Abstract
Objective
In functional electrophysiological imaging, signals are often contaminated by interference that can be of considerable magnitude compared to the signals of interest. This paper proposes a novel algorithm for removing such interferences that does not require separate noise measurements.
Approach
The algorithm is based on a dual definition of the signal subspace in the spatial- and time-domains. Since the algorithm makes use of this duality, it is named the dual signal subspace projection (DSSP). The DSSP algorithm first projects the columns of the measured data matrix onto the inside and outside of the spatial-domain signal subspace, creating a set of two preprocessed data matrices. The intersection of the row spans of these two matrices is estimated as the time-domain interference subspace. The original data matrix is projected onto the subspace that is orthogonal to the interference subspace
Main results
The DSSP algorithm is validated first by using the computer generated data, and then by using two sets of real biomagnetic data: SCEF data measured from a healthy volunteer and MEG data from a patient with a vagus nerve stimulator.
Significance
The proposed DSSP algorithm is effective for removing overlapped interference in a wide variety of biomagnetic measurements.
1. Introduction
Functional electrophysiological imaging can be achieved using a variety of modalities including electroencephalography (EEG), magnetoencephalography (MEG) or magnetospinography (MSG). In all of these modalities, measurement signals are often contaminated by interference that can be of considerable magnitude compared to the signals of interest. Although quite a few algorithms have been developed to deal with such interference, these algorithms often rely on availability of separate measurements that capture the statistical properties of the interference [1][2][3]. Therefore, if such separate measurements are not available, those existing algorithms would not be effective for removing overlapped interference. We present two examples of such cases in this paper: imaging of the spinal cord electrophisiological activitiy and magnetoencephalographic (MEG) imaging for epilepsy patients implanted with vagus nerve stimulators.
There has been growing interest in dynamic source imaging of spinal cord electrophysiological activity using its evoked magnetic field. In spite of the fact that spinal cord disorders are very common[4], there are no effective methods for accurate diagnosis of spinal cord lesions, primarily because spinal cord abnormalities found in a patient’s anatomical image (such as MRI or X-ray images) often do not correspond with the patient’s clinical symptoms. Thus, functional imaging of spinal cord nerve activity can be a promising diagnostic tool for spinal cord disorders.
Biomagnetometers optimized for measuring the spinal cord evoked magnetic field (SCEF) have been developed[5][6][7], along with efficient source reconstruction algorithms suited to functional spinal cord imaging[8][9]. However, one serious problem, the removal of large stimulus-induced artifacts1, remains to be solved. Such artifacts exist for 8–10 ms immediately after the stimulus onset. These artifacts overlie the SCEF signal, and can distort imaging results of spinal cord activity as shown in our experiments described in Sections 3.1 and 4.
Similarly, in MEG measurements, extremely large artifacts sometimes contaminate the measured data and it can be quite difficult to visualize the underlying signal of interest within this artifact. One striking example is in MEG recordings obtained from epilepsy patients implanted with vagal nerve stimulators (VNS) where the artifact from the stimulator and the lead-wires can completely contaminate the recordings such that it is extremely difficult to see interictal epileptiform activity or stimulus evoked responses from patients’ primary sensory cortices.
This paper proposes a novel algorithm to remove these artifacts from biomagnetic measurements. The algorithm is based on the two kinds of definitions of the signal subspace, as the spatial-domain signal subspace and the time domain signal subspace. Since the algorithm makes use of this duality, it is named the dual signal subspace projection (DSSP) algorithm. The DSSP algorithm first projects the columns of the measured data matrix onto the inside and outside of the spatial-domain signal subspace, creating two “projected” data matrices.
The intersection of the row spans of those two “projected” matrices is then taken to be an estimate of the time-domain interference subspace, and artifact removal is carried out on the basis of this estimated interference subspace. In this paper, the theory behind the DSSP algorithm is provided in sufficient details to explain why the interference subspace can be estimated in such a manner. The proposed algorithm is first validated by using the computer generated data that simulates the SCEF measurements and by using the SCEF data taken from a healthy volunteer.
It should be mentioned that estimating the interference subspace as the intersection between two time-domain subspaces was first proposed in [10]. That method is called temporal signal space separation (tSSS). However, there is a key difference between tSSS and DSSP in that tSSS uses the vector spherical harmonics expansions, which are not used in DSSP. The application of the tSSS method, therefore, is limited to magnetoencephalography (MEG) applications in which the data is acquired by an array of sensors that are arranged on a surface of a sphere-like helmet. The tSSS method is, therefore, not applicable to our SCEF measurements, in which the sensors are arranged on a nearly-flat surface.
In contrast, the proposed DSSP algorithm is a versatile algorithm. Its application is not limited to the artifact removal in SCEF measurements, and it is applicable to a wide variety of biomagnetic data. To demonstrate this versatility, we present the results of applying the algorithm to MEG data acquired from a patient with a vagus nerve stimulator. Although the MEG data contained a large amount of interference generated from the stimulator, primary somatosensory cortices were successfully localized by interference removal using the proposed DSSP algorithm.
2. Dual signal subspace projection (DSSP)
2.1. Data Model
We assume that the biomagnetic measurements are conducted by using an array of M sensors. We denote the output of the mth sensor at time t by bm(t) and the column vector containing the outputs of all sensors is b(t): b(t) = [b1, …, bM(t)]T. The b(t) is called the data vector. We use a data model in which the measured data consists of the signal magnetic field generated from signal sources of interest, the interference magnetic field generated from interference sources, and the sensor noise. That is, the data vector is expressed as
| (1) |
where bs(t), the signal vector, represents the signal magnetic field, bI(t), the interference vector, represents the interference magnetic field, and ε, the noise vector, represents the sensor noise. The spatio-temporal data matrices of b(t), bs(t), and bI (t) are, respectively, denoted by B, BS, and Bj, such that
| (2) |
| (3) |
| (4) |
where the data acquisition takes place at t1, …, tK and M < K is assumed. The M × K spatio-temporal data matrix B is modeled as
| (5) |
where Bε is a matrix whose columns consist of the noise vectors ε at t1, …, tK. The spatio-temporal data model in Eq. (5) is used in our analysis.
2.2. Sensor lead field and signal subspace
Let us assume that a unit-magnitude source exists at r. When the source is oriented in the x, y, and z directions, the corresponding outputs of the jth sensor are, respectively, denoted , , and . We define an M × 3 matrix L(r) in which the jth row is equal to the row vector [, , ]. This matrix L(r), which is called the lead field matrix, represents the sensitivity of the whole sensor array at the location of r.
Let us assume that, in total, Q sources exist. Their locations are denoted by r1, …, rQ, their orientations by η1, …, ηQ2, and their intensities by s1(t), …, SQ(t). Then, the signal vector bs(t) is expressed as
| (6) |
where lp = L(rp)ηp, and lp is called the lead field vector of the pth source. The equation above indicates that the signal vector bs(t) is expressed as a linear combination of the lead field vectors. In other words, the vector bs(t) lies within the span of the lead field vectors, l1, …, lQ, i.e.,
| (7) |
This subspace, span{l1, …, lq}, is referred to as the (spatial-domain) signal subspace[11].
In general, the signal subspace is unknown because the source locations and the source orientations are unknown. However, a subspace that approximates the signal subspace can be obtained from an augmented lead field over the source space, which is the region where a source may exist. To derive the augmented lead field, we define voxels over a source space. Denoting the locations of the voxels by r1, …, rN, the augmented lead field matrix over all voxel locations is defined as
| (8) |
It is clear that (ignoring the voxel discretization error) the column span of F includes the signal subspace, that is,
| (9) |
The column span of F is called the pseudo signal subspace in this paper, and the projector onto the pseudo signal subspace can be derived by applying eigenvalue decomposition to FFT, such that3
| (10) |
We assume that the eigenvalues γ1, …, γζ are distinctively larger than the other eigenvalues. Then, the set of eigenvectors {e1, …, eζ} is an orthonormal basis of the column span of the augmented lead field. Accordingly, the pseudo signal subspace projector P is derived as
| (11) |
and the following relationship holds:
| (12) |
2.3. Time-domain signal and interference subspaces
Now we define the time-domain signal subspace, which is different from the spatial domain signal subspace defined in Section 2.2. For the arguments on the time-domain signal subspace, we denote row vectors consisting of the signal-source time courses by sp: sp = [sp(t1), …, sp(tK)] (p = 1, …, Q). We assume that Q is smaller than the number of sensors M. This assumption is called the low-rank signal assumption. Also, the vectors s1, …, sQ are assumed to be linearly independent. Using Eqs. (3) and (6), we have the following relation
| (13) |
Here, are the elements of the lead field vector .
The row vector whose elements form the jth row of the matrix BS is denoted . Equation (13) then implies the relation,
| (14) |
Namely, is expressed as a linear combination of the vectors s1, …, sQ. The row vector thus lies within the span of these vectors. That is,
| (15) |
This subspace, span{s1, …, sQ}, is defined as the time-domain signal subspace, and denoted :
| (16) |
It can be shown under the low-rank signal assumption that is equal to the row span of BS, i.e.,
| (17) |
The proof is presented in Section 8.1 in the Appendix.
Let us define the time-domain interference subspace, which plays a key role in the proposed algorithm. We express the interference vector as
| (18) |
where P is the number of interference sources. Their intensities are denoted by σj(t) (j = 1, …, P), and their lead field vectors by ξj (j = 1, …, P). We define row vectors consisting of the interference-source time courses as σp: σp = [σp(t1), …, σp(tK)] (p = 1, …, P). The vectors σ1, …, σP are assumed to be linearly independent. The span of σ1, …, σP is defined to be the time domain interference subspace, and is denoted by , i.e. . The row vector whose elements form the jth row of the matrix BI is denoted by . Using the same arguments used to derive Eq. (14), we can derive the relation,
| (19) |
where are the elements of the lead field vector . It is clear that is expressed as a linear combination of the vectors σ1, …,σP and that the vector lies within the interference subspace (i.e., ). Under the low-rank signal assumption (P + Q < M), it can be shown that is also equal to .
2.4. Finding the time-domain interference subspace
In the proposed DSSP algorithm, the sensor measurements B are first projected onto the inside and the outside of the pseudo signal subspace. Let us define Bin and Bout as the projections of B onto the inside and the outside of the pseudo signal subspace,
| (20) |
| (21) |
where I is the identity matrix. Using Eqs. (5) and (12), we have,
| (22) |
| (23) |
Note here that, due to the “dull” cut-off property4 of the projector P, neither PBI nor (I − P)BI becomes negligibly small.
We now analyze the subspace spanned by the row vectors of Bin. We can write
| (24) |
where . The equation above indicates that although the projector P modifies the lead field vectors, it never changes the time courses of the inter-ference sources. Therefore, defining to be the jth row vector of the matrix PBI, we can show
| (25) |
where are the elements of the modified lead field vector . This equation indicates that the row vector of the matrix PBI lies within the time-domain interference subspace, i.e., . We denote a row vector of Bin by and the span of by . Then, taking the relations and into consideration, Equation (22) implies that
| (26) |
where the span of the row vectors of PBε is denoted by . The proof of (26) is presented in Section 8.3 in the Appendix.
We next analyze the subspace spanned by the row vectors of Bout. We can write
| (27) |
where . Using the same arguments employed to derive Eq. (25) and denoting the jth row vector of the matrix (I − P)BI by , we can show
| (28) |
where are the elements of the modified lead field vector . This equation indicates that the relation holds, and the row span of PBI is equal to .
Thus, denoting the jth row vector of Bout by and the span of by , Equation (23) implies that is given by
| (29) |
where is the subspace spanned by the row vectors of (I − P)Bε. Equations (26) and (29) imply that the intersection of the two subspaces and is the time-domain interference subspace :
| (30) |
if holds5 where ∅ indicates the empty set. It is actually straight-forward to show , because the relationship
holds. This means that the rows of PBε are orthogonal to the rows of (I−P)Bε. In the equation above, we assume that where ρ2 is the power of the sensor noise.
2.5. Derivation of interference-subspace projector and removal of interference
In this subsection, we present an algorithm to derive an orthonormal basis of . We first extract and from Bin and Bout, respectively by applying singular value decomposition:
| (31) |
and
| (32) |
For the two sets of singular values, distinctively large singular values are denoted by φ1, …,φμ and ϕ1, …,ϕv. We can then derive the subspaces and such that
| (33) |
| (34) |
Additional arguments, presented in Section 6, lead to the conclusion that the interference subspace is equal to the intersection of span{u1, …, uμ} and span{v1, …, vv}, That is, .
The procedure used to find the intersection is described below. According to [12], an orthonormal basis of the intersection is obtained as a set of the principal vectors whose principal angles are equal to zero. To find those principal vectors, we first define
| (35) |
| (36) |
Singular-value decomposition of a matrix UTV is then performed, and the results are expressed as
| (37) |
where Y and Z are matrices whose columns consist of singular vectors. In the equation above, we use the fact that μ > v. Equation (37) indicates that the singular values of the matrix UTV are equal to the cosines of the principal angles between span{u1, …, uμ} and span{v1, …, vv}. The intersection has the property that the principal angles are equal to zero. Thus, by observing the relation,
the dimension of is determined to be r.
The principal vectors are then obtained either as the first r columns of the matrix UY or the first r columns of the matrix VZ. Defining the first r columns of UY as ψ1, …, ψr, the relation
| (38) |
holds and the vectors ψ1, …, ψr form an orthonormal basis for . The interference removal is thus carried out by projecting the measured data B onto the subspace orthogonal to the interference subspace. Defining a matrix G by
| (39) |
the interference removal can be accomplished by right multiplying B by (I − GGT). That is, the interference-removed measurements are obtained as
| (40) |
3. Computer Simulation
3.1. Spinal cord evoked field (SCEF) measurements distorted by large artifacts
Computer simulations were carried out to investigate the validity of the proposed algorithm. We first simulated spinal cord evoked field (SCEF) measurements distorted by large stimulus-induced artifacts. For data generation, we assumed the sensor array of a 120-channel biomagnetometer [5][7], which has been specifically developed for SCEF measurements. The biomagnetometer is equipped with 40 vector sensors6, which are arranged at 8 × 5 measurement locations covering a 14 cm×9 cm area.
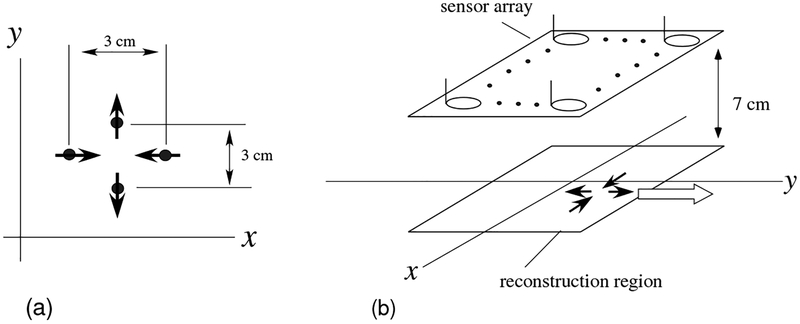
In this computer simulation, we used a source model consisting of four equal-intensity current vectors; such a source model has been found to be physiologically plausible in our previous studies [8]. The model is shown in Fig. 1(a). In this model, the two sources aligned along the y axis are intercellular sources propagating in the nerve axon, representing two anti-directional current dipoles [14], called the leading and the trailing dipoles [15][16]. The other two sources aligned along the x axis represent the volume current. The volume current flows from the extracellular milieu to the site between the two dipole sources in the nerve axon. In this computer simulation, the distance between the two dipole sources, as well as that between the two volume current sources, was set to 3 cm.
Figure 1:
(a) The source model used in the computer simulation. The model consists of four sources in which each pair of sources is 3 cm apart. (b) Schematic drawing of the source-sensor geometry used in the computer simulation. The four sources, depicted by the four solid arrows, travel along the y axis with a speed of 60 cm/s in the direction indicated by the white arrow. The source space is a two-dimensional plane (−8 ≤ x ≤ 8 cm, −6 ≤ y ≤ 6 cm), which is located 7 cm below the sensor array.
The SCEF sources move with a speed of 40–80 cm/s. In this computer simulation, we assumed that four sources traveled along the y direction (from −y to +y) with a speed of 60 cm/s. The source-sensor geometry used in the computer simulation is shown in Fig. 1 (b). The magnetic field generated from the four moving sources was computed, and a plot of the simulated measurements of all 120 sensors is shown in Fig. 2(a). Here, we used no conductor models, and the forward solution was computed using the well-known Biot-Savart law, which is derived based on the quasi-static approximation of Maxwell’s equations. The time was set at zero when the y coordinate of the leading dipole was equal to −30 cm. The field contour map at a latency of 5 ms is shown in Fig. 3(a).
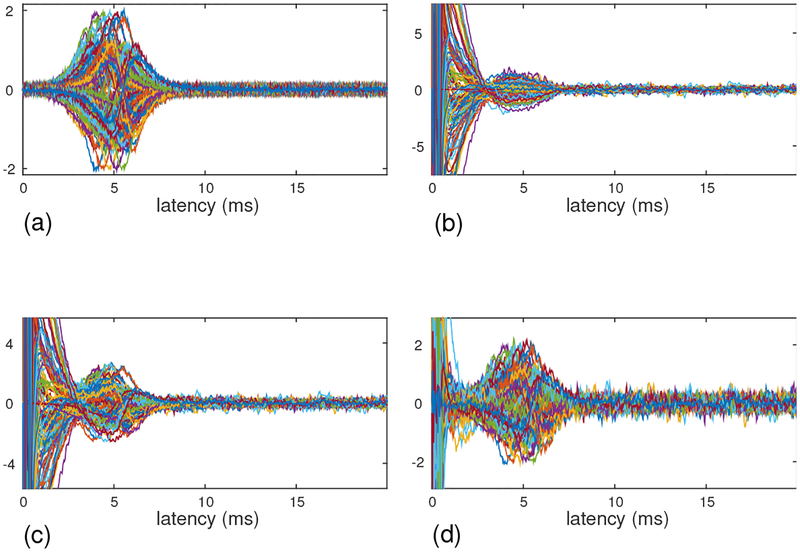
Figure 2:
(a) The sensor time courses of the signal magnetic field, computed assuming that four sources travel along the y axis with the speed of 60 cm/s. (b) The sensor time courses of the artifact-only measurement, which was taken with the stimulus electrode positioned a few centimeters away from the median nerve of the subject. (c) The sensor time courses of the combined (signal plus artifact) results. (d) The sensor time courses of the artifact-removed results. The ordinates of these plots show relative values of field intensity. (The values are common for all four plots.)
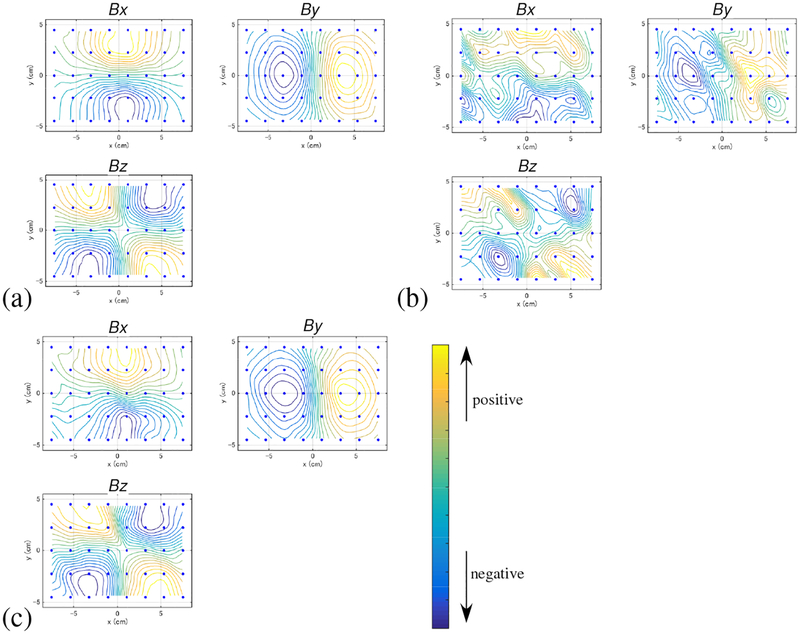
Figure 3:
The field contour maps at the latency of 5 ms for (a) the signal-only sensor time courses, (b) the combined (signal plus artifact) sensor time courses, and (c) the artifact-removed sensor time courses. The upper-left, upper-right, and the lower-right panels in each figure show, respectively, the plots of the x, y, and z components of the magnetic field (denoted Bx, By, and Bz). (Note that the x and y components are the tangential components. The z component is the normal component.) The color of the contours indicates the relative intensity of the magnetic field according to the color bar. The small filled circles indicate the 40 locations of the sensors.
The artifact data was obtained from artifact-only measurements, in which we applied exactly the same stimulus as used in measuring the SCEF, with a stimulus electrode positioned a few centimeters away from the median nerve of a subject [17]. Since the location of the electrode was not on the nerve, stimuli did not induce the nerve activity but elicited only the artifacts. The artifact-only data is shown in the lower-left panel of Fig. 2(b).
The results of adding the artifact data onto the computer-generated signal are shown in Fig. 2(c), and the field contour map of the artifact-contaminated data at a latency of 5 ms is shown in Fig. 3(b). Here, the interference-to-signal ratio (ISR) was set at 12. The ISR is defined as the ratio of ǁBIǁF/ǁBSǁF where ǁXǁF indicates the Frobenius norm of a matrix X. Both the sensor time courses and the contour map show that a significant amount of distortion arises due to contamination by the artifacts.
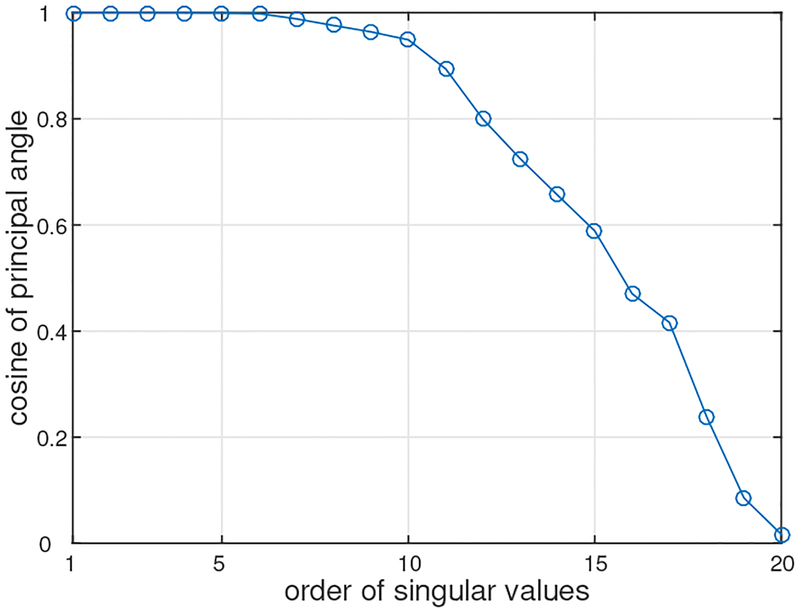
We then applied the DSSP algorithm to this artifact-contaminated data. Here, a two-dimensional region (−8 ≤ x ≤ 8 cm, −6 ≤ y ≤ 6 cm), which is located 7 cm below the sensor plane, was defined as the source space. The augmented lead field was computed with a 0.5-cm voxel grid over this source space. A plot of the singular values cos(θj) in Eq. (37) is shown in Fig. 4. The plot shows that the first six singular values are very close to 1. Thus, the dimension of the intersection, r, was determined to be six by thresholding the singular values at 0.99. The artifact-removed sensor time courses are shown in Fig. 2(d). A field contour map of the artifact-removed data at a latency of 5 ms is shown in Fig. 3(c). Comparing between Figs. 2 and 3, we can see that the distortion from the artifacts is significantly reduced.
Figure 4:
The singular values cos(θj) in Eq. (37) versus their order j.
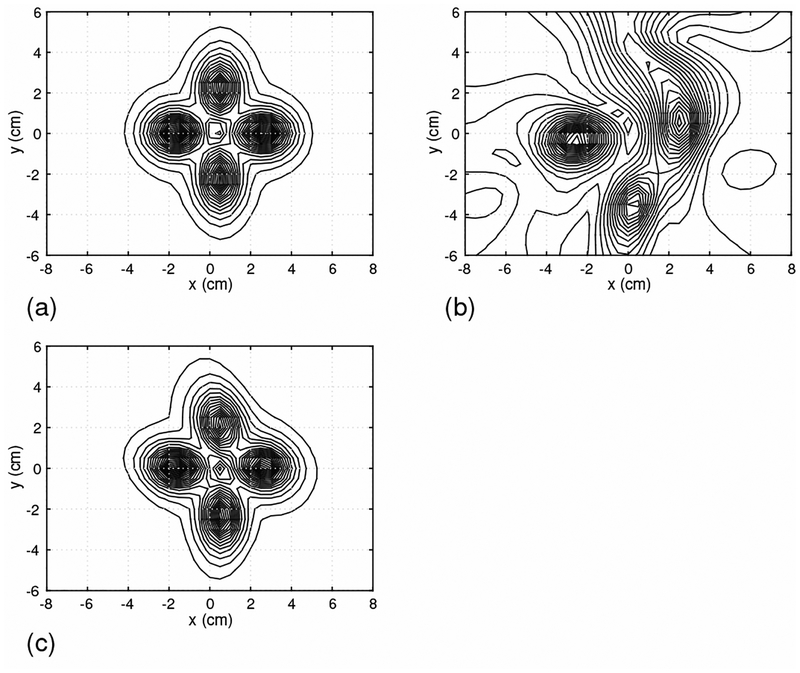
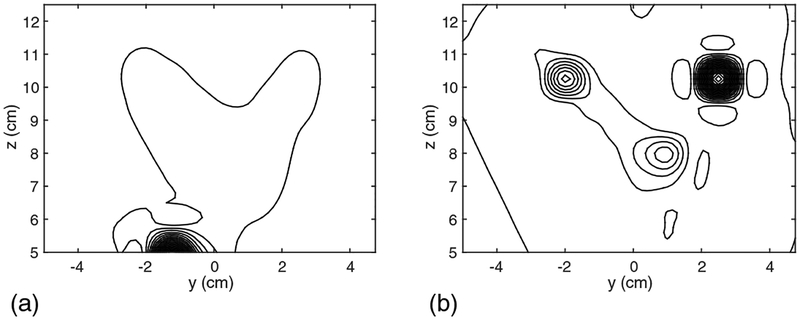
We next assessed the effectiveness of the DSSP algorithm on the basis of source reconstruction results. Time-point by time-point source reconstruction is essential for imaging spinal cord activity because of the rapid movements of the sources. We therefore used the recursively applied null-steering (RENS) beamformer algorithm [9][18] because of its applicability to single time-point data. A snapshot of the source image at 5 ms latency is shown in Fig. 5. In this figure, the results in (a) are obtained by using the artifact-free sensor data in Fig. 3(a). The results from the artifact-contaminated sensor data in Fig. 3(b) are shown in Fig. 5(b). Due to the presence of the interference, the reconstruction results are significantly distorted. The results from the artifact-removed sensor data in Fig. 3(c) are shown in Fig. 5(c). In these results, the four sources are clearly reconstructed and the results are very close to the artifact-free case in Fig. 5(a), demonstrating the effectiveness of the proposed method.
Figure 5:
The source reconstruction results obtained using the field maps at 5 ms, which are shown in Fig. 3. (a) The source image from the signal-only data in Fig. 3(a). (b) The source image from the signal-plus-artifact data in Fig. 3(b). (c) The source image from the artifact-removed data in Fig. 3(c).
3.2. MEG measurement overlapped with large interference magnetic field
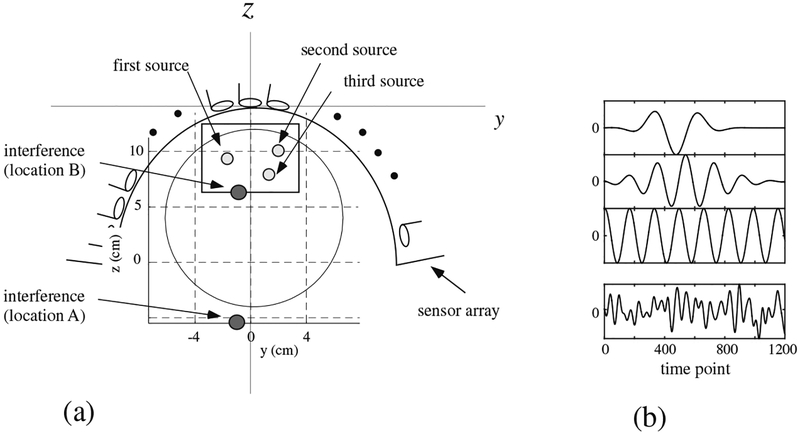
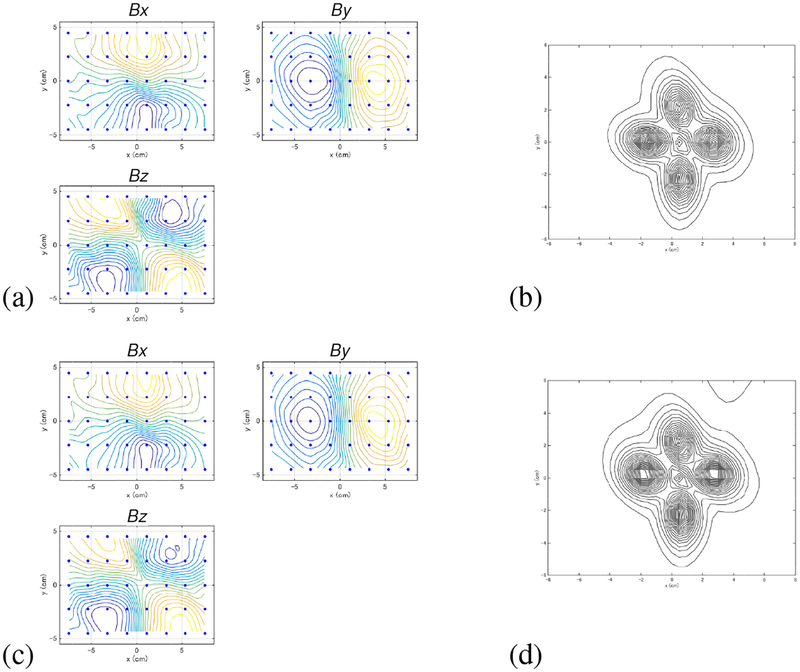
We next present a computer simulation on MEG measurements in which the data was overlapped with a large interference caused near the source space. A sensor alignment of the 275-channel whole-head sensor array from the Omega™ (VMS Medtech, Coquitlam, Canada) neuromagnetometer was used. The coordinate system and source-sensor configuration used in the computer simulation are depicted in Fig. 6(a). A vertical plane (x = 0 cm) was assumed at the middle of the sensor array, and three sources were assumed to exist on this plane. The coordinates of the sources are (0, −2,10.3), (0, 2.5,10.3), and (0,1, 7.3) cm. The time courses assigned to the three sources are shown in the top three panels in Fig. 6(b).
Figure 6:
(a) The coordinate system and source-sensor configuration used in the computer simulation in Section 3.2. Small three circles show the locations of the signal sources. The filled circles with the labels of”interference (location A)” and “interference (location B)” indicate the two locations of the interference source. The square indicates the y-z cross section of the source space. The large circle indicates the sphere used for the forward calculation. (b) The assumed time courses of the first, the second, and the third sources are shown respectively in the first, the second and the third panels from the top. The bottom panel shows the assumed time course of the interference source. The ordinate indicates the normalized, relative intensity and the abscissa indicates the time points.
We put a single interference source shown as a filled circle with the label “interference (location A)” in Fig. 6(a). As shown in this figure, the interference source was fairly close to the signal sources and it was located just outside the head near the head surface. The coordinate of the interference source was (−1, −1, −6) cm. This interference simulated the noise caused from some types of brain stimulator. In Section 5, we actually present experiments in which the MEG data was contaminated by the noise from patient’s vagus nerve stimulator (VNS). The time course of the interference source is presented in the bottom panel of Fig. 6(b). We generated interference magnetic field with the interference-to-signal ratio (ISR) equal to 100 where the ISR is defined as ǁBIǁ/||BSǁ The interference magnetic field was computed and overlapped onto the signal magnetic field computed from the activities of the three signal sources.
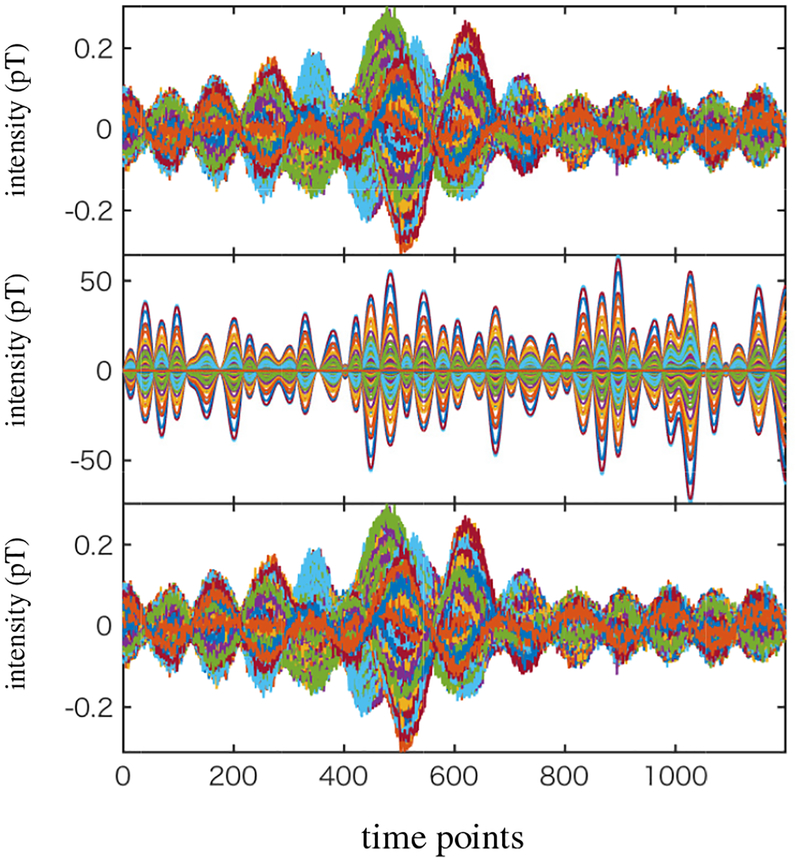
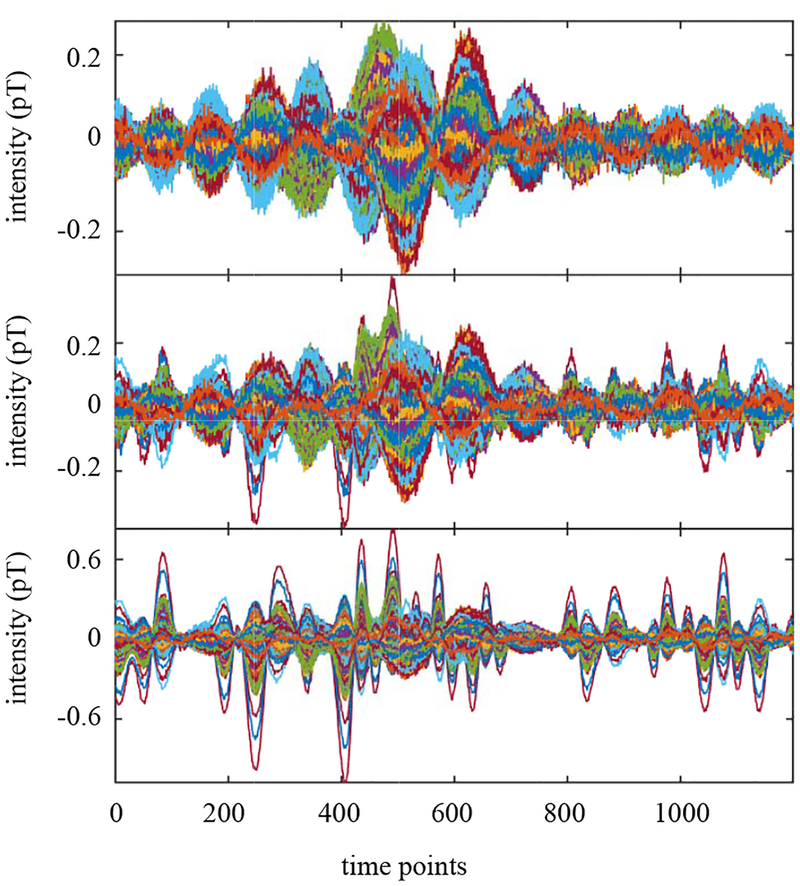
To generate the magnetic fields, source activities were projected to the sensor time courses through the lead field, which is obtained using the homogeneous spherical head model [19] with the center of the sphere set to (0, 0, 4) cm. The spatio-temporal data with 1200 time points was generated. In Fig. 7, the time courses of the signal magnetic field (plus sensor noise) are shown in the top panel. The time courses of interference-overlapped magnetic field are shown in the middle panel. Since the interference magnetic field is 100 times stronger than the signal magnetic field, the sensor time courses in the bottom panel are dominated by the interference magnetic field.
Figure 7:
The sensor data corresponding to the signal magnetic field plus sensor noise (top). The interference-added sensor data (middle). The interference-to-signal ratio was set equal to 100 in which the ISR is defined by the ratio ǁBIǁ/ǁBSǁ. (The time courses of the sensor data in this panel is dominated by the interference-source time course, due to the large ISR.) The interference-removed sensor time courses (bottom).
We set the source space to a region of −4.5 ≤ x ≤ 4.5 cm, −5 ≤ x ≤ 5 cm and 5 ≤ z ≤ 13 cm. This region approximately covers the whole brain and includes the locations of the three sources, but it does not include the location of the interference source. The augmented lead field was computed using a 0.5 cm voxel grid over this region. We then applied the DSSP algorithm for removing the interference. The dimension of the interference subspace r was determined to be 1 by thresholding the cosine of the principal angle (Eq. (37)) by the value of 0.99. The interference-removed sensor time courses are shown in the bottom panel in Fig. 7, which shows that the interference is nearly completely removed.
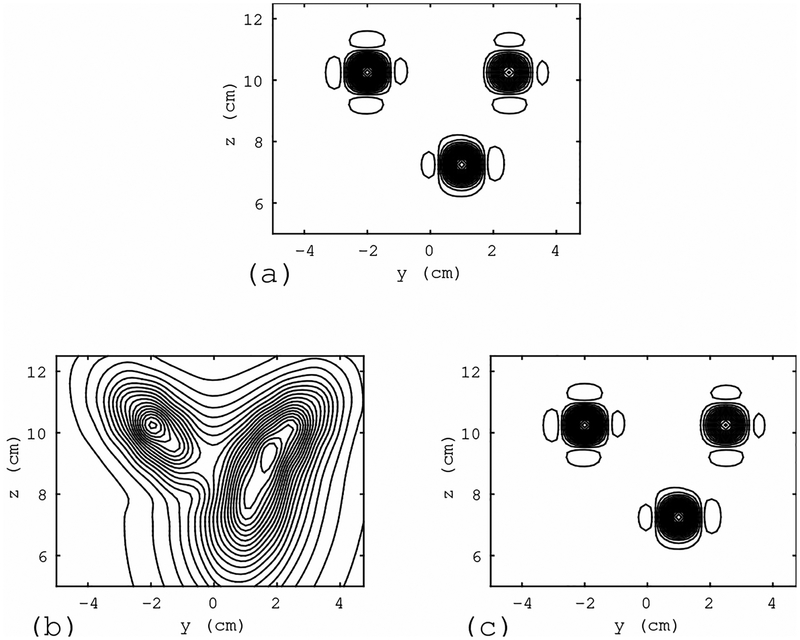
We next evaluate the effect of interference removal based on the quality of reconstructed source images. The adaptive beamformer algorithm[11] was applied for source reconstruction from the sensor data shown in Fig. 7. The results are shown in Fig. 8. In Fig. 8(a), the source reconstruction results obtained from the interference-free magnetic field (the top panel of Fig. 7) are shown. Three sources can clearly be observed. The reconstruction results obtained using the interference-overlapped magnetic field (the middle panel of Fig. 7) are shown in Fig. 8(b). Due to the overlap of the large interference, the results contain significant distortion. The reconstruction results obtained using the artifact-removed sensor data (the bottom panel of Fig. 7) are shown in Fig. 8(c) in which the distortion is nearly completely removed, demonstrating the effectiveness of the proposed method.
Figure 8:
Results of source reconstruction obtained using the sensor data in Fig. 7. (a) Results obtained from the interference-free magnetic field (the top panel of Fig. 7). (b) Results using the interference-overlapped magnetic field (the middle panel of Fig. 7). (c) Results obtained using the interference-removed sensor data (the bottom panel of Fig. 7). The reconstruction region was set equal to the source space −4.5 ≤ x ≤ 4.5 cm, −5 ≤ x ≤ 5 cm, and 5 ≤ z ≤ 13 cm. The source distribution on the plane x = 0 cm is shown. The filled circle with the label”interference (location A)” in Fig. 6(a) indicates the location of the interference source in this simulation.
As shown in the computer simulation above, the DSSP algorithm allows interference sources to be fairly close to the source space. However the prerequisite exists that the interference sources are located outside the source space. We performed computer simulation in which this prerequisite was not fulfilled. We here put a single interference source at (−1, −1,5), exactly on the border of the source space. This location is indicated by a filled circle with the label “interference (location B)” in Fig. 6(a). The results of source reconstruction are shown in Fig. 9 where the results without interference removal are shown in (a) and the results with the removal in (b). These results clearly indicate that the DSSP algorithm can still remove the influence of interference but the removal is achieved at the sacrifice of significant amount of signal source intensity.
Figure 9:
Results of source reconstruction when the interference source put at (−1,−1,5), which is just on the border of the source space. (a) Results obtained without the interference removal. (b) Results obtained with the DSSP interference removal. The reconstruction region (the source space) was set equal to the source space −4.5 ≤ x ≤ 4.5 cm, −5 ≤ x ≤ 5 cm, and 5 ≤ z ≤ 13 cm. The source distribution on the plane x = 0 cm is shown. The filled circle with the label”interference (location B)” in Fig. 6(a) indicates the location of the interference source in this simulation.
4. Experiments using SCEF measurements from a healthy volunteer
4.1. SCEF measurements
A 120-channel biomagnetometer[5][7] was used for measuring a human SCEF7. The subject was a healthy male volunteer. The experiment was approved by the ethics committee of Tokyo Medical and Dental University.
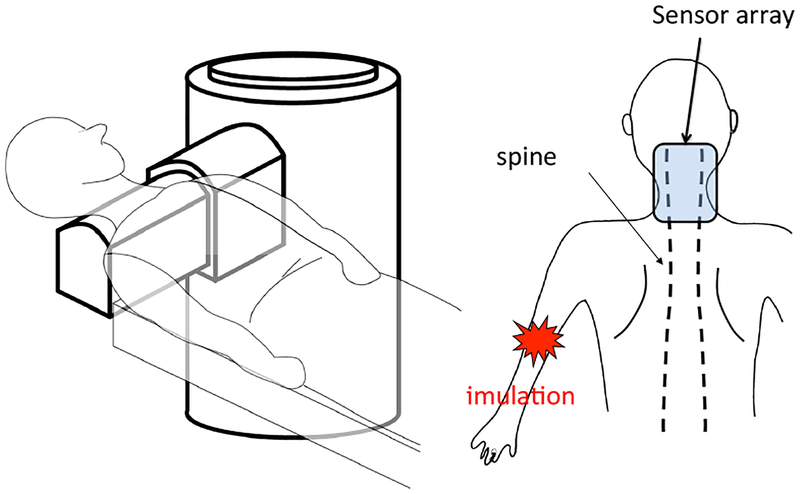
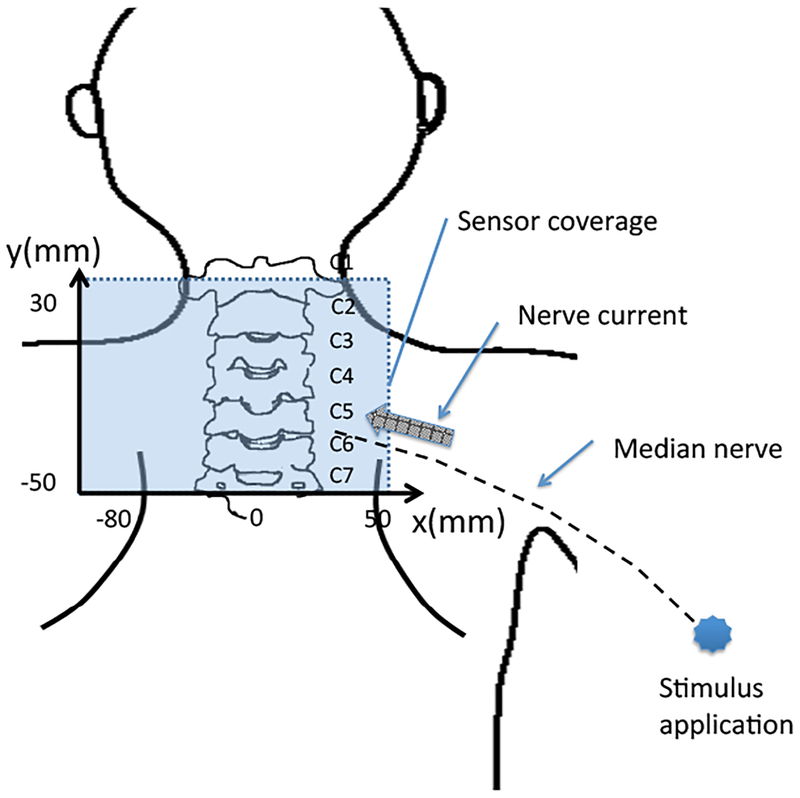
The experimental setup is schematically shown in Fig. 10. As depicted here, the cryostat of the biomagnetometer has a cylindrical body with a protrusion, and this protrusion contains sensors directed upward. The subject lies down in the supine position, and the subject’s lower neck is positioned on the upper surface of the protrusion of the cryostat. A stimulus current was applied to the subject’s median nerve near his elbow. The stimulus with an intensity of 10 mA and a duration of 0.3 ms was repeated 2000 times at a repetition rate of 4 Hz. The data acquisition was performed with a sampling frequency of 40 kHz. An analog bandpass filter with a bandwidth of 100–5000 Hz was applied. The signal was averaged across all 2000 measured trials.
Figure 10:
Schematic illustration of the experimental setup for measuring a spinal cord evoked field. The subject lies down in the supine position with the lower neck placed on the upper surface of the protrusion of the cryostat; the protrusion contains 40 vector sensors directed upward, covering a 14 × 9 cm area. The sensors are arranged at 8 × 5 measurement locations. A stimulus current is applied to the subject’s median nerve near the elbow
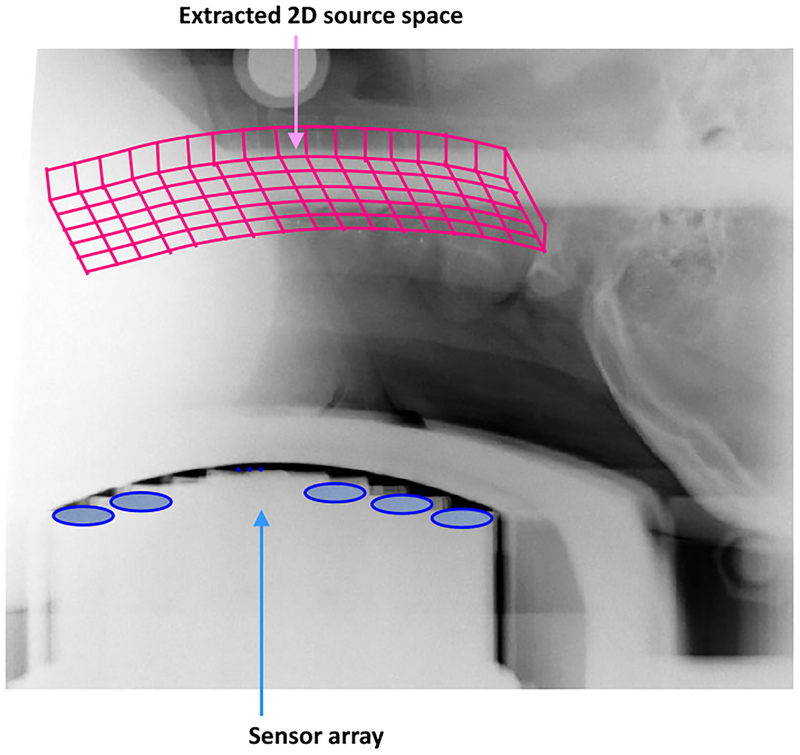
An X-ray image covering the subject’s neck and the sensors of the biomagnetometer was obtained to identify the location of the spinal cord. The reconstruction region was determined to be a curved plane containing the spinal cord. An X-ray image with the extracted 2-D reconstruction region is shown in Fig. 11.
Figure 11:
X-ray image covering the subject neck and the sensors, used for determining a plane containing the spinal cord. This 2D plane is defined as the source space and the source reconstruction is performed on this plane.
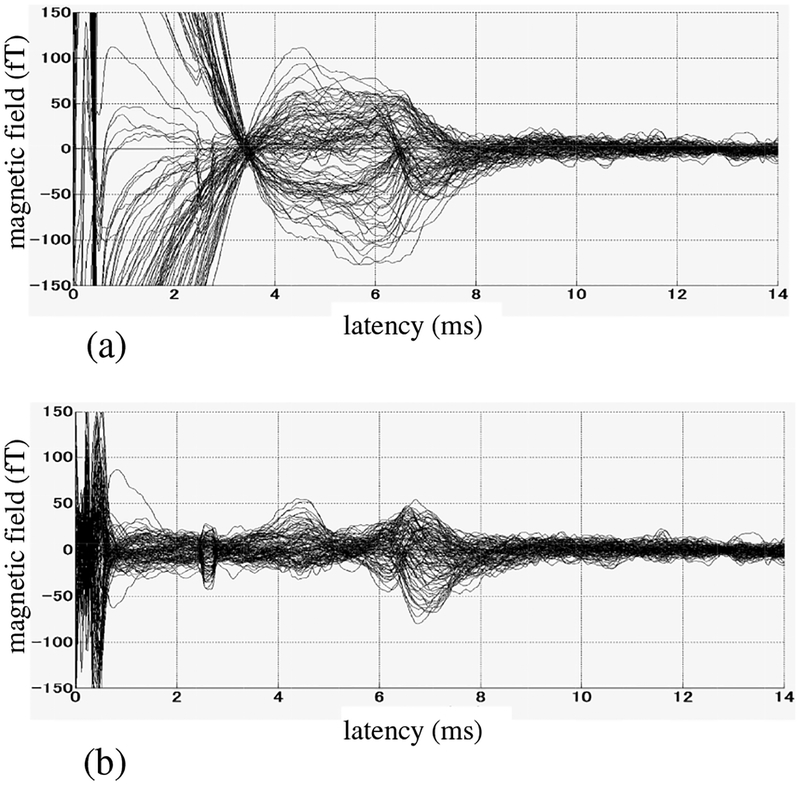
The measured SCEF, averaged over 2000 trials, is shown in Fig. 12(a). The electric stimulus was given at a latency of 0 ms. Large artifacts are observed particularly in the data before 4 ms, although the peaks between 4 and 8 ms, which are caused by the spinal cord nerve activity, can still be observed. We applied the proposed DSSP algorithm to remove these artifacts. The 2-D reconstruction region was used as the source space over which the augmented lead field matrix was computed. The results are shown in Fig. 12(b). Here, the artifacts are significantly reduced and the SCEF signal, consisting of peaks between 4 and 8 ms, is clearly observed.
Figure 12:
(a) The original sensor time courses. The results averaged across 2000 trials are shown, (b) Artifact-removed sensor time courses. The reconstruction plane shown in Fig. 11 was used as the source space over which the augmented lead field matrix was computed. The dimension of the intersection, r, was set at three, which was determined by applying the 0.99 thresholding criterion. The fairly sharp peak near 2.5 ms is due to a hardware problem, and should be disregarded.
4.2. Validation based on reconstruction of spinal cord nerve activity
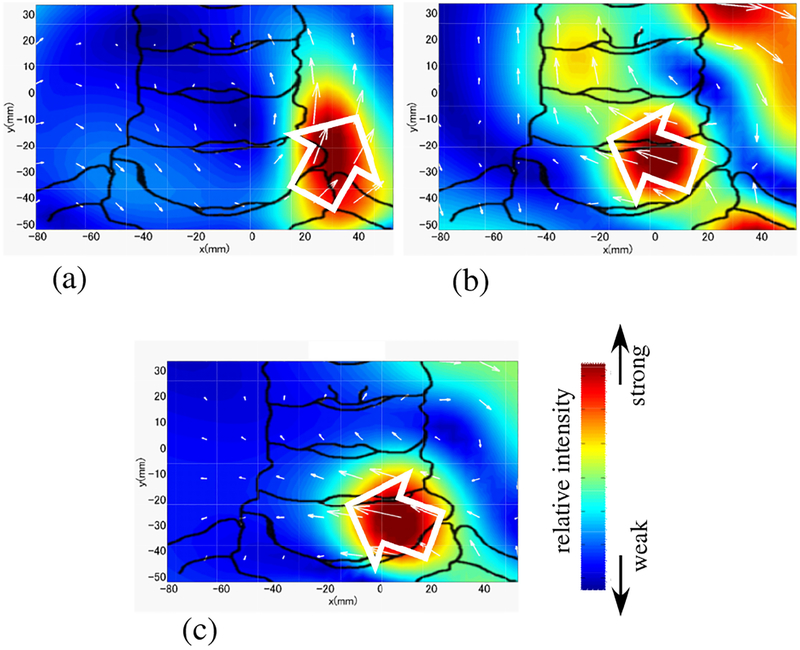
The spinal cord source activity was reconstructed by using the RENS beam-former. Schematic illustration in Fig. 13 shows relative positions of the subject’s neck and median nerve with respect to the reconstruction region, which had an area of 16 × 12 cm with voxel dimensions of 0.5 cm in the x and y directions. The reconstructed source images at a latency of 5.8 ms are shown in Fig. 14. In this figure, the source image from the original artifact-contaminated sensor data is shown in (a). The image from the artifact-removed data is shown in (b). Both images show the leading dipoles but their directions (indicated by the white arrows) are significantly different.
Figure 13:
Schematic illustration of the reconstruction region, which is indicated by the square. Relative positions of the subject’s neck and median nerve are shown. The arrow indicates a typical orientation of the current vector of the leading dipole flowing from the peripheral nerve into the spinal cord when subject’s left median nerve is stimulated.
Figure 14:
Results of source reconstruction when a stimulus applied to the subject’s left median nerve. (a) Source image at the latency of 5.8 ms obtained from the original sensor data in Fig. 12(a). (b) Source image at the latency of 5.8 ms from the artifact-removed data in Fig. 12(b). (c) Source image at 9.85 ms obtained with the stimulus applied to the median nerve near the subject’s wrist. (This image was obtained without using DSSP artifact removal.) The white arrows indicate the directions of the leading dipoles. The relative intensity of the reconstructed source is color-coded according to the color bar. The sketch of the spine was drawn from the overlaid X-ray image used for aligning the sensor coordinates to the subject’s neck position.
To determine which results are physiologically more plausible, we used a source image obtained with the stimulus applied at the median nerve near the subject’s wrist; the image is shown in Fig. 14(c). The signal obtained with wrist stimulation is known to be less affected by artifacts. This is because, with the stimulation near the subject’s wrist, it takes 4–5 ms more for the nerve activation to reach the neck region, and thus the spinal cord signal is much less contaminated by stimulus-induced artifacts due to their rapid decay. Therefore, the results from the wrist stimulation should serve as the physiological”ground truth” for the comparison. The leading dipoles have transverse (the negative x) components in Figs. 14(b) and (c), while the direction is almost upward in (a), i.e., the current vector has almost no x component in (a).
Since the subject’s left median nerve is stimulated, the nerve activity is known to propagate from the left median nerve into the spinal cord near the fourth vertebra (c4). Thus, it should be more plausible that the current vector has a transverse, negative x component, as depicted in Fig. 13. Accordingly, considering the fact that the leading dipoles in (b) and (c) have such negative x components, we can draw the conclusion that the results in Fig. 14(b) are physiologically more plausible than those in Fig. 14(a).
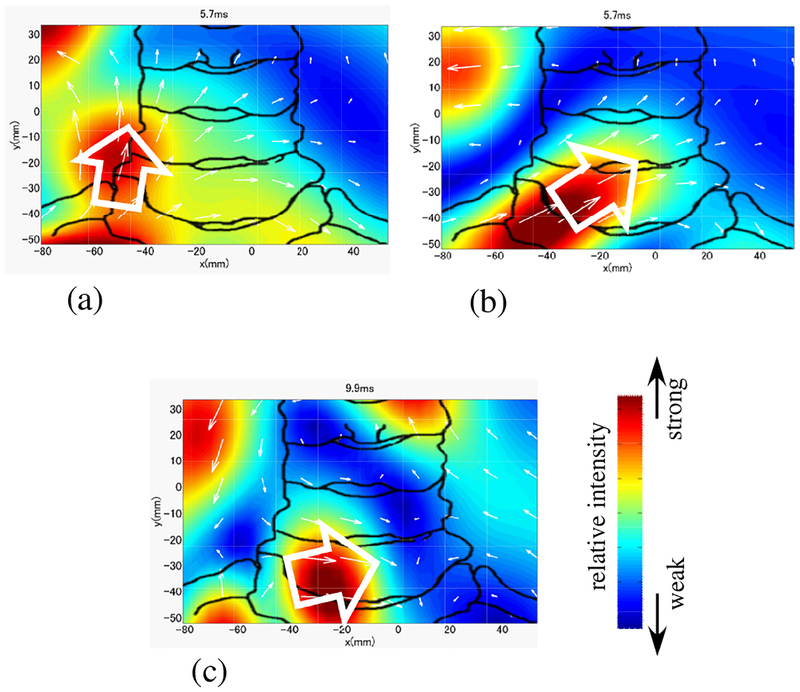
We have conducted measurements exactly the same as the ones described above but with right median nerve stimulation. The source images from these measurements are shown in Fig. 15. Here, the source image from the original artifact-contaminated sensor data is shown in (a). The image from the artifact-removed data is shown in (b). Both results show the leading dipole but the direction of the leading dipole (indicated by the white arrow) is again different between these two images. The source image obtained with the stimulus applied at the right median nerve near subject’s wrist is shown in (c). In this figure, the current vector should have a transverse, positive x component since the x component of the source should be opposite to that in the left median nerve stimulation. The images in Fig. 15(b) and (c) show that the dipoles have the expected positive x components. Therefore, we can again conclude that the results in (b) are physiologically more plausible than those in (a), which show the source vector with an almost upward direction. It should be noted that the DSSP algorithm can be used with any type of source reconstruction methods. In these experiments, the RENS beamformer was used for reconstructing the spinal cord activity, primarily because it gives the spatial resolution better than other existing methods8 applicable to single time point data.
Figure 15:
Results of source reconstruction when a stimulus applied to the subject’s right median nerve. (a) Source image at the latency of 5.8 ms obtained from the original sensor data in Fig. 12(a). (b) Source image at the latency of 5.8 ms from the artifact-removed data in Fig. 12(b). (c) Source image at 9.85 ms obtained with the stimulus applied to the median nerve near the subject’s wrist. See Fig. 14 for legend.
5. Experiments using MEG data measured from a patient with a vagus nerve stimulator
The application of the DSSP algorithm is not limited to artifact removal in SCEF measurements, and the algorithm can be applied to a wide variety of biomagnetic measurements. To demonstrate the versatility of the algorithm, we present results from applying the DSSP algorithm to MEG data obtained from a patient with a vagus nerve stimulator (VNS). Generally, MEG data from VNS patients contains a large amount of interference generated from the stimulator located near the patient’s chest area. The measurements were conducted using the 275-channel whole-head sensor array of the Omega™ (VMS Medtech, Coquitlam, Canada) neuromagnetometer.
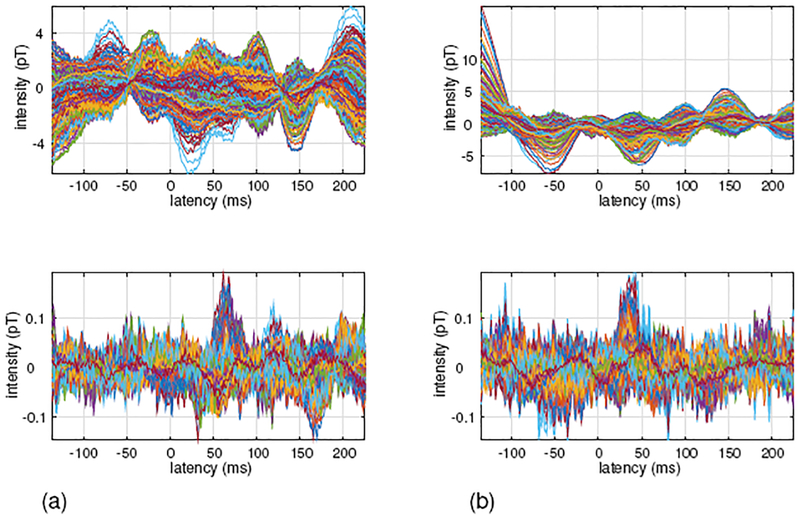
In the upper panel of Fig. 16(a), the somatosensory MEG data measured with tactile stimulation applied to the patient’s left index finger is shown. Since the tactile stimulation was used, a large peak should be observed around the latency of 50 ms. However, such a peak is not observed due to the presence of interference from the VNS stimulator.
Figure 16:
Somatosensory MEG data obtained from a patient with a vagus nerve stimulator (VNS). (a) Sensor time courses when the tactile stimulus was applied to the subject’s left index finger. (b) Sensor time courses when the tactile stimulus was applied to the subject’s right index finger. Original sensor time courses are shown in the upper panels, and sensor time courses processed by the proposed DSSP algorithm for interference removal are shown in the lower panels.
The sensor time course with the interference removed by the proposed DSSP method is shown in the lower panel of Fig. 16(a). Here, a peak around the latency of 50 ms is clearly observed. The MEG data taken from the same patient with the tactile stimulation applied to the patient’s right index finger is shown in Figure 16(b). The upper panel shows the original sensor time courses and the lower panel shows the sensor time courses processed with the DSSP algorithm. Again, the sensor time courses processed by the DSSP algorithm show a peak around the latency of 50 ms, but such a peak is not observed in the original sensor time courses.
The source localization results are shown in Fig. 17. The results for the case of the left index finger stimulation are shown in Fig. 17(a), and those for the right index finger stimulation are shown in Fig. 17(b). In both cases, the source activity is successfully localized near the primary somatosensory area in the contra-lateral hemisphere. These reconstruction results show that the DSSP algorithm removes the influence of the VNS stimulator and enables the mapping of the primary somatosensory cortices, demonstrating that the DSSP algorithm can be used for the interference removal in MEG. It should be mentioned that, without interference removal, a single strong fake source was detected near the center of the head for the both cases of left- and right-index finger stimulations, although these results are not shown.
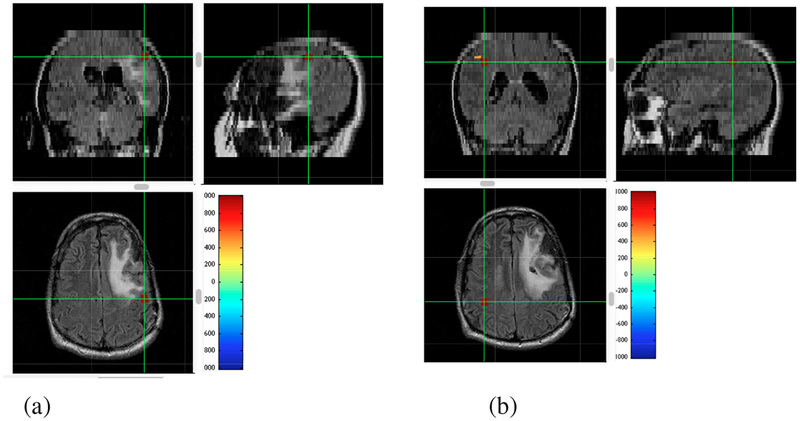
Figure 17:
Results of source localization using the interference-removed sensor time courses shown in the bottom panels of Fig. 16. The relative voxel intensity is color-coded according to the color-bar, and overlaid onto the patient’s MRI. The cross hair indicates the point with the maximum reconstruction intensity, and three cross sectional MR images at this point are shown. (a) Results from the data taken with left index finger stimulation. (b) Results from the data taken with right index finger stimulation.
6. Discussion and summary
Since equations (33) and (34) are important in the derivation of the proposed method, we should further clarify the meaning of these equations. Theoretically, if Q and P are known and μ and v are respectively set to Q + P and P, the following relationships hold:
| (41) |
| (42) |
The proof can be found, for example, in [23] or [11]9. Therefore, we could directly derive the interference subspace using Eq. (42) if we knew the number of interference sources P.
However, information on the number of sources is difficult to obtain in real-life applications. In addition, the singular values in Eqs. (31) and (32) may not have sharp cutoffs due to a low signal-to-noise ratio (SNR) condition or an insufficient number of time points. Accordingly, there is always considerable ambiguity in determining μ and v, the threshold values of the singular value spectra. A general strategy for such cases is to overestimate μ and v. However, span{u1, …, uμ} and span{v1, …, vv} then unavoidably contain a part of the noise subspace. Thus, to be exact with overestimated μ and v, Eqs. (33) and (34) should be rewritten as
| (43) |
| (44) |
where is the part of that is contained in span{u1, …, uμ} and the part of that is contained in span{v1, …, vv}.
The arguments above indicate that the choices of the parameters μ and v are not crucial for implementing the algorithm, because the excess dimensions of span{u1, …, uμ} and span{v1, …, vv} do not affect the extraction of the interference subspace, since the intersection between span{u1, …, uμ} and span{v1, …, vv} is computed. In all the experiments described in previous sections, the parameters μ and v were always set at 20. In the computer simulation in Section 3.1, the field map and the reconstructed source image were recomputed with μ and v set to 40. The results are shown in Fig. 18(a) and (b), which are almost the same as the results obtained with μ and v set at 20 (shown in Fig. 3(c) and Fig. 5(c)).
Figure 18:
(a) The field contour maps at the latency of 5 ms for the artifact-removed results when μ and v are set at 40. (b) The source reconstruction results using the field maps shown in (a). (c) The field contour maps at the latency of 5 ms for the artifact-removed results when r was set at 12 (μ and v were set at 20). (d) The source reconstruction results using the field maps shown in (c).
Unlike μ and v, the choice of the dimension of the interference subspace, r, can be crucial in the implementation of the algorithm. The influence of overestimating r is analyzed as follows. Even when the parameter r is overestimated, since the interference time-course vectors still lie within the column span of G, the orthogonal projector (I − GGT) suppresses the interference BI. Therefore, only when the projector (I − GGT) also suppresses the signal of interest, the overestimation becomes problematic. This scenario happens if the excess dimensions of the interference subspace include a part of the signal subspace.
Part of the signal subspace may be included in the estimated interference subspace when the distance between the signal subspace, span{s1, …, sQ}, and the interference subspace, span{σ1, …, σP}, is small, i.e., when the two subspaces are close. The distance between the two subspaces can be defined using the subspace angles between the two subspaces [24], and if all the inner products between the basis vectors are small, the distance between the two subspaces is large. If some of these inner products have large values, the distance between the two subspaces can be small.
Therefore, when the time courses of the signal sources sp (p = 1, …,Q) and those of the interference sources σp (p = 1, …, P) are very different and the correlations between them are small, the distance between the signal and interference subspaces is large. In such cases, the overestimation of r may not be problematic, because the excess columns of G are nearly orthogonal to the signal time-course vector sp (p = 1, …, Q), so the projector (I − GGT) hardly affects BS. The results of overestimating r at 12 in our computer simulation are shown in Fig. 18(c) and (d). Comparison of these results with the results with r = 6 (Fig. 3(c) and Fig. 5(c)) shows that overestimation of r causes no serious problems. This result can be explained by the arguments above, because in our computer simulation, sp and σp were very different.
Many algorithms have been proposed by our group and others for removing artifacts and interferences from sensor measurements. One method, called common-mode subspace projection (CSP), was reported by our group as a conference proceedings paper [17]. The CSP algorithm uses a part of the DSSP algorithm, and estimates the interference subspace as an intersection between the row spans of two spatio-temporal data matrices. It is, however, different from the DSSP algorithm in that the CSP algorithm requires the artifact-only measurements to be conducted separately from the signal measurements. The proposed DSSP algorithm does not require such separate, additional measurements, and thus the total measurement time for the DSSP algorithm is half of that for the CSP method, which is advantageous in clinical environments, because the burden for a patient is nearly proportional to the total time for measurements.
Signal space projection (SSP) is a well-known method for interference removal in MEG and EEG measurements[25][26]. In the SSP method, the (spatial-domain) interference subspace projector PI is first computed, and the sensor data b(t) is then projected onto the subspace orthogonal to the interference subspace by computing (I − PI)b(t). In order to derive PI, the SSP method requires the information on the locations of interference sources. Therefore, the method cannot be applied to the artifact removal problem in the spinal cord imaging, because the spatial locations of the artifacts cannot be determined. The proposed DSSP algorithm, in contrast, does not require such information, and this fact, in general, makes the DSSP algorithm much more useful than the SSP method, because such information is unavailable in many real-life applications.
For interferences from brain stimulators such as the VNS interference, the location of the interference source may be predetermined. However, even in this case, a very accurate determination is needed to implement SSP when the interference is much stronger than the signal. This fact can be seen using our computer simulation in Section 3.2. We applied SSP to the interference-overlapped data shown in the top panel of Fig. 7. The results are shown in Fig. 19. In this figure, the top panel shows the results obtained when the exact location of the interference source is used. The middle and bottom panels, respectively, show the results obtained when the location of the interference source has 1 mm and 3 mm errors. These results show that even a 1 mm localization error affects results of interference removal, suggesting that SSP requires a very accurate localization of an interference source. This fact may make the application of SSP to removing large artifacts (such as the VNS interference) somewhat impractical.
Figure 19:
Results of applying the signal space projection (SSP) to the interference-overlapped data shown in the top panel of Fig. 7. Top panel shows the results obtained when the accurate location of the interference source is available. The moddle and the bottom panels, respectively, show results obtained when the location has 1 mm and 3 mm errors.
In summary, this paper has proposed a novel algorithm to remove large interferences overlapped on biomagnetic data. Since the algorithm makes use of the duality in defining the signal subspace, it is named dual signal subspace projection (DSSP). We have presented a detailed mathematical description of the proposed DSSP algorithm, which we have validated by computer simulation and two kinds of biomagnetic data: spinal cord data obtained from a healthy volunteer and MEG data taken from a patient with a vagus nerve stimulator. The proposed DSSP algorithm is suited to remove interferences from sources located fairly close to the boundary of the source space. The eye-blink and cardiac artifacts encountered in MEG measurements are also such interferences. The investigation on applying the DSSP algorithm to the removal of these artifacts is planned and results will be published.
7. Acknowledgements
This work has been supported by grants from the Japanese Ministry of Education, Science, Culture and Sports (26282149, 26462231) and by a grant from SECOM Science and Technology Foundation.
8. Appendix
8.1. The row span of BS is equal to
Here, we prove that, under the low-rank signal assumption (Q < M), is equal to the row span of BS. That is,
| (45) |
Equation (45) is equivalent to claiming that any row vector can be expressed as a linear combination of the row vectors, . The claim is proved as follows. Since , x can be expressed as a linear combination of sp:
| (46) |
If this vector x is also expressed as a linear combination of , we have
| (47) |
Substituting Eq. (14) into (47) and using Eq. (46), we get
| (48) |
Comparing the coefficients of the vector sp in the left- and right-hand sides of the equation above, we have a set of Q linear equations:
Under the low-rank signal assumption M > Q, for any arbitrary set of γ1, … γQ, we have a set of α1, …, αM that fulfills the above equations. Therefore, Eq. (47) always holds for any , and thus is equal to .
Using the same arguments, it can be shown that the row span of BI, , is equal to the interference subspace . Also, both the row span of , , and the row span of , , are equal to .
8.2. Cutoff property of the pseudo signal subspace projector
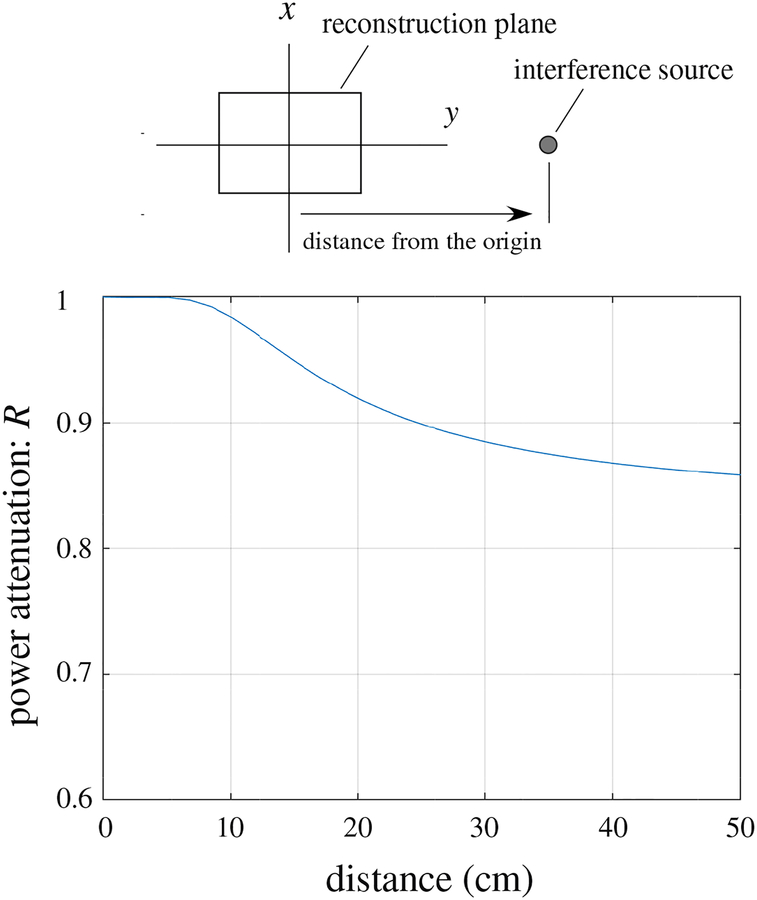
Here, we present an example of the cutoff property of the pseudo signal subspace projector. The example was derived assuming the source-sensor geometry used in our computer simulation where the augmented lead field was computed over the two-dimensional source space defined by −8 ≤ x ≤ 8 cm, −6 ≤ y ≤ 6 cm, and z = 0 cm. The sensors are arranged on a plane of z = 7 cm. Assuming that a source generating interference signal existed at r. the ratio R = ǁPlI(r)||2/||lI(r)||2 was computed where lI(r) indicates the lead field vector of the interference source. The ratio R expresses the decrease of the total power of the interference after processing by the projector P. In this example, the source is located on the y axis (i.e., r = (0, y, 0)), and the configuration of the source space and the interference source is shown in the upper panel of Fig. 20. The power decrease ratio R versus the y coordinate, which is the distance of the interference source from the origin, is shown in the lower part of Fig. 20.
Figure 20:
Cutoff property of the pseudo signal subspace projector P computed using the geometry in our computer simulation. The ratio R = ǁPlI(r)ǁ2/ǁlI(r)ǁ2 is plotted with respect to y, the distance from the center of the source space. The upper panel shows the relative locations of the source space and the interference source.
The plot shows that the cutoff property of the pseudo signal subspace projector P is not sharp. It passes almost 100% of the signal power if the interference source is located within the source space (y ≤ 6 cm) and attenuates only 5–20 % of the signal power when the source is located in the vicinity of the source space (10 ≤ y ≤ 50 cm). The proposed DSSP algorithm first applies the pseudo subspace projector P to create Bin and I − P to create Bout. Both Bin and Bout should contain significant amount of the interference, because of this “dull” cutoff of the pseudo signal subspace projector P. The DSSP algorithm makes use of this property of P and detects the interference subspace by finding common components in the row spans of Bin and Bout.
8.3. Proof of Eq. (26)
Here, we present a proof of Eq. (26) in its general form. That is, we define two subspaces, and , with basis vectors x1, …, xD and y1, …, yD′, respectively. We show that, if and , the relationship
holds. The proof is straightforward. Since x ∈ span{x1, …, xD} and y ∈ span{y1, …, iD′}, we can write
| (49) |
Therefore, we get
| (50) |
Footnotes
Although the exact causes of these artifacts are still unknown, we speculate that they are caused by combined effects of stimulus-induced body electric currents and transient responses of receiver electronics.
The orientation ηj is a 3 × 1 column vector consisting of its x, y, and z components
The matrix FFT is called the Gram matrix.
An example of the cut-off property of P computed assuming the source-sensor geometry used in our computer simulation is shown in Section 8.2 in the Appendix.
If , denoting , we have and the intersection contains an additional noise-related space.
A vector sensor can measure the radial component and two tangential components of the magnetic field. The details of the sensor are reported in [13].
The sensor array of this biomagnetometer was used for the data generation in our computer simulation.
Equation (41) can be proved, for example, by replacing B in Section 13.1 of [11] with and by following the same arguments in that section.
References
- [1].Nagarajan SS, Attias HT, Hild KE, and Sekihara K, “A probabilistic algorithm for robust interference suppression in bioelectromagnetic sensor data,” Statistics in Medicine, vol. 26, pp. 3886–910, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nagarajan SS, Attias HT, Hild II KE, and Sekihara K, “A graphical model for estimating stimulus-evoked brain responses from magnetoen-cephalography data with large background brain activity,” NeuroImage, vol. 30, no. 2, pp. 400–416, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].de Cheveigné A and Parra LC, “Joint decorrelation, a versatile tool for multichannel data analysis,” Neuroimage, vol. 98, pp. 487–505, 2014. [DOI] [PubMed] [Google Scholar]
- [4].Shinomiya K, Furuya K, Sato R, Okamoto A, Kurosa Y, and Fu-chioka M, “Electrophysiologic diagnosis of cervical OPLL myelopathy using evoked spinal cord potentials,” Spine, vol. 13, pp. 1225–1233, 1998. [DOI] [PubMed] [Google Scholar]
- [5].Adachi Y, Oyama D, Kawabata S, Sekihara K, Haruta Y, and Ue-hara G, “Magnetospinography: Instruments and application to functional imaging of spinal cords,” IEICE transactions on electronics, vol. 96, no. 3, pp. 326–333, 2013. [Google Scholar]
- [6].Kawabata S, Komori H, Mochida K, Ohkubo H, and Shinomiya K, “Visualization of conductive spinal cord activity using a bio-magnetometer,” Spine, vol. 27, pp. 475–479, 2003. [DOI] [PubMed] [Google Scholar]
- [7].Adachi Y, Kawai J, Miyamoto M, Ogata H, Tomori M, Kawabata S, Sato T, and Uehara G, “A SQUID system for measurement of spinal cord evoked field of supine subjects,” IEEE Trans. Appl. Supercond, vol. 19, pp. 861–866, 2009. [Google Scholar]
- [8].Sato T, Adachi Y, Tomori M, Ishii S, Kawabata S, and Sekihara K, “Functional imaging of spinal cord electrical activity from its evoked magnetic field,” Biomedical Engineering, IEEE Transactions on, vol. 56, no. 10, pp. 2452–2460, 2009. [DOI] [PubMed] [Google Scholar]
- [9].Kumihashi I and Sekihara K, “Array-gain constraint minimum-norm spatial filter with recursively updated gram matrix for biomagnetic source imaging,” Biomedical Engineering, IEEE Transactions on, vol. 57, no. 6, pp. 1358–1365, 2010. [DOI] [PubMed] [Google Scholar]
- [10].Taulu S and Simola J, “Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements,” Phys. Med. Biol, vol. 51, pp. 1759–1768, 2006. [DOI] [PubMed] [Google Scholar]
- [11].Sekihara K and Nagarajan SS, Adaptive spatial filters for electromagnetic brain imaging. Berlin, Heidelber: Springer-Verlag, 2008. [Google Scholar]
- [12].Golub GH and Van Loan CF, Matrix computations, vol. 3 The Johns Hopkins University Press, 2012. [Google Scholar]
- [13].Adachi Y, Kawai J, Miyamoto M, Uehara G, Kawabata S, Okubo H, Fukuoka Y, and Komori H, “Three dimensionally configured squid vector gradiometers for biomagnetic measurement,” Superconductor Science and Technology, vol. 16, no. 12, p. 1442, 2003. [Google Scholar]
- [14].Wikso JP, “Biomagnetic sources and their models,” in Advances in Biomagnetism (Williamson SJ et al. , eds.), (New York: ), pp. 1–18, Plenum Press, 1989. [Google Scholar]
- [15].Fukuoka Y, Komori H, Kawabata S, Ohkubo H, Shinomiya K, and Terasaki O, “Imaging of neural conduction block by neuromagnetic recording,” Clin. Neurophysiol, vol. 113, pp. 1985–1992, 2002. [DOI] [PubMed] [Google Scholar]
- [16].Fukuoka Y, Komori H, Kawabata S, Ohkubo H, and Shinomiya K, “Visualization of incomplete conduction block by neuromagnetic recording,” Clin. Neurophysiol, vol. 115, pp. 2113–2122, 2004. [DOI] [PubMed] [Google Scholar]
- [17].Watanabe T, Kawabata Y, Ukegawa D, Kawabata S, Adachi Y, and Sekihara K, “Removal of stimulus-induced artifacts in functional spinal cord imaging,” in Engineering in Medicine and Biology Society (EMBC), 2013 35th Annual International Conference of the IEEE, pp. 3391–3394, IEEE, 2013. [DOI] [PubMed] [Google Scholar]
- [18].Sekihara K and Nagarajan SS, Electromagnetic brain imaging: A Bayesian perspective. Berlin, Heidelber: Springer-Verlag, 2015. [Google Scholar]
- [19].Sarvas J, “Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem,” Phys. Med. Biol, vol. 32, pp. 11–22, 1987. [DOI] [PubMed] [Google Scholar]
- [20].Hämäläinen MS and Ilmoniemi RJ, “Interpreting measured magnetic fields of the brain: Estimates of current distributions,” Tech. Rep TKK-F-A559, Helsinki University of Technology, 1984. [Google Scholar]
- [21].Hämäläinen MS and Ilmoniemi RJ, “Interpreting magnetic fields of the brain: minimum norm estimates,” Med. & Biol. Eng. & Comput, vol. 32, pp. 35–42, 1994. [DOI] [PubMed] [Google Scholar]
- [22].Pascual-Marqui RD, “Standardized low resolution brain electromagnetic tomography (sLORETA): technical details,” Methods and Findings in Experimental and Clinical Pharmacology, vol. 24, pp. 5–12, 2002. [PubMed] [Google Scholar]
- [23].Scharf LL, Statistical Signal Processing: detection, estimation, and time series analysis. New York: Addison-Wesley Publishing Company, 1991. [Google Scholar]
- [24].Meyer CD, Matrix Analysis and Applied Linear Algebra. Philadelphia, PA: Society for Industrial and Applied Mathematics, 2000. [Google Scholar]
- [25].Uusitalo M and Ilmoniemi R, “Signal-space projection method for separating meg or eeg into components,” Medical and Biological Engineering and Computing, vol. 35, no. 2, pp. 135–140, 1997. [DOI] [PubMed] [Google Scholar]
- [26].Nolte G and Curio G, “The effect of signal-space projection on dipole localization accuracy,” IEEE Trans. Biomed. Eng, vol. 46, pp. 400–408, 1999. [DOI] [PubMed] [Google Scholar]