Abstract
The canonical Wnt/β-catenin signaling pathway plays multiple roles during Xenopus gastrulation, including posteriorization of the neural plate, patterning of the mesoderm, and induction of the neural crest. Wnt signaling stabilizes β-catenin, which then activates target genes. However, few targets of this signaling pathway that mediate early developmental processes are known. Here we sought to identify transcriptional targets of the Wnt/β-catenin signaling pathway using a genome-wide approach. We selected putative targets using the criteria of reduced expression upon zygotic Wnt knockdown, β-catenin binding within 50kb of the gene, and expression in tissues that receive Wnt signaling. Using these criteria, we found 21 novel direct transcriptional targets of Wnt/β-catenin signaling during gastrulation and in addition have identified putative regulatory elements for further characterization in future studies.
Keywords: Transcriptional targets, Wnt/β-catenin signaling, Xenopus gastrulation, Posterior neural tube development, Mesoderm patterning, Neural crest induction
Introduction
Extracellular signaling pathways regulate the patterning and specification of tissues during development, with the same pathway often being deployed to serve many different functions. For example, in the gastrulating Xenopus embryo, canonical Wnt signaling functions to posteriorize the neural plate (reviewed in Niehrs, 2004), pattern the developing mesoderm (reviewed in Harland, 2004), and contribute to neural crest induction (reviewed in Pegoraro and Monsoro-Burq, 2012 and Groves and LaBonne, 2014). To better understand the mechanisms by which Wnt signaling patterns and specifies these different tissues, this study takes a genome-wide and unbiased approach to discover new target genes of this pathway as well as their putative regulatory regions during Xenopus gastrulation.
The canonical Wnt signaling pathway has a single mediator of transcriptional activation, β-catenin, which simplifies identification of regulatory regions. The canonical Wnt signaling pathway is initiated when Wnt extracellular ligands bind to low-density lipoprotein receptor-related protein 5/6 (LRP 5/6) and Frizzled (Fz) receptors (Hikasa and Sokol, 2013). As a consequence, GSK3, which normally phosphorylates β-catenin to target it for destruction, is inhibited, allowing for stabilization of β-catenin and its subsequent translocation into the nucleus, where it binds to TCF/LEF transcription factors (Clevers, 2006; Kim et al., 2013). This binding releases Groucho from TCF/LEF and recruits transcriptional co-activators to initiate transcription of target genes (Roose et al., 1998). There are many antagonists of this pathway, one of which is Dickkopf-1 (Dkk). Dkk is a secreted protein that binds to LRP5/6, inhibiting Wnt ligands from forming a canonical signaling complex (Bafico et al., 2001; Mao et al., 2002; Semënov et al., 2001).
In this study we sought to identify direct transcriptional targets of the canonical Wnt/β-catenin signaling pathway by taking advantage of the recent release of X. laevis genome assemblies and annotations (Xenbase.org: Karpinka et al., 2015; Session et al., 2016 (submitted)). We used three criteria to create a list of target genes. First, a direct transcriptional target must have reduced expression in embryos in which we have inhibited Wnt signaling (Park et al., 2012; Gentsch et al., 2013; Chiu et al., 2014; Schuijers et al., 2014). Second, target genes in our candidate list must have a β-catenin binding site within 50 kb of the gene (Park et al., 2012; Gentsch et al., 2013; X. Zhang et al., 2013; Schuijers et al., 2014; Chiu et al., 2014). Third, direct target genes must also be expressed in a tissue where Wnt signaling is known to occur (Gentsch et al., 2013). Finally, we validated genes that fulfilled all prior criteria by quantifying individual gene expression in uninjected control versus Wnt knockdown embryos by qPCR as well as in situ hybridization. This approach allows us not only to identify the target genes of the Wnt/β-catenin signaling pathway, but also to start characterizing putative enhancers of these target genes.
Results
Transcriptome analysis of Wnt knockdown embryos reveals candidate canonical Wnt/β-catenin target genes involved in mesoderm and neural development.
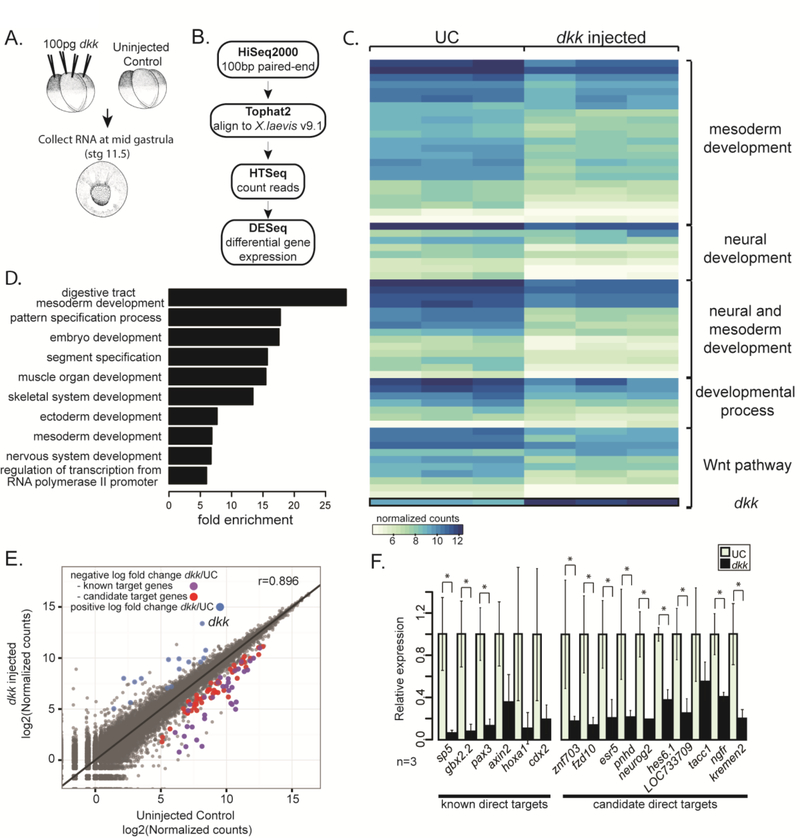
To determine the targets of canonical Wnt/β-catenin signaling that mediate neural and mesodermal patterning at mid gastrula stages, we compared transcriptomes of uninjected control and dkk-injected embryos at stage 11.5. Dkk, a secreted Wnt antagonist, inhibits canonical Wnt signaling at the mid-gastrula stage but does not interfere with the maternal role of Wnt in specifying dorsal mesoderm (Glinka et al., 1998; Brott and Sokol, 2002). We confirmed that embryos injected with 100 pg dkk display enlarged heads and cement glands, indicating that we used a sufficient dose to inhibit Wnt signaling (Glinka et al., 1998) (Sup. Fig. 1A). Uninjected control and dkk-injected embryos were collected at mid gastrula (stage 11.5) from three different mating pairs. After confirming substantial knockdown of known target genes cdx2, hoxa1, and axin2 in single embryos (data not shown) by RT-qPCR of a fraction of the RNA, the remaining RNA was used for transcriptome sequencing (GSE77365) (Fig. 1A).
Figure 1. Expression screen for transcriptional targets of Wnt signaling during gastrulation.
(A) Schematic of experiment. 100pg dkk was injected animally into all blastomeres of 4-cell staged X. laevis embryos. mRNA was extracted from single embryos at stage 11.5 (mid gastrula) and used to make Illumina TruSeq RNA sequencing libraries. (B) Schematic pipeline of RNAseq analysis. (C) Heat map of normalized counts for differentially expressed (DE) genes between three single uninjected control (UC) and three dkk-injected embryos grouped based on categories from GO analysis. (D) Differentially expressed genes were analyzed for enrichment of PANTHER GOslim terms. Fold enrichment is expressed as the observed fraction of genes in GO term category divided by the expected fraction of genes in GO term category. (E) log2 of normalized counts plotted. Uninjected control (UC) counts on the x-axis and dkk-injected counts on the y-axis. Blue dots have a significant (p-adjusted≤0.05) positive log2 fold change in dkk-injected embryos, purple and red dots have a significant negative log2 fold change in dkk-injected embryos. The purple dots are known direct target genes of Wnt signaling and the red dots are candidate target genes of Wnt signaling. (F) qPCR data from single embryos in 3 independent experiments validating selected known and candidate target genes (n=3). Error bars indicate standard deviation and significance was calculated by Student’s t-test (*=p<0.05).
To identify differentially expressed genes, we followed a standard pipeline for analysis (Fig. 1B) using the recently released X. laevis version 9.1 genome assembly (Anders and Huber, 2010; Anders et al., 2015; Session et al., 2016 (submitted) ). Ninetyseven genes were significantly altered in expression (Sup. Table 1). The majority of differentially expressed genes (n=82, 85%) were inhibited in dkk-injected embryos compared to uninjected controls, suggesting that most of the differentially expressed genes might be directly activated by β-catenin (Fig. 1C, E, and Sup. Fig. 2). Indeed, around 30% (n=27/97) of these genes have been previously reported to be canonical Wnt targets (Table 1, Sup. Table 1) (Hikasa and Sokol, 2013). Importantly, the differentially expressed genes found here are enriched for GO terms that suggest involvement in mesoderm and ectoderm/neural development, supporting a role for Wnt signaling in mesoderm and neural patterning at gastrula stages (Fig. 1C,D, and Sup. Fig. 1C). To validate the RNAseq results, expression levels of a set of both known and candidate target genes were tested by qPCR, comparing expression in uninjected control embryos to those injected with 100pg dkk (Fig. 1F). In each case, the target genes showed reduced expression compared to uninjected control (Fig. 1F).
Table 1.
List of characterized direct targets and candidate direct targets of Wnt/β-catenin. Genes in bold are proposed candidate direct target genes. One homeolog per gene is listed and only those genes that are properly annotated in X. laevis annotation v1.8 are included in this list.
| Wnt targets during Xenopus gastrulation | Reference |
|---|---|
| sp5 ------------- | Weidinge et al. 2005 |
| hoxdl ------------- | In der Rieden et al. 2010 |
| znf703 | |
| gbx2.1 ------------- | |
| Li et al. 2009 | |
| gbx2.2 | |
| Li et al. 2009 | |
| xarp | |
| esr5 | |
| pax3 ------------- | |
| Gargett et al. 2012 | |
| axin2 ------------- | |
| Jho et al. 2002 | |
| myf5 ------------- | |
| Shi et al. 2002 | |
| hes5 like | |
| fxd10 | |
| msgn1 ------------- | |
| Wittler et al. 2007 | |
| hoxa1 ------------- | |
| In der Rieden et al. 2010 | |
| pnhd | |
| tp63 | |
| hoxb1 ------------- | |
| In der Rieden et al. 2010 | |
| ventxl.1 ------------- | |
| Ramel, M. C. 2004 | |
| cdx2 ------------- | |
| Zhao et al. 2013 | |
| cdx1 | |
| Prinos et al. 2001 | |
| t | |
| Yamaguchi et al. 1999 | |
| msx2 | |
| Hussein et al. 2003 | |
| irx3 ------------- | |
| Janssens et al. 2010 | |
| vegt | |
| myo3b | |
| tacc1 | |
| tcf7 | |
| ngfr | |
| kremen2 | |
| hes6.1 | |
| mtx2 | |
| zadh2 |
Genome-wide binding pattern of β-catenin revels candidate enhancers of Wnt/β-catenin signaling.
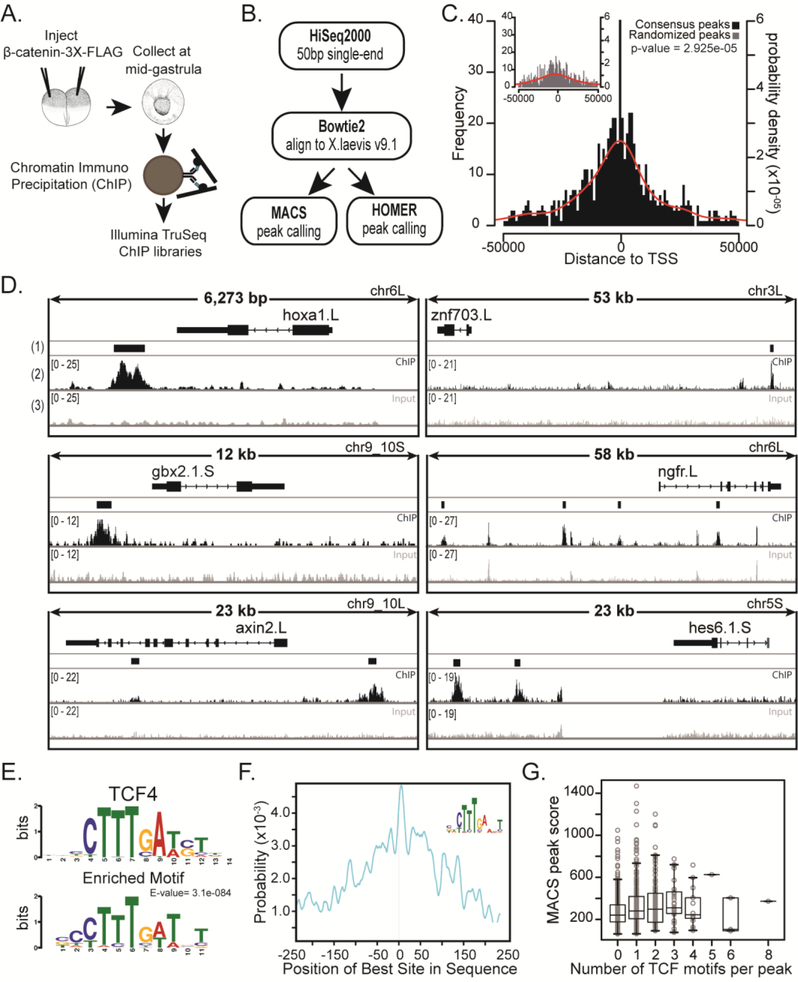
To further investigate whether the candidate target genes from the transcriptome analysis are direct transcriptional targets of β-catenin, we reasoned that a target gene would have a β-catenin binding region within 50kb of the gene. Therefore, we used a triple FLAG-tagged β-catenin and anti-FLAG antibody for Chromatin Immunoprecipitation followed by sequencing (ChIPseq) (GSE77365). Embryos were injected with 500 pg of mRNA encoding a C-terminally FLAG-tagged β-catenin, which, as previously shown, does not induce a secondary axis or posteriorization when injected animally at the two-cell stage (Goentoro and Kirschner, 2009; Young et al., 2014) (Fig. 2A). Previously, Yost et al., (1996) showed that mRNA encoding full-length β-catenin is much less potent in induction of secondary axes than mRNA encoding a protein that removed the N-terminal phosphorylation site. The wild type protein is also very unstable compared to the mutant. The inference is that most ectopically expressed full-length β-catenin is turned over, to restore near physiological levels of the tagged protein under the conditions we used. Expression of the FLAG-tagged β-catenin was confirmed by Western blot (Sup. Fig. 3A) and mid-gastrula (stage 11.5) embryos were collected for ChIP. We made three Illumina libraries from independent chromatin immunoprecipitates, and three controls from input chromatin (chromatin before immunoprecipitation). Fifty base sequencing reads were mapped to the new X. laevis version 9.1 genome and peaks were called using both MACS and Homer peak callers (Y. Zhang et al., 2008; Heinz et al., 2010). The final set of peaks used in this study consisted of those peaks present in all three libraries and found significant by both peak callers (Fig. 2B, Sup. Fig. 3B). We found that the final set of 855 peaks were located an average distance of 2929 bp upstream of all transcription start sites (TSS) compared to 9131 bp upstream of TSS for randomized peaks (Fig. 2C), suggesting an enrichment of β-catenin binding closer to genes than to random genomic locations. In addition, we looked at H3K27ac marks from X. laevis stage 10.5 ChIPseq data at peak locations. While this data (http://veenstra.ncmls.nl/trackhub.htm) represents the chromatin architecture at an earlier stage, we can see that many of the peaks are enriched by inspection for H3K27ac, suggesting that these peaks may act as enhancers (data not shown).
Figure 2. Chromatin Immunoprecipitation and sequencing (ChIPseq) of a FLAG-tagged β-catenin at mid gastrula identifies β-catenin bound regions.
(A) Schematic of experiment. 500 pg of mRNA encoding triple FLAG-tagged β-catenin was injected animally into both blastomeres of a 2-cell stage embryo. Chromatin was crosslinked at mid gastrula, sonicated, and used to make Illumina TruSeq libraries. (B) Pipeline of ChIPseq analysis. 50 bp single-end reads were aligned to X. laevis genome version 9.1 using Bowtie2. Peaks were called with MACS and HOMER. Common peaks were identified by both peak callers in all three biological replicates. (C) Histogram of distances from peak to transcription start site (TSS). Inset: Common peaks were randomly distributed along the genome to make Randomized Peaks (see methods). Red line is the probability density for distances. For ChIP peaks the mean distance to TSS = - −2929 bp and for randomized peaks the mean distance to TSS = −9131 bp. Samples are significantly different with a p-value = 2.925e-05 (Kolmogorov-Smirnov test). Note: Only peaks within 50kb from TSS are shown. (D) IGV browser views of ChIPseq coverage at previously identified target genes and new candidate targets. (1) The Consensus Peak track shows the consensus peaks from three replicate experiments. (2) Read pile-up coverage from a single ChIPseq replicate sample. We note that the width of the peak displayed depends on the overall length of DNA represented, such that peaks appear narrow in 50kb windows. (3) Read pile-up coverage from a single Input replicate sample. (E) Peaks were extended by 250bp from the middle nucleotide and submitted to MEMEchip. Position Weight Matrix of TCF4 motif and most enriched motif found in consensus peaks. E-value=3.1e-084. (F) Distribution of enriched motif in consensus peaks. (G) Box plot representing number of TCF4 motifs in a peak versus the MACS score (MACS score = −10*log10pvalue) of that peak.
Individual peaks were further validated by ChIP qPCR by comparing fold change of signal between uninjected controls and embryos injected with 500pg β-catenin-3xFLAG mRNA or embryos co-injected with 500pg β-catenin-3xFLAG and either 100 or 400pg dkk (Sup. Fig. 3C). With each increase in dose of dkk, the signal of each selected peak was reduced (Sup. Fig. 3C), confirming that the signal for each peak was a result of Wnt dependent stabilization of β-catenin and subsequent binding of β-catenin to chromatin.
To confirm previously identified Wnt/β-catenin responsive regulatory regions, and verify the β-catenin-3xFLAG construct, we used the Integrative Genomics Viewer (IGV) to visualize peaks near known direct target genes (Robinson et al., 2011) (Fig. 2D, Sup. Fig. 4). Hoxa1, Gbx2, Axin2, Sp5 and Cdx1 are all demonstrated direct targets and showed binding profiles similar to those that were previously published (Prinos et al., 2001; Jho et al., 2002; Weidinger et al., 2005; Li et al., 2009; In der Rieden et al., 2010) (Fig. 2D, Sup. Fig. 4). Importantly, many new candidate genes have ChIP peaks within 50 kb of their coding DNA sequence (CDS) (Table 2). Binding landscapes vary such that some peaks are upstream, some are downstream, of CDS and there is often more than one binding site per gene (Fig. 2D).
Table 2.
21 candidate direct targets of Wnt/β-catenin are both differentially expressed in RNAseq experiment and have β-catenin ChIPseq peaks within 50kb of CDS. For full list see Supplemental Table 1.
| Common Name | Rank | V1.8 annotation | baseMean | log2 Fold Change UC/dkk | adjusted P-value | # of peaks within 50 kb of gene | |
|---|---|---|---|---|---|---|---|
| 1 | znf703 | 3 | znf703.L | 602.7 | −2.862 | 2.38E-23 | 1 |
| 9 | znf703.S | 1033.374 | −2.29 | 2.32E-19 | 2 | ||
| 2 | esr5 | 7 | Xelaev18000894m.g | 503.763 | −2.356 | 9.77E-19 | 1 |
| 3 | hes-like | 13 | Xelaev18035791m.g.L | 375.868 | −2.081 | 5.99E-13 | 2 |
| 4 | fzd10 | 15 | fzd10.S | 757.614 | −1.952 | 2.87E-08 | 3 |
| 28 | fzd10.L | 1514.842 | −1.619 | 1.75E-05 | 2 | ||
| 5 | xarp like | 19 | Xetrov90019363m.L | 37.315 | −1.874 | 1.78E-07 | 4 |
| 20 | xarp-like.S | 2207.046 | −1.791 | 1.38E-14 | 5 | ||
| 6 | pnhd | 24 | pnhd.S | 1713.472 | −1.722 | 1.10E-07 | 2 |
| 26 | pnhd.L | 2318.014 | −1.645 | 6.53E-15 | 1 | ||
| 7 | tp63 | 27 | tp63.S | 262.359 | −1.644 | 1.14E-05 | 2 |
| 8 | uncharacterized | 30 | LOC101731931.S | 156.616 | −1.603 | 1.09E-08 | 2 |
| 9 | hes-like | 45 | Xelaev18035790m.g.L | 128.309 | −1.317 | 0.001072 | 2 |
| 10 | hes-like | 48 | LOC733709.L | 240.457 | −1.297 | 0.000673 | 2 |
| 11 | vegt | 52 | Xelaev18010507m.g.S | 4630.587 | −1.25 | 0.00049 | 2 |
| 12 | myo3b | 53 | myo3b-like.1.L | 19.822 | −1.239 | 0.00946 | 5 |
| 13 | tacc1 | 54 | tacc1-like.L | 392.711 | −1.228 | 0.002117 | 2 |
| 14 | tcf7 | 59 | tcf7.S | 505.298 | −1.189 | 7.99E-05 | 1 |
| 15 | ngfr | 67 | ngfr.L | 3846.596 | −1.126 | 0.001554 | 4 |
| 16 | hes-like | 68 | Xelaev18035789m.g.L | 140.925 | −1.117 | 0.019785 | 2 |
| 17 | kremen2 | 70 | kremen2.S | 194.401 | −1.114 | 0.038376 | 1 |
| 18 | uncharacterized | 74 | Xetrov90018470m.S | 1078.929 | −1.086 | 0.000136 | 1 |
| 19 | hes6.1 | 76 | hes6.1.S | 1887.573 | −1.069 | 3.25E-05 | 4 |
| 20 | mtx2 | 77 | mtx2.S | 95.887 | −1.062 | 0.008915 | 5 |
| 21 | zadh2 | 78 | zadh2.L | 69.105 | −1.059 | 0.044797 | 1 |
β-catenin does not directly bind DNA, but rather binds to the transcription factors TCF/LEF (Clevers and van de Wetering, 1997; Clevers, 2006; Schuijers et al., 2014). We confirmed that indeed, the TCF/LEF motif was enriched in the peaks, using MEMEChIP (Machanick and Bailey, 2011). Taking the middle nucleotide of each peak and extending 250bp on either side, we found that the TCF4 motif is the most significantly enriched of any identifiable motif, and is centrally positioned within each peak (Machanick and Bailey, 2011) (Fig. 2E,F). In addition, peaks with more TCF motifs per peak have slightly larger peak scores as called by MACS, suggesting a slight increase in binding of β-catenin when there are more TCF transcription factors bound to DNA (Fig. 2G).
In addition to the TCF motif being enriched in the consensus peaks, there are other motifs that are highly enriched (Sup. Fig. 5). A motif resembling a ZNF281 motif is the second most highly enriched motif followed by a motif for Zic1/3/4 (Sup. Fig. 5). Because β-catenin is thought to use the TCF/LEF family of transcription factors exclusively as a binding partner, this suggests a recurring set of other transcription factors cooperate in regulating Wnt/β-catenin target genes at this stage (Schuijers et al., 2014).
Candidate direct target genes have both a β-catenin binding site as well as reduced expression in dkk-injected embryos.
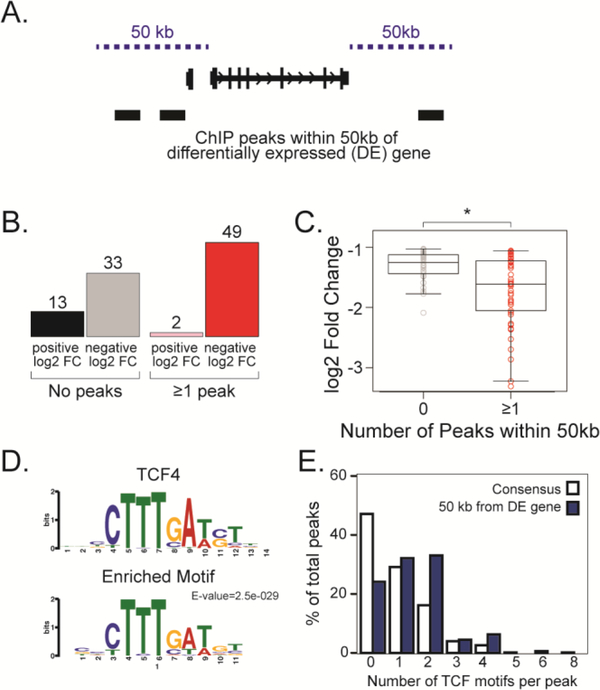
To determine which genes have both a β-catenin binding site as well as reduced expression in dkk-injected embryos, we asked how many ChIP peaks lie within 50 kb of each differentially expressed gene from the RNAseq data (Fig. 3A, Table 2). Here we use the coding DNA sequence (CDS) instead of the transcription start site (TSS) to find peaks both 50kb upstream of the TSS and 50kb downstream of the last coding nucleotide. The tetraploid genome of X. laevis frequently has two homeologs for each gene, therefore here we count each homeolog individually. Of the differentially expressed genes, 53% (n=51/97) had at least 1 peak within 50 kb of the CDS, and of these, 96% (n=49/51) were inhibited by DKK, as would be expected for β-catenin direct targets (Fig. 3B). This is probably an underestimate because our stringent peak calling methods exclude the weaker peaks. Regardless, all genes with 2 or more peaks within 50 kb of a differentially expressed gene are inhibited by DKK, and those genes with at least 1 peak within 50 kb have more significantly reduced expression than those with 0 peaks (Fig. 3C).
Figure 3. Candidate Wnt direct target genes have both a negative log2 fold change in dkk-injected embryos and a ChIP peak within 50 kb of the coding DNA sequence.
(A) Differentially expressed (DE) genes were designated as candidate direct target genes if they had a ChIPseq peak within 50 kb from the coding DNA sequence (CDS). (B) Number of DE genes that have either no peaks or at least one peak within 50 kb from CDS. (C) Log2 fold change (y-axis) of DE gene plotted as a function of number of peaks within 50 kb (x-axis). Genes that have at least one peak have a significantly greater negative log2 fold change than genes with zero peaks. p < 0.05. (D) A TCF motif is enriched in the set of peaks within 50 kb of DE genes as determined by MEMEChIP. E-value=2.5e-029. (E) Percentage of peaks with TCF motif. Blue is peaks within 50 kb of DE gene; white represents all consensus peaks.
A new motif analysis of only the peaks within 50 kb of differentially expressed genes revealed that the TCF4 motif is still enriched in this subset of peaks (Fig. 3E). Further, the percentage of peaks with 1 or more TCF motifs increased when using this subset; whereas around 55% of all peaks have at least one TCF motif, this increased to 75% of peaks within 50 kb of differentially expressed genes (Fig. 3F).
Expression pattern of candidate direct target genes is similar to wnt8 expression at mid gastrula.
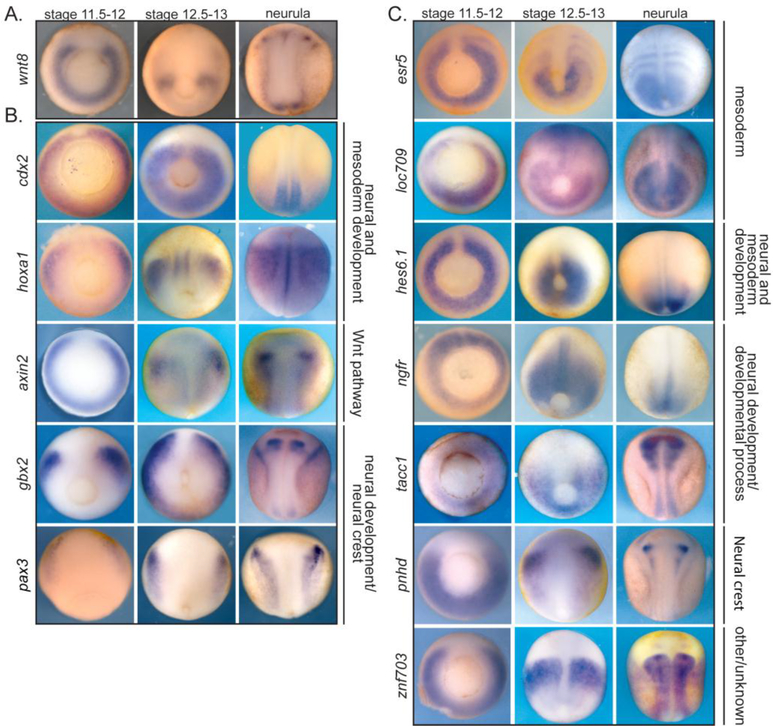
Although Wnt8 is a secreted molecule and its protein expression domains may exceed the bounds of the mRNA expression, Wnt/β-catenin candidate target genes should be expressed in similar or overlapping domains to that of wnt. Of the known direct target genes, most show very similar expression domains, in many cases resembling the horseshoe pattern of wnt8 (Smith and Harland, 1991; Christian and Moon, 1993; Harland and Gerhart, 1997; In der Rieden et al., 2010) (Fig. 4A). For example, the direct targets cdx1, cdx2, hoxa1, axin2, and gbx2 are all expressed in the ventral and lateral marginal zone at stage 11.5 with expression domains slightly more anterior to that of wnt8 (Fig. 4A,B, Sup. Fig. 6). Despite their similar expression patterns, these four target genes are involved in development and patterning of two different tissue types (the mesoderm and neurectoderm) and go on to have very different expression patterns at later neurula stages (Fig 4B).
Figure 4. In situ hybridization shows that the expression patterns of many characterized and candidate Wnt/β-catenin direct target genes resemble wnt8 expression.
Genes are grouped based on functional role. (A) Expression of wnt8 at different developmental stages. (B) Expression of a few characterized direct target genes of Wnt/β-catenin at different developmental stages. (C) Expression of selected candidate direct target genes. Stage 11.5–12 and 12.5–13 is blastopore view with dorsal up. Neurula stage is dorsal view with anterior up.
Like the previously demonstrated direct targets of Wnt signaling, many of the candidate direct target genes, regardless of their functions at later developmental stages, have a similar expression patterns at stage 11.5 (Fig. 4C). While the size of the dorsal midline gap and the thickness of the horseshoe expression domain vary, the general pattern is consistent with these genes being direct targets of Wnt/β-catenin signaling. At later stages the expression pattern of these candidate targets varies significantly, as for the known direct targets. For example, esr5 is expressed in developing somites, whereas hes6.1 is expressed in the posterior mesoderm (Fig. 4C). The varied expression of both candidate and direct target genes after gastrulation further confirms that our approach identified target genes of Wnt signaling during gastrulation, though their mode of regulation after gastrulation may differ.
Validation of Wnt-dependent target gene expression by in situ hybridization
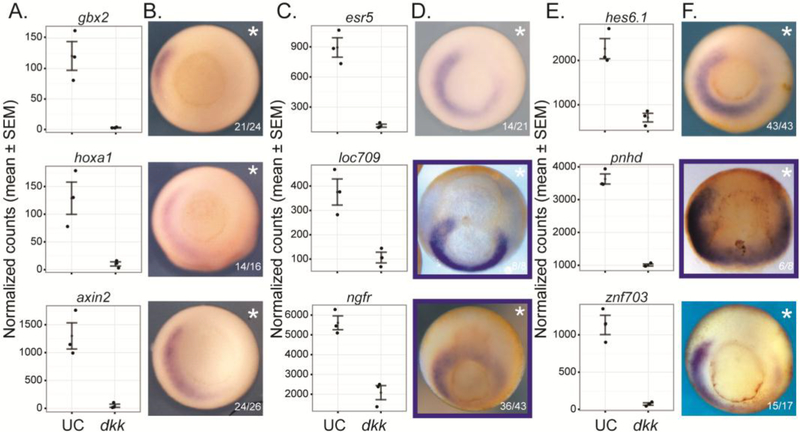
To determine whether the expression pattern of candidate genes was dependent on Wnt signaling, we injected dkk into the 2 right blastomeres of a 4-cell embryo. While RNAseq reports the global loss of expression upon Wnt knockdown (Fig. 5A,C,E), in situ hybridization can reveal tissue type-specific changes in expression of each candidate gene resulting from dkk injection (Fig. 5B,D,F). For both known (Fig. 5A,B, Sup. Fig. 7) and candidate (Fig. 5C-F, Sup. Fig. 7) target genes, there are varying degrees of loss of expression on the injected side, with the ventral domain of expression being most resistant to the manipulation. While some genes such as znf703 show nearly complete loss of gene expression on the dkk-injected side, other genes such as esr5 do not show such a dramatic loss of expression (Fig. 5C-F). Variation in the location of expression changes may be attributed to co-regulation of target genes by other signaling pathways or transcription factors. For example, esr5, a gene expressed in the paraxial mesoderm and important for developing somites, is also regulated by Notch signaling (Jen et al., 1999).
Figure 5. Expression of Wnt/β-catenin target genes is reduced by dkk-injection.
(A,C,E) Normalized counts from RNAseq data comparing uninjected control (UC) and dkk-injected embryos for selected known (A) and candidate (C,E) target genes at stage 11.5 (mid-gastrula). (B,D,F) In situ hybridization showing expression pattern of known (B) and candidate (D, F) target genes. Embryos were injected in the right (asterisk) 2 blastomeres at the 4-cell stage with a total of 100 pg dkk. Blue box indicates that embryo was cleared in either benzyl benzoate:benzoic acid (BB:BA) or benzyl benzoate alone (BB; to reduce transparency and visualize the archenteron). Blastopore view with dorsal up, except pnhd is dorsal view with anterior up.
Discussion
Canonical Wnt signaling has been extensively studied in the context of development and cancer, but while some of the direct transcriptional targets of this signaling pathway have previously been documented, our study reveals many novel direct target candidates. Furthermore, by examining both the transcriptional responses to Wnt knockdown and binding locations of β-catenin, we have identified not only potential direct targets but also potential enhancer regions that regulate these genes.
Our unbiased genome-wide approach was able to detect subtle changes in gene expression to discover new target genes of the Wnt pathway. The known direct target genes axin2, gbx2, pax3, sp5, cdx1, cdx2, hoxa1, hoxd1, and irx3 have the largest and most significant log2 fold changes from our RNAseq data (Table 2, Sup. Table 1). Thus, these large changes in expression upon Wnt manipulation made these genes more readily identifiable, compared to the new candidate targets discovered here.
Further, many of the previously characterized Wnt/β-catenin responsive enhancers that regulate these genes are verified in our list of candidate enhancers. For example, a Wnt responsive regulatory element for cdx1 has been characterized immediately 5’ to the TSS in developing mouse embryos, as well as a 5’ promoter proximal sp5 element in zebrafish (Prinos et al., 2001; Weidinger et al., 2005). We see both of these elements called as significant peaks in our ChIPseq data. In fact, a common feature of many of the known Wnt responsive regulatory elements is that they are near promoter sites, as this is the most easily identified region. Again, our unbiased genome-wide approach was able to detect potential regulatory elements as far as 50kb away from CDS (Fig. 2C), and added new and potentially more sensitive targets to genes such as axin2, where the documented enhancer immediately 5’ to the gene (Jho et al., 2002) showed an increase in immunoprecipitation, but was not called as significant, compared to a larger ChIP peak further 5’ to the gene.
Many of the known and candidate target genes of the canonical Wnt signaling pathway during gastrulation are expressed in a domain resembling the wnt8 expression domain around the blastopore (Fig. 4A). Wnt is a secreted ligand and can act as a morphogen, signaling several cell diameters away so that we would expect that some of the target gene expression could extend modestly beyond the wnt-expressing cells (Zecca et al., 1996; Mii and Taira, 2009). In fact, while many of the genes resemble the expression pattern of wnt, there are differences in expression. For example, pax3 and gbx2, two genes involved in neural patterning as well as neural crest specification, are expressed more anteriorly than wnt and most of the other target genes (Tour et al., 2002; Li et al., 2009; Garnett et al., 2012) (Fig 4). We also see differences in the size of the gap in expression across the organizer (Fig.4). In addition to wnt8, wnt3a is expressed in the paraxial mesoderm of the early gastrulating Xenopus embryo and has been demonstrated to posteriorize the neural plate via activation of meis3 (Elkouby et al., 2010).
If these genes are all targets of Wnt signaling mediated by β-catenin, then why are there differences in their expression domains? Three mutually nonexclusive hypotheses may explain this: 1.) There are differences in the binding landscape of β-catenin. Some genes such as ngfr have many β-catenin binding sites nearby, while other genes such as znf703 only have one binding site much further away. 2.) These genes are also being regulated by other signaling pathways or transcription factors. Fgf and RA signaling are known to have overlapping roles in caudalization of the neural plate (Blumberg et al., 1997; Cox and Hemmati-Brivanlou, 1995; Kengaku and Okamoto, 1995; Lamb and Harland, 1995; Y. Chen et al., 2001; Kudoh et al., 2002). Indeed, these pathways share many of the same direct targets; for example, RA and Wnt converge on the cdx1 promoter (Prinos et al., 2001). Other pathways such as BMP signaling pathway help regulate genes expressed at the neural plate border that are important for neural crest development (LaBonne and Bronner-Fraser, 1998; Kléber et al., 2005). The upstream promoter of msx2, for example, is synergistically and directly activated by BMP and Wnt signaling (Hussein et al., 2003). By examining other motifs that are enriched in the β-catenin binding regions we can start to predict which factors may be required or permissive for expression of these Wnt target genes. For example, the motif for Zic1, a transcription factor important for neuroectodermal differentiation, is enriched in the β-catenin binding regions identified in our ChIP experiment and therefore might serve to co-regulate a subset of genes required for neural differentiation (Mizuseki et al., 1998; Aruga and Mikoshiba, 2011). 3.) These genes are each integrating repressive inputs in their regulatory landscape, and thus the threshold at which the repressors outcompete the activators determine the extent of the expression domain (H. Chen et al., 2012).
In conclusion, our genome-wide approach combining RNAseq and ChIPseq data has identified new candidate direct target genes of the canonical Wnt/β-catenin signaling pathway. The direct transcriptional target genes identified here are down-regulated when zygotic Wnt signaling is inhibited with DKK1 and have β-catenin ChIP peaks within 50 kb of the gene’s CDS. In addition, these genes are expressed in a similar domain to wnt8 mRNA expression during gastrula stages in X. laevis. The identification of putative regulatory regions via ChIP-seq provides the basis for future studies in understanding how these target genes are regulated such that they are expressed in a context dependent manner.
Materials and Methods
Embryo Culture
X.laevis embryos were obtained (Sive et al., 2010) and staged (Nieuwkoop and Faber, 1967) as described previously. All controls are uninjected controls (UC).
DNA constructs and RNA synthesis
The triple FLAG epitope tagged X. laevis β-catenin (β-catenin-3X FLAG) was generated from a single FLAG epitope β-catenin, described previously (Young et al., 2014), using the restriction enzymes, AvrII and NotI and the double stranded oligonuclotide:
GCATCCTAGGAGATTACAAGGATGACGACGATAAGGACTATAAGGACGATG ATGACAAGGACTACAAAGATGATGACGATAAATAAGCGGCCGCAAGGCC.
The sequence for the triple FLAG epitope is underlined, and the start of each single FLAG is in bold.
Capped RNAs were synthesized using mMessage mMachine (Ambion). DKK1 pCS2 (Glinka et al., 1998) was digested with NotI and β-catenin-3X FLAG pCS108 was digested with AscI. Both were transcribed with SP6 RNA polymerase. All RNAs were injected in 5nl or 10nl bursts along with mCherry RNA to serve as a tracer.
Whole-mount in situ hybridization
Embryos were stained by in situ hybridization as described (Harland, 1991). Probe sequences for axin2, esr5, loc709, ngfr, hes6.1, tacc1, pnhd, znf703, irx3, frzd10 and ngn2 are available on request.
RNAseq
100pg dkk1 mRNA was injected into all blastomeres of a 4-cell stage embryo. Total RNA was extracted from three single dkk-injected embryos and three single uninjected control (UC) embryos at stage 11.5 (mid gastrula) using Trizol (Invitrogen). Illumina TruSeq RNA sequencing libraries were made from single embryos resulting in a total of three libraries made from uninjected control (UC) embryos and three libraries made from dkk-injected embryos. Each of the replicate pairs (uninjected control and DKK injected) came from the same mating pair. For quantitative PCR (qPCR) analysis of candidate target genes, cDNA was made using 1ug of total RNA using iScript (Bio-Rad) and reactions were amplified using a CFX96 light cycler (Bio-Rad) with SsoAdvanced Universal SYBR Green supermix (Bio-Rad). Data shown is from RNA extracted from single embryos from 3 independent experiments (n=3). All samples were normalized to control embryos and elongation factor 1a1 (eef1a1) was used as internal control.
RNAseq analysis
100bp paired-end sequencing reads (Illumina HiSeq2000) were aligned to the X. laevis genome (assembly version 9.1) using Tophat2.1.0 (Trapnell et al., 2009). Read counts were generated using htseq-count (Anders et al., 2015) and the X. laevis genome annotation v1.8. Differential expression (DE) analysis was performed using DESeq (Anders and Huber, 2010) with an adjusted p value < 0.05 and log2 fold change > 1 cutoffs. DE genes were analyzed for enrichment of GO terms using PANTHER GOslim (Mi et al., 2012).
Chromatin Immunoprecipitation (ChIP)
500pg of either single-FLAG or triple-FLAG C-terminus tagged β-catenin was injected into both blastomeres of X. laevis embryos at the 2-cell stage. Embryos were collected at stage 11.5 and processed for ChIP as previously described (Blythe et al., 2009; Wills et al., 2014) using an anti-FLAG antibody (Sigma F3165). Sequencing libraries were made from immunoprecipitates of between 200–500 embryos and input chromatin (chromatin before immunoprecipitation) using Illumina TruSeq ChIP prep kit per manufacturer’s instructions. Antibodies used in Supplemental Figure 3A: FLAG (Sigma F3165), beta Actin (GeneTex GT5512).
ChIPseq analysis
50 bp single-end sequencing reads were aligned to the X. laevis genome (assembly version 9.1) using bowtie2. Peaks were called using both MACS (Y. Zhang et al., 2008) and Homer (Heinz et al., 2010). Final peaks were taken as those called by both peak callers and the common peaks reported in Figure 2 were significant in each of the three replicate experiments. Peaks and read pile-ups were visualized with Integrative Genomics Viewer (IGV) (Robinson et al., 2011). De novo motif analysis was performed with MEMEchip (Machanick and Bailey, 2011). Distance from peak to transcription start site was calculated using closestBed and randomized sample was made using shuffleBed from the bedtools suite (Quinlan and Hall, 2010).
Candidate gene selection
Differentially expressed (DE) genes from the RNAseq analysis with a ChIPseq peak within 50kb up or downstream of the gene body (CDS) were considered candidate target genes. Analysis was performed using custom scripts in Rstudio and python.
Supplementary Material
Highlights.
21 novel transcriptional targets of Wnt/β-catenin are identified.
Targets are identified by combining transcriptome analysis as well as genome wide binding of β-catenin.
Most transcriptional targets display similar expression pattern to that of wnt8 during gastrulation.
Acknowledgements
We thank Cameron Exner and Ryan Morrie for editing; Albert Wu for help with cloning; James Hart, Darwin Dichman and Sofia Medina-Ruiz for computational advice.
Funding
This work was supported by a National Institutes of Health grants [GM42341 and HD065705] to R.M.H.
Footnotes
Competing Interests
The authors declare no competing financial interests.
Accession Numbers
The Gene Expression Omnibus accession number for the ChIP-seq and RNA- seq datasets reported in this paper is GSE77365.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anders S, Huber W, 2010. Differential expression analysis for sequence count data. Genome Biol 11, R106. doi:10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders S, Pyl PT, Huber W, 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi:10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aruga J, Mikoshiba K, 2011. Role of BMP, FGF, calcium signaling, and Zic proteins in vertebrate neuroectodermal differentiation. Neurochem. Res 36, 1286–1292. doi:10.1007/s11064-011-0422-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA, 2001. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol 3, 683–686. doi:10.1038/35083081 [DOI] [PubMed] [Google Scholar]
- 5.Blumberg B, Bolado J, Moreno TA, Kintner C, Evans RM, Papalopulu N, 1997. An essential role for retinoid signaling in anteroposterior neural patterning. Development 124, 373–379. [DOI] [PubMed] [Google Scholar]
- 6.Blythe SA, Reid CD, Kessler DS, Klein PS, 2009. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev. Dyn 238, 1422–1432. doi:10.1002/dvdy.21931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brott BK, Sokol SY, 2002. Regulation of Wnt/LRP Signaling by Distinct Domains of Dickkopf Proteins. Mol. Cell. Biol 22, 6100–6110. doi:10.1128/MCB.22.17.6100-6110.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Xu Z, Mei C, Yu D, Small S, 2012. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149, 618–629. doi:10.1016/j.cell.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Pollet N, Niehrs C, Pieler T, 2001. Increased XRALDH2 activity has a posteriorizing effect on the central nervous system of Xenopus embryos. Mech. Dev 101, 91–103. [DOI] [PubMed] [Google Scholar]
- 10.Chiu WT, Charney Le R, Blitz IL, Fish MB, Li Y, Biesinger J, Xie X, Cho KWY, 2014. Genome-wide view of TGF /Foxh1 regulation of the early mesendoderm program. Development 141, 4537–4547. doi:10.1242/dev.107227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christian JL, Moon RT, 1993. Interactions between Xwnt-8 and Spemann organizer signaling pathways generate dorsoventral pattern in the embryonic mesoderm of Xenopus. Genes Dev. 7, 13–28. [DOI] [PubMed] [Google Scholar]
- 12.Clevers H, 2006. Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480. doi:10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 13.Clevers H, van de Wetering M, 1997. TCF/LEF factor earn their wings. Trends Genet. 13, 485–489. [DOI] [PubMed] [Google Scholar]
- 14.Cox WG, Hemmati-Brivanlou A, 1995. Caudalization of neural fate by tissue recombination and bFGF. Development 121, 4349–4358. [DOI] [PubMed] [Google Scholar]
- 15.Elkouby YM, Elias S, Casey ES, Blythe SA, Tsabar N, Klein PS, Root H, Liu KJ, Frank D, 2010. Mesodermal Wnt signaling organizes the neural plate via Meis3. Development 137, 1531–1541. doi:10.1242/dev.044750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnett AT, Square TA, Medeiros DM, 2012. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development 139, 4220–4231. doi:10.1242/dev.081497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentsch GE, Owens NDL, Martin SR, Piccinelli P, Faial T, Trotter MWB, Gilchrist MJ, Smith JC, 2013. In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep 4, 1185–1196. doi:10.1016/j.celrep.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C, 1998. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357–362. doi:10.1038/34848 [DOI] [PubMed] [Google Scholar]
- 19.Goentoro L, Kirschner MW, 2009. Evidence that fold-change, and not absolute level, of beta-catenin dictates Wnt signaling. Mol. Cell 36, 872–884. doi:10.1016/j.molcel.2009.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groves AK, LaBonne C, 2014. Developmental Biology. Dev. Biol 389, 2–12. doi:10.1016/j.ydbio.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harland R, Gerhart J, 1997. Formation and function of Spemann’s organizer. Annu. Rev. Cell Dev. Biol 13, 611–667. doi:10.1146/annurev.cellbio.13.1.611 [DOI] [PubMed] [Google Scholar]
- 22.Harland RM (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695. [DOI] [PubMed] [Google Scholar]
- 23.Harland RM. Dorsoventral Patterning of the Mesoderm In: Stern CD, editor. Gastrulation: from cells to embryo. Cold Spring Harbor, NY: CSHL Press; 2004. p. 373–88. [Google Scholar]
- 24.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, 2010. Simple Combinations of LineageDetermining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589. doi:10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hikasa H, Sokol SY, 2013. Wnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol 5, a007955. doi:10.1101/cshperspect.a007955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussein SM, Duff EK, Sirard C, 2003. Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J. Biol. Chem 278, 48805–48814. doi:10.1074/jbc.M305472200 [DOI] [PubMed] [Google Scholar]
- 27.In der Rieden PMJ, Vilaspasa FL, Durston AJ, 2010. Xwnt8 directly initiates expression of labial Hox genes. Dev. Dyn 239, 126–139. doi:10.1002/dvdy.22020 [DOI] [PubMed] [Google Scholar]
- 28.Janssens S, Denayer T, Deroo T, Van Roy F, Vleminckx K, 2010. Direct control of Hoxd1 and Irx3 expression by Wnt/beta-catenin signaling during anteroposterior patterning of the neural axis in Xenopus. Int. J. Dev. Biol 54, 1435–1442. doi:10.1387/ijdb.092985sj [DOI] [PubMed] [Google Scholar]
- 29.Jen WC, Gawantka V, Pollet N, Niehrs C, Kintner C, 1999. Periodic repression of Notch pathway genes governs the segmentation of Xenopus embryos. Genes Dev. 13, 1486–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jho E-H, Zhang T, Domon C, Joo C-K, Freund J-N, Costantini F, 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol 22, 1172–1183. doi:10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD, 2015. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 43, D756–D763. doi:10.1093/nar/gku956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kengaku M, Okamoto H, 1995. bFGF as a possible morphogen for the anteroposterior axis of the central nervous system in Xenopus. Development 121, 3121–3130. [DOI] [PubMed] [Google Scholar]
- 33.Kim S-E, Huang H, Zhao M, Zhang X, Zhang A, Semonov MV, MacDonald BT, Zhang X, Garcia Abreu J, Peng L, He X, 2013. Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science 340, 867–870. doi:10.1126/science.1232389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kléber M, Lee H-Y, Wurdak H, Buchstaller J, Riccomagno MM, Ittner LM, Suter U, Epstein DJ, Sommer L, 2005. Neural crest stem cell maintenance by combinatorial Wnt and BMP signaling. J. Cell Biol 169, 309–320. doi:10.1083/jcb.200411095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudoh T, Wilson SW, Dawid IB, 2002. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development 129, 4335–4346. [DOI] [PubMed] [Google Scholar]
- 36.LaBonne C, Bronner-Fraser M, 1998. Neural crest induction in Xenopus: evidence for a two-signal model. Development 125, 2403–2414. [DOI] [PubMed] [Google Scholar]
- 37.Lamb TM, Harland RM, 1995. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development 121, 3627–3636. [DOI] [PubMed] [Google Scholar]
- 38.Li B, Kuriyama S, Moreno M, Mayor R, 2009. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development 136, 3267–3278. doi:10.1242/dev.036954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machanick P, Bailey TL, 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. doi:10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C, 2002. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 417, 664–667. doi:10.1038/nature756 [DOI] [PubMed] [Google Scholar]
- 41.Mi H, Muruganujan A, Thomas PD, 2012. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386. doi:10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mii Y, Taira M, 2009. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083–4088. doi:10.1242/dev.032524 [DOI] [PubMed] [Google Scholar]
- 43.Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y, 1998. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development 125, 579–587. [DOI] [PubMed] [Google Scholar]
- 44.Niehrs C, 2004. Regionally specific induction by the Spemann–Mangold organizer. Nat Rev Genet 5, 425–434. doi:10.1038/nrg1347 [DOI] [PubMed] [Google Scholar]
- 45.Nieuwkoop PD and Faber J (1967). Normal Table of Xenopus Laevis (Daudin). New York: Garland Publishing. [Google Scholar]
- 46.Park J-S, Ma W, O’Brien LL, Chung E, Guo J-J, Cheng J-G, Valerius MT, McMahon JA, Wong WH, McMahon AP, 2012. Six2 and Wnt Regulate Self-Renewaland Commitment of Nephron Progenitors through Shared Gene Regulatory Networks. DEVCEL 23, 637–651. doi:10.1016/j.devcel.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pegoraro C, Monsoro-Burq AH, 2012. Signaling and transcriptional regulation in neural crest specification and migration: lessons from xenopus embryos. WIREs Dev Biol 2, 247–259. doi:10.1002/wdev.76 [DOI] [PubMed] [Google Scholar]
- 48.Prinos P, Joseph S, Oh K, Meyer BI, Gruss P, Lohnes D, 2001. Multiple pathways governing Cdx1 expression during murine development. Dev. Biol 239, 257–269. doi:10.1006/dbio.2001.0446 [DOI] [PubMed] [Google Scholar]
- 49.Quinlan AR, Hall IM, 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi:10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramel MC, 2004. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development 131, 3991–4000. doi:10.1242/dev.01277 [DOI] [PubMed] [Google Scholar]
- 51.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, 2011. Integrative genomics viewer. Nature Publishing Group; 29, 24–26. doi:10.1038/nbt0111-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H, 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395, 608–612. doi:10.1038/26989 [DOI] [PubMed] [Google Scholar]
- 53.Schuijers J, Mokry M, Hatzis P, Cuppen E, Clevers H, 2014. Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. EMBO J. 33, 146–156. doi:10.1002/embj.201385358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Semënov MV, Tamai K, Brott BK, Kühl M, Sokol S, He X, 2001. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr. Biol 11, 951–961. [DOI] [PubMed] [Google Scholar]
- 55.Session A, Genome evolution in the allotetraploid frog Xenopus laevis. Nature. submitted -place holder until published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi D-L, Bourdelas A, Umbhauer M, Boucaut J-C, 2002. Zygotic Wnt/β-Catenin Signaling Preferentially Regulates the Expression of Myf5 Gene in the Mesoderm of Xenopus. Dev. Biol 245, 124–135. doi:10.1006/dbio.2002.0633 [DOI] [PubMed] [Google Scholar]
- 57.Sive HL, Grainger RM and Harland RM (2010). Early Development of Xenopus Laevis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 58.Smith WC, Harland RM, 1991. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell 67, 753–765. [DOI] [PubMed] [Google Scholar]
- 59.Tour E, Pillemer G, Gruenbaum Y, Fainsod A, 2002. Gbx2 interacts with Otx2 and patterns the anterior-posterior axis during gastrulation in Xenopus. Mech. Dev 112, 141–151. [DOI] [PubMed] [Google Scholar]
- 60.Trapnell C, Pachter L, Salzberg SL, 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi:10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT, 2005. The Sp1-Related Transcription Factors sp5 and sp5-like Act Downstream of Wnt/β-Catenin Signaling in Mesoderm and Neuroectoderm Patterning. Current Biology 15, 489–500. doi:10.1016/j.cub.2005.01.041 [DOI] [PubMed] [Google Scholar]
- 62.Wills AE, Gupta R, Chuong E, Baker JC, 2014. Chromatin immunoprecipitation and deep sequencing in Xenopus tropicalis and Xenopus laevis. Methods 66, 410–421. doi:10.1016/j.ymeth.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittler L, Shin E-H, Grote P, Kispert A, Beckers A, Gossler A, Werber M, Herrmann BG, 2007. Expression of Msgn1 in the presomitic mesoderm is controlled by synergism of WNT signalling and Tbx6. EMBO Rep. 8, 784–789. doi:10.1038/sj.embor.7401030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP, 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 13, 3185–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT, 1996. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 10, 1443–1454. [DOI] [PubMed] [Google Scholar]
- 66.Young JJ, Kjolby RAS, Kong NR, Monica SD, Harland RM, 2014. Spalt-like 4 promotes posterior neural fates via repression of pou5f3 family members in Xenopus. Development 141, 1683–1693. doi:10.1242/dev.099374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zecca M, Basler K, Struhl G, 1996. Direct and long-range action of a wingless morphogen gradient. Cell 87, 833–844. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Peterson KA, Liu XS, McMahon AP, Ohba S, 2013. Gene Regulatory Networks Mediating Canonical Wnt Signal Directed Control of Pluripotency and Differentiation in Embryo Stem Cells. Stem Cells. doi:10.1002/stem.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS, 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9, R137. doi:10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao T, Gan Q, Stokes A, Lassiter RNT, Wang Y, Chan J, Han JX, Pleasure DE, Epstein JA, Zhou CJ, 2013. -catenin regulates Pax3 and Cdx2 for caudal neural tube closure and elongation. Development 141, 148–157. doi:10.1242/dev.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.