, et al. Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget 2017; 8:71924–71932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian J, et al. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep 2013; 30:877–889. [DOI] [PubMed] [Google Scholar]
- 23.Bargiela-Iparraguirre J, Prado-Marchal L, Fernandez-Fuente M, Gutierrez-Gonzalez A, Moreno-Rubio J, Munoz-Fernandez M, et al. CHK1 expression in Gastric Cancer is modulated by p53 and RB1/E2F1: implications in chemo/radiotherapy response. Sci Rep 2016; 6:21519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casimiro MC, Crosariol M, Loro E, Li Z, Pestell RG. Cyclins and cell cycle control in cancer and disease. Genes Cancer 2012; 3:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005; 24:2776–2786. [DOI] [PubMed] [Google Scholar]
- 26.Ooi A, Oyama T, Nakamura R, Tajiri R, Ikeda H, Fushida S, et al. Gene amplification of CCNE1, CCND1, and CDK6 in gastric cancers detected by multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Hum Pathol 2017; 61:58–67. [DOI] [PubMed] [Google Scholar]
- 27.Hunt KK, Keyomarsi K. Cyclin E as a prognostic and predictive marker in breast cancer. Semin Cancer Biol 2005; 15:319–326. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, Bandla S, Ye J, Xia Y, Que J, Luketich JD, et al. Cyclin E involved in early stage carcinogenesis of esophageal adenocarcinoma by SNP DNA microarray and immunohistochemical studies. BMC Gastroenterol 2014; 14:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama K, Rahman MT, Rahman M, Nakamura K, Ishikawa M, Katagiri H, et al. CCNE1 amplification is associated with aggressive potential in endometrioid endometrial carcinomas. Int J Oncol 2016; 48:506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama N, Nakayama K, Shamima Y, Ishikawa M, Katagiri A, Iida K, et al. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer 2010; 116:2621–2634. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn E, Bahadirli-Talbott A, Shih Ie M. Frequent CCNE1 amplification in endometrial intraepithelial carcinoma and uterine serous carcinoma. Mod Pathol 2014; 27:1014–1019. [DOI] [PubMed] [Google Scholar]
- 32.Fang D, Huang S, Su SB. Cyclin E1-CDK 2, a potential anticancer target. Aging 2016; 8:571–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res 2009; 15:1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Josefsberg Ben-Yehoshua L, Beider K, Shimoni A, Ostrovsky O, Samookh M, Peled A, et al. Characterization of cyclin E expression in multiple myeloma and its functional role in seliciclib-induced apoptotic cell death. PLoS ONE 2012; 7:e33856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SH, Ho JN, Jin H, Lee SC, Lee SE, Hong SK, et al. Upregulated expression of BCL2, MCM7, and CCNE1 indicate cisplatin-resistance in the set of two human bladder cancer cell lines: T24 cisplatin sensitive and T24R2 cisplatin resistant bladder cancer cell lines. Investig Clin Urol 2016; 57:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia W, Prokopenko SN. Cyclin E at the centre of an identity crisis. Nat Cell Biol 2005; 7:3–5. [DOI] [PubMed] [Google Scholar]
- 37.Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol 2008; 180:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]


