Abstract
PqqE is the first enzyme in the biosynthetic pathway of the redox cofactor pyrroloquinoline quinone (PQQ), catalyzing the formation of a carbon–carbon bond in the precursor peptide PqqA. PqqE is a radical S-adenosyl-l-methionine (SAM) (RS) enzyme, a family of enzymes that use the reductive cleavage of a [4Fe–4S] cluster-bound SAM molecule to generate a 5′-deoxyadenosyl radical. This radical is then used to initiate an array of reactions that otherwise would be unlikely to occur. PqqE is a founding member of a subset family of RS enzymes that, additionally to the SAM [4Fe–4S] cluster, have a SPASM domain containing additional, auxiliary Fe–S clusters.
Most radical SAM enzymes are highly sensitive to oxygen, which destroys their Fe–S clusters. This can pose several limitations when working with these enzymes, since most of the work has to be done under anaerobic conditions. Here, we summarize the methods developed in our lab for the expression and purification of PqqE. We also highlight the several methods we have used for the characterization of the enzyme.
1. INTRODUCTION
Pyrroloquinoline quinone (4,5-dihydro-4,5-dioxo-1H-pyrrolo[2,3-f] quinolone-2,7,9-tricarboxilic acid, PQQ) is a quinone-containing noncovalently bound redox cofactor first described in 1964 as the “new prosthetic group” of a Bacterium anitratum glucose dehydrogenase (Hauge, 1964). It is used by a wide variety of bacterial enzymes, mainly alcohol and sugar dehydrogenases in the nonglycolytic substrate oxidation for the production of ATP (Duine, 1999).
Quinocofactors are derived from canonical amino acid side chains in either a protein or a peptide, by posttranslational modifications and, besides the peptide-derived PQQ, include the protein-derived trihydroxyphenylalanine quinone (TPQ), tryptophan tryptophylquinone (TTQ), lysine tyrosyl quinone (LTQ), and cysteine tryptophylquinone (CTQ) (Klinman & Bonnot, 2014).
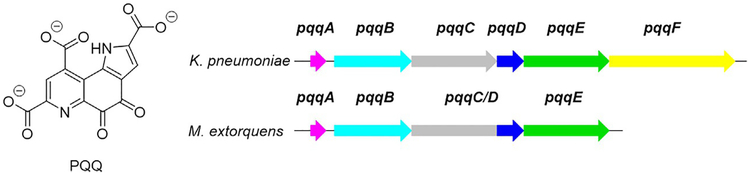
PQQ is a ribosomally synthesized and posttranslationally modified peptide (RiPP), being derived from the peptide precursor PqqA. The enzymes involved in the biosynthesis of PQQ are encoded by genes arranged in the pqq operon, with five of the genes—pqqA, pqqB, pqqC, pqqD, and pqqE—being obligatory for PQQ production (Shen et al., 2012). pqqF, encoding a protease, may also be present—in the operon or elsewhere in the genome—but it is not consistently conserved (Fig. 1). The PQQ molecule derives from the evolutionarily conserved glutamate and tyrosine side chains within the ribosomally produced peptide substrate PqqA, encoded by pqqA (Velterop et al., 1995).
Fig. 1.
The structure of PQQ and the pqq operons in K. pneumoniae and M. extorquens (Mex). Note that in Mex, pqqD is fused to the C-terminal of pqqC, while in K. pneumoniae PqqC and PqqD are encoded by separate genes. In K. pneumoniae the gene encoding the PqqF protease is located in the operon, unlike in Mex.
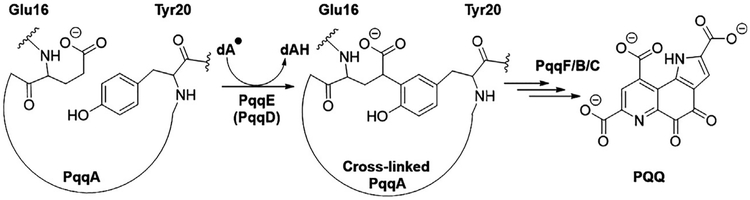
The biosynthesis of PQQ from PqqA starts with the formation of a new carbon–carbon bond between the γ-carbon of the glutamate and the C5 of the tyrosine ring, catalyzed by the radical S-adenosyl-L-methionine (SAM) (RS) enzyme PqqE (Barr et al., 2016) (Fig. 2).
Fig. 2.
Simplified scheme of the PQQ biosynthetic pathway. PqqE catalyzes the first step in PQQ biosynthesis. The specific involvement of PqqF and PqqB within the pathway is under investigation. The roles of PqqA, PqqC, PqqE, and PqqD are now well established (see text).
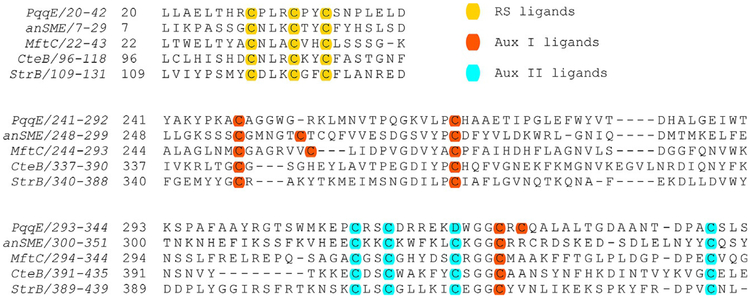
PqqE uses a [4Fe–4S] cluster located in its N-terminal domain to reductively cleave SAM, initiating a free radical cascade that ultimately leads to the formation of the new carbon–carbon bond, creating a cross-linked product. PqqE belongs to a subset of RS enzymes that additionally contain a SPASM domain in their C-terminal (RS-SPASM) comprised of most generally two auxiliary Fe–S clusters that are also essential for the net reaction (Grell, Goldman, & Drennan, 2015). PqqE has been proposed to contain the typical RS site (Wecksler et al., 2009) as well as two auxiliary Fe–S cluster-binding sites, designated AuxI and AuxII (Barr et al., 2016; Saichana et al., 2017). What distinguishes PqqE from other SPASM proteins is the fact that it contains a unique Cx2Cx5Dx21C motif with the ligand aspartate (D) in AuxII, while other SPASM proteins contain a conserved Cx(2–4)Cx5Cx(21–22)C, with four cysteines in their AuxII site. Additionally there is a CxC motif present in AuxI of PqqE, a feature observed in both [4Fe–4S] and [2Fe–2S] containing proteins (Banci et al., 2013; Wagner, Koch, Ermler, & Shima, 2017; Fig. 3).
Fig. 3.
Partial sequence alignment of SPASM proteins. The protein sequences near the Fe–S clusters in PqqE in Methylobacterium extorquens, anaerobic sulfatase-maturating enzyme (anSME) in Clostridium perfringens, mycofactocin radical SAM maturase (MftC) in Mycobacterium tuberculosis, sactisynthase (CteB) in Clostridium thermocellum, and a radical SAM enzyme in biosynthesis of streptide (StrB) Streptococcus thermophiles. Note that the assignment of Aux I and Aux II ligands for anSME and CteB is based on crystal structures in the database (PDB: 4K37 and PDB: 5WHY) (Goldman et al., 2013; Grove et al., 2017). The assignment of PqqE, MftC, and StrB is predicted based on the structure model created by SWISSMODEL (http://swissmodel.expasy.org/).
PqqD, a small protein, acts as a chaperone to deliver PqqA to PqqE, as shown by the formation of a PqqA–PqqD complex and its interaction with PqqE (Latham, Iavarone, Barr, Juthani, & Klinman, 2015). The hypothesis is that PqqD shields PqqA from premature proteolytic cleavage catalyzed by cellular proteases.
The cross-linked precursor to PQQ is hypothesized to be cleaved by a protease, which in some cases is encoded by the gene pqqF, and the resulting molecule is later acted on by PqqB, a metallo-β-lactamase enzyme, ultimately yielding 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid, AHQQ (Bonnot, Iavarone, & Klinman, 2013; Magnusson et al., 2004). Finally, the well-studied PqqC, a cofactorless synthase, catalyzes the eight-electron oxidation and ring cyclization of AHQQ to PQQ (Magnusson et al., 2004).
This chapter focuses on the proteins acting in the initial step of PqqA cross-linking, with an emphasis on enzymes from Methylobacterium extorquens (Mex). PqqE from different sources has different tolerances to oxygen, thus the expression and purification methods differ based on the strain from which the enzyme is being purified (Barr et al., 2016; Latham et al., 2015; Saichana et al., 2016; Wecksler et al., 2009). Most radical SAM enzymes are highly sensitive to oxygen due to the oxidation of their Fe–S clusters, but the Mex enzyme is quite tolerant, making it easier to express and purify under normal lab conditions (Saichana et al., 2016).
2. PROTEIN EXPRESSION, PURIFICATION, AND RECONSTITUTION
The initial attempts to express and purify recombinant PqqE, from Klebsiella pneumoniae, were done under aerobic conditions (Wecksler et al., 2009). In these conditions the protein was restricted to inclusion bodies and the purification procedure included a protein refolding step prior to chromatographic purification. The protein obtained was >95% pure and was reconstituted with Fe2+/Fe3+ and S2− ions, under anaerobic conditions and in the presence of dithiothreitol (DTT). Although the reconstituted protein exhibited the reddish-brown color characteristic of the Fe–S presence, the EPR spectral characteristics were not in accordance with a radical SAM enzyme and a different approach for enzyme purification had to be pursued (Wecksler et al., 2009). Growth and induction under anaerobic conditions yielded soluble protein but subsequent aerobic purification resulted in an enzyme with EPR spectra similar to the one grown and purified aerobically. Thus, growth and purification of K. pneumoniae PqqE must be done in strict anaerobic conditions (see protocol in Sections 2.2.1 and 2.3).
Mex PqqE, on the other hand, is much more tolerant to oxygen, and it can be expressed under aerobic conditions (Barr et al., 2016; Saichana et al., 2016), although it was initially expressed under anaerobic conditions (Latham et al., 2015).
All anaerobic work must be done in a controlled atmosphere chamber (glovebox) with <5ppm oxygen (Barr et al., 2016). The buffers to be used in anaerobic work are prepared under normal lab conditions and then purged with argon. The reagent solutions are either prepared like the buffers or the powder reagent is brought to the glovebox, weighed, and reconstituted with the anaerobic buffers.
All steps involving work with bacterial cell hosts are done in sterile conditions, using autoclaved media and autoclaved or filter (0.2 μm)-sterilized solutions.
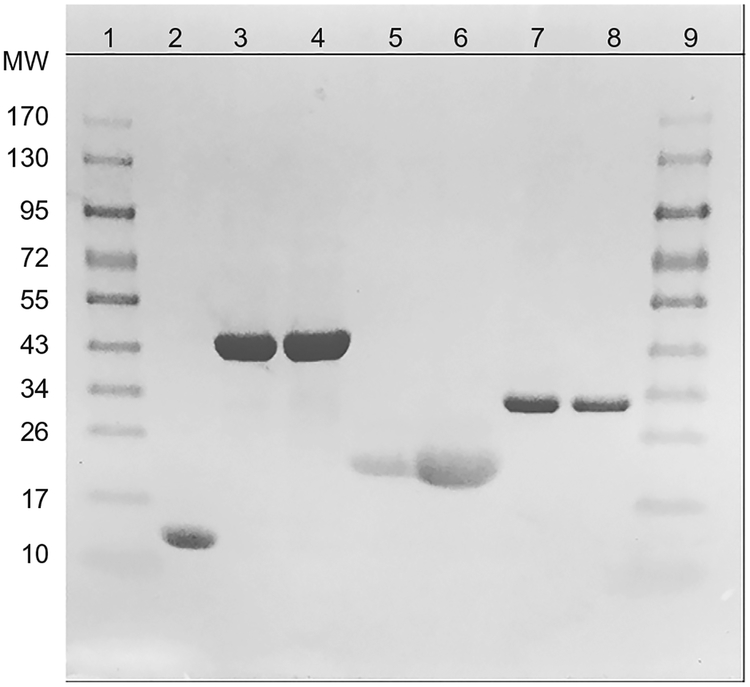
Fig. 4 shows a typical SDS-PAGE result for Mex proteins expressed and purified as described in this section.
Fig. 4.
SDS-PAGE gel of proteins expressed and purified in this work. The lanes are as follows: (1),(9) protein ladder; (2) Mex PqqD; (3),(4) Mex PqqE; (5),(6) A. vinelandii FldA;(7),(8) Mex FNR.
2.1. pqqE Cloning Strategies
The gene encoding PqqE is amplified from genomic DNA by regular molecular biology techniques, using a set of primers designed in order to insert the required restrictions sites. PqqE has been expressed and purified as a His6-tag construct (either N-terminal or C-terminal), thus the vectors pET28 (Novagen) have been used for cloning purposes (Barr et al., 2016; Latham et al., 2015; Wecksler et al., 2009). The pET cloning system uses a strong T7 promoter, facilitating a robust overexpression of the protein encoded by the cloned gene. The plasmids are then transformed into host competent cells—E. coli XL1 Blue or E. coli TOP10 (Invitrogen)—isolated and sequenced. Once the sequence is confirmed, the plasmid harboring the correct construct is cotransformed into E. coli BL21(DE3) (Stratagene) with vectors expressing the E. coli suf ABCDSE operon (pPH151) genes, which encodes proteins involved in Fe–S cluster repair and assembly (Nachin, Loiseau, Expert, & Barras, 2003). The vectors can also be cotransformed in E. coli Rosetta 2 (DE3) (Novagen), which contain rare codons that are seldom used in E. coli (Saichana et al., 2016). The pET vector contains a kanamycin (Kan) resistance gene, while the pPH151 is resistant to chloramphenicol (Cam); thus, the transformed strain is Kan and Cam resistant and can be grown in selective media with both antibiotics.
2.2. PqqE Overexpression
Depending on the bacterial source of PqqE, the protein expression must be done anaerobically (e.g., K. pneumoniae) or it can be done in aerobic conditions (e.g., Mex). A summary of PqqE expression techniques is shown in Table 1.
Table 1.
Methods for Expression of PqqE in Anaerobic (K. pneumoniae or Mex) or Aerobic (Mex) Conditions
| Anaerobic (Latham et al., 2015; Wecksler et al., 2009) | Aerobic (Barr et al., 2016) | |
|---|---|---|
| Heterologous protein | His6-PqqE or PqqE-His6 | His6-PqqE |
| Vectors | pET28a or pET24b (Novagen) | pET28a (Novagen) |
| Vector hosts | E. coli BL21(DE3) (withpPH151) (Stratagene) | E. coli BL21 (DE3) (with pPH51) (Stratagene) |
| Starter cultures/inoculum | Cells grown aerobically overnight at 37°C | Cells grown aerobically overnight at 37°C |
| Growth medium | LBKan50Cam50 (Klbp) TBKan50Cam50 + 50 mM fumarate + 50 μM Fe(III) citrate (Mex) |
TBKan50Cam50 + 100 μM Fe(III) citrate |
| Induction | OD600 = 0.1 with 100 μM IPTG (Klbp) + 5mg/L Fe(NH4)2(SO4)2·6(H2O) OD600 = 0.6 with 400 μM IPTG (Mex) |
OD600 = 0.1 with 100 μM IPTG + 50 μM cysteine |
| Expression conditions | 18°C, overnight (Klbp) 19°C, 12h (Mex) |
20°C, 18h |
2.2.1. Growth and Expression of K. pneumoniae PqqE in Anaerobic Conditions (Wecksler et al., 2009)
Select a single isolated colony of E. coli BL21 (DE3) on agar plates of Luria–Bertolli (LB) medium containing 50μg/mL Kan and Cam antibiotics (LBKan50Cam50).
Use this colony to inoculate a flask with 100mL of LBKan50Cam50 media and grow overnight at 37°C, 250rpm.
Use 20mL of the overnight culture to inoculate 2L of the same medium in Erlenmeyer flasks with a rubber septum and purge the head space with argon.
Grow at 30°C, 220rpm until reaching an OD600 of 0.1 and induce the expression by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 100 μM. Simultaneously, add Fe(NH4)2(SO4)2 6 (H2O) to a final concentration of 5mg/L, for the formation of Fe–S clusters.
Purge the head space with argon and allow the cultures to grow anaerobically overnight at 18°C, 220rpm.
Harvest by centrifugation, freeze the cell paste in liquid nitrogen, and store at −80°C until use.
2.2.2. Growth in Aerobic Conditions and Expression of Mex PqqE in Anaerobic Conditions (Latham et al., 2015)
Select a single isolated colony of E. coli BL21 (DE3) on LBKan50Cam50 plates.
Prepare a starter culture by inoculating a flask with 100mL of terrific broth (TB) media with 50 μg/mL Kan and 50 μg/mL Cam (TBKan50Cam50) and growing overnight at 37°C, 250rpm.
Use 20mL of the overnight culture to inoculate Erlenmeyer flasks containing 2L of the same medium and grow in the same conditions until OD600 reaches 0.6.
Add 50mM fumarate, 50 μM Fe(III) citrate, stopper the flasks, and purge the head space with argon to transition from aerobic to anaerobic conditions.
Allow cells to grow for 30 min in anaerobic conditions before inducing protein expression with 400 μM IPTG.
Incubate cells anaerobically at 19°C for 12h.
Harvest the cells by centrifugation and freeze as described in Section 2.2.1.
2.2.3. Growth and Expression of Mex PqqE in Aerobic Conditions (Barr et al., 2016)
Select a single isolated colony of E. coli BL21 (DE3) on LBKan50Cam50 plates and prepare a starter culture like described in Section 2.2.2.
Use 20mL of the overnight culture to inoculate Erlenmeyer flasks containing 2L of TBKan50Cam50 with 4g/L glycerol as carbon source, supplemented with 100 μM Fe(III) citrate or ammonium Fe(III) citrate.
Allow the cultures to grow at 31°C, 220rpm for 4h and then decrease the temperature to 20°C.
One hour later induce protein expression by adding IPTG to a final concentration of 100 μM. Simultaneously, add cysteine as a source of sulfur, to a final concentration of 50 μM.
Grow the cells overnight in the same conditions, harvest by centrifugation, and freeze and store as described previously.
2.2.4. Expression of 57Fe-Labeled PqqE to Be Used in Mössbauer Spectroscopy
Protein samples to be used in Mössbauer spectroscopy must be labeled with 57Fe, and, for that purpose, the cultures are grown in the presence of labeled Fe(III) citrate (Barr et al., 2016).
Prepare a 50-mL starter overnight culture as described previously.
Prepare 20mM 57Fe citrate by reacting 117mg iron oxide (Cambridge Isotopes) with 1mL 10 M HCl at 80°C. Dilute the formed 57FeCl3 10-fold with 40mM citric acid.
Prepare a solution of 5g OmniPur® casamino acids (Sigma-Aldrich) in 10mL water and chelate the free metals by incubating the solution with2.5g of Chelex-100 chelating ion-exchange resin (Bio-Rad) for 1h.
Prepare flasks with 1L of M9 medium and supplement with 1mL of 1000× trace minerals solution (Sigma-Aldrich), the 10mL casamino acids solution (5g/L final concentration), and the 1mL 20mM 57Fe citrate (20 μM final concentration). Sterilize by autoclaving. Add Kan and Cam to final concentrations of 50 μg/mL.
Inoculate the flasks and follow the procedure described in Section 2.2.3. When inducing with IPTG, supplement with pyridoxine HCl and cysteine to final concentrations of 10 and 100 μM, respectively.
Follow the protocol described for unlabeled PqqE (Section 2.2.3).
As a control, express the protein using the same protocol but with regular Fe(III) citrate as the iron source.
2.3. PqqE Purification
The expressed His6-PqqE is purified by affinity chromatography from a nickel column. The purification is simplified by the fact that the protein has a reddish-brown color due to the presence of Fe–S clusters; thus, the PqqE-containing fractions are easily identified by their color (Barr et al., 2016; Wecksler et al., 2009).
Purification of the K. pneumoniae His6 tag-PqqE is performed under strict anaerobic conditions in an inert atmosphere glovebox (Wecksler et al., 2009), while Mex PqqE can be purified in anaerobic (Barr et al., 2016; Latham et al., 2015) or aerobic conditions (Saichana et al., 2016). The steps used in anaerobic and aerobic purifications are identical; the only difference being that aerobic purification is done in normal lab conditions, while anaerobic purification is done in a glovebox using degassed solutions and buffers. The following protocol is for anaerobic purification. In all steps aliquots (10–50 μL) should be collected for SDS-PAGE analysis. A summary of PqqE purification procedures is shown in Table 2.
Table 2.
Methods for Purification of K. pneumoniae or M. extorquens PqqE
| K. pneumoniae (Wecksler et al., 2009) | M. extorquens (Barr et al., 2016; Latham et al., 2015) | |
|---|---|---|
| Cell pellet disruption | Lysis with BugBuster (Novagen) in 50mM Tris pH 7.9, 1 mM DTT, 300mM NaCl, 10mM imidazole, 5μL benzonase nuclease; centrifugation 15,000 × g, 20 min, 4°C | Lysis with BugBuster (Novagen) in 50 mM Tris pH 7.9, 300mM NaCl, 1mM TCEP, 30mM imidazole, 5μL benzonase nuclease; 15,000 × g, 20min, 4°C |
| Chromatography | (i) Ni-NTA column (12 in. long, 2.5 in wide) equilibrated with 50 mM Tris pH 7.9, 1mM DTT, 300mM NaCl, 10mM imidazole. Washed with 100 mL of the same buffer, then with 100mL of 25 mM imidazole in buffer, followed by 100mL of 50 mM imidazole in buffer. Eluted with 200mM imidazole in buffer. Fractions selected based on color, pooled and concentrated to 5 mL by ultrafiltration (Amicon Ultra 30K membrane) (ii) Gel filtration in PD-10 column equilibrated with 50 mM Tris pH 7.9, 1mM DTT, 300 mM NaCl to remove imidazole. Protein collected off the column and concentrated to ~10 mg/mL by ultrafiltration (Amicon Ultra 30K membrane) |
(i) His-Trap FF column (Novagen) equilibrated with 50 mM Tris pH 7.9, 300mM NaCl, 1mM TCEP, 30 mM imidazole buffer. Washed with the same buffer and eluted with 300 mM imidazole in buffer. Fractions pooled and concentrated to 2.5 mL by ultrafiltration (Amicon Ultra 30K membrane) (ii) Gel filtration in PD-10 columns equilibrated with 50mM Tris pH 7.9, 300mM NaCl, 10% (v/v) glycerol |
| Yield | 0.5–1.5mg/L LB medium | 18mg/L TB medium |
| Reconstitution | Not possible, the protein precipitates | Chemical reconstitution with Na2S and ammonium Fe(III) citrate |
| Fe and S content | 10.4 ± 0.9 mol Fe and 7.0 ± 1.0mol S/monomer | 7–10 mol Fe/monomer (as-purified) 13 mol Fe and 12.2 mol S/monomer (reconstituted) |
All procedures are done in anaerobic conditions, in a glovebox.
Prepare and degas all buffers needed for the purification: Lysis buffer—50mM Tris buffer pH 7.9, 300 mM NaCl, 30 mM imidazole, 1 mM DTT or TCEP, 10% (v/v) glycerol; Wash buffer—50 mM Tris buffer pH 7.9, 300 mM NaCl, 40 mM imidazole, 1 mM DTT or TCEP, 10% (v/v) glycerol; Elution buffer—50 mM Tris buffer pH 7.9, 300 mM NaCl, 300 mM imidazole, 10% (v/v) glycerol; Storage buffer—50 mM Tris pH7.9, 300 mM NaCl, 10% (v/v) glycerol.
Defrost the frozen cell pellets on ice and resuspend in five times the mass of cell paste volume of lysis buffer.
Lyse by the addition of 0.2% (w/v) CHAPS or 1× BugBuster (Novagen), 0.1mg/mL lysozyme (final concentrations), and 5μL benzonase nuclease, for 30min, following the manufacturer’s guidelines.
Transfer the lysates to anaerobic centrifuge tubes (Beckman) and centrifuge at 15,000 × g, 15min, 4°C.
Reintroduce the centrifuge tubes into the glovebox and collect the supernatant.
Equilibrate a Nickel column (Ni-NTA resin or His-Trap FF column (GE Healthcare)) with cold lysis buffer. Load the supernatant into the column manually with a pipette or using a peristaltic pump.
Upon protein binding, wash the column with three volumes of cold wash buffer.
Elute the protein with cold elution buffer. Collect and pool all the reddish-brown fractions.
Continuing in the glove box, concentrate by centrifugal ultrafiltration in an Amicon Ultra 30K filtration device (Millipore).
To remove the imidazole, pass the concentrated protein sample through a PD-10 column previously equilibrated with 50 mM Tris pH 7.9, 300 mM NaCl, and 10% (v/v) glycerol.
If needed, concentrate the protein again using centrifugal ultrafiltration as described previously.
Aliquot in sealed vials, flash-freeze in liquid nitrogen, and store in a liquid nitrogen dewar.
Untagged PqqE needed for SPR protein interaction assays (Section 3.4.2) is obtained by cleavage of the His-tag with a thrombin kit (Novagen), according to the manufacturer’s instructions.
2.4. Chemical Reconstitution of the As-Purified PqqE
K. pneumoniae PqqE purified anaerobically cannot be reconstituted with excess of iron and sulfide since the protein precipitates upon Fe2+/Fe3+ and S2− addition (Wecksler et al., 2009). As-purified Mex PqqE can contain incomplete Fe–S clusters. Thus, for activity assays and the majority of spectroscopic studies, the enzyme was routinely reconstituted with iron and sulfide. The protein reconstitution is done under anaerobic conditions, in a glovebox with <5ppm O2 (Barr et al., 2016). The 57Fe-labeled PqqE (described in Section 2.2.4) is reconstituted following a similar protocol, but using solutions (except for ammonium Fe(III) sulfate) treated with Chelex-100 chelating ion-exchange resin (Bio-Rad) for 30min, to minimize the iron contamination.
Prepare 100mM fresh stock solutions of DTT, ammonium Fe(III) citrate, and Na2S in 1 M Tris pH 7.9 buffer: 15.4mg DTT/1mL buffer, 24.3mg ammonium Fe(III) citrate/1mL buffer, and 24mg Na2S/1mL buffer.
Add 1.5 μL of DTT stock solution into 1mL 100 μM PqqE sample while stirring.
Then add 1μL of the ammonium Fe(III) citrate stock solution to the sample and stir slowly for 15min at room temperature.
Finally add 1μL of the Na2S stock solution to the sample and stir slowly for 15 more min at room temperature. Repeat steps 3 and 4 four times.
Pass the sample through a PD-10 column preequilibrated with storage buffer (see Section 2.3) to remove excess of iron and sulfide.
Flash-freeze in liquid nitrogen and store in a liquid nitrogen dewar.
2.5. Cloning, Expression, and Purification of Flavodoxin Reductase and Flavodoxin A
Because the PqqE activity in vitro requires the presence of a flavodoxin A (FldA) and flavodoxin reductase (FNR)-based reduction system (other reductants lead to an uncoupled cleavage of the cosubstrate SAM) (Barr et al., 2016), these proteins are also expressed and purified in our lab. Because no FldA could be identified in the Mex genome, FldA was amplified from the Azotobacter vinelandii ATCC478 genome.
2.5.1. Mex FNR
All the buffers used are the ones described in Section 2.3 for PqqE purification but without DTT/TCEP.
Amplify FNR from Mex AM1 genomic DNA and clone into the pET28a plasmid.
Transform the plasmid into E. coli BL21 (DE3) competent cells and plate in LBKan50 agar plates.
Pick up a single, isolated colony to inoculate 50mL of LBKan50 medium and incubate at 37°C overnight, at 250rpm.
Use 10mL of the overnight culture to inoculate 1L of fresh LBKan50 medium, grow in the same conditions, and when OD600 reaches 0.5 induce the expression by adding IPTG to a final concentration of 500μM.
Simultaneously add riboflavin to a final concentration of 100μM and express the protein overnight at 31°C, 220rpm.
Harvest cells by centrifugation and freeze the cell pellets at −80°C or proceed to purification.
The protein is purified at 4°C using a Ni-NTA column (Thermo Fisher Scientific).
Add 20mL of lysis buffer and FAD to a final concentration of 10 mM to the cell pellet obtained from 1L of culture.
Sonicate the cell suspension on ice, with cycles of 20s on and 30s off, for a total on time of 2min.
Centrifuge the cell lysate at 16,000 × g for 30min at 4°C.
Load the supernatant (cell-free extract) in the column.
Wash the column with three-column volumes of washing buffer.
Elute the protein using three-column volumes of elution buffer and collect 15mL bright yellow fractions.
Pool the fractions and concentrate by centrifugal ultrafiltration using a 15-mL Amicon Ultra 3K filter device (Millipore).
To remove the imidazole, pass the concentrated protein sample through a PD-10 column (GE Healthcare) previously equilibrated with storage buffer and collect the bright yellow protein fractions.
Flash-freeze 200μL aliquots and store at −80°C.
2.5.2. A. vinelandii FldA
All the buffers used are the ones described in Section 2.3 for PqqE purification.
Amplify FldA from the A. vinelandii genomic DNA using an appropriate set of primers and clone into the pGEX-6p-1 plasmid (GE Healthcare).
Transform the plasmid into E. coli BL21 (DE3) competent cells and select positive transformants in LB agar plates with 100 μg/mLAmp (LBAmp100).
Use a single, isolated colony to inoculate 50mL of LBAmp100 medium and incubate overnight at 37°C, 250rpm.
Use 10mL of overnight culture to inoculate 1L of fresh LBAmp100 medium, grow in the same conditions, and induce with IPTG to a final concentration of 500μM, when OD600 is 0.5.
Simultaneously add FMN to a final concentration of 50μM and express the protein overnight at 31°C, 220rpm.
Harvest the cells by centrifugation and freeze the cell pellets at −80°C or proceed to purification.
Purify the protein by affinity chromatography in a glutathione Sepharose column (GE Healthcare) as follows.
Add 20mL of storage buffer to the cell pellet obtained from 1L of culture.
Prepare the cell-free extract as described in Section 2.5.1 for cells expressing FNR.
Equilibrate the 5mL glutathione-Sepharose column with 50 mM Tris–HCl pH 7.9, 300 mM NaCl, and load the dark-greenish supernatant.
Wash with three-column volumes of storage buffer and discard the flow-through.
Elute the protein with a 30-mL of a 15-mM solution of glutathione (GSH) in storage buffer.
To remove the GST-tag add 40μL PreScission protease (GE Healthcare) to the ~15mL of eluted protein solution and shake on a nutator (Clay Adams brand) for 12h at 4°C.
Concentrate the protein sample to 5mL using an Amicon Ultra 10K filtration device (Millipore).
Run the sample in a size-exclusion column HiPrep 26/60 Sephacryl S-200 High Resolution (GE Healthcare) equilibrated with storage buffer, outfitted to an AKTA FPLC (GE Healthcare) using a flow rate of 0.5mL/min (this purification step must be performed at 4°C).
Run aliquots of the fractions with protein in SDS-PAGE and identify the ones containing untagged FldA.
Pool the fractions with untagged FldA and concentrate the protein solution to 3 mM using an Amicon Ultra 3K membrane (Millipore).
Aliquot into 50μL samples, flash-freeze in liquid nitrogen, and store at −80°C.
2.6. Cloning, Expression, and Purification of PqqCD and PqqD
For activity assays and interaction experiments of PqqE with PqqD and PqqA, Mex PqqD was also cloned, expressed, and purified in our lab. In vivo Mex PqqD is N-terminally fused to PqqC, and this fusion was amplified from the Mex genome as a His6-PqqCD (Section 2.6.1) (Latham et al., 2015) and used for interaction assays (Section 3.4.2). Additionally PqqD was expressed and purified as a single protein, either as a His6-PqqD (Latham et al., 2015), for interaction assays, or as untagged PqqD (Barr et al., 2016), for activity assays.
2.6.1. His6-PqqCD
All the buffers used are the ones described in Section 2.3 for PqqE purification. PqqCD is amplified from Mex AM1 genomic DNA and cloned into the pET28a plasmid. The expression and purification procedures are similar to the ones described for FNR (Section 2.5.1), except that the protein was expressed at 19°C and the lysis buffer does not contain FAD.
2.6.2. His6-PqqD
The sequence of a truncation of PqqD not containing the PqqC part (amino acids 280–372 of the PqqCD sequence) was obtained as described by Latham et al. (2015). This sequence was amplified from the Mex AM1 genome and cloned into pET28a. The expression and purification procedures are similar to the ones described in Section 2.6.1.
2.6.3. Untagged PqqD
Amplify PqqD from Mex AM1 genomic DNA and clone into the pGEX-6p-1 plasmid (GE Healthcare).
Transform into E. coli BL21 (DE3) competent cells and plate in LBAmp100 agar plates.
Prepare a starter culture by inoculating 50mL LBAmp100 medium with a single, isolated colony from the LBAmp100 plate, and incubate overnight at 37°C, 220rpm.
Use 10mL of overnight culture to inoculate 1L of fresh LBAmp100 medium, grow in the same conditions until OD600 is 0.5, and induce the expression by adding IPTG to a final concentration of 500μM.
Incubate the cultures overnight at 19°C, 220rpm to allow the protein to express.
Harvest the cells by centrifugation, freeze, and store at −80°C until use.
Purify the protein in a glutathione-Sepharose column followed by size-exclusion chromatography using the same methodology described in Section 2.5.
3. PROTEIN CHARACTERIZATION
3.1. Determination of Protein Concentration and Purity Evaluation
The purity of the PqqE, FNR, FldA, FNR, PqqCD, and PqqD preparations during the purification process is monitored by SDS-PAGE using the method of Laemmli (1970).
The concentration of protein is determined by the Bradford assay (Bradford, 1976) and its comparison with the absorbance at 280nm (A280), as described by Barr et al. (2016). Correct protein quantification is very important to determine the number of Fe–S clusters per polypeptide. Thus, a conversion factor relating protein determined by the Bradford assay, which relies on a standard curve usually prepared with a different protein (e.g., BSA), with that determined by absorbance at 280nm, must be calculated. This is done using the following procedure (Barr et al., 2016):
Denature the protein by incubating for 1h with 6 M guanidinium HCl, and 0.1 M citric acid to destroy the Fe–S clusters and chelate the free iron.
Pass the protein through a PD-10 column to remove the citric acid.
Determine the protein concentration by measuring the absorbance at 280nm and determine the molar extinction coefficient using the method of Pace, Vajdos, Fee, Grimsley, and Gray (1995). Alternatively, a theoretical molar extinction coefficient of 57,870M−1cm−1 can be calculated in ProtParam, assuming that all Cys residues are reduced (Gasteiger et al., 2005).
Determine the concentration of protein in the same sample using the Bradford assay with a BSA standard curve linearized by dividing the absorbance at 590nm by the absorbance at 450nm (Zor & Selinger, 1996).
Compare the values obtained by the two methods and determine the conversion factor.
3.2. Characterization of Fe–S Clusters
3.2.1. Iron and Sulfide Quantification
The catalysis of PqqE depends on the correct incorporation of the Fe–S clusters that are formed in the protein either during expression or by chemical reconstitution after purification. It is necessary to quantitatively determine the total content of iron and sulfide for each batch of expressed and purified PqqE. We have adopted the method of Crack, Green, Thomson, and Le Brun (2014) for both determinations.
Iron content determination
Since the as-purified PqqE generally contains roughly 10 iron molecules per monomer (Barr et al., 2016; Latham et al., 2015), the protein should be diluted 10-fold prior to the assays. It is also worth noting that the use of screwed cap vials and the cooling process are necessary in order to avoid the change of concentration due to the water loss by evaporation during the heating process.
Add 100μL of PqqE sample into screwed cap tubes in triplicate—tubes #1–3.
Set up a control vial by adding 100μL of storage buffer (see Section 2.3) to tube #4.
Add 100μL of each concentration of iron ICP-MS standard solution (Sigma)—0, 25, 50, 100, 150, 200, and 350μM—to tubes #5–11.
Add 100μL of 22% (v/v) HNO3 to each tube.
Incubate the tubes in a dry bath at 90°C for 30min.
Cool the samples to room temperature and spin in a microcentrifuge at 12,000×g for 1min.
Neutralize the samples by adding 600μL of 7.5% (w/v) ammonium acetate to each tube and mix by vortexing.
Reduce the samples by adding of 100μL of 12.7% (w/v) freshly made ascorbic acid and mix by vortexing.
Add 100μL of 10 mM ferrozine solution and mix by vortexing.
Spin the samples in a microcentrifuge at 12,000×g for 1min.
Transfer the solutions into disposable cuvettes and read the absorbance at 562nm on the UV–vis spectrometer (Cary 50 spectrophotometer with a spectral bandwidth 1.5nm).
Prepare a standard curve using the solutions with different concentrations of standard iron (tubes #5–11) and calculate the total iron content in the protein solution (tubes #1–3).
Calculate the iron/monomer of PqqE based on the protein concentration of the same PqqE sample that is used in the total iron content measurement (see Section 3.1).
Repeat the experiment individually three times and calculate the average iron content of the protein sample.
Sulfide content determination
Add 200μL of an appropriately diluted PqqE sample into tubes #1–3.
Prepare a control vial by adding 200μL of storage buffer (see Section 2.3) in tube #4.
Prepare a 2-mM sulfide stock solution by dissolving 240mg of sodium sulfide nonahydrate (Na2S 9H2O) in 500mL 10 mM NaOH.
Prepare sulfide standard solutions by dilution of the 2 mM stock to 0, 10, 50, 100, 150, 200, and 250μM. Use a 10-mM NaOH solution as a blank control.
Add 200μL of the sulfide standard solutions prepared in step 2 to tubes #5–11.
Immediately add 600μL of freshly prepared 1% (w/v) zinc acetate into each tube.
Add 50μL of 12% (v/v) NaOH into each tube.
Cap the tubes well and mix by inversion. Incubate at room temperature for 15min
Centrifuge the tubes at 16,000×g for 1min.
Add 150μL of freshly prepared 0.1% (w/v) N,N-dimethyl-p-phenylenediamine dihydrochloride in 5M HCl.
Add 150μL of freshly prepared 10 mM FeCl3 in 1M HCl.
Vortex the solutions to mix and incubate at room temperature for 30min.
Transfer the solutions into disposable cuvettes and read the absorbance at 670nm on UV–vis spectrometer (Cary 50 spectrophotometer, spectral bandwidth 1.5nm).
Prepare a standard curve using standard solutions of sodium sulfide prepared in step 2 (tubes #5–11) and use it to calculate the total sulfide content in the protein samples (tubes #1–3).
Calculate the sulfide/monomer of PqqE based on the protein concentration of the same PqqE sample used in the total sulfide content measurement (see Section 3.1).
Repeat the experiment individually three times and calculate the average sulfide content of the protein sample.
3.2.2. EPR Characterization
EPR spectroscopy has been successfully used in the characterization of the Fe–S clusters in structure–function studies of radical SAM enzymes (Stich, Myers, & Britt, 2014). EPR spectra can provide useful information, such as the identity of Fe–S clusters, the protein environment around the clusters, and the quantity/quality of the iron and sulfur in the protein sample. Without the addition of reducing reagent, the oxidized [3Fe–4S] cluster presented in the as-purified/reconstituted PqqE can be easily detected by EPR at 40K, while the oxidized iron in the [4Fe–4S] clusters is EPR silent (Pandelia, Lanz, Booker, & Krebs, 2015). Therefore, the sample preparation for characterization of PqqE using EPR spectroscopy involves the use of both, the oxidized and the reduced form of enzyme.
The sodium dithionite (DTH) solution must be freshly prepared anaerobically, inside the glovebox, using an aliquot of DTH powder. The commercially available DTH is usually less than 85% pure, and the purity decreases after long-term storage, even if in the glovebox. In this case, it is necessary to determine the quality of DTH using a redox titration method (McKenna, Gutheil, & Song, 1991). If highly concentrated PqqE is needed for better spectroscopy quality, the concentration step is done during reconstitution. Protein precipitated during the reconstitution steps is removed by centrifugation at 14,000×g for 10min.
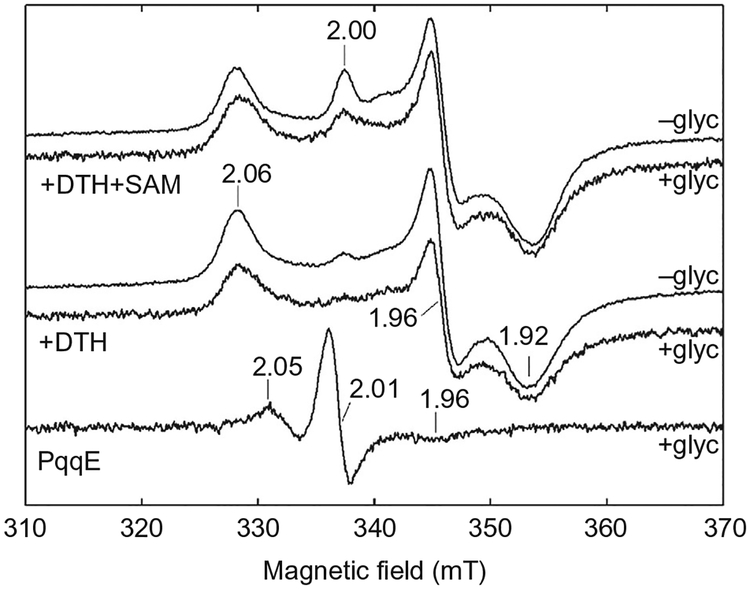
The representative procedure described here is focused on the comparative Fe–S cluster characterization of K. pneumoniae PqqE and the PqqE–SAM complex, with the control being as-isolated enzyme in the absence of SAM/DTH (Fig. 5). The EPR spectrometer used was a Bruker ECS106 X-band spectrometer with a TE102 cavity (ER4102ST) resonating at 9.5GHz, equipped with an Oxford ESR900 liquid helium cryostat in conjunction with an ITC503 temperature controller.
Fig. 5.
A representative continuous wave X-band EPR spectrum of K. pneumoniae PqqE samples in various conditions. The signals are as follows, from top to bottom: dithionite (DTH)-reduced PqqE with a 10-fold excess of SAM (with and without glycerol), DTH-reduced PqqE (with and without glycerol), and as-isolated PqqE with glycerol. Adapted from Wecksler, S. R., Stoll, S., Tran, H., Magnusson, O. T., Wu, S. P., King, D., et al. (2009). Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry, 48(42), 10151–10161. Copyright © 2009 American Chemical Society.
Mix a 10-fold excess of DTH solution (freshly prepared in 50 mM Tris, 300 mM NaCl pH 7.9, under anaerobic conditions) with 100μM PqqE in 50 mM Tris pH 7.9, 300 mM NaCl with or without 20% (v/v) glycerol by stirring the reaction mixture at room temperature in the glovebox for 10min. Alternatively, a fivefold excess of DTH can be used with an incubation time extended to 30min.
For the experiments with PqqE–SAM, after the reduction reaction, add a 10-fold excess of a 10 mM SAM solution in water (prepared as described in Zhang & Klinman, 2015) to the reduced PqqE solution and incubate for approximately 5min.
Aliquot 300μL of the DTH-reduced PqqE sample into an X-band EPR tube.
Prepare the control samples by adding only PqqE or reduced PqqE (prepared in step 1) to EPR tubes.
Seal the tube with a rubber stopper in the glovebox and fast-freeze the sample in liquid nitrogen immediately after removal from the glovebox.
The spectrometer settings are set as follows: EPR conditions: temperature, 40K; microwave power, 6.3mW; microwave frequency, 9.480GHz.
Determine the spectra of the PqqE samples at 40K.
Spectra are simulated using the EasySpin MATLAB® package.
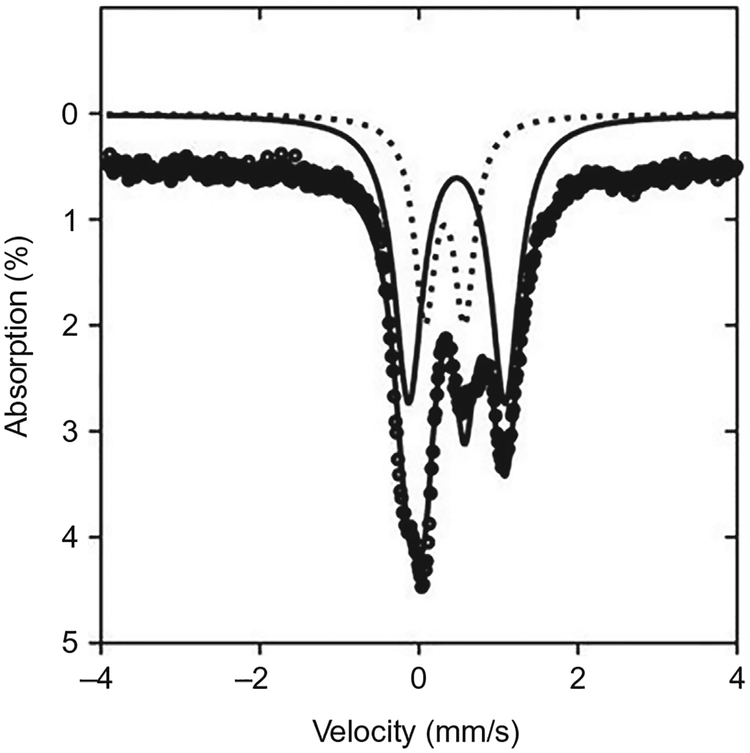
3.2.3. Mössbauer Spectroscopy
Compared to EPR, Mössbauer spectroscopy provides valuable information about the total iron present either in Fe–S clusters or adventitiously bound, without the need of prior reduction of the protein. Based on the parameters provided by the spectra, one can distinguish the different iron oxidation state and the [Fe–S] species in a mixed protein environment. This is especially useful for characterization of the clusters in PqqE, which has been found to contain a mix of [2Fe–2S] and [4Fe–4S] clusters (Barr et al., 2016; Saichana et al., 2016; Fig. 6). By analyzing proteins with different Fe–S site knockouts, one may also be able to identify the source of certain signals in the Mössbauer spectra. Here, we provide a general method for quantitatively determining the different iron species in the Fe–S clusters of as-purified WT PqqE labeled with 57Fe (prepared as described in Section 2.2.4). We used a Mössbauer spectrometer (See Co., Edina, MN) connected to a Janis Research Co. cryostat (Wilmington, MA).
Fig. 6.
Illustrative zero-field Mössbauer spectrum of as-purified Mex 57Fe-PqqE at 4K. Black circles show the raw data, which have been offset from the axis by 0.5% for ease of interpretation. These data are decomposed into 4Fe–4S (solid line) and 2Fe–2S (dotted line) components. Adapted from Barr, I., Latham, J. A., Iavarone, A. T., Chantarojsiri, T., Hwang, J. D., & Klinman, J. P. (2016). Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. Journal of Biological Chemistry, 291(17), 8877–8884. Copyright ©2016 by American Society for Biochemistry and Molecular Biology.
Add the purified and reconstituted 57Fe-labeled PqqE solution into the Teflon sample holder (thickness 0.2in.) in the glovebox with O2<1ppm.
Place the sample holder snuggly in the sample rod holder and wrap in Kapton® Tape prior to introduction into the spectrometer.
Collect spectra and analyze with the WMOSS software package (Prisecaru, 2009–2016).
3.3. Activity
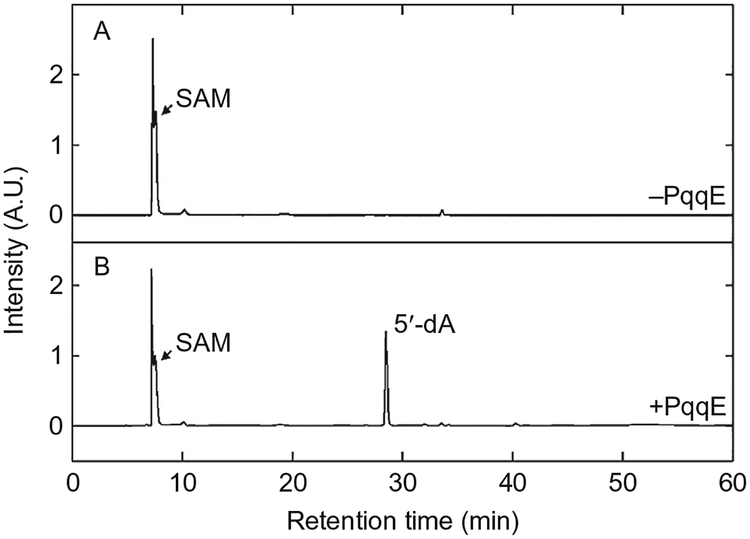
3.3.1. SAM Cleavage Activity
The PqqE-catalyzed PqqA cross-linking reaction is initiated by the generation of a SAM-derived radical, concomitant with oxidation of the adjacent RS [4Fe–4S]+ cluster to [4Fe–4S]2+. This can lead to peptide (PqqA) cross-linking (Section 3.3.2) or uncoupled cleavage to yield an equivalent of 5′-deoxyadenosine (5′-dA). The formation of 5′-dA can be easily quantified by HPLC. In this experiment, we used a Beckman HPLC equipped with a diode array detector and operated by 32 Karat version 8.0 software. The column used was a reverse-phase C18 Jupiter® 4μm Proteo 90Å, 250mm × 4.60mm C18 column.
Set up reactions in the glovebox with O2<0.8ppm.
Prepare 100 mM DTH in 50 mM Tris pH 7.9, 1 mM DTT, 300 mM NaCl, 20% (v/v) glycerol.
Add 89μL PqqE stock solution in the same buffer into tubes #1–8.
Add 1μL 100 mM DTH into each sample.
Leave the samples in the glovebox at room temperature for 10min to complete the reduction.
Add 10μL 5 mM SAM into each sample.
Quench the reactions at various time points (1min–16h) by addition of 5μL 88% (v/v) formic acid per 100μL of the sample.
Take the quenched reactions out of the glovebox and centrifuge at 16,000×g for 1min.
Inject 100μL of each sample into HPLC for 5′-dA determination; the conditions are: 1mL/min flow rate, 0.05% (v/v) formic acid, 1.0% (v/v) acetonitrile in water for 30min, followed by a linear gradient from 1% to 100% acetonitrile over 10min. Spectra are recorded at 215 and 260nm (Fig. 7).
Prepare a 5′-dA standard curve by analyzing solutions with different concentrations—0, 5, 10, 50, 100, 150, 200, and 250μM, in the same conditions.
Fig. 7.
LC-MS analysis of SAM cleavage reaction by K. pneumoniae DTH-reduced PqqE.(A) LC-MS elution profile (monitored at 260nm) of a control sample without PqqE.(B) LC-MS elution profile of an anaerobic reaction mixture containing PqqE, SAM, and DTH. Adapted from Wecksler, S. R., Stoll, S., Tran, H., Magnusson, O. T., Wu, S. P., King, D., et al. (2009). Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry, 48(42), 10151–10161. Copyright © 2009 American Chemical Society.
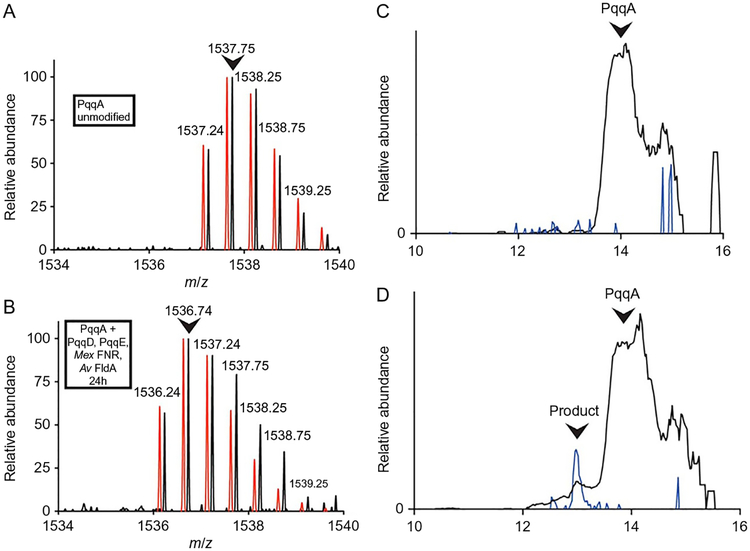
3.3.2. PqqA Modification Assay
The SAM cleavage reaction described in Section 3.3.1 provides insight about the catalytic activity of the radical SAM [4Fe–4S]+ cluster that generates the active radical in the initial steps of the PqqE reaction, but it does not reflect the catalytic efficiency of PqqE to perform the biologically active peptide modification reaction. The direct measurement of the PqqE bioactivity can be determined by assaying the production of the modified peptide by LC-MS. The cross-linked peptide product has a lower retention time compared to the unmodified PqqA in LC and can be identified from the m/z in the MS spectra.
The reaction mixture must contain both the peptide chaperone PqqD and an appropriate reduction system, in this instance FldA/FNR/NADPH (Barr et al., 2016). While the ratio of each component can be further optimized for better PqqA conversion, the FNR concentration should always be lower than the FldA concentration, due to the FldA-binding competition between PqqE and FNR, which is expected to reduce the yield of the cross-linked product.
In the following protocol, we used an Agilent 1200 LC with a C4 column (Restek) connected in-line with a Thermo LTQ-Orbitrap-XL mass spectrometer equipped with an electrospray ionization source and operated in the positive ion mode.
PqqA (with a nonconserved cysteine mutated to serine for increased stability) was custom synthesized and obtained from CPC Scientific.
All solutions and enzymes used in this protocol are prepared in storage buffer. Stock solutions are: 100μM PqqE, 1 mM PqqA, 3 mM PqqD, 5 mM SAM, 10 mM NADPH, 2 mM FNR, and 2 mM FldA. All concentrations mentioned in the following protocol are final concentrations in the mixtures.
Mix and incubate 1 mM NADPH, 5μM FNR, and 20μM FldA in tube #1 in the glovebox for 30min.
Mix 50μM PqqE and 500μM SAM in tube #2 in the glovebox.
Mix 50μM PqqD and 50μM PqqA in tube #3 in the glovebox.
Mix the reaction mixtures from the three tubes and complete the volume to 200μL with 50 mM Tris pH 7.9, 300 mM NaCl, 10% (v/v) glycerol.
Incubate at room temperature for 16 h and quench the reaction by addition of 10μL 88% (v/v) formic acid.
Centrifuge the quenched reaction mixture at 14,000×g for 5min and dilute 20μL of the supernatant to 200μL.
Apply 5μL of the sample onto LC-MS and elute the sample with a linear gradient 20%–70% water (2% formic acid)/acetonitrile (2% formic acid).
Analyze the sample peak containing the PqqA and PqqA cross-linked product using the Xcalibur software (version 2.0.7, Thermo). The extent of cross-linking of the PqqA peptide is determined by integrating extracted ion chromatograms for the [M+2H]2+ ions of cross-linked and unmodified forms of PqqA (occurring at m/z = 1536.7 and 1537.7, respectively) (Fig. 8).
Fig. 8.
LC-MS analysis of modification of PqqA by PqqE and PqqD. (A) Initial ion mass envelope of unmodified PqqA with two charge state. (B) Mass envelope of a minor peak, eluting 1min earlier, seen following a 24-h reaction under anaerobic conditions. A noticeable shift in mass by 2Da is observed, consistent with cross-linking of residues in PqqA. The calculated mass envelopes for PqqA (A) and modified PqqA (B) are shown in red and slightly offset for ease of comparison. The most abundant ions are indicated by the arrows, and these were used to quantify the relative amount of modified PqqA.(C) Chromatograph showing the elution profile of 1537.7 (black) and 1536.7 (blue) ions in an unreacted PqqA sample. (D) Chromatograph showing the elution profile of 1537.7 (black) and 1536.7 (blue) ions in a 24-h reaction mixture; a small peak containing cross-linked PqqA is seen to elute earlier than the unreacted PqqA. Adapted from Barr, I., Latham, J. A., Iavarone, A. T., Chantarojsiri, T., Hwang, J. D., & Klinman, J. P. (2016). Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. Journal of Biological Chemistry, 291(17), 8877–8884. Copyright ©2016 by American Society for Biochemistry and Molecular Biology.
3.4. Protein Interaction Studies
3.4.1. Nanoelectrospray Ionization Mass Spectrometry of Native Proteins
In the study of the interaction between PqqE with other proteins in the PQQ biosynthesis, nanoelectrospray ionization mass spectrometry (NanoESI-MS) provides a direct measurement of the binding ratio of PqqE to other proteins in the pathway. The procedure for the determination of the binding ratio in the PqqE/D complex in solution is described here. Based on the ions of PqqE monomer, PqqD monomer, and the m/z of the PqqE/D complex, one can determine the number of PqqE and PqqD in the PqqE/D complex. The instrument used was a Quadrupole time-of-flight (Q-TOF) mass spectrometer equipped with a nano-Z-spray nano-ESI source (Q-TOF Premier, Waters, Beverly, MA).
Exchange the purified PqqE and PqqD (in 50 mM Tris, 300 mM NaCl pH 7.9 buffer) into 25 mM ammonium bicarbonate buffer pH 7.9 using preequilibrated PD-10 columns (GE Healthcare).
Prepare the PqqD and PqqE complex sample by mixing 10μM PqqE and 50μM PqqD in ammonium bicarbonate buffer pH 7.9.
Load 10μL of the complex sample into the nano-ESI emitter using a gas-tight syringe.
Initiate the electrospray by increasing the DC potential. Set up the instrument parameters as previously described by Beeson, Iavarone, Hausmann, Cate, and Marletta (2011).
Record mass spectra as previously described (Wecksler et al., 2010).
Collect the MS data of the intact proteins and the complex and process using MassLynx software (version 4.1, Waters) (Fig. 9).
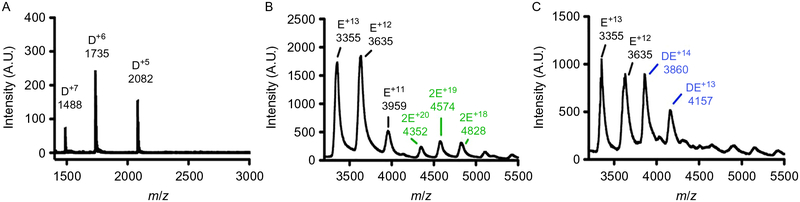
Fig. 9.
Nano-Q-TOF-ESI-MS characterization of the PqqD:PqqE complex. (A) The Mex PqqD native mass spectrum shows a monomer population with a molecular weight of 10.4kDa calculated from the charge-state distribution. (B) The native mass spectrum of Mex PqqE shows two sets of charge-state distributions with masses of 43.6 and 86.9kDa, indicating that both the monomeric and dimeric species are present. (C) In the presence of Mex PqqD, a 54.0-kDa charge-state distribution is formed corresponding to a 1:1 Mex PqqE:PqqD complex. Adapted from Latham, J. A., Iavarone, A. T., Barr, I., Juthani, P. V., & Klinman, J. P. (2015). PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. Journal of Biological Chemistry, 290(20), 12908–12918. Copyright ©2015 by American Society for Biochemistry and Molecular Biology.
3.4.2. Surface Plasmon Resonance
For determination of the dissociation constants between PqqD and PqqE, and PqqA and PqqCD fusion, surface plasmon resonance (SPR) measurements and isothermal titration calorimetry (ITC) analysis have both been applied. However, the ITC measurements require highly concentrated protein samples due to the relatively high Kd of PqqD and PqqE (Latham et al., 2015). Therefore, SPR gives more accurate measurements in this system.
In the following experiment, we used a Biacore T100 SPR (Biacore, GE Healthcare). In order to reduce the buffer effect on the measurements, all the solutions should be exchanged to the same stock buffer prior to the determinations. For the PqqCD/PqqA interaction, His6-PqqCD is immobilized on the Ni-NTA chip and the PqqA solution is injected as the analyte. In the study of PqqD/E interaction, His6-PqqD is immobilized, and native PqqE is injected as the analyte. For both experiments, the analyte is serially diluted seven- to ninefold, depending on the number of injections in the experiment. A careful wash must be done after each cycle so that the binding capacity is kept optimal throughout the entire experiment.
Interaction of PqqCD and PqqA
Exchange His6-PqqCD and PqqA (both in 50 mM Tris 300 mM NaCl pH 7.9 buffer) into HBS-P buffer (GE Healthcare) and determine the concentration.
In the Biacore T100 SPR leave Flow cell 1 with HBS-P buffer as the reference channel.
Immobilize His6-PqqCD by injecting 10nM on the NTA chip (Biacore, GE Healthcare) at room temperature.
Inject the serial dilutions of native PqqA solution for 30s at 30μL/min followed by a 60-s dissociation phase.
After each cycle (15 injections), regenerate the surface injecting 300 mM EDTA, followed by 500μM NiCl2 and 3 mM EDTA.
Calculate the binding affinities from steady-state analysis of the maximum steady-state response.
Analyze the data by plotting the time and response units using the Prism 5 Software (Prism) (Fig. 10).
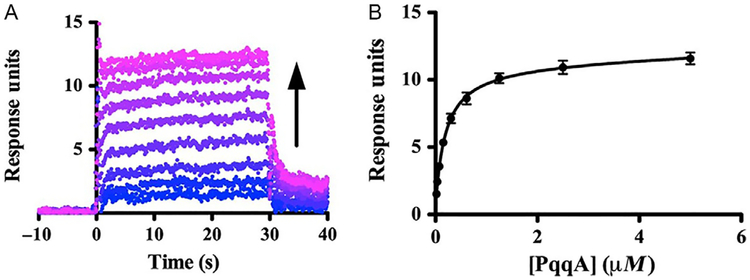
Fig. 10.
Binding of PqqCD and PqqA observed by SPR. (A) Representative dataset (Mex His6-PqqCD as ligand and Mex PqqA as analyte) showing that the SPR response increases as Mex PqqA concentration increases (blue to violet). (B) The steady-state fits were used to calculate the dissociation constant (error bars represent the standard deviation of three independent experiments). Adapted from Latham, J. A., Iavarone, A. T., Barr, I., Juthani, P. V., & Klinman, J. P. (2015). PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. Journal of Biological Chemistry, 290(20), 12908–12918. Copyright ©2015 by American Society for Biochemistry and Molecular Biology.
Interaction of PqqD and PqqE
The protocol is identical to the previous one, with His6-PqqD replacing His6-PqqCD and untagged PqqE substituting PqqA as the analyte.
4. SUMMARY AND CONCLUSIONS
PQQ, although only produced by bacteria, has been widely recognized as an important nutrient for growth and development in plant and animals (Mure, 2004). PQQ exhibits growth-promoting activity, anti-diabetic effects, antioxidant, and neuroprotection properties (Akagawa, Nakano, & Ikemoto, 2015). Despite the significant importance of PQQ to human health, the details of the biosynthetic pathway of this molecule have been, until recently, largely unknown. Recent studies by our group revealed that a key step in the PQQ biosynthesis involves the formation of a C–C bond between two amino acids in the peptide substrate PqqA, catalyzed by the radical SAM enzyme PqqE in complex with the chaperone PqqD (Barr et al., 2016; Latham, Barr, & Klinman, 2017; Latham et al., 2015). This reaction is both complicated and fascinating not only because it is an O2-sensitive reaction occurring in an aerobic bacteria (Mex) but also due to the fact that it requires precise redox control and interaction among multiprotein complexes. The chaperone protein PqqD is most generally encoded by an individual gene pqqD in the gene cluster (e.g., K. pneumoniae operon), whereas in Mex it is fused to the C-terminus of the pqqC gene. In the latter case, PqqD that is fused to the C-terminus of PqqC may only bind PqqE transiently during the cross-linking of PqqA, dissociating prior to the downstream catalytic steps required for PQQ production. This contrasts with the other RiPP proteins such as CteB where the peptide-binding domain is fused to the N-terminus of the radical SAM protein (Grove et al., 2017). For the activity studies of PqqA cross-linking described herein, PqqD was cloned from the Mex PqqCD gene and expressed as a separate protein.
Even though the O2 tolerance is relatively high for Mex PqqE, it is necessary to purify the enzyme under strict anaerobic conditions, to preserve the Fe–S clusters essential for the enzyme activity. With regard to the activity assays, DTH can be used for examination of the SAM cleavage activity. However, a biological reducing system, such as FldA/FNR/NADPH, is essential for successful modification of PqqA (Barr et al., 2016). While a Mex FldA/FNR reducing pair would have been ideal for assaying the Mex PqqE reaction, A. vinelandii FldA was used in our studies due to the inability of obtaining the FldA from Mex. The requirement to use chemically reconstituted PqqE to catalyze the PqqA modification is almost certainly related to the fact that PqqE requires three intact Fe–S centers of varying structures and stabilities for substrate turnover. The exact nature of the two auxiliary clusters is an ongoing and exciting direction to research on PqqE structure–function (Barr et al., 2016; Saichana et al., 2017).
ABBREVIATIONS
- 5′-dA
5′-deoxyadenosine
- AHQQ
3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid
- Amp
ampicillin
- Cam
chloramphenicol
- DTH
sodium dithionite
- DTT
dithiothreitol
- FldA
flavodoxin A
- FNR
flavodoxin reductase
- GSH
glutathione
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- ITC
isothermal titration calorimetry
- Kan
kanamycin
- Klbp
Klebsiella pneumoniae
- LB
Luria–Bertani medium
- LBAmp100
Luria–Bertani medium with 100μg/mL ampicillin
- LBKan50Cam50
Luria–Bertani medium with 50μg/mL kanamycin and 50μg/mL chloramphenicol
- Mex
Methylobacterium extorquens
- Nano-ESI-MS
nanoelectrospray ionization mass spectrometry
- PQQ
pyrroloquinoline quinone (4,5-dihydro-4,5-dioxo-1H-pyrrolo[2,3-f] quinolone-2,7,9-tricarboxilic acid)
- RiPPs
ribosomally synthesized and posttranslationally modified peptides
- RS
radical SAM
- SAM
S-adenosyl-l-methionine
- SPR
surface plasmon resonance
- TB
terrific broth
- TBKan50Cam50
terrific broth with 50μg/mL kanamycin 50μg/mL chloramphenicol
- TCEP
Tris(2-carboxyethyl)phosphine
REFERENCES
- Akagawa M, Nakano M, & Ikemoto K (2015). Recent progress in studies on the health benefits of pyrroloquinoline quinone. Bioscience, Biotechnology, and Biochemistry, 80(1), 13–22. [DOI] [PubMed] [Google Scholar]
- Banci L, Ciofi-Baffoni S, Mikolajczyk M, Winkelmann J, Bill E, & Pandelia ME (2013). Human anamorsin binds [2Fe-2S] clusters with unique electronic properties. Journal of Biological Inorganic Chemistry, 18(8), 883–893. [DOI] [PubMed] [Google Scholar]
- Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, & Klinman JP (2016). Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. Journal of Biological Chemistry, 291(17), 8877–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson WT, Iavarone AT, Hausmann CD, Cate JH, & Marletta MA (2011). Extracellular aldonolactonase from Myceliophthora thermophila. Applied and Environmental Microbiology, 77(2), 650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnot F, Iavarone AT, & Klinman JP (2013). Multistep, eight-electron oxidation catalyzed by the cofactorless oxidase, PqqC: Identification of chemical intermediates and their dependence on molecular oxygen. Biochemistry, 52(27), 4667–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Crack JC, Green J, Thomson AJ, & Le Brun NE (2014). Techniques for the production, isolation, and analysis of iron-sulfur proteins. Methods in Molecular Biology, 1122, 33–48. [DOI] [PubMed] [Google Scholar]
- Duine JA (1999). The PQQ story. Journal of Bioscience and Bioengineering, 88(3), 231–236. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. (2005). Protein identification and analysis tools on the ExPASy server In Walker JM (Ed.), The proteomics protocols handbook (pp. 571–607). New York: Humana Press. [Google Scholar]
- Goldman PJ, Grove TL, Sites LA, McLaughlin MI, Booker SJ, & Drennan CL (2013). X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proceedings of the National Academy of Sciences of the United States of America, 110(21), 8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell TA, Goldman PJ, & Drennan CL (2015). SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. Journal of Biological Chemistry, 290(7), 3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove TL, Himes PM, Hwang S, Yumerefendi H, Bonanno JB, Kuhlman B, et al. (2017). Structural insights into thioether bond formation in the biosynthesis of sactipeptides. Journal of the American Chemical Society, 139(34), 11734–11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauge JG (1964). Glucose dehydrogenase of Bacterium anitratum: An enzyme with a novel prosthetic group. Journal of Biological Chemistry, 239, 3630–3639. [PubMed] [Google Scholar]
- Klinman JP, & Bonnot F (2014). Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chemical Reviews, 114(8), 4343–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685. [DOI] [PubMed] [Google Scholar]
- Latham JA, Barr I, & Klinman JP (2017). At the confluence of ribosomally synthesized peptide modification and radical S-adenosylmethionine (SAM) enzymology. Journal of Biological Chemistry, 292(40), 16397–16405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Iavarone AT, Barr I, Juthani PV, & Klinman JP (2015). PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. Journal of Biological Chemistry, 290(20), 12908–12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson OT, Toyama H, Saeki M, Rojas A, Reed JC, Liddington RC, et al. (2004). Quinone biogenesis: Structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proceedings of the National Academy of Sciences of the United States of America, 101(21), 7913–7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna CE, Gutheil WG, & Song W (1991). A method for preparing analytically pure sodium dithionite. Dithionite quality and observed nitrogenase-specific activities. Biochimica et Biophysica Acta, 1075(1), 109–117. [DOI] [PubMed] [Google Scholar]
- Mure M (2004). Tyrosine-derived quinone cofactors. Accounts of Chemical Research, 37(2),131–139. [DOI] [PubMed] [Google Scholar]
- Nachin L, Loiseau L, Expert D, & Barras F (2003). SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. The EMBO Journal, 22(3), 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, & Gray T (1995). How to measure and predict the molar absorption coefficient of a protein. Protein Science, 4(11), 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandelia ME, Lanz ND, Booker SJ, & Krebs C (2015). Mössbauer spectroscopy of Fe/S proteins. Biochimica et Biophysica Acta, 1853(6), 1395–1405. [DOI] [PubMed] [Google Scholar]
- Prisecaru I (2009–2016). WMOSS4 Mössbauer spectral analysis software. Retrieved from www.wmoss.org.
- Saichana N, Tanizawa K, Pechousek J, Novak P, Yakushi T, Toyama H, et al. (2016). PqqE from Methylobacterium extorquens AM1: A radical S-adenosyl-lmethionine enzyme with an unusual tolerance to oxygen. Journal of Biochemistry, 159(1), 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saichana N, Tanizawa K, Ueno H, Pechoušek J, Novák P, & Fr ebortová J (2017). Characterization of auxiliary iron–sulfur clusters in a radical S-adenosylmethionine enzyme PqqE from Methylobacterium extorquens AM1. FEBS Open Bio, 7, 1864–1879. Retrieved from https://doi.org/10.1002/2211-5463.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YQ, Bonnot F, Imsand EM, RoseFigura JM, Sjolander K, & Klinman JP (2012). Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry, 51(11), 2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stich TA, Myers WK, & Britt RD (2014). Paramagnetic intermediates generated by radical S-adenosylmethionine (SAM) enzymes. Accounts of Chemical Research, 47(8), 2235–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velterop JS, Sellink E, Meulenberg JJ, David S, Bulder I, & Postma PW (1995). Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway. Journal of Bacteriology, 177(17), 5088–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Koch J, Ermler U, & Shima S (2017). Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science, 357(6352), 699–703. [DOI] [PubMed] [Google Scholar]
- Wecksler SR, Stoll S, Iavarone AT, Imsand EM, Tran H, Britt RD, et al. (2010). Interaction of PqqE and PqqD in the pyrroloquinoline quinone (PQQ) biosynthetic pathway links PqqD to the radical SAM superfamily. Chemical communications (Cambridge), 46(37), 7031–7033. [DOI] [PubMed] [Google Scholar]
- Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu SP, King D, et al. (2009). Pyrroloquinoline quinone biogenesis: Demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry, 48(42), 10151–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, & Klinman JP (2015). High-performance liquid chromatography separation of the (S,S)- and (R,S)-forms of S-adenosyl-L-methionine. Analytical Biochemistry, 476, 81–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zor T, & Selinger Z (1996). Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Analytical Biochemistry, 236(2), 302–308. [DOI] [PubMed] [Google Scholar]