Abstract
Neurotrauma frequently results in neuropathic pain. Our earlier studies revealed that peripheral neurotrauma-induced fragmentation of the myelin basic protein (MBP), a major component of the myelin sheath formed by Schwann cells, initiates a pain response from light touch stimuli (mechanical allodynia) in rodents. Here, we identified the cyclin-dependent kinase 5 (CDK5), as an intracellular interactor of MBP in Schwann cells. The algesic peptide fragment of MBP directly associated with CDK5. When complexed with its p25 co-activator, CDK5 phosphorylated the conserved MBP sequence. The expressed MBP fragment colocalized with CDK5 in Schwann cell protrusions. Roscovitine, an ATP-competitive CDK5 inhibitor, disrupted localization of the expressed MBP peptide. Mutations in the evolutionary conserved MBP algesic sequence resulted in the interference with intracellular trafficking of the MBP fragment and kinase activity of CDK5 and diminished pain-like behavior in rodents. Our findings show that MBP fragment amino acid sequence conservation determines its interactions, trafficking and pro-nociceptive activity. Because CDK5 activity controls both neurogenesis and nociception, the algesic MBP fragment may be involved in the regulation of the CDK5 functionality in pain signaling and post-injury neurogenesis in vertebrates.
Keywords: proteolysis, CDK5, MBP, neuropathic pain, neurodegeneration, myelin, pain, Schwann cells
Neurotrauma induces fragmentation of myelin basic protein (MBP) and results in neuropathic pain. Here, we identified that the algesic MBP fragment directly associated with CDK5. Mutations in the evolutionarily conserved MBP peptide sequence interfered with the CDK5 kinase activity and diminished pain-like behavior. Our findings suggest that the algesic MBP fragment may be involved in pain signaling regulation.
Introduction
Mechanosensitive axons of the peripheral nervous system (PNS) are insulated by the myelin sheath, a multilamellar membrane that permits propagation of action potential from mechanical stimulation [1]. Schwann cells form the myelin sheath by enwrapping their cell membranes around mechanosensitive afferent axons [2]. Demyelination in PNS injury is thought to contribute to severe neuropathic pain states, including pain from light touch, or mechanical allodynia [1, 3]. Myelin basic protein (MBP), a major component of the myelin sheath, is an intrinsically unstructured cationic protein that interacts with anionic lipids and cytoskeletal proteins. These interactions regulate myelin compaction and structural assembly of the axon-glia unit [4]. The invariant α-helix motif 87-VVHFF-91 domain of MBP, conserved across species, is essential to T cell receptor recognition [5].
We previously reported that physical injury in the PNS microenvironment stimulates the protease-mediated cleavage of MBP and the release of fragments comprising the invariant α-helix motif. These fragments accumulate at the paranodal/nodal regions of the damaged myelinated fibers. A single injection of synthetic peptides comprising this MBP sequence into an intact nerve is sufficient to produce robust mechanical allodynia sustained for weeks in rodents [3, 6–8]. However, the molecular identity of cellular proteins that interact with the algesic MBP peptides was unknown. Here, we investigated the cellular interactions and the role of the amino acid sequence conservation in its relationship with the cell components that are involved in the peptide trafficking mechanisms in Schwann cells.
The present study provides the experimental evidence that the algesic MBP peptides directly bind CDK5. CDK5 is an intracellular serine/threonine kinase and a global orchestrator of neuronal cytoskeletal dynamics, including sensory PNS neurons [9]. CDK5 deregulation contributes to neurodegeneration [10–15] and pain signaling [16, 17]. CDK5 phosphorylates a diverse list of substrates, including TRPV1 and NMDA receptors, implicating its activity in a broad range of cellular processes during pain states [16–18]. CDK5 is bound to the cell membrane via its interactions with the p39 and p35 non-cyclin activator proteins [19, 20]. Cleavage of the p39 and p35 by calpain results in the N-end truncated p29 and p25 activators. As a result, the stable CDK5 complex with either p29 or p25 and with the elevated activity is trafficked from the membrane to the cytoplasm and the nucleus. Our findings established that the reciprocal interaction between CDK5 and the algesic MBP peptides represents a novel molecular mechanism of intracellular MBP trafficking and depends on the MBP sequence conservation. The sequence conservation of the algesic MBP peptide is essential in the development of neuropathic pain.
Results
Synthetic MBP epitope internalizes and localizes to cell projections in cultured Schwann cells
The algesic MBP84-104 cryptic epitope is released in the nerve post-injury as a result of proteolysis of MBP. In our earlier studies, the injection of the main algesic, autoimmune MBP84-104 epitope that is hidden in the native MBP fold, into an intact nerve was sufficient to induce robust, long-lasting allodynia in rodents [3, 6–8]. Peptides representing MBP epitope region MBP80-99 and MBP84-104 exhibit a high level of amino acid conservation among evolutionary distant animal species (Fig. 1A, Table 1). Notably, the highly conserved motif 87-VVHFF-91 representing an alpha-helix [21] was present in both peptide species. The MBP epitope region is encoded by exons 3-4 of all four classic MBP splicing variant, and exons 6-7 of variant 1 of the Golli MBP in humans (Fig. 1B).
Fig. 1. A highly conserved cryptic MBP fragment.
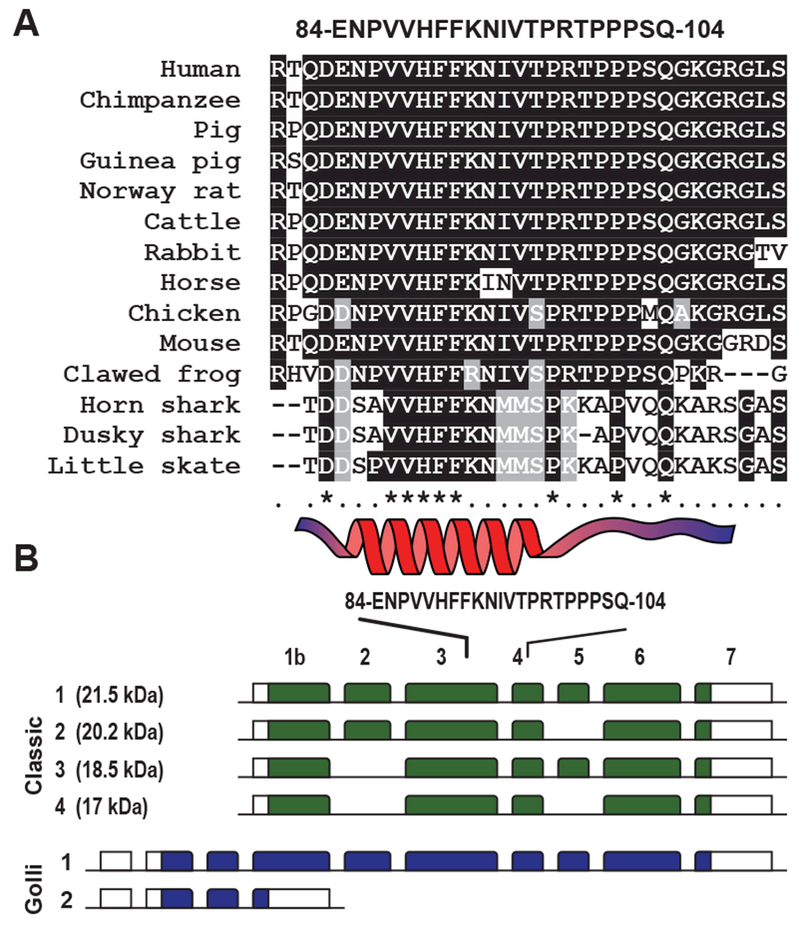
(A) Sequence alignment of the evolutionary conserved central region of MBP. Human (Homo sapiens), orangutan (Pongo abelii), chimpanzee (Pan troglodytes), Rhesus macaque (Macaca mulatta), mouse (Mus musculus), rat (Rattus norvegicus), mole-rat (Heterocephalus glaber), pig (Sus scrofa), guinea pig (Cavia porcellus), dog (Canis lupus), cattle (Bos taurus), panda (Ailuropoda melanoleuca), horse (Equus caballus), rabbit (Oryctolagus cuniculus), chicken (Gallus gallus), clawed frog (Xenopus laevis), horn shark (Heterodontus francisci), ghostshark (Callorhinchus milii), dusky shark (Carcharhinus obscurus) and skate (Leucoraja erinacea). Conserved and semi-conserved residues are shaded black and gray, respectively. Consensus sequence indicates conserved (*) and semi-conserved (.) residues. The invariant α-helix motif is shown at the bottom. (B) Schematic presentation of the gene structure of 4 classic and 2 Golli MBPs. Exons are indicated in green (classic MBPs) and blue (Golli MBP) boxes. The position of the MBP84-104 coding sequence is shown on top.
Table 1. Synthetic MBP peptide sequences.
pT, Phospho-Threonine.
| Peptide | Amino acid sequence |
|---|---|
| MBP84-104-WT | ENPVVHFFKNIVTPRTPPPSQ |
| MBP84-104-H89G | ENPVVGFFKNIVTPRTPPPSQ |
| MBP84-104-SCR | SHVPFNTEQPFPVNVPKPRIT |
| MBP84-104-pT96 | ENPVVHFFKNIVpTPRTPPPSQ |
| MBP84-104-pT99 | ENPVVHFFKNIVTPRpTPPPSQ |
| MBP84-104-T96A/T99A | ENPVVHFFKNIVAPRAPPPSQ |
| MBP80-99-FITC/TMR | RTQDENPVVHFFKNIVTPRT |
We initially screened whether the synthetic MBP peptides can specifically bind receptors on the surface of Schwann cells, the dominant cell type and thus, the likely source for MBP interactors in MBP-injected nerves [8]. The synthetic fluorescently labeled MBP peptide MBP80-99 was administered extracellularly to the primary rat Schwann cell cultures. After 30 min incubation, cells were fixed and fluorescent signals were recorded using in situ epi-fluorescent microscopy. Using a FITC-labeled MBP80-99-FITC peptide, we detected its specific association with the cell periphery, including in cell projections/lamellipodia (Fig. 2A). This specific association was not observed with the scramble MBP80-99-SCR-FITC-labeled peptide.
Fig. 2. In situ localization of algesic MBP fragments in Schwann cells.
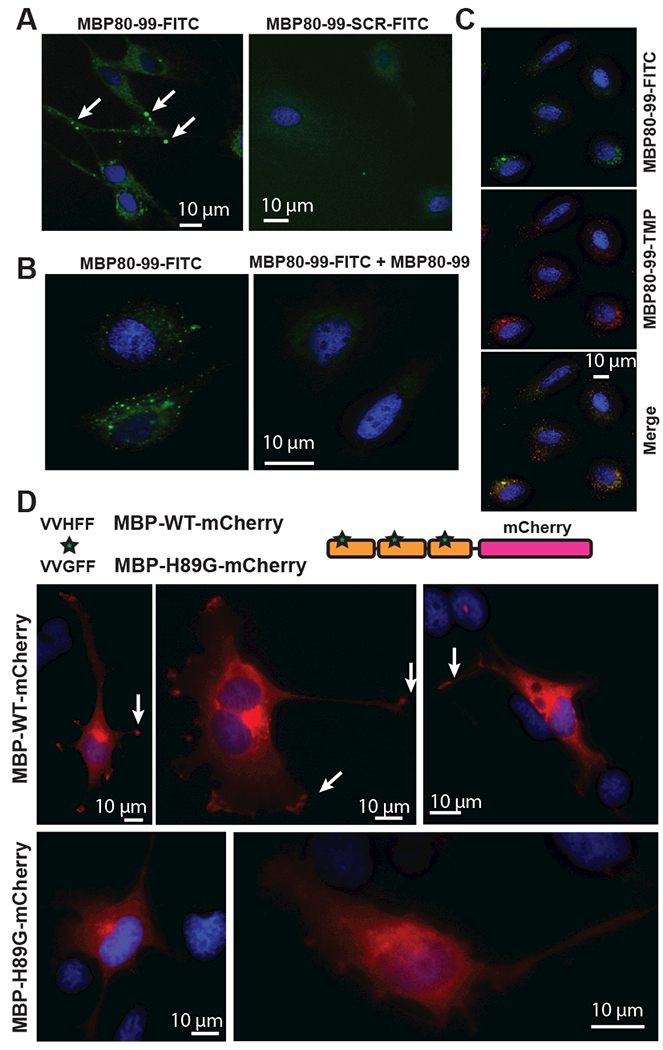
(A) Left panel, Localization of MBP80-99-FITC (green) in projections (lamellipodia) of Schwann cells. Peptides were added to cell culture medium (10 μM) and incubated for 1h. Increase of signal intensity is marked by arrows. Right panel, the scramble MBP80-99-SCR-FITC peptide was incubated and visualized under the identical experimental conditions. (B) Competition titration of MBP80-99-FITC peptide (green) using unlabeled MBP80-99 peptide. Left panel, 25 μM MBP80-99-FITC (green). Right panel, Cells incubated with both 25 μM MBP80-99-FITC, and 100 μM MBP80-99 for 1 h. (C) Colocalization of MBP80-99 peptide labelled with FITC (top panel, green) or TMR (middle panel, red), merged images (bottom panel). (D) The mCherry-tagged wild-type (WT) and mutant (H89G) MBP constructs with three MBP68-102 repeats and the C-terminal mCherry are shown on the top. H89G substitution (asterisk) was inserted in each of the MBP68-102 repeats. Schwann cells expressing MBP-WT-mCherry (red) or MBP-H89G-mCherry (red). Arrows indicate signal localization in lamellipodia of Schwann cells. Blue, DAPI.
The competitive titration of MBP80-99-FITC using a 4-fold excess of unlabeled MBP80-99 peptide reduced the signal intensity thus supporting the specific nature of cell projection binding of MBP80-99-FITC (Fig. 2B). The signal specificity was further confirmed by co-incubation of FITC- and Tetramethylrhodamine (TMR) labeled MBP80-99 peptides. The FITC- and TMR-specific signals juxtaposed at cell periphery loci (Fig. 2C).
Trafficking of the algesic MBP peptide to Schwann cell lamellipodia relies on MBP amino acid sequence conservation
To identify the potential cellular interactors of the internalized algesic MBP peptide, we designed a specialized expression construct that encoded the three head-to-tail linked MBP68-102 sequences C-terminally tagged with mCherry (MBP-WT-mCherry, Fig. 2D, Table 2).Based on the well-established knowledge by others, we used the MBP-tri-peptide-mCherry constructs, rather than a mono-peptide construct, to achieve a better protein stability and to signal readout, as well as to directly monitor the tagged construct in the cell milieu. We have also designed the mutant construct that deviated from the highly conserved in vertebrates central alpha-helix 84-95 portion of MBP (residues are numbered according to human MBP, GenBank NP_001020261). In the mutant construct, amphoteric Gly-89 substituted for the basic His-89 (MBP-H89G-mCherry). Both constructs were expressed in rat Schwann cells and the subcellular localization of the constructs was then recorded. The MBP-WT-mCherry construct was consistently trafficked to the Schwann cell lamellipodial protrusions involved in myelination (Fig. 2D). In contrast, the lamellipodia-specific signals could not be observed in cells expressing the H89G mutant. We concluded that the cell protrusion-directed trafficking of the algesic MBP fragment occurs via amino acid sequence-specific binding to a cellular interactor molecule(s).
Table 2.
The sequence of the MBP-mCherry constructs containing three head-to-tail MBP68-102 sequences.
| Construct | Amino acid sequence |
|---|---|
| MBP-WT-mCherry | MAHYGSLPQKSHGRTQDENPVVHFFKNIVTPRTPPPAHYGSLPQKSHGRTQDENPVVHFFKNIVTPRTPPPAHYGSLPQKSHGRTQDENPVVHFFKNIVTPRTPPP |
| MBP-H89G-mCherry | MAHYGSLPQKSHGRTQDENPVVGFFKNIVTPRTPPPAHYGSLPQKSHGRTQDENPVVGFFKNIVTPRTPPPAHYGSLPQKSHGRTQDENPVVGFFKNIVTPRTPPP |
Expression of MBP peptide activates pain-related signaling pathways
To determine which signaling pathways became altered in response to the MBP-WT-mCherry expression in Schwann cells the genome-wide transcriptome analysis was conducted. Naive Schwann cells were used as a control. RNA sequencing (RNA-seq) was performed using NextSeq 500 DNA sequencer (Illumina), RNA-seq read data were mapped to the R. norvegicus genome (version Rn5) and normalized using Cufflinks software. To predict the effect of MBP-WT-mCherry on the regulation of canonical signaling pathways the gene expression data (log2(FPKM) ≥ 0.01) were analyzed using IPA (Qiagen) (Fig. 3). We compared the activated canonical pathways in Schwann cells to our previously published gene expression data [7, 8]. The latter were obtained after the intrasciatic injection of the MBP84-104-WT peptide in female rats (n=6). In 7 days after the intrasciatic peptide injection the specimens of sciatic nerves, dorsal root ganglia and dorsal spinal cord were collected and the coding RNAs were analyzed by the genome-wide transcriptional profiling using microarray (GEO dataset GSE34868).
Fig. 3. Regulation of signaling pathways in Schwann cells in response to the expression of MBP-WT-mCherry.
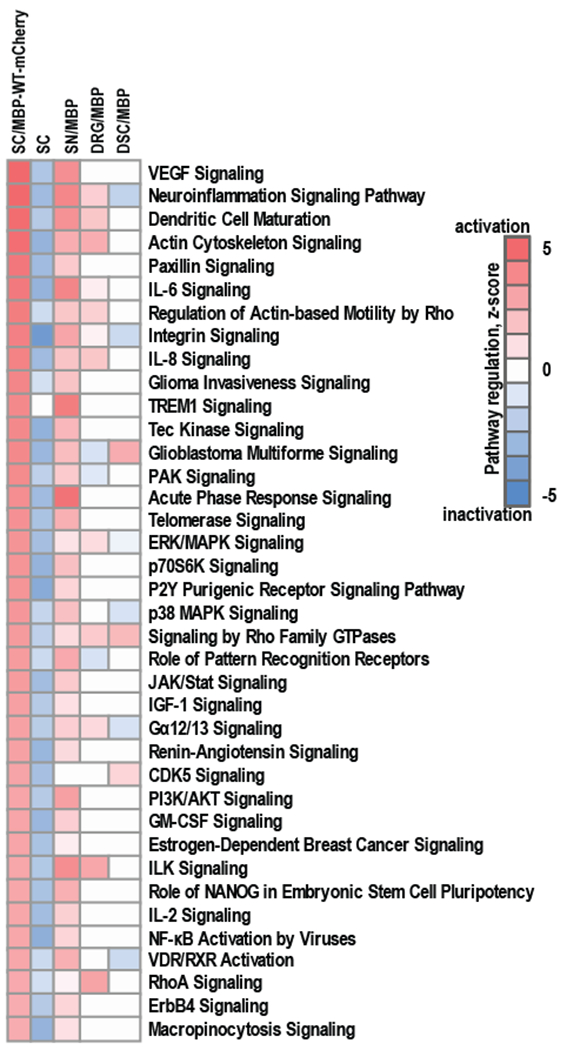
RNA-seq was conducted on RNAs from Schwann cells expressing MBP-WT-mCherry (SC/MBP-WT-mCherry). Naive Schwann cells were used as a control (SC). Whole transcriptome sequencing was conducted using NextSeq 500 instrument (Illumina). RNA-seq read data were mapped to R. norvegicus genome (version Rn5) and normalized using Cufflinks software. RNA-seq data (Log2(FPKM) ≥ 0.01) were analyzed using Ingenuity IPA knowledgebase (Qiagen, version 2017) to predict the regulation of signaling pathways. RNA-seq data were compared to gene expression data from our previously published study [7, 8] in which MBP84-104 peptide was intrasciatically injected in female rats and following 7 days specimen of ipsilateral sciatic nerves (SN/MBP), dorsal root ganglia (DRG/MBP) and dorsal spinal cord (DSC/MBP) were collected, pooled (n=6/animal group) and total RNAs were profiled using gene expression microarrays (GEO ID GSE34868). Color scales represent predictive activation (red) or inactivation (blue) of signaling pathways based on their z-scores.
Our comparative system biology analysis (Fig. 3) indicated that at least 38 signaling pathways activated in both Schwann cells expressing MBP84-104-WT-mCherry and rat nerves injected with the peptide. These pathways included VEGF signaling, neuroinflammation signaling, paxillin signaling, IL-6, IL-8 and IL-2 signaling, p38/MAPK, CDK5 signaling, purinergic receptor signaling, and others. These data suggest that Schwann cells expressing MBP-WT-mCherry exhibit the characteristics that are comparable to our MBP-mediated pain models in rodents.
The MBP peptide specifically interacts with CDK5
We used a mCherry antibody to pull-down the MBP-WT-mCherry and MBP-H89G-mCherry constructs and the associated proteins from the lysates of the respective transfected Schwann cells. Intact Schwann cells were used as a control. To determine the identity of the co-pulled-down proteins, the precipitated material was subjected to the LC-MS analysis using a linear trap quadrupole mass-spectrometer (LTQ-MS). Seventeen unique proteins were present in the pulled-down the MBP-WT-mCherry samples relative to the control (Table 3), including CDK5, actin-related protein 2/3 complex subunit 1A (ARPC1A) [22], plakoglobin (JUP) [23] and testin (TES) that are known to be associated with cell junctions, focal adhesions and cell protrusions-lamellipodia.
Table 3. Proteins specifically immunoprecipitated using the MBP-WT-mCherry construct in Schwann cells.
Coverage, the percentage of the protein amino acid sequence covered by the peptides identified by mass spectrometry analysis.
| ID | Gene name | Description | Coverage, % |
|---|---|---|---|
| Q03114 | CDK5 | Cyclin-dependent-like kinase 5 | 7.5 |
| Q2LAP6 | TES | Testin | 5.5 |
| Q9R1A7 | NR1I2 | Nuclear receptor subfamily 1 group I member 2 | 5.1 |
| Q2KN99 | SPECC1L | Cytospin-A | 4.7 |
| Q91XJ0 | SS18L1 | Calcium-responsive transcription coactivator | 4.5 |
| P38659 | PDIA4 | Protein disulfide-isomerase A4 | 4 |
| Q6L711 | HABP2 | Hyaluronan-binding protein 2 | 3.8 |
| Q2MJT0 | MEF2A | Nuclear receptor subfamily 1 group I member 2 | 3.6 |
| Q8R5I4 | RASGRP4 | RAS guanyl-releasing protein 4 | 3.5 |
| Q99PD4 | ARPC1A | Actin-related protein 2/3 complex subunit 1A | 3 |
| P23785 | GRN | Granulins | 2.9 |
| Q6P0K8 | JUP | Junction plakoglobin | 2.7 |
| P47819 | GFAP | Glial fibrillary acidic protein | 2.62 |
| D4A7T3 | BRDT | Bromodomain testis-specific protein | 2 |
| P02563 | MYH6 | Myosin-6 | 2 |
| A2VD12 | PBXIP1 | Pre-B-cell leukemia transcription factor-interacting protein 1 | 1.7 |
| Q8CFM6 | STAB2 | Stabilin-2 (Fragment) | 1 |
We evaluated the co-pulled-down samples by immunoblotting using the CDK5, ARPC1A, JUP, and TES antibodies. The samples were positive for the presence of CDK5 (Fig. 4A), while other four proteins were undetectable (data not shown). Next, we pulled-down the MBP-H89G-mCherry construct and compared side-by-side the level of CDK5 in the WT and H89G precipitated samples. Our results indicated that the H89G mutant exhibited the reduced ability to bind to CDK5 (Fig. 4A).
Fig. 4. MBP peptide-CDK5 Interactions.
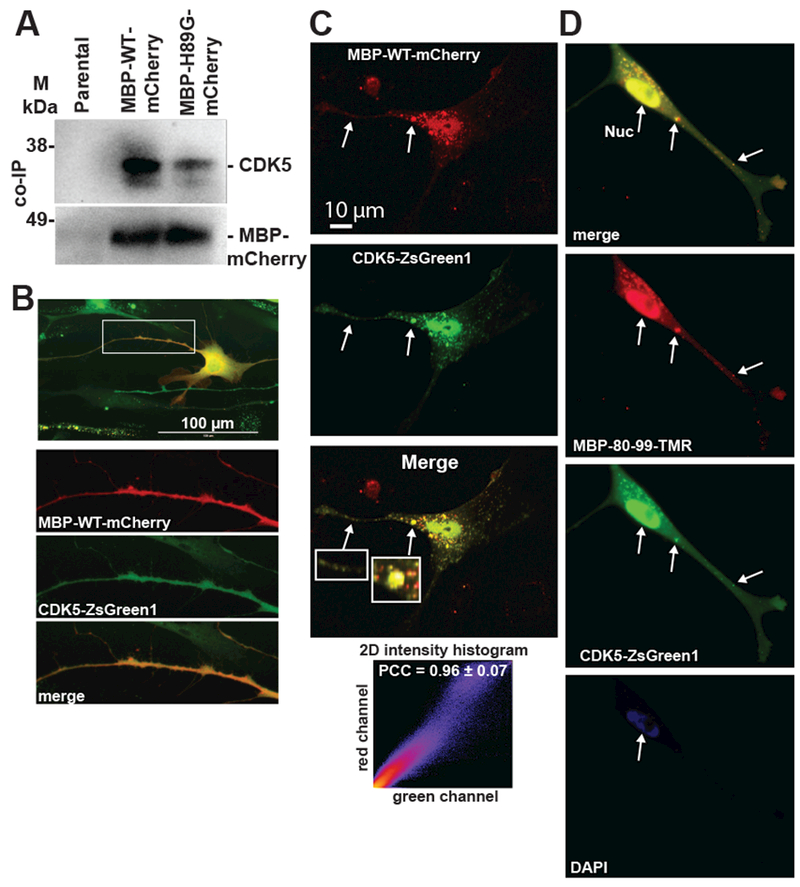
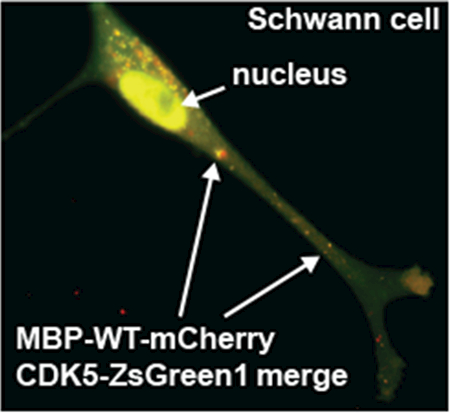
(A) Immunoprecipitation of endogenous CDK5 in Schwann cells expressing the MBP-WT-mCherry and MBP-H89G-mCherry constructs. Proteins were co-immunoprecipitated using a mCherry antibody and analyzed by immunoblotting using anti-CDK5 and anti-mCherry antibodies. Parental Schwann cells were used as a control. M – pre-stained protein marker. (B,C) Schwann cells co-expressing the MBP-WT-mCherry (red) and CDK5-ZsGreen1 (green) constructs. (B) Enlarged images of red, green and merged channels correspond to a Schwann cell protrusion indicated by a rectangle. (C) Fluorescence intensity histogram and Pearson’s correlation coefficient (PCC) indicate the level of co-localization of the constructs in cell protrusions (5 areas). Arrows indicate the overlapping signals. (D) MBP80-99-TMR (10 μM, red) was extracellularly administered to Schwann cells expressing the CDK5-ZsGreen1 (green) construct and incubated for 1h. Overlapping signals are marked by arrows. DAPI, blue.
CDK5 and MBP colocalize in Schwann cells
We co-expressed MBP-WT-mCherry and the human CDK5 fused to a ZsGreen1 tag (CDK5-ZsGreen1) in cultured Schwann cells. A significant level of colocalization of both constructs was detected in cell projections (Fig. 4B, C, Movie). In addition, we observed a significant localization of MBP-WT-mCherry to cell nuclei. The expressed MBP-WT-mCherry alone did not result in accumulation of fluorescent signals in cell nuclei. Because CDK5 protein is known to localize in both cell periphery and nuclear/perinuclear regions [24, 25] we proposed that the exogenously overexpressed CDK5 is responsible for nuclear trafficking of MBP-WT-mCherry. We incubated Schwann cells expressing CDK5-ZsGreen1 with extracellularly administered TMR-labeled MBP80-99 peptide. Using confocal imaging we observed colocalization of CDK5 and MBP80-99-TMR specific signals in both cell protrusions and cell nuclei (Fig. 4D) consistent with the co-expression data (Fig. 4B,C). We concluded that the extracellular MBP peptide can be internalized by Schwann cells and CDK5 binds both extracellularly administered and the expressed MBP peptide species.
CDK5 phosphorylates the algesic MBP peptides
The algesic MBP sequence comprises multiple potential phosphorylation sites sharing a consensus motif Thr/Ser-Pro [26, 27]. The kinase substrate prediction suggested that Thr-96 and Thr-99 can be targeted by multiple Thr/Ser-specific protein kinases, including mitogen-activated protein (MAP) kinases and CDKs (Fig. 5A) [26]. CDK5 was predicted to phosphorylate the Thr-96, Thr-99 and Ser-166 (Fig. 5B). Using LTQ-MS of proteins in the MBP-WT-mCherry pull-down sample, we detected the NIVpTPRTPPPAHYGSLPQK fragment with phosphorylated Thr-96 (data not shown). Based on these data, we hypothesized that CDK5 binds to the MBP peptides and then phosphorylates either Thr-96 or Thr-99, or both.
Fig. 5. Predicted phosphorylation sites of MBP conserved central region.
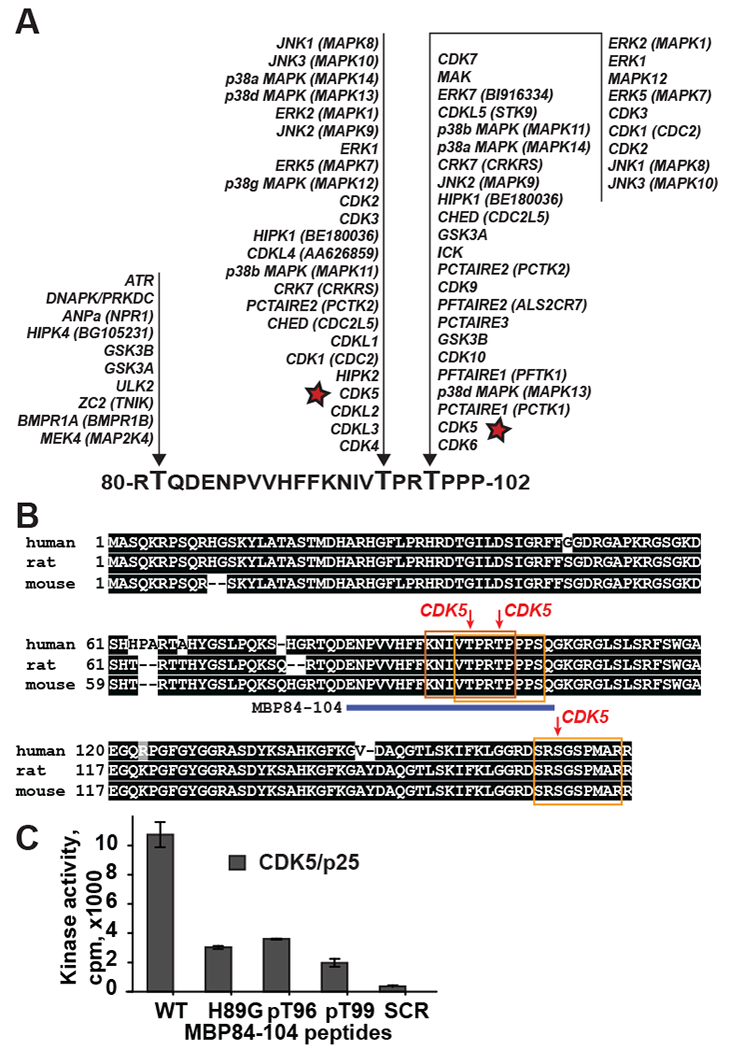
(A) Predicted protein kinases that phosphorylate threonine residues in the MBP 80-102 region. Asterisk, CDK5 (PhosphoNET, www.kinexus.ca). (B) Predicted CDK5 phosphorylation sites (red arrows) in human (NP_001020261, MBP isoform 3), rat (NP_001020463, MBP isoform 2) and mouse (NP_001020426, MBP isoform 3) MBP sequences. Consensus sequences are indicated by yellow boxes. Predictions were made using NetPhos tool (http://www.cbs.dtu.dk/services/NetPhos-2.0). Only sites with high confidence scores (>0.5) are shown. MBP84-104 peptide sequence is shown as a blue bar. Conserved residues are shaded black. (C) CDK5/p25 kinase complexes phosphorylate the MBP84-104-WT, -H89G, -pT96, -pT99 peptides but not the MBP84-104-SCR peptide. Bars correspond to the mean normalized cpm (n=3; **, p≤0.005).
To test if the cryptic algesic peptides are CDK5 substrates, the MBP84-104-WT, MBP84-104-H89G and MBP84-104-SCR synthetic peptides were co-incubated with the purified CDK5/p25 and the level of phosphorylation was then measured by the incorporation of γ−33P. We also used the MBP84-104-WT peptides with synthetic phospho-threonine (pT) at either 96 or 99 positions to determine which individual Thr residue is phosphorylated. The MBP84-104-WT peptide but not the scrambled peptide was efficiently phosphorylated by CDK5/p25 (Fig. 5C). The H89G mutation reduced the level of the peptide phosphorylation at least 2-fold suggesting that the amino acid conservation of the invariant motif 87-VVHFF-91 is important for the CDK5 interactions with the algesic MBP. The level of CDK5-dependent phosphorylation of the MBP84-104-WT peptides with either pT96 or pT99, was reduced more than 2-fold relative to the unmodified peptide. These data implied that both Thr-96 and Thr-99 in the algesic peptide were phosphorylated by CDK5 with a roughly similar efficacy.
Roscovitine disrupts the MBP peptide-CDK5 interactions
Roscovitine is a broad-range purine inhibitor of CDKs, including CDK5, through its direct competition at their ATP-binding site [28, 29]. Based on our modeling data, we hypothesized that roscovitine would interfere with the CDK5-MBP peptide interactions. To test this model, we co-incubated roscovitine for 2 h with Schwann cells co-expressing the MBP-WT-mCherry and CDK5-ZsGreen1 constructs. In the absence of roscovitine, the CDK5 and the MBP peptide immunoreactivity overlapped, especially in the nucleus and cell protrusions (Fig. 6A). In the presence of roscovitine, the MBP peptide immunoreactivity was detected in the cell protrusions, but not in the nucleus. In turn, cell protrusions became free of CDK5 fluorescence which persisted in the nucleus. These findings implied that roscovitine effectively blocked the MBP peptide-CDK5 complex and liberated MBP peptide-free CDK5 from Schwann cell protrusions, yet had no effect on CDK5 translocation to the nucleus.
Fig. 6. Specificity of MBP peptide-CDK5 interactions.

(A) Roscovitine affects cellular co-localization of the MBP-WT-mCherry with CDK5. Schwann cells co-expressing the CDK5-ZsGreen1 (green) and MBP-WT-mCherry (red) constructs. Cells were treated with 10 μM roscovitine for 2 h. White and purple arrows point to the fluorescence in the cell protrusions and nuclei, respectively. Green arrow indicates a nucleus of a cell expressing CDK5-ZsGreen1 alone. Bottom left, PCC indicates the degree of co-localization of MBP- and CDK5-specific fluorescence in cell protrusions (Pr) and nuclei (Nuc) in the control and roscovitine-treated cells (n = 5 areas). Bottom right, Fluorescence intensity of the red and green signals in the control and roscovitine-treated cells (n = 5 areas; **, p≤0.005, *** ≤0.0005). (B) H89G substitution in the MBP peptide affects the kinase activity of CDK5/p25. Boxplot shows the kinase activity of CDK5/p25 in the presence of 25, 50 or 100 μM of the MBP84-104-WT, -H89G, -pT96 and -pT99 peptides.. Kinase activity was measured by the conversion of ATP to ADP. Significant differences were calculated using ANOVA with post-hoc Tukey-HSD test (*, p≤0.05, **, p≤0.005, *** ≤0.0005, n.s. – non significant). (C) Molecular modeling of the MBP84-104-WT peptide bound to CDK5. The predicted position of His-89 is shown by an arrow. Electrostatic map of CDK5 is based on the X-ray structure of the CDK5/p25 complex. Red and blue colors indicate positively and negatively charged residues, respectively.
ATP hydrolysis by CDK5 is affected by His-89
Similar to others [30], we measured ATP to ADP conversion catalyzed by CDK5/p25 in the presence of MBP84-104-WT, MBP84-104-H89G, MBP84-104-SCR and MBP84-104-WT with either synthetic pT96 or pT99. Except MBP84-104-H89G, the effect of the peptides on CDK5 activity was insignificant. In contrast, the mutant peptide considerably reduced the ATP to ADP conversion suggesting the vital importance of the conserved 87-VVHFF-91 motif in the physiological regulation of the CDK5 function (Fig. 6B).
We hypothesized that the conserved 87-VVHFF-91 motif of the CDK5-bound algesic MBP84-104 wild-type peptide is positioned near the ATP-binding site. Mutations in its conserved sequence led to the interference with the ATP to ADP conversion. To model the CDK5 complex with the ENPVVHFFKNIVpTPRTPPPSQ peptide, we used the structures of CDK5/p25 (PDB:3o0g) [31] and the MBP fragment bound to HLA-DR2 (PDB:1vf1) [32]. The fold of the pT96 region in the CDK5 active site was modeled based of the positions of the substrate peptides in the active site of CDK2 [33, 34]. The CDK2 structure was aligned with CDK5 (PDB:3o0g) and then the positions of the main chain of the MBP peptide were aligned with the CDK2 substrate peptides. This approach allowed us to define the relative positions of the P3 and P4 residues (K and N in the MBP peptide, respectively) and roughly assess the general fold of the peptide bound to the CDK5 globule (Fig. 6C). Accordingly, the peptide chain including P7 His-89 is localized near the negatively charged ATP-binding site of CDK5. Alterations of the MBP peptide sequence may alter the peptide fold that potentially interferes with ATP binding in the CDK5 ATP-binding pocket and consequently inhibit the ATP to ADP conversion.
His-89 alters the algesic potency of the MBP peptide
To determine if the His-89 mutation affected not only the trafficking pathway of the MBP84-104 peptide but also its algesic potency, the three synthetic peptides – the wild-type (MBP84-104-WT), the H89G mutant (MBP84-104-H89G), the scrambled peptide (MBP84-104-SCR) and T96A/T99A dual mutant (MBP84-104-T96A/T99A)– were each administered by intra-fascicular injection into sciatic nerve in rats (Fig. 7A). The animals were then subjected to von Frey testing for 7 days. The withdrawal thresholds in von Frey tests in the ipsilateral paw of rats that received the MBP84-104-WT peptide dropped within day 1 post-injection, indicative of the robust mechanical allodynia (Fig. 7B). Withdrawal thresholds remained low in animals injected with MBP84-104-WT peptide for the entire 7-day test period, especially when compared with the scrambled peptide that was inactive in our von Frey tests (p≤0.05). Notably, the pain-inducing efficacy of a MBP84-104-T96A-T99A dual mutant peptide was reduced on days 2 to 7 as compared to the wild-type peptides. A single H89G mutation was sufficient to significantly reduce the pain-inducing efficacy of the peptide. Apart from the mechanical paw hypersensitivity behavior, the rats demonstrated no additional behavioral alterations, consistent with our previous data [7, 8].
Fig. 7. Algesic potency of MBP peptides.
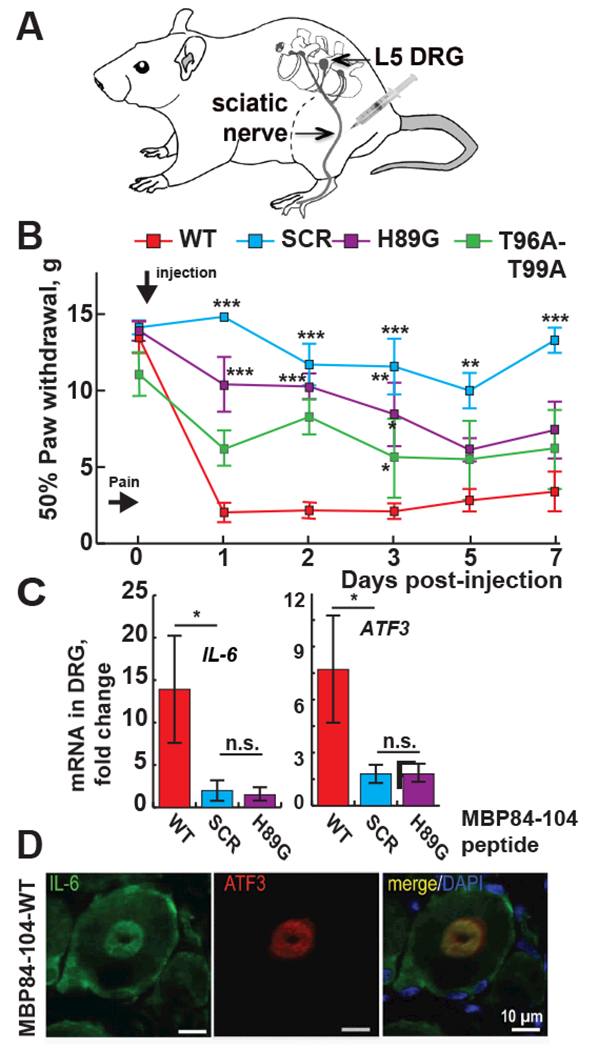
(A) A schematic drawing of the peptide injection into naïve sciatic nerve. (B) The H89G and -T96A/T99A mutations diminish the MBP-induced allodynia, by von Frey testing of paw withdrawal thresholds (n=6/group). Mean gram force (g) ± SEM on days 1, 2, 3, 5 and 7 post-injection of MBP84-104-WT, -SCR, -H89G and -T96A/T99A. *, p≤0.05; **, p≤0.005; ***, p≤0.0005 relative to -WT. (C) RT-PCR of Il-6 and Atf3 genes in L5 DRG. The mean relative mRNA levels of n=3/group were normalized to Gapdh and compared to naïve nerve. Solid bars indicate fold-change relative to the naive nerve samples. Statistical analyses were done using 2-way ANOVA with Bonferroni post-hoc test (*, p<0.05). Standard errors are indicated. (D) Immunostaining of IL-6 (green) and ATF3 (red) in DRG after intra-sciatic MBP84-104-WT injection. DAPI (blue).
To corroborate the von Frey findings, we determined the expression levels of the interleukin 6 (IL-6, or interferon β2) [35] and activating transcription factor 3 (ATF3) [36] in the respective lumbar 5 dorsal root ganglia (L5 DRG) containing the sensory neuronal cell bodies [7]. At day 9 post-injection both Il6 and Atf3 genes were induced following the injection of MBP84-104-WT but not the scramble or H89G peptides (Fig. 7C). In agreement with our von Frey tests, an increase of intraganglionic IL-6 and ATF3 was observed only in the MBP84-104-WT samples (Fig. 7D), but not in the H89G mutant and scramble peptide samples (data not shown).
Localization of MBP peptides in PNS
In the PNS, Schwann cells myelinate the nerves and synthesize MBP. Schwann cells are the main immunoreactive cell type for the algesic MBP peptide post-injury and the main cell type in the MBP-injected nerve [6, 8]. Furthermore, the MBP68-102 peptide translocates toward the paranode/node of Ranvier of the Schwann cell-axon unit, as shown in fibers teased out from the nerve at day 1 post-axotomy (Fig. 8A), in agreement with our previous report [6]. Our comparative transcriptional profiling determined elevated expression of the CDK family genes, including CDK5, in nerve microenvironment at day 1 post-axotomy (Fig. 8B and GEO dataset GSE34868) corroborating the earlier reports of CDK5 expression in the PNS [9, 37]. Myelinating Schwann cells of characteristic crescent morphology produce both the MBP68-102 peptide and CDK5 in nerve at day 1 post-axotomy (Fig. 8C). Based on our in vitro and in vivo data, we propose a model of CDK5-dependent translocation of the algesic MBP to the Schwann cell lamelopodia towards the paranodes and nodes of Ranvier (Fig. 8D), the sites of pain-related ion channel clustering [38].
Fig. 8. Localization of CDK5 and algesic MBP peptides in PNS.
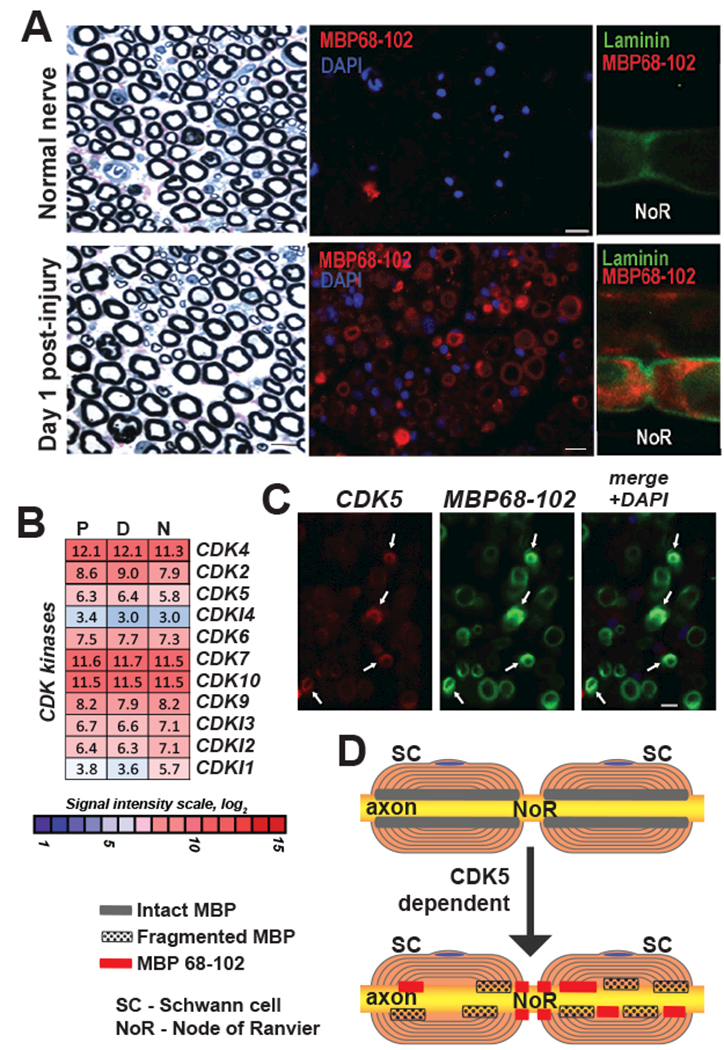
(A) Laminin (green) and MBP (MBP68-102) (red) dual-immunostaining in teased fibers, distal segment, at day 1 post-axotomy. NoR, node of Ranvier. DAPI (blue). (B) Expression of the CDK family genes in the proximal (P) and distal (D) segments of the rat sciatic nerve at day 1 post-axotomy. Heatmap represents normalized intensity values (p<0.05). Color inset shows the signal intensity log2 scale. N, intact nerve. (C) Dual-immunostaining of CDK5 (red) and MBP (MBP68-102) (green) in distal nerve sections at day 1 post-axotomy. Arrows indicate sites of co-localization. Blue, DAPI. (D) A hypothesis schematic of CDK5 dependent distribution of algesic MBP peptides in PNS. SC, Schwann cells, NoR, node of Ranvier. Intact and fragmented MBP, and MBP68-102 are indicated by gray, dotted and red rectangles, respectively.
Together, our data for the first time demonstrate the direct interactions between CDK5 and the highly conserved, pro-nociceptive and auto-antigenic MBP84-104 sequence. Our results suggest that the algesic function of the degraded MBP peptide species in PNS is CDK5-dependent. The reciprocal nature of this relationship is evidenced from the ability of the kinase to phosphorylate MBP84-104, as the amino acid sequence conservation is required to sustain ATP hydrolysis regulated by CDK5.
Discussion
Normally, pain serves as an alarm of impending danger. But an injury in the PNS leading to a damage of the myelin sheath may contribute to a chronic state of neuropathic pain termed mechanical allodynia [1, 3]. MBP is a component of the myelin sheath synthesized by Schwann cells. After nerve injury, the upregulated protease activity cleaves MBP [3, 6, 8]. While the released proteolytic fragments activate the pro-nociceptive machinery, the underlying molecular mechanisms remained elusive. We previously reported that intrasciatic injections of the synthetic peptides corresponding to the 68-102 sequence of MBP that is hidden in the native intact MBP fold reproduce long-lasting mechanical allodynia [3, 6–8]. This MBP region folds into amphipathic α-helix and exhibits remarkable conservation in vertebrates (Fig. 1A). The corresponding MBP fragments are known primary immunodominant epitopes in multiple sclerosis [39]. The 89-HFFK-92 pocket is essential to T cell receptor recognition [5]. However, Schwann cells produce the algesic MBP peptides prior to T cell recruitment to the nerve and the development of the MBP-induced pain is only partially depended on T cells as shown using T cell-deficient nude rats [8]. Phosphorylation of full-length MBP by CDK5 and other protein kinases has been reported previously [40–42].Threonine residues Thr-96 and Thr-99 can be phosphorylated by kinases including ERK1 and ERK2 [43]. Furthermore, other residues, such as His-89 and Lys-92, also may undergo phosphorylation to yield difficult to detect acid-labile phosphoramidates [44–46].
Here, we report that the wild-type peptides derived from the algesic MBP sequence directly interact with CDK5 in Schwann cells. CDK5 is a unique CDK family enzyme known to orchestrate cytoskeletal dynamics in the nervous system and a well-established component in pain signaling processes [16, 17, 47]. CDK5 phosphorylates and regulates the activity of a number of major substrates central to nociception, including N-methyl-D-aspartate (NMDA) [48], P/Q-type voltage-dependent calcium channel (VDCC) [49], transient receptor potential vanilloid receptor-1 (TRPV1) [50], potassium voltage-gated channel Kv2.1 [51] and ATP-gated ion channels [52, 53]. Aberrant phosphorylation by CDK5/p25 is involved in pathological neurodegenerative states including Alzheimer’s disease, brain injuries, Parkinson’s disease and ALS [11–15]. We demonstrated that CDK5 bound to the algesic MBP sequence and then phosphorylated both Thr residues. The MBP binding to CDK5 greatly affected both its cell compartmentalization and enzymatic activity. However, whether this molecular event affects the functions of MBP or CDK5 in pain versus in normal development warrants future investigation.
The selective trafficking to the Schwann cell protrusions and algesic activity were repressed in the mutant MBP peptides, highlighting the importance of the amino acid sequence conservation of the MBP hidden epitope. Schwann cells release signals that guide the assembly of multi-protein cell adhesion, ion channels, and scaffolding complexes into the distinct longitudinal myelin domains [38]. MBP has a well-established role in myelin compaction by regulation of the multi-protein and lipid cell adhesions of the opposing Schwann cell membranes [4]. Our experimental results also support and extend the earlier observations by others [54] who identified the phosphorylation-dependent active nuclear trafficking of MBP.
Inherently, myelinating Schwann cells are the source of myelin proteins in the PNS, including MBP [2]. Physical disruption of the Schwann cell-axon unit and its plasma and myelin membranes caused by nerve injury releases myelin peptides. The algesic MBP84-104 peptide is primarily localized in Schwann cells in vivo [6, 8] consistent with its efficient internalization in cultured Schwann cells. The endogenous MBP peptide does not co-localize with macrophages in the post-injury nerve [6] implying that the peptide escapes the macrophage-mediated phagocytosis. Inhibition of matrix metalloproteinases both blocks the MBP epitope release and diminishes pain caused by nerve injury [3, 6, 8].
The myelin lamellae terminate at the Schwann cell cytoplasm-filled paranodal loops in a close proximity to action potential propagation, ionic fluxes and extensive phosphorylation/dephosphorylation of the molecular machinery at the paranode and node of Ranvier, the sites of pain-related ion channel clustering [2, 38]. The algesic MBP fragments are selectively internalized by mechanosensory A-fibers (inherently myelinated) and not nociceptive C-fibers (inherently unmyelinated), producing pain from mechanical but not thermal stimulation [6–8]. Both MMPs and the algesic MBP peptides translocate toward the paranode/node of Ranvier regions in response to nerve injury [8, 55]. The algesic MBP peptides accumulation in the Schwann cell protrusions in vitro and the paranodal/nodal regions following nerve trauma is likely to be central to their algesic action [8, 56].
In the nodal/paranodal area, the site of the CDK5-MBP binding, the energy demand is high. To support extensive ionic fluxes and action potential propagation, the energy is commonly released in the form of ATP. Our data indicate that CDK5 promotes hydrolysis of ATP more efficiently using the H89G mutant peptide as substrate compared to the wildtype peptide. Our modeling suggests that the H89G mutant fold may interfere with the ATP binding. In turn, the original peptide does not interfere with the ATP binding and, as a result, is efficiently phosphorylated. These data imply that conservation of this sequence region of MBP is important. Conversely, if this conservation is abolished, the mutant MBP sequence may interfere with the ATP binding to CDK5 and CDK5 function in general.
Our data suggest that the binding mode of the algesic MBP fragments involves regions proximal to the ATP-binding site of CDK5. Given the two-substrate kinetics, it is plausible that the algesic MBP peptide binding to CDK5 would affect the efficiency of ATP conversion to ADP. ATP alters nociceptive sensitivity via the ATP-gated P2X receptor cation channels localized at the site of nerve injury [57]. Our results also imply that mutations that potentially destroy the invariant helical structure of the 68-102 MBP region may lead to the fold that affects both the kinase activity of CDK5 and the algesic ability of MBP. Conservation of this MBP region in vertebrates may protect the vital CDK5 functionality in neurogenesis or nociception. In sum, our study identified CDK5 as an intracellular target of the specific immunodominant, algesic fragments of MBP. Whether or not this interaction contributes to pain facilitation warrants investigation, but it clearly controls cellular trafficking of the algesic MBP fragments. Sequence conservation of the algesic 68-102 MBP peptides in vertebrates defines the molecular mechanism in its functional regulation of neuropathic pain.
Materials and Methods
Reagents and antibodies
Reagents were purchased from Thermo Fisher Scientific unless indicated otherwise. The following antibodies were used: rabbit polyclonal anti-mCherry (Abcam, Cat. No. ab183628); mouse monoclonal anti-CDK5 clone 1H3 (Cell Signaling Technologies, Cat. No. 12134); rabbit polyclonal CDK5 (Bioss Antibody, Cat. No. bs-0559R); rabbit polyclonal anti-laminin antibody (Sigma, Cat. No. L9393), goat polyclonal anti-IL-6 (R&D Systems, Cat. No. AF506, rabbit polyclonal ATF3 (Santa Cruz Biotechnology, Cat. No. SC-188, rabbit polyclonal anti-degraded MBP (Millipore, Cat. No. AB5864) and mouse monoclonal MBP antibodies (Biolegend, Cat. No. 808401). Roscovitine (Cayman Chemical) was dissolved (10 mM) in DMSO and diluted to 10 μM in culture medium prior to use [29] or delivered at 25 mg/kg in DMSO [58] in vivo. Peptides (Table 1) were synthesized by GenScript and purified using HPLC-chromatography. DNA oligonucleotides and synthetic DNA were generated by Integrated DNA Technologies.
Cloning of expression constructs
Synthetic cDNA encoding three MBP68-102 fragments (Table 2) and synthetic DNA encoding the mCherry tag [59] were joined and cloned into the linearized pCpGfree-vitroBmcs vector (Invivogen) using isothermal assembly (New England Biolabs). The wild type and H89G mutant pAC-MBP-WT-mCherry and pAC-MBP-H89G-mCherry expression plasmids, respectively, were next constructed and their identity was validated by DNA sequencing. The signal peptide (MKTIIALSYIFCLVFAD) of the influenza hemagglutinin (hsp) [60] was added to the N-terminus of both the wild-type and mutant sequences by DNA synthesis producing pAC-hsp-MBP-WT-mCherry and pAC-hsp-MBP-H89G-mCherry expression constructs, respectively. CDK5 cDNA (GenBank NM_004935.2) was amplified using primers CDK5-GA-F (TAC AGT AGC TTC CAA GTC ACC ATG CAG AAA TAC GAG AAA CTG GAA) and CDK5-GA-R (GTG CAC TAG ATC TTA GTA CTC CGG GCG GAC AGA AGT CGG AGA). CDK5 and a synthetic DNA fragment encoding the C-terminal ZsGreen1 (fluorescent protein derived from Zoanthus sp. reef coral [61]) were assembled and cloned into the linearized pCpGfree-vitro Bmcs vector using isothermal assembly. All constructs were validated by DNA sequencing.
Cell culture
Primary Schwann cells were isolated from sciatic nerves of postnatal (day 1-3) Sprague-Dawley rats [62]. Cells were purified from fibroblasts using 10 μM cytosine arabinoside (AraC), an anti-fibronectin Thy1.1 antibody, and the rabbit complement [63]. Schwann cell purity was confirmed by S100B staining [62]. Schwann cells (purity >99%) were plated in a 75 cm2 flask coated with 50 μg/ml poly-D-lysine (EMD Millipore) in DMEM containing 1 g/L glucose, 10% FBS, 100 μg/ml primocin (Invivogen), 21 μg/ml bovine pituitary extract and 4 μM Forskolin (EMD Millipore). Cells (90-100% confluent) were used in our experiments at passages 3-10. Cultures were routinely maintained at 37°C and 5% CO2.
Transfection of Schwann cells
Cells were grown to 80-90% confluency before transfection. Cells (1×106) were transfected using the recombinant DNA (1 μg) and Lipofectamin 3000. Cells were analyzed after 48 h post-transfection. Positive cells were enriched by passaging in the medium containing 10 μg/ml blasticidin S (Invivogen).
RNA-seq and system biology analysis
Total RNA was extracted from Schwann cells using TRI reagent (Zymo Research) and purified using Direct-zol microRNA system (Zymo Research). The RNA purity was estimated by measuring the OD260/280 ratio using a ND-1000 spectrophotometer (ThermoFisher Scientific). RNA integrity was determined using 2100 Bioanalyser (Agilent Technologies). Poly(A) RNA was enriched using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). Ribosomal RNA was depleted using NEBNext rRNA Depletion reagents (New England Biolabs). A NEBNext Ultra Directional II RNA Library Preparation Kit for Illumina (New England Biolabs) was used to obtain barcoded cDNA libraries from total RNA. Barcoded libraries were pooled and the High Output v2 kit (IlluminaO) was employed to prepare the sequencing libraries. Libraries were sequenced on a NextSeq 500 DNA sequencer (Illumina).
The reads data were processed using BaseSpace software (Illumina). Reads were aligned to R. norvegicus genome (version Rn5) using STAR aligner [64] with default settings. Statistical data processing and differential transcript expression was determined using the Cufflinks package (http://cufflinks.cbcb.umd.edu/). Global differential expression analysis was conducted using the previously published workflow [65]. Biological pathway analysis and comparison were performed using Ingenuity Pathway Analysis (Qiagen). RNA-seq datasets were deposited in the GEO database (GSE107020).
Immunoprecipitation
Cells (5×106) were grown in a 100 mm dish to reach a 80-90% confluency. Culture medium was removed, cells were washed in ice-cold PBS and then lysed for 10 min on ice in 1 ml Lysis/IP Buffer (20 mM Tris HCl, pH 8.0, 150 mM NaCl, 1% Triton X100, 2 mM EDTA) supplemented with Complete Ultra Protease Inhibitors (Roche). Cells were collected using a cell scraper and centrifuged (11,000 g; 10 min, 4°C) to remove cell debris. Cell lysates were incubated with a rabbit polyclonal mCherry antibody (10 μg; Abcam) for 16 h at 4°C with gentle agitation. Protein G magnetic bead slurry (50 μl) was pre-washed in Lysis/IP Buffer, added to the samples and the samples were incubated for 1 h at 4°C with gentle agitation. Magnetic beads were washed 6 times with 1 ml Lysis/IP Buffer supplemented with 300 mM NaCl. The bound proteins were eluted for 10 min at 70°C in 50 μl lithium dodecyl sulphate (LDS) sample buffer supplemented with 50 mM DTT.
Mass spectroscopy
Protein samples were homogenized by sonication and extracted at ambient temperature for 1 h in 100 mM Tris-HCl, pH 8.0, containing 8 M urea and the protease and phosphatase inhibitor cocktails, and the insoluble material was removed by centrifugation 16,000 g for 15 min. The supernatant samples (at least 0.5 mg total protein each) were reduced (10 mM tris(2-carboxyethyl) phosphine, 37°C, 30 min), alkylated (20 mM iodoacetamide, 37°C, 40 min in the dark), and digested using Modified Trypsin, Mass Spectrometry Grade (Promega; 1:100 w/w ratio; 37 °C, 16-18 h). The samples were desalted using a SepPack cartridge, dried using a SpeedVac and re-suspended in 0.1 ml 5% formic acid. The resulting peptides were separated into 24 fractions using an offline Michrom MDLC pump (Michrom) with a Michrom Strong Cation Exchange column. The 1/10 aliquot of each peptide fraction was analyzed using an LTQ-Orbitrap XL mass-spectrometer and a 15 cm Michrom Magic C18 column coupled with a low-flow Michrom ADVANCED device. The data were analyzed by Sorcerer Enterprise v.3.5 software (Sage-N Research) using the ipi.Rat.v3.56 protein database. 57 Da and 16 Da were added to cysteines and methionines to identify carboxyamidomethylated and oxidated residues, respectively. The search results were sorted, filtered, and statistically analyzed using a trans-proteomic pipeline (TPP) (Institute for Systems Biology, Seattle, WA) with a 90% minimum probability score and an error rate ≤2%. An additional search was performed using a Prolucid search algorithm with a DTASelect function via an Integrated Proteomics Pipeline (IP2) server. Relative levels of the proteins in the samples were then analyzed using IP2 for a Label-Free differential peptide/protein analysis. The proteome data were analyzed using Ingenuity IPA 8.7 software (Qiagen).
Immunoblotting
Proteins were separated on NuPAGE 4-12% Bis-Tris Protein Gels (Life Technologies) and transferred onto a PVDF membrane using an OWL Hep-1 dry blotting system. The membranes were blocked for 1 h at 4°C in SuperBlock in TBS/Tween and incubated 16 h with the primary antibodies diluted with SuperBlock in TBS/Tween. Membranes were washed in TBS/Tween and incubated for 1 h with respective HRP-conjugated secondary antibody (Cell Signaling Technologies). Immunostaining was developed using SuperSignal West Dura substrate and visualized using ChemiDoc XRS+ imaging system with Image Lab software (Bio Rad). SeeBlue Plus2 pre-stained marker was used to visualize protein molecular weight.
Immunocytochemistry
Cells were seeded in wells of poly-D-lysine coated 96-well glass-bottom Sensoplate (Greiner Bio-One) and grown to 70% confluency. Cells were fixed for 10 min in 2% formaldehyde/PBS (Tousimis) at 25°C, permeabilized in 0.1% Triton X-100/PBS and blocked in 4% BSA in PBS. Cells were incubated for 1 h with the respective primary antibodies followed by incubation (30 min; 25°C) with species-specific secondary antibody. Cells were mounted on slides using the Prolong Gold reagent (Life Technologies) containing 4′,6-diamidino-2-phenylindole (DAPI). Confocal microscopy was conducted using a LSM 710 NLO Zeiss Multiphoton Laser Point scanning confocal microscope equipped with a multi-photon Mai-Tai laser HB – DeepSee system (690-1024 nm). Images were acquired using ZEN software (Zeiss). Alternatively, a Spinning Disc confocal microscope DSU-IX81 (Olympus) was used and images were captured using Slidebook software (Olympus). Images were further processed using ImageJ (http://www.macbiophotonics.ca) and combined in Photoshop CC (Adobe System). Signal co-localization was determined using Coloc2 utility (http://imagej.net/coloc_2).
Protein phosphorylation assays
Reactions were performed at ambient temperature in a 25 μl reactions consisting of 10 nM of the kinase/activator complex (Kinexus) and 500 μM peptide subtrates in the kinase assay buffer (Kinexus). The reactions were initiated by the addition of 1.25 nmole of [γ−33P] ATP (Perkin Elmer). After 30 min reactions were terminated by spotting 10 μl of the mixtures onto a multiscreen phosphocellulose P81 plate (EMD Millipore). The multiscreen phosphocellulose P81 plate was washed 3 times for 15 min each in 1% H3PO4. The radioactivity was counted in the presence of scintillation fluid in a Trilux scintillation counter (Perkin Elmer).
ATP hydrolysis assay
Reactions (50 μl each) were performed at ambient temperature using Universal Fluorimetric Kinases Assay system (Sigma). Each reaction consisted the CDK5/p25 complex (10 nM each; Sigma) and the substrate peptides (25-100 μM each). The reactions were initiated by the addition of 50 μM ATP. ADP accumulation was recorded in using a microplate reader. Enzyme kinetics data were processed using GraphPad Prism (GraphPad Software).
Animal models
Female Sprague-Dawley rats (8-10 weeks old, 200-225 g) were obtained from Envigo Labs and housed in a temperature-controlled room (∼22 °C), on a 12-h light/dark cycle with free access to food and water. The procedure and testing were conducted during the light cycle. Under isoflurane anesthesia, the common sciatic nerve was exposed unilaterally at the mid-thigh level. A single intrasciatic bolus injection of the MBP peptides (60 μg in 5 μl PBS each) was performed into a nerve fascicle using a 33-gauge needle on a Hamilton syringe. In a separate animal cohort, nerves were transected using surgical scissors to produce axotomy. Sciatic nerve segments (injection site; the proximal and distal to axotomy) and L5 DRG were collected for the quantitative RT-PCR and immunostaining analyses. Tissues from naïve animals were used as a control. Animals were sacrificed using Beuthanasia IP (Schering-Plough Animal Health). All animal procedures were performed in agreement with the PHS Policy on Humane Care and Use of Laboratory Animals and the protocol approved by the Institutional Animal Care and Use Committee at the VA San Diego Healthcare System.
Von Frey test
Sensitivity to non-noxious mechanical stimuli was measured using the up-and-down method [66] by an investigator unaware of the animal groups. The plantar surface of the hind paw was stimulated using calibrated von Frey filaments (Stoelting). Baseline measurements were done for three consecutive days before and then up to daily after the MBP peptide injection. Stimuli were applied for 2 s with a 0.4-15.0 g buckling force to the mid paw plantar surface with ascending filament stiffness until a paw withdrawal response occurred. Stimuli were separated by several-second intervals or until the animal was calm with both hind paws placed on the grid. The consecutive way of applying filaments was continued until six responses were recorded. The 50% threshold was calculated as described [66].
Nerve immunostaining
In cryoprotected, OCT-embedded, 4% paraformaldehyde-fixed 10-μm-thick DRG sections, non-specific binding was blocked using 5% donkey serum in 0.3% Triton X-100 for 30 min at ambient temperature. Teased nerve fibers were prepared from desheathed sciatic nerves at day 1 post-axotomy, distal segments. Nerve bundles were separated using forceps, incubated for 1 h in PBS substituted with 5% fish skin gelatin and 0.1% Triton X-100 (PBS/Gelatin/Triton). Individual fibers were teased out using 0.20- to 0.22-mm acupuncture needles (Vinco, Oxford Medical Supplies, Kingham, UK) and dried. The slides were incubated with a primary antibody (16-18 h at 4 °C) followed by the species-specific Alexa 488-conjugated secondary antibody (green; 1 h, ambient temperature), a second primary antibody (16-18 h, 4 °C), and the species-specific Alexa 594-conjugated secondary antibody (red; 1 h, ambient temperature). The slides were rinsed in PBS and mounted using the Slowfade Gold antifade reagent containing DAPI. Signal specificity was confirmed by omitting the primary antibody. The images were acquired using a Leica DMRB microscope and Openlab software (PerkinElmer).
RT-PCR
Total RNA was extracted from DRG using TRIzol (Invitrogen) and purified on an RNeasy mini column (Qiagen). The RNA purity was estimated by measuring the OD260/280 ratio. The samples were treated with RNase-free DNAse I (Qiagen). cDNA was synthesized using a SuperScript first-strand RT-PCR kit (Invitrogen). Real-time RT-PCR was conducted using MX3005P qPCR System (Agilent) in 25 μl reactions containing Taqman Universal PCR Master Mix (Ambion), cDNA (50 ng), specific forward and reverse primers (900 nM each) and Taqman probes (200-300 nM) for Il-6 and Atf3 mRNAs [7] with a one-step program: 95°C, 10 min; 95°C, 30 s; 60°C, 1 min for 50 cycles. Duplicate samples without cDNA (a no template control) showed no contaminating DNA. Normalization to GAPDH and relative mRNA quantification were calibrated to naïve samples using the 2 (-Delta Delta C(T)) method [67]. The fold-change calculations were performed using MxPro qPCR software (Agilent) [8, 68].
Data analyses
Statistical analyses were performed using GraphPad Prism (GraphPad Software) by analyses of variance (ANOVA) for repeated measures, followed by the Bonferroni post-hoc test, unless specified otherwise. p≤0.05 values were considered significant.
Supplementary Material
Acknowledgements
We thank Drs. S.L. Pelech, S. Hong, W. Choi and J. Ko for discussions and technical assistance. Funding was provided by the grants from the National Institutes of Health RO1 DE022757 (to VIS and AYS) and the Department of Veterans Affairs 5I01BX000638 (to VIS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Abbreviations
- MBP
myelin basic protein
- CDK5
cyclin-dependent kinase 5
- PNS
peripheral nervous system
Footnotes
Database
The novel RNA-seq datasets were deposited in the GEO database under the accession number GSE107020.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Devor M (2009) Ectopic discharge in Abeta afferents as a source of neuropathic pain, Exp Brain Res. 196, 115–28. [DOI] [PubMed] [Google Scholar]
- 2.Pereira JA, Lebrun-Julien F & Suter U (2012) Molecular mechanisms regulating myelination in the peripheral nervous system, Trends Neurosci. 35, 123–34. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi H, Chattopadhyay S, Kato K, Dolkas J, Kikuchi S, Myers RR & Shubayev VI (2008) MMPs initiate Schwann cell-mediated MBP degradation and mechanical nociception after nerve damage, Mol Cell Neurosci. 39, 619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boggs JM (2006) Myelin basic protein: a multifunctional protein, Cell Mol Life Sci. 63, 1945–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendz GL, Moore WJ & Martenson RE (1986) NMR studies of myelin basic protein. XIII. Assignment of histidine residues in rabbit, bovine and porcine proteins, Biochim Biophys Acta. 871, 156–66. [DOI] [PubMed] [Google Scholar]
- 6.Hong S, Remacle AG, Shiryaev SA, Choi W, Hullugundi SK, Dolkas J, Angert M, Nishihara T, Yaksh TL, Strongin AY & Shubayev VI (2017) Reciprocal relationship between membrane type 1 matrix metalloproteinase and the algesic peptides of myelin basic protein contributes to chronic neuropathic pain, Brain Behav Immun. 60, 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko JS, Eddinger KA, Angert M, Chernov AV, Dolkas J, Strongin AY, Yaksh TL & Shubayev VI (2016) Spinal activity of interleukin 6 mediates myelin basic protein-induced allodynia, Brain Behav Immun. 56, 378–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Shiryaev SA, Chernov AV, Kim Y, Shubayev I, Remacle AG, Baranovskaya S, Golubkov VS, Strongin AY & Shubayev VI (2012) Immunodominant fragments of myelin basic protein initiate T cell-dependent pain, J Neuroinflammation. 9, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ino H, Ishizuka T, Chiba T & Tatibana M (1994) Expression of CDK5 (PSSALRE kinase), a neural cdc2-related protein kinase, in the mature and developing mouse central and peripheral nervous systems, Brain Res. 661, 196–206. [DOI] [PubMed] [Google Scholar]
- 10.Tarricone C, Dhavan R, Peng J, Areces LB, Tsai LH & Musacchio A (2001) Structure and regulation of the CDK5-p25(nck5a) complex, Mol Cell. 8, 657–69. [DOI] [PubMed] [Google Scholar]
- 11.Lee KY, Clark AW, Rosales JL, Chapman K, Fung T & Johnston RN (1999) Elevated neuronal Cdc2-like kinase activity in the Alzheimer disease brain, Neurosci Res. 34, 21–9. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen MD, Lariviere RC & Julien JP (2001) Deregulation of Cdk5 in a mouse model of ALS: toxicity alleviated by perikaryal neurofilament inclusions, Neuron. 30, 135–47. [DOI] [PubMed] [Google Scholar]
- 13.Shukla V, Skuntz S & Pant HC (2012) Deregulated Cdk5 activity is involved in inducing Alzheimer’s disease, Arch Med Res. 43, 655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith PD, Crocker SJ, Jackson-Lewis V, Jordan-Sciutto KL, Hayley S, Mount MP, O’Hare MJ, Callaghan S, Slack RS, Przedborski S, Anisman H & Park DS (2003) Cyclin-dependent kinase 5 is a mediator of dopaminergic neuron loss in a mouse model of Parkinson’s disease, Proc Natl Acad Sci U S A. 100, 13650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yousuf MA, Tan C, Torres-Altoro MI, Lu FM, Plautz E, Zhang S, Takahashi M, Hernandez A, Kernie SG, Plattner F & Bibb JA (2016) Involvement of aberrant cyclin-dependent kinase 5/p25 activity in experimental traumatic brain injury, J Neurochem. 138, 317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pareek TK & Kulkarni AB (2006) Cdk5: a new player in pain signaling, Cell Cycle. 5, 585–8. [DOI] [PubMed] [Google Scholar]
- 17.Wang GQ, Cen C, Li C, Cao S, Wang N, Zhou Z, Liu XM, Xu Y, Tian NX, Zhang Y, Wang J, Wang LP & Wang Y (2015) Deactivation of excitatory neurons in the prelimbic cortex via Cdk5 promotes pain sensation and anxiety, Nat Commun. 6, 7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhavan R & Tsai LH (2001) A decade of CDK5, Nat Rev Mol Cell Biol. 2, 749–59. [DOI] [PubMed] [Google Scholar]
- 19.Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P & Tsai LH (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration, Nature. 402, 615–22. [DOI] [PubMed] [Google Scholar]
- 20.Su SC & Tsai LH (2011) Cyclin-dependent kinases in brain development and disease, Annu Rev Cell Dev Biol. 27, 465–91. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed MA, De Avila M, Polverini E, Bessonov K, Bamm VV & Harauz G (2012) Solution nuclear magnetic resonance structure and molecular dynamics simulations of a murine 18.5 kDa myelin basic protein segment (S72-S107) in association with dodecylphosphocholine micelles, Biochemistry. 51, 7475–87. [DOI] [PubMed] [Google Scholar]
- 22.Egile C, Rouiller I, Xu XP, Volkmann N, Li R & Hanein D (2005) Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions, PLoS Biol. 3, e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber GF, Bjerke MA & DeSimone DW (2012) A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration, Dev Cell. 22, 104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ino H & Chiba T (1996) Intracellular localization of cyclin-dependent kinase 5 (CDK5) in mouse neuron: CDK5 is located in both nucleus and cytoplasm, Brain Res. 732, 179–85. [DOI] [PubMed] [Google Scholar]
- 25.Ito Y, Asada A, Kobayashi H, Takano T, Sharma G, Saito T, Ohta Y, Amano M, Kaibuchi K & Hisanaga S (2014) Preferential targeting of p39-activated Cdk5 to Rac1-induced lamellipodia, Mol Cell Neurosci. 61, 34–45. [DOI] [PubMed] [Google Scholar]
- 26.Safaei J, Manuch J, Gupta A, Stacho L & Pelech S (2011) Prediction of 492 human protein kinase substrate specificities, Proteome Sci. 9 Suppl 1, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudrabhatla P KJ, Zheng YL, Amin ND, Kesavapani S, Pant HC (2008) Cyclin-Dependent Kinase 5 (Cdk5) Modulates Signal Transduction Pathways Regulating Neuronal Survival in Cyclin Dependent Kinase 5 (Cdk5) (Ip Nancy Y., T L-H., ed) pp. 69–90, Springer. [Google Scholar]
- 28.De Azevedo WF, Leclerc S, Meijer L, Havlicek L, Strnad M & Kim SH (1997) Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine, Eur J Biochem. 243, 518–26. [DOI] [PubMed] [Google Scholar]
- 29.Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG & Moulinoux JP (1997) Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5, Eur J Biochem. 243, 527–36. [DOI] [PubMed] [Google Scholar]
- 30.Enke DA, Kaldis P & Solomon MJ (2000) Kinetic analysis of the cyclin-dependent kinase-activating kinase (Cak1p) from budding yeast, J Biol Chem. 275, 33267–71. [DOI] [PubMed] [Google Scholar]
- 31.Ahn JS, Radhakrishnan ML, Mapelli M, Choi S, Tidor B, Cuny GD, Musacchio A, Yeh LA & Kosik KS (2005) Defining Cdk5 ligand chemical space with small molecule inhibitors of tau phosphorylation, Chem Biol. 12, 811–23. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Li H, Martin R & Mariuzza RA (2000) Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins, J Mol Biol. 304, 177–88. [DOI] [PubMed] [Google Scholar]
- 33.Bao ZQ, Jacobsen DM & Young MA (2011) Briefly bound to activate: transient binding of a second catalytic magnesium activates the structure and dynamics of CDK2 kinase for catalysis, Structure. 19, 675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook A, Lowe ED, Chrysina ED, Skamnaki VT, Oikonomakos NG & Johnson LN (2002) Structural studies on phospho-CDK2/cyclin A bound to nitrate, a transition state analogue: implications for the protein kinase mechanism, Biochemistry. 41, 7301–11. [DOI] [PubMed] [Google Scholar]
- 35.De Jongh RF, Vissers KC, Meert TF, Booij LH, De Deyne CS & Heylen RJ (2003) The role of interleukin-6 in nociception and pain, Anesth Analg. 96, 1096–103, table of contents. [DOI] [PubMed] [Google Scholar]
- 36.Seijffers R, Mills CD & Woolf CJ (2007) ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration, J Neurosci. 27, 7911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terada M, Yasuda H, Kogawa S, Maeda K, Haneda M, Hidaka H, Kashiwagi A & Kikkawa R (1998) Expression and activity of cyclin-dependent kinase 5/p35 in adult rat peripheral nervous system, J Neurochem. 71, 2600–6. [DOI] [PubMed] [Google Scholar]
- 38.Poliak S & Peles E (2003) The local differentiation of myelinated axons at nodes of Ranvier, Nat Rev Neurosci. 4, 968–80. [DOI] [PubMed] [Google Scholar]
- 39.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL & Hafler DA (1994) Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones, J Exp Med. 179, 279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant NJ, Coates PJ, Woods YL, Bray SE, Morrice NA, Hastie CJ, Lamont DJ, Carey FA & Sutherland C (2015) Phosphorylation of a splice variant of collapsin response mediator protein 2 in the nucleus of tumour cells links cyclin dependent kinase-5 to oncogenesis, BMC Cancer. 15, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosales JL, Sarker K, Ho N, Broniewska M, Wong P, Cheng M, van der Hoorn FA & Lee KY (2007) ODF1 phosphorylation by Cdk5/p35 enhances ODF1-OIP1 interaction, Cell Physiol Biochem. 20, 311–8. [DOI] [PubMed] [Google Scholar]
- 42.Veeranna, Shetty KT, Takahashi M, Grant P & Pant HC (2000) Cdk5 and MAPK are associated with complexes of cytoskeletal proteins in rat brain, Brain Res Mol Brain Res. 76, 229–36. [DOI] [PubMed] [Google Scholar]
- 43.Sanghera JS, Aebersold R, Morrison HD, Bures EJ & Pelech SL (1990) Identification of the sites in myelin basic protein that are phosphorylated by meiosis-activated protein kinase p44mpk, FEBS Lett. 273, 223–6. [DOI] [PubMed] [Google Scholar]
- 44.Smith LS, Kern CW, Halpern RM & Smith RA (1976) Phosphorylation on basic amino acids in myelin basic protein, Biochem Biophys Res Commun. 71, 459–65. [DOI] [PubMed] [Google Scholar]
- 45.Ciesla J, Fraczyk T & Rode W (2011) Phosphorylation of basic amino acid residues in proteins: important but easily missed, Acta Biochim Pol. 58, 137–48. [PubMed] [Google Scholar]
- 46.Klumpp S & Krieglstein J (2009) Reversible phosphorylation of histidine residues in proteins from vertebrates, Sci Signal. 2, pe13. [DOI] [PubMed] [Google Scholar]
- 47.Cheung ZH & Ip NY (2012) Cdk5: a multifaceted kinase in neurodegenerative diseases, Trends Cell Biol. 22, 169–75. [DOI] [PubMed] [Google Scholar]
- 48.Li BS, Sun MK, Zhang L, Takahashi S, Ma W, Vinade L, Kulkarni AB, Brady RO & Pant HC (2001) Regulation of NMDA receptors by cyclin-dependent kinase-5, Proc Natl Acad Sci U S A. 98, 12742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomizawa K, Ohta J, Matsushita M, Moriwaki A, Li ST, Takei K & Matsui H (2002) Cdk5/p35 regulates neurotransmitter release through phosphorylation and downregulation of P/Q-type voltage-dependent calcium channel activity, J Neurosci. 22, 2590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utreras E, Prochazkova M, Terse A, Gross J, Keller J, Iadarola MJ & Kulkarni AB (2013) TGF-beta1 sensitizes TRPV1 through Cdk5 signaling in odontoblast-like cells, Mol Pain. 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerda O & Trimmer JS (2011) Activity-dependent phosphorylation of neuronal Kv2.1 potassium channels by CDK5, J Biol Chem. 286, 28738–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coddou C, Sandoval R, Castro P, Lazcano P, Hevia MJ, Rokic M, Hall B, Terse A, Gonzalez-Billault C, Kulkarni AB, Stojilkovic SS & Utreras E (2017) Cyclin-dependent kinase 5 modulates the P2X2a receptor channel gating through phosphorylation of C-terminal threonine 372, Pain. 158, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nair A, Simonetti M, Fabbretti E & Nistri A (2010) The Cdk5 kinase downregulates ATP-gated ionotropic P2X3 receptor function via serine phosphorylation, Cell Mol Neurobiol. 30, 505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedraza L, Fidler L, Staugaitis SM & Colman DR (1997) The active transport of myelin basic protein into the nucleus suggests a regulatory role in myelination, Neuron. 18, 579–89. [DOI] [PubMed] [Google Scholar]
- 55.Nishihara T, Remacle AG, Angert M, Shubayev I, Shiryaev SA, Liu H, Dolkas J, Chernov AV, Strongin AY & Shubayev VI (2015) Matrix metalloproteinase-14 both sheds cell surface neuronal glial antigen 2 (NG2) proteoglycan on macrophages and governs the response to peripheral nerve injury, J Biol Chem. 290, 3693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shubayev VI, Strongin AY & Yaksh TL (2016) Role of myelin auto-antigens in pain: a female connection, Neural Regen Res. 11, 890–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jarvis MF (2010) The neural-glial purinergic receptor ensemble in chronic pain states, Trends Neurosci. 33, 48–57. [DOI] [PubMed] [Google Scholar]
- 58.Sallam H, Jimenez P, Song H, Vita M, Cedazo-Minguez A & Hassan M (2008) Age-dependent pharmacokinetics and effect of roscovitine on Cdk5 and Erk1/2 in the rat brain, Pharmacol Res. 58, 32–7. [DOI] [PubMed] [Google Scholar]
- 59.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE & Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein, Nat Biotechnol. 22, 1567–72. [DOI] [PubMed] [Google Scholar]
- 60.Guan XM, Kobilka TS & Kobilka BK (1992) Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide, J Biol Chem. 267, 21995–8. [PubMed] [Google Scholar]
- 61.Richards B, Zharkikh L, Hsu F, Dunn C, Kamb A & Teng DH (2002) Stable expression of Anthozoa fluorescent proteins in mammalian cells, Cytometry. 48, 106–12. [DOI] [PubMed] [Google Scholar]
- 62.Brockes JP, Fields KL & Raff MC (1979) Studies on cultured rat Schwann cells. I. Establishment of purified populations from cultures of peripheral nerve, Brain Res. 165, 105–18. [DOI] [PubMed] [Google Scholar]
- 63.Shubayev VI, Angert M, Dolkas J, Campana WM, Palenscar K & Myers RR (2006) TNFalpha-induced MMP-9 promotes macrophage recruitment into injured peripheral nerve, Mol Cell Neurosci. 31, 407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner, Bioinformatics. 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, Lun AT & Smyth GK (2016) From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline, F1000Res. 5, 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaplan SR, Bach FW, Pogrel JW, Chung JM & Yaksh TL (1994) Quantitative assessment of tactile allodynia in the rat paw, J Neurosci Methods. 53, 55–63. [DOI] [PubMed] [Google Scholar]
- 67.Livak KJ & Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods. 25, 402–8. [DOI] [PubMed] [Google Scholar]
- 68.Chernov AV, Dolkas J, Hoang K, Angert M, Srikrishna G, Vogl T, Baranovskaya S, Strongin AY & Shubayev VI (2015) The calcium-binding proteins S100A8 and S100A9 initiate the early inflammatory program in injured peripheral nerves, J Biol Chem. 290, 11771–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


