Highlights
-
•
dsRNA production was growth-associated according to Luedeking-Piret model.
-
•
dsRNA/biomass yield was 0.06 g g−1, the same value in bacth and fed-batch culture.
-
•
dsRNA productivity was 37% higher in fed-batch fermentation.
-
•
Production of dsRNA occurred while the bacteria were actively multiplying.
Keywords: dsRNA production, Escherichia coli HT115, Fed-batch, Luedeking-Piret kinetics
Abstract
Double-stranded RNA can induce interference processes. The specificity of this system raises the possibility of using dsRNA for therapeutic applications targeting viral diseases. Escherichia coli HT115 (DE3) has been widely used to produce dsRNA; however, the kinetics of dsRNA production and the relationship between dsRNA and biomass remain unknown. Our aims were to study the kinetics of dsRNA production and to improve dsRNA productivity with fed-batch technology. The results revealed that the production of dsRNA was growth-associated. In batch fermentation, the dsRNA/biomass yield remained close to 0.06 g·g−1, with a maximum productivity of 11.1 mg l−1 h−1 at 10 h of culture. In fed-batch fermentation, the yield was 0.06 g g−1, with a maximum dsRNA productivity of 15.2 mg l−1 h−1 at the end of the feed (12 h). Therefore, to increase the production of dsRNA, it is necessary to enhance the biomass that produces the recombinant nucleic acid.
1. Introduction
Double-stranded RNA (dsRNA) is involved in various biological processes, one of which is RNA interference (RNAi), wherein dsRNA triggers the sequence-specific degradation of messenger RNA [1]. This mechanism silences the expression of specific genes based on homologous sequences [2]. Several new treatments based on RNAi are currently being developed, including treatments directed at controlling viral diseases in intensive animal farming systems such as aquaculture [[3], [4], [5], [6]]. Notably, several of these high-scale treatments use bacteria for dsRNA production. The use of recombinant bacteria to produce dsRNA, as bacterial factories, is an effective strategy because of the ease of handling and the high growth rate of bacteria, in addition to their capacity to retain plasmids [7]. Escherichia coli HT115 (DE3) possesses all of the aforementioned characteristics in addition to containing the T7 polymerase-encoding gene [[8], [9], [10], [11], [12], [13]] and can thus generate dsRNA from dual T7 promoter-containing plasmids. Moreover, this bacterial strain has a mutation in the RNAase III-encoding gene [8,9] that confers characteristics that are suitable for its use as a dsRNA-producing factory.
The use of batch cultures under controlled conditions allows for the study of bacterial growth and dsRNA production kinetics. Some products, such as proteins or acetic acid, are associated with microbial growth. However, other bacterial products, such as xanthan and lactic acid, are formed when bacterial growth decelerates or stops [14,15]. To date, although numerous studies have used E. coli HT115 (DE3) to produce dsRNA, the kinetics of dsRNA production and their relationship to the growth of the bacteria remain unknown. This information would allow for the development of more efficient dsRNA bacterial factories, maximize the productivity of dsRNA and expand the application of this technology. The aims of this work were to study the kinetics of dsRNA production in E. coli HT115 (DE3) and to improve dsRNA productivity in fed-batch fermentation.
2. Materials and methods
2.1. Bacteria strain and media
Escherichia coli HT115 (DE3) RNAase III deficient strain (kindly donated by the Cold Spring Harbor Laboratory, NY, USA) containing the double T7-promoter plasmid L4440 with one 480-bp segment (construct L4440-s3, developed in our laboratory) was used for dsRNA production. The bacteria were grown in batches in flasks with LB broth (10 g tryptone·l−1, 5 g yeast extract·l−1 and 10 g NaCl·l−1) supplemented with 100 μg ampicillin·ml−1 and 12.5 μg tetracycline·ml−1 as selection markers.
2.2. Batch fermentation and dsRNA production
dsRNA production was conducted in a 5 l bioreactor at pH 7, 37 °C, a 300-rpm agitation rate, and 1 VVM aeration for 12 h. To evaluate the growth of E. coli, the OD620nm was recorded every hour during bacterial growth and is expressed as the biomass concentration (g·l−1) based on a linear model. When the culture was in its second hour of growth, the dsRNA production was induced using a 25-mM lactose pulse [16].
2.3. dsRNA extraction and quantification
Total nucleic acid was extracted from the biomass 2 h post-induction (at 4 h of bacterial growth) and every 2 h thereafter as previously described [5,8,9,17] using the one-step protocol of Posiri et al. [13] with modifications. The bacterial pellet was resuspended in 5 ml of 70% v/v ethanol in PBS, incubated at 4 °C for 5 min and collected by centrifugation at 10,000 g for 10 min at 4 °C. Next, the bacterial pellet was resuspended in 1 ml of 150 mM NaCl and incubated at 4 °C for 1 h. The centrifugation was then repeated for 10 min. The supernatant was collected, and the genetic material was allowed to precipitate at −20 °C overnight in absolute ethanol. Afterwards, the supernatant was centrifuged for 30 min at 10,000 g at 4 °C, and the formation of a white pellet was observed [5,13]. The purified dsRNA was quantified as described by García et al. [5].
2.4. Fed-batch fermentation
A fed-batch culture with an exponential feeding flow at a constant rate (0.27 h−1) in a 3 l bioreactor was utilized. Fermentation began with an initial volume of 2 l and ended after 12 h (9 h of feeding) with a volume of 2.8 l. The conditions were the same as those of the batch culture, i.e., LB broth supplemented with 100 μg ampicillin·ml−1 and 12.5 μg tetracycline·ml−1, pH 7, 37 °C, 300 rpm agitation rate, and 1 VVM aeration. At 2 h, the culture was induced with 25 mM lactose, then, at the 3rd hour of culture, a feed of 0.016 l·h−1 began and increased exponentially at a constant rate of 0.27 h−1 to a maximum flow of 0.235 l·h−1 at the end of fermentation. The system was fed with culture media and antibiotics concentrated 2x to maintain a permanent saturation condition in the system. Furthermore, the system included 25 mM lactose to maintain a constant inductor concentration in the reactor.
2.5. Modeling and statistical analysis
All experiments were performed in triplicate. The statistical analyses were performed using one-way ANOVA to compare the dsRNA data in time, and p < 005 was considered statistically significant. The data were analyzed using GraphPad Prism 5.01 (GraphPad Software, San Diego, USA). The fit of the culture data and the modeling were performed using MATLAB (MATLAB R2016a, MathWorks, USA).
3. Results and discussion
3.1. dsRNA production in batch culture
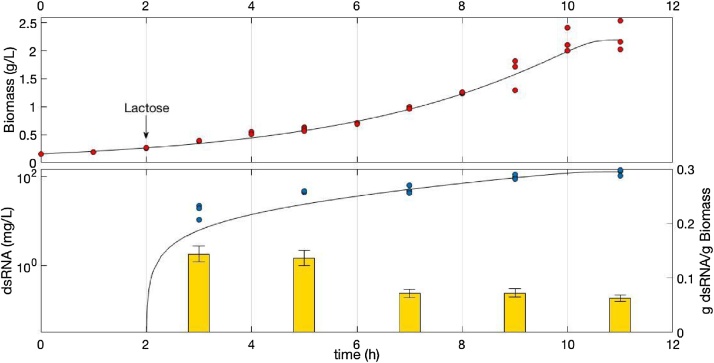
The bacterial growth in the batch culture is illustrated in Fig. 1a. A significant biomass increase was observed at 12 h. A bacterial growth model involving Monod kinetics was used to calculate a specific growth rate of 0.27 ± 0.03 h−1. At the end of fermentation, a final biomass concentration of 2.2 ± 0.2 g·l−1 and a final dsRNA concentration of 110 mg l−1 were observed. The dsRNA was visualized as one band of approximately 500 bp in a 1% w/v agarose gel (Fig. 2). The literature regarding the specific growth rate of E. coli strains grown in the same conditions indicates that this parameter should be higher, i.e., approximately 0.5 h−1 [14,18]. This difference may be explained by the double T7 promoter in the plasmid and the RNAase III deficiency of E. coli HT115, which slow the turnover of nucleic acid within the bacterial cell. Strong expression systems, such as the T7 system, have been described to tend to exceed the metabolic capacity of the bacterial host [19]. Furthermore, the plasmid has been referred to as a molecular parasite; hence, increased replication of the plasmid of interest may cause stress to the host because not only is replication increased but the transcriptional and translational machinery are also challenged and soon run out of metabolic building blocks and energy [18,20].
Fig. 1.
Production of dsRNA in batch culture of E. coli HT115 (DE3) induced with lactose at 2nd hour of fermentation. a Bacterial growth curve in batch culture, the line is a model using Monod kinetics (μmax 0.27 h−1, K 0.1 g·l−1, r2 0.97). b dsRNA production in batch culture, the line is a growth-associated model using Luedeking-Piret kinetics (α 90, β 0, r2 0.96). Columns are dsRNA/biomass yield.
Fig. 2.
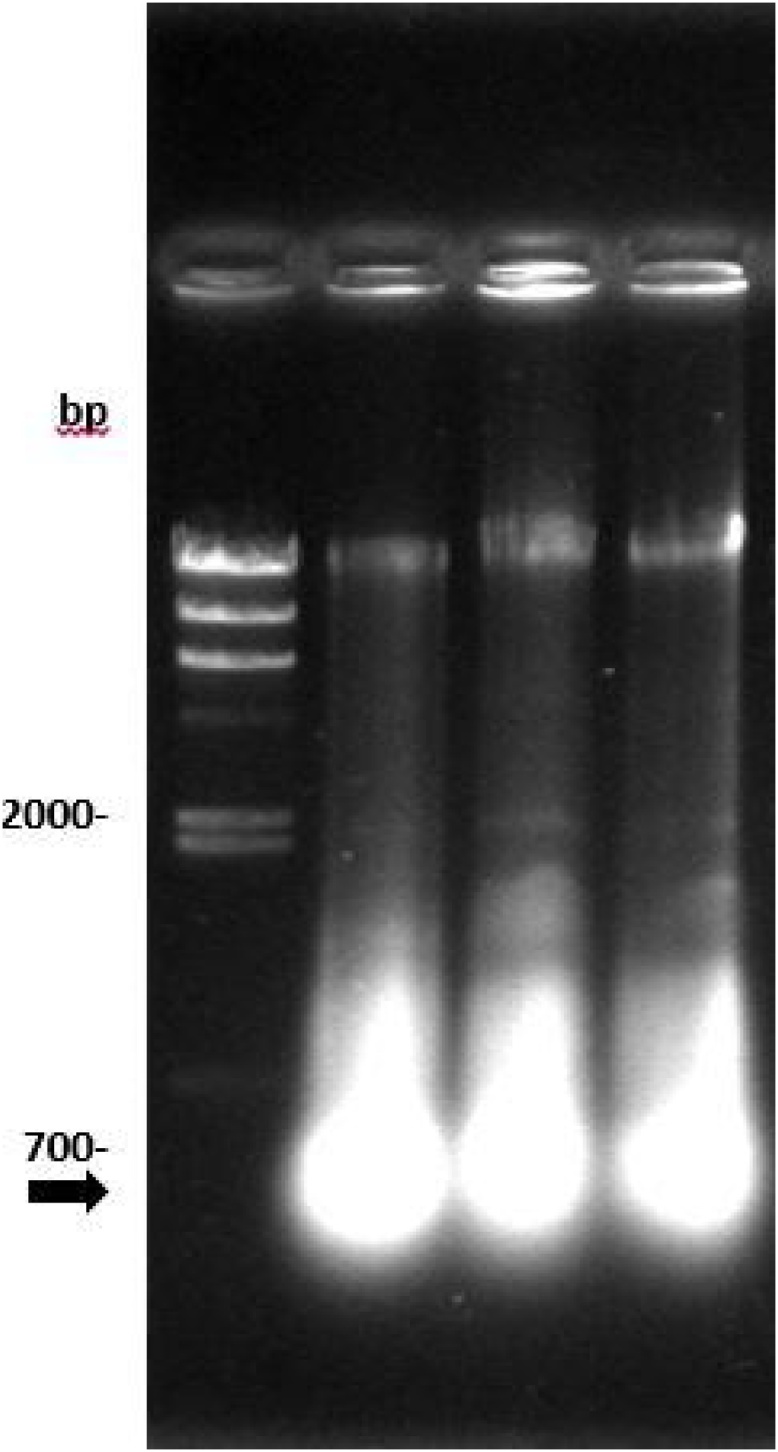
Agarose gel electrophoresis of dsRNA expressed in 12 h batch culture of E. coli HT115 (DE3) induced with lactose. Lane 2, 3 and 4 are triplicates. Arrow indicate dsRNA band that resolved at ∼480 bp.
3.2. Relationship between bacterial growth and dsRNA production
To understand the relationship between dsRNA production and biomass during bacterial growth, we studied the dsRNA production kinetics in batch culture under controlled conditions. To define this kinetics, a dsRNA/biomass yield was estimated as the ratio of the total mass of dsRNA produced and the total mass of bacteria at a certain time of cultivation [14]. This yield allowed for the determination of whether the amount of dsRNA produced by each bacterial cell varied depending on the stage of the microbial growth curve and thus indicated whether the dsRNA production was growth-associated, partially growth-associated, or not growth-associated [15]. The yield results of the batch culture (Fig. 1b) revealed that there were no significant differences in the dsRNA/biomass yields at the different times of fermentation, and the yields exhibited a constant value of approximately 0.06 g·g−1 from 6 to 12 h of fermentation (4 to 10 post-induction) and a maximum productivity of 11.1 mg l−1 h−1 at 10 h of culture (8 h post-induction). Thammasorn et al. [11] recently reported a yield of 1.08 μg dsRNA·1 × 109 CFU−1 with the same strain and culture media. This yield is similar to our results when the equivalent units are used; the calculated Thammasorn’s yield corresponds to 0.02 g·g−1. These results contrast with the results of Posiri et al. [13] in the same conditions because these authors originally reported a yield of 45 μg dsRNA for each 1 OD600 E. coli HT115 (DE3), which can be standardized to 1·10-4 g·g−1, i.e., a 600-fold lower value. This significant difference in dsRNA/biomass yield could be associated with the stress of the host cell as proposed by Grabherr & Bayer [19] and Diaz Ricci & Hernández [18]. Furthermore, Posiri et al. [13] used one strain to host several plasmids to produce different dsRNA, whereas in our protocol and that of Thammasorn et al. [11], each strain harbored one plasmid to produce one dsRNA [11,18,20].
We used a Luedeking-Piret product synthesis model to represent and fit the dsRNA production data in batch culture [14,15]. The dsRNA production model in batch fermentation is described by a mass balance for the dsRNA concentration with respect to time (t):
where x is the biomass concentration, and α and β are the growth-associated, and non-growth associated coefficients, respectively. The fit of the dsRNA concentration data to the model (Fig. 1b) revealed that the dsRNA production was growth-associated, probably because all of the biochemical reactions required for dsRNA production are directly associated with microbial growth [14]. Therefore, the best strategy is a fed-batch culture with exponential feeding, which can maximize the amount of produced biomass and consequently increase the amount of dsRNA. The time at which the culture was induced with lactose was relevant; synthesizing recombinant dsRNA at the beginning of the culture may not be advantageous because dsRNA is sensitive to nucleases and thus will be more susceptible to degradation before the final harvest [10,12].
3.3. Fed-batch fermentation for dsRNA production
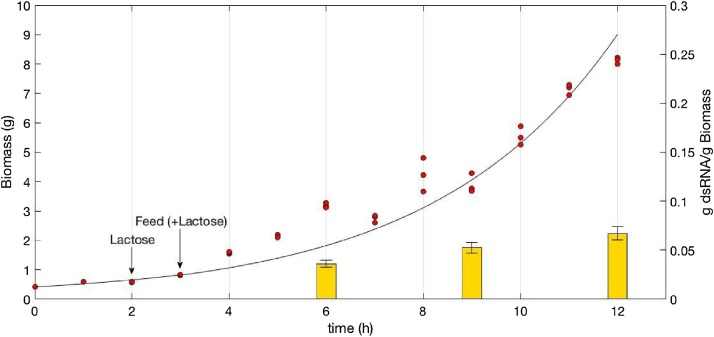
The results of the 12 h fed-batch fermentation (Fig. 3) revealed a final biomass of 8.2 ± 0.5 g (3 g·l−1), a final dsRNA concentration of 182 mg l−1 and a maximum productivity of 15.2 mg l−1 h−1 at the end of fed-batch culture. This productivity was 37% greater than that obtained in the batch culture due to the greater amount of biomass achieved in this fermentation system. Improving productivity is important if the produced dsRNA is to be used on an industrial scale, for example, as an antiviral therapy in intensive animal farming. There were no significant differences in the dsRNA/biomass yields at the different times of culture; the dsRNA/biomass yield was constantly approximately 0.06 g·g−1 as observed over the 4th hour of fermentation, which is the same value that resulted from the batch culture. This observation is coincident with the dsRNA/biomass yield reported by Thammasorn et al. [11], i.e., a batch fermentation value of 0.02 g·g−1 and a fed-batch value of 0.03 g·g−1. Our results for the batch and fed-batch cultures resulted in the same yield value, which reinforces the idea that the production of dsRNA was growth-associated. Indeed, the fed-batch culture, which was fed by an exponentially stream of fresh media, never caused growth limitation, which enabled us to conclude that dsRNA production occurred while the bacteria were actively multiplying.
Fig. 3.
Production of dsRNA in fed-batch culture of E. coli HT115 induced with lactose at 2nd hour of fermentation. Bacterial growth curve in fed-batch with exponential feeding, the line is a model using Monod kinetics (μmax 0.27 h−1, F0 0.0187 l·h−1, r2 0.95). Columns are dsRNA/biomass yield.
4. Conclusions
Our results demonstrate that dsRNA production is growth-associated according to the Luedeking-Piret kinetic model. Thus, to increase the production of dsRNA, it is necessary to maximize the biomass that produces the nucleic acid. The dsRNA production associated with bacterial growth results in an increase in dsRNA productivity of 37% when using a fed-batch fermentation system in the same conditions. Further optimization of the production of dsRNA in fed-batch fermentation systems will allow for the use of this technology in therapeutic applications for viral diseases in biomedicine, veterinary medicine and large-scale production systems, such as aquaculture.
Acknowledgments
This research was funded by Fondecyt1171129 and FONDEFDI17I10185 from CONICYT.
References
- 1.Plasterk R.H. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 2.Leung R., Whittaker P. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol. Ther. 2005;107:222–239. doi: 10.1016/j.pharmthera.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papić L., García K., Romero J. Progress and limitations of dsRNA strategies in the control of viral diseases in aquaculture. Lat. Am. J. Aquat. Res. 2015;43(3):388–401. [Google Scholar]
- 4.Reshi M., Wu J., Wang H., Hong J. RNA interference technology used for the study of aquatic virus infections. Fish Shellfish Immunol. 2014;40:14–23. doi: 10.1016/j.fsi.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 5.García K., Ramirez-Araya S., Díaz A., Reyes-Cerpa S., Espejo R., Higuera G., Romero J. Inactivated E. coli transformed with plasmids that produce dsRNA against infectious salmon anemia virus hemaglutinin show antiviral activity when added to infected ASK cells. Front. Microbiol. 2015;6:300. doi: 10.3389/fmicb.2015.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poynter S.J., DeWitte-Orr S.J. Length-dependent innate antiviral effects of double-stranded RNA in the rainbow trout (Oncorhynchus mykiss) cell line, RTG-2. Fish Shellfish Immunol. 2015;46(2):557–565. doi: 10.1016/j.fsi.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotech. 2006;72(2):211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 8.Timmons L., Court D., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 9.Ongvarraspone C., Roshorm Y., Panyim S. A simple and cost effective method to generate dsRNA for RNAi studies in invertebrates. Sci. Asia. 2007;33:35–39. [Google Scholar]
- 10.Yin G., Sun Z., Liu N., Zhang L., Song Y., Zhu C., Wen F. Production of double-stranded RNA for interference with TMV infection utilizing a bacterial prokaryotic expression system. Appl. Microbiol. Biotechnol. 2009;84:323. doi: 10.1007/s00253-009-1967-y. [DOI] [PubMed] [Google Scholar]
- 11.Thammasorn T., Sangsuriya P., Meemetta W., Senapin S., Jitrakorn S., Rattanarojpong T., Saksmerprome V. Large-scale production and antiviral efficacy of multi-target double-stranded RNA for the prevention of white spot syndrome virus (WSSV) in shrimp. BMC Biotechnol. 2015;15:110. doi: 10.1186/s12896-015-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenllado F., Martínez-García B., Vargas M., Díaz-Ruíz J. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 2003;3 doi: 10.1186/1472-6750-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posiri P., Ongvarrasopone C., Panyim S. A simple one-step method for producing dsRNA from E. coli to inhibit shrimp virus replication. J Virol Meth. 2013;188(1-2):64–69. doi: 10.1016/j.jviromet.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 14.Bailey J.E., Ollis D.F. McGaw-Hill; New York: 1986. Biochemical Engineering Fundamentals. [Google Scholar]
- 15.Garnier A., Gaillet B. Analytical solution of Luedeking-Piret equation for batch fermentation obeying Monod growth kinetics. Biotechnol. Bioeng. 2015;112(12):2468–2474. doi: 10.1002/bit.25669. [DOI] [PubMed] [Google Scholar]
- 16.Viitanen M., Vasala A., Neubauer P., Alatossava T. Cheese whey-induced high cell density production of recombinant proteins in Escherichia coli. Microb. Cell Fact. 2003;2(1):1. doi: 10.1186/1475-2859-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solis C.F., Santi-Rocca J., Perdomo D., Weber C., Guillén N. Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression. PLoS One. 2009;4(12):e8424. doi: 10.1371/journal.pone.0008424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz Ricci J.C., Hernández M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000;20(2):79–108. doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- 19.Grabherr R., Bayer K. Impact of targeted vector design on ColE1 plasmid replication. Trends Biotechnol. 2002;20(6):257–260. doi: 10.1016/s0167-7799(02)01950-9. [DOI] [PubMed] [Google Scholar]
- 20.Patnaik P.R. An evaluation of models for the effect of plasmid copy number on bacterial growth rate. Biotechnol. Lett. 2000;22(1):1719–1725. [Google Scholar]