Abstract
Autophagy captures intracellular components and delivers them to lysosomes where they are degraded and recycled to sustain metabolism and to enable survival in starvation1–5. Acute, whole-body deletion of the essential autophagy gene Atg7 in adult mice causes a systemic metabolic defect manifested by starvation intolerance and gradual loss of white adipose tissue (WAT), liver glycogen, and muscle mass1. Cancer cells also benefit from autophagy. Deletion of essential autophagy genes impairs the metabolism, proliferation, survival and malignancy of spontaneous tumors in autochthonous cancer models6,7. Acute, systemic deletion of Atg7 or acute, systemic expression of a dominant-negative ATG4b in mice induces greater regression of Kras-driven cancers than tumor-specific autophagy deletion, suggesting a role for host autophagy in promoting tumor growth1,8. Here we show that host-specific Atg7 deletion impairs growth of multiple different allografted tumors, although not all tumor lines were sensitive to host autophagy status. Host autophagy loss was associated with reduction in circulating arginine and the sensitive tumor cells lines were arginine auxotrophs due to lack of expression of the enzyme argininosuccinate synthase (ASS1). Serum proteomic analysis identified the arginine-degrading enzyme Arginase I (ARG1) in the circulation of Atg7-deficient hosts, and in vivo arginine metabolic tracing demonstrated degradation of serum arginine to ornithine. ARG1 is predominantly expressed in liver and can be released from hepatocytes into the circulation. Liver-specific Atg7 deletion produced circulating ARG1, and reduced serum arginine and tumor growth. Deletion of Atg5 in the host similarly regulated circulating arginine and tumorigenesis, demonstrating specificity to autophagy function. Dietary supplementation of Atg7-deficient hosts with arginine partially restored circulating arginine levels and tumor growth. Thus, defective host autophagy leads to release of ARG1 from liver and degradation of circulating arginine essential for tumor growth, identifying a novel metabolic vulnerability of cancer.
To validate whether host autophagy promotes tumor growth, we tested the growth of an autophagy-competent C57Bl/6J isogenic BrafV600E/+, Pten−/−, Cdkn2−/− mouse melanoma cell line (YUMM 1.1) in C57Bl/6J host mice without (Atg7+/+) and with (Atg7Δ/Δ) conditional, whole body Atg7 deficiency (Fig. 1a). YUMM 1.1 tumors were significantly smaller when grown on Atg7Δ/Δ compared to Atg7+/+ hosts (Fig. 1b), demonstrating that host autophagy promoted tumor growth. Examination of additional autophagy-competent isogenic C57Bl/6J BrafV600E/+, Pten−/−, Cdkn2−/− YUMM 1.3 melanoma, carcinogen-induced MB49 urothelial carcinoma, and KrasG12D/+, p53−/− 71.8 non-small-cell lung cancer cell lines revealed a similar requirement for host autophagy for tumor growth (Extended data Fig. 1a, c, e). The decreased tumor growth observed in Atg7Δ/Δ hosts was associated with decreased proliferation. In some tumor types, there was also increased apoptosis (Fig. 1c and Extended data Fig. 1b, d, f). Host autophagy was, however, not required for the growth of autophagy-competent isogenic C57Bl/6J BrafV600E/+, Pten−/−, Cdkn2−/− YUMM 1.7 and 1.9 melanoma cell lines (Extended data Fig. 2a-d), indicating that dependency on host autophagy is common, but there are tumor-specific adaptation mechanisms.
Fig. 1: Host autophagy promotes growth of arginine auxotrophic tumors.
a, Experimental design to induce host mice with conditional whole-body Atg7 deletion (Atg7Δ/Δ) and wild type controls (Atg7+/+) with which to assess tumor growth. Ubc-CreERT2/+;Atg7+/+ and Ubc-CreERT2/+;Atg7flox/flox mice were injected with TAM to delete Atg7 and were then injected subcutaneously with tumor cells. Tumor growth was monitored over 3 weeks. b, Comparison of tumor weight between Atg7+/+ (n=5) and Atg7Δ/Δ hosts (n=8). Data are mean +/− S.E.M (****p<0.0001). c, IHC quantification of Ki67 and active Caspase-3 positive cells in tumors from Atg7+/+ and Atg7Δ/Δ hosts. Data are mean +/− S.E.M (***p<0.001). d, Serum metabolites with log2 fold change cut-offs of >1 or <−1 between Atg7+/+ (n=17) and Atg7Δ/Δ (n=17) hosts obtained by LC-MS with p<0.05. e, Illustration of the arginine metabolism. ASS1, Argininosuccinate synthase 1; ASL, Argininosuccinate lyase; ARG1, Arginase 1; NOS, Nitric oxide synthetase; OTC, Ornithine transcarbamylase. f, YUMM 1.1 proliferation in vitro in medium containing different percentages of arginine. Cell density was measured every 2 hours using IncuCyte. Data are representative of 3 independent experiments performed in duplicate. g, Western blotting showing expression of ASS1, ASL and OTC in tumors from Atg7+/+ and Atg7Δ/Δ hosts (n=4 each) representative of 3 independent experiments. Kidney was used as a control tissue for ASS1 and ASL while liver was used for OTC. Actin was used a loading control. In all figures, n= number of mice.
The melanoma cell lines dependent on host autophagy for tumor growth are derived from genetically engineered mouse models of cancer and thereby have a low mutation burden and neoantigen load and fail to provoke an efficient T-cell response9. Nonetheless, autophagy modulates a variety of immune mechanisms that could underlie defective tumor growth on autophagy-deficient hosts. Atg7Δ/Δ hosts did not modify infiltration of YUMM 1.1 tumors with CD3, CD4 or CD8 positive cells (Extended data Fig. 2e). Depletion of CD4+ and CD8+ T-cells modestly increased tumor growth in Atg7+/+ hosts but did not significantly rescue growth in Atg7Δ/Δ hosts (Extended data Fig. 2f). Thus, despite relative increased fraction of myeloid-derived suppressor cells and CD8+ T cells in Atg7Δ/Δ hosts (Extended data Fig. 2g), the decreased tumor growth on Atg7Δ/Δ hosts was not due to induction of an anti-tumor T-cell response.
Autophagy supports metabolism by recycling cargo to provide anabolic and catabolic substrates6. This metabolic recycling function of autophagy promotes mammalian survival during fasting1–3, and tumor cell survival to nutrient limitation4,10,11. One major source of tumor nutrients is from the host blood supply. Accordingly, we tested whether circulating nutrients provided by host autophagy were required for tumor growth. Metabolite profiling of serum from Atg7+/+ and Atg7Δ/Δ hosts identified 12 metabolites that were decreased and 7 that were increased with autophagy knockout (Fig. 1d, Supplementary Table 1 and 2). Serum arginine was strikingly downregulated in Atg7Δ/Δ compared to Atg7+/+ hosts (−2.37 log2 fold change) (Fig. 1d), confirming previous results1.
Arginine is a non-essential amino acid derived from the diet, de novo synthesis and protein turnover, and is important for mTOR activation12, ammonia detoxification through the urea cycle, as well as the synthesis of proteins, creatine, polyamines and nitric oxide (NO)13. It is long known that some human cancers silence expression of ASS1 resulting in arginine auxotrophy14. Without ASS1, cancer cells are unable to synthesize arginine from citrulline and are dependent on exogenous arginine15,16. ASS1 silencing prevents consumption of aspartate by the urea cycle, increasing the availability of this amino acid, which is required for pyrimidine biosynthesis and can become limiting in hypoxia (Fig. 1e)17,18. These findings suggested that low circulating arginine may underlie defective tumor growth on autophagy-deficient hosts.
To determine their requirement for exogenous arginine, YUMM 1.1, 1.3, 1.7, 1.9, MB49 and 71.8 cells were tested for growth without and with arginine. Proliferation was blocked in vitro in complete medium with the sole absence of arginine, and this was not associated with cell death. Growth rates increased with increased percentage of arginine in the medium demonstrating arginine auxotrophy (Fig. 1f and Extended data Fig. 3a). YUMM 1.1 tumors were tested for lack expression of enzymes involved in arginine biosynthesis: ASS1, Argininosuccinate Lyase (ASL) that converts citrulline to arginine, and Ornithine Transcarbamylase (OTC) that converts ornithine to citrulline (Fig. 1e). As previously shown for melanoma19,20, irrespective of use of Atg7+/+ and Atg7Δ/Δ hosts, tumors lacked ASS1 and OTC expression explaining arginine auxotrophy (Fig. 1g). In contrast to tumors, both Atg7+/+ and Atg7Δ/Δ hosts express ASS1, ASL or OTC in liver and kidney suggesting that they are capable of arginine synthesis (Extended data Fig. 3b). Consistent with the YUMM 1.1 and the literature, YUMM 1.7 tumors which grew on Atg7Δ/Δ hosts also lacked expression of ASS1 and OTC, suggesting a mechanism of intrinsic resistance independent of arginine auxotrophy in a subset of tumor cell lines (Extended data Fig. 3c).
To determine how circulating arginine is depleted in Atg7Δ/Δ hosts, we examined the serum proteome by nano LC-MS/MS, identifying 19 proteins which were downregulated and 32 which were upregulated upon loss of Atg7 (Fig. 2a and Supplementary Table 3). Among proteins upregulated in Atg7Δ/Δ hosts serum was ARG1 (2.43 log2 fold change) (Fig. 2b). ARG1 is expressed in liver where it degrades arginine to ornithine. Appearance of ARG1 in serum without altered levels in liver in Atg7Δ/Δ hosts was confirmed by western blotting (Fig. 2c). Serum NO levels were not modified, suggesting that serum arginase did not alter the arginine availability for NO synthesis (Extended data Fig. 3d). Serum arginase activity in vitro was increased as shown by greater 13C6-arginine degradation to 13C5-ornithine in serum from Atg7Δ/Δ hosts (Extended data Fig. 4a). To determine how Atg7 deficiency altered arginine metabolism in vivo, we infused Atg7+/+ and Atg7Δ/Δ hosts with 13C6-15N4-labeled arginine for 3h (Fig. 2d)21. To analyze 13C and 15N enrichment, serum was collected at different times during infusion, and tumors, kidney and liver were collected at the end of the infusion (3h). Atg7Δ/Δ host serum showed decreased arginine (12C, 13C615N4 and 13C515N2) associated with increased ornithine (12C and 13C515N2), indicating degradation of circulating arginine to ornithine (Fig. 2e and Extended data Fig. 4b). Atg7Δ/Δ host kidney showed decreased arginine levels with no change in ornithine or citrulline; no difference was observed in arginine, citrulline or ornithine levels in liver of Atg7Δ/Δ compared to Atg7+/+ hosts (Extended data Fig. 4c-d). Tumors from Atg7Δ/Δ hosts also displayed decreased arginine (12C, 13C615N4 and 13C515N2) and increased ornithine levels (12C and 13C515N2) (Fig. 2f). These results confirm that in ASS1-deficient tumors dependent on exogenous arginine, arginine is depleted, consistent with insufficient circulating arginine in Atg7Δ/Δ hosts.
Fig. 2: Serum ARG1 levels increase in Atg7Δ/Δ hosts and deplete circulating arginine.
a, Comparison of serum proteins between Atg7+/+ and Atg7Δ/Δ hosts (n=5 each) obtained by nano LC-MS/MS with corrected p<0.05. b, Proteins with log2 fold change cut-offs of >1 or <−1 between Atg7+/+ and Atg7Δ/Δ hosts. c, Western blotting showing expression of ARG1 in serum and liver from Atg7+/+ and Atg7Δ/Δ hosts. *p<0.05 compared to Atg7+/+ hosts. Data are representative of 2 independent experiments. Actin and transferrin were used as loading controls. d, Illustration of the 13C615N4-arginine tracer labelling pattern. e, Concentration (μΜ) of arginine, citrulline, and ornithine in serum from Atg7+/+ and Atg7Δ/Δ hosts (n=3 and 4, respectively) after infusion with 13C615N4-arginine. Data are mean +/− S.E.M. f, Concentration (nmol/g) of arginine, citrulline, and ornithine in tumor from Atg7+/+ and Atg7Δ/Δ hosts (n=2 each) after infusion with 13C615N4-arginine. Data are mean (***p<0.001 by Two-way ANOVA test).
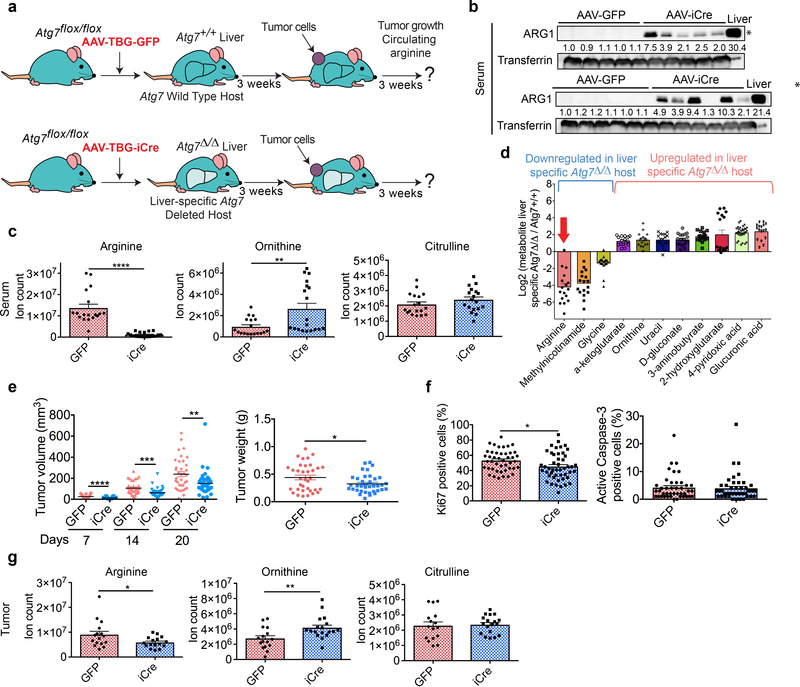
During inflammation, injury and liver disease, ARG1 is released from hepatocytes into the circulation leading to arginine depletion22. Atg7Δ/Δ hosts have steatosis1 and liver-specific deletion of Atg5 or Atg7 is associated with liver damage2,23,24. Accordingly, we hypothesized that ARG1 is released into the circulation following deletion of Atg7 in liver. To test this hypothesis, we deleted Atg7 specifically in the liver and examined arginine and ARG1 levels in the circulation, and tumor growth (Fig. 3a). Injection of AAV-TBG-iCre vector efficiently deleted Atg7 in liver but not in others organs such as brain and kidney (Extended data Fig. 5a-c). As expected, liver-specific Atg7 deletion led to histopathologic changes in liver cells without affecting other tissues (Extended data Fig. 5d). As seen in Atg7Δ/Δ hosts, liver-specific Atg7Δ/Δ hosts showed increased serum ARG1 (Fig. 3b), with reduced arginine and increased ornithine (Fig. 3c) and no change in NO levels (Extended data Fig. 5e). Liver-specific Atg7 deletion also modified levels of other circulating metabolites, with 18 increased and 4 decreased in liver-specific Atg7Δ/Δ compared to Atg7+/+ hosts (Supplementary Table 4 and 5). Some of these circulating metabolites (e.g. 4-pyridoxic acid, D-gluconate and glucuronic acid) were also altered in Atg7Δ/Δ hosts suggesting that their dysregulation had a liver-specific origin (Fig. 1d and Extended data Fig. 5f). Serum arginine was downregulated in liver-specific Atg7Δ/Δ compared to Atg7+/+ hosts (Fig. 3d) as previously shown in Atg7Δ/Δ hosts (Fig. 1d). The weight and volume of melanoma tumors (YUMM 1.1) were significantly decreased in liver-specific Atg7Δ/Δ compared to Atg7+/+ hosts (Fig. 3e), which was associated with decreased proliferation and no change in apoptosis (Fig. 3f). Tumors from liver-specific Atg7Δ/Δ hosts had decreased arginine and increased ornithine, but to a lesser extent than the tumors from Atg7Δ/Δ hosts, which may explain why the decreased tumor growth in liver-specific Atg7Δ/Δ hosts was not as dramatic as with Atg7Δ/Δ hosts (Fig. 3g). In liver-specific Atg7Δ/Δ hosts, autophagy in the microenvironment may locally feed the tumor with amino acids, as previously shown in pancreatic cancer and in Drosophila tumors25,26. These results suggest that Atg7 deletion in the liver is responsible for ARG1 release into the circulation leading to depletion of circulating arginine and decreased tumor growth.
Fig. 3: Atg7 deletion in liver increases serum ARG1, and decreases serum arginine and tumor growth.
a, Experimental design to induce liver-specific deletion of Atg7. Atg7flox/flox mice were injected (tail vein) with AAV-TBG-GFP or AAV-TBG-iCre to delete Atg7 in liver and were injected subcutaneously with tumor cells. Tumor growth was monitored over 3 weeks. b, Western blotting showing expression of ARG1 in serum from Atg7+/+ and liver-specific Atg7Δ/Δ hosts (n=11 each). *p<0.05 compared to Atg7+/+ hosts. Data are representative of 2 independent experiments. Transferrin was used as a loading control. c, Serum arginine, ornithine and citrulline levels in Atg7+/+ and liver-specific Atg7Δ/Δ hosts (n=18 each) obtained by LC-MS. Data are mean +/− S.E.M (**p<0.01, ****p<0.0001). d, Serum metabolites with log2 fold change cut-offs of >1 or <−1 between Atg7+/+ and liver-specific Atg7Δ/Δ hosts (n=17 each) obtained by LC-MS with p<0.05. Data are mean +/− S.E.M. e, Comparison of tumor volume and weight between Atg7+/+ (n=17) and liver-specific Atg7Δ/Δ hosts (n=19). Data are mean +/− S.E.M (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). f, IHC quantification of Ki67 and active Caspase-3 positive cells in tumors from Atg7+/+ and liver-specific Atg7Δ/Δ hosts. Data are mean +/− S.E.M (*p<0.05). g, Tumor arginine, ornithine and citrulline levels in Atg7+/+ (n=16) and liver-specific Atg7Δ/Δ (n=16) hosts obtained by LC-MS. Data are mean +/− S.E.M (*p<0.05, **p<0.01).
To determine if the degradation of circulating arginine by ARG1 in Atg7Δ/Δ hosts was due to loss of autophagy, we examined mice with conditional deletion of Atg5. Whole-body conditional deletion of Atg5 also introduced ARG1 into the circulation and decreased serum arginine, and tumor growth was also decreased on these Atg5Δ/Δ hosts (Extended data Fig. 6). Similar to liver-specific deletion of Atg7, liver-specific deletion of Atg5 led to histopathologic liver changes with increased circulating ARG1 and reduced arginine (Extended data Fig. 7), confirming that modulation of circulating arginine and tumorigenesis was autophagy-dependent.
We next tested whether dietary arginine supplementation can rescue tumor growth in Atg7Δ/Δ hosts (Fig. 4a). Dietary arginine supplementation was able to partially increase serum arginine levels in Atg7Δ/Δ hosts and did not modify ornithine or citrulline levels (Fig. 4b). This increased circulating arginine promoted growth and proliferation of melanoma cell lines YUMM 1.1 and 1.3 in Atg7Δ/Δ compared to Atg7+/+ hosts (Fig. 4c-d and Extended data Fig. 8a-c), confirming that limiting circulating arginine can curtail tumor growth.
Fig. 4: Dietary arginine supplementation rescues tumor growth in Atg7Δ/Δ hosts.
a, Experimental design to perform arginine supplementation and induce conditional whole-body Atg7 deletion to assess YUMM 1.1 tumor growth. Ubc-CreERT2/+;Atg7+/+ and Ubc-CreERT2/+;Atg7flox/flox mice were supplied with supplementary dietary arginine (0 or 1%). Seven days later, TAM was injected to delete Atg7 and mice were injected subcutaneously with tumor cells. Tumor growth was monitored over 3 weeks. b, Serum arginine, ornithine and citrulline in Atg7+/+ (n=5) and Atg7Δ/Δ (n=6) hosts with or without arginine supplementation, obtained by LC-MS. Data are mean +/− S.E.M (*p<0.05, **p<0.01, ***p<0.001). c, Comparison of tumor weight between Atg7+/+ (n=13), Atg7+/+ + 1% arginine (n=13), Atg7Δ/Δ (n=13) and Atg7Δ/Δ + 1% arginine (n=14) hosts. Data are mean +/− S.E.M (**p<0.01, ***p<0.001, ****p<0.0001). d, IHC quantification of Ki67 and active Caspase-3 positive cells in tumors from Atg7+/+ and Atg7Δ/Δ hosts with or without arginine supplementation. Data are mean +/− S.E.M (***p<0.001, ****p<0.0001). e, Model of host autophagy promoting tumor growth.
In summary, autophagy in the liver prevents the release of ARG1 and the degradation of circulating arginine important for the growth of arginine auxotrophic tumors (Fig. 4e). As some tumor cells auxotrophic for arginine in vitro were capable of growth on Atg7Δ/Δ hosts, this suggests the existence of adaptation mechanisms27. Recent work demonstrated that autophagy in the local tumor microenvironment can provide amino acids that promote tumor growth25,26. Our work demonstrates that host autophagy also sustains a circulating amino acid (arginine) essential for tumor growth. This finding underscores the importance of understanding the sensitivity of ASS1-deficient tumors to arginine deprivation therapy28 with or without autophagy inhibition29. As tumor nutrients are mainly derived from the host circulation, restricting essential tumor nutrients in the circulation, as done with asparaginase treatment, is a form of cancer therapy ripe to be exploited further30.
METHODS
Mice.
All animal care and treatments were carried out in compliance with Rutgers University Institutional Animal Care and Use Committee guidelines (IACUC). Mice for conditional whole-body Atg7 deletion (C57Bl/6J Ubc-CreERT2/+;Atg7flox/flox) were engineered with floxed alleles of Atg7 (Atg7flox/flox)2 and a transgene expressing the Tamoxifen (TAM)-regulated Cre recombinase fusion protein under the control of the ubiquitously expressed ubiquitin C promoter (Ubc)31 as previously described1. Acute deletion of Atg7 throughout the mouse is obtained after TAM injection1. TAM (T5648, Sigma) was suspended at a concentration of 20 mg/ml, in a mixture of 98% sunflower seed oil and 2% ethanol and 200 μl per 25 g of body weight was injected intraperitoneally into 8 to 10 weeks old male Ubc-CreERT2/+;Atg7+/+ or Ubc-CreERT2/+;Atg7flox/flox mice once per day for 4 days to generate cohorts of Atg7 deleted (Atg7Δ/Δ) and wild type (Atg7+/+) control host mice. To assess the consequence of acute Atg7 deletion on tumorigenesis of C57Bl/6J isogenic male tumor cells, one week post TAM treatment, YUMM 1.1 (1 × 106 cells), 1.3 (2 × 106 cells), 1.7 (0.1 × 106 cells), 1.9 (1 × 106 cells), 71.8 (1 × 106 cells) or MB49 (0.25 × 106 cells) cells were resuspended in 100 μL PBS and injected subcutaneously into the dorsal flanks of mice. Three weeks post cell injection, mice were sacrificed and serum and tumors were collected. The maximal tumor volume (1700 mm3) permitted by Rutgers University IACUC was never exceeded. For arginine supplementation, 1% arginine (A8094, Sigma) in drinking water was given to the mice a week before TAM and throughout the experiment.
Mice for conditional whole-body Atg5 deletion (C57Bl/6J Ubc-CreERT2/+;Atg5flox/flox) were engineered with floxed alleles of Atg5 (Atg5flox/flox)32 and a transgene expressing the Tamoxifen (TAM)-regulated Cre recombinase fusion protein under the control of the ubiquitously expressed ubiquitin C promoter (Ubc)31. Acute deletion of Atg5 throughout mice was obtained after TAM injection (200 μl of TAM per 25 g of body weight injected intraperitoneally into 8 to 10 week old male Ubc-CreERT2/+;Atg5+/+ or Ubc-CreERT2/+;Atg5flox/flox mice once a week for 4 weeks) to generate cohorts of Atg5 deleted (Atg5Δ/Δ) and wild type (Atg5+/+) control host mice.
Liver-specific Atg7 and Atg5 deletion was achieved by injected an Adeno Associated Virus (AAV)-thyroxine binding globulin (TBG) promoter-Cre recombinase vector (AAV-TBG-iCre, Vector Biolabs) to Atg7flox/flox and Atg5flox/flox mice. An AAV-TBG promoter-GFP vector (AAV-TBG-GFP, Vector Biolabs) was injected to Atg7flox/flox and Atg5flox/flox mice as a control. 1.5 × 1011 genome copy (g.c) of either AAV-TBG-iCre or AAV-TBG-GFP vectors in 100μl PBS was injected into the tail vein of 8 to 10 week old male Atg7flox/flox and Atg5flox/flox mice to generate liver-specific Atg7Δ/Δ or Atg5Δ/Δ and Atg7+/+ or Atg5+/+ control mice, respectively. Three weeks post injection, YUMM 1.1 cells (1 × 106 cells) were resuspended in 100 μL PBS and injected subcutaneously into the dorsal flanks of the liver-specific Atg7Δ/Δ and Atg7+/+ control mice. Tumor growth was monitored daily. Tumor volume was calculated with the following formula: volume = π/6 x L x W x H. Three weeks post cell injection, mice were sacrificed and liver, kidney, brain, serum and tumors were collected.
Cell lines.
Cell culture.
Cell lines were authenticated using whole exome sequencing. YUMM 1.1, 1.3, 1.7 and 1.9 cells derived from BrafV600E/+, Pten−/−, Cdkn2−/− C57Bl/6J mouse melanomas were generated previously33 and cultured in Dulbecco’s minimum essential medium and Ham’s F12 (DMEM-F12) (10–092-CV, Corning) supplemented with 10% fetal bovine serum (FBS) (F0926, Sigma) in a 5% CO2 incubator at 37°C. Mouse lung cancer cell line 71.8 was derived from p53−/−, KrasG12D/+ mouse lung tumors previously11 and the MB49 cell line34 was provided by the Ratliff laboratory and cultured in Roswell Park Memorial Institute medium (RPMI) (11875–093, Gibco). Cells were tested negative for mycoplasma contamination.
Cell proliferation in arginine-deficient medium.
YUMM 1.1, 1.3, 1.7, 1.9, 71.8 and MB49 cell were seeded at a density of 15,000 cells per well in 24-well plates. The following day, cells were washed with phosphate-buffered saline (PBS) (14190–144, Gibco) and cultured in arginine-free DMEM-F12 (DFL27, Caisson Labs) or arginine-free RPMI (R1780, Sigma) supplemented with 10% dialyzed FBS (89986, Thermo Scientific) and an increasing percentage of arginine from 2.5 to 100%. Growth was assessed using an IncuCyte ZOOM™ with images of the proliferative cells recorded every 2 hours for a total duration of 6 days.
Metabolite analysis by LC-MS.
Metabolite extraction for LC-MS.
Metabolites from 10 μl serum samples were first extracted with 40 μl of ice-cold methanol. The mixture was allowed to sit at −20°C for 20 min, and then centrifuged at 16,000×g for 10 min at 4°C. Supernatants were transferred to clean tubes and pellets were extracted again with 200 μl 40:40:20 methanol:acetonitrile:H2O. The mixture was allowed to sit on ice for 10 min, and then centrifuged at 16,000×g for 10 min at 4°C. Supernatants were combined with the first extraction, resulting in roughly 240 μl of extract. Extracts were further processed with Phree® Phospholipid Removal 1mL Tube (Phenomenex) according to the manufacturer’s instructions. The final extract was stored at −80°C until analyzed by LC-MS. To extract metabolites from the tissues and tumors, samples (25 mg) were first pulverized using a Cryomill (Retsch) in liquid nitrogen at 25Hz for 2 min. Extraction was performed by adding −20°C 40:40:20 methanol:acetonitrile:water with 0.5% formic acid solution (500 μl) to the ground samples, followed by vortexing and centrifugation at 16,000×g for 10 min at 4°C. The supernatants were transferred to clean tubes and the pellets were extracted again by repeating the previous step. The supernatant was then combined with the first extract. 500 μl of extract was neutralized with 44 μl of 15% NH4HCO3 solution and centrifuged at 16,000×g for 10 min at 4°C to remove protein precipitate. 300 μl of supernatant was removed to clean tubes and stored at −80°C until analyzed by LC-MS.
LC-MS analysis.
LC−MS analysis of the extracted metabolites was performed on a Q Exactive PLUS hybrid quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific) coupled to hydrophilic interaction chromatography (HILIC). The LC separation was performed on UltiMate 3000 UHPLC system with an XBridge BEH Amide column (150 mm × 2.1 mm, 2.5 μM particle size, Waters) with the corresponding XP VanGuard Cartridge. The liquid chromatography used a gradient of solvent A (95%:5% H2O:acetonitrile with 20mM ammonium acetate, 20mM ammonium hydroxide, pH 9.4), and solvent B (20%:80% H2O:acetonitrile with 20 mM ammonium acetate, 20 mM ammonium hydroxide, pH 9.4). The gradient was 0 min, 100% B; 3 min, 100% B; 3.2 min, 90% B; 6.2 min, 90% B; 6.5 min, 80% B; 10.5 min, 80% B; 10.7 min, 70% B; 13.5 min, 70% B; 13.7 min, 45% B; 16 min, 45% B; 16.5 min, 100% B. The flow rate was 300 μl/min. Injection volume was 5 μl and column temperature 25°C. The MS scans were in negative ion mode with a resolution of 70,000 at m/z 200. The automatic gain control (AGC) target was 3 × 106 and the scan range was 75−1000. In order to increase metabolome coverage, the samples were also analyzed with a secondary LC-MS method, which involves two separate instrument platforms covering both positive charged and negative charged metabolites. Negative charged metabolites were analyzed via reverse-phase ion-pairing chromatography coupled to an Exactive orbitrap mass spectrometer (ThermoFisher Scientific). The mass spectrometer was operated in negative ion mode with resolving power of 100,000 at m/z 200, scanning range being m/z 75–1000. The LC method has been described previously35, using a Synergy Hydro-RP column (100 mm × 2 mm, 2.5 μm particle size, Phenomenex) with a flow rate of 200 μL/min. The LC gradient was 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; 25 min, 0% B. Solvent A is 97:3 water:methanol with 10 mM tributylamine and 15 mM acetic acid; solvent B is methanol. Positive charged metabolites were analyzed on a Q Exactive Plus mass spectrometer coupled to Vanquish UHPLC system (ThermoFisher Scientific). The mass spectrometer was operated in positive ion mode with resolving power of 140,000 at m/z 200, scanning range being m/z 75– 1000. The LC separation was achieved on an Agilent Poroshell 120 Bonus-RP column (150 × 2.1 mm, 2.7 μm particle size). The gradient was 0 min, 50 μL/min, 0.0%B; 6 min, 50 μL/min, 0% B; 12 min, 200 μL/min, 70% B; 14 min, 200 μL/min, 100 %B; 18 min, 200 μL/min, 100% B; 19 min, 200 μL/min, 0% B; 24 min, 200 μL/min, 0% B; 25 min, 50 μL/min, 0% B. Solvent A is 10mM ammonium acetate + 0.1% acetic acid in 98:2 water:acetonitrile and solvent B is acetonitrile36. Metabolite features were extracted in MAVEN v70737 with the labeled isotope specified and a mass accuracy window of 5 ppm. For the 13C-15N arginine infusions, the isotope natural abundance and impurity of labeled substrate was corrected using a matrix-based algorithm.
Labelled arginine infusion.
For jugular vein catheterization, the procedure was modified from work previously described38. In brief, Atg7Δ/Δ and Atg7+/+ mice were anesthetized using isoflurane carried by oxygen, followed by placement of a central venous catheter (polyurethane tubing, 1F in O.D.) (SAI Infusion Technologies) into the right jugular vein. A minimal amount of blood was carefully withdrawn to verify the catheter patency. Afterwards, the saline solution in the catheter was replaced by heparin/glycerol catheter lock solution (SAI Infusion Technologies). The proximal end of the catheter was then tunneled subcutaneously, exited between the shoulder blades, and properly secured. A fully recovered surgical mouse was placed in a plastic harness (SAI Infusion Technologies), and the catheter was connected to an infusion pump (New Era Pump System, Inc) through a mouse tether and swivel system (Instech Laboratories). Arginine isotope tracer (13C615N4, CNLM-539-H-PK, Cambridge Isotope Laboratories) was dissolved in sterile saline and infused at a rate of 3.5 nmol/g/min (0.1 μl/g/min) for 3 hours. Infusion rate was determined using turnover flux calculations21. Mice were sacrificed after infusion for serum, tumor, liver and kidney analysis by LC-MS. The isotope natural abundance and impurity of labeled substrate was corrected using a matrix-based algorithm. The construction of the purity matrix and C/N joint correction matrix is similar to AccuCor39. For calculation of the circulating amino acid concentration, the average ion counts from the Atg7+/+ mice were normalized to the amino acid concentration measured previously40. The amino acid concentration in the Atg7Δ/Δ mice were calculated proportionally.
Arginase activity assay.
To follow conversion of arginine to ornithine, 15 μl serum samples were added to 5 μl of 9.7 mM MnCl2, 5 μl of 360 mM pH 9.7 glycine and 5 μl of 300 μM 13C6-arginine followed by incubation at 37°C for 0, 5, 20, 60 or 120 min. 870 μl 40:40:20 methanol:acetonitrile:water with 0.5% formic acid solution were added to stop the reaction; the mixture was allowed to sit on ice for 10 min. The extract was neutralized with 40 μl of 15% NH4HCO3 solution and centrifuged at 16,000×g for 10 min at 4°C. 500 μl of supernatant was removed to clean tubes and stored at −80°C until analyzed by LC-MS. The LC−MS analysis was performed on the Q Exactive PLUS mass spectrometer coupled to UltiMate 3000 UHPLC system with an XBridge BEH Amide column (150 mm × 2.1 mm, 2.5 μM particle size, Waters, Milford, MA) with the corresponding XP VanGuard Cartridge. The liquid chromatography used a 6-min isocratic elution of 28% solvent A (95%:5% H2O:acetonitrile with 20mM ammonium acetate, 20 mM ammonium hydroxide, pH 9.4) and 72% solvent B (20%:80% H2O:acetonitrile with 20 mM ammonium acetate, 20 mM ammonium hydroxide, pH 9.4). The flow rate was 300 μl/min. Injection volume was 5 μl and column temperature 25°C. The MS scans were in negative ion mode with a resolution of 70,000 at m/z 200. The automatic gain control (AGC) target was 3 × 106 and the scan range was 75−1000. Metabolite features were extracted in MAVEN v707 with the labeled isotope specified and a mass accuracy window of 5 ppm.
Proteomic Analysis by LC-MS.
Technical duplicate of pooled serum samples from both Atg7+/+ (n=5) and Atg7Δ/Δ (n=5) mice were processed in parallel. Two different methods were also used to reduce the amount of the major serum proteins to allow detection of rarer components: AlbuVoid™ (Biotech Support Group, LLC) was used to deplete albumin while the Agilent multiple affinity removal spin cartridge mouse 3 system (Mars3) was used to remove albumin, IgG, and transferrin following the manufacturer’s protocol. Untreated or depleted sera were loaded onto NuPage™ 10% Bis-Tris Gel (Invitrogen), run a short distance into the gel, and proteins reduced, alkylated and digested with trypsin as described41. Digests were analyzed by nano LC-MS/MS using a Dionex Ultimate 3000 RLSC nano System interfaced with Q Exactive HF (Thermofisher). Peptides were loaded onto a self-packed 100 μm x 2 cm trap (Magic C18AQ, 5 μm 200 Å, Michrom Bio resources, Inc.) and washed with Buffer A (0.1% trifluoroacetic acid) for 5 min with a flow rate of 10 μl/min. The trap was brought in-line with the analytical column (self-packed Magic C18AQ, 3 μm 200 Å, 75 μm x 50 cm) and fractionated at 300 nl/min using a segmented linear gradient of 4–15% B in 30 min (A: 0.2% formic acid; B: 0.16% formic acid/80% acetonitrile), 15–25%B in 40min, 25–50% in 44 min and 50–90%B in 11 min. Mass spectrometry data was acquired using a data-dependent acquisition procedure with each cycle consisting of a MS1 scan (resolution 120,000) followed MS/MS scans (HCD relative collision energy 27%, resolution 30,000) of the 20 most intense ions using a dynamic exclusion duration of 20 sec. The raw data was converted into MASCOT Generic Format (MGF) using Proteome Discover 2.1 (ThermoFisher) and searched against the Ensemble mouse database and a database of common laboratory contaminants (http://www.thegpm.org/crap/) using a local implementation of the global proteome machine (GPM Fury)42. Peptide spectrum matches were assigned to genes using BioMart Ensembl tables. To estimate differential abundances of proteins, data from all LC-MS runs were combined (neat, Albivoid-depleted, and Mars3-depleted for each of the four samples). For mouse proteins having 10 or more spectral counts, differential expression was estimated using the QLSpline option of the QuasiSeq package (https://cran.r-project.org/web/packages/QuasiSeq/index.html)43. Data is presented as thresholded log2 fold change of Atg7Δ/Δ/Atg7+/+ with adjusted p-values <0.05. p-values were adjusted using Holm correction using the “p.adjust” function in the base R package (https://cran.r-project.org). The raw mass spectrometry data have been deposited in the MassIVE repository, entry MSV000082879.
Enzymatic assays.
Serum nitric oxide levels were determined with the nitric oxide assay kit (ab65328, Abcam).
Histology.
Mouse tissues were fixed in 10% buffer formalin solution overnight and then transferred to 70% ethanol for paraffin-embedded sections. Tissue sections were deparaffinized, rehydrated and boiled for 45 min in 10 mM pH 6 Citrate buffer. Slides were blocked in 10% goat serum for an hour and then incubated at 4˚C overnight with primary antibody against Ki67 (1:200, Ab15580, Abcam), active Caspase-3 (1:300, #9661, Cell Signaling), CD3 (1:100, Ab16669, Abcam), CD4 (1:1,000, Ab183685, Abcam) and CD8 (1:100, 14–0808-82, Invitrogen). The following day, tissue sections were incubated with biotin-conjugated secondary antibody for 15min (Vector Laboratories), 3% hydrogen peroxide for 5 min, horseradish peroxidase streptavidin for 15 min (SA-5704, Vector Laboratories) and developed by 3,3-diaminobenzidine (Vector Laboratories) followed by hematoxylin staining (3536–16, Ricca). Sections were then dehydrated, mounted in Cytoseal 60 mounting medium (8310, Thermo Scientific) and analyzed using Nikon Eclipse 80i microscope. For quantification of IHC, at least 10 images containing a minimum of 100 cells were analyzed at 60X magnification for each genotype.
Western Blotting.
Tissues and tumor samples were grounded in liquid nitrogen, lysed in Tris lysis buffer (50 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 0.1% NP40, 5 mM MgCl2, 10% glycerol), separated on 12.5% SDS-PAGE gel and then transferred on PVDF membrane (Millipore). Membranes were blocked with 5% non-fat milk for 1 hour and probed overnight at 4˚C with antibodies against ASS1 (1:1,000, Ab170952, Abcam), ASL (1:500, sc-374353, Santa Cruz), OTC (1:500, sc-515791, Santa Cruz), ARG1 (1:500, sc-271430, Santa Cruz), ATG7 (1:2,000, A2856, Sigma), transferrin (1:1,000, sc-22597, Santa Cruz), ATG5 (1:1,500, Ab108327, Abcam) and β-actin (1:5,000, A1978, Sigma). Immunoreactive bands were detected using peroxidase-conjugated antibody (GE Healthcare) and enhanced chemiluminescence detection reagents (NEL105001EA, Perkin Elmer) and were analyzed using the ChemiDoc XRS+ system (Biorad). Protein levels were quantified using the Image Lab v6.0.1 software. Antibodies were validated with the use of positive and negative control following manufacturer’s protocol.
T cell depletion and flow cytometry.
A week post TAM and every 5 days, 200 μg of CD4 (clone GK1.5; BE003–1, BioXCell) and CD8 (clone 2.43; BE0061, BioXCell) antibodies were injected intraperitoneally into Atg7Δ/Δ and Atg7+/+ mice. Two days after the first antibody injection, YUMM 1.1 (1 × 106 cells) cells were resuspended in 100 mL PBS and injected subcutaneously into the dorsal flanks of the mice. Three weeks post cell injection, mice were sacrificed and tumors and spleen were collected. Tumors were homogenized in PBS in a gentleMACS Octo Dissociator (Miltenyi Biotec Inc.) according to manufacturer’s protocol, and passed through a 70 mm cell restrainer. Spleens were grounded with a rubber grinder through steel mesh, treated with ACK Lysis Buffer to remove erythrocytes and passed through a 70 mm cell restrainer. Nonspecific binding of antibodies to cell Fc receptors was blocked using 20 mL/107 cells of FcR blocker (Miltenyi Biotec Inc). Cell surface immunostaining was performed with the following antibodies (1:200) : CD11c-PE-eFluor610 (clone N418, 61–0114-82), CD4-APC (clone GK1.5, 17–0041-82) CD3-AF700 (clone 17A2, 56–0032-82) and CD11b-APC-Cy7 (clone M1/70, A15390) (eBioscience); and CD45-FITC (clone 30-F11, 103107), MHC-II-BV605 (clone M5/114.15.2, 107639), Ly6G-BV650 (clone 1A8, 127641) and CD8-BV785 (clone 53.67, 100749) (BioLegend). Aqua Live/Dead (Invitrogen) was included to determine live cells. After staining of surface markers, cells were fixed and permeabilized using Transcription Factor staining kit and stained with FoxP3-eFluor450 (eBioscience). Cell staining data were acquired using a LSR-II flow cytometer (BD Biosciences, BD FACS Diva v2 software) and analyzed with FlowJo v10 software (Tree Star). Live lymphocytes were gated using forward scatter area (FSC-A) versus side scatter area (SSC-A), followed by FSC-A versus forward scatter height (FSC-H), SSC-A versus side scatter height (SSC-H) plots, forward scatter width (FSC-W) versus side scatter width (SSC-W), and Aqua Live/Dead. Populations were gated as follows: CD45 (%CD45+ of total live lymphocytes), CD3 (%CD3+ of CD11b-, CD11c-, CD45+), CD8 (%CD8+ of CD3), CD4 (%CD4+ of CD3), Treg (%FoxP3+ of CD4), DC (%CD11c+ of MHC-II+, CD45+), MDSC (%Ly6G, CD11b+ of MHC-II-, CD45+).
Antibodies for western blotting, flow cytometry and immunohistochemistry were validated with the use of positive and negative control (gene knock-outs and through the use of control tissues and cell lines) and following manufacturer’s protocol.
Statistics.
All statistical analyses were performed with Prism v7 software using two-sided Student’s t-test, unless specified otherwise. The sample size was chosen in advance on the basis of common practice of the described experiment and is mentioned for each experiment. No statistical methods were used to predetermine sample size. Each experiment was conducted with biological replicates and repeated multiple times. All attempts at replication were successful and no data were excluded. Mice were randomly allocated to experimental groups and the investigators were not blinded during the experiments and outcome assessment.
Extended Data
Extended data Fig. 1: Host autophagy promotes growth of different tumor cell types.
a-c-e, Comparison of tumor weight between Atg7+/+ (n=5) and Atg7Δ/Δ (a, n=4; c, n=5; e, n=4) hosts after injection of 1.3 (a), MB49 (c) or 71.8 (e) cells. Data are mean +/− S.E.M (*p<0.05, **p<0.01. b-d-f, IHC quantification of Ki67 and active Caspase-3 positive cells in tumors from Atg7+/+ and Atg7Δ/Δ hosts. Data are mean +/− S.E.M (*p<0.05, ***p<0.001, ****p<0.0001).
Extended data Fig. 2: Immune response is not involved in decreased tumor growth observed in Atg7Δ/Δ hosts.
a-c, Comparison of tumor weight between Atg7+/+ (n=5) and Atg7Δ/Δ (a, n=5; c, n=6) hosts after injection of 1.7 (a), 1.9 (c) cells. Data are mean +/− S.E.M. b-d, IHC quantification of Ki67 and active Caspase-3 positive cells in 1.7 (b) and 1.9 (d) tumors from Atg7+/+ and Atg7Δ/Δ hosts. Data are mean +/− S.E.M. e, Representative IHC pictures and quantification of CD3, CD4 and CD8 positive cells in tumors from Atg7+/+ and Atg7Δ/Δ hosts. Data are mean +/− S.E.M. f, Comparison of tumor volume and weight between Atg7+/+ (n=10), Atg7+/+ + CD4/CD8 antibody depletion (n=15), Atg7Δ/Δ (n=7) and Atg7Δ/Δ + CD4/CD8 antibody depletion (n=8) hosts. Data are mean +/− S.E.M (*p<0.05, ****p<0.0001). g, Immune components fold change between Atg7+/+ and Atg7Δ/Δ with or without antibody depletion (n=5 each). Treg, T regulatory cells; DC, dendritic cells; MDSC, myeloid-derived suppressor cells. Data are mean +/− S.E.M (***p<0.001, ****p<0.0001) by Two-way ANOVA test.
Extended data Fig. 3: Tumor cells are arginine auxotrophs.
a, YUMM 1.3, 71.8, MB49 and YUMM 1.7, 1.9 proliferation in vitro in medium containing different percentage of arginine. Cell density was measured every 2 hours using the IncuCyte. Data are representative of 3 independent experiments performed in duplicate. b, Western blotting showing expression of ASS1, ASL and OTC in kidney and liver from Atg7+/+ and Atg7Δ/Δ hosts. *p<0.05 compared to Atg7+/+ hosts. Data are representative of 3 independent experiments. Actin was used as a loading (Kidney ASL, Liver OTC) and processing (Kidney ASS1, Liver ASS1 and ASL) control. c, Western blotting showing expression of ASS1, ASL and OTC in YUMM 1.7 tumors from Atg7+/+ and Atg7Δ/Δ hosts. Data are representative of 2 independent experiments. Actin was used as a loading (OTC) and processing (ASS1, ASL) control. d, Analysis of serum nitric oxide levels in Atg7+/+ (n=11) and Atg7Δ/Δ (n=9) hosts. Data are mean +/− S.E.M.
Extended data Fig. 4: Atg7 deletion increases serum arginine degradation but does not modify arginine metabolism in kidney and liver.
a, Serum 13C6-arginine and 13C5-ornithine in Atg7+/+ and Atg7Δ/Δ hosts (n=3 each) overtime. Data are mean +/− S.E.M. b, Concentration (μΜ) of arginine, citrulline, and ornithine in serum from Atg7+/+ (n=3) and Atg7Δ/Δ hosts (n=4) after infusion with 13C615N4-arginine. c-d, Concentration (nmol/g) of arginine, citrulline, and ornithine in kidney (c) and liver (d) from Atg7+/+ and Atg7Δ/Δ hosts (n=2 each) after infusion with 13C615N4-arginine. Data are mean (**p<0.01 by Two-way ANOVA test).
Extended data Fig. 5: Liver-specific Atg7 deletion lead to liver cell enlargement without affecting other tissues.
a-b-c, Western blotting showing expression of Atg7 in liver (n=11 each) (a), brain (n= 9 and 11, respectively) (b) and kidney (n=10 each) (c) from Atg7+/+ and liver-specific Atg7Δ/Δ hosts. *p<0.05 compared to Atg7+/+ hosts. Data are representative of 2 independent experiments. Actin was used as a loading control. d, Representative H&E tissue staining from Atg7+/+ and liver-specific Atg7Δ/Δ hosts. Pictures are representative of 2 independent experiments. e, Analysis of serum nitric oxide levels in Atg7+/+(n=13) and liver-specific Atg7Δ/Δ (n=15) hosts. Data are mean +/− S.E.M. f, Comparison of serum metabolites significantly regulated in Atg7Δ/Δ and liver-specific Atg7Δ/Δ hosts (n=17 each, p<0.05).
Extended data Fig. 6: Atg5 deletion increased serum ARG1, decreased serum arginine and tumor growth.
a, Experimental design to induce host mice with conditional whole-body Atg5 deletion (Atg5Δ/Δ) and wild type controls (Atg5+/+) with which to assess tumor growth. Ubc-CreERT2/+;Atg5+/+ and Ubc-CreERT2/+;Atg5flox/flox mice were injected with TAM at 8 to 10 weeks of age to delete Atg5 and create Atg5+/+ and Atg5Δ/Δ hosts. Mice were then injected subcutaneously with tumor cells and tumor growth was monitored over 3 weeks. b, Comparison of tumor weight between Atg5+/+ (n=4) and Atg5Δ/Δ hosts (n=3). Data are mean +/− S.E.M (**p<0.01). c, IHC quantification of Ki67 and active Caspase-3 positive cells in tumors from Atg5+/+ and Atg5Δ/Δ hosts. Data are mean +/− S.E.M (****p<0.0001). d, Western blotting showing expression of ARG1 in serum from Atg5+/+ (n=3), Atg5Δ/Δ (n=4) and Atg7Δ/Δ (n=3) hosts. *p<0.05 compared to Atg5+/+ hosts. Transferrin was used as a loading control e, Serum arginine, ornithine and citrulline levels in Atg5+/+ (n=4) and Atg5Δ/Δ (n=3) hosts obtained by LC-MS. Data are mean +/− S.E.M (*p<0.05, **p<0.01).
Extended data Fig. 7: Liver-specific Atg5 deleted hosts present liver cell enlargement, increased serum ARG1 and decreased serum arginine.
a, Experimental design to induce liver-specific deletion of Atg5. Atg5flox/flox mice were injected (tail vein) with AAV-TBG-GFP or AAV-TBG-iCre at 8 to 10 weeks of age to delete Atg5 in liver and create Atg5+/+ and liver-specific Atg5Δ/Δ hosts, respectively. b, Western blotting showing expression of Atg5 in liver, brain and kidney from Atg5+/+ and liver-specific Atg5Δ/Δ hosts (n=6 each). *p<0.05 compared to Atg5+/+ hosts. Actin was used as a loading control c, H&E tissue staining from Atg5+/+ and liver-specific Atg5Δ/Δ hosts (n=6 each). d, Western blotting showing expression of ARG1 in serum from Atg5+/+ and liver-specific Atg5Δ/Δ hosts (n=6 each). *p<0.05 compared to Atg5+/+ hosts. Transferrin was used as a loading control e, Serum arginine, ornithine and citrulline levels in Atg5+/+ and liver-specific Atg5Δ/Δ hosts (n=6 each) obtained by LC-MS. Data are mean +/− S.E.M (***p<0.001, ****p<0.0001).
Extended data Fig. 8: Dietary arginine supplementation rescues YUMM 1.3 tumor growth in Atg7Δ/Δ hosts.
a, Serum arginine, ornithine and citrulline in Atg7+/+ (n=5), Atg7+/+ + 1% arg (n=5), Atg7Δ/Δ (n=6) and Atg7Δ/Δ + 1% arg (n=6) hosts, obtained by LC-MS. Data are mean +/− S.E.M (*p<0.05, **p<0.01). b, Comparison of YUMM 1.3 tumor weight between Atg7+/+ and Atg7Δ/Δ (n=5 each) hosts with or without arginine supplementation. Data are mean +/− S.E.M. (**p<0.01, ****p<0.0001). c, IHC quantification of Ki67 and active Caspase-3 positive cells in tumors from Atg7+/+ and Atg7Δ/Δ hosts with or without arginine supplementation. Data are mean +/− S.E.M (**p<0.01, ****p<0.0001).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants: R01CA130893, R01CA193970 (to EW), R01CA163591 (to EW and JDR), K22CA190521 (to JYG), R50CA211437 (to WL), R01CA193970 (to JMM), P30CA072720 (Rutgers Cancer Center), and S10OD016400 (Rutgers Biological Mass Spectrometry Facility), and the V Foundation for Cancer Research (to JMM). Laura Poillet-Perez received support from a postdoctoral fellowship from the New Jersey Commission for Cancer Research (DHFS16PPC034). We thank the Rutgers Cancer Institute Metabolomics Service Shared Resource, the Rutgers-New Brunswick/Robert Wood Johnson Medical School Biological Mass Spectrometry Facility for mass spectrometry analysis and Dirk Moore for the statistical analysis of the proteomics data. Illustrations used in Fig. 4e are licensed under the Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/ or send a letter to Creative Commons, PO Box 1866, Mountain View, CA 94042, USA.
Footnotes
Supplementary Information is available in the online version of the paper.
Data availability. All data are available from the authors upon reasonable request. Source Data for Figs 1d, 2b, 3d are provided with the online version of the paper. The raw mass spectrometry data have been deposited in the MassIVE repository, entry MSV000082879.
The other authors declare no competing financial interests.
References
- 1.Karsli-Uzunbas G et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4, 914–927, doi:10.1158/2159-8290.CD-14-0363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komatsu M et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169, 425–434, doi:10.1083/jcb.200412022 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuma A et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036, doi:10.1038/nature03029 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Guo JY et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev 30, 1704–1717, doi:10.1101/gad.283416.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada Y, Sekito T & Ohsumi Y Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol 279, 73–84 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Kimmelman AC & White E Autophagy and Tumor Metabolism. Cell Metab 25, 1037–1043, doi:10.1016/j.cmet.2017.04.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaravadi R, Kimmelman AC & White E Recent insights into the function of autophagy in cancer. Genes Dev 30, 1913–1930, doi:10.1101/gad.287524.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang A et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer Discov, doi:10.1158/2159-8290.CD-17-0952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res 30, 428–435, doi:10.1111/pcmr.12591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strohecker AM et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov 3, 1272–1285, doi:10.1158/2159-8290.CD-13-0397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JY et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27, 1447–1461, doi:10.1101/gad.219642.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chantranupong L et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164, doi:10.1016/j.cell.2016.02.035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris SM Jr. Arginine metabolism: boundaries of our knowledge. J Nutr 137, 1602S–1609S (2007). [DOI] [PubMed] [Google Scholar]
- 14.Delage B et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J Cancer 126, 2762–2772, doi:10.1002/ijc.25202 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Dillon BJ et al. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer 100, 826–833, doi:10.1002/cncr.20057 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Patil MD, Bhaumik J, Babykutty S, Banerjee UC & Fukumura D Arginine dependence of tumor cells: targeting a chink in cancer’s armor. Oncogene 35, 4957–4972, doi:10.1038/onc.2016.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabinovich S et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383, doi:10.1038/nature15529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagamani SC & Erez A A metabolic link between the urea cycle and cancer cell proliferation. Mol Cell Oncol 3, e1127314, doi:10.1080/23723556.2015.1127314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feun LG et al. Negative argininosuccinate synthetase expression in melanoma tumours may predict clinical benefit from arginine-depleting therapy with pegylated arginine deiminase. Br J Cancer 106, 1481–1485, doi:10.1038/bjc.2012.106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam TL et al. Recombinant human arginase inhibits the in vitro and in vivo proliferation of human melanoma by inducing cell cycle arrest and apoptosis. Pigment Cell Melanoma Res 24, 366–376, doi:10.1111/j.1755-148X.2010.00798.x (2011). [DOI] [PubMed] [Google Scholar]
- 21.Hui S et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118, doi:10.1038/nature24057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris SM Jr. Arginases and arginine deficiency syndromes. Curr Opin Clin Nutr Metab Care 15, 64–70, doi:10.1097/MCO.0b013e32834d1a08 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamura A et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25, 795–800, doi:10.1101/gad.2016211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu M et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163, doi:10.1016/j.cell.2007.10.035 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Sousa CM et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483, doi:10.1038/nature19084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katheder NS et al. Microenvironmental autophagy promotes tumour growth. Nature 541, 417–420, doi:10.1038/nature20815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer JC et al. Arginine Deprivation Inhibits the Warburg Effect and Upregulates Glutamine Anaplerosis and Serine Biosynthesis in ASS1-Deficient Cancers. Cell Rep 18, 991–1004, doi:10.1016/j.celrep.2016.12.077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yau T et al. A phase 1 dose-escalating study of pegylated recombinant human arginase 1 (Peg-rhArg1) in patients with advanced hepatocellular carcinoma. Invest New Drugs 31, 99–107, doi:10.1007/s10637-012-9807-9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen W et al. A novel and promising therapeutic approach for NSCLC: recombinant human arginase alone or combined with autophagy inhibitor. Cell Death Dis 8, e2720, doi:10.1038/cddis.2017.137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koprivnikar J, McCloskey J & Faderl S Safety, efficacy, and clinical utility of asparaginase in the treatment of adult patients with acute lymphoblastic leukemia. Onco Targets Ther 10, 1413–1422, doi:10.2147/OTT.S106810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruzankina Y et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126, doi:10.1016/j.stem.2007.03.002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara T et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889, doi:10.1038/nature04724 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Meeth K, Wang JX, Micevic G, Damsky W & Bosenberg MW The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res 29, 590–597, doi:10.1111/pcmr.12498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Summerhayes IC & Franks LM Effects of donor age on neoplastic transformation of adult mouse bladder epithelium in vitro. J Natl Cancer Inst 62, 1017–1023 (1979). [PubMed] [Google Scholar]
- 35.Lu W et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem 82, 3212–3221, doi:10.1021/ac902837x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papazyan R et al. Physiological Suppression of Lipotoxic Liver Damage by Complementary Actions of HDAC3 and SCAP/SREBP. Cell Metab 24, 863–874, doi:10.1016/j.cmet.2016.10.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melamud E, Vastag L & Rabinowitz JD Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82, 9818–9826, doi:10.1021/ac1021166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan L et al. Dysregulation of bile acid homeostasis in parenteral nutrition mouse model. Am J Physiol Gastrointest Liver Physiol 310, G93–G102, doi:10.1152/ajpgi.00252.2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su X, Lu W & Rabinowitz JD Metabolite Spectral Accuracy on Orbitraps. Anal Chem 89, 5940–5948, doi:10.1021/acs.analchem.7b00396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sailer M et al. Increased plasma citrulline in mice marks diet-induced obesity and may predict the development of the metabolic syndrome. PLoS One 8, e63950, doi:10.1371/journal.pone.0063950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sleat DE et al. Mass spectrometry-based protein profiling to determine the cause of lysosomal storage diseases of unknown etiology. Mol Cell Proteomics 8, 1708–1718, doi:10.1074/mcp.M900122-MCP200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beavis RC Using the global proteome machine for protein identification. Methods Mol Biol 328, 217–228, doi:10.1385/1-59745-026-X:217 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Lund SP, Nettleton D, McCarthy DJ & Smyth GK Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol 11, doi:10.1515/1544-6115.1826 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.