SUMMARY
Tcrb locus V(D)J recombination is regulated by positioning at the nuclear periphery. Here, we used DamID to profile Tcrb locus interactions with the nuclear lamina at high resolution. We identified a lamina-associated domain (LAD) border composed of several CTCF-binding elements that segregates active non-LAD from inactive LAD regions of the locus. Deletion of the LAD border causes an enhancer-dependent spread of histone H3 lysine 27 acetylation from the active recombination center into recombination center-proximal LAD chromatin. This is associated with a disruption to nuclear lamina association, increased chromatin looping to the recombination center, and increased transcription and recombination of recombination center-proximal gene segments. Our results show that a LAD and LAD border are critical components of Tcrb locus gene regulation and suggest that LAD borders may generally function to constrain the activity of nearby enhancers.
In Brief
Chen et al. identify a Tcrb locus lamina-associated domain border that constrains the activity of the Tcrb enhancer. Deletion of the border causes enhancer-dependent loss of nuclear lamina association, spreading of H3K27 acetylation, and elevated transcription and VDJ recombination of gene segments in affected chromatin.
Graphical Abstract
INTRODUCTION
The association of chromatin with the nuclear periphery plays an essential role in gene regulation. The nuclear periphery consists of the inner nuclear membrane, nuclear membrane proteins, and the nuclear lamina (NL), a filamentous network of A-, C-, and B-type lamin proteins (Dobrzynska et al., 2016). Regions of chromatin in molecular contact with the NL, termed lamina-associated domains (LADs), have been defined by DamID, a profiling technology that utilizes adenine methylation of DNA to label chromatin localized to the NL (Guelen et al., 2008; van Steensel and Henikoff, 2000). The mapping of LADs in human and mouse cell lines by DamID has shown that approximately one-third of the genome is organized into LADs, which typically span 0.1–10 Mb in size (Guelen et al., 2008). LADs and non-LADs are not static subnuclear compartments but can interconvert upon cell differentiation or activation (Peric-Hupkes et al., 2010; Robson et al., 2017). DNA sequence composition (Bian et al., 2013; Meuleman et al., 2013; Zullo et al., 2012), histone modifications (Bian et al., 2013; Towbin et al., 2012), and transcription factors (Harr et al., 2015; Zullo et al., 2012) have all been implicated as molecular regulators of LAD formation.
The nuclear periphery is generally considered to be transcriptionally repressive (van Steensel and Belmont, 2017). LADs have a lower gene density than non-LADs, and genes within LADs are either transcriptionally inactive or expressed at lower levels than non-LAD genes (Guelen et al., 2008; Peric-Hupkes et al., 2010). Transcriptional suppression at the NL is mediated directly, by repressors like heterochromatin protein 1 (HP1) and histone deacetylase 3 (HDAC3), which interact with the NL or NL-associated proteins (Poleshko et al., 2017; Polioudaki et al., 2001; Somech et al., 2005; Zullo et al., 2012), and indirectly, by the exclusion of transcription factors and transcriptional machinery from the nuclear periphery (Yao et al., 2011).
The adaptive arm of the mammalian immune system is composed of T and B lymphocytes that recognize antigens using diverse arrays of T and B cell receptors, respectively, with each lymphocyte bearing a receptor of unique specificity. Complete antigen receptor genes are not encoded in the germline but are instead generated by a process of somatic recombination termed V(D)J recombination (Schatz and Ji, 2011). V(D)J recombination mediates the assembly of variable (V), diversity (D), and joining (J) gene segments at antigen receptor loci and depends on the activity of recombination activating gene 1 and 2 proteins (hereafter referred to as RAG) and DNA repair proteins.
Antigen receptor gene assembly is regulated by multiple mechanisms. V(D)J recombination is limited to developing lymphocytes by tight control of RAG gene expression. In addition, antigen receptor gene segments are selectively made available for recombination by regulated transcription from nearby promoter elements (Abarrategui and Krangel, 2006, 2007; Yancopoulos and Alt, 1985). This transcription removes repressive histone modifications in favor of activating histone modifications and promotes chromatin remodeling needed for RAG binding (Bevington and Boyes, 2013; Gopalakrishnan et al., 2013; Ji et al., 2010; Maman et al., 2016). High-level RAG binding is directed to D and J segments to form the recombination center (RC) (Schatz and Ji, 2011). Because antigen receptor loci can span megabases along the linear genome, large-scale changes in chromatin conformation and looping are required to bring distal V gene segments into spatial proximity of the RC for recombination (Bossen et al., 2012). These changes in looping and conformation are dependent on the activity of architectural proteins like CTCF (Chen et al., 2015; Guo et al., 2011; Shih et al., 2012) and transcription factors like Pax5 (Ebert et al., 2011) and YY1 (Medvedovic et al., 2013). V(D)J recombination is also regulated by subnuclear positioning, with antigen receptor loci generally localized with the NL or repressive heterochromatin compartments during developmental stages when the loci are inactive and repositioned to the nuclear interior to set the stage for recombination (Kosak et al., 2002; Schlimgen et al., 2008).
The mouse T cell receptor (TCR) β locus (Tcrb) encodes one chain of the TCRαβ heterodimer. This locus includes Vβ, Dβ, Jβ, and Cβ gene segments along with multiple trypsinogen genes that are inactive in the T cell lineage. These trypsinogen genes are organized into two discrete chromatin regions of 150 and 250 kb that flank the major cluster of 21 functional Vβ gene segments and segregate the Vβ gene segments from the Dβ -Jβ RC. The RC also contains the only known Tcrb locus enhancer, Eβ. Tcrb recombination is a two-step process that takes place in the CD4−CD8−double-negative (DN) stage of thymocyte development, with Dβ-to-Jβ recombination followed by Vβ-to-DJβ recombination. Vβ-to-DJβ recombination is tightly regulated to ensure that only one of the two Tcrb alleles in a cell undergoes functional recombination, a process known as allelic exclusion (Brady et al., 2010).
The Tcrb locus is unique among antigen receptor loci in that it localizes to the nuclear periphery during the developmental stage (DN) in which it undergoes recombination. Prior work showed that Tcrb alleles associate frequently and stochastically with the NL in DN thymocytes and that high-frequency association persists during the subsequent CD4+CD8+ double-positive (DP) stage of development, when Tcrb recombination has concluded (Schlimgen et al., 2008). Analysis of DN thymocytes revealed that Vβ-to-DJβ recombination of NL-localized Tcrb alleles was suppressed compared to alleles localized to the nuclear interior, and this suppression was hypothesized to contribute to Tcrb allelic exclusion (Chan et al., 2013). Suppression may be mediated, in part, by sequestration of Tcrb alleles from RAG proteins.
Here, we defined the organization of the Tcrb locus at the NL and identified a LAD border that separates the Vβ gene segments and trypsinogen genes from the RC. Deletion of the LAD border disrupts NL association of 300 kb of Tcrb locus chromatin. This disruption is accompanied by changes in histone modification, long-distance chromatin contacts, transcription, and recombination, that primarily affect RC-proximal genes. Therefore, we demonstrate that the LAD border is a core regulatory element of the Tcrb locus.
RESULTS
Tcrb Locus Organization at the NL
To obtain a high-resolution map of Tcrb-NL interactions, we conducted DamID. To do so, we transduced the mouse DP thymocyte cell line VL3-3M2 (Groves et al., 1995) with constructs encoding either a Dam-Lamin B1 fusion protein or un-fused Dam and assayed adenine methylation of cellular DNA. VL3-3M2 cells bear an in-frame VDJβ rearrangement on one Tcrb allele and a DJβ rearrangement on the second allele (Figures 1A and 1B). Because the VDJβ-rearranged allele joins Trbv3 to Trbd1 by deletional recombination, the 500-kb region between these gene segments is represented in a monoallelic fashion on the DJβ-rearranged allele. Thus, the DJβ-rearranged allele (hereafter referred to as the experimental allele) can serve as a valuable substrate for analysis and manipulation. Moreover, because VL3-3M2 cells express RAG proteins and support V(D)J recombination on extrachromosomal substrates (Williams et al., 2001), these cells offer a potentially valuable experimental system to test the relationship between NL association and Vβ-to-DJβ recombination at the endogenous Tcrb locus.
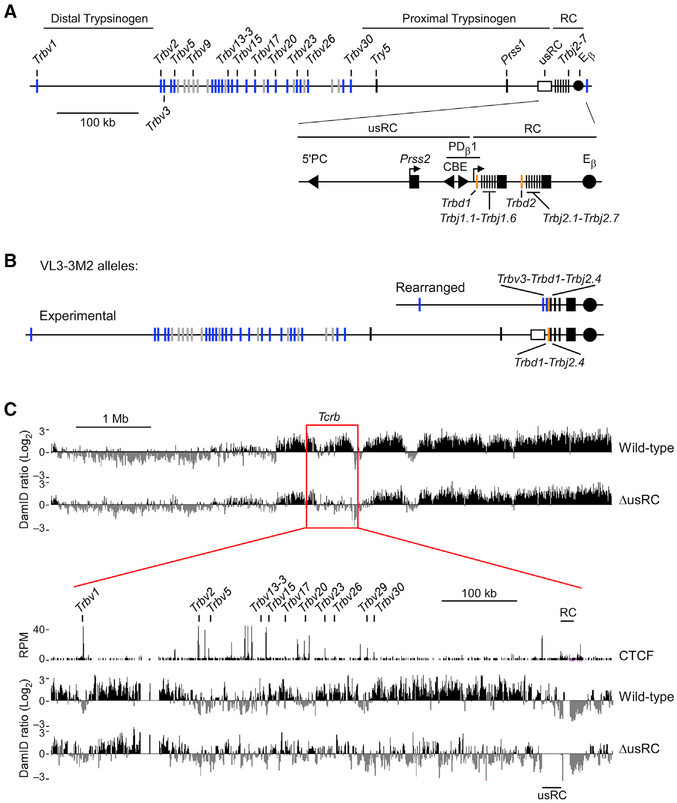
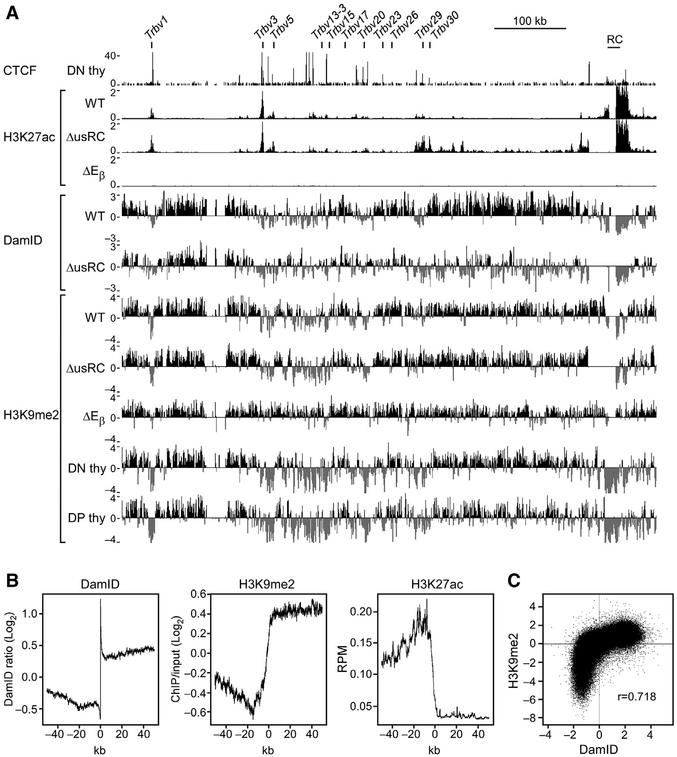
Figure 1. Characterization of Tcrb-NL Association in Wild-Type VL3-3M2 and ΔusRC.
(A) Scale schematic of the Tcrb locus indicating positions of Vβ gene segments, trypsinogen regions, and regulatory elements. The 3′ end of the locus is magnified to show elements within the recombination center (RC) and the lamina-associated domain (LAD) border upstream of the RC (usRC). Filled triangles denote the position and orientation of relevant CTCF-binding elements (CBEs). The filled circle denotes the Tcrb enhancer (Eβ). Functional Vβ gene segments and pseudo-genes are indicated in blue and gray, respectively.
(B) Tcrb alleles in the VL3-3M2 cell line. The rearranged allele bears a Trbv3-Trbd1-Trbj2.4 rearrangement while the experimental allele bears a Trbd1-Trbj2.4 rearrangement.
(C) DamID-seq profiles of the Tcrb locus in wild-type and ΔusRC. Experimental values are expressed as the log2 ratio of Dam-Lamin B1 over unfused Dam. Reads were merged from two independent replicates. CTCF ChIP-seq data are from GEO: GSE41743 (Shih et al., 2012). CTCF ChIP-seq values are expressed as reads per million.
See also Figure S1.
High-throughput sequencing coupled with DamID (DamID-seq) was used to generate a high-resolution map of Tcrb-NL interactions. We observed that, compared to the unfused Dam control, Dam-Lamin B1 signals were generally enriched throughout the Tcrb locus and were highest at the RC-proximal and RC-distal trypsinogen regions, consistent with these regions constituting LADs (wild-type, Figure 1C). The cluster of Vβ gene segments that lies between the trypsinogen regions exhibited a mixed character, wherein RC-proximal Vβ gene segments were associated with the NL and RC-distal ones were generally free of the NL. In contrast, a well-defined border between the RC and the RC-proximal trypsinogen region segregated the RC into a non-LAD (Figure 1C).
The LAD Border Upstream of the RC Insulates Tcrb Chromatin from the Influence of Eβ
We assessed the mechanisms enforcing segregation of the Tcrb locus into LAD and non-LAD chromatin regions by characterizing the sharp LAD border immediately upstream of the RC. The 28-kb region spanning the LAD border contains previously characterized regulatory elements (Figure 1A; Majumder et al., 2015). The 3′end of this region, immediately adjacent to the RC, contains elements of the Trbd1 promoter PDβ 1, including a pair of CTCF-binding elements (CBEs), which function as an insulator that blocks the spread of activating histone modifications from the RC. The 5′end of this region, farthest from the RC, is marked by the CBE 5′PC, which functions as a tether that brings distal Vβ segments into proximity of the RC. The trypsinogen gene Prss2, normally silent in T-lineage cells, is found between 5′PC and PDβ 1. Using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, we created a derivative VL3-3M2 cell line that deleted this 28-kb region on the experimental allele, taking care that core PDβ 1 promoter elements were not disrupted and that PDβ 1 transcription was minimally affected (Figure S1). We designated this 28-kb region “upstream of the recombination center” (usRC) and the VL3-3M2 derivative cell line bearing a deletion of this region ΔusRC. DamID-seq analysis of ΔusRC revealed that the RC-proximal trypsinogen genes and two most RC-proximal Vβ genes, Trbv29 and Trbv30, showed substantial reductions in NL association, whereas Vβ gene segments spanning Trbv23 to Trbv26 had more modest reductions in NL association (Figure 1C). In contrast, the remainder of the locus, as well as surrounding chromatin, was largely unchanged (Figure 1C). Analysis of DamID by qPCR (DamID-qPCR) recapitulated the results of DamID-seq with respect to the alternating regions of NL association on the wild-type allele as well as a highly significant loss of NL association in the RC-proximal region in ΔusRC (Figures 2A and 2B).
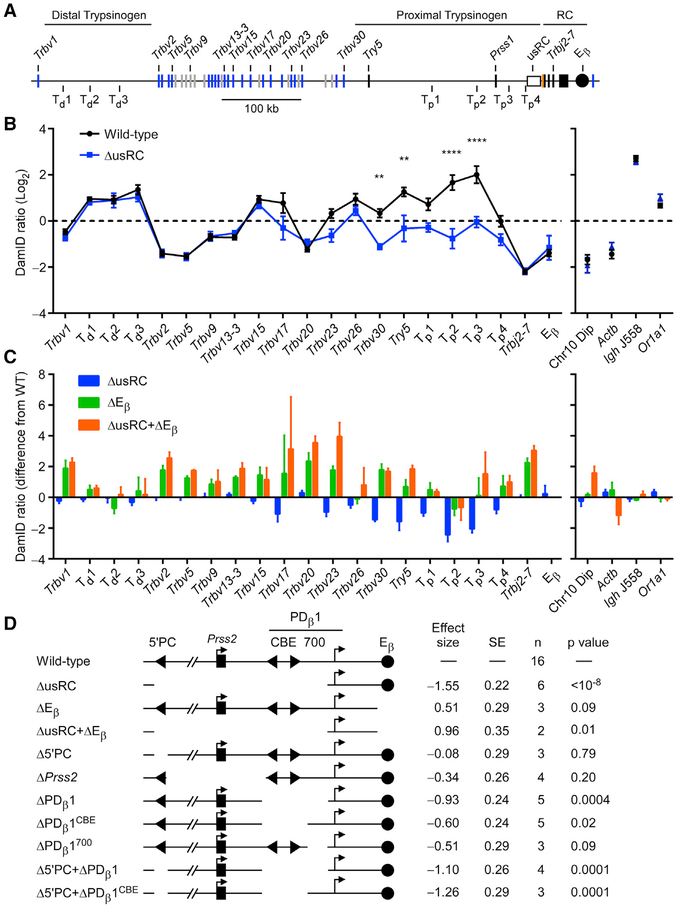
Figure 2. Dissection of usRC LAD Border Function.
(A) Scale schematic of the Tcrb locus indicating sites analyzed by qPCR. Td and Tp refer to sites within the RC-distal and RC-proximal trypsinogen regions, respectively.
(B) DamID-qPCR profiles of the Tcrb locus (left) and non-Tcrb control loci (right). Control loci were chosen from regions that showed consistently high or low signals in DamID-seq. The data represent the mean and SE of 16 (wild-type) or 6 (ΔusRC) independent experiments. **p < 0.01 and ****p < 0.0001, by two-way ANOVA with Holm-Sidak’s multiple-comparisons test.
(C) DamID-qPCR difference maps relative to wild-type VL3-3M2. The graph shows the difference in DamID-qPCR signals from wild-type at each site analyzed. Data for wild-type and ΔusRC were transformed from Figure 2B.
(D) Effects of Tcrb locus deletions on NL association of the RC-proximal region. Clonal derivative VL3-3M2 cell lines bearing the indicated deletions were analyzed by DamID-qPCR. Overall effect sizes and p values were calculated for six RC-proximal sites spanning Trbv30 to Tp4, using a restricted maximum-likelihood model, which accounted for DamID signal, genotype, and batch effects. Positive effect sizes indicate greater NL association; negative effect sizes indicate reduced NL association. Data for wild-type, ΔusRC, ΔEβ and ΔusRC+ΔEβ were from the same dataset that was used to generate Figure 2C.
See also Figure S1 and Table S1.
We hypothesized that the RC is positioned away from the NL due to the activity of the Tcrb enhancer, Eβ, and that this activity is normally confined to the RC by the usRC LAD border. To test this, we assessed the effects of Eβ deletion. In these experiments, we plotted differences in DamID-qPCR signals relative to wild-type VL3-3M2 cells. ΔEβ exhibited an increase in NL association at the RC (as indicated by Trbj2–7) and at all Vβ gene segments (Figure 2C). Minimal effects were observed on either the proximal or distal trypsinogen regions. The effects of Eβ deletion on NL association were generally opposite in nature to the effects of usRC deletion. To determine the functional relationship between usRC and Eβ, we created an additional line bearing deletions in both elements (ΔusRC+ΔEβ) and observed a Tcrb-NL association pattern similar to that of the ΔEβ line (Figure 2C). Because the deletion of usRC diminishes RC-proximal chromatin-NL associations only in the presence of Eβ, we conclude that usRC functions as a boundary to modulate the activity of Eβ and prevent encroachment of non-LAD chromatin into the RC-proximal trypsinogen and Vβ regions. However, usRC does not inhibit the impact of Eβ on more distal Vβ gene segments, which is likely to occur by a distinct mechanism.
Multiple Elements Contribute to usRC Function
To dissect the functional elements contributing to the LAD border activity of usRC, we created several VL3-3M2 lines bearing smaller deletions (Figure 2D; Table S1). We used DamID-qPCR data to calculate overall effect sizes and p values relative to wild-type for the region spanning Trbv30 to Tp4, which showed the greatest change in ΔusRC. Among Δ5′PC, ΔPrss2, and ΔPDβ 1, only ΔPDβ1 displayed an effect size approaching that of ΔusRC. Further dissection of PDβ1 into a 5′region containing both CBEs (ΔPDβ1CBE) and a 700-bp 3′region containing a binding site for the transcription factor Sp1 (ΔPDβ1700) (Sikes et al., 1998) revealed that both segments contribute to PDβ1 activity. The greatest effect sizes were obtained by the combined deletion of 5′PC with either PDβ1 (Δ5′PC+ΔPDβ1) or PDβ1CBE (Δ5′PC+ΔPDβ1CBE). We therefore conclude that the three elements 5′PC, PDβ1CBE, and PDβ1700 function cooperatively to enforce a boundary that prevents Eβ-mediated positioning of the RC-proximal region of the Tcrb locus into the nuclear interior.
usRC Suppresses RC-Proximal Transcription
We asked whether the boundary function of usRC influenced transcriptional activation across the Tcrb locus. The two most proximal Vβ gene segments (Trbv29 and Trbv30) and two genes in the proximal trypsinogen region (Try5 and Prss1) showed substantial transcriptional upregulation in ΔusRC compared to wild-type (Figures 3A and 3B). However, there was no transcriptional upregulation of the RC-distal Vβ gene segments Trbv3, Trbv5, and Trbv13–2 on the experimental allele. Notably, all detected transcription was dependent on Eβ, as transcription was completely abrogated in ΔEβ and ΔusRC+ΔEβ (Figure 3B). Thus, similar to NL association, usRC blocks the effects of Eβ on RC-proximal promoters but fails to block more distal effects.
Figure 3. usRC Protects RC-Proximal Genes from Eβ-Dependent Transcriptional Activation.
(A) Scale schematic of the Tcrb locus indicating gene segments assayed.
(B–D) Transcription of Vβ gene segments and RC-proximal trypsinogen genes were analyzed by RT-qPCR. (B) Effects of usRC and Eβ. (C) Dissection of usRC. (D) Effects of compound usRC deletions. Vβ primers only detect transcripts originating from unrearranged gene segments on the experimental allele. Expression values are normalized to Actb, with Actb expression set to 4 on a log10 scale. The data represent the mean and SE of two to eight independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, by two-way ANOVA with Holm-Sidak’s multiple-comparisons test. Number sign, not detectable. The same data for wild-type and ΔusRC were plotted in all three panels.
We also analyzed transcription in the VL3-3M2 lines bearing smaller deletions within usRC. Among the lines bearing deletions of individual elements, transcriptional upregulation was restricted to Try5 and only observed in ΔPDβ1 (Figure 3C). Among the lines with compound deletions, Δ5′PC+ΔPDβ1CBE largely mirrored ΔPDβ1, whereas Δ5′PC+ΔPDβ1 had a transcriptional profile very similar to ΔusRC (Figure 3D).
usRC Suppresses RC-Proximal V(D)J Recombination
Since VL3-3M2 cells express RAG and can support V(D)J recombination (Williams et al., 2001), we asked whether deletion of usRC impacted Tcrb recombination. As an initial assessment, we analyzed recombination in genomic DNA (gDNA) from wild-type, ΔusRC, ΔPDβ1, and Δ5′PC+ΔPDβ1 by qPCR. Relative to wild-type, we detected 15- to 150-fold increases in Trbv29 and Trbv30 rearrangement on the experimental allele in ΔusRC and Δ5′PC+ΔPDβ1 (Figure 4A). However, the detection and quantification of rearrangement events was challenging due to the low rate of recombination of the experimental allele in wild-type VL3-3M2 cells and non-standardized culture periods during which recombination events would have accumulated.
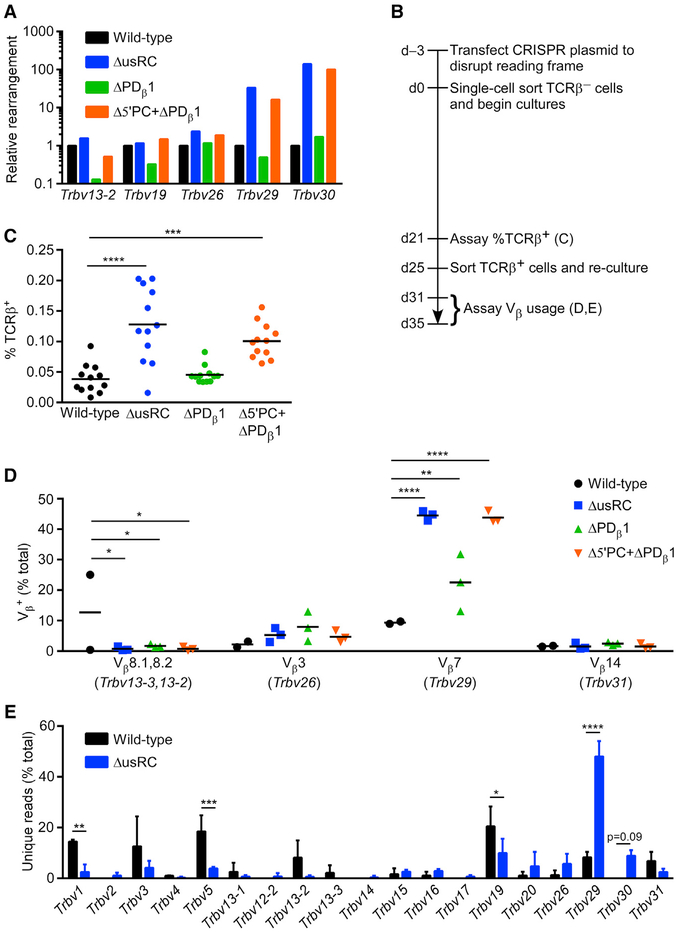
Figure 4. Changes in Tcrb Recombination Frequency and Repertoire upon usRC Deletion.
(A) Relative Tcrb rearrangement in unsynchronized populations of VL3-3M2 cell lines analyzed by qPCR of gDNA. qPCR values were initially normalized to Cd14 in each sample, and normalized values for each Vβ rearrangement in VL3-3M2 mutant cell lines were expressed relative to wild-type, which was set to 1 (n = 1).
(B) Timeline for the disruption of the rearranged allele in VL3-3M2 and its derivatives and analysis of the subsequent accumulation of Tcrb rearrangements in synchronized cultures.
(C) Flow-cytometric analysis of Tcrb recombination in clonal cultures of TCRβ− cells grown for 21 days (n = 12). Horizontal lines denote the mean in each genotype. ***p < 0.001 and ****p < 0.0001, by non-parametric one-way ANOVA with Dunn’s multiple-comparisons test.
(D) Flow-cytometric analysis of Vβ usage in TCRβ+ cells obtained from synchronized cultures of wild-type (n = 2) and mutants (n = 3) by sorting at 25 days followed by expansion in culture. Another wild-type sample in which TCRβ+ cells arose predominantly from an early replacement rearrangement on the rearranged allele was excluded from analysis. Horizontal lines denote the mean in each genotype. *p < 0.05, **p < 0.01, and ****p < 0.0001, by two-way ANOVA with Holm-Sidak’s multiple-comparisons test.
(E) Tcrb repertoire analyzed by high-throughput sequencing of gDNA isolated from TCRβ+ cells analyzed in (D). Vβ usage is plotted as the percentage of unique reads (n = 206 and 209 for wild-type; n = 162, 216, and 240 for ΔusRC) representing rearrangement events on the experimental allele. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, by two-way ANOVA with Holm-Sidak’s multiple-comparisons test.
See also Figure S2.
To circumvent the abovementioned problems, we devised a controlled system in which cells with an initial DJβ rearrangement on the experimental allele were cultured for a defined time period prior to assaying Vβ-to-DJβ recombination (Figure 4B). VL3-3M2 cells have one functional Tcrb and one functional Tcra rearrangement and express surface αβ TCRs detectable by a pan-TCRβ monoclonal antibody (mAb). We used the CRISPR/Cas9 system to disrupt the coding sequence on the rearranged Tcrb allele in order to generate surface TCRβ− derivatives of the VL3-3M2 lines. This allowed us to reliably detect cells with newly generated in-frame recombination events on the experimental allele by staining for the appearance of TCRβ+ cells. After expanding single-cell sorted TCRβ− cells for 21 days in culture, we assayed the rate of recombination in wild-type VL3-3M2, ΔusRC, ΔPDβ1, and Δ5′PC+ΔPDβ1 by flow-cytometric analysis. ΔusRC and Δ5′PC+ΔPDβ1 displayed 2.5- to 3-fold increases in Tcrb recombination frequency compared to wild-type (Figure 4C). To analyze the Tcrb repertoire in greater detail, we sorted TCRβ+ cells that arose in these cultures and analyzed Vβ usage by flow cytometry using Vβ-specific mAbs. We observed a modest increase in the usage of Vβ7 (encoded by the RC-proximal Trbv29 gene segment) in ΔPDβ1 and dramatic increases in usage in ΔusRC and Δ5′PC+ΔPDβ1 (Figure 4D). We could not assay recombination of the RC-proximal Trbv30 gene segment in this manner because a mAb that recognizes its protein product is not available. To analyze Tcrb repertoires in a more comprehensive fashion, we quantified Vβ usage in TCRβ+ wild-type and ΔusRC cultures by high-throughput sequencing. Unique rearrangements to the DJβ gene segment on the experimental allele exhibited a very strong bias toward the usage of Trbv29 and, to a lesser extent, Trbv30, in ΔusRC compared to wild-type (Figure 4E). Because the number of unique sequences represented a substantial proportion of the number of TCRβ+ cells seeded, the RC-proximal bias in Vβ usage reflected multiple unique recombination events, rather than the outgrowth of a limited number of clones that had rearranged early and expanded throughout the 21-day time course.
In addition to recovering recombination events that utilized the DJβ gene segment on the experimental allele, we also recovered an unexpected class of recombination events that restored TCRβ expression to TCRβ− VL3-3M2 cells. These appeared to be direct Vβ-to-Jβ rearrangements that used Trbv1 or Trbv2 with Trbj2–5 or Trbj2–7. Based on their limited Vβ usage, these events were most likely to have occurred on the rearranged rather than the experimental allele, replacing the CRISPR/Cas9-inactivated Trbv3-Trbd1-Trbj2–4 rearrangement by joining an upstream Vβ to a downstream Jβ. Direct Vβ-to-Jβ rearrangement is not observed in vivo, even though it is formally permissible according to the 12/23 rule (Bassing et al., 2000; Sleckman et al., 2000). To independently verify this result, we performed qPCR to detect the original rearrangement and the most heavily represented Vβ-to-Jβ “replacement rearrangement” (Trbv2-Trbj2–5) in pre-sorted (mostly TCRβ−) and sorted TCRβ+ populations. In two wild-type and three ΔusRC samples, the original Trbv3-Trbd1-Trbj2–4 rearrangement was abundant both pre- and post-sort, with the replacement rearrangement detected at low levels in the post-sorted TCRβ+ populations (Figure S2A). However, in the remaining wild-type sample, we observed a substantial depletion of the original rearrangement with a proportional increase in the replacement rearrangement. Consistent with this result, two-thirds of the sorted TCRβ+ cells in this sample expressed surface Vβ4 (encoded by Trbv2) (Figure S2B). Notably, sequence analysis of this wild-type sample identified only a single highly abundant Trbv2-Trbj2–5 sequence and only three unique rearrangements on the experimental allele. This indicated that a single replacement rearrangement had occurred early in the culture period, thus compromising the selection for functional rearrangements on the experimental allele. Hence, we excluded this highly unusual sample from the analyses in Figures 4D and 4E.
usRC Limits Proximal Vβ-RC Looping
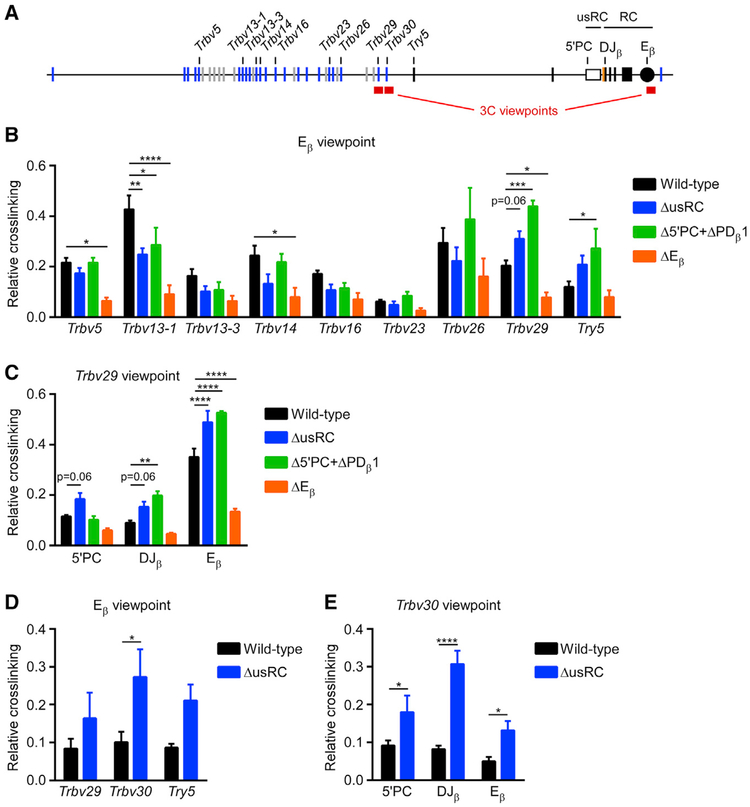
To better understand the basis for the effects of usRC on transcription and recombination, we analyzed the spatial organization of the Tcrb locus using chromosome conformation capture (3C). We initially used Eβ as a viewpoint to test interactions between the Eβ-containing HindIII fragment and other HindIII fragments in the locus. The Eβ-containing HindIII fragment interacted broadly across the Tcrb locus in wild-type cells, and these interactions were diminished by an average of 60% in ΔEβ, showing that Eβ is an essential regulator of Tcrb locus conformation (Figures 5A and 5B). Compared to wild-type, ΔusRC and Δ5′PC+ΔPDβ1 exhibited significant increases in Eβ interactions with RC-proximal Trbv29 and Try5. This was accompanied by significant decreases in interactions with the RC-distal Trbv13–1 gene segment and a similar trend at several other RC-distal sites (Figure 5B). In accordance with these results, the use of Trbv29 as a viewpoint revealed increased interactions with Eβ, 5′PC and the DJβ-rearrangement in the same cell lines (Figure 5C). Similar 3C experiments testing interactions among BglII fragments revealed elevated Trbv30 interactions with Eβ, 5′PC, and DJβ (Figures 5D and 5E). Therefore, we conclude that Eβ acts as a key regulatory element that mediates the interactions of Vβ gene segments with the RC and that usRC functions to balance these interactions by reducing the frequency of interactions with RC-proximal targets and increasing the frequency of interactions with RC-distal targets.
Figure 5. Looping of the RC to RC-Proximal Chromatin Is Suppressed by usRC.
(A) Scale schematic of the Tcrb locus indicating positions of 3C viewpoint and test fragments analyzed.
(B) 3C analysis of long-distance interactions between test HindIII fragments and the Eβ-containing HindIII viewpoint fragment. Results for different ligation products were normalized to their abundance in a digested and religated BAC standard and were expressed relative to the frequency of ligation with an Eβ nearest neighbor fragment. The data represent the mean and SE of five to six independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, by two-way ANOVA with Holm-Sidak’s multiple-comparisons test.
(C) 3C analysis of HindIII fragments with the Trbv29 viewpoint (n = 5–6).
(D) 3C analysis of BglII fragments with the Eβ viewpoint (n = 4–5).
(E) 3C analysis of BglII fragments with the Trbv30 viewpoint (n = 4–5).
usRC Prevents the Spread of H3K27ac from the RC
To further explore the mechanism for usRC effects on NL association, transcription, and recombination, we analyzed histone modifications across the Tcrb locus. Histone H3 lysine 27 acetylation (H3K27ac) is enriched at actively transcribed genes and their enhancers (Creyghton et al., 2010; Tie et al., 2009). Chromatin immunoprecipitation sequencing (ChIP-seq) analysis revealed substantial enrichment of H3K27ac at the RC in wild-type VL3-3M2 cells, with small peaks of H3K27ac detected at Vβ segments (Figure 6A). A large peak detected at Trbv3 most likely represents a signal from the rearranged allele. Similar analysis of ΔEβ cells revealed complete depletion of H3K27ac across the entire Tcrb locus (Figure 6A). Thus, Eβ is absolutely required for Tcrb locus transcription and deposition of H3K27ac in VL3-3M2 cells. Notably, ΔusRC displayed elevated H3K27ac that extended from the RC, across the RC-proximal trypsinogen region and Vβ segments Trbv29 and Trbv30, terminating at a CBE located 8.5 kb upstream of Trbv29 (Figure 6A). Hence, elevated H3K27ac was tightly correlated with portions of the locus that dissociated from the NL and displayed elevated transcription and Vβ-to-DJβ recombination.
Figure 6. usRC Blocks the Spread of H3K27ac from the RC.
(A) ChIP-seq and DamID-seq profiles of the Tcrb locus. CTCF ChIP-seq data are from GEO: GSE41743 (Shih et al., 2012). DamID-seq data are identical to Figure 1C. CTCF and H3K27ac ChIP-seq values are expressed as reads per million. H3K9me2 ChIP-seq values are expressed as the log2 ratio of bound H3K9me2 over input. Reads were merged from two independent replicates.
(B) Feature profiles of LAD borders. LAD borders were identified genome-wide, and average signals across LAD borders were graphed across 100 kb centered on the LAD border. Non-LADs are oriented on the left, and LADs are oriented on the right.
(C) Correlation between H3K9me2 ChIP-seq and DamID-seq in wild-type VL3-3M2. Reads were placed into 10-kb bins. Pearson correlation coefficient is indicated.
See also Figures S3 and S4.
Histone H3 lysine 9 dimethylation (H3K9me2) is enriched at the nuclear periphery and overlaps with LADs genome-wide (Kind et al., 2013; Peric-Hupkes et al., 2010; Poleshko et al., 2017; Wen et al., 2009). Disruption of the methyltransferases that deposit H3K9me2 by knockdown or inhibition has been shown to cause genes in LADs to move away from the nuclear periphery (Bian et al., 2013; Harr et al., 2015; Kind et al., 2013; Towbin et al., 2012). Analysis of wild-type VL3-3M2 cells confirmed the correlation between H3K9me2 and DamID at the Tcrb locus (Figure 6A). Consistent with increased NL association, ΔEβ cells showed increased H3K9me2 across the Vβ array and the RC. However, despite the substantially reduced NL association of the RC-proximal region in ΔusRC, we detected no loss of H3K9me2 in ΔusRC compared to wild-type (Figure 6A).
Further analysis of wild-type VL3-3M2 cells confirmed that LAD borders were demarcated by a sharp transition from H3K9me2 to H3K27ac genome-wide (Figure 6B), and that DamID and H3K9me2 signals were strongly correlated (Figure 6C). However, a fraction of the genome displayed low DamID signals despite enrichment for H3K9me2 (Figure 6C; Figure S3, red box). This observation, together with finding that usRC deletion perturbs NL association with no loss of H3K9me2, agrees with the literature indicating that H3K9me2 is necessary but not sufficient for NL association (Bian et al., 2013; Harr et al., 2015; Kind et al., 2013; Towbin et al., 2012).
In this regard, some studies have revealed enrichment of H3K27 trimethylation (H3K27me3) within LADs (Guelen et al., 2008; Harr et al., 2015), although this observation is not universal (Bian et al., 2013; Kind et al., 2015). It has been hypothesized that H3K27me3 maintains the integrity of LADs near LAD borders (Harr et al., 2015). Because elevated H3K27ac should erase H3K27me3, we hypothesized that the extended region of H3K27ac in ΔusRC disrupts NL association in the RC-proximal portion of the locus by reducing H3K27me3. To test this hypothesis, we performed H3K27me3 ChIP-qPCR at various sites across the RC-proximal region of the Tcrb locus, including 5′PC, which was shown to be H3K27me3-enriched in mouse embryonic fibroblasts (Simon et al., 2013). However, relative to H3K27me3-enriched control loci (Ins1 and Olfr446), we detected only low levels of H3K27me3 at all sites tested across the Tcrb locus in wild-type VL3-3M2 cells (Figure S4). In addition, there were no reductions of H3K27me3 in ΔusRC, although we did detect a trend toward increased H3K27me3 in ΔEβ (Figure S4). Therefore, the NL association of the Tcrb locus is likely regulated by H3K9me2 and H3K27ac but is independent of H3K27me3.
Because it has not been possible to perform DamID in vivo, we assayed H3K9me2 in primary DN and DP thymocytes as a surrogate measure of NL association. Primary DN and DP thymocytes had very similar H3K9me2 profiles, with high signals in both trypsinogen regions and depletion of H3K9me2 at the RC and across large sections of the Vβ array (Figure 6A). These H3K9me2 profiles are broadly similar to the H3K9me2 profile obtained in wild-type VL3-3M2, although there is a greater depletion of H3K9me2 across the Vβ array in primary DN and DP thymocytes. We conclude that the or ganization of the Tcrb locus is likely to be similar in wild-type VL3-3M2 cells and primary DN and DP thymocytes, with the Tcrb locus anchored to the NL by the proximal and distal trypsinogen regions. This configuration suggests that Vβ and RC looping from the NL facilitates transcriptional activation and recombination of Tcrb gene segments (Figure 7).
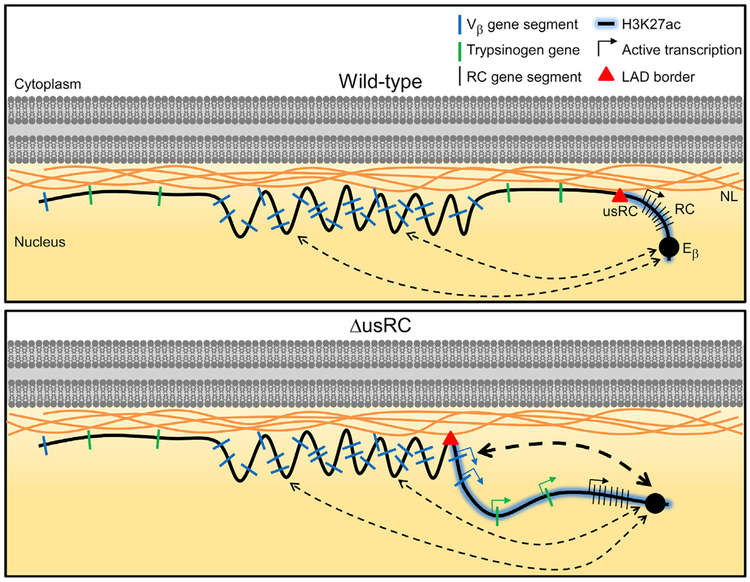
Figure 7. Model of usRC LAD Border Function in VL3-3M2.
Tcrb locus organization at the NL is depicted for wild-type VL3-3M2 and ΔusRC. Dotted lines indicate looping interactions with the RC; line thicknesses represent relative frequencies of looping interactions.
DISCUSSION
In this study, we described the LAD organization of the Tcrb locus and identified a LAD border, termed usRC, which segregates Vβ gene segments and trypsinogen genes from the RC. In wild-type VL3-3M2 cells, the RC is positioned away from the nuclear periphery in an Eβ-dependent manner, and usRC insulates upstream genes from Eβ to ensure NL positioning and suppression. Deletion of usRC caused H3K27ac to spread from the RC into the RC-proximal portion of the Tcrb locus, causing proximal Vβ gene segments and trypsinogen genes to dissociate from the NL and interact more frequently with the RC (Figure 7). These changes in Tcrb locus structure and localization resulted in greatly elevated transcription and recombination of the two most RC-proximal Vβ gene segments and transcriptional activation of normally silent trypsinogen genes. Thus, our results show that a Tcrb locus LAD and LAD border are critical components of Tcrb locus gene regulation.
Multiple elements contribute to LAD border function. The 28-kb usRC LAD border region contains three CBEs: 5′PC and two CBEs upstream of PDβ1. Deletion of all three CBEs (Δ5′PC+ΔPDβ 1CBE) was required to obtain an effect on NL association similar to that of ΔusRC. However, transcriptional upregulation at Trbv29, Trbv30, Try5, and Prss1 in Δ5′PC+ΔPDβ 1CBE was 3- to 8-fold lower than in ΔusRC. Maximal transcriptional upregulation required the additional deletion of PDβ 1700 (Δ5′PC+ΔPDβ1). In fact, Δ5′PC+ΔPDβ1 phenocopied ΔusRC with regard to NL association, Tcrb locus conformation, transcription, and recombination. Because transcriptional activation did not accompany NL dissociation in all experimental genotypes, we believe that dissociation from the NL occurs prior to transcriptional activation.
Tcrb locus LADs and the usRC LAD border have properties typical of LADs and LAD borders genome-wide. LAD borders are enriched for CTCF binding and active promoters oriented away from the LAD (Guelen et al., 2008). usRC has a similar organization and our functional dissection of this LAD border indicates that the CBEs and promoter elements cooperate to mediate LAD border activity. LADs have been shown to be enriched for H3K9me2 along their entire length, and this modification is thought to be essential for anchoring to the NL (Harr et al., 2015; Kind et al., 2013; Peric-Hupkes et al., 2010; Poleshko et al., 2017; Towbin et al., 2012; Wen et al., 2009). Our analysis of wild-type VL3-3M2 cells confirmed the correlation between DamID and H3K9me2 signals, both genome-wide and at the Tcrb locus. However, we were surprised that the deletion of usRC had no effect on Tcrb locus H3K9me2. Instead, we observed a propagation of H3K27ac from the RC into LAD chromatin, suggesting that usRC protects the upstream LAD from invasion of non-LAD chromatin. Although H3K27ac spreads for hundreds of kilobases from the RC upon the deletion of usRC, it stops abruptly at a CBE located 8.5 kb upstream of Trbv29, skipping two other CBEs located more proximal to the RC. The mechanism by which H3K27ac disrupts NL association remains to be determined, as our results do not indicate a role for H3K27me3. H3K27me3 was shown to be essential for NL association at several other loci (Harr et al., 2015).
A recent study described how non-coding RNA (ncRNA) transcription at the Bcl11b enhancer caused Bcl11b gene activation along with release of the surrounding 2 Mb of chromatin from the NL (Isoda et al., 2017). ncRNA transcription was shown to promote DNA demethylation, binding of CTCF and cohesin, and the formation of chromatin loops, which were hypothesized to sequester the Bcl11b locus in a transcriptionally active compartment away from the NL. By comparison, our study provides an example in which boundary deletion causes chromatin release from the NL due to the unrestrained influence of enhancer activity in neighboring chromatin. Similar to Bcl11b, dissociation of Tcrb from the NL is associated with increased enhancer looping. The spread of H3K27ac from the Tcrb RC to adjacent chromatin may link directly to the process of loop extrusion (Barrington et al., 2017; Merkenschlager and Nora, 2016), by which long-range Eβ contacts may be established upon deletion of the usRC boundary. We suggest that loop extrusion, coupled with enhancer-dependent chromatin modifications, dissociates Tcrb chromatin from the NL and facilitates gene segment transcription within the dissociated region. As noted previously, we believe that increased gene segment transcription likely occurs after dissociation from the NL. Such transcription may function subsequently to reinforce and stabilize chromatin localization to the nuclear interior. We do not know whether there may be additional ncRNAs that serve critical roles in loop extrusion, chromatin modifications, and dissociation from the NL.
It is intriguing that Tcrb locus transcription is completely dependent on Eβ in VL3-3M2 cells, whereas in primary DN thymocytes, Eβ deletion abrogates transcription within the RC but has only modest effects on the transcription of Vβ gene segments (Bouvier et al., 1996; Majumder et al., 2015; Mathieu et al., 2000). Because VL3-3M2 is a DP thymocyte cell line, this result may reflect a natural transition from Eβ-independent to Eβ-dependent transcription of Vβ gene segments during thymocyte development. We note that usRC suppresses transcriptional effects of Eβ on RC-proximal Vβ gene segments and trypsinogen genes but fails to suppress Eβ-dependent activation of more distal Vβ gene segments. This suggests that Eβ may influence the transcription of proximal and distal gene segments by different mechanisms. We propose that usRC poses no intrinsic barrier to Eβ-mediated long-distance looping to upstream genes and gene segments, a conclusion that is consistent with our 3C studies. Instead, we suggest that usRC suppresses Eβ contacts with more proximal sites that would otherwise be generated by loop extrusion.
usRC bears structural and functional similarities to clusters of intergenic CBEs found proximal to the RCs in other antigen receptor loci, including IGCR1 in the Igh locus (Guo et al., 2011; Jain et al., 2018; Lin et al., 2015; Qiu et al., 2018), INT1–2 in the Tcra-Tcrd locus (Chen et al., 2015), and Cer-Sis in the Igk locus (Xiang et al., 2011, 2013). Deletion of these CBEs caused locus conformational changes and modified repertoires so that they were biased toward the usage of RC-proximal V gene segments. Such repertoire changes have been variously attributed to changes in long-distance interactions among rearranging gene segments, long-distance interactions among transcriptional regulatory elements, or RAG tracking from the RC (Hu et al., 2015; Jain et al., 2018). These mechanisms are equally applicable in the usRC-deleted Tcrb locus. However, the Tcrb locus is unique among antigen receptor loci in its high-level NL association, and usRC plays a unique role in maintaining LAD integrity. Therefore, it is likely that dissociation from the NL is a key upstream event for repertoire perturbation in ΔusRC VL3-3M2 cells.
Vβ-to-DJβ recombination occurs in DN thymocytes but is then suppressed in DP thymocytes to enforce allelic exclusion. Prior three-dimensional fluorescence in situ hybridization (3D-FISH) analysis of the Tcrb locus indicated high NL association in primary DN and DP thymocytes (Schlimgen et al., 2008). This association appeared to be stochastic, with each Tcrb allele associating with the NL independently of the other allele. Because Vβ-to-DJβ rearrangement is suppressed on NL-associated alleles in DN thymocytes (Chan et al., 2013), we proposed that NL association reduces the efficiency of Vβ-to-DJβ recombination and diminishes the likelihood of both alleles attempting recombination in a similar time frame. Although we were unable to perform DamID in primary thymocytes developing in vivo due to the toxicity of the transduced Dam protein, our 3D-FISH studies (Chan et al., 2013; Schlimgen et al., 2008), together with the similar H3K9me2 profiles in VL3-3M2, DN and DP thymocytes, suggest that the Tcrb locus LAD organization of the VL3-3M2 cell line is likely to be applicable to DN and DP thymocytes in vivo. Nevertheless, we must remain cautious that chromatin regulation in VL3-3M2 may not perfectly mimic that in primary thymocytes. With this caveat in mind, we propose that the usRC LAD border functions in vivo to keep the proximal trypsinogen region anchored to the NL in a suppressed state, insulated from the RC. This in turn may limit the activation and rearrangement of Trbv29 and Trbv30, thereby suppressing their contribution to the Tcrb repertoire and enforcing their allelic exclusion.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LEAF-purified anti-mouse CD3ε (145–2C11) | Biolegend | Cat# 100331; RRID: AB_1877073 |
| PE anti-mouse CD4 (GK1.5) | Biolegend | Cat# 100408; RRID: AB_312693 |
| APC anti-mouse CD8α (53–6.7) | Biolegend | Cat# 100712; RRID: 100712 |
| Brilliant Violet 421 anti-mouse TCRb (H57–597) | Biolegend | Cat# 109230; RRID: 2562562 |
| Anti-mouse TCR Vβ screening panel | BD Biosciences | Cat# 557004; RRID: AB_647180 |
| Anti-H3K9me2 | abcam | Cat# ab1220; RRID: AB_449854 |
| Anti-H3K27ac | abcam | Cat# ab4729; RRID: AB_2118291 |
| Anti-H3K27me3 | Millipore | Cat# 07–449; RRID: AB_310624 |
| Normal rabbit IgG control antibody | R&D Systems | Cat# AB-105-C; RRID: AB_354266 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 7-amino-actinomycin D | Biolegend | Cat# 420404 |
| Trizol | Ambion | Cat# 15596–026 |
| DNase I (RNase-free) | New England Biolabs | Cat# M0303 |
| BglII | New England Biolabs | Cat# R0144 |
| DpnI | New England Biolabs | Cat# R0176 |
| DpnII | New England Biolabs | Cat# R0543 |
| HindIII | New England Biolabs | Cat# R0104 |
| T4 DNA ligase | New England Biolabs | Cat# M0202 |
| T4 DNA ligase | Roche | Cat# 10481220001 |
| Hexadimethrine bromide | Sigma-Aldrich | Cat# H9268 |
| Benzamidine hydrochloride hydrate | Sigma-Aldrich | Cat# B6506 |
| Phenylmethanesulfonyl fluoride | Sigma-Aldrich | Cat# P7626 |
| Protein A agarose/salmon sperm DNA slurry | Millipore | Cat# 16–157 |
| Micrococcal nuclease | New England Biolabs | Cat# M0247 |
| Proteinase K, Biotechnology Grade | VWR | Cat# 0706 |
| Sodium butyrate | Sigma-Aldrich | Cat# 303410 |
| Paraformaldehyde aqueous solution, 16% | Electron Microscopy Sciences | Cat# 15710 |
| Glycine | Sigma-Aldrich | Cat# G7126 |
| Critical Commercial Assays | ||
| iScript cDNA Synthesis Kit | Bio-Rad | Cat# 170–8891 |
| QuantiFast SYBR Green PCR Kit | QIAGEN | Cat# 204057 |
| Cell Line Nucleofector Kit V | Amaxa | Cat# VCA-1003 |
| Advantage 2 Polymerase Mix | Takara | Cat# 639201 |
| QIAquick PCR Purification Kit | QIAGEN | Cat# 28106 |
| LightCycler 480 Probes Master | Roche | Cat# 4902343001 |
| NEBNext End Repair Module | New England Biolabs | Cat# E6050 |
| NEBNext Ultra DNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7370 |
| DNA Clean and Concentrator 5 | Zymo Research | Cat# D4003 |
| immunoSEQ mouse TCRb sequencing | Adaptive Biotechnologies | N/A |
| Deposited Data | ||
| DamID-seq | This paper | GEO: GSE116954 |
| H3K27ac ChIP-seq | This paper | GEO: GSE116954 |
| H3K9me2 ChIP-seq | This paper | GEO: GSE116954 |
| CTCF ChIP-seq | Shih et al., 2012 | GEO: GSE41743 |
| Experimental Models: Cell Lines | ||
| M. musculus VL3-3M2 cell line | Groves et al., 1995 | N/A |
| M. musculus VL3-3M2 derivative cell lines | This paper | Table S1 |
| H. sapiens BOSC23 cell line | Pear et al., 1993 | RRID: CVCL_4401 |
| Experimental Models: Organisms/Strains | ||
| M. musculus 129 strain Rag2 −/− | Shinkai et al., 1992 | RRID: IMSR_TAC:rag2 |
| Oligonucleotides | ||
| Primers and probes for qPCR (all assays) | This paper | Table S2 |
| DamID adaptor AdRt CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA | Vogel et al., 2007 | N/A |
| DamID adaptor AdRb TCCTCGGCCG | Vogel et al., 2007 | N/A |
| DamID primer AdR-PCR GGTCGCGGCCGAGGATC | Vogel et al., 2007 | N/A |
| Recombinant DNA | ||
| BAC: RP23–416M23 | BACPAC Resources Center | Cat# RP23–416M23 |
| BAC: RP23–31E15 | BACPAC Resources Center | Cat# RP23–31E15 |
| BAC: RP23–238C12 | BACPAC Resources Center | Cat# RP23–238C12 |
| pX458 | Ran et al., 2013 | Addgene plasmid #48138 |
| pkat2ampac | Finer et al., 1994 | N/A |
| pSMGV Dam-V5 | Reddy et al., 2008 | N/A |
| pSMGV Dam-V5-LMNB1 | Reddy et al., 2008 | N/A |
| Software and Algorithms | ||
| CRISPR design tool | Ran et al., 2013 | http://crispr.mit.edu/ |
| LADetector | Harr et al., 2015 and this paper | https://github.com/thereddylab/pyLAD |
| Bowtie | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| R | R Core Team, 2018 | https://www.r-project.org/ |
| Bioconductor in R | Huber et al., 2015 | https://www.bioconductor.org/ |
| R package: DNAcopy | Seshan and Olshen, 2018 | https://bioconductor.org/packages/release/bioc/html/DNAcopy.html |
| R package: dada2 | Callahan et al., 2016 | https://bioconductor.org/packages/release/bioc/html/dada2.html |
| R package: QuasR | Gaidatzis et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/QuasR.html |
| R package: Rsamtools | Morgan et al., 2018 | https://bioconductor.org/packages/release/bioc/html/Rsamtools.html |
| R package: mosaics | Chung et al., 2018 | https://bioconductor.org/packages/release/bioc/html/mosaics.html |
| R package: genomation | Akalin et al., 2015 | https://bioconductor.org/packages/release/bioc/html/genomation.html |
| Graphpad Prism | GraphPad Software | https://www.graphpad.com/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael S. Krangel (krang001@mc.duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Rag2–/–mice were of strain 129 background (Shinkai et al., 1992). Mice were housed in a specific-pathogen-free facility managed by the Duke University Division of Laboratory Animal Resources. Mice of both sexes were included in all experiments; no differences on the basis of sex were noted. All mice were handled in accordance with protocols approved by the Duke University Institutional Animal Care and Use Committee.
VL3-3M2 cell line
This study utilized the VL3-3M2 cell line (Groves et al., 1995), which was derived from a radiation leukemia virus-induced C57BL6/Ka thymic lymphoma. Derivative VL3-3M2 cell lines were generated by CRISPR/Cas9-mediated mutation.
METHOD DETAILS
Mice
Mice were generally sacrificed at 4 weeks of age. DN thymocytes were obtained directly from Rag2–/– mice, while DP thymocytes were obtained from Rag2–/– mice that were injected intraperitoneally with 150 μg of anti-CD3ε antibody (145–2C11, Biolegend) 10 days prior to sacrifice.
Cell culture
VL3-3M2 cells were maintained in RPMI 1640 with L-glutamine (GIBCO) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products), 50 U/mL penicillin-streptomycin (GIBCO) and 55 μM 2-mercaptoethanol (GIBCO). BOSC23 cells were maintained in DMEM (GIBCO) supplemented with 10% FBS (Gemini Bio-Products) and 50 U/mL penicillin-streptomycin. Cells were cultured at 37°C in 5% CO2 in a humidified incubator.
Flow cytometry and cell sorting
VL3-3M2 cells were stained with the following mAbs and reagents from Biolegend: CD4-PE (GK1.5), CD8α-APC (53–6.7), BV421-TCRβ (H57–597) and 7-amino-actinomycin D (7AAD). Repertoire analysis was performed by staining with the following FITC-conjugated mAbs from the Mouse Vβ TCR screening panel (BD Biosciences): Vβ3 (JK25), Vβ4 (KT4), Vβ7 (TR310), Vβ8.1/8.2 (MR5–2) and Vβ14 (14–2). Flow cytometric data acquisition was performed on a FACSCanto II (Becton Dickinson). Cell sorting was performed on an Astrios (Beckman Coulter), MoFlo XDP (Beckman Coulter) or FACSDiVa (Becton Dickinson).
CRISPR/Cas9 plasmid construction and transfection
CRISPR/Cas9 guides were designed using the Zhang lab CRISPR design tool available at http://crispr.mit.edu/ (Ran et al., 2013). Guides were cloned into pX458 according to the Zhang lab general cloning protocol. For transient transfection into VL3-3M2 cells, 2 μg of plasmid was mixed with 106 cells in 100 μL of Cell Line Nucleofector Kit V solution (Amaxa) and electroporation was performed using program X-001. Transfected cells were allowed to expand for three days before single cell sorting into 96-well plates. Clones were expanded for nine days before gDNA was harvested for screening by PCR. Initial screening was performed on column and row pools, after which prospective clones were expanded for a second round of PCR screening followed by Sanger sequencing of the PCR product. Cell lines bearing compound deletions were generated by two rounds of this process. ΔusRC+ΔEβ was created using ΔEβ as a base, Δ5′PC+ΔPDβ 1CBE was created using ΔPDβ 1CBE as a base, and Δ5′PC+ΔPDβ1 was created using ΔPDβ1 as a base. Mutated sequences (with indicated guides) are listed in Table S1.
Retroviral packaging and transduction
The BOSC23 cell line (Pear et al., 1993), a derivative of the HEK293T cell line, was grown to 80%–90% confluency in 10 cm tissue culture-treated dishes and transfected with 10 μg of retroviral vector and 5 μg of packaging plasmid pkat2ampac (Finer et al., 1994) using calcium phosphate. Media was replaced one day later and viral supernatant harvested the following day. VL3-3M2 cells were transduced by spin-infection. In brief, 106 VL3-3M2 cells were resuspended in 1 mL of viral supernatant and 10 μg of hexadimethrine bromide (Sigma-Aldrich) and placed into a 48-well tissue culture-treated plate. Spin-infection was performed by centrifugation at 1500 g for 90 minutes at 32°C. Following spin-infection, the viral supernatant was removed and replaced with fresh culture medium.
RNA isolation and analysis
RNA was isolated using Trizol (Ambion). Following purification, RNA was treated with DNase I (New England Biolabs) and cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed using the QuantiFast SYBR Green PCR kit (QIAGEN) on a Roche Lightcycler 480 using the following program: 5 min at 95°C, followed by 45 cycles of 10 s at 95°C and 30 s at 62°C. RT-qPCR primers detecting Vβ gene segment transcription were designed to span the RSS to only detect transcripts originating from unrearranged gene segments. qPCR primers and probes (if applicable) for all qPCR-based assays are listed in Table S2. Experimental values were normalized to values for Actb in each sample.
DamID
VL3-3M2 cells were transduced with pSMGV Dam-V5 (unfused Dam control) or pSMGV Dam-V5-LMNB1 (Dam-Lamin B1) (Reddy et al., 2008) and grown for three days before sorting and expansion of GFP+ cells. gDNA was harvested and purified by phenol-chloroform extraction and isopropanol precipitation. Subsequent steps followed the protocol described in Vogel et al. (2007). In brief, 2.5 μg of gDNA was digested with 10 U of DpnI (New England Biolabs) in 10 μL overnight at 37°C, followed by heat inactivation at 80°C for 20 min. The DpnI-digested product was ligated to 40 pmol of double-stranded adaptor dsAdR, consisting of the annealed oligonucleotides AdRt: CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA and AdRb: TCCTCGGCCG, with 400 U of T4 DNA ligase (New England Biolabs) in 20 μL for 2 h at 16°C, followed by heat inactivation at 65°C for 10 min. Unmethylated fragments were digested with 10 U of DpnII (New England Biolabs) in 50 μL for 1 h at 37°C, followed by heat inactivation at 65°C for 20 min. 20% of the sample was utilized for PCR amplification using Advantage 2 polymerase mix (Takara) with 62.5 pmol of AdRPCR primer: GGTCGCGGCCGAGGATC using the following program: 68°C for 10 min, 94°C for 1 min, 65°C for 5 min, 68°C for 15 min, 3 cycles of 94°C for 1 min, 65 C for 1 min, 68°C for 10 min, and 18 cycles of 94°C for 1 min, 65°C for 1 min, 68°C for 2 min. The PCR product was purified using the QIAquick PCR Purification Kit (QIAGEN).
For DamID-qPCR, qPCR analysis was performed by SYBR-based qPCR with conditions identical to those used for RT-qPCR. For DamID-seq, the DamID PCR product was randomized by sonication following end repair and ligation. Specifically, 1–2 μg DamID material was subjected to end repair using the NEBNext End Repair Module (New England Biolabs) and cleaned up using DNA Clean and Concentrator −5 (Zymo Research). Cleaned up DNA was ligated with 5U T4 DNA ligase (Roche) per 1 μg material in 20 μL to generate a library of large DNA fragments. This DNA library was diluted to 200 μL in H2O, sonicated for one hour in 1.5 mL DNA LoBind tubes (Eppendorf) using a Bioruptor (Diagenode), then transferred to 1.5 mL TPX tubes (Diagenode) for four rounds of 10 min sonication to yield fragments of 150–300 bp in size. Samples were transferred to new TPX tubes between each round of sonication to minimize loss of DNA fragments. The sonicated DNA was transferred to 1.5mL DNA LoBind tubes, ethanol precipitated and resus-pended in 10 μL of 10 mM Tris-HCl pH 8.0. Samples were quantified using Qubit and sequencing libraries were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs). In independent experiments, we obtained 100 bp single end reads using an Illumina HiSeq 2500 or 150 bp paired end reads (with one end discarded) using an Illumina HiSeq 4000.
DamID-seq analysis
DamID-seq reads were processed using LADetector (available at https://github.com/thereddylab/pyLAD), an updated Python implementation of LADetector described in Harr et al. (2015) with incorporated sequence mapping. pyLAD was run with the parameters “–multimapping–quality-trim–unalignable–count-overlaps–seed 5.” Reads were quality trimmed using a sliding window quality score average over 3 bases and a minimum score cutoff of 30. This was followed by trimming any matching overlap between read-ends and sequencing or DamID adaptor-primer sequence. Reads containing a DamID adaptor-primer sequence were split and adaptor-primer sequence removed. All resulting reads greater than 20 bp were aligned to mm9 using Bowtie (Langmead et al., 2009) with parameters “—tryhard–best–m 1.” Unaligned reads had 10 bases trimmed from the 5′ end and remapped, and the resultant unmapped reads were trimmed 10 bases from the 3′ end and remapped. Total aligned reads were assigned to DpnI bins, with reads straddling bin boundaries counting toward both. Prior to scoring, a value of 0.5 was added to bins with no reads. Bins falling in unaligned regions were removed prior to analysis. DamID scores were calculated for all non-zero bins as the log2 ratio of Dam-Lamin B1 over unfused Dam. Scores were partitioned using circular binary segmentation using the DNAcopy package in R (Seshan and Olshen, 2018). LADs were classified as regions > 100 kb in size of positive signal, allowing for smaller regions of negative signal < 10 kb in size.
ChIP
107 VL3-3M2 cells or primary thymocytes were washed in phosphate buffered saline supplemented with 5 mM EDTA, 500 μM spermidine and 150 μM spermine, followed by the same solution lacking EDTA. Cells were lysed by incubation for 5 min on ice in 250 mM sucrose, 80 mM NaCl, 10 mM Tris-HCl pH 8.0, 6 mM MgCl2, 1 mM CaCl2, 500 μM spermidine, 150 μM spermine, containing either0.2% v/v NP-40 (for VL3-3M2 cells) or 0.02% v/v NP-40 (for primary thymocytes). Samples were centrifuged at 750 g for 5 min at 4°C and the pellet was washed in 200 μl of digestion buffer (250 mM sucrose, 10 mM NaCl, 10 mM Tris-HCl pH 8.0, 3 mM MgCl2, 1 mM CaCl2). The pellet was then resuspended in 200 μl of digestion buffer and 200 gel units of micrococcal nuclease (New England Biolabs) was added for a 10 min incubation at 37°C. The reaction was terminated by addition of 300 μl 10 mM Tris-HCl pH 8.0, 5 mM EDTA. After centrifugation at 18000 g for 10 min at 4°C, supernatants were harvested and Triton X-100 added to a final concentration of 1% v/v. After checking digestion efficiency on a gel, chromatin was pre-cleared by addition of 50 ml protein A agarose/salmon sperm DNA slurry (Millipore) and mixing for 1 h at 4°C. Supernatant corresponding to 5 × 106 cells was incubated overnight at 4°C with 5 μg of antibody specific for H3K9me2 (Abcam), H3K27ac (Abcam), H3K27me3 (Millipore) or control IgG (R&D Systems). Pulldown was performed by adding 50 μl protein A agarose/salmon sperm DNA slurry (Millipore) and mixing for 1 h at 4°C. The slurry was washed twice with 1 mL of each of the following wash buffers: 167 mM NaCl, 16.7 mM Tris-HCl pH 8.0, 1.2 mM EDTA, 1.1% v/v Triton X-100, 0.01% w/v SDS; 300 mM NaCl, 20 mM Tris-HCl pH 8.0, 2 mM EDTA, 1% v/v Triton X-100, 0.1% w/v SDS; 50 mM Tris-HCl pH 8.0, 0.25 M LiCl, 0.5% v/v NP-40, 0.5% w/v sodium deoxycholate; 10 mM Tris-HCl pH 8.0, 1 mM EDTA. Elution was performed by resuspending the slurry in 100 μl of 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.3% w/v SDS and 1 mg/mL proteinase K (VWR) overnight at 65 C. Elution buffer was set aside and a second elution performed using 0.5 M NaCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA. Eluates were combined and volumes were adjusted to 500 μl with 10 mM Tris-HCl pH 8.0, 1 mM EDTA. DNA was purified by phenol-chloroform extraction and isopropanol precipitation and resuspended in H2O. In addition to the listed components, all buffers used before elution also contained 10 mM sodium butyrate (Sigma-Aldrich), 100 μM PMSF (Sigma-Aldrich) and 100 μM benzamidine (Sigma-Aldrich). qPCR analysis of H3K27me3 was performed by SYBR-based qPCR with conditions identical to those used for RT-qPCR.
ChIP-seq
Library preparation and high-throughput sequencing were performed by the Duke Center for Genomic and Computational Biology Core Facility. 50 bp single end reads were obtained using an Illumina HiSeq 4000. Analysis was primarily performed using the Bio-conductor set of packages in R (Huber et al., 2015; R Core Team, 2018). Demultiplexed.fastq files were trimmed using the fastqFilter command in the dada2 package (Callahan et al., 2016) with the parameters “truncLen = 0, trimLeft = 0, maxN = 0, minQ = 0, rm.phix = TRUE.” The trimmed files were aligned to the mm9 genome using the qAlign command in the QuasR package (Gaidatzis et al., 2015; Langmead et al., 2009), a wrapper for Rbowtie with the parameters “aligner = “Rbowtie,” maxHits = 1, paired = NULL.” .bam files were merged using the mergeBam command in the Rsamtools package (Morgan et al., 2018). To generate ChIP-seq tracks, reads were placed into 200 bp bins using the constructBins command in the mosaics package (Chung et al., 2018) with the parameters “fragLen = 150, binSize = 200, capping = 50, PET = FALSE.” Reads that corresponded to chromosome Y, chromosome M or un-mapped parts of the chromosome were excluded. Bins were assembled into tracks in the .bedgraph format using a custom script. For H3K27ac tracks, bins were normalized to reads per million. H3K9me2 tracks were normalized for read count relative to the input sample and a value of 1 was added to bins with no reads. Log2 ratios of the H3K9me2 sample over input were then plotted. Correlation graphs between DamID-seq and H3K9me2 ChIP-seq were constructed by placing reads into 10 kb bins using the constructBins command as described above, except changing “binSize = 10000.” Matching bins were plotted and the Pearson’s correlation coefficient obtained using the cor function in R. To align the features surrounding LAD borders, we first defined the regions extending 50 kb upstream and downstream of the 5′LAD borders. These regions were then aligned using the plotMeta command in the genomation package (Akalin et al., 2015).
3C
5 × 106 VL3-3M2 cells were crosslinked in 8 mL of RPMI 1640 (GIBCO) containing 10% v/v FBS (Gemini Bio-Products) and 2% w/v paraformaldehyde (Electron Microscopy Sciences) for 10 min at 20°C. Crosslinking was terminated by the addition of 1 mL 1.25M glycine (Sigma-Aldrich). Fixed cells were pelleted by centrifugation and cytoplasm removed by incubation in 5 mL 10 mM Tris-HCl pH 8.0, 10 mM NaCl, 0.2% v/v NP-40, 1 mM PMSF (Sigma-Aldrich) and 1 mM benzamidine (Sigma-Aldrich) for 10 min on ice. Nuclei were pelleted by centrifugation and resuspended in 450 mL 1.1x restriction enzyme buffer (buffer 2 for HindIII or buffer 3 for BglII) and 7.5 mL of 20% w/v SDS was added for 1 h incubation at 37°C. 50 μL of 20% v/v Triton X-100 was then added for 1 h at 37°C, and 200 U HindIII or BglII (New England Biolabs) was added for overnight digest at 37°C. Reactions were then supplemented with an additional 200 U of restriction enzyme for a 6 h incubation. After heat inactivation and sequential incubation with SDS and Triton X-100 as above, the samples were diluted to 7 mL in 1x T4 DNA ligase buffer, and 4000 U T4 DNA ligase (New England Biolabs) was added for overnight incubation at 16°C. Reactions were then supplemented with an additional 4000 U T4 DNA ligase for a 6 h incubation. Samples were then digested by addition of proteinase K (VWR) to 1 mg/mL for overnight incubation at 65°C, and DNA was purified by phenol/chloroform extraction followed by isopropanol precipitation. 3C products were quantified by a Taqman-based qPCR strategy using the LightCycler 480 probes master mix (Roche) on a Roche Lightcycler 480 using the following program: 5 min at 95°C, followed by 48 cycles of 10 s at 95°C and 30 s at 62°C. Values were normalized to a bacterial artificial chromosome (BAC) standard generated from RP23–416M23, RP23–31E15 and RP23–238C12 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute) and a second normalization step was performed by normalizing values to the nearest neighbor fragment.
Tcrb repertoire analysis
Initial assessment was performed by Taqman-based qPCR of gDNA isolated from unsynchronized cells using PCR conditions identical to those for 3C. Readings were normalized to Cd14. For the timed recombination system, each VL3-3M2 line was transfected with a pX458 derivative targeting the VDJβ junction on the rearranged allele and three days later, TCRβ GFP+ cells were single cell sorted into 96-well plates. For each cell line, twelve clones were cultured for 21 days and recombination was scored by flow cytometry to quantify TCRβ+ cells. To enrich for cells with functional rearrangements, we chose three clones from each line which displayed recombination frequencies closest to the median and sorted TCRβ+ cells. The Tcrb repertoire was assessed by flow cytometric staining using Vβ-specific mAbs (BD Biosciences) and by high-throughput sequencing of rearrangement events using the immunoSEQ service (Adaptive Biotechnologies). Verification of the direct Vβ-to-Jβ rearrangements was performed by SYBR-based qPCR with conditions identical to those used in measuring transcription. Readings were normalized to Cd14.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise specified, statistical analysis was carried out by two-way ANOVA with adjustment for multiple comparisons using GraphPad Prism software. Differences with adjusted p values of < 0.05 were considered significant. The number of biological replicates (n) is listed in the legend of each figure. For DamID-qPCR, effect sizes and significance were calculated by fitting a model of the form:
Where yijk is the DamID signal for the i-th locus, j-th VL3-3M2 line and k-th run. The model explains variation in expression levels relative to a baseline μ, which represents the average expression level at the Tp4 locus in wild-type cells. The terms αi show the difference in expression at other loci (relative to baseline) in wild-type cells. The terms βj show the effect (average difference in expression) for other lines relative to wild-type. The assumption is that this effect is constant across all loci. The terms bk represent the random effect of run/batch, assumed to have a zero mean Gaussian distribution. Finally εijk is unexplained/measurement error, also assumed to have a zero mean Gaussian distribution but independent of the batch effect. The model was fit to observed data for all runs but restricted to loci lying between Trbv30 and Tp4, both inclusive, using the method of restricted maximum likelihood (REML).
DATA AND SOFTWARE AVAILABILITY
The accession number for DamID-seq and ChIP-seq reported in this paper is GEO: GSE116954.
Supplementary Material
Highlights.
A lamina-associated domain (LAD) border controls Tcrb nuclear lamina (NL) association
The LAD border consists of CTCF-binding elements and an active promoter
Deletion of the LAD border causes regional loss of NL association
Disrupted NL association results in increased transcription and VDJ recombination
ACKNOWLEDGMENTS
We thank the members of the Duke Cancer Institute Flow Cytometry Shared Resource Facility for help with cell sorting, Dr. Y. Zhuang for valuable suggestions, and Danielle Dauphars and Catherine Lewis for comments on the manuscript. This work was supported by the NIH (R01 AI49934 to M.S.K. and R21 AG050132 to K.L.R.). T.R.L. was partially supported by NIH grant T32 GM007445. K.R.C. was partially supported by the Duke Biostatistics Core, which is funded by the National Center for Advancing Translational Science (UL1TR001117).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.10.052.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abarrategui I, and Krangel MS (2006). Regulation of T cell receptor-a gene recombination by transcription. Nat. Immunol 7, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Abarrategui I, and Krangel MS (2007). Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO J 26, 4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Franke V, Vlahoviček K, Mason CE, and Schübeler D (2015). Genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 31, 1127–1129. [DOI] [PubMed] [Google Scholar]
- Barrington C, Finn R, and Hadjur S (2017). Cohesin biology meets the loop extrusion model. Chromosome Res 25, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Alt FW, Hughes MM, D’Auteuil M, Wehrly TD, Woodman BB, Gärtner F, White JM, Davidson L, and Sleckman BP (2000). Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature 405, 583–586. [DOI] [PubMed] [Google Scholar]
- Bevington S, and Boyes J (2013). Transcription-coupled eviction of histones H2A/H2B governs V(D)J recombination. EMBO J 32, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Khanna N, Alvikas J, and Belmont AS (2013). β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol 203, 767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen C, Mansson R, and Murre C (2012). Chromatin topology and the regulation of antigen receptor assembly. Annu. Rev. Immunol 30, 337–356. [DOI] [PubMed] [Google Scholar]
- Bouvier G, Watrin F, Naspetti M, Verthuy C, Naquet P, and Ferrier P (1996). Deletion of the mouse T-cell receptor b gene enhancer blocks alpha-beta T-cell development. Proc. Natl. Acad. Sci. USA 93, 7877–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady BL, Steinel NC, and Bassing CH (2010). Antigen receptor allelic exclusion: an update and reappraisal. J. Immunol 185, 3801–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EA, Teng G, Corbett E, Choudhury KR, Bassing CH, Schatz DG, and Krangel MS (2013). Peripheral subnuclear positioning suppresses Tcrb recombination and segregates Tcrb alleles from RAG2. Proc. Natl. Acad. Sci. USA 110, E4628–E4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Carico Z, Shih HY, and Krangel MS (2015). A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires. Nat. Immunol 16, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Kuan PF, Welch R, and Keles S (2018). MOSAiCS (model-based one and two sample analysis and inference for ChIP-seq). R package, version 2.16.0 http://groups.google.com/d/forum/mosaics_user_group.
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynska A, Gonzalo S, Shanahan C, and Askjaer P (2016). The nuclear lamina in health and disease. Nucleus 7, 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, and Busslinger M (2011). The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34, 175–187. [DOI] [PubMed] [Google Scholar]
- Finer MH, Dull TJ, Qin L, Farson D, and Roberts MR (1994). kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood 83, 43–50. [PubMed] [Google Scholar]
- Gaidatzis D, Lerch A, Hahne F, and Stadler MB (2015). QuasR: quantification and annotation of short reads in R. Bioinformatics 31, 1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Majumder K, Predeus A, Huang Y, Koues OI, Verma-Gaur J, Loguercio S, Su AI, Feeney AJ, Artyomov MN, and Oltz EM (2013). Unifying model for molecular determinants of the preselection Vβ repertoire. Proc. Natl. Acad. Sci. USA 110, E3206–E3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, and Guidos CJ (1995). In vitro maturation of clonal CD4+CD8+ cell lines in response to TCR engagement. J. Immunol 154, 5011–5022. [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, and van Steensel B (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951. [DOI] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. (2011). CTCF-binding elements mediate control of V(D)J recombination. Nature 477, 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, and Reddy KL (2015). Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol 208, 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, and Alt FW (2015). Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell 163, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, et al. (2017). Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171, 103–119.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Ba Z, Zhang Y, Dai HQ, and Alt FW (2018). CTCF-binding elements mediate accessibility of RAG substrates during chromatin scanning. Cell 174, 102–116.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, and Schatz DG (2010). The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, and van Steensel B (2013). Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178–192. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. (2015). Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163, 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, and Singh H (2002). Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296, 158–162. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SG, Guo C, Su A, Zhang Y, and Alt FW (2015). CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D) J recombination. Proc. Natl. Acad. Sci. USA 112, 1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder K, Koues OI, Chan EA, Kyle KE, Horowitz JE, Yang-Iott K, Bassing CH, Taniuchi I, Krangel MS, and Oltz EM (2015). Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J. Exp. Med 212, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maman Y, Teng G, Seth R, Kleinstein SH, and Schatz DG (2016). RAG1 targeting in the genome is dominated by chromatin interactions mediated by the non-core regions of RAG1 and RAG2. Nucleic Acids Res 44, 9624–9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Hempel WM, Spicuglia S, Verthuy C, and Ferrier P (2000). Chromatin remodeling by the T cell receptor (TCR)-β gene enhancer during early T cell development: implications for the control of TCR-β locus recombination. J. Exp. Med 192, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, Sun Q, Huis In ’t Veld PJ, Guo C, Yoon HS, et al. (2013). Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity 39, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, and Nora EP (2016). CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet 17, 17–43. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, and van Steensel B (2013). Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Pagès H, Obenchain V, and Hayden N (2018). Rsamtools: binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package, version 1.30.0 http://bioconductor.org/packages/release/bioc/html/Rsamtools.html.
- Pear WS, Nolan GP, Scott ML, and Baltimore D (1993). Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko A, Shah PP, Gupta M, Babu A, Morley MP, Manderfield LJ, Ifkovits JL, Calderon D, Aghajanian H, Sierra-Pagan JE, et al. (2017). Genome-nuclear lamina interactions regulate cardiac stem cell lineage restriction. Cell 171, 573–587.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos PA, Singh PB, Giannakouros T, and Georgatos SD (2001). Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep 2, 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Kumari G, Gerasimova T, Du H, Labaran L, Singh A, De S, Wood WH 3rd, Becker KG, Zhou W, et al. (2018). Sequential enhancer sequestration dysregulates recombination center formation at the IgH locus. Mol. Cell 70, 21–33.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: a language and environment for statistical computing (R Foundation for Statistical Computing) https://www.R-project.org/.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, and Singh H (2008). Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247. [DOI] [PubMed] [Google Scholar]
- Robson MI, de Las Heras JI, Czapiewski R, Sivakumar A, Kerr ARW, and Schirmer EC (2017). Constrained release of lamina-associated enhancers and genes from the nuclear envelope during T-cell activation facilitates their association in chromosome compartments. Genome Res 27, 1126–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, and Ji Y (2011). Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol 11, 251–263. [DOI] [PubMed] [Google Scholar]
- Schlimgen RJ, Reddy KL, Singh H, and Krangel MS (2008). Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nat. Immunol 9, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan VE, and Olshen A (2018). DNAcopy: DNA copy number data analysis. R package, version 1.52.0 https://www.bioconductor.org/packages/release/bioc/html/DNAcopy.html.
- Shih HY, Verma-Gaur J, Torkamani A, Feeney AJ, Galjart N, and Krangel MS (2012). Tcra gene recombination is supported by a Tcra enhancerand CTCF-dependent chromatin hub. Proc. Natl. Acad. Sci. USA 109, E3493–E3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. (1992). RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867. [DOI] [PubMed] [Google Scholar]
- Sikes ML, Gomez RJ, Song J, and Oltz EM (1998). A developmental stage-specific promoter directs germline transcription of D β J β gene segments in precursor T lymphocytes. J. Immunol 161, 1399–1405. [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, and Lee JT (2013). High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman BP, Bassing CH, Hughes MM, Okada A, D’Auteuil M, Wehrly TD, Woodman BB, Davidson L, Chen J, and Alt FW (2000). Mechanisms that direct ordered assembly of T cell receptor β locus V, D, and J gene segments. Proc. Natl. Acad. Sci. USA 97, 7975–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, and Gal-Yam EN (2005). The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci 118, 4017–4025. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, and Harte PJ (2009). CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, and Gasser SM (2012). Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947. [DOI] [PubMed] [Google Scholar]
- van Steensel B, and Belmont AS (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, and Henikoff S (2000). Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol 18, 424–428. [DOI] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, and van Steensel B (2007). Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc 2, 1467–1478. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, and Feinberg AP (2009). Large his-tone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet 41, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Grandal I, Vesprini DJ, Wojtyra U, Danska JS, and Guidos CJ (2001). Irradiation promotes V(D)J joining and RAG-dependent neoplastic transformation in SCID T-cell precursors. Mol. Cell. Biol 21, 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhou X, Hewitt SL, Skok JA, and Garrard WT (2011). A multi-functional element in the mouse Igk locus that specifies repertoire and Ig loci subnuclear location. J. Immunol 186, 5356–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Park SK, and Garrard WT (2013). Vk gene repertoire and locus contraction are specified by critical DNase I hypersensitive sites within the Vk-Jk intervening region. J. Immunol 190, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, and Alt FW (1985). Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40, 271–281. [DOI] [PubMed] [Google Scholar]
- Yao J, Fetter RD, Hu P, Betzig E, and Tjian R (2011). Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev 25, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, and Singh H (2012). DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.