1. Introductory Remarks
The focus of this review is assembly of biologically important systems. We are excluding protein-ligand binding and will present a number of case studies that deal exclusively with self-assembly. In a way this topic goes back to the roots of ion mobility spectrometry-mass spectrometry (IMS-MS) as developed at UCSB. The broad scope of that work has recently been reviewed (1) and self-assembly has played a key role in IMS studies at UCSB from the beginning. The first in depth application of IMS-MS to any system was to the self-assembly of carbon in carbon arcs 25 years ago. (2–5) Carbon clusters were a very hot topic at the time, especially the formation of Fullerenes (6,7) which were being touted as a new form of carbon and lead to a Nobel Prize for Smalley, Curl and Kroto in 1996. The primary contribution of the Bowers group was the determination of the structural evolution as the carbon clusters increased in size and the determination of the mechanism of Fullerene formation for sizes larger than 30 atoms. Specifically, they showed that carbon grew from linear structures to rings to Fullerenes and that Fullerenes arose from the annealing of large carbon planar ring systems (2,4,8,9).
Not long after the carbon cluster studies, the Bowers group initiated studies on the structure and assembly of biological molecules (10,11) but the real breakthrough came with the development of a new electrospray IMS instrument (12). The results from this instrument, and a later higher resolution instrument with an ultra-soft ion source (13), will provide most of the data for the case studies coming next. Before we discuss these case studies, however, a few general points need to be emphasized. The first of these is to understand the type of assembly that is occurring in solution and how best to capture it using IMS-MS. One of the requirements is to pay attention to the natural (native) charge state the biological system under study has in solution. For example, our first case study will be on the amyloid β-protein systems that are responsible for Alzheimer’s disease. We started on this problem in 2003 but we had no experience dealing with rapidly assembling biological systems. Hence we took our Aβ samples and used positive ion nano ESI and tuned to maximize signal. We felt we had to do this since our primary target, Aβ42 would assemble so fast it rapidly clogged out spray tip shutting down the experiment. After more than a year of experiments we had no useful, reproducible data. We then decided to back up and rethink our approach. We realized that Aβ42 had a minus 3 charge in solution at physiological pH. We decided we had to learn to work with this system using negative ion electrospray so that we could directly sample both monomer structures and assembled oligomers in their native charge states. Negative nano ESI was rarely used in those days and it took us a year to figure out how to obtain usable and reproducible signal. The beautiful results we obtained (14) will be summarized in our first Case Study later in this paper.
The second general point to be made is “softness”. If you are interested in studying the self-assembly of a biological system in which the oligomers are weakly bound non-covalent complexes you absolutely must eliminate all factors that energize these complexes during the IMS-MS measurement. Both of the nano ESI instruments that were used to record the data discussed in case studies 1 through 3 (12,13) are “soft” but our high-resolution instrument is “ultra-soft” (13). In part this is a lucky accident since we built the source to be at essentially the same pressure as the drift tube (10 to 15 torr) with no intervening segments that could energize the oligomers captured from solution. More will be said about the physical principles that underlie “softness” below.
A third crucial point is insuring that the oligomer distributions and the structures of the individual, size selected oligomers that we analyze by IMS correspond to those originally formed in solution. This is an interesting issue of sufficient importance that it deserves an entire Annual Review article on its own and, in part, is related to both of the points made above as will be apparent shortly. Most of the effort put into this topic to date has focused on individual peptides or proteins (see ref. (15) for a recent example and good summary of the literature). Hence, even though our emphasis here is on assemblies it is important to comment on this long standing issue of monomer folding since “solution” structures have been observed for even smallish monomer peptides like Aβ42 (16) and human isle amyloid polypeptide (hIAPP), a 37-residue hormone involved with type 2 diabetes (17).
One final point: IMS methods are ideally suited for the kinds of studies discussed here. Traditional spectroscopic methods are poorly matched to the study of solutions with multiple oligomers present that are in a state of dynamic growth. Such methods yield average results for all oligomers present in solution. IMS-MS on the other hand can directly sample solution oligomer distributions and size selected oligomer structures since these vary in mass-to-charge ratio (m/z) and in shape (cross section). The power of IMSMS was first demonstrated in the analysis of carbon arcs as discussed earlier. This commentary focuses on the non-covalent assembly of amino acids and peptides, systems at the heart of the amyloid process that plays a central role in a number of devastating diseases (18–20). In the case studies that follow we will start with the largest example (the 42 residue Aβ42 peptide), followed with several “model peptide systems (5 to 11 residues), and finish with assembly of individual amino acids. While this ordering may seem counter intuitive, it first highlights an important system responsible for Alzheimer’s disease and follows with our efforts to more fundamentally and mechanistically understand the amyloid process on a molecular level. First, however a more detailed discussion of instrumental “softness” and why it is required if we are to study solution structures of any type and hence relate our results to structural biology. We will focus these remarks on monomeric peptide/protein folding since this topic is of widespread interest. However virtually all of the points we make are equally valid for the assembly of these systems with differences pointed out where appropriate.
2. Softness of IMS-MS measurements: the ability to study biologically relevant structures of proteins and their assemblies.
How closely the structure of biological ions measured by IMS-MS methods reflects the biologically relevant structure present in solution is currently an active area of research.
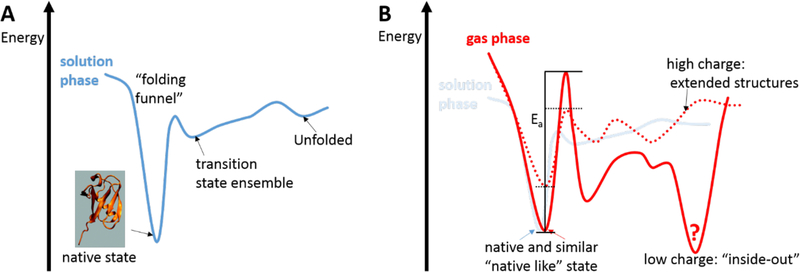
Protein folding in solution has been subject of an overwhelming body of research in the past three decades and many excellent reviews and perspectives are available (21–25). Figure 1a depicts a simplified picture of the energy landscape that governs protein folding in solution. This energy landscape involves a funneled energy landscape comprising an ensemble of compact, folded native protein structures, an ensemble of transition state conformations, and unfolded structures. The free energy difference between the folded and unfolded conformations is typically on the order of 5–20 kcal/mol. The driving force for formation of the folded, native conformations are interactions between the various functional groups present in the protein and between these groups and the water solvent (26,27). At neutral pH, proteins and peptides carry a number of ionic groups, including the carboxylate (R-CO2−), guanidinium (Gd+), and ammonium (R-NH3+) groups. These groups strongly bind several water molecules on the order of ~10 to 15 kcal/mol (28). The binding enthalpy of each single water molecule is thus comparable to the free energy required to unfold a native protein structure. By contrast, hydrophobic side chains of e.g. leucine or phenyl-alanine residues do not favorably interact with water. As a consequence, the enthalpy of protein conformations is minimized when ionic/hydrophilic groups are exposed to water on the protein’s surface while hydrophobic side chains cluster together to form a hydrophobic core on the interior of protein. When proteins do expose hydrophobic patches to the solvent, these patches often serve as inter-protein binding-sites resulting in a “hydrophobic collapse” and aggregation of the protein (29), a process that is the focus of the Case Studies discussed below in this Review. In the absence of solvent, ionic groups with localized charges become ‘charge-solvated’ by other groups in the protein through hydrogen-bonded interactions (30,31). Hence, in a solvent-free environment, a “natively” folded bare protein is expected to bury polar groups in its interior while exposing hydrophobic residues on its surface (32). Such folded, gas-phase structures of biomolecules are often referred to as “inside-out” structures (32). However, the charged side chains in a metastable, desolvated solution type structure, as often encountered in ESI, will “self-solvate” with any side chains in its immediate area with little change in backbone structure due to the high barriers of backbone rearrangement and formation of the “inside out” structure (15,32–39). Since these interactions depend on the charges on the amino acid residues, it is important to choose the polarity of the ESI process to match the native solution charge state of the protein (complex) under investigation in order to capture the native solution like structures using IMS-MS. For the case of Aβ40 and Aβ42, this issue had turned out to be pivotal, as will be detailed further below.
Figure 1. Simplified protein folding landscape (A) in solution and (B) in the absence of solvent.
(A) In the presence of solvent, protein folding is governed by a funneled energy landscape comprising an ensemble of compact, folded native protein structures, an ensemble of transition state conformations, and unfolded structures. The driving force for formation of the folded, native conformations is to minimize the enthalpy by exposing hydrophilic groups to the solvent (water) while burying hydrophobic side chains in its interior (“hydrophobic core). (B) In the absence of solvent, protein structures turn “inside-out” by exposing hydrophobic patches on the protein surface and charge-solvating ionic groups. For low charge-states, a significant activation barrier can separate the protein solution structure from the “inside-out” structure, making it possible that solution structures can be kinetically trapped in “soft” IMS-MS measurements.
Consider a peptide or protein sprayed using ESI that is gently desolvated without changes its native structure. Ions formed from ESI are expected to be subthermal due to evaporative cooling (40) and hence must become energetically activated if they are to structurally rearrange. Ion activation can occur in an IMS-MS experiment when translational energy is converted into internal (vibrational) energy via inelastic collisions between the ion and buffer gas particles. This collisional heating process reaches a steady state between activating and de-activating collisions and the internal energy distribution of the ion population can be characterized by an (effective) internal temperature Tint, which increases with increasing energy transfer per collision (41–43). Basic physics (44) indicate that the amount of translational energy that is converted into internal energy through collisions, δE is given in Eqn (1)
| (1) |
where Elab is the translational energy of an analyte ion with mass m in the laboratory frame, mbg the mass of a buffer gas particle, and γ he inelasticity parameter of the collision that depends on the densities of states of the buffer gas particle and the analyte ion and on the strength of the ion-neutral interaction potential. A second important factor in Eqn. (1) is the mass mbg of the buffer gas particle. For heavy analytes with mass m ≫ mbg, energy transfer occurs approximately seven times more effectively in collisions with nitrogen (mbg = 28 Da) than helium (mbg = 4 Da). Hence, helium is a much “softer” buffer gas for ion mobility spectrometry experiments than the more commonly used nitrogen. This increased “softness” of helium is one reason why helium buffer gas is used for any of the structural study of biological analyte ions at UCSB (12,13) that we discuss in the following sections in this Review. At FSU, nitrogen buffer gas is used as our trapped IMS measurement (45,46) requires a constant flow of buffer gas through the instrument which would be prohibitively costly with helium.
Finally, Eqn. (1) reveals that the increase in internal energy δE of the analyte ion depends decisively on the translational (kinetic) energy Elab of the analyte ion. The kinetic energy of an ion in an ion mobility experiment depends on the energy uptake from the electric field between two collisions with the buffer gas, ΔElab In traditional drift tube IMS instruments, ions are accelerated between two ion-neutral collisions by a uniform DC electric field (47,48). Under these drift conditions, the gain in translational energy ΔElab of an ion from the electric field EDC between two collisions with neutrals is given in Eqn (2)
| (2) |
where m and q are the mass and charge of the analyte ion, Δv is the velocity gained by the analyte ion from the acceleration by the force EDC = qEDCin the electric field EDC within time interval δt between two collisions. A radially confining radio-frequency (RF) field—present in the trapped IMS setup (45,46) used at FSU and the traveling wave instruments of Waters (49)—contributes to the acceleration of the analyte ion between two ion-neutral collisions. Tolmachev and Smith have shown (43) that the mean kinetic energy of an ion induced by an RF-field within a single RF period increases quadratically with charge q and RF-field strength ERF. Thus, on a qualitative level, the energy uptake from the RF-field and that from a DC-field are similar: the higher the strength of the RF-confining field, the more an ion is accelerated.
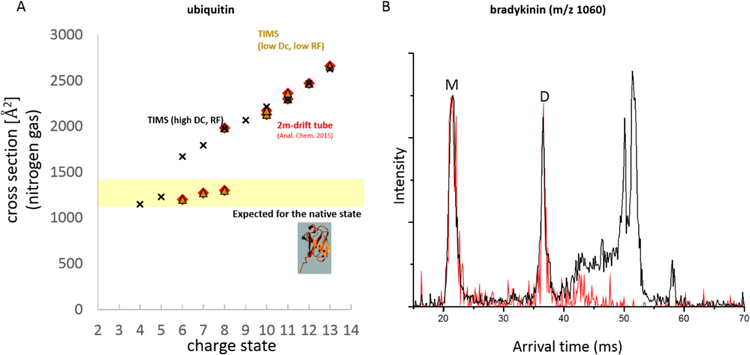
Since δt depends on the pressure in the IMS tube, Eqn (2) tells us that to prevent effective ion heating the pressure should be fairly high so to have many, but low-energy, collisions. Measurements in the drift tubes at UCSB are typically carried out in helium gas at pressures in the order of 4–15 torr (12,13). Such pressures correspond to mean free paths between collisions of approximately 0.4 μm to 0.1 μm. At FSU, where we carry out measurements at ~3 mbar in nitrogen buffer gas (50,51), the mean free path is in the order of 0.6 μm. In travelling-wave instruments, which are typically operated at 0.5mbar to 1mbar (49), mean free paths are on the order of 3.6 μm, resulting in energy transfer that is ~36 and ~1237 more effective than in our TIMS setup at FSU and the high-resolution drift tube at UCSB, respectively. An example of how this can affect the measured cross sections, and hence structures, of the small protein ubiquitin is given in Fig (2).
Figure 2. Observation of ion heating processes in IMS-MS measurement.
(A) Cross sections recorded for various charge states of ubiquitin in the drift tube at UCSB (red diamond), soft-tuned (yellow triangle), and untuned (black cross) trapped IMS systems. Charge states +6, +7, and +8 measured on the UCSB drift tube and the soft-tuned trapped IMS system at FSU are consistent with the solution structure of ubiquitin while solution structures are not retained when the trapped IMS system is not carefully “soft”-tuned. (Note that cross sections of “inside-out” structures for charge states +4 or +5 are similar to the solution structure; see reference (55) for details.) Ubiquitin cross sections were taken from references (53), (50), and (54). (B) Trapped IMS spectra for m/z 1060 of bradykinin with and without mass selection in the quadrupole (red and black traces, respectively) comprises monomers and oligomers. Differences in the IMS spectra demonstrate that complicated ion optics between the IMS measurement and mass analysis/ion detection can obscure the interpretation of IMS sepctra when peptide or protein oligomers are present. (S. R. Kirk and C. Bleiholder, unpublished).
Figure 2A compares cross sections recorded on different instruments for several ubiquitin charge states to the cross section expected for the ubiquitin x-ray structure (52). The data recorded at UCSB (15,53) show low charge states with cross sections consistent with the x-ray structure and high charge states with much larger cross sections that correspond to denatured ubiquitin ions. The data recorded on the trapped IMS system at FSU agrees essentially numerically with these data but only after careful “soft-tuning” of the operating conditions by down tuning the DC and RF electric fields as well as the source conditions (needle potential, desolvation gas temperature) (50). The cross sections of charge states 6,7, and 8 are significantly larger—indicative of substantial collision-induced unfolding of ubiquitin ions during the measurement—when high DC and RF fields are used and the ESI desolvation gas is heated in excess of 370K in a trapped IMS device (54). Charge states 4 and 5, which are not generally observed under native conditions (50,53), should not be considered indicative of the ubiquitin solution structure as cross sections of their “inside-out” gas-phase structures are not distinguishable from that of the ubiquitin solution structure (55).
Here we have discussed energizing collisions only in the IMS cell. Of course the entire instrument must be extremely soft, not just the IMS cell, when studying non-covalent assemblies of peptides. An illustrative example is m/z 1060 of bradykinin. The instruments at UCSB show monomer (1/1), dimer (2/2), and trimer (3/3) species (12,13). In these instruments, mass analysis occurs in a quadrupole directly attached to the IMS drift tube, effectively preventing any reactions of the assemblies to take place between IMS and mass analysis. In the trapped IMS system used at FSU ions traverse a quadrupole and a collision cell between IMS and mass analysis (45,46,50,51). We find that the presence of triply-charged bradykinin trimers depends significantly on the energy used to injected the assemblies into the collision cell (not shown). Further, spectra recorded with and without precursor ion selection in the quadrupole can differ substantially when the collision energy isn’t carefully tuned (Figure 2B). These observations underscore that great care must be taken to avoid energizing analyte ions in the source, the transport region before the IMS cell and following the cell before mass analysis and ion detection. In the instruments used to obtain the data discussed in the following Case Studies, all of these sections provide negligible ion activation, which is what made the studies possible even though the details will not be pointed out in the narratives.
3. Case 1: Amyloid β-protein and Alzheimer’s disease (AD)
Alzheimer’s is a complex disease that has resisted every attempt to both understand its detailed mechanism and to establish even a minimally effective therapy. It is finally being recognized as a priority in the USA and elsewhere due to the aging of our population and due to the current and projected human and financial cost (~$1,000,000,000,000/year by 2050) (56). The literature dealing with various aspects of AD is vast and cannot even be summarized here. Our focus will be on the molecular basis of AD, especially how IMS-MS can make unique contributions to our understanding of it. For a very long time AD was felt to be an amyloid disease (20) with research emphasis on the terminal fibrillar constructs produced by the amyloid cascade. However, about the turn of the 21st century the community began to realize that the initial oligomers formed in the amyloid cascade may be the most important, not the terminal fibrils themselves (56). It was this realization that sparked our interest since IMS-MS is uniquely suited to the study of dynamic oligomer formation but incapable of studying the terminal fibrils.
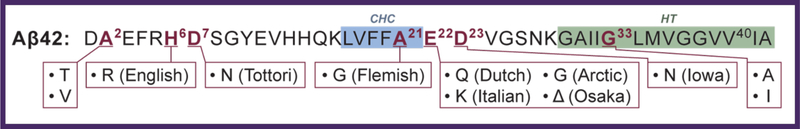
Our efforts began with a collaboration with Prof. David Teplow in 2003 (initially at Harvard Medical School but now at UCLA), a collaboration that has born much fruit and that continues to this day. In addition to providing abundant and ultrapure Aβ samples of many varieties, Dave was essential in bringing us up to speed on the outstanding issues in AD, particularly those we could attack using IMSMS methods. Of crucial importance is the fact that AD appears to be primarily caused by early oligomers of Amyloid β-protein, peptides that are cleaved from the much larger amyloid precursor protein (APP) that is over 700 residues long. The primary cleavage products are Aβ40 and Aβ42 shown below in Scheme I.
Scheme 1.
Aβ40 makes up about 90% of the cleavage products and Aβ42 9%; however the plaques found in AD patients are formed primarily from Aβ42 monomers (57,58). Early work indicated that Aβ40 and Aβ42 oligomers assembled by different mechanisms (59) but it was not understood why Aβ42 was so much more neurotoxic than Aβ40 (60) nor were the two assembly mechanisms understood. Perhaps most important, while Aβ oligomers were implicated in the aetiology of AD there was little or no information on the identity, formation mechanism and structure of the neurotoxic agent(s). Hence, our initial goals were to determine the assembly details for Aβ40 and Aβ42, to determine the identity and structure of the toxic agent(s), and to develop possible therapeutic strategies for treatment of AD. A summary of our initial results and mechanistic conclusions is given in Figure 3.
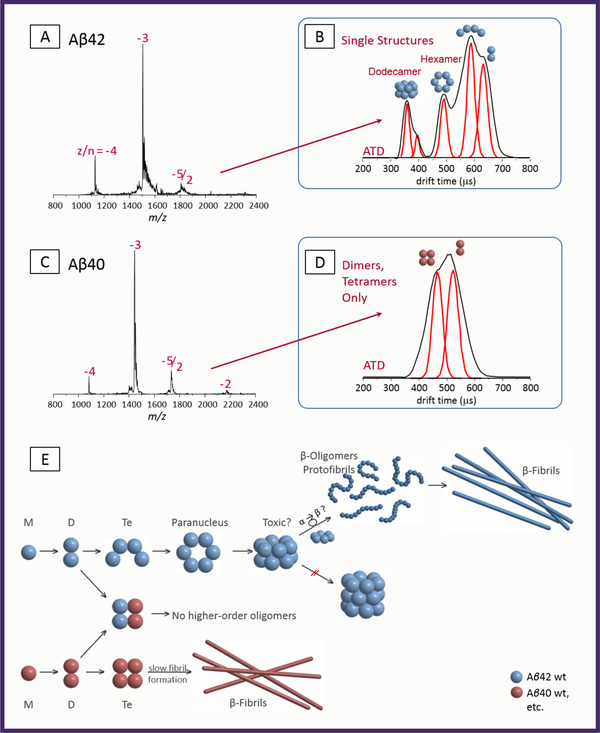
Figure 3. Summary of amyloid β-protein structures and reaction mechanisms.
A and C, Mass spectra of Aβ42 and Aβ40 respectively at 10 uM concentration. B and D, arrival time distributions for the −5/2 peak of Aβ42 and Aβ40 repectively. The model structures given over the various ATD features are taken from ref (61). The mechanism in E summarizes the differences in reactivity for Aβ42 and Aβ40 and qualitatively represents the current understanding for Amyloid β-protein assembly.
On the top left of the figure are negative ion mass spectra of 10 μM solutions of Aβ42 and Aβ40. From these data alone it would be difficult to say much about either species. However, the arrival time distributions (ATDs) at the top right of the figure are much more revealing. These are taken for the z/n = – 5/2 mass spectral peaks and hence must represent even numbered oligomers (n is the oligomer number and z is the charge). The assignments of the features in the ATDs have been established elsewhere (14,61). For Aβ42 the – 5/2 peak contains dimer, tetramer, hexamer and dodecamer while for Aβ40 only dimer and tetramer are observed. Coarse grain structures are given above each of the features in the ATDs. These were obtained by comparing the experimentally measured cross section with cross sections obtained from model structures (61). Of importance is the fact the tetramer of Aβ42 is an open structure which is able to add a dimer to form hexamer while the tetramer for Aβ40 is a closed structure that could add a dimer only with difficulty. These tetramer structures are pivotal in determining the oligomer assembly of the two alloforms and consequently the aetiology of AD. The Aβ42 hexamer is planar and cyclic while the dodecamer is composed of two stacked hexamers. Of interest is the fact a third hexamer doesn’t stack which probably indicates the hydrophobic cores of the individual hexamers composing the dodecamer interact leaving relatively non-bonding top and bottom surfaces. Also of interest is the fact that no octamer is observed. Consideration of these facts, and others available to us at the time, lead to the assembly mechanism given at the bottom of the figure.
One interesting point is the fact that mixed tetramers of Aβ42 and Aβ40 do not further oligomerized (62). This result implies that Aβ40 is effective in inhibiting Aβ42 assembly and thus is over all protective against AD (see below). It is also interesting that only the 2×2 tetramer is observed (62) indicating that oligomer growth occurs primarily by dimer addition supporting the mechanism given in Figure 3. There is support for the dodecamer as the primary neurotoxic agent in AD from mouse model studies (63,64): Transgenic mice with the human APP gene were found to develop memory loss before plaques were detected. The dominant Aβ agent found was termed Aβ56* as it was approximately 56 kD in mass which is the approximate mass of the Aβ42 dodecamer.
While the IMS-MS data appear to be unambiguous, it is important to verify them by independent means and to gain further insight into the fibril formation mechanism. In this regard we have recently published work in collaboration with Professor Steve Buratto at UCSB on the Aβ-peptide systems using ultra high resolution atomic force microscopy with the data summarized in Figure 4 (65).
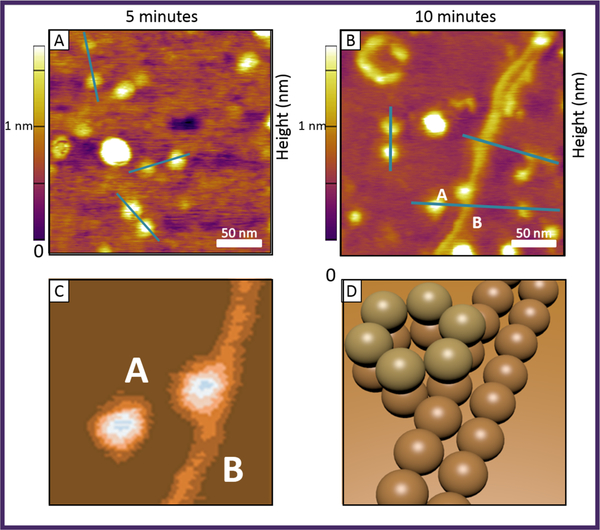
Figure 4. Ultra-high resolution atomic force microscopy of Aβ42 at a concentration of 1 μM drop cast on a freshly cleaved mica surface.
Panel A is at a solution incubation time of 5 minutes and panel B at a solution incubation time of 10 minutes. The blue lines indicate features that are characterized by line cuts to establish their height and width (see text). Panel C shows a blow up of the dodecamer features in region labeled A/B in the 10 minute image. The long filament is growing out of the lower hexamer of the dodecamer in this image (see text). A cartoon of the dodecamer-filament connection is shown in panel D. The images are reconstructed from reference (65).
The top two images show data taken from a 1 μM solution of Aβ42 drop cast onto a freshly cleaved mica surface following solution incubation times of 5 and 10 minutes. The 5 min image shows primarily circular shaped objects, which line cuts indicate are either 0.75 or 1.5 nm high and 10 to 15 nm in diameter. These dimensions are fully consistent with cross sections measured for the hexamers and docamers using IMS-MS and the structures given in Figure 3 (14,61). In the 10 min image an important new feature is observed; a long (> 200 nm) filament like species 0.75 nm in height and 10 nm in width, consistent with the expected dimensions of pre-protofibrils previously reported (66). The bottom left image in Figure 4 is a blowup of a segment of the filament shown in the 10 min image. It is apparent the filament appears to be growing out of the bottom hexamer of a dodecamer as depicted in the cartoon in the bottom right of Figure 4. A search of the surface indicated all such filaments are associated only with dodecamers similar to what is shown in Figure 4. While the dodecamer is becoming accepted as an important neurotoxic agent in AD, it now appears it is also the seed assembly for initiation of fibril growth, which is currently viewed as protective in AD. Hence the dodecamer appears to be the central figure in the assembly of Aβ42 and possibly in the aetiology of the disease.
The story is very different for Aβ40 (65). In this instance no hexamers or dodecamers are detected by AFM at early times, only smaller oligomers. At longer times (> 30 minutes) highly branched filaments appear that are 0.75 nm in height and 10 nm in width. At longest times these filaments compete with large spherical objects similar to those also observed in Aβ42 at long times. The results given by both the IMSMS and AFM studies clearly answer the questions about the very different assembly of Aβ40 and Aβ42 as well as why Aβ42 is so much more toxic than Aβ40. Further the results provide evidence for the first time that the Aβ56* toxic agent observed in mouse model studies is in fact an Aβ42 dodecamer composed of stacked, planar, cyclic Aβ42 hexamers. This information could not have been obtained by other means currently available.
Familial Alzheimer’s disease
The primary structure of Aβ42 given in Scheme I has notations where various mutations in the APP gene have been observed. These are often associated with a geographic region where the mutation has been found to occur in abundance. Formally each mutation leads to a somewhat different outcome and as such can have quite different symptoms from sporadic Alzheimer’s disease caused by wild type Aβ, often occurring with an earlier onset date (sporadic AD is known to ubiquitously begin close to age 65). For example, in a recent newspaper (June 29, 2016) it was reported that the iconic women’s basketball coach Pat Summit had died at age 64 from early onset Alzheimer’s disease. At UCSB, we have initiated studies on many of the mutants listed in Scheme I (67,68) but space limitation doesn’t let a detailed discussion be made of the results. What we have found so far, however, is that every mutant has profound effects on the assembly of both Aβ40 and Aβ42 leading to very different oligomer distributions and structures. Perhaps most important is the observation that the A2V and E22G mutations allow Aβ40 to form dodecamers of stacked cyclic hexamers and other mutations allow hexamers of Aβ40 to be formed. Since Aβ40 is 10 times more abundant than Aβ42 such mutations may be very dangerous indeed. We continue to pursue these systems including AFM measurements to confirm what we see in our IMS-MS studies and to investigate the onset of fibril formation in each system.
4. Case 2: Peptide Assembly and the Amyloid Paradigm
An amyloid is a proteinaceous, extracellular deposit with β-sheet structure. Consequently, since AD patients are known to have such extracellular plaques, an early hypothesis for AD was the Amyloid Cascade Hypothesis (20). In recent years it has become much more likely that early oligomers are responsible for AD yet the name “amyloid β-protein” has become synonymous with the Aβ40 and Aβ42 peptides believed to be involved in AD (see Case Study 1). The Amyloid Paradigm is different from the Amyloid Cascade Hypothesis. In the Amyloid Paradigm the focus is on the initial steps in the assembly process. It is widely accepted that amyloid is formed from association of (perhaps misfolded) peptides or proteins that at some size range undergo a transition to a β-sheet assembly followed by rapid addition of monomer to form pre-fibrils, fibrils and (possibly) plaques. Historically traditional methods could be used to study isolated monomers (various spectroscopies) and the terminal fibrils (x-ray, NMR) but the interesting and important intermediate species could not be studied using these methods. However, IMSMS can be used to follow oligomer growth with a classic example given in Case Study 1. Hence, there is a possibility that the amyloid process can be followed from monomer to eventual β-sheet dominated structures. Here we show the first example of the observation of the complete amyloid process using model peptides extracted from larger amyloid forming systems.
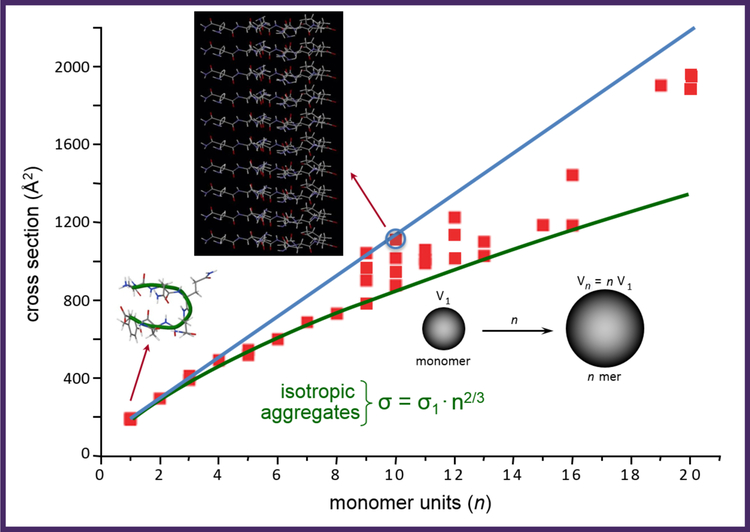
The data for the assembly of the peptide NNQQNY are shown in Figure 5 (69). This peptide was chosen because it is known to form fibrils under certain controlled conditions (70). The mass spectrum of NNQQNY clearly shows extensive assembly with oligomers to n = 20 readily observed. ATDs of each peak were taken and cross sections obtained for each feature in the ATDs. A plot of cross section versus oligomer number is shown in Figure 5. The green line is the prediction for isotropic growth. This curve follows the equation σn = n2/3σ1 where σn is the cross section of the nth oligomer, and assumes the nth oligomer has a volume n-times the volume of the monomer. What is apparent from the plot is that NNQQNY assembles in an isotropic manner through n = 8 but beginning at n = 9 significant positive deviations from the isotropic curve occur. The blue line is the prediction of cross section growth of oligomers with structures taken from the x-ray data (70). The structures of the monomer and the β-sheet decamer (taken from the x-ray data) are shown in Figure 5. Amazingly the cross section of the most extended n = 10 oligomer observed in the IMS-MS experiment essentially exactly fits the structure found in the x-ray data. A variety of cross sections (structures) are observed between n = 9 and n = 16 but above n = 16 only β-sheet structures are found. The NNQQNY system represents the first observation of the complete amyloid paradigm assembly process from monomer to fully β-sheet oligomers.
Figure 5. A plot of collision cross section against the number of monomers in each oligomer for the peptide NNQQNY.
The data points are given as the orange squares. The green line is the isotropic growth curve given by the formula in the figure. The blue line is taken from the cross sections of the β-sheet oligomers taken from the x-ray data of reference (70). The structures of the monomer and the β-sheet decamer are given. The cross section of the β-sheet decamer is given by the blue circle, in excellent agreement with the experimental data point. The cross section data were taken from reference (69).
In the same IMS study (69) the assembly of the opioid YGGFL was also reported. In this instance only isotropic growth was observed from monomer to the largest oligomer (n = 18). This result is consistent with the fact that YGGFL forms 3-dimensional isotropic crystals and not β-sheet fibrils (71). Hence not all systems form fibrils and the question remains as to why some do and why some don’t under biological assembly conditions.
There have been several approaches developed to attempt to predict the probability a particular primary structural sequence will assemble into β-sheet oligomers/fibrils (72–74). Perhaps the most successful of these is termed PASTA (72), an algorithm developed from a statistical analysis of β-sheet regions of folded proteins found in the protein data bank (75). The Bowers group has systematically mutated NNQQNY and YGGFL (76,77) and compared their assembly results with predictions of PASTA and other algorithms (76,77). The results indicate the various algorithms do not do a good job predicting the changes observed in assembly due to mutation of the peptides. More will be said on this topic in Case Study 3 on Amino Acid Assembly.
Two additional points on peptide assembly will be briefly discussed. First the Eisenberg group put forth a creative suggestion that there may be metastable assembly of peptides and proteins into specific structures that would be conducive to cell membrane disruption. They termed these structures “cylindrins”. They provided evidence that a specific eleven residue fragment of α-B crystalline assembles into a cylindrical β-barrel structure composed of 6 monomers aligned in off register anti-parallel β-sheets (78). They chose this fragment for a variety of reasons but, practically speaking, the most important was its relatively slow assembly rate that allowed them to isolate and crystalize the hexamer and to obtain its structure using xray methods.
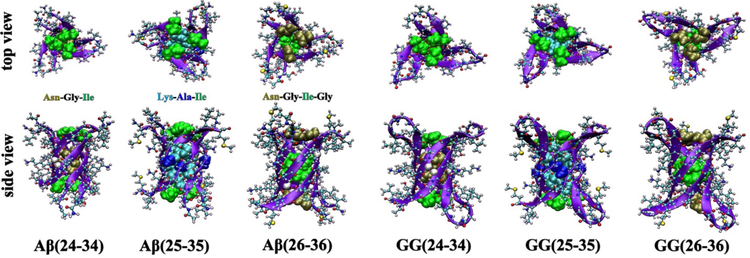
These are exciting and potentially important results. In a collaborative effort with the Eisenberg group we chose to apply the IMS-MS technique to a series of eleven residue fragments of the Aβ42 peptide: Aβ(24–34), Aβ(25–35) and Aβ(26–36). The Aβ(25–35) peptide fragment is known to exist in the brain and to be cytotoxic (79) and has been studied by our group previously for different purposes (80). The great advantage of IMS-MS is that essentially any system can be studied and individual oligomers analyzed regardless of how complex the mixture of oligomers in solution. Most systems do not lend themselves to the isolation/crystallization of specific oligomers in a dynamic assembly environment. While space does not allow the details to be given here, our efforts on all three systems were successful and the combination of IMS-MS and high level molecular modeling confirmed that cylindrins were formed by the hexamers of all 3 systems (46). Also, following Eisenberg’s lead, the 11 residue Aβ fragments were attached head to tail by -GG- linkers and the trimers of these 24 residue peptides also readily formed cylindrins (81). The structures of all six systems are given in Figure 6.
Figure 6. Cylindrin models of single-repeat Aβ hexamers and tandem-repeat GG trimers.
Each peptide chain is shown as a violet β-strand in CPK representation. The side chains inside the cylindrin cavities are shown in space filling representation. Both top views and side views are given for each system. The models are reconstructed from reference (81).
These results are very encouraging and indicate that the cylindren/β-barrel structure proposed by Eisenberg is worthy of further research to see how general it is.
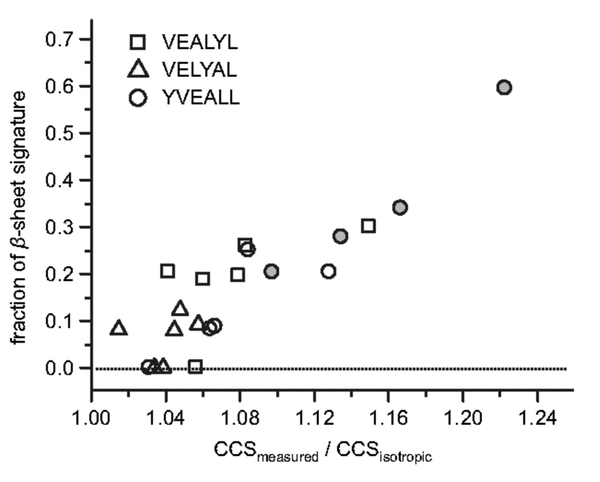
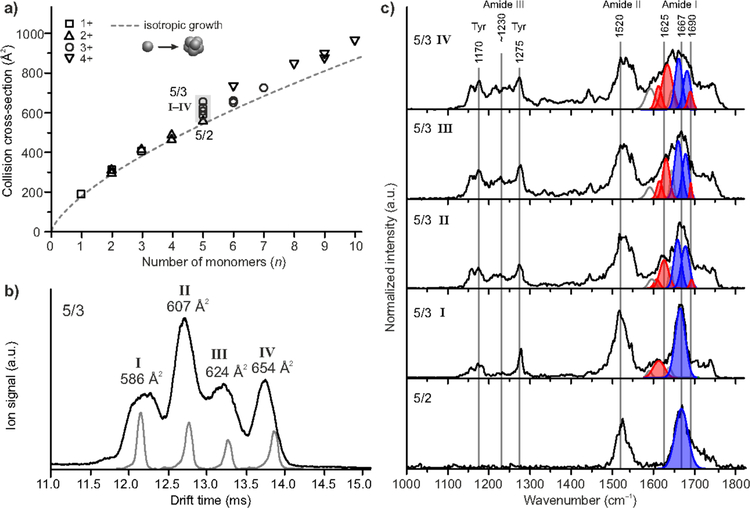
Finally, the growth of β-sheet content as peptides assemble leads to cross sections larger than isotropic as n increases. It should be possible to observe this growth using infrared spectroscopy as there are characteristic β-sheet absorbtions in the Amide I infrared band. What is needed is a high resolution IMS system (13) for conformational selection coupled to an MS for mass selection coupled to a free electron laser/MS detector for spectroscopy of the shape and mass selected oligomers. Such a system has been assembled in the lab of Gert von Helden at the Fritz Haber Institute in Berlin in a collaborative effort of the von Helden, Bowers and Kevin Pagel (Free University in Berlin) groups (82). An initial study has been undertaken of the VEALYL fragment of Insulin that prior work has suggested forms β-sheet oligomers at relatively small oligomer sizes (69). Two scrambled sequences of VEALYL were included; YVEALL and VELYAL. IMS-MS studies indicated the first of these behaved similar to VEALYL, with early oligomers more extended than expected from isotropic assembly, and that the second assembled in an isotropic manner and hence could act as a negative standard. Again space constraints prevent the details from being given here. It was found that β-sheet onset was spectroscopically observed for both VEALYL and YVEALL with onset at n = 4 extending to n = 9 (the largest oligomer studied) but no β-sheet absorptions were observed for VELYAL in accordance of expectations from both IMS-MS and TEM results (83). This direct spectroscopic observation of β-sheet onset in oligomer growth is potentially a very important result and suggests that this type of investigation may be possible on systems of direct biological importance. In addition a direct correlation of the spectroscopic results with IMS measured cross sections was observed: The greater the positive deviation above the isotropic line the greater the percentage of β-sheet observed spectroscopically directly vetting the IMS-MS method as a way to determine β-sheet content as peptides and proteins assemble (Figure 7). Key results for the onset of β-sheet in oligomers of the YVEALL peptide are given in Figure 8.
Figure 7. Fraction of β-sheet in the amide I band of VEALYL (squares), VELYAL (triangles), and YVEALL (circles) oligomers as a function of relative deviation in collision cross section from the isotropic growth model.
Solid circles denote the polymorph pentamers (5/2 and 5/3, I–IV) of YVEALL, which are shown in Figure 6. Reconstructed from reference (83).
Figure 8. Ion mobility spectrometry and conformer-selected IR spectroscopy of YVEALL oligomers.
(a) Collision cross sections of YVEALL oligomers as a function of the number of monomers n. The dashed line denotes isotropic growth. The statistical error of the measured cross sections is less than 1% and smaller than the symbol size. (b) The ATD of the triply protonated pentamer (5/3) for which multiple conformers with distinct cross sections are observed. The narrow peaks depicted in gray correspond to the portions of the oligomer distribution that were selected for IRMPD-spectroscopic analysis. (c) IRMPD spectra of the doubly-protonated pentamer (5/2) and the drift-time selected species (I–IV) of the triply protonated YVEALL pentamers (5/3). The amide I region (1600–1700 cm−1) was fitted by multiple Gaussian peaks. The blue and red Gaussians represent IR bands corresponding to turn-like (1660–1685 cm−1) and β-sheet structures (1610–1640 cm–1 and ~1690 cm−1), respectively. For details about the fitting procedure see reference (83) from which the figure was reconstructed.
These results indicate the IMS-MS/FEL technique developed in the FHI in Berlin has a bright future in the analysis of the secondary structure of peptides and proteins and their assemblies adding a very powerful compliment to the ability of IMS-MS methods to directly measure tertiary structure of monomers and the quaternary structure of assemblies (36,83).
5. Case Study 3: Amino Acid Assembly
The fundamental building blocks of peptides and proteins are amino acids of which there are 20 that are commonly found in living systems. The focus here will be on the non-covalent assembly of AAs, the resulting assembly structures and whether these structures are relevant to disease and to peptide/protein aggregation. The amino acid serine has historically been the most studied since early work by Cooks and coworkers indicated the serine octamer was a “magic number” cluster and it had a strong propensity for forming chirally pure structures (84,85). This observation stimulated great interest with other research groups focusing on the determination of the structure of the serine octamer (38,86–88) but the detailed structure remains undetermined to the present time. There have been reports of various levels of assembly of other AAs including arginine (89), tryptophan (90), proline (91), tyrosine (92) and phenylalanine (93–95). However, until recently there had been little effort to understand the non-covalent assembly of AA oligomers based on their chemical and physical properties, nor had this information been applied to help understand the aggregation of peptides and proteins (96). This latter point will be briefly addressed below. First, however, the assembly of Phe will be discussed.
Gazit and coworkers (93) were the first to investigate the assembly of non-covalent Phe oligomers under conditions similar to those common for patients suffering from the disease of phenylketonuria (PKU). They observed formation of amyloid like fibrils and using molecular dynamics simulations at high pH proposed a ladder like mechanism for forming the fibrils. In addition, they showed that Phe was cytotoxic at high concentrations and that they developed antibodies specific to Phe oligomers. This excellent and exciting work stimulated our interest in amino acid assembly and we chose phenylalanine as the first system to study (95).
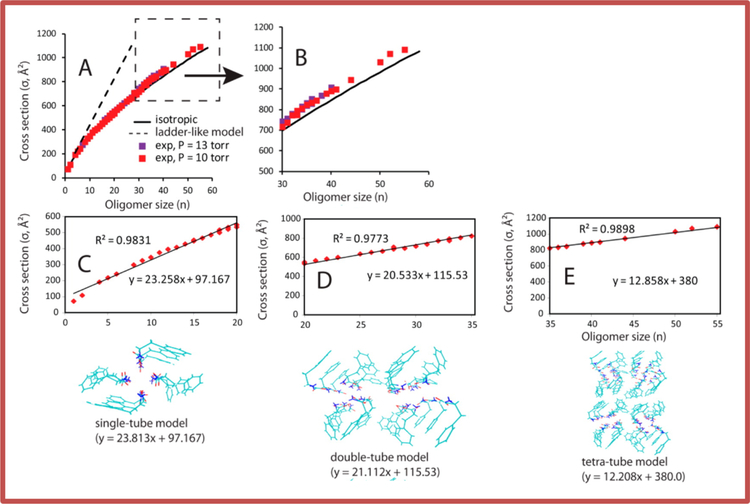
At similar concentrations used by Gazit (5 mM) in our soft nano ESI instrument, we observed Phe oligomers to at least n = 60. The cross sections of the oligomers were measure and plotted versus the oligomer number n (Figure 9 top part) and compared to the prediction of isotropic assembly.
Figure 9. Plots of collision cross section versus oligomer number for the self-assembly of phenylalanine oligomers.
The dashed linear line in A is the predicted growth pattern of the ladder mechanism of reference 60. The black solid line is the isotropic growth line. Panel B is a blow up of panel A for n > 30. The data points in panels C, D, and E are from panel A. The solid linear lines are the cross sections generated from the various pore structures beneath the panels: single pore for panel C, double pore for panel D, and tetra-pore for panel E. The R2 values for the fits of the models to the data are given. The figure was reconstructed from reference (96).
While early assembly appeared isotropic the fit was systematically off and above n = 30 experimental cross sections were clearly larger than isotropic. The ladder mechanism of Gazit and coworkers did not fit the data, increasing too fast with n (Figure 9 top left). [As an aside, IMS data taken at pH 11 did show a minor component with cross sections close to those predicted from the ladder mechanism (95)]. Fortunately, as we were considering how to interpret our data, Hansmann and coworkers published results of MD simulations of Phe assembly in pure water at pH 7. They found a pore like zwitterionic structure to be dominant. This structure is composed of 4 molecules in a plane with charged termini focused to the inside and the phenyl groups on the outside. Layers of these planes then stacked, stabilized by both ionic bonding in the interior and π-stacking of the phenyl groups on the exterior. Comparison of calculated cross sections of the model structures with the IMS experimental data gave reasonable agreement up to about n = 20. However, as n increased the model structures systematically overestimated the experimental cross sections. Our solution was to build first a double pore set of model structures starting with the single pore model of Hansmann and then a tetra pore model as n increased above n = 36 (Figure 9 bottom). The fit with experiment for the “growing” pore models is excellent, as shown in the middle portion of Figure 9.
These are very surprising results. It is unusual for biological systems to assemble with the hydrophobic part on the outside and the hydrophilic part on the inside. However, in Phe oligomers this does seem to be the case. As a consequence, such oligomers are well suited for insertion into the cellular plasma membrane, possibly causing leakage across it. Further it makes sense that oligomers, not fibrils, be the neurotoxic agents in PKU since they are so much more mobile than fibrils. In addition, oligomers are more likely to form than fibrils at the concentrations of Phe in blood of the PKU patient. Finally, as noted in Case Study 1, oligomers and not fibrils are now widely believed to be the toxic agents in Alzheimer’s disease and many other amyloid related diseases. PKU appears to be fitting into this scenario.
One final comment will be made on the possible utility of AA assembly. As noted in Case Study 2 there is need to find a useful algorithm to predict amyloid propensity from stretches of primary structure in peptides and proteins. There have been several attempts to develop such an algorithm (72–74) but all have been unsuccessful when compared to data for peptide amyloid formation (76,77). In observing assembly of several AAs we noticed that hydrophilic AAs tended to assembly more compactly than isotropic and hydrophobic more extended than isotropic (96). Given this unexpected observation we developed a simple algorithm based on these tendencies. We have begun testing the algorithm by constructing peptides of the tested AAs and predicting how they would assemble. From this very limited set of data we found the new algorithm performed admirably, accurately predicting not only the propensity for amyloid formation but also resistance to amyloid formation resulting in 3-d crystal formation instead (96). Work is continuing to further test and extend the reliability of this algorithm. What can be said at present, however, is that it appears the best way to establish what an AA will do when sequenced in a peptide or protein is to see how it interacts with itself without tethering it to other AAs.
6. Acknowledgements
The support of the National Science Foundation under grants CHE-1301032 and CHE-1565941, The Air Force Office of Scientific Research under grant FA9550–11-0113 and the National Institutes of Health under grant 1R01AG047116–01 is gratefully acknowledged (MTB).
7 References
- 1.Bowers MT. Ion mobility spectrometry: A personal view of its development at UCSB. Int J Mass Spectrom. 2014; 370:75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helden G von Hsu M-T, Kemper PR Bowers MT. Structures of carbon cluster ions from 3 to 60 atoms: Linears to rings to fullerenes. J Chem Phys 1991; 95(5):3835. [Google Scholar]
- 3.Helden G von, Hsu MT, Gotts N, Bowers MT. Carbon cluster cations with up to 84 atoms: structures, formation mechanism, and reactivity. J Phys Chem 1993; 97(31):8182–8192. [Google Scholar]
- 4.Gert von Helden Nigel G. Gotts, Bowers Michael T.. Experimental Evidence for the Formation of Fullerenes by Collisional Heating of Carbon Rings in the Gas Phase. Nature. 1993; 363:60–63. [Google Scholar]
- 5.Hunter J, Fye J, Jarrold MF. Annealing C60+: Synthesis of Fullerenes and Large Carbon Rings. Science. 1993; 260:784–786. [DOI] [PubMed] [Google Scholar]
- 6.Kroto HW, Heath JR, O’Brien SC, Curl RF, Smalley RE. C60: Buckminsterfullerene. Nature. 1985; 318:162–164. [Google Scholar]
- 7.Kraetschmer W, Fostiropoulis K, Huffman DR. The infrared and ultraviolet absorption spectra of laboratory-produced carbon dust: evidence for the presence of the C60 molecule. Phys Chem Fuller Repr Collect 1993; 1:23. [Google Scholar]
- 8.Helden G von Gotts NG, Bowers MT. Annealing of carbon cluster cations: rings to rings and rings to fullerenes. J Am Chem Soc 1993; 115(10):4363–4364. [Google Scholar]
- 9.Gotts NG, Helden G von, Bowers MT. Carbon cluster anions: Structure and growth from C5− to C62−. Int J Mass Spectrom Ion Process 1995; 149:217–229. [Google Scholar]
- 10.Wyttenbach T, Helden G von, Bowers MT. Gas-phase conformation of biological molecules: Bradykinin. J Am Chem Soc 1996; 118(35):8355–8364. [Google Scholar]
- 11.Wyttenbach T, Bowers MT. Gas phase conformations of biological molecules: the hydrogen/deuterium exchange mechanism. J Am Soc Mass Spectrom. 1999; 10(1):9–14. [Google Scholar]
- 12.Wyttenbach T, Kemper PR, Bowers MT. Design of a new electrospray ion mobility mass spectrometer. Int J Mass Spectrom 2001; 212(1):13–23. [Google Scholar]
- 13.Kemper PR, Dupuis NF, Bowers MT. A new, higher resolution, ion mobility mass spectrometer. Int J Mass Spectrom 2009; 287(1–3):46–57. [Google Scholar]
- 14.Bernstein SL, Wyttenbach T, Baumketner A, et al. Amyloid ß-Protein: Monomer Structure and Early Aggregation States of Aß42 and Its Pro19 Alloform. J Am Chem Soc 2005; 127(7):2075–2084. [DOI] [PubMed] [Google Scholar]
- 15.Wyttenbach T, Bowers MT. Structural Stability from Solution to the Gas Phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility-Mass Spectrometry Environment. J Phys Chem B 2011; 115(42):12266–12275. [DOI] [PubMed] [Google Scholar]
- 16.Baumketner A, Bernstein S, Wyttenbach T, et al. Amyloid beta-protein monomer structure: A computational and experimental study. Protein Sci 2006; 15(3):420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupuis NF, Wu C, Shea J-E, Bowers MT. Human Islet Amyloid Polypeptide Monomers Form Ordered ß-hairpins: A Possible Direct Amyloidogenic Precursor. J Am Chem Soc 2009; 131(51):18283–18292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier JJ, Kayed R, Lin C-Y, et al. Inhibition of human IAPP fibril formation does not prevent beta-cell death: evidence for distinct actions of oligomers and fibrils of human IAPP. AJP Endocrinol Metab. 2006; 291(6):E1317–E1324. [DOI] [PubMed] [Google Scholar]
- 19.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J Neurosci Res 2002; 69(5):567–577. [DOI] [PubMed] [Google Scholar]
- 20.Hardy J, Selkoe DJ. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science. 2002; 297(5580):353–356. [DOI] [PubMed] [Google Scholar]
- 21.Dill KA. Dominant forces in protein folding. Biochemistry (Mosc). 1990; 29(31):7133–7155. [DOI] [PubMed] [Google Scholar]
- 22.How Haran G., when and why proteins collapse: the relation to folding. Curr Opin Struct Biol 2012; 22(1):14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruebele M Protein folding: the free energy surface. Curr Opin Struct Biol 2002; 12:161–168. [DOI] [PubMed] [Google Scholar]
- 24.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012; 338(6110): 1042–1046. [DOI] [PubMed] [Google Scholar]
- 25.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol 2004; 14(1):70–75. [DOI] [PubMed] [Google Scholar]
- 26.Lazar GA, Desjarlais JR, Handel TM. De novo design of the hydrophobic core of ubiquitin. Protein Sci 1997; 6(6):1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson EC, Lazar GA, Desjarlais JR, Handel TM. Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin. Structure. 1999; 7(8):967–976. [DOI] [PubMed] [Google Scholar]
- 28.Wyttenbach T, Bowers MT. Hydration of biomolecules. Chem Phys Lett 2009; 480(1–3): 1–16. [Google Scholar]
- 29.Stefani M Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta BBA - Mol Basis Dis 2004; 1739(1):5–25. [DOI] [PubMed] [Google Scholar]
- 30.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev 2005; 24(4):508–548. [DOI] [PubMed] [Google Scholar]
- 31.Wyttenbach T, Paizs B, Barran P, et al. The Effect of the Initial Water of Hydration on the Energetics, Structures, and H/D Exchange Mechanism of a Family of Pentapeptides: An Experimental and Theoretical Study. J Am Chem Soc 2003; 125(45):13768–13775. [DOI] [PubMed] [Google Scholar]
- 32.Meyer T, Gabelica V, Grubmüller H, Orozco M. Proteins in the gas phase. Wiley Interdiscip Rev Comput Mol Sci 2013; 3(4):408–425. [Google Scholar]
- 33.Badman ER, Hoaglund-Hyzer CS, Clemmer DE. Monitoring Structural Changes of Proteins in an Ion Trap over ~ 10–200 ms: Unfolding Transitions in Cytochrome c Ions. Anal Chem 2001; 73(24):6000–6007. [DOI] [PubMed] [Google Scholar]
- 34.Breuker K, McLafferty FW. Stepwise evolution of protein native structure with electrospray into the gas phase, 10−12 to 102 s. Proc Natl Acad Sci 2008; 105(47):18145–18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warnke S, Helden G von, Pagel K. Protein Structure in the Gas Phase: The Influence of Side-Chain Microsolvation. J Am Chem Soc 2013; 135(4):1177–1180. [DOI] [PubMed] [Google Scholar]
- 36.Seo J, Hoffmann W, Warnke S, Bowers MT, Pagel K, von Helden G. Retention of Native Protein Structures in the Absence of Solvent: A Coupled Ion Mobility and Spectroscopic Study. Angew Chem Int Ed 2016; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silveira JA, Fort KL, Kim D, et al. From Solution to the Gas Phase: Stepwise Dehydration and Kinetic Trapping of Substance P Reveals the Origin of Peptide Conformations. J Am Chem Soc 2013; 135(51):19147–19153. [DOI] [PubMed] [Google Scholar]
- 38.Counterman AE, Clemmer DE. Magic Number Clusters of Serine in the Gas Phase. J Phys Chem B 2001; 105(34):8092–8096. [Google Scholar]
- 39.Koeniger SL, Merenbloom SI, Sevugarajan S, Clemmer DE. Transfer of Structural Elements from Compact to Extended States in Unsolvated Ubiquitin. J Am Chem Soc 2006; 128(35): 11713–11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S-W, Freivogel P, Schindler T, Beauchamp JL. Freeze-Dried Biomolecules: FT-ICR Studies of the Specific Solvation of Functional Groups and Clathrate Formation Observed by the Slow Evaporation of Water from Hydrated Peptides and Model Compounds in the Gas Phase. J Am Chem Soc 1998; 120(45):11758–11765. [Google Scholar]
- 41.Laskin J, Futrell JH. Collisional activation of peptide ions in FT-ICR mass spectrometry. Mass Spectrom Rev 2003; 22(3):158–181. [DOI] [PubMed] [Google Scholar]
- 42.McLuckey SA, Goeringer DE. Slow heating methods in tandem mass spectrometry. J Mass Spectrom 1997; 32(5):461–474. [Google Scholar]
- 43.Tolmachev AV, Vilkov AN, Bogdanov B, PÄsa-Tolic L, Masselon CD, Smith RD. Collisional activation of ions in RF ion traps and ion guides: The effective ion temperature treatment. J Am Soc Mass Spectrom 2004; 15(11):1616–1628. [DOI] [PubMed] [Google Scholar]
- 44.Hoxha A, Collette C, De Pauw E, Leyh B. Mechanism of Collisional Heating in Electrospray Mass Spectrometry: Ion Trajectory Calculations. J Phys Chem A. 2001; 105(31):7326—7333. [Google Scholar]
- 45.Femandez-Lima FA, Kaplan DA, Park MA. Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev Sci Instrum 2011; 82(12):126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Lima F, Kaplan DA, Suetering J, Park MA. Gas-phase separation using a trapped ion mobility spectrometer. Int J Ion Mobil Spectrom 2011; 14(2–3):93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langevin MP. Ann Chim Phys. 1905; 8:245–288. [Google Scholar]
- 48.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. New York: Wiley; 1988. [Google Scholar]
- 49.Pringle SD, Giles K, Wildgoose JL, et al. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom 2007; 261(1):1–12. [Google Scholar]
- 50.Liu FC, Kirk SR, Bleiholder C. On the structural denaturation of biological analytes in trapped ion mobility spectrometry - mass spectrometry. The Analyst. 2016; 141(12):3722–3730. [DOI] [PubMed] [Google Scholar]
- 51.Bleiholder C Towards measuring ion mobilities in non-stationary gases and non-uniform and dynamic electric fields (I). Transport equation. Int J Mass Spectrom 2016; 399–400:1–9. [Google Scholar]
- 52.Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ. Three-dimensional structure of ubiquitin at 2.8 A resolution. Proc Natl Acad Sci 1985; 82(11):3582–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bleiholder C, Johnson NR, Contreras S, Wyttenbach T, Bowers MT. Molecular Structures and Ion Mobility Cross Sections: Analysis of the Effects of He and N 2 Buffer Gas. Anal Chem 2015; 87(14):7196–7203. [DOI] [PubMed] [Google Scholar]
- 54.Benigni P, Marin R, Molano-Arevalo JC, et al. Towards the analysis of high molecular weight proteins and protein complexes using TIMS-MS. Int J Ion Mobil Spectrom [Internet]. 2016. [cited 2016 Jul 25]; . Available from: http://link.springer.com/10.1007/s12127-016-0201-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laszlo KJ, Munger EB, Bush MF. Folding of Protein Ions in the Gas Phase after Cation to Anion Proton Transfer Reactions (CAPTR). J Am Chem Soc 2016; . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alzheimer’s Association. Changing the trajectory of Alzheimer’s disease: A national imperative [Internet] http://alz.org/alzheimersdiseasetrajectory.asp Available from: http://alz.org/alzheimersdiseasetrajectory.asp
- 57.Suzuki N, Cheung TT, Cai XD, et al. An increased percentage of long amyloid beta protein secreted by familial amyloid beta protein precursor (beta APP717) mutants. Science. 1994; :1336–1340. [DOI] [PubMed] [Google Scholar]
- 58.Gravina SA, Ho LB, Eckman C, et al. Amyloid beta-protein (Abeta) in Alzheimer’s disease brain: Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Abeta40 or Abeta42(43). J Biol Chem 1995; 270:7013–7016. [DOI] [PubMed] [Google Scholar]
- 59.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid ß-protein (Aß) assembly: Aß40 and Aß42 oligomerize through distinct pathways. Proc Natl Acad Sci 2003; 100(1):330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dahlgren KN, Arlene M. Manell, W. Blaine Stine, Lorinda K. Baker, Grant A. Krafft, Mary Jo LaDu. Oligomeric and Fibrillar Species of Amyloid-beta Peptides Differentially Affect Neuronal Viability. J Biol Chem 2002; 277(35):32046–32053. [DOI] [PubMed] [Google Scholar]
- 61.Bernstein SL, Dupuis NF, Lazo ND, et al. Amyloid-P protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem 2009; 1(4):326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray MM, Bernstein S, Nyugen V, Condron MM, Teplow D., Bowers MT. Amyloid beta-protein: Abeta40 inhibits Abeta42 oligomerization. J Am chem Soc 2009; 131:6316–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesné S, Koh MT, Kotilinek L, et al. A specific amyloid-P protein assembly in the brain impairs memory. Nature. 2006; 440(7082):352–357. [DOI] [PubMed] [Google Scholar]
- 64.Cheng IH, Scearce-Levie K, Legleiter J, et al. Accelerating Amyloid-P Fibrillization Reduces Oligomer Levels and Functional Deficits in Alzheimer Disease Mouse Models. J Biol Chem 2007; 282(33):23818–23828. [DOI] [PubMed] [Google Scholar]
- 65.Economou NJ, Giammona MJ, Do TD, et al. Amyloid P-Protein Assembly and Alzheimer’s Disease: Dodecamers of AP42, but Not of AP40, Seed Fibril Formation. J Am Chem Soc 2016; 138(6): 1772–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB. Amyloid P-protein fibrillogenesis detection of a protofibrillar intermediate. J Biol Chem 1997; 272(35):22364–22372. [DOI] [PubMed] [Google Scholar]
- 67.Gessel MM, Bernstein S, Kemper M, Teplow DB, Bowers MT. Familial Alzheimer’s Disease Mutations Differentially Alter Amyloid P-Protein Oligomerization. ACS Chem Neurosci 2012; 3(11):909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng X, Liu D, Roychaudhuri R, Teplow DB, Bowers MT. Amyloid P-Protein Assembly: Differential Effects of the Protective A2T Mutation and Recessive A2V Familial Alzheimer’s Disease Mutation. ACS Chem Neurosci 2015; 6(10):1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to P-sheet in amyloid fibril formation. Nat Chem 2011; 3(2):172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sawaya MR, Sambashivan S, Nelson R, et al. Atomic structures of amyloid cross-P spines reveal varied steric zippers. Nature. 2007; 447(7143):453–457. [DOI] [PubMed] [Google Scholar]
- 71.Smith GD, Griffin JF. Conformation of [Leu5] enkephalin from X-ray diffrachartion: Features important for recognition at the opiate receptor,. Science. 1978; 199:1214–1216. [DOI] [PubMed] [Google Scholar]
- 72.Trovato A, Chiti F, Maritan A, Seno F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLoS Comput Biol. 2006; 2(12):e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “Aggregation- prone” and “Aggregation-susceptible” Regions in Proteins Associated with Neurodegenerative Diseases. J Mol Biol 2005; 350(2):379–392. [DOI] [PubMed] [Google Scholar]
- 74.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci 2010; 107(8):3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berman HM, Henrick K, Nakamura H. Announcing the world wide protein data bank. Nat Struct Biol 10:980. [DOI] [PubMed] [Google Scholar]
- 76.Do TD, Economou NJ, LaPointe NE, et al. Factors That Drive Peptide Assembly and Fibril Formation: Experimental and Theoretical Analysis of Sup35 NNQQNY Mutants. J Phys Chem B. 2013; 117(28):8436–8446. [DOI] [PubMed] [Google Scholar]
- 77.Do TD, LaPointe NE, Sangwan S, et al. Factors That Drive Peptide Assembly from Native to Amyloid Structures: Experimental and Theoretical Analysis of [Leu-5]-Enkephalin Mutants. J Phys Chem B. 2014; 118(26):7247–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu C, Zhao M, Jiang L, et al. Out-of-register -sheets suggest a pathway to toxic amyloid aggregates. Proc Natl Acad Sci 2012; 109(51):20913—20918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pike CJ, Walencewicz-Wasserman AJ, Kosmoski J, Cribbs DH, Glabe CG, Cotman CW. Structure-Activity Analyses of ß-Amyloid Peptides: Contributions of the ß25–35 Region to Aggregation and Neurotoxicity. J Neurochem 1995; 64:253–265. [DOI] [PubMed] [Google Scholar]
- 80.Bleiholder C, Do TD, Wu C, et al. Ion Mobility Spectrometry Reveals the Mechanism of Amyloid Formation of Aß(25–35) and Its Modulation by Inhibitors at the Molecular Level: Epigallocatechin Gallate and Scyllo -inositol. J Am Chem Soc 2013; 135(45):16926–16937. [DOI] [PubMed] [Google Scholar]
- 81.Do TD, LaPointe NE, Nelson R, et al. Amyloid ß-Protein C-Terminal Fragments: Formation of Cylindrins and ß-Barrels. J Am Chem Soc 2016; 138(2):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Warnke S, Baldauf C, Bowers MT, Pagel K,Helden G von. Photodissociation of Conformer-Selected Ubiquitin Ions Reveals Site-Specific Cis / Trans Isomerization of Proline Peptide Bonds. J Am Chem Soc 2014; 136(29):10308–10314. [DOI] [PubMed] [Google Scholar]
- 83.Seo J, Hoffman W, Huang X, et al. An infrared spectorscopy approach to follow beta-sheet formation in peptide amyloid assemblies. Nat Chem in press. [DOI] [PubMed] [Google Scholar]
- 84.Koch KJ, Zhang D, Cooks RG, Gozzo FC, Eberlin MN. Serine octamer metaclusters: formation, structure elucidation and implications for homochiral polymerization. Chem Commun 2001; (18): 1854–1855. [DOI] [PubMed] [Google Scholar]
- 85.Cooks RG, Zhang D, Koch KJ, Gozzo FC, Eberlin MN. Chiroselective Self-Directed Octamerization of Serine: Implications for Homochirogenesis. Anal Chem 2001; 73(15):3646–3655. [DOI] [PubMed] [Google Scholar]
- 86.Julian RR, Hodyss R, Kinnear B, Jarrold MF, Beauchamp JL. Nanocrystalline Aggregation of Serine Detected by Electrospray Ionization Mass Spectrometry: Origin of the Stable Homochiral Gas-Phase Serine Octamer. J Phys Chem B. 2002; 106(6):1219–1228. [Google Scholar]
- 87.Schalley CA, Weis P. Unusually stable magic number clusters of serine with a surprising preference for homochirality. Int J Mass Spectrom 2002; 221(1):9–19. [Google Scholar]
- 88.Spencer EAC, Ly T, Julian RR. Formation of the serine octamer: Ion evaporation or charge residue? Int J Mass Spectrom 2008; 270(3):166–172. [Google Scholar]
- 89.Julian RR, Hodyss R, Beauchamp JL. Salt Bridge Stabilization of Charged Zwitterionic Arginine Aggregates in the Gas Phase. J Am Chem Soc 2001; 123(15):3577—3583. [DOI] [PubMed] [Google Scholar]
- 90.Feketeová L, Khairallah GN, Brunet C, et al. Fragmentation of the tryptophan cluster [Trp9–2H]2-induced by different activation methods. Rapid Commun Mass Spectrom 2010; 24(22):3255–3260. [DOI] [PubMed] [Google Scholar]
- 91.Counterman AE, Clemmer DE. Anhydrous Polyproline Helices and Globules. J Phys Chem B. 2004; 108(15):4885—4898. [Google Scholar]
- 92.Shaham-Niv S, Adler-Abramovich L, Schnaider L, Gazit E. Extension of the generic amyloid hypothesis to nonproteinaceous metabolite assemblies. Sci Adv 2015; 1(7):e1500137–e1500137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adler-Abramovich L, Vaks L, Carny O, et al. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat Chem Biol 2012; 8(8):701–706. [DOI] [PubMed] [Google Scholar]
- 94.Reches M, Gazit E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science. 2003; 300:625–627. [DOI] [PubMed] [Google Scholar]
- 95.Do TD, Kincannon WM, Bowers MT. Phenylalanine Oligomers and Fibrils: The Mechanism of Assembly and the Importance of Tetramers and Counterions. J Am Chem Soc 2015; 137(32): 10080–10083. [DOI] [PubMed] [Google Scholar]
- 96.Do TD, Almeida NEC de, LaPointe NE, Chamas A, Feinstein SC, Bowers MT. Amino Acid Metaclusters: Implications of Growth Trends on Peptide Self-Assembly and Structure. Anal Chem 2016; 88(1):868–876. [DOI] [PubMed] [Google Scholar]