Abstract
Acamprosate is an FDA-approved medication for the treatment of alcoholism that is unfortunately only effective in certain patients. Although acamprosate known to stabilize the hyper-glutamatergic state in alcoholism, pharmacological mechanisms of action in brain tissue remains unknown. To investigate the mechanism of acamprosate efficacy, we employed a pharmacoproteomics approach using an animal model of alcoholism, type 1 equilibrative nucleoside transporter (ENT1) null mice. Our results demonstrated that acamprosate treatment significantly decreased both ethanol drinking and preference in ENT1 null mice compared to that of wild-type mice. Then, to elucidate acamprosate efficacy mechanism in ENT1 null mice, we utilized label-free quantification proteomics comparing both genotype and acamprosate treatment effect in the nucleus accumbens (NAc). We identified 1,040 protein expression changes in the NAc among 3,634 total proteins detected. Our proteomics and western blot result demonstrated that acamprosate treatment decreased EAAT expression implicating stabilization of the hyper-glutamatergic condition in ENT1 null mice. Pathway analysis suggested that acamprosate treatment in ENT1 null mice seems to rescue glutamate toxicity through restoring of RTN4 and NF-Kb medicated neuroimmune signaling compared to wild-type mice. Overall, pharmacoproteomics approaches suggest that neuroimmune restoration is potential efficacy mechanism in the acamprosate treatment of certain sub-populations of alcohol dependent subjects.
Keywords: Pharmacoproteomics, Label-free Proteomics, Alcoholism, Acamprosate, Bioinformatics
Introduction
Recent data from the National Institute of Health indicate that alcohol use disorders (AUDs) affected 7.2% of Americans in 2012, making it a significant public health concern, however, pharmacological treatments of AUDs are still challenging. Acamprosate has been widely used since the Food and Drug Administration (FDA) approved the medication for treatment of AUD in 2004. Acamprosate is known to increase the time to relapse owing to its ability to reduce hyper-glutamatergic condition in the brain, preventing serious alcohol withdrawal symptoms (1).
Although acamprosate is effective for many patients, not all patients respond to treatment the same way (2), which has led to doubts about the overall efficacy of the drug. One six month study performed in the United Kingdom showed no difference in the effects of acamprosate versus a placebo (3), as did another performed at US family medicine clinics (4). These studies seem to be in contention with others that illustrate the effect of acamprosate in reducing alcohol craving and withdrawal symptoms (5). These contradicting studies suggest that acamprosate may only be effective in some patients, indicating that the identification of responders and non-responders to acamprosate is necessary to treat the right patient with the right drug. In addition, identifying the subset of patients who are responsive to acamprosate would help uncover the mechanism of action of acamprosate.
Using brain magnetic resonance spectroscopy (MRS), it was found that alcohol-dependent patients have elevated glutamate levels compared to healthy controls in the anterior cingulate cortex (6, 7). Levels of glutamate in the anterior cingulate were reduced in patients in response to acamprosate (6, 7), while those treated with placebo showed a trend towards an increase in glutamate levels in the anterior cingulate cortex (6, 7). Moreover, glutamate levels in the anterior cingulate cortex are positively associated with alcohol craving (6, 7), suggesting that acamprosate may be able to reduce alcohol craving.
By using animal models, the pharmacological action of acamprosate in the brain can be more easily manipulated. The equilibrative nucleoside transporter (ENT1) gene has been associated with alcohol related phenotypes and increased susceptibility of ethanol withdrawal behavior (8). Mice lacking ENT1 (ENT1−/− mice) demonstrate hyper-glutamatergic condition in the nucleus accumbens (NAc) and consume more ethanol than wild-type mice (9, 10). Acamprosate treatment seems to be effective in reducing ethanol drinking behaviors in ENT1−/− mice, not wild-type mice (10). Moreover, our previous brain MRS study demonstrated that acamprosate treatment significantly reduced glutamate levels in the NAc of ENT1−/− mice compared to the saline-treated group, an effect that was not observed in wild-type mice (10). However, the molecular mechanism how acamprosate treatment attenuates the symptom of hyper-glutamatergic condition, which is related to decreased alcohol drinking in ENT1−/− mice is not unveiled.
In this study, we investigated NAc protein expression changes in response to acamprosate treatment using pharmacoproteomics. This approach enabled us to identify a subpopulation of patients with AUDs who could potentially benefit from acamprosate treatment. To demonstrate a possible mechanism involved in acamprosate efficacy, we employed a label-free proteomics and bioinformatics approach including heat-map analysis, principal component analysis (PCA), and ingenuity pathway analysis (IPA) to compare molecular responses between genotypes.
MATERIALS AND METHODS
Animals.
We used 8–16 weeks old male ENT1 knockout (ENT1−/−) mice. Mice were housed (4–5 mice per group) in standard Plexiglas cages under a 12 h light (>500 lux)/dark (<0.5 lux) cycle with lights on at 6:00 AM. Male mice used for all experiments were housed at a constant temperature (24 ± 0.5°C) and humidity (60 ± 2%). Food and water was provided ad libitum. The animal care and handling procedures were both approved by the Mayo Clinic College of Medicine Institutional Animal Care and Use Committees in accordance with NIH guidelines.
Chronic Ethanol Administration Using Two-bottle Choice Self-administration.
To administer ethanol voluntarily, a two-bottle choice drinking experiment was performed as previously described (9, 10). During the experiment, mice were given one week to acclimate to individual housing conditions and handling. Mice were (n = 9 ~ 10 per group) then given 24 h access to two bottles, one containing plain tap water and the other containing an ethanol solution. The concentration of ethanol was raised every 4 days, increasing from 3 to 6 to 10 % (v/v) ethanol in tap water (Figure 1A). To examine the effect of acamprosate, both ENT1 +/+ mice and ENT1 –/– mice were given saline or acamprosate beginning on day 18. The acamprosate-treated mice were given 200 mg/kg (i.p.) acamprosate every 12 hours for five days after the mice steadily consumed 10% ethanol in a two-bottle choice drinking experiment. Throughout self-administration experiments, both alcohol/water intake and body weight were measured every 2 days to calculate average daily ethanol consumption (g/kg/day) and ethanol preference (%; ethanol solution consumption/total fluid consumption × 100). Blood acamprosate levels were measured using LC-MS/MS to verify acamprosate administration (11).
Figure 1.
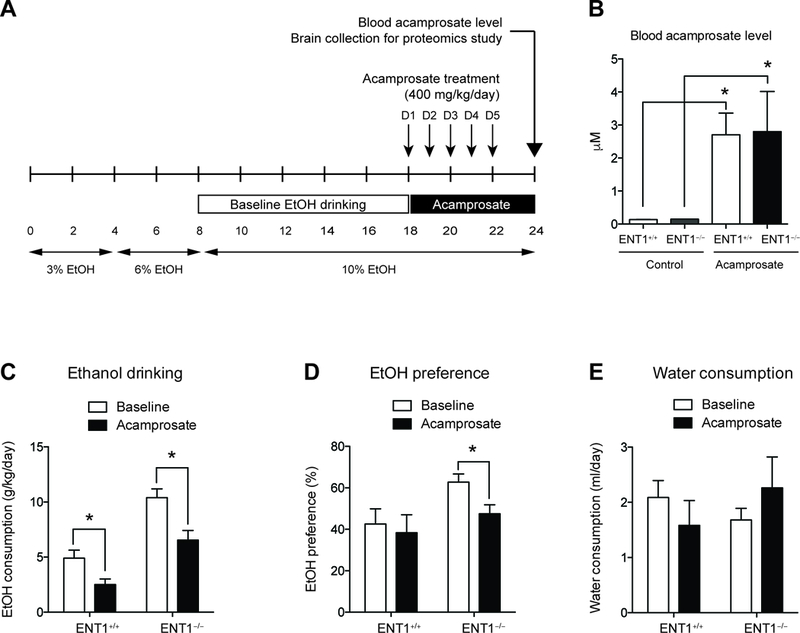
Acamprosate treatment in ENT1−/− mice during two-bottle choice alcohol administration. A. Schematic representation of the experimental paradigm. Mice were given 18 days to acclimate to a 10% (v/v) ethanol concentration. Then mice were injected with either saline or acamprosate twice per day (2 times x 200mg/kg, i.p.) for the next 5 days during the two-bottle choice self-administration (n = 9 ~10). B. There was no difference in the blood acamprosate levels between genotypes. C. Effect of Acamprosate treatment in alcohol consumption. Alcohol consumption (g/kg/day) is reduced in both genotypes with acamprosate treatment. D. Ethanol preference (%) is significantly reduced in in ENT1 –/– mice. E. Water drinking is similar between genotypes during acamprosate treatment. All data are presented as mean ± SEM.
Brain Tissue Sample Preparation.
Both ENT1+/+ mice and ENT1−/− mice were subjected to rapid CO2 inhalation to induce unconsciousness, followed by decapitation and subsequent harvesting of brain for isolation of the NAc from both hemispheres under a surgical microscope (n = 4 per treatment in each genotype). The extracted tissue was snap-frozen on dry ice and stored at −80°C until it was processed for SDS-PAGE (Bio-Rad Criterion system). The NAc from each mouse was homogenized using Pellet Pestle (Fisher) in a Cell-lytic MT mammalian tissue extraction reagent (Sigma-Aldrich) containing 50 mM Tris buffer (pH 7.4), 2 mM EDTA, 5 mM EGTA, 0.1% SDS protease inhibitor cocktail type I (Roche) and II (Sigma). Whole NAc tissue lysates were then centrifuged at 500 g at 4°C and supernatants were collected. Protein concentration from each replicate supernatant was quantified using the Bradford protein assay (Bio-Rad). Tissue samples were loaded at 30 μg proteins/lane and separated in MOPS buffer via electrophoresis in a 4–12% Bis-Tris poly-acrylamide gel (Criterion, Bio-Rad, Hercules CA, USA) at 70 V for 20 min, followed by 140 V for 80 min.
Label-free Proteomics Using 1D-SDS PAGE.
Each lane in 1D Gel was divided into 6 sections using a Precision Plus Kaleidoscope standard (Bio-Rad) to indicate prominent proteins bands common to each lane as section boundaries. Gel sections were then excised and the resulting fractions were incubated in 200 mM Tris for 30 min followed by destaining with 50 mM Tris/50%acetronitrile for 1–2 hr. Fractions were then dehydrated with 100% acetonitrile until gel pieces appeared opaque. Destaining was repeated a second time, followed by reduction with 20 mM Dithiothreitol (Sigma) in 50 mM Tris for 1 hr at 60°C. Gel fraction dehydration and alkylation was done with 100% acetonitrile and 40 mM iodoacetamide (Sigma) in 50 mM Tris for 1 hr at room temperature. Samples were rehydrated in 25 mM Tris and subjected to a final dehydration step prior to trypsin digest. Samples were mixed with 0.2 to 0.3 μg of trypsin (Promega) in 20 mM Tris/0.0002% zwittergent 3–16 overnight at 37°C. Trypsin was inactivated and peptides were extracted by adding 2% trifluoroacetic acid to the digest for 30 min followed by the addition of acetonitrile for an additional 30 min. The supernatant was removed and saved, followed by addition of acetonitrile a third time to the gel fractions for 30 min. The gel fractions were then added to the saved supernatant. These fractions were dried under vacuum and stored at −20°C.
NanoLC Tandem Mass Spectrometry (MS/MS).
Dried peptide extracts were reconstituted in an acidic aqueous solution of 0.2% formic acid for analysis by nano-scale liquid chromatography interfaced to tandem mass spectrometry (nLC-MS/MS). Peptide mixture was loaded onto an Optipak 0.25 ul cartridge (Optimize Technologies, Oregon City, OR) custom packed with Michrom Magic C8 (Michrom BioResource, Auburn, CA) using an Eksigent nanoLC-Ultra 2D with AS3 autosampler. Peptides were separated by reversed-phase LC using a 75μm i.d. fused silica column, packed in-house with 37 cm of 3 μm Magic C18 (Michrom BioResource, Auburn, Ca) with a gradient of 2–40% B over 60 minutes with a mobile phase flow rate of 300 nL/min. Mobile phase A contained water, acetonitrile (98/2 by volume), whereas mobile phase B was composed of acetonitrile, isopropanol, water (80/10/10 by volume). Both mobile phase A and B were acidified using 0.2% formic acid. Eluting peptides were analyzed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) configured to measure peptides within the molecular weight range of 360–2000 m/z at a resolving power of 70K (FMHM, m/z 200) survey scans (MS1), followed by isolating top 15 most abundant ions for higher energy C-trap dissociation scans (MS2) using an isolation window of 2 Da at a resolving power of 17.5K and a 45 second exclusion duration (12).
Data Processing and Label-free Data Analysis.
Data files were imported into the Rosetta Elucidator software (Seattle, WA) in which a relative quantitative label-free pipeline was used for analysis (13, 14). We utilized a label-free peptide MS1 intensity-based method for finding differentially expressed proteins between experimental groups. Quantification was detected such that m/z, retention time, and MS1 peak intensity data were extracted across pooled replicates and samples. Peptide sequencing on features selected with charge state <4 and >1, peak confidence score >0.6, peak time score > 0.8, and peak m/z score >0.9, was initiated within Elucidator using Mascot (v2.2, Matrix Science) with mouse Swiss-Prot database (release-2012_02) appended with common contaminants and a reverse decoy database. Mascot search parameters were set with peptide precursor tolerance of 10 ppm, ms/ms tolerance of 0.6 Da, 2 missed cleavages, and variable modifications carbamidomethyl C, oxidation M, and propionamide C. Annotation was performed using the Peptide Prophet implementation in Elucidator. All proteins were identified at least one unique peptide with a probability score of > 95%. The table of peptides and peptide intensities was exported in excel. Then, detection of differentially expressed peak ratios was performed with ANOVA utilizing a signature p value decoy error rate of no more than 5%. The annotation was assigned across aligned sample features so that relative quantitation of corresponding protein identifications across sample groups could be compared (15–17).
Heat Map Analysis, Principal Component Analysis, and Ingenuity Pathway Analysis.
Heat maps were drawn to determine the expression patterns of significantly up- or down-regulated protein at each time point and to compare the expression levels with the other time points, using R version 2.15.1 and the R packages. PCA can reduce the dimensionality of a data set consisting of a large number of interrelated variables, whereas retaining as much as possible of the variation present in the data set. PCA was conducted as an “unsupervised” analysis to clarify the variance among proteomics data from NAc samples using R. To clarify the variances among samples, proteomics data were calculated using a Q-mode PCA package ‘prcomp’of R. The proportion of variance and factor loading were also calculated. To classify the protein functionally, we used IPA where we entered the genes whose fold changes were more than 1.2-fold and P values were less than 0.05. IPA shows possible networks involved in microarray profiles by the IPA Network Generation Algorithm. Proteins were clustered and classified by the IPA Network Generation algorithm and the networks were ranked by the network score. The network score was calculated based on the tight-tailed Fisher’s Exact Test that takes into account: 1) the number of network eligible molecules in the network, 2) size of the network, 3) the total number of network eligible molecules in the given dataset, and 4) the number of molecules in the IPA database that could potentially be included in the networks. In the networks, solid and dashed lines indicated direct and indirect interactions, respectively. Direct interactions require the two molecules make direct physical contact with each other; there is no intermediate step. Indirect interactions do not require that there is physical contact between the two molecules, such as a signaling cascade instead of the two molecules making physical contact with each other (18).
Western Blot Analysis.
NAc tissues were homogenized in a solution containing 50 mM Tris buffer (pH 7.4), 2 mM EDTA, 5 mM EGTA, 0.1% SDS, protease inhibitor cocktail (Roche, Indianapolis, IN), and phosphatase inhibitor cocktail type I and II (Sigma). Homogenates were centrifuged at 500 g for 15 min and supernatants were collected. Proteins were analyzed using Bradford protein assay (BioRad, Hercules, CA). Proteins were separated by 4–12% NuPAGE Bis Tris gels at 130 V for 2 h, transferred onto PVDF membranes at 30 V for 1 h (Thermo Fisher Scientific, Waltham, MA), and incubated with antibodies against EAAT2 (Santa Cruz, Dallas, TX; SC-365634; 1:500), CAP1 (Santa Cruz, Dallas, TX; SC-376286; 1:500), RTN4 (Santa Cruz, Dallas, TX; SC-271878; 1:500), pERK; phospho-p44/42MAPK (Cell Signaling, Danvers, MA; 9106; 1:500), ERK; p44/42MAPK (Cell Signaling, Danvers; 9102; 1:500), and GAPDH (Millipore, Burlington, MA; MAB374; 1:2000). Chemiluminescent bands were detected on an Image Station and quantified using NIH Image J software.
Statistical Analysis.
The data was shown as mean with standard error of the mean (SEM). To detect statistical differences between acamprosate treatment and genotype, we conducted two-way ANOVA (SigmaStat vs. 3.1, SYSTAT Software, Point Richmond, CA). Criterion for statistical significance was p < 0.05.
RESULTS
Acamprosate treatment in ENT1–/– mice during ethanol self-administration.
To investigate the pharmacological effect of acamprosate in the NAc of animals that model responders to acamprosate treatment, we designed a pharmacoproteomics study using ENT1 –/– mice. A previous brain MRS study revealed that ENT1 –/– mice show decreased glutamate levels in the NAc after chronic acamprosate treatment during a two-bottle choice ethanol self-administration (10). Therefore, we profiled changes in the proteome in response to acamprosate in the NAc of ENT1–/– mice and ENT1+/+ mice using a label-free quantification proteomics approach. Mice were pre-exposed to a two-bottle choice ethanol self-administration experiment for 18 days and then treated with acamprosate for 5 days during 5 additional days of ethanol consumption (Figure 1A). To validate the acamprosate administration, the blood acamprosate level was measured using LC-MS/MS, and it showed no significant differences between genotypes (Figure 1B). Then, we examined whether Acamprosate treatment reduces ethanol consumption or preference using two-bottle choice ethanol self-administration experiment. Before Acamprosate treatment, ENT1 –/– mice showed increased baseline ethanol consumption (Figure 1C) and ethanol preference (Figure 1D) compared with wild-type mice. During the Acamprosate treatment session, both ENT1 +/+ mice and ENT1 –/– mice demonstrated significantly decreased ethanol consumption. Two-way ANOVA indicated a significant effect of genotype [ F(1,30) = 38.6, p <0.0001] and acamprosate treatment [ F(1, 30) = 16.6, p <0.01]. For ethanol preference compared to total liquid intake, two-way ANOVA showed significant effect of acamprosate treatment [ F(1,30) = 5.76, p <0.05], while there is no effect of genotype (Figure 1D). Water consumption was similar between genotypes during acamprosate treatment (Figure 1E). These results are consistent with previous finding that acamprosate reduces ethanol preference in ENT1 –/– mice.
Large-scale proteomics analysis of the NAc in response to acamprosate treatment in ENT1 null mice
Then, to elucidate proteome changes in the NAc of ENT1 –/– mice, we employed pharmacoproteomics approach. NAc tissue (AP: 0.98 ~ 1.54 mm) was isolated from both ENT1 +/+ mice and ENT1 –/– mice with micro-punch, and then processed for protein lysate (Figure 2A). Using label-free proteomics, proteins from each NAc tissue (4 per each group) were separated by 1D-SDS PAGE and each lane was pre-fractionated into 6 pieces for label-free proteomics using LC-MS/MS (Figure 2B). We identified 3,634 total proteins in the NAc (Table S1). 1,040 protein expression changes were found in response to acamprosate treatment and/or genotype. As shown in Figure 2C, we categorized protein expression change having a statistical significance value of p < 0.05 to identify significantly altered proteins. Although ethanol consumption itself can modify protein expression in the NAc (19, 20), here, the measurable changes in protein expression were due to acamprosate treatment, while changes induced by ethanol would be consistent with saline treated group. 5% of protein expression was altered by both lack of ENT1 and acamprosate treatment (Figure 2C-a), while 3.5 % of protein expression was changed by ENT1 genotype without an effect of acamprosate treatment (Figure 2C-b). Strikingly, 20% of proteins responded to acamprosate treatment only, which could imply that drug treatment goes beyond that of genotype differences (Figure 2C-c). 71.5% of proteins showed no significant response to acamprosate treatment or genotype (Figure 2C-d). These data suggest that pharmacological intervention induced dramatic changes in protein expression in the NAc, and it is possible that a mechanism could be identified in ENT1–/– mice that is associated with a positive acamprosate treatment response.
Figure 2.
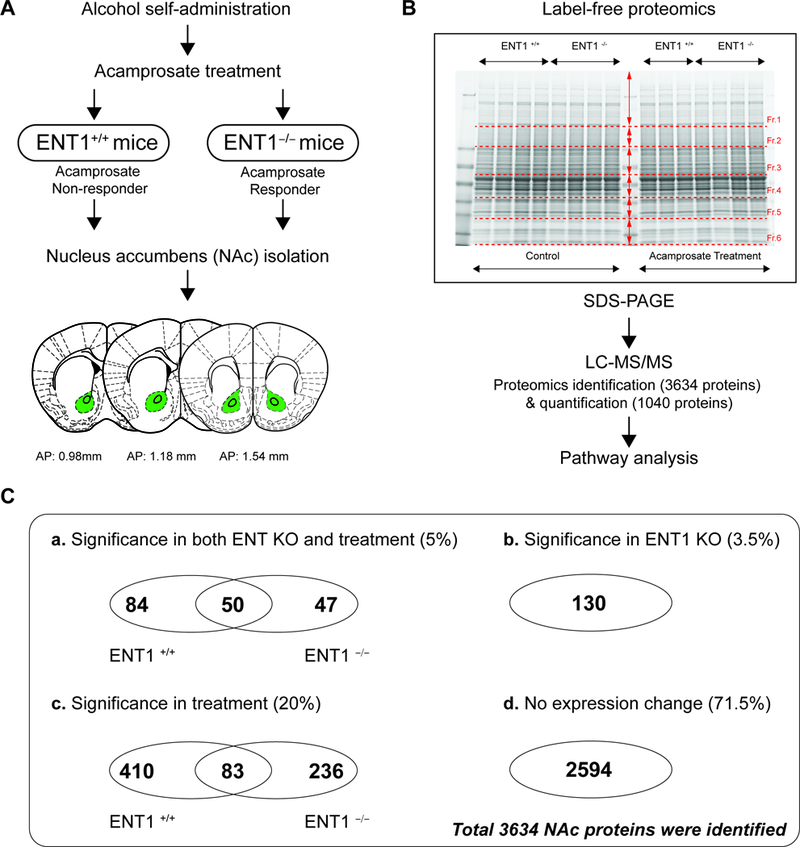
Pharmacoproteomics strategy for acamprosate treatment using label-free proteomics. A. Pharmacoproteomics workflow for NAc of ENT1 +/+ mice and in ENT1 –/– mice. Brain coordinates for NAc tissue collection. B. Label-free proteomics approach to identify acamprosate efficacy mechanism. Accumbal proteins were separated using SDS PAGE (n = 4 per group). C. Pharmacoproteomics analysis in the NAc of ENT1−/− mice in response to acamprosate treatment. (a) 181 proteins were changed in response to both genotype and acamprosate treatment. Among 181 proteins, 50 proteins were changes in both genotypes. (b) 130 proteins were changed by genotype only. (c) 729 proteins were changed by acamprosate treatment only. Among 729 proteins, 83 proteins were changes in both genotypes. (d) 2594 proteins were not changed by either genotype or treatment.
PCA and heat map analysis of protein profiles in response to acamprosate treatment in ENT1 null mice.
To determine whether acamprosate treatment during alcohol administration alters NAc protein expression in ENT1 –/– mice, we conducted principal component analysis (PCA) on the 1040 altered proteins in the NAc. In this dataset, three principal components (PC) were automatically identified, and their proportion ratios were calculated as PC1 (63%), PC2 (35%), and PC3 (2%) (Figure 3A). The ratio scatterplot for each condition (genotype and drug treatment) identified a significant effect of acamprosate treatment between genotypes (Figure 3A). The ENT1 –/– mice treated with acamprosate (green dot) showed significant differences according to PC1 compared to that of drug treatment in wild type mice (orange dot). While acamprosate treatment in ENT1 +/+ mice (blue dot) responded to PC2, representing a distinctive effect of acamprosate treatment in both genotypes (Figure 3B). The ratio scatterplot for the total 1040 protein also identified significant proteins projected over the first two principal components (Figure 3C). The mean value of the data was circled, and everything more than three standard deviations away from the mean was labeled individually as outliers (Figure 3C). The results demonstrated that JAK3 positively responds to PC1; and BAP29, Q3TJ49, A3KGK6, ANXA5, HYI, Q3U1N3, and CAP1 positively respond to PC2.
Figure 3.
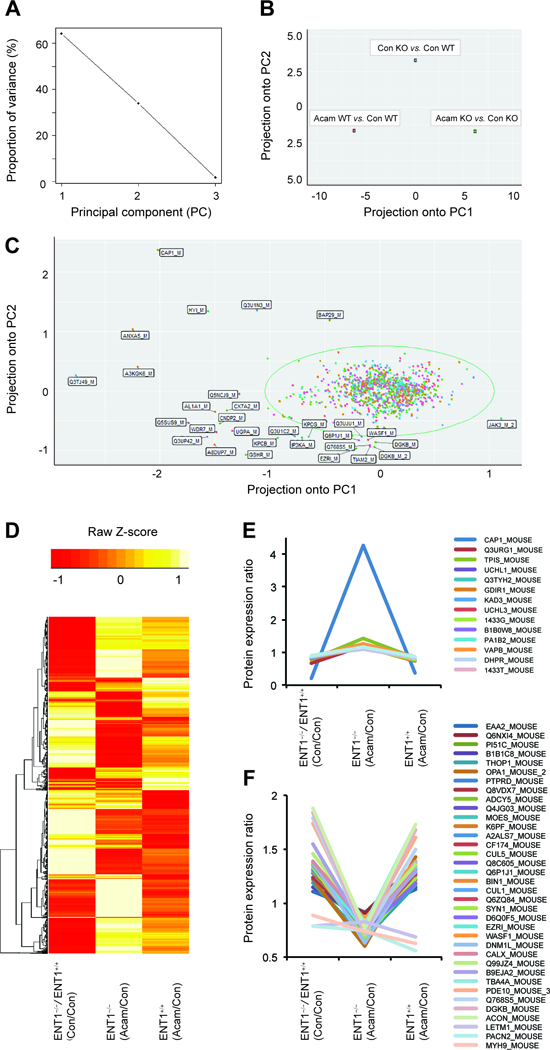
PCA analysis of protein expression in response to acamprosate treatment and deletion of ENT1. A. Ratio proportion of variance line graph identifies the 60% of proportion of variance of the first principal components. B. Acamprosate treatment in ENT1−/− mice shows significant projection in the first principal component (PC1) in ratio scatterplot. C. Ratio scatterplot for the total 1040 proteins. Labeled proteins indicate outliers having more than 3 standard deviations away from the mean. D. Heat map for altered protein expression in response to acamprosate treatment and deletion of ENT1 gene. Values were normalized by Z-Score E. Significantly increased protein expression by acamprosate treatment in ENT1−/− mice. F. Significantly decreased protein expression by acamprosate treatment in ENT1−/− mice.
To further profile protein expression changes between genotypes and acamprosate treatment, a heat map represented by normalized Z-score for expression of the 1040 proteins was generated (Figure 3D). Fifty proteins showed significant altered protein expression at baseline between genotypes as well as in response to acamprosate treatment between genotypes (Table 1). Adenylyl cyclase related proteins (ADCY5 and CAP1), PKC related proteins (PI51C PACN2, BIBIC8, and DGKB), and synaptic vesicle related proteins (Q768S5, EAAT2, SYN1, and VAPB) showed significant differences in their expression level between ENT1 +/+ mice and ENT1 –/– mice. Since acamprosate treatment in ENT1 –/– mice showed the most significant response in our PCA analysis (Figure 3B), clusters for 50 significantly altered proteins in ENT1 –/– mice were selected for the heat map (Table 1). Among the 50 proteins, 14 proteins were increased (Figure 3E) and 36 proteins were decreased (Figure 3F) by acamprosate treatment in ENT1 –/– mice. Interestingly, acamprosate treatment in ENT1 –/– mice significantly reverses those protein expressions by induced by lack of ENT1, while acamprosate treatment in ENT1 +/+ mice shows similar profiles to that of ENT1 deletion. These proteins may play a pivotal role in glutamate regulation in the NAc and suggest the pharmacological action of acamprosate and how it may prevent alcohol drinking.
Table 1.
50 NAc proteins altered by both ENT1 genotype and acamprosate treatment.
| Protein Name | Gene | KO vs. WT | KO vs. KO | WT vs. WT | |||
|---|---|---|---|---|---|---|---|
| Con vs. Con | Acam vs. Con | Acam vs. Con | |||||
| Ratio | p-value | Ratio | p-value | Ratio | p-value | ||
| Myc box-dependent-interacting protein 1 | BIN1_MOUSE | 1.316 | 8.43E-09 | 0.799 | 0.004 | 1.29 | 6.16E-07 |
| Diacylglycerol kinase beta | DGKB_MOUSE | 1.789 | 1.68E-08 | 0.71 | 7.05E-04 | 1.502 | 7.33E-05 |
| Rabphilin-3A | Q768S5_MOUSE | 1.838 | 2.13E-07 | 0.721 | 0.008 | 1.684 | 3.51E-06 |
| MKIAA1758 protein | B9EJA2_MOUSE | 1.548 | 1.92E-06 | 0.748 | 0.007 | 1.295 | 0.047 |
| Msn protein | MOES_MOUSE | 1.456 | 2.61E-06 | 0.828 | 0.002 | 1.354 | 9.80E-04 |
| Wiskott-Aldrich syndrome protein family member 1 | WASF1_MOUSE | 1.744 | 3.87E-06 | 0.759 | 0.012 | 1.61 | 1.88E-04 |
| Ezrin | EZRI_MOUSE | 1.879 | 7.97E-06 | 0.768 | 0.033 | 1.733 | 0.001 |
| Putative uncharacterized protein | Q8C605_MOUSE | 1.391 | 1.16E-05 | 0.802 | 1.20E-04 | 1.351 | 0.002 |
| Q6P1J1_MOUSE Crmp1 protein | Q6P1J1_MOUSE | 1.743 | 4.31E-05 | 0.802 | 0.049 | 1.729 | 0.002 |
| Isoform 2 of Dynamin-like 120 kDa protein | OPA1_MOUSE_2 | 1.28 | 1.01E-04 | 0.849 | 0.017 | 1.282 | 0.007 |
| Phosphofructokinase | Q8VDX7_MOUSE | 1.304 | 1.33E-04 | 0.837 | 0.003 | 1.307 | 1.92E-04 |
| Hypoxanthine guanine phosphoribosyl transferase 1 | B1B0W8_MOUSE | 0.812 | 2.07E-04 | 1.154 | 0.017 | 0.735 | 2.95E-05 |
| Adenylyl cyclase-associated protein | CAP1_MOUSE | 0.206 | 2.66E-04 | 4.267 | 0.03 | 0.389 | 0.029 |
| Glial high affinity glutamate transporter | EAA2_MOUSE | 1.109 | 2.79E-04 | 0.914 | 0.004 | 1.125 | 0.018 |
| LETM1 and EF-hand domain-containing protein 1 | LETM1_MOUSE | 1.188 | 3.36E-04 | 0.671 | 9.93E-11 | 1.168 | 0.005 |
| Dynein cytoplasmic 1 intermediate chain 2 | D6Q0F5_MOUSE | 1.32 | 3.51E-04 | 0.785 | 0.008 | 1.344 | 0.001 |
| Protein kinase C epsilon type | B1B1C8_MOUSE | 1.379 | 3.60E-04 | 0.855 | 0.045 | 1.388 | 0.013 |
| Calnexin | CALX_MOUSE | 1.371 | 6.20E-04 | 0.749 | 0.002 | 1.332 | 0.004 |
| Aconitate hydratase, mitochondrial | ACON_MOUSE | 1.336 | 6.66E-04 | 0.678 | 6.31E-05 | 1.228 | 0.009 |
| Rab15 | Q3TYH2_MOUSE | 0.668 | 0.002 | 1.21 | 0.009 | 0.722 | 0.029 |
| Thimet oligopeptidase | THOP1_MOUSE | 1.296 | 0.002 | 0.85 | 0.014 | 1.301 | 0.018 |
| 14–3-3 protein theta | 1433T_MOUSE | 0.898 | 0.002 | 1.088 | 0.025 | 0.834 | 5.97E-04 |
| cGMP 3’,5’-cyclic phosphodiesterase 10A | PDE10_MOUSE | 1.357 | 0.003 | 0.734 | 4.51E-04 | 1.189 | 0.041 |
| Adenylate cyclase type 5 | ADCY5_MOUSE | 1.149 | 0.003 | 0.836 | 8.99E-04 | 1.174 | 0.022 |
| Rap1 GTPase-activating protein | A2ALS7_MOUSE | 1.227 | 0.003 | 0.816 | 0.006 | 1.263 | 0.002 |
| Receptor-type tyrosine-protein phosphatase delta | PTPRD_MOUSE | 1.319 | 0.003 | 0.842 | 0.037 | 1.349 | 0.01 |
| Tubulin polymerization-promoting protein | Q3URG1_MOUSE | 0.777 | 0.004 | 1.43 | 0.001 | 0.846 | 0.007 |
| Synapsin-1 | SYN1_MOUSE | 1.238 | 0.004 | 0.787 | 0.001 | 1.218 | 0.007 |
| Phosphatidylinositol-4-phosphate 5-kinase gamma | PI51C_MOUSE | 1.15 | 0.004 | 0.857 | 0.006 | 1.153 | 0.007 |
| Rapgef2 protein | Q6NXI4_MOUSE | 1.146 | 0.007 | 0.903 | 0.035 | 1.366 | 9.48E-10 |
| Putative uncharacterized protein | DNM1L_MOUSE | 1.29 | 0.008 | 0.752 | 1.54E-05 | 1.233 | 0.042 |
| 14–3-3 protein gamma | 1433G_MOUSE | 0.914 | 0.009 | 1.162 | 0.002 | 0.861 | 0.005 |
| 6-phosphofructokinase, muscle type | K6PF_MOUSE | 1.289 | 0.01 | 0.823 | 0.015 | 1.298 | 0.005 |
| Dihydropteridine reductase | DHPR_MOUSE | 0.879 | 0.01 | 1.128 | 0.031 | 0.824 | 0.022 |
| Putative uncharacterized protein | Q99JZ4_MOUSE | 0.787 | 0.012 | 0.748 | 0.001 | 0.559 | 1.27E-06 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1_MOUSE | 0.832 | 0.012 | 1.213 | 0.011 | 0.757 | 2.87E-05 |
| Cullin 5 | CUL5_MOUSE | 1.292 | 0.015 | 0.804 | 0.04 | 1.32 | 0.006 |
| Uncharacterized protein C6orf174 homolog | CF174_MOUSE | 1.335 | 0.015 | 0.809 | 0.045 | 1.314 | 0.019 |
| Rho GDP-dissociation inhibitor 1 | GDIR1_MOUSE | 0.872 | 0.017 | 1.206 | 0.003 | 0.863 | 0.017 |
| Putative uncharacterized protein | CUL1_MOUSE | 1.277 | 0.019 | 0.79 | 0.003 | 1.298 | 0.007 |
| MKIAA0617 protein | Q6ZQ84_MOUSE | 1.295 | 0.021 | 0.788 | 0.018 | 1.273 | 0.013 |
| Vesicle-associated membrane protein B | VAPB_MOUSE | 0.861 | 0.021 | 1.136 | 0.019 | 0.784 | 0.004 |
| AMP phosphotransferase, mitochondrial | KAD3_MOUSE | 0.859 | 0.023 | 1.204 | 0.01 | 0.841 | 0.003 |
| Platelet-activating factor acetylhydrolase IB subunit | PA1B2_MOUSE | 0.896 | 0.031 | 1.137 | 0.015 | 0.822 | 0.003 |
| Tubulin alpha-4A chain | TBA4A_MOUSE | 0.885 | 0.031 | 0.737 | 0.029 | 0.623 | 5.69E-10 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L3 | UCHL3_MOUSE | 0.864 | 0.035 | 1.2 | 0.024 | 0.826 | 0.025 |
| Myosin-9 | MYH9_MOUSE | 1.214 | 0.039 | 0.601 | 1.00E-04 | 1.427 | 2.46E-07 |
| HECT, UBA and WWE domain containing 1 | Q4JG03_MOUSE | 0.785 | 0.042 | 0.829 | 0.004 | 0.686 | 0.011 |
| Protein kinase C and casein kinase substrate | PACN2_MOUSE | 1.39 | 0.044 | 0.631 | 0.02 | 1.382 | 0.015 |
| Triosephosphate isomerase | TPIS_MOUSE | 0.872 | 0.05 | 1.251 | 0.004 | 0.863 | 0.003 |
p < 0.05 compared with control treatment by unpaired two-tailed t-test.
Acamprosate restores glutamatergic protein expressions in ENT1 –/– mice.
We previously reported that the hyper-glutamatergic state in the NAc promotes excessive ethanol seeking in ENT1 –/– mice. Acamprosate is known to stabilize glutamatergic imbalance, so we sought to identify the restoration mechanism of glutamate neurotransmission mediated by acamprosate in ENT1 –/– mice. Among the proteins significantly altered by both genotype and acamprosate treatment, we validated EAAT2, CAP1, RTN4, and ERK protein expression changes between ENT1 +/+ mice and ENT1 –/– mice using western blot (Figure 4A). Consistent with our proteomics data, EAAT2 expression was increased in the NAc of ENT1 –/– mice (Figure 4B). Interestingly, acamprosate treatment significantly decreased EAAT2 expression in ENT1 –/– mice compared to wild-type mice, which suggest that acamprosate treatment rescues extracellular glutamate uptake mediated by EAAT2 (Figure 4C). In addition, we identified altered protein expression related to synaptic plasticity such as CAP1, RTN4 and ERK pathway in the baseline of ENT1 –/– mice. Decreased RNT4 and ERK expressions in the NAc were normalized by acamprosate treatment, which are consistent with proteomics results (Figure 4B and 4C). However, CAP1 demonstrated opposite expression patterns between proteomics and western blot results, which may be induced by fragmentation or post-translational modification. Acamprosate treatment could normalize CAP1 expression in ENT1 –/– mice as we observed in proteomics results.
Figure 4.
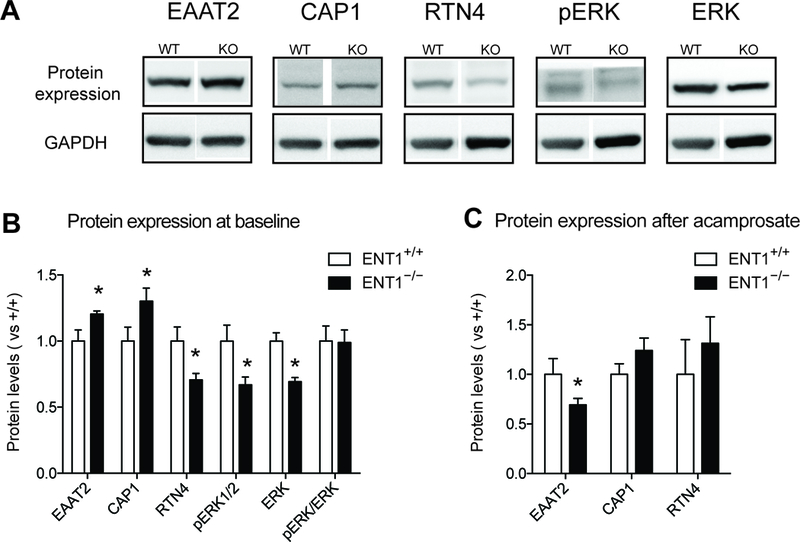
Validation of altered glutamatergic and synaptic molecules in the NAc of ENT1−/− mice. A. Western Blot showing altered protein expression changes between ENT1+/+ mice and ENT1−/− mice. B. EAAT2 [ t(8) =2.34, p <0.05] and CAP1 [ t(8) =2.28, p <0.05] expressions were significantly increase in the NAc of ENT1−/− mice, while RTN4 [ t(8) =2.50, p <0.05], pERK [ t(8) =2.46, p <0.05], and ERK [ t(8) =4.33, p <0.005], were decreased (n = 5). C. After acamprosate treatment, EAAT expression in the ENT1−/− mice were significantly decreased [ t(11) =2.30, p <0.05], while the other proteins showed no genotype differences (n = 6 ~ 7). All data are presented as mean ± SEM.
Pathway analysis of protein profiles in response to acamprosate treatment
To gain more insight into the intracellular mechanism of acamprosate treatment in ENT1 –/– mice, ingenuity pathway analysis (IPA) pathway analysis was carried out to map the biological processes. Including the 50 proteins (Table 1) that showed a difference in basal expression and that significantly responded to acamprosate treatment in both genotypes, 84 proteins and 47 proteins significantly responded to acamprosate treatment in ENT1 +/+ mice and ENT1 –/– mice, respectively (Figure 3A). IPA results of the significant proteins demonstrated that a signaling network related to tissue morphology and inflammation response were significantly different in both ENT1 +/+ mice (score 26; 14 proteins) and ENT1 –/– mice (score 26; 14 proteins) (Figure 5A). This approach newly identified a p53 and interleukin-related pathway that could possibly be involved in acamprosate response in ENT1 –/– mice, whereas these profiles were in the opposite direction in ENT1 +/+ mice. Our results may suggest that glutamate excitotoxicity caused by alcoholism is related to neuroimmune systems regulated by p53 and interleukin signaling, and acamprosate treatment could possibly alleviate the cell damage response in ENT1 –/– mice specifically (Figure 5A).
Figure 5.
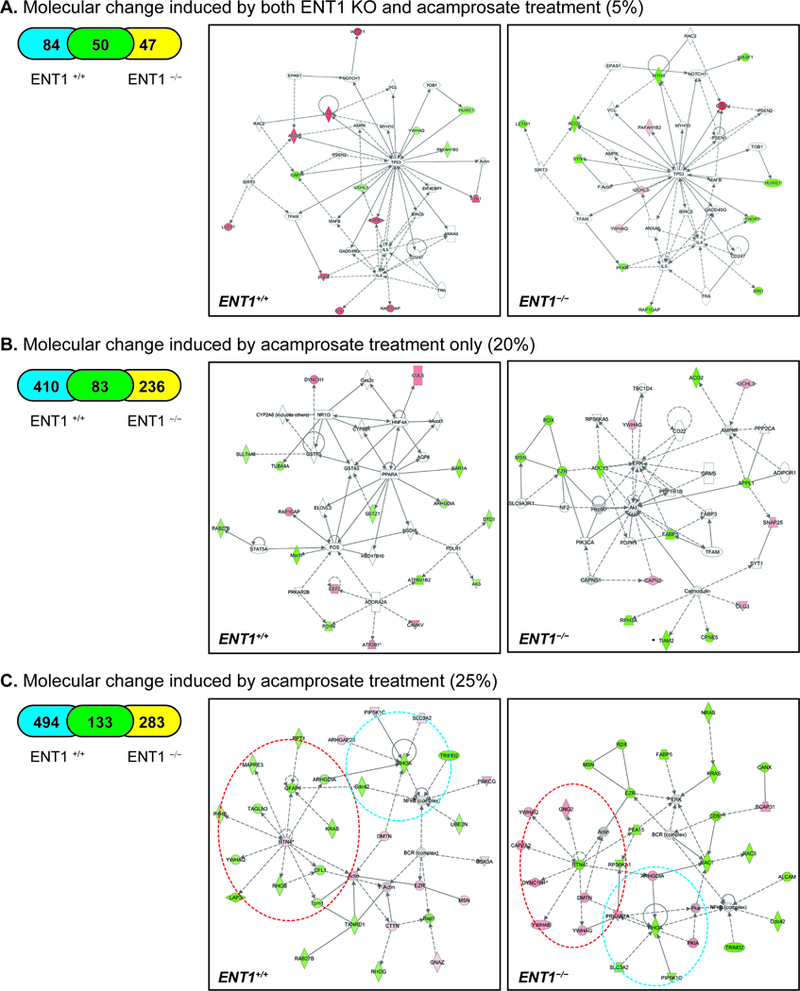
Summary of IPA pathway analysis and representative network in the NAc of ENT1–/– mice. A. Significantly changed network by both genotype and acamprosate treatment B. Significantly changed network by acamprosate treatment only. C. Significantly changed network by acamprosate treatment with genotype. Blue circle represents RhoA cluster, while red circle indicates RTN4 network. Orange color indicates up-regulated protein, while green color indicates down-regulated proteins with statistical significance. Grey indicates protein was detected but did not show statistical significance. A color gradient indicates an identified protein complex consisting of significantly altered proteins and proteins that did not show statistical significance. (*p < 0.05 by t-test for mean protein expression change measured by label-free proteomics). Table 1. 50 accumbal proteins altered by both ENT1 genotype and acamprosate treatment.
Next, we investigated the protein profile responding to acamprosate treatment only (Figure 3C), since acamprosate treatment led to the most protein expression changes (20%). This illustrates the strength of pharmacological intervention, which goes beyond that of the genotype effect induced by the deletion of ENT1. Of 729 proteins, the expression of 410 proteins was changed in ENT1 +/+ mice, while 236 proteins were altered in ENT1−/− mice after acamprosate treatment (Figure 5B). IPA analysis suggests that acamprosate treatment increases ADORA2A signaling, which decreases FOS in ENT1 +/+ mice. This pathway is related to molecular transport and lipid metabolism (score 26; 17 proteins). Although ENT1+/+ mice demonstrated more changes in protein expression after acamprosate treatment, ENT1−/− mice showed decreased activation of the AKT, AMPK, ERK, and calmodulin (CaM) pathways related to cellular assembly (score 25; 15 proteins).
Identification of RTN4 and RHOA clusters related to acamprosate treatment
Finally, all of the proteins altered by acamprosate treatment were analyzed by IPA pathway analysis. A total of 910 proteins among 1040 proteins showed significant changes in response to acamprosate treatment, and IPA pathway analysis identified that cellular assembly and organization were involved in acamprosate treatment in both ENT1 –/– mice (score 38; 30 proteins) and ENT1 –/– mice (score 44; 30 proteins). RTN4 and RHOA signaling showed significant profile differences between ENT1 +/+ mice and ENT1 –/– mice, while NF-κB-related signaling was decreased in both genotypes (Figure 5C). RTN4, named Nogo-A, acts as a developmental neurite growth regulatory factor involved in synaptic plasticity restriction (21–24). Interestingly, reduced expression of RTN4 is known to lead to decreased motivational behavior, which may correlate to decreased ethanol drinking in ENT1−/− mice in response to acamprosate (25). The IPA results show that decreased RTN4 expression in the in ENT1−/− mice negatively regulates RTN4 network protein expression, including YWHAQ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation), GNG2 (guanine nucleotide binding protein, subunit gamma 2), CAPZA2 (F-actin-capping protein subunit alpha-2), DYNC1H1 (cytoplasmic dynein 1 heavy chain 1), YWHAB (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta), and YWHAG (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma). YWHAQ is an adapter protein implicated in the regulation of signaling pathways, and it negatively regulates the kinase activity of PDPK1 (3-phosphoinositide-dependent protein kinase) (26) and PKC signaling (27). GNG2 is involved in various transmembrane signaling cascades, and both the gamma and beta chains are needed for GTPase activity (28). CAPZA2 binds the ends of actin filaments in a calcium-dependent manner, blocking subunit exchange (29). DYNC1H1 is a motor for the intracellular retrograde motility of vesicles and organelles along microtubules (30). YWHAB is involved in neuronal apoptosis, while YWHAG interacts with protein kinase C (PKC) (31). On the other hand, the RTN4 network was decreased in ENT1 +/+ mice, implicating a difference in cytoskeletal and microtubule formation. GFAP (glial fibrillary acidic protein) is a cell-specific marker that distinguishes astrocytes from other glial cells in development. PPT1 (palmitoyl protein thioesterase 1) removes thioester-linked fatty acyl groups from cysteine residues in lysosomal degradation (32). KRAS has intrinsic GTPase activity, playing an important role in cell proliferation. MAPRE3 (microtubule-associated protein RP/EB family member 3) binds the plus end of microtubules and regulates the movement of the cytoskeleton (33). TAGLN3 (transgelin-3) is a transformation-sensitive actin cross-linking protein in fibroblasts and smooth muscle, which may be an early indicator of transformation (34). LAP3 (leucine aminopeptidase 3) catalyzes the removal of unsubstituted N-terminal amino acids from peptides and may also be involved in the processing of intracellular proteins in the brain (35). RhoB (Rho-related binding GTP-binding protein) serves as a microtubule-dependent signal required for contractile ring formation in cytokinesis (36).
Another group of proteins altered by acamprosate treatment was the RHOA network, which also is involved in synaptic plasticity (37–39). There were four proteins identified in both groups related to the amount of RHOA: SLC3A2 (solute carrier family 3, member 2), PIP5K1C (phosphatidylinositol-4-phosphate 5-kinase type 1 gamma), DMTN (dematin actin-binding protein), and ARHGDIA (Rho GDP dissociation inhibitor alpha). SLC3A2 and PIP5K1C were decreased in the ENT1−/− mice, but increased in the ENT1+/+ mice; while ARHGDIA was increased in the ENT1−/− mice and decreased in the ENT1+/+ mice. Since RHOA showed decreased expression in both groups, it could be inferred that the actions of ARGHDIA counter the effects of PIP5K1C and SLC3A2. Moreover, SLC3A2 comprises the light subunit of a large amino acid transporter (LAT) and is important for the transport of LAT to the plasma membrane (40). PIP5K1C catalyzes the phosphorylation of phosphatidylinositol 4-phosphate (PtdIns4P) to form phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) (41). This reaction product contributes to secretion and vesicular trafficking, both of which are important for presynaptic neuronal release of neurotransmitters. DMTN, which acts as an inhibitor of RHOA, is a membrane-cytoskeleton-associated protein with F-actin-binding activity that induces F-actin stabilization. ARHGDIA regulates the GDP/GTP exchange reaction of Rho proteins (including RhoA) by inhibiting GDP dissociation. It sequesters the Rho proteins in the cytoplasm, protecting them from degradation (42).
Moreover, acamprosate treatment seems to decrease the NF-κB network in both genotypes, implicating a change in neuroimmune function. This finding is consistent with the finding that acamprosate treatment altered p53 and interleukin-related pathway. The ERK pathway was also sensitive to the effect of acamprosate treatment in the ENT1−/− mice (Figure 5C), which was not observed in in ENT1+/+ mice. Our findings indicate that RTN4, ERK, RHOA, and NF-κB signaling is significantly associated with a positive outcome of acamprosate treatment in ENT1–/– mice. Although it is well known that acamprosate acts as a partial antagonist of NMDAR and attenuates hyper-glutamatergic condition in the NAc (10, 43), our study is the first report demonstrating intracellular signaling associated with acamprosate efficacy.
DISCUSSION
Our study provides the possible mechanism of RTN4, ERK, RHOA, and NF-κB involved in the positive outcome of acamprosate treatment in ENT1–/– mice. Although it is well known that acamprosate acts as a partial antagonist of NMDAR and attenuates glutamate increases in the NAc, our study is the first report demonstrating possible synaptic plasticity target genes associated with acamprosate efficacy using a mouse model. Notably, in ENT1–/– mice, the RTN4 level was significantly decreased after acamprosate treatment during ethanol administration, whereas there was increased RTN4 expression in wild-type littermates. In addition, RTN4 plays a pivotal role in regulating a protein cluster in both genotypes, suggesting that acamprosate changes the synaptic plasticity-related network in opposing fashion based on genotype. The hyper-glutamatergic condition caused by the deletion of ENT1 may result in neuroimmune signaling changes that are normalized by acamprosate.
Pharmaco-omics, including pharmacogenomics and pharmacometabolomics, is a general trend of contemporary pharmacological research suggesting the use of blood biomarkers for individualized medicine strategies. These biomarkers could be useful in predicting therapeutic outcomes, but the sensitivity and specificity have yet to be determined. Although proteomics is underused for biomarker application due to the complexity of the proteome, including post-translational modification and proteolysis, it is an essential tool to measure the changes in protein contents in a target tissue. Pharmacoproteomics enables us to study the mechanism of acamprosate action in a target brain tissue in an alcoholism animal model. The variant of the ENT1 gene is highly associated with ethanol withdrawal in humans, and ENT1–/– mice showed an alcohol-related hyper-glutamatergic condition by microdialysis and brain MRS approaches. In this study, the same ethanol administration and acamprosate treatment paradigm for a previous MRS study was utilized to elucidate underlying glutamate excitotoxicity in the NAc (10). The protein expression change in the NAc of ENT1–/– mice elucidates the effect of Acamprosate in hyper-glutamatergic condition and ethanol self-administration.
Acamprosate is thought to modulate NMDA receptor (44), Glycine receptor (45, 46), or mGluR5 receptor (47) in the NAc; however this based on physiological findings. Therefore, our study was designed to instead focus on the identification of molecular signaling, which provides better understanding of acamprosate action. In our previous studies, we showed that excitatory amino acid transporter 2 (EAAT2) expression in the NAc of ENT1–/– mice plays an essential role in supporting the hyper-glutamatergic condition and excess alcohol seeking in mice (9). Consistently, our pharmacoproteomics results demonstrate that ethanol exposure increases EAAT2 expression, which uptakes more extracellular glutamate into the glia, in ENT1–/– mice and following Acamprosate treatment significantly decreases EAAT2 expression in ENT1–/– mice (Figure 3 and 4). Therefore, decreased EAAT2 expression by acamprosate treatment could be an important biomarker for validating our ENT1–/– mouse-model system and hyper-glutamatergic regulation in alcohol drinking. Since NF-kB transcription activity regulates EAAT2 expression, acamprosate may strengthen the neuroimmune response by alleviating excitotoxicity, which may reflect less expression of NF-kB and EAAT2 in acamprosate-treated ENT1–/– mice.
Recent advances in proteomics methodologies serve as powerful platforms for hypothesis generation or discovery of novel targets involved in pharmacological actions. The advantages of employing a label-free proteomics approach shows increased proteome coverage, decreased experimental steps for stable isotope labeling, enhanced ability to detect various types of post-translational modification on proteins, and the ability for single experiment relative comparisons of expression changes in multiple biological samples and technical replicates (48). However, pharmacological treatment and genotype comparison in brain proteomic analysis can be challenging, due to highly complex and interconnected heterogeneous neural and glial cell populations, in addition to computing and validating the large amounts of data generated (49). It must also be noted that label-free proteomics are less strictly quantitative compared to targeted MRM analysis (50). In addition, the sample size for this investigative study was quite small (4 mice per group) with a short duration of treatment (5 days). Based on these realities, we strengthened the validity of our findings by measurements of blood acamprosate level and by confirmation of protein expression using western blotting. Then, we utilized PCA and IPA bioinformatics analysis to facilitate our understanding of the interest and reference protein data. We hope that our label-free approach to the pharmacological response in a mouse brain tissue lysate will contribute to the standardization of clinical proteomics approaches as an example for robust quantitative analysis
Overall, our study provides a possible mechanism for examining proteome response and pursuing multiple comparisons as a result of genotype and acamprosate treatment using PCA and IPA protein network analysis. Separation using PCA and cluster analysis was carried out to identify protein sub-groups, and heat map analysis was conducted to distinguish significant clusters from the total proteome. In addition, pathway analysis using IPA suggest that there is great potential for candidate protein network analysis, as noted here, in the acamprosate treatment of certain sub-populations of alcohol dependent patients. Future research will focus on the interpretation of pharmacoproteomics findings as related to the pathophysiology of alcohol use disorder.
Supplementary Material
Statement of significance of the study.
Pharmaco-omics, including pharmacogenomics, pharmacoproteomics, and pharmacometabolomics, is a general trend of contemporary pharmacological research suggesting the use of biomarkers for individualized medicine strategies. These drug efficacy biomarkers could be useful in predicting therapeutic outcomes, but the sensitivity and specificity have yet to be determined. Although proteomics is underused for biomarker application due to the complexity of the proteome, such as post-translational modification, it is an essential tool to measure the changes in protein contents in a target tissue. In this study, pharmacoproteomics enables us to study the mechanism of acamprosate action in a target brain tissue in an alcoholism animal model. Moreover, our label-free proteomics revealed that acamprosate’s effect is possibly regulated by EAAT2 dependent glutamatergic regulation as well as RTN4 mediated neuroimmune pathway. Therefore, pharmacoproteomics approaches suggest that there is great potential for candidate protein network analysis, as noted here, in the acamprosate treatment of certain sub-populations of alcohol dependent subjects.
ACKNOWLEDGEMENTS
This project was supported by a NARSAD Young Investigator Award from Brain & Behavior Research Foundation and BRF Seed Funding from Biomedical Research Foundation to HWN, the Samuel C. Johnson for Genomics of Addiction Program at Mayo Clinic in parts by a grant from the National Institutes of Health (NIH) to DSC (AA018779).
List of abbreviations:
- ENT1
equilibrative nucleoside transporter
- NAc
nucleus accumbens
- AUD
alcohol use disorders
- PCA
principal components analysis
- IPA
ingenuity pathway analysis
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
Footnotes
CONFLICT OF INTEREST
All authors declare no conflict of interest.
References
- 1.Maisel NC; Blodgett JC; Wilbourne PL; Humphreys K; Finney JW, Meta-analysis of naltrexone and acamprosate for treating alcohol use disorders: when are these medications most helpful? Addiction 2013, 108, (2), 275–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morley KC; Teesson M; Reid SC; Sannibale C; Thomson C; Phung N; Weltman M; Bell JR; Richardson K; Haber PS, Naltrexone versus acamprosate in the treatment of alcohol dependence: A multi-centre, randomized, double-blind, placebo-controlled trial. Addiction 2006, 101, (10), 1451–62. [DOI] [PubMed] [Google Scholar]
- 3.Chick J; Howlett H; Morgan MY; Ritson B, United Kingdom Multicentre Acamprosate Study (UKMAS): a 6-month prospective study of acamprosate versus placebo in preventing relapse after withdrawal from alcohol. Alcohol and Alcoholism 2000, 35, (2), 176–187. [DOI] [PubMed] [Google Scholar]
- 4.Berger L; Fisher M; Brondino M; Bohn M; Gwyther R; Longo L; Beier N; Ford A; Greco J; Garbutt JC, Efficacy of acamprosate for alcohol dependence in a family medicine setting in the United States: a randomized, double-blind, placebo-controlled study. Alcohol Clin Exp Res 2013, 37, (4), 668–74. [DOI] [PubMed] [Google Scholar]
- 5.Jonas DE; Amick HR; Feltner C; Bobashev G; Thomas K; Wines R; Kim MM; Shanahan E; Gass CE; Rowe CJ; Garbutt JC, Pharmacotherapy for adults with alcohol use disorders in outpatient settings: a systematic review and meta-analysis. JAMA 2014, 311, (18), 1889–900. [DOI] [PubMed] [Google Scholar]
- 6.Umhau JC; Momenan R; Schwandt ML; Singley E; Lifshitz M; Doty L; Adams LJ; Vengeliene V; Spanagel R; Zhang Y; Shen J; George DT; Hommer D; Heilig M, Effect of acamprosate on magnetic resonance spectroscopy measures of central glutamate in detoxified alcohol-dependent individuals: a randomized controlled experimental medicine study. Arch Gen Psychiatry 2010, 67, (10), 1069–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frye MA; Hinton DJ; Karpyak VM; Biernacka JM; Gunderson LJ; Feeder SE; Choi DS; Port JD, Anterior Cingulate Glutamate Is Reduced by Acamprosate Treatment in Patients With Alcohol Dependence. J Clin Psychopharmacol 2016, 36, (6), 669–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JH; Karpyak VM; Biernacka JM; Nam HW; Lee MR; Preuss UW; Zill P; Yoon G; Colby C; Mrazek DA; Choi DS, Functional role of the polymorphic 647 T/C variant of ENT1 (SLC29A1) and its association with alcohol withdrawal seizures. PLoS One 2011, 6, (1), e16331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam HW; Lee MR; Zhu Y; Wu J; Hinton DJ; Choi S; Kim T; Hammack N; Yin JC; Choi DS, Type 1 equilibrative nucleoside transporter regulates ethanol drinking through accumbal N-methyl-D-aspartate receptor signaling. Biol Psychiatry 2011, 69, (11), 1043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MR; Hinton DJ; Wu J; Mishra PK; Port JD; Macura SI; Choi DS, Acamprosate reduces ethanol drinking behaviors and alters the metabolite profile in mice lacking ENT1. Neurosci Lett 2011, 490, 90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam HW; Karpyak VM; Hinton DJ; Geske JR; Ho AM; Prieto ML; Biernacka JM; Frye MA; Weinshilboum RM; Choi DS, Elevated baseline serum glutamate as a pharmacometabolomic biomarker for acamprosate treatment outcome in alcohol-dependent subjects. Transl Psychiatry 2015, 5, e621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayers-Ringler JR; Oliveros A; Qiu Y; Lindberg DM; Hinton DJ; Moore RM; Dasari S; Choi DS, Label-Free Proteomic Analysis of Protein Changes in the Striatum during Chronic Ethanol Use and Early Withdrawal. Front Behav Neurosci 2016, 10, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neubert H; Bonnert TP; Rumpel K; Hunt BT; Henle ES; James IT, Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J Proteome Res 2008, 7, (6), 2270–9. [DOI] [PubMed] [Google Scholar]
- 14.Wiener MC; Sachs JR; Deyanova EG; Yates NA, Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem 2004, 76, (20), 6085–96. [DOI] [PubMed] [Google Scholar]
- 15.Elias JE; Gygi SP, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007, 4, (3), 207–14. [DOI] [PubMed] [Google Scholar]
- 16.Keller BO; Wang Z; Li L, Low-mass proteome analysis based on liquid chromatography fractionation, nanoliter protein concentration/digestion, and microspot matrix-assisted laser desorption ionization mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2002, 782, (1–2), 317–29. [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI; Keller A; Kolker E; Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003, 75, (17), 4646–58. [DOI] [PubMed] [Google Scholar]
- 18.Omura S; Kawai E; Sato F; Martinez NE; Chaitanya GV; Rollyson PA; Cvek U; Trutschl M; Alexander JS; Tsunoda I, Bioinformatics multivariate analysis determined a set of phase-specific biomarker candidates in a novel mouse model for viral myocarditis. Circ Cardiovasc Genet 2014, 7, (4), 444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell RL; Kimpel MW; Rodd ZA; Strother WN; Bai F; Peper CL; Mayfield RD; Lumeng L; Crabb DW; McBride WJ; Witzmann FA, Protein expression changes in the nucleus accumbens and amygdala of inbred alcohol-preferring rats given either continuous or scheduled access to ethanol. Alcohol 2006, 40, (1), 3–17. [DOI] [PubMed] [Google Scholar]
- 20.McBride WJ; Schultz JA; Kimpel MW; McClintick JN; Wang M; You J; Rodd ZA, Differential effects of ethanol in the nucleus accumbens shell of alcohol-preferring (P), alcohol-non-preferring (NP) and Wistar rats: a proteomics study. Pharmacol Biochem Behav 2009, 92, (2), 304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aloy EM; Weinmann O; Pot C; Kasper H; Dodd DA; Rulicke T; Rossi F; Schwab ME, Synaptic destabilization by neuronal Nogo-A. Brain Cell Biol 2006, 35, (2–3), 137–56. [DOI] [PubMed] [Google Scholar]
- 22.Blochlinger S; Weinmann O; Schwab ME; Thallmair M, Neuronal plasticity and formation of new synaptic contacts follow pyramidal lesions and neutralization of Nogo-A: a light and electron microscopic study in the pontine nuclei of adult rats. J Comp Neurol 2001, 433, (3), 426–36. [DOI] [PubMed] [Google Scholar]
- 23.Zemmar A; Weinmann O; Kellner Y; Yu X; Vicente R; Gullo M; Kasper H; Lussi K; Ristic Z; Luft AR; Rioult-Pedotti M; Zuo Y; Zagrebelsky M; Schwab ME, Neutralization of Nogo-A enhances synaptic plasticity in the rodent motor cortex and improves motor learning in vivo. J Neurosci 2014, 34, (26), 8685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrinovic MM; Hourez R; Aloy EM; Dewarrat G; Gall D; Weinmann O; Gaudias J; Bachmann LC; Schiffmann SN; Vogt KE; Schwab ME, Neuronal Nogo-A negatively regulates dendritic morphology and synaptic transmission in the cerebellum. Proc Natl Acad Sci U S A 2013, 110, (3), 1083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enkel T; Berger SM; Schonig K; Tews B; Bartsch D, Reduced expression of nogo-a leads to motivational deficits in rats. Front Behav Neurosci 2014, 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato S; Fujita N; Tsuruo T, Regulation of kinase activity of 3-phosphoinositide-dependent protein kinase-1 by binding to 14–3-3. J Biol Chem 2002, 277, (42), 39360–7. [DOI] [PubMed] [Google Scholar]
- 27.Robinson K; Jones D; Patel Y; Martin H; Madrazo J; Martin S; Howell S; Elmore M; Finnen MJ; Aitken A, Mechanism of inhibition of protein kinase C by 14–3-3 isoforms. 14–3-3 isoforms do not have phospholipase A2 activity. Biochem J 1994, 299 ( Pt 3), 853–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pronin AN; Gautam N, Interaction between G-protein beta and gamma subunit types is selective. Proc Natl Acad Sci U S A 1992, 89, (13), 6220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Y; Tang X; Vitriol E; Chen G; Zheng JQ, Actin capping protein is required for dendritic spine development and synapse formation. J Neurosci 2011, 31, (28), 10228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirokawa N; Niwa S; Tanaka Y, Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 2010, 68, (4), 610–38. [DOI] [PubMed] [Google Scholar]
- 31.Van Der Hoeven PC; Van Der Wal JC; Ruurs P; Van Dijk MC; Van Blitterswijk J, 14–3-3 isotypes facilitate coupling of protein kinase C-zeta to Raf-1: negative regulation by 14–3-3 phosphorylation. Biochem J 2000, 345 Pt 2, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verkruyse LA; Hofmann SL, Lysosomal targeting of palmitoyl-protein thioesterase. J Biol Chem 1996, 271, (26), 15831–6. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa H; Koyama K; Murata Y; Morito M; Akiyama T; Nakamura Y, EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene 2000, 19, (2), 210–6. [DOI] [PubMed] [Google Scholar]
- 34.Fan L; Jaquet V; Dodd PR; Chen W; Wilce PA, Molecular cloning and characterization of hNP22: a gene up-regulated in human alcoholic brain. J Neurochem 2001, 76, (5), 1275–81. [DOI] [PubMed] [Google Scholar]
- 35.Hui KS; Saito M; Hui M, A novel neuron-specific aminopeptidase in rat brain synaptosomes. Its identification, purification, and characterization. J Biol Chem 1998, 273, (47), 31053–60. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto J; Kikuchi A; Ueda T; Ohga N; Takai Y, A GTPase-activating protein for rhoB p20, a ras p21-like GTP-binding protein--partial purification, characterization and subcellular distribution in rat brain. Brain Res Mol Brain Res 1990, 8, (2), 105–11. [DOI] [PubMed] [Google Scholar]
- 37.Briz V; Zhu G; Wang Y; Liu Y; Avetisyan M; Bi X; Baudry M, Activity-dependent rapid local RhoA synthesis is required for hippocampal synaptic plasticity. J Neurosci 2015, 35, (5), 2269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iida J; Ishizaki H; Okamoto-Tanaka M; Kawata A; Sumita K; Ohgake S; Sato Y; Yorifuji H; Nukina N; Ohashi K; Mizuno K; Tsutsumi T; Mizoguchi A; Miyoshi J; Takai Y; Hata Y, Synaptic scaffolding molecule alpha is a scaffold to mediate N-methyl-D-aspartate receptor-dependent RhoA activation in dendrites. Mol Cell Biol 2007, 27, (12), 4388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang HG; Lu FM; Jin I; Udo H; Kandel ER; de Vente J; Walter U; Lohmann SM; Hawkins RD; Antonova I, Presynaptic and postsynaptic roles of NO, cGK, and RhoA in long-lasting potentiation and aggregation of synaptic proteins. Neuron 2005, 45, (3), 389–403. [DOI] [PubMed] [Google Scholar]
- 40.Lin CH; Lin PP; Lin CY; Lin CH; Huang CH; Huang YJ; Lane HY, Decreased mRNA expression for the two subunits of system xc(−), SLC3A2 and SLC7A11, in WBC in patients with schizophrenia: Evidence in support of the hypo-glutamatergic hypothesis of schizophrenia. J Psychiatr Res 2016, 72, 58–63. [DOI] [PubMed] [Google Scholar]
- 41.Wright BD; Loo L; Street SE; Ma A; Taylor-Blake B; Stashko MA; Jin J; Janzen WP; Frye SV; Zylka MJ, The lipid kinase PIP5K1C regulates pain signaling and sensitization. Neuron 2014, 82, (4), 836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gee HY; Saisawat P; Ashraf S; Hurd TW; Vega-Warner V; Fang H; Beck BB; Gribouval O; Zhou W; Diaz KA; Natarajan S; Wiggins RC; Lovric S; Chernin G; Schoeb DS; Ovunc B; Frishberg Y; Soliman NA; Fathy HM; Goebel H; Hoefele J; Weber LT; Innis JW; Faul C; Han Z; Washburn J; Antignac C; Levy S; Otto EA; Hildebrandt F, ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest 2013, 123, (8), 3243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinton DJ; Lee MR; Jacobson TL; Mishra PK; Frye MA; Mrazek DA; Macura SI; Choi DS, Ethanol withdrawal-induced brain metabolites and the pharmacological effects of acamprosate in mice lacking ENT1. Neuropharmacology 2012, 62, (8), 2480–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomek SE; Lacrosse AL; Nemirovsky NE; Olive MF, NMDA Receptor Modulators in the Treatment of Drug Addiction. Pharmaceuticals (Basel) 2013, 6, (2), 251–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chau P; Hoifodt-Lido H; Lof E; Soderpalm B; Ericson M, Glycine receptors in the nucleus accumbens involved in the ethanol intake-reducing effect of acamprosate. Alcohol Clin Exp Res 2010, 34, (1), 39–45. [DOI] [PubMed] [Google Scholar]
- 46.Chau P; Soderpalm B; Ericson M, The mGluR5 antagonist MPEP elevates accumbal dopamine and glycine levels; interaction with strychnine-sensitive glycine receptors. Addict Biol 2011, 16, (4), 591–9. [DOI] [PubMed] [Google Scholar]
- 47.Blednov YA; Harris RA, Metabotropic glutamate receptor 5 (mGluR5) regulation of ethanol sedation, dependence and consumption: relationship to acamprosate actions. Int J Neuropsychopharmacol 2008, 11, (6), 775–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakolitsa C; Kumar A; McMullan D; Krishna SS; Miller MD; Carlton D; Najmanovich R; Abdubek P; Astakhova T; Chiu HJ; Clayton T; Deller MC; Duan L; Elias Y; Feuerhelm J; Grant JC; Grzechnik SK; Han GW; Jaroszewski L; Jin KK; Klock HE; Knuth MW; Kozbial P; Marciano D; Morse AT; Nigoghossian E; Okach L; Oommachen S; Paulsen J; Reyes R; Rife CL; Trout CV; van den Bedem H; Weekes D; White A; Xu Q; Hodgson KO; Wooley J; Elsliger MA; Deacon AM; Godzik A; Lesley SA; Wilson IA, The structure of the first representative of Pfam family PF06475 reveals a new fold with possible involvement in glycolipid metabolism. Acta Crystallogr Sect F Struct Biol Cryst Commun 2010, 66, (Pt 10), 1211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craft GE; Chen A; Nairn AC, Recent advances in quantitative neuroproteomics. Methods 2013, 61, (3), 186–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wegler C; Gaugaz FZ; Andersson TB; Wisniewski JR; Busch D; Groer C; Oswald S; Noren A; Weiss F; Hammer HS; Joos TO; Poetz O; Achour B; Rostami-Hodjegan A; van de Steeg E; Wortelboer HM; Artursson P, Variability in Mass Spectrometry-based Quantification of Clinically Relevant Drug Transporters and Drug Metabolizing Enzymes. Mol Pharm 2017, 14, (9), 3142–3151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


