Abstract
The analysis of animal models of neurological disease has been instrumental in furthering our understanding of neurodevelopment and brain diseases. However, animal models are limited in revealing some of the most fundamental aspects of development, genetics, pathology, and disease mechanisms that are unique to humans. These shortcomings are exaggerated in disorders that affect the brain, where the most significant differences between humans and animal models exist, and could underscore failures in targeted therapeutic interventions in affected individuals. Human pluripotent stem cells have emerged as a much-needed model system for investigating human-specific biology and disease mechanisms. However, questions remain regarding whether these cell-culture-based models are sufficient or even necessary. In this review, we summarize human-specific features of neurodevelopment and the most common neurodevelopmental disorders, present discrepancies between animal models and human diseases, demonstrate how human stem cell models can provide meaningful information, and discuss the challenges that exist in our pursuit to understand distinctively human aspects of neurodevelopment and brain disease. This information argues for a more thoughtful approach to disease modeling through consideration of the valuable features and limitations of each model system, be they human or animal, to mimic disease characteristics.
Keywords: modeling, human, species, pluripotent stem cells, cerebral cortex, evolution, neural, brain, intellectual disability, developmental disorders
Main Text
Introduction
Animal models, particularly rodent models, are essential for determining the molecular mechanisms of gene regulation and have been instrumental in our understanding of neurodevelopment and disorders. However, the advancement of imaging, genomic, and genetic technologies has increasingly revealed human-specific aspects of neural development, genetics, pathology, and disease mechanisms. Fundamental differences between humans and animal models raise questions about the value of information gained primarily from animal models, particularly widely used rodents. In addition, the failure of clinical trials that have been based on animal studies has further highlighted the limitations of animal-model-focused human disease research.
Analysis of human-specific characteristics of the brain is hindered by the difficulty in acquiring developing and diseased human brain tissue. Human pluripotent human stem cells (hPSCs) provide an important alternative for studying the development and function of brain cells. To facilitate brain disease modeling, human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) provide paradigms for defining human-specific biology and identifying disease mechanisms. However, questions remain about the utility and value of these cell-culture-based models and how best to move the technology forward. In this review, we summarize human-specific features of neurodevelopment and neurodevelopmental diseases, present the gaps between animal models and human diseases, demonstrate how human stem cell models can bridge some of these gaps, and discuss the challenges for further improvement.
Unique Characteristics of Human Brian Development
Conserved Characteristics among Humans and Animal Models
The neocortex in mammals is involved in higher cognitive functions, such as sensory perception, generation of motor commands, spatial reasoning, conscious thought, and in humans, language. The higher-order functions that distinguish humans from rodents are accomplished through complex and evolutionarily emergent differences in cortical expansion, arealization, and connectivity. Yet, specific fundamental aspects of gross neuroanatomy, neuronal function, and principles of initial neocortical development are similar in all mammals, and thus our understanding of these aspects of brain development come largely from studying animal models.
The neuroepithelium of the neural tube is the origin of the entire central nervous system. Cell genesis during cortical development follows an intrinsic time sequence, which begins with the development of neurons, followed by astrocytes and then oligodendrocytes. The cortex forms in an “inside-out” manner whereby deep layers (layers 4–6) form before upper layers (layers 1–3).1, 2, 3 This sequence of events is largely conserved in animals.4
Two major classes of neurons populate the mammalian cortex: excitatory and inhibitory neurons. Complex neuronal information processing depends on precise spatial and temporal coordination of the activity of principal excitatory neurons that constitute the majority of neurons in the brain. Such coordination is provided by network oscillations that synchronize activity within local cortical circuits. Inhibitory interneurons, although constituting less than 20% of cortical neurons, form extensive local connections, such that each interneuron innervates thousands of principal neurons, enabling them to exert decisive forms of control over large neuronal assemblies. The interactions between excitatory and inhibitory neurons are critical in generating and regulating network oscillations, synchronizing the activity of principal neurons, and setting time windows for synaptic integration. Thus, these interactions drive circuit activity5, 6 and switch networks between different states of activities.7, 8 The balance between the actions of excitatory and inhibitory neurons is critical in all mammals in that it enables information on the mechanisms of neuronal function gained from rodents to be applied to humans.
Proper brain functioning relies on the correct number and placement of excitatory and inhibitory neurons. This is a remarkably complex process because each type of neuron has a different developmental origin. Excitatory glutamatergic projection neurons arise from progenitor cells in the dorsal ventricular zone; these migrate to their proper cortical layer destinations via radial migration.2, 9, 10, 11, 12 In contrast, inhibitory GABAergic interneurons develop from progenitors in the ganglionic eminences of the ventral forebrain and migrate first radially in the ventricular zone and then tangentially to the cortex.13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 Correct formation, and thus function, of the cortex relies on synchronization of these two developmental programs. Comparison of human fetal brain tissue with information gleaned from animal models suggests that basic steps in excitatory and inhibitory neuron development are similar in mammals.28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40 These results imply that mechanisms of cortical neuron development in rodents are relevant to humans.
These specific shared aspects of neocortical development in humans and mammals suggest that knowledge gained through animal modeling can be attributed to human development. Yet, emerging evidence makes it clear that the development of the human neocortex includes additional mechanisms to form the highly complex structure that can carry out higher-order functions, and it is in these higher-order functions from which disorders of intellectual disability often arise.
Human-Specific Characteristics of Brain Development
Despite the similarity between humans and rodents, there are a number of human-specific characteristics of neocortical development, including cortical expansion, protracted time of development, and genetics. A consequence of these differences between humans and rodent models is that not all aspects of human neurological diseases can be recapitulated in animal models, leading to misinterpretation of data and potentially failed clinical trials.
Cortical Expansion. The most obvious difference between rodent and human brains is the much larger size of the human cortex. The evolutionary expansion of the cerebral cortex is reliant on increased cell numbers, which is in turn dependent on an increase in the numbers of neural progenitors.41, 42, 43, 44, 45, 46 Specific cellular and molecular characteristics of cortical neural progenitors that are unique to primates drive this expansion. Neural progenitor cells, specifically radial glia that give rise to neurons, undergo more symmetric divisions in primates, enabling the generation of more neural progenitors. Further, these neurogenic radial glia (oRGs) translocate to an additional outer subventricular zone (oSVZ), where they undergo mitosis to provide vastly more proliferative neuronal progenitors that, in turn, differentiate into more neurons.26, 27, 47, 48, 49 Although oRGs have been found in rodents, they are rare in number, and their contribution to rodent brain development remains unclear.
Molecular mechanisms that regulate the expansion of the human cortex have been gleaned primarily from global transcriptomic analyses of the developing human brain. A human-specific gene, ARHGAP11B (MIM: 616310), encoding Rho GTPase activating protein 11B, promotes basal progenitor amplification and neocortex expansion and might have contributed to the evolutionary expansion of the human neocortex.50, 51 Other suggested molecular regulators include non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), that have undergone dynamic evolutionary changes.52, 53, 54, 55 Further, DNA methylation is dynamic during human brain development.56 Thus, the ability to accurately model the expansion of the human cortex requires recapitulation of these critical molecular regulators, and so accurate modeling of cortical expansion is not possible in rodents.
Protracted Time of Development. The development of the human brain occurs over a prolonged time course in comparison with that of animal models.4 The gestational period is 40 weeks in humans and 3 weeks in rodents. The bulk of human brain formation occurs prenatally. The neural tube is completely formed by embryonic day 27 (gestational age of 6 weeks) in humans and embryonic day 10 in mice. Human brains thus have 34 weeks, versus 11 days in mice, to develop before birth. The major period of generation and migration of cortical neurons in humans occurs from 10 to 25 weeks of gestation.2, 43, 55 Cortical neurons generated in the ventricular zone, primarily in the oSVZ, reach their destination in the cortical plate by both radial and non-radial migration.25, 57, 58, 59 Newly born neurons are added to the cortical plate in an “inside-out” fashion and differentiate to form the cell-specific layers of the cortex.60, 61 Although most neuroblasts have been generated by the 16th gestational week, many neurons have not yet reached their final destination.2 Furthermore, medium and small neurons, as well as glia, can continue to be produced after 16 weeks of gestation,2 and neurogenesis continues at low levels after birth into old age.62 Glial genesis continues throughout infancy and early childhood.55, 63, 64 Infancy is also characterized by the formation of synaptic connections, including outgrowth of dendrites and axons. Structural and molecular reorganization of cells and synapses continues into early adolescence.43, 55 This long-drawn-out timing of neurodevelopment provides opportunities for mechanisms to increase complexity in the human brain, and this is difficult to model in rodents.
Increased Complexity. Although the functions of excitatory and inhibitory cortical neurons are conserved, interneurons in humans have distinct features. Recent evidence indicates that the human cortex contains more diverse types of interneurons and that humans have greater reliance on interneurons than other mammals.32 Interneuron progenitors arise from the ventral forebrain in the ganglionic eminences (GEs), although there is dispute about whether interneurons in humans also arise from the dorsal forebrain.65, 66, 67, 68, 69, 70, 71 The medial GE is the primary source of interneurons in mice, and its development is well characterized. The medial GE gives rise to 70% of the total cortical interneurons in mice, but recent data suggest a greater contribution from caudal GE progenitors in primates.72, 73 Human cortical neuron development integrates unique mechanisms, including prolonged neurogenesis and migration,74, 75 to generate interneurons.27, 32, 47, 76, 77, 78 Thus, developmental mechanisms that regulate interneuron development in primates are more complex and could lead to the generation of different classes and/or proportions of interneuron subtypes than in rodents.79 In fact, a new human interneuron subtype was recently identified with features never described in rodents.80
Unique Genetics. Comparisons of DNA between human and animal models reveal only subtle differences in protein-coding regions. However, transcribed non-protein-coding sequences, including many species of non-coding RNA, are significantly less conserved than protein-coding mRNAs.44, 63, 81, 82, 83 Thus, differences in regulatory elements, non-coding genes, and limited human-specific genes, as well as human-specific gene removal, can contribute significantly to phenotypic differences between human and animal brains.
An increase in comparative genomic analyses between the human or primate brain and the rodent brain in the last 10 years has led to reports of different expression patterns of the same genes in humans and mice.44, 55, 81, 82, 84, 85 Some of these differences can be accounted for by chromosome-level regulation, given the divergent chromosomal organization of human and mouse genes.86 Differences in chromosomal architecture can impede the ability to genetically model disorders that are due to large changes in chromosomes, including aneuploidies (e.g., trisomy 21, also known as down syndrome [DS (MIM: 190685)]) and duplication syndromes (e.g., chromosome 15q11–q13 duplication syndrome [MIM: 608636]). Several examples of divergent regulation of the same genes have recently emerged.87, 88 A number of studies have identified human accelerated regions (HARs): short, evolutionarily conserved DNA sequences that have acquired significantly more DNA substitutions than expected in the human lineage since our divergence from chimpanzees.89, 90 Many of the HARs, such as GLI2 (GLI family zinc finger 2 [MIM: 165230]) and NPAS3 (neuronal PAS domain protein 3 [MIM: 609430]), are enhancers near genes and have important functions in development. For example, Prabhakar et al. identified cis-regulatory elements that have undergone human-specific accelerated evolution, and one of them, human-accelerated conserved non-coding sequence 1 (HACNS1), leads to enhanced gene expression in limbs and potentially affects digit development and dexterity.91, 92 The impact of HARs in human neural development is currently unknown. In addition, recent evidence has revealed uniquely human enhancers that might drive human cortical neurogenesis.93, 94 Johnson et al. surveyed the transcriptome of 13 brain regions, including prefrontal cortical regions, of mid-gestation human brains and discovered genes differentially expressed and differentially spliced within subregions of the neocortex.44 Interestingly, a significant number of differentially expressed genes, including forkhead box P2 (FOXP2 [MIM: 605317]), are involved in speech and language and have undergone human-specific accelerated evolution.44 Besides genomic regulatory elements, differences in untranslated regions (UTRs) of mRNAs can lead to differential gene regulation. In the developing human cortex, nitric oxide synthase 1 (NOS1 [MIM: 163731]) mRNA is bound by fragile X mental retardation protein (FMRP), and its protein translation is regulated by FMRP. However, NOS1 mRNAs in rodent and other non-primate mammals lack a G-quartet motif for FMRP binding, and so such regulation is absent.44, 84, 95 The authors hypothesize that such differential post-transcriptional regulation could be responsible for reduced amounts of NOS1 in individuals with human fragile X syndrome (FXS [MIM: 300624]); such differential regulation is not found in FXS mouse models. Relevant to neuron function and neural disorders, Qiu et al. compared activity-dependent gene expression in neurons differentiated from either human or mouse ESCs and found that basal neuronal gene expression is less divergent than depolarization-triggered gene expression.88 For example, human ETS proto-oncogene 2 (ETS2 [MIM:164740]) is induced more strongly and rapidly than mouse Ets2. This difference could be due to the presence of a human-specific AP-1 transcription factor binding site in the human ETS2 promoter but not in the mouse Ets2 promoter. The differential regulation and expression of this gene is particularly relevant to DS because ETS2 is encoded on human chromosome 21, and so its expression and downstream consequences could be very different between humans and mouse models. Thus, an increasing number of examples of divergent gene expression between rodents and humans could lead to differences in function and relevance to disease.
Genome-wide approaches have identified a small number of human-specific genes that are likely to be important for neurodevelopment.50 Some of these human-specific genes result from alternative splicing that is highly enriched in developing human brains.44 For example, Kallikrein-related peptidase 8 (KLK8, also known as neuropsin [MIM: 605644]) encodes a secreted-type serine protease that is involved in synaptogenesis, neurite outgrowth, and plasticity in the hippocampus and neocortex. KLK8 is involved in learning and memory and has a long spliced form that is expressed only in humans (not in non-human primates) and produces human-specific type II KLK8.96 Other human-specific genes could be products of human-specific duplications or variants of otherwise evolutionarily conserved genes. ARHGAP11B is a human-specific duplication of ARHGAP11A (MIM: 610589), is highly expressed in radial glia, and promotes cell proliferation and cortical expansion.50 SLIT-ROBO Rho GTPase activating protein 2 (SRGAP2 [MIM: 606524]) has two human-specific duplications, SRGAP2B and SRGAP2C. It has been shown that SRGAP2C inhibits SRGAP2, leading to increased dendritic spine density.97 In addition to duplications, human-specific gene removal (deletion) also occurs. One example is an Alu-mediated frameshift mutation of the gene encoding the enzyme cytidine monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) hydroxylase (CMAH, named CMAHP for the pseudogene in humans [MIM: 603209]). Whereas other mammals, including our closest relative (chimpanzee), express functional CMAH to convert Neu5Ac to Neu5Gc, humans do not, leading to an accumulation of amino acid Neu5Ac in the human brain.98 The functional impact of such gene loss is unclear, and at least one study has shown that a lack of Neu5Gc prevents malaria parasite infection.99 Other examples of gene loss, although not human specific, could also contribute to differences between humans and animal models. For example, primates (including humans) and guinea pigs have gained mutations in the gene encoding gulonolactone oxidase (GULOP), which produces ascorbic acid (vitamin C).100 Humans and some primates have lost the enzymatic activity of urate oxidase (UOX), which catalyzes the oxidation of uric acid to allantoin during primate evolution, thus predisposing us to conditions including gouty arthritis and renal stones.101 How human-specific gene addition or removal contributes to the differences observed between rodent and human neurodevelopment remains unclear. These studies further bolster the idea that studying animal models alone will yield incomplete information about human neural development and function.
Even evolutionarily conserved genes can have different functions in humans and animal models. One example is FOXP2, the only gene that has been firmly linked to speech and language development in humans through gene identification of individuals with language disorders (MIM: 602081).102, 103 FOXP2 has gone through accelerated evolution in humans. FOXP2 encodes a transcription factor and is among the top 5% of highly conserved genes among mammals, such that only three amino acids differ between the human and mouse proteins, and only two amino acids differ between the human protein and those of other primates.104 In mice, FOXP2 has been found to regulate lung development, and Foxp2-null mice die before adulthood as a result of severe motor and respiratory defects.105 However, mice with FOXP2 variants that cause language disorders in humans do not exhibit apparent vocalization deficits, suggesting differential functions of FOXP2 between humans and mice.106, 107, 108, 109 In fact, chromatin immunoprecipitation analysis demonstrates that human FOXP2 has distinct transcriptional targets compared with those of its counterparts in rodents and chimpanzees.110 Therefore, the two amino acid substitutions in FOXP2 could affect its specificity for transcriptional targets that are uniquely important for human language development. Recapitulation of specific human gene mutations does not, therefore, predictably recapitulate gene function in animal models.
A large portion of both human and mouse genomes is transcribed into non-protein-coding RNAs. A comparison of orthologous human and mouse gene pairs has revealed that the sequence, number, and length of exons are highly conserved, but both the sequence and the length of introns vary significantly between species (e.g., human genes have larger introns).111 The functional significance of this divergence in introns remains unclear, but non-coding RNA genes are sometimes embedded within introns.112 In fact, both the human and mouse genomes encode a large number of non-coding RNAs that include both lncRNAs that are over 200 bases long112, 113 and small RNAs that are 20–30 nucleotides in length.114 Unlike protein-coding genes, lncRNAs are less conserved. Approximately one-third of lncRNAs are primate specific.115 Also, a number of conserved miRNAs seem to play similar functions in both humans and rodents. For example, both MIR9-1 (MIM: 611186) and MIR124-1 (MIM: 609327) are enriched in neurons and promote neural stem cell (NSC) differentiation into neurons in multiple species.116 On the other hand, human-specific miR-941 is highly expressed in hPSCs, and its expression decreases upon differentiation.117 The functional importance of this miRNA in human-specific traits remains unclear, but the fact that it targets hedgehog- and insulin-signaling-pathway genes that are important for brain development suggests that it could have a critical function in neural development. As with protein-coding genes, the impact of human-specific RNA species on neural development and function remains unclear but expands the known differences between humans and mice.
Together, expansion and complexity of the human cortex, at both the cellular and genomic levels, most likely lead to human-specific aspects of disease when they go awry. Thus, some aspects of human neurological diseases cannot be recapitulated in animal models.
Human-Specific Genetic Characteristics of Neurodevelopmental Disorders
Mistakes in any of the unique aspects of human brain development summarized above lead to neurodevelopmental disorders that generally result in intellectual impairment. Two of the most prevalent genetic causes of intellectual disability in humans are FXS and DS. We will use these two disorders as examples to discuss human-specific traits of neurodevelopmental disorders and the ability to investigate their causes through model systems.
Unique Genetics
Although both FXS and DS are categorized as genetic neurodevelopmental disorders, they represent two ends of the spectrum in terms of genetic mechanisms: single-gene mutation (Figure 1) versus whole-chromosome duplication (Figure 2).
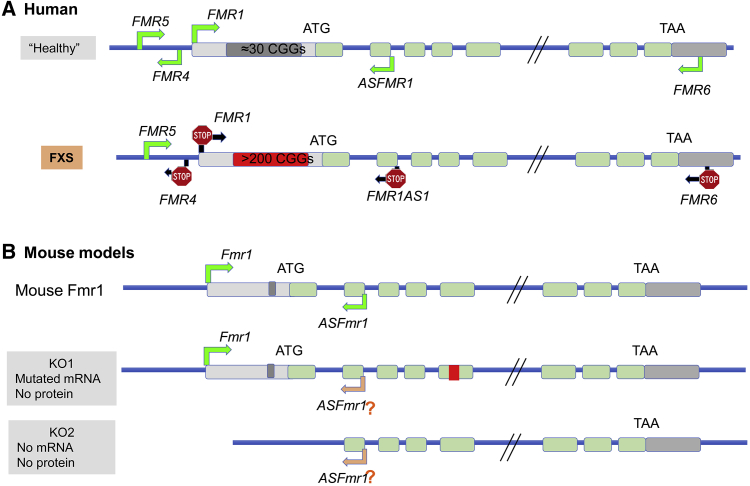
Figure 1.
The Genetics of Human FXS versus Mouse Models of FXS
(A) The human FMR1 locus contains several primate-specific lnRNAs, FMR4, FMR5, and FMR6. ASFMR1 exists in both humans and mice. In FXS-affected individuals with a full mutation, most of these lncRNAs are also silenced.
(B) Mouse models of FXS have been created through the insertion of a neomycin cassette into exon 5 (KO1) or the deletion of the promoter and exon 1 (KO2), leading to a lack of protein production in both KO1 and KO2. ASFmr1 expression is unknown in mouse models.
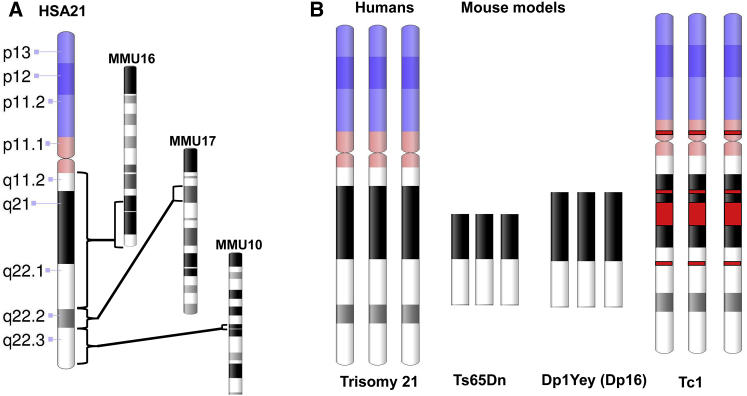
Figure 2.
Comparison of Human Chromosome 21 and Mouse Models
(A) Genes in human chromosome 21 have orthologous regions on three mouse chromosomes.
(B) Schematic of chromosome 21 genes that are represented in three mouse models: Ts65Dn (the most widely studied mouse model), Dp16, and Tc1 (a transchromosomic mouse line).118, 119, 120
The causal mutation in FXS affects a single gene, FMR1 (MIM: 309550; Xq27.3), whose protein-coding sequence is highly conserved among species: the murine homolog is 97% homologous to the human amino acid sequence.121, 122 However, human FMR1 has a large CGG repeat region that is absent in mouse Fmr1. The expansion of this CGG repeat in the 5′ UTR of human FMR1 leads to a chromosomal fragile site on the X chromosome123 and epigenetic silencing of FMR1, resulting in FXS. The number of CGG repeats in human FMR1 is highly polymorphic (the mode peak is 30 or 31), and more than 90% of the human population has fewer than 40 CGG repeats.123, 124 When the CGG repeat number exceeds 200, the CGGs become methylated and FMR1 is transcriptionally silenced. Interestingly, when CGG repeats are expanded to 55–200, FMR1 transcription is enhanced and leads to a pathological condition called fragile X pre-mutation. Individuals with pre-mutation have an increased risk of developing fragile-X-related primary ovarian insufficiency (FXPOI) and fragile-X-associated tremor and ataxia syndrome (FXTAS) in older age.125 In addition, the CGG repeats in individuals with pre-mutation are unstable and tend to expand further during meiosis in the germline, leading to increased risk of FXS as a result of repeat expansion in the next generation. Despite extensive efforts, it remains unclear how the variable CGG-repeat length in FMR1 affects FMR1 transcription and why having more than 200 repeats leads to DNA methylation and gene shutdown.
Mouse Fmr1 has very few (about six) CGG repeats at the 5′ UTR, and no CGG expansion has been observed in any experimental mouse lines, thus limiting the utility of the mouse for studying the mechanisms of FMR1 shutdown. Several mouse models of fragile X pre-mutation have been created through the insertion of human CGG repeat (∼100) sequences into the 5′ regions of mouse Fmr1. Although these mice exhibit some deficits found in human individuals with pre-mutation, such as reduced cognitive ability, increased oxidative stress, RNA-induced toxicity, and instability of repeats, the CGG repeats do not expand into full mutation in the germline as they do in humans.126 Therefore, mouse FXS models rely on Fmr1 deletion, a completely different mechanism than the human mutation.
The protein product of FMR1 and Fmr1, FMRP, is an RNA-binding protein that binds to specific mRNAs to control their stability, localization, and protein translation. The function of FMRP in human neuronal development and how a lack of FMRP causes the characteristics in FXS-affected individuals are largely unknown. FMRP’s targets and function might differ between humans and rodents, leading to different phenotypes when the protein is not expressed. An example has been demonstrated by FMRP binding and translational regulation of human NOS1 mRNA,95 but not of mouse Nos1 mRNA. However, a comprehensive identification of FMRP targets in human neurons has not yet been realized, defining a large gap in our knowledge of the similarities and differences of FMRP function between mice and humans.
Importantly, the human FMR1 locus also produces several human-specific lncRNAs. Among them, FMR4 (MIM: unavailable), FMR1AS1 (MIM: 300805), and FMR6, but not FMR5, are silenced in FXS.127 The functions of these lncRNAs are largely unclear, although FMR4 has been shown to promote neural progenitor proliferation.128 Therefore, it is currently unknown whether human FXS pathology is a result of the deletion of FMRP alone or the silencing of several coding and non-coding genes. Answering these questions will require human experimental models that have the full repertoire of human FMR1 components.
DS (also known as trisomy 21) is a complex multigene disorder and the most common viable human aneuploidy.129 Neurological consequences include intellectual disability and early-onset Alzheimer disease (AD). The complexity of an additional chromosome provides a daunting problem for deciphering the underlying mechanisms, especially genotype-phenotype investigations. Further, the variability of characteristics among DS-affected individuals suggests that genetic, environmental, and stochastic influences are likely to play a role in defining the characteristics of DS.130, 131, 132
Chromosome 21 (HSA21), comprising an estimated 225–400 genes, is the smallest human chromosome,133, 134 which could explain why trisomy of this chromosome and not others (e.g., 13 and 18) is compatible with life and results in only mild to moderate intellectual disability. The short arm (p) of HSA21 is very small and highly repetitive, and its loss has only rarely been reported to result in significant disease phenotypes. Well-studied genes on the long arm (q) of HSA21 include genes that encode cell-adhesion molecules (e.g., DS cell-adhesion molecule [DSCAM (MIM: 602523)]), signaling molecules and kinases (e.g., dual-specificity tyrosine phosphorylation-regulated kinase 1A [DYRK1A (MIM: 600855)]), and transcription factors (e.g., ETS2 [MIM: 164740]). Several genes on chromosome 21 have been implicated in DS and other disorders: familial AD (amyloid beta precursor protein (APP [MIM: 104760]), familial amyotrophic lateral sclerosis (superoxide dismutase 1 [SOD1 (MIM: 147450)]), and a predisposition for leukemia (runt-related transcription factor 1 [RUNX1 (MIM: 151385)] and GATA binding protein 1 [GATA1 (MIM: 305371)]). Using rare partial trisomies, human studies have tried to pinpoint the critical or smallest HSA21 region whose triplication might confer specific attributes of DS.135, 136, 137, 138, 139, 140 However, some phenotypic characteristics might map outside the minimum critical region, and it is likely that the combination of genes and their downstream effects on other genes result in the characteristics of DS.
Genes on HSA21 are encoded across three mouse chromosomes, and mouse DS models that contain various combinations of orthologous HSA21 genes have been generated118, 119, 141, 142, 143, 144, 145, 146 (Figure 2). Trisomy 16 mice were originally the most complete HSA21 trisomy model,147, 148, 149 but their utility is limited because fetuses do not survive as live-born animals, and so analysis is limited to prenatal development. In the most widely used mouse model, Ts65Dn,150 a large trisomic segment of mouse chromosome 16 contains most regions orthologous to HSA21. Mouse models Ts1Cje and Ts2Cje contain smaller regions.143, 144 The mouse strain Dp1Yey (also known as Dp16) contains the largest number of orthologous chromosome 16 genes. In addition, a “transchromosomic” mouse model that contains most of HSA21 has been developed.120 Although these varied mouse models of DS contain substantial portions of HSA21 gene content, these genes lack the same regulation as human genes. As discussed above, the regulation of genes and their expression can vary in a human context. Thus, none of the mouse models are trisomic for all HSA21 orthologs, some have additional triplicated mouse genes, and none have human-specific gene regulation, so the full impact of human trisomy 21 has not been realized in mouse models. Although mouse models have provided a wealth of information about DS, it is crucial to examine the effects of the full trisomy 21 in the context of human cells, where human-relevant genetics and development come into play.
Success and Limitations of Animal Models for Neurodevelopmental Disorders
FXS- and DS-affected individuals exhibit distinct yet overlapping phenotypes. FXS-affected individuals have mild to severe cognitive impairment, increased incidence of childhood seizures, reduced motor coordination, and heightened anxiety. DS-affected individuals also have cognitive impairment (although it is often milder and includes deficits in different domains) and motor dysfunction characterized by hypotonia and a small seizure risk. Language, a human trait, is affected in both syndromes. These characteristics of both FXS and DS are due to affected higher-order functions that are largely mediated by the cerebral cortex and are unique to humans. Thus, models of these disorders must account for the varied human-specific aspects of cortical development, cortical expansion, human genetics, and genetic regulation.
Modeling Neurological Symptoms and Behavioral Deficits
Neurodevelopmental disorders encompass a broad spectrum of phenotypes with potential contributions from a large number of genes.151, 152, 153 Although mice and humans are 80 million years apart in evolution, most protein-coding genes are highly conserved; therefore, mice have been used extensively for modeling human diseases. Behavioral tests of rodents have been used for modeling human deficits with both successes and limitations.142, 154, 155, 156, 157 Symptoms of social impairment can be evaluated through interactions, preference, and recognition of other mice. The communication between mice can be assessed by their responses to vocalizations or olfactory cues and their interactions. Repetitive behavior and resistance to change in routine can be observed in stereotypical behavior or in reversal learning tasks. Cognition is standardly evaluated in rodent models of neurological disorders via the assessment of memory and learning through novel-object recognition, the Morris water maze, fear conditioning, and passive avoidance tests. These tests can define memory differences due to specific disease-mutation modeling but still cannot completely model human-specific aspects of cognition, including language and conscience. The hope is that these measurements can be used for assessing specific gene contributions to neurodevelopmental disorders and evaluating potential drug candidates in a pre-clinical model system.
Fragile X Syndrome. Animal models of FXS have been developed in Drosophila, zebrafish, mice, and rats. Extensive behavioral modeling has been done primarily with mouse models of FXS.158 In the most widely used mouse model (Fmr1 knockout [KO]), an insertion in exon 5 of Fmr1 produces a mutant Fmr1 mRNA but not FMRP159 (Figure 1). Another Fmr1-mutant mouse line (Fmr1 KO2) has been generated by germline deletion of the floxed promoter and the first exon of Fmr1, and neither mRNA nor protein is produced.160 Ever since its creation, the Fmr1-KO mouse model has been extensively tested with the hope that it will faithfully recapitulate human FXS phenotypes. Human male FXS-affected individuals have macroorchidism, an elongated face, prominent ears, and flexible joints. Fmr1-KO mice indeed have enlarged testes; however, joint flexibility has not been effectively assessed in mouse models.161 Dense-surface modeling techniques have been used for assessing the facial features of human FXS and Fmr1 KO.162 Fmr1-KO mice do not show an increased anterior-posterior length of the skull nor increased length of the frontal bone. However, Fmr1-KO mice exhibit an increased vertical dimension of the mandible skull, increased width of the premaxilla, and nasal bone differences, which could resemble the more oval face and wide nose of FXS-affected individuals. Seizure is prevalent in neurodevelopmental disorders. About 10%–20% of FXS-affected individuals have childhood seizures, but these seizures are generally infrequent and can be controlled by medication. Fmr1-KO mice exhibit susceptibility to audiogenic seizures triggered by loud sounds. The mouse model exhibits transient and mild seizure phenotypes that could be similar to human FXS, but the seizures occur only transiently in young mice and only in certain mouse strains.
The core behavioral deficits of human FXS-affected individuals include cognitive deficits, increased anxiety, hyperactivity, social phobia, and repetitive behaviors. Although FXS is primarily a single-gene disorder, FXS-affected individuals display variable degrees of severity. Because FMR1 is an X-linked gene, the differences in clinical phenotypes between males and females are expected, and in fact, FXS-affected females tend to have less severe disability as a result of compensation by the second X chromosome. However, even males with the full mutation exhibit heterogeneous characteristics. Both the size of the CGG expansion and the extent of CGG methylation vary significantly among males with the full mutation. Furthermore, mosaic CGG methylation and partial FMR1 inactivation have been detected in somatic cells of some individuals.163 Therefore, genetic differences lead to different levels and patterns of FMRP expression and are responsible for additional variability in clinical presentations and severity of this single-gene disorder. This heterogeneity is somewhat recapitulated in FXS mouse models that are developed in different genetic backgrounds.164, 165
Fmr1-KO mice exhibit several human FXS-related behavioral deficits, but not others (for a comprehensive review of behavioral studies of FXS mice, please see Kazdoba et al.157). FXS-affected individuals have impaired working memory, short-term visual memory, visual-spatial abilities, sequential information processing, executive function, and attention. Several mouse behavioral tests have been used for assessing these complex cognitive functions in Fmr1-KO mice. Although some studies have shown that Fmr1-KO mice are impaired in passive avoidance, contextual fear conditioning, delayed trace conditioning, the Morris water maze, novel-object recognition, and novel-location tests, other laboratories have found no change or even opposite results (Figure 3 and Table 1).
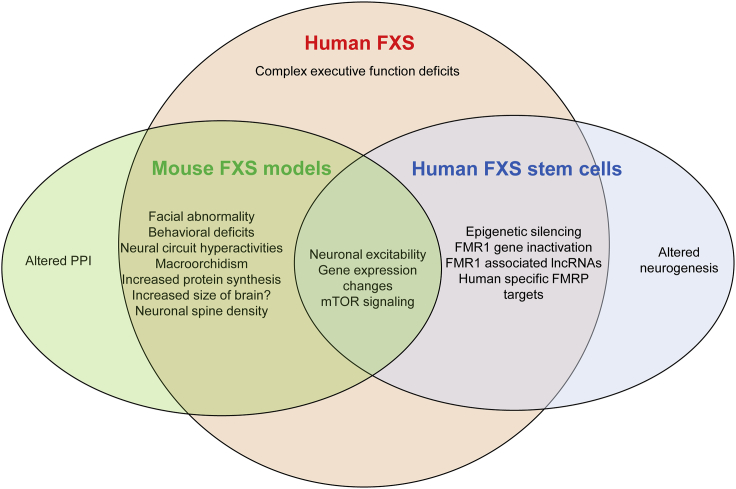
Figure 3.
Human FXS Neurological and Behavioral Deficits that Are Represented in Mouse Models and/or Human Stem Cell Models
Although FXS-affected individuals also exhibit reduced pre-pulse inhibition (PPI), mouse models show either increased or no change in PPI.
Table 1.
Comparison of Characteristics of FXS-Affected Individuals and Model Systems
| FXS-Affected Individuals | Rodent Models | Human Stem Cell Models | |
|---|---|---|---|
| Physical symptoms | facial abnormality (long face, prominent ears) | some mild facial abnormality | ND |
| flexible joints | ND | ND | |
| gastrointestinal issues | ND | ND | |
| macroorchidisma | macroorchidisma | ND | |
| facial abnormality (long face, prominent ears) | some mild facial abnormality | ND | |
| Behavioral symptoms | reduced PPI | increased, decreased, or no change in PPI | ND |
| severe cognitive deficits (working memory, short-term memory, executive functioning, attention) | some mild cognitive deficits and variable resultsb | ND | |
| motor deficits | ND | ND | |
| hyperactivity | variable resultsb | ND | |
| significant anxiety, social anxiety | variable resultsb | ND | |
| compulsive behaviorsa | impaired reversal learning, marble buryinga | ND | |
| childhood seizuresa | audiogenic seizure in juvenile micea | ND | |
| Brain and neuronal deficits | increased brain size | both embryonic and adult NPCs have increased proliferation | ND |
| ND | embryonic NSCs have increased neuronal differentiation; adult NSCs have reduced neuronal differentiation | reduced neuronal differentiation, no difference, variable results | |
| ND | reduced dendritic complexitya | reduced dendritic complexitya | |
| increased spine densitya,c | increased spine densitya,c | ND | |
| Neuronal functional deficits | neural circuit hyperactivitya | neural circuit hyperactivitya | ND |
| neuronal hyperexcitabilityd | neuronal hyperexcitabilityd | neuronal hyperexcitabilityd | |
| ND | reduced synaptic pruning | ND | |
| ND | impaired LTD | ND | |
| ND | impaired LTP | ND | |
| ND | impaired development of GABAergic inhibitory neurons | ND | |
| Genetic basis | FMR1 CGG expansion and gene silencinga | Fmr1 deletion in exon 5 (KO1 produces mutant mRNA and no protein) or exon 1 (KO2 produces no mRNA or protein)e | FMR1 CGG expansion and gene silencinga |
| mosaic gene silencinga | ND | mosaic gene silencinga | |
| human lncRNAs FMR4 and FMR6 inactivateda | ND | human lncRNAs within FMR1 locusa | |
| FMR1AS1 inactivateda | ND | FMR1AS1 inactivateda | |
| Molecular deficits | increased protein synthesisa | increased protein synthesisa | ND |
| altered mTOR signalingd | altered mTOR signalingd | altered mTOR signalingd | |
| elevated PI3K activitiesa | elevated PI3K activitiesa | ND | |
| ND | signaling pathways (e.g., APP, ERK, IGF, and P53) | ND | |
| Molecular targets | gene expression changese | gene expression changese | gene expression changese |
| some FMRP targets (e.g., Map1b)d | some FMRP targets (e.g., Map1b)d | some FMRP targets (e.g., Map1b)d | |
| human-specific FMRP targets (e.g., NOS1)a | ND | human-specific FMRP targets (e.g., NOS1)a |
The following abbreviations are used: LTD, long-term depression; LTP, long-term potentiation; ND, no data.
Consistent between FXS-affected individuals and only one of the experimental models.
Large variability in behavioral deficits was observed. See Kazdoba et al.157 for a review.
Spine density has only been shown in specific brain regions and in very few human cases.
Consistent among FXS-affected individuals and both experimental models.
Not consistent between experimental models and FXS-affected individuals.
FXS-affected individuals exhibit significant anxiety despite the genetic background. Anxiety can be assessed in rodents with the open-field activity test, dark-light exploration, and the elevated plus maze. These tests are non-invasive and are based on the natural tendency of mice to seek dark and protected locations. However, the mouse models display increased, decreased, or no change in anxiety depending on the genetic background, age, methods, and labs tested.155, 157 Compulsive and repetitive behaviors are frequently seen in FXS-affected individuals. Fmr1-KO mice have shown altered reverse learning. The marble-burying test has been used to show compulsive burying of strange objects in mice.155 Fmr1-KO mice in some backgrounds have been shown to bury more marbles.166 Impairment of social behavior in FXS is largely due to social anxiety. The social-interaction results of mouse models are mixed—some studies have shown decreased anxiety, whereas others have found no difference in social interaction in Fmr1-KO mice. A recent study has shown that Fmr1-KO mice might have trouble recognizing strangers in a social-novelty test.167, 168 Whether this is a result of learning or social ability remains to be validated. Thus, the behavioral phenotypes that are relevant to FXS-affected individuals are not robustly recapitulated in FXS mouse models.
Many FXS-affected individuals are hyperactive and have impaired sustained attention. They have difficulty performing difficult tasks requiring switching and inhibiting responses. Fmr1-KO mice have not shown impaired attention in the five-choice serial-reaction-time task. However, analyses of open-field activity have shown increased locomotor activity in some studies, but not others.157 Sensorimotor gating is impaired in many neurodevelopmental disorders, including autism and FXS, which can be assessed with pre-pulse inhibition (PPI) tests for both rodents and humans. FXS-affected individuals are more sensitive to pure tones, as are Fmr1-KO mice. The severity of PPI deficits correlates with the severity of impaired attention and cognition in FXS. However, the PPI results from mice have been confusing because most studies have found that PPI is enhanced in Fmr1-KO mice, contradictory to the reduced PPI observed in FXS-affected individuals.
The assessment of language impairment is difficult in rodents. Ultrasonic vocalization emitted by pups separated from their mother has been used for assessing language in rodents; however, the findings are inconsistent. Importantly, it is not clear how relevant rodent vocalization is to human language.
Altogether, it is not clear whether the variable results observed in FXS mouse models reflect variability in the disease phenotypes that are being modeled or whether these mouse models simply fail to recapitulate human disease phenotypes.
Down Syndrome. Non-neural DS features, including hallmark craniofacial characteristics, heart malformations, and leukemia, have been modeled in some of the DS animal models.169, 170, 171, 172, 173, 174 DS is characterized by learning, memory, and language abnormalities that result in mild to moderate intellectual disability.175, 176, 177, 178, 179, 180, 181, 182 Additional neurological hallmarks of DS include motor dysfunction and hypotonia178, 183, 184, 185 and predisposition to AD pathology.178, 186, 187, 188, 189 In recent years, research in DS has been increasingly focused on the degeneration and dementia hallmarks of AD in DS. DS and neurodegeneration are reviewed in detail in other recent reviews.190, 191, 192 Here, we focus on neurodevelopment, although the two are probably linked.
Cognitive disability in DS-affected individuals suggests that hippocampus-dependent mechanisms are affected but that other cortical regions and cerebellum contribute. Hippocampus-dependent learning and motor learning are impaired in DS mouse models, as shown in behavioral tests and electrophysiology (Figure 4 and Table 2).193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208 Yet, thorough analyses of all the different trisomic mouse models are still underway, and results are not consistent among models.142, 145, 196, 204, 209 Importantly, the different mouse models have differing or no phenotypes that might link to DS disease characteristics.210
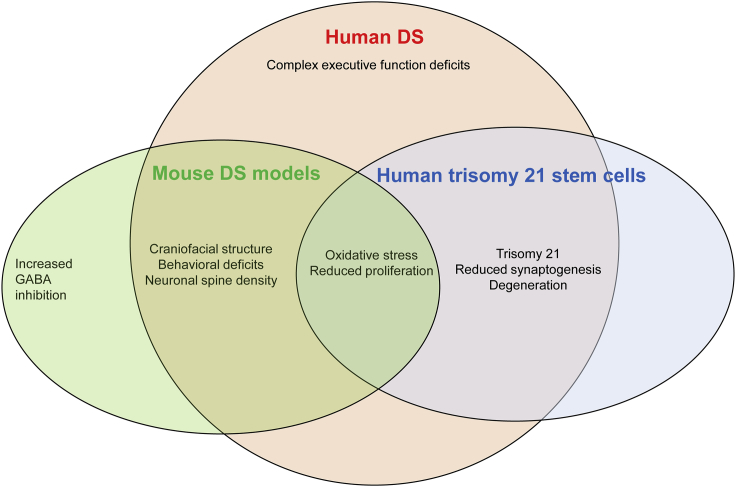
Figure 4.
Human DS Neurological and Behavioral Deficits that Are Represented in Mouse Models and/or Human Stem Cell Models
Increased GABAergic inhibition has not yet been shown in human DS.
Table 2.
Comparison of Characteristics of DS-Affected Individuals and Model Systems
| DS-Affected Individuals | Rodent Models | Human Stem Cell Models | |
|---|---|---|---|
| Physical symptoms | altered craniofacial structurea | altered craniofacial structurea | ND |
| short stature | ND | ND | |
| Behavioral symptoms | severe cognitive deficits (learning and memory, executive function) | some cognitive deficits and variable resultsb | ND |
| Brain and neuronal deficits | decreased brain size | ND | reduced neuronal differentiation, no difference, variable results |
| reduced excitatory and inhibitory neurons | increased inhibitory neuronsc | reduced inhibitory neurons | |
| dendritic spine defects | dendritic spine defects | ND | |
| Alzheimer disease pathologyd | increased amyloid beta proteind | increased amyloid beta proteind | |
| Neuronal functional deficits | oxidative stressd | oxidative stressd | oxidative stressd |
| decreased neurogenesis and proliferation | variable increased and decreased neurogenesisb | decreased neurogenesis and proliferation | |
| ND | increased inhibition | ND | |
| decreased myelinationa | decreased myelinationa | ND | |
| Genetic basis | trisomy 21a | each animal model has different combinations of orthologous HSA21 genes; some have triplicated non-HSA21 genesc | trisomy 21a |
| Molecular deficits and targets | upregulation of some HSA21 genes; downstream consequences on whole genomea | different sets of genes; potentially different downstream consequencesc | upregulation of some HSA21 genes; downstream consequences on whole genomea |
The following abbreviation is used: ND, no data.
Consistent between DS-affected individuals and only one of the experimental models.
Large variability in behavioral deficits was observed. See Herault et al.142 for a review.
Not consistent between experimental models and DS-affected individuals.
Consistent among DS-affected individuals and both experimental models.
Although the behavioral phenotypes of some of the DS mouse models could be reminiscent of cognitive characteristics of DS-affected individuals, structural and molecular mechanisms that underlie the phenotypes are different.
Modeling Structural and Cellular Deficits
Fragile X Syndrome. Whether FXS-affected individuals have an enlarged brain is unclear because larger brain size has been found in some affected individuals but not others.211 Compared with those of wild-type mice, the brains of Fmr1-KO mice show no significant changes in size, weight, or overall number of cells.159 A number of studies have assessed the size of the brain and major brain structures in FXS-affected individuals by using magnetic resonance imaging (MRI) and revealed abnormalities including reduced cerebellar vermis, increased hippocampal volume, larger volume of the caudate nucleus, and enlarged ventricles, although these findings are not consistent across studies.212, 213 Fmr1-KO mice have also been analyzed by MRI. Although an early analysis found no significant change in gross structure,214 two later studies found reduced volumes in the deep cerebellar nuclei215 and increased white-matter volume.216 In these studies, no change in brain volume was found. Because Fmr1-KO mice exhibit mild facial structural changes,162 changes in regional brain structures and cell number could be too subtle for MRI detection.
Neurons have complex morphological development that is critical for their function. A highly cited finding is that neurons in post-mortem brain tissue from humans with FXS have long, thin dendritic spines. This observation was first reported in apical dendrites of pyramidal neurons of the isocortex in one FXS-affected male and was later confirmed in neocortical neurons of three more individuals.217, 218, 219 A subsequent quantitative analysis comparing the layer V pyramidal neurons of temporal visual cortices of three FXS-affected individuals with that of three control individuals corroborated the spine phenotypes.220 However, the limited analyses, in terms of both types of neurons and numbers of subjects, limit the potential generalizability of this finding to phenotypical modeling in animal models. Nevertheless, because this dendritic-spine abnormality is one of the only neuronal deficits identified in human neurons, spine analysis has been widely carried out to identify “human FXS-individual-relevant phenotypes” in Fmr1-mutant mouse models221 despite the fact that the spine phenotype has been inconsistent in animal models.222 More analysis of human neurons and the identification of additional neuronal deficits in human FXS neurons are critically needed to guide animal studies and to expand our understanding of the impact of FXS on the brain.
Electroencephalography (EEG) analysis has revealed that FXS-affected individuals exhibit hyperexcitability in the neocortex and altered functional connectivity, suggesting impaired excitatory and inhibitory balance.223, 224, 225, 226 The Fmr1-KO mouse model enables cellular physiology analysis that is impossible in humans. Altered EEG has also been found in Fmr1-KO mice, and it has been used as a readout for pre-clinical assessment.227 These studies have shown that Fmr1-KO mice exhibit altered synaptic plasticity and a disrupted critical period for neuronal maturation (see the review by Contractor et al.228). Validation of these morphological and functional neuronal deficits in human models is needed.
Down Syndrome. Post-mortem observations and volumetric MRI studies have indicated that people with DS, in contrast to those with FXS, have reduced brain volumes.229, 230, 231, 232, 233, 234 Fewer neurons in the developing cerebral cortex of DS-affected individuals have been consistently reported over the last 70 years.229, 230, 231, 232, 233, 235, 236, 237, 238, 239 Cortical neuron deficiency appears near the end of cortical neurogenesis (>23 weeks of gestation)238, 240 and, according to limited histopathological analyses, is area, cell-type, and age specific. Data suggest that primarily small, granular, presumably GABAergic neurons in layers II and IV of the cortex are affected.239 Additional hypocellularity has been reported in other brain regions, including the hippocampus and cerebellum,241, 242, 243 during development, suggesting a general defect in neurogenesis in DS.244
Impaired neurogenesis has been reported in DS mouse models241, 245, 246, 247 but does not consistently decrease the number of neurons in the mouse brain to recapitulate the reduced number of neurons in the brains of DS-affected individuals. For example, increased interneuron progenitor proliferation and increased production of interneurons is found in T65Dn,248 in contrast to observations of fewer interneurons present in human DS cortex. Further, at least one mouse model, Dp16, that contains the largest number of triplicated orthologous HSA21 genes has no detectable neurodevelopmental defects.210 Neuronal structure is also altered in the brains of DS-affected individuals, such that neurons show fewer and shorter dendritic spines.229, 240, 249, 250, 251, 252, 253, 254, 255 A reduced number of spines has been reported in DS mouse models, although the morphology varies among models.209, 256, 257, 258 These results highlight the inability of DS mouse models to recapitulate the most established neuroanatomical characteristics of DS-affected individuals.
Modeling Molecular Mechanisms
Fragile X Syndrome. Extensive studies using animal models have revealed several significant molecular functions of FMRP. The most prominent function of FMRP is regulating protein synthesis and acting as a brake for metabotropic glutamate-receptor-dependent protein syntheses. In the absence of FMRP, exaggerated mGluR5 signaling leads to enhanced protein synthesis and changes in synaptic plasticity. According to this mGluR5 theory,259 a number of pharmacological preclinical studies targeting mGluR5 have been performed and shown remarkable effects in correcting Fmr1-KO mice. Subsequent large human clinical trials were carried out and were able to successfully alleviate disease symptoms.158 Another group of favorite drug targets for FXS is GABA receptors and drugs to enhance GABAergic signaling. Some of these drugs were found to correct behaviors in Fmr1-KO mice, and hence clinical trials were moved forward, but they did not show a successful response in FXS-affected individuals.161, 260 Although the failure of these trials could be due to many reasons, including differences in drug bioavailability, drug metabolism, and methods of delivery and outcome assessment, there was a clear lack of validation in human experimental systems before the trials. In fact, only one study assessed mGluR5 in humans and found only mild increases with marginal significance (p = 0.058) in the density and amount of mGluR in post-mortem prefrontal cortex.261 Importantly, the activity of mGluR5 has not been assessed in human FXS neurons, and so it remains unclear whether pathways targeted for therapy in FXS mouse models are the correct targets for human disease.
Several FMRP-regulated downstream pathways have been discovered, and the most well-known mechanistic target is rapamycin (mTOR) signaling.262, 263 However, a critical role of mTOR signaling has not been validated in human FXS. Many signaling pathways, including mitogen-activated protein kinase 1 (MAP2K1) signaling and Wingless-type MMTV integration site family (WNT) signaling, have been shown to alter mTOR signaling in Fmr1-KO mice. Targeting these pathways has had remarkable rescue effects in mice, but none of these changes have been validated in human neurons.158 Therefore, it is unclear whether any of these pathways are important mechanisms underlying FXS in humans.
Down Syndrome. Most lines of research to identify molecular mechanisms underlying DS have focused on the premise that specific genes located on chromosome 21 are responsible for DS symptoms and that these genes might exert direct effects that lead to manifestations of DS or have secondary effects on other genes. Thus, intense focus on the contribution of a few individual HSA21 genes to regulating aspects of DS pathology has been carried out with DS mouse models. In particular, DYRK1A, a kinase that plays a role in many signaling pathways (including those involved in neural development), has been linked to behavioral deficits in mouse models and has been targeted as a potential therapeutic candidate.264, 265, 266 More recently, the role of DYRK1A has been implicated in the proliferation and differentiation of human neural progenitors.267 Regulator of calcineurin 1 (RCAN1) regulates calcineurin—so its overexpression might affect neural development—and it has been linked to aspects of DS.268, 269, 270, 271 One chromosome 21 gene, USP16, has been shown to be a controller of stem cells in both animals and humans and can affect the proliferation and differentiation of many cell types that are affected in DS.272 Most of these studies do not analyze the role of genes in the context of trisomy 21 but instead provide good data for each chromosome 21 gene product’s roles in neural development.
The predominant candidate mechanism that has emerged from mouse model studies as underlying intellectual disability in DS is an imbalance between excitation and inhibition in the cortex, specifically over-inhibition. Excitation-inhibition imbalance is a mechanism commonly proposed in disorders characterized by intellectual disability and cortical dysfunction. The establishment of this phenotype in mouse models has led to the targeting of over-inhibition in the cortex by various drugs, including GABA agonists and epigallocatechin-3-gallate.273, 274, 275, 276, 277 It is important to note that in the Ts65Dn mouse model, the one on which therapeutic targets and clinical trials have been based, 25% of trisomic genes are not human chromosome 21 orthologs and 45% of human chromosome 21 orthologs are not trisomic.118, 141 Importantly, the over-inhibition phenotype has not been validated in human DS brain tissue or neurons, and the fact that the human DS brain has fewer interneurons suggests that over-inhibition might not underlie cortical dysfunction in DS-affected individuals.
The examples of data from mouse models of the two most common genetic causes of intellectual disability highlight important knowledge gained but also reflect differences, inconsistencies, and limited (sometimes contradictory) associations with disease characteristics. Many biochemical or structural differences identified in animal models have not been validated in humans and therefore might not be of physiological relevance to human disease. Therapeutic targets that have not been validated in affected individuals are being pursued. So, the impact of these therapeutic interventions could be limited to treatments that reverse mouse phenotypes rather than disease characteristics. Thus, human models are critical to identifying therapeutic targets that will be more likely to benefit affected persons. To do so, we need to better understand foundations of human development and developmental disorders through the use of human tissue, cells, and models.
The Advantages of Human Stem Cell Models
Human Experimental Models for Neurodevelopmental Disorders
Various peripheral cell types, including lymphocytes, amniocytes, and fibroblasts, have been used for studying molecular aspects of neurological disease. The primary advantage of these cells is their accessibility in humans. These peripheral cells can also serve as critical platforms for identifying biomarkers for early diagnosis if characteristics of these cells are shared with neural cells or, more likely, if strong correlations between neural function and specific peripheral cell phenotypes are defined. However, in the context of understanding the causes of neurodevelopmental disorders, cells of the developing cortex are dynamic and have cell-type-specific characteristics that cannot be represented by these surrogate cells. Thus, studying human neural cells is critical to providing insight into the foundations of neurodevelopmental disorders.
Post-mortem Brain Tissues. Post-mortem brain tissues have long been a major experimental source for analyses to inform our understanding of neurological diseases. The fact that the bulk of the formation of the human brain occurs prenatally necessitates study of the fetal period to understand human cortical development. The use of human post-mortem fetal tissue is limited by access, ethical and sometimes political controversy, and static time points. Yet, primary fetal tissue provides a critical opportunity to view human brain development and thus remains the gold standard by which to compare other model systems. In fact, fetal tissue samples were essential for recent transcriptomic analysis of normal human brain development and identifying differences in cortical development in DS.64
Some of the technical, but not societal, limitations of post-mortem human fetal tissue can be overcome to some extent by forcing expansion of precursor cells within the tissue. Human neural progenitor cells (hNPCs) provide a relatively pure population of neural cells that can be used for studying early brain formation. Exposure of primary fetal cortical tissue to mitogens causes the formation of aggregates of progenitor cells, termed “neurospheres.”278 Neurospheres contain uncommitted and restricted neural progenitor cells. Progenitors within the neurospheres divide when cultured in the presence of growth factors for extended periods of time and differentiate into neurons and glia in the absence of growth factors.279, 280, 281, 282 Yet, there are significant limitations of using hNPCs to study neural development. First, the isolation of cells from tissue, as in any primary-tissue in vitro paradigm, removes them from their developmental niche and therefore provides information about the potential of hNPCs but not necessarily what the cells would do in vivo. hNPCs differentiate into limited neuronal subtypes because they are derived from fetal tissue at a single time point, approximately 20 gestational weeks. hNPCs are not very valuable for studying synaptic development because synaptically active cortical neurons have not been successfully generated from hNPCs. Therefore, the utility of hNPCs for studying human neural development and neurodevelopmental disorders is limited.
Despite these limitations, critical information about neurodevelopmental disorders, particularly DS, has been gleaned from studies of hNPCs. The ability to diagnose DS prenatally and to terminate a pregnancy has allowed for access to DS fetal tissue. Although hNPCs are derived from a specific developmental time point, this particular time point has proved to be useful for identifying differences in DS cortical development, such as reduced neuron generation283, 284, 285 and increased astrocyte generation,286, 287 suggesting a possible change in developmental timing during cortical development in DS.
hPSCs overcome many limitations of primary brain tissue and are an important tool for studying the development of specific cell types, including neural cells.288, 289, 290, 291, 292, 293, 294 hPSCs, including hESCs derived from embryos and hiPSCs derived from somatic cells, have tremendous potential for human disease modeling and therapeutic development. Because hPSCs are pluripotent, meaning they have the ability to differentiate into any cell type in humans, they can be used to generate disease-relevant cell types, tissues, and organs to unveil cellular and molecular events underlying normal and abnormal neural development. In addition, because hPSCs are non-transformed human cells, they can be used to elucidate neurological disease pathogenesis in a human genetic background. Additionally, pure populations of specific neural cell types derived from hiPSCs can be used for the rapid screening of pharmaceutical compounds to accelerate drug discovery and advance treatments. The power of hPSCs to model human brain development lies in the ability to generate specific neural cell types in vitro over a long period of time that corresponds to in vivo development and thus recapitulate many of the developmental steps. Methods of generating specific neuronal and glial subtypes from hPSCs that follow in vivo developmental principles and timing have been developed.29, 30, 31, 36, 40, 292, 294, 295, 296, 297, 298, 299, 300, 301, 302
A critical opportunity for hPSCs is their use in studying the developmental course of genetic disorders. hESCs can be derived from embryos that have been found to carry genetic mutations after pre-implantation genetic diagnosis (PGD).303 By building upon studies of ESCs, the establishment of iPSC technology in humans289, 291 has resolved many difficult issues facing hESCs and revolutionized our ability to study human development and diseases. To date, hiPSCs can be derived from not only skin fibroblasts but also more accessible lymphocytes, hair follicle cells, and dental pulp cells, for example. Characteristics of a given genetic disorder can be defined at the molecular level with hPSCs that either carry the genetic defect (derived from affected individuals) or have been genetically engineered to carry the disease mutation. Unlike their mouse counterparts, hPSCs have been difficult to modify genetically. Gene editing of hPSCs is typically achieved by random transgene insertion due to inefficiency in homologous recombination and cellular cloning, and such transgenes are often downregulated upon stem cell differentiation, especially during differentiation to functional neurons. Technical difficulties in genetically modifying hPSCs have significantly impeded neuroscience research involving hPSCs. Recent technological advances enable more efficient genetic hPSC modifications, including homologous recombination based on zinc fingers, transcription-activator-like effector nucleases, and CRISPR-Cas systems.
Human Stem Cells for Modeling Neurodevelopmental Disorders
The unique ability of hPSCs to model human brain development has facilitated their use in defining human-specific aspects of cortical development and defining differences that occur in neurodevelopmental disorders. Human-derived hiPSCs have been generated for modeling both single-gene mutations and complex mutations that lead to intellectual disability in humans. PSC models of neurodevelopmental disorders caused by mutations in single genes have been established and include those for Rett syndrome (MECP2 [MIM: 300005]),304, 305, 306, 307, 308 tuberous sclerosis (TSC1 [MIM: 605284]),309, 310, 311 and FXS.312, 313, 314, 315, 316 More complex genetic disorders have benefitted from the ability to retain the chromosomal defect in hiPSCs for disorders that result in deletions or duplications of chromosomal regions, such as 15q11.2 microdeletion syndrome (MIM: 615656),317 Prader-Willi syndrome (MIM: 608636),318, 319 and DS.320, 321, 322, 323, 324 Some phenotypes in neural cells derived from these cells link to disease symptomology, whereas other phenotypes do not, and there is variability in the phenotypes in different reports.
Fragile X Syndrome. Both hESC and iPSC models of FXS have been developed.312, 313, 314, 315, 316 FXS hESCs are derived from embryos that are determined to have the full mutation by PGD.325, 326 Analyses of neurons derived from these cells confirm that the epigenetic mutation is preserved during reprogramming in the case of hiPSCs and during differentiation. The phenotypes of neural progenitor cells differentiated from these cells vary among reports; some have provided evidence that neurogenesis is aberrant,316 whereas others have reported that neurogenesis is unaffected.312, 327 As discussed above, FXS-affected individuals have grossly normal brain formation, so neurogenesis does not appear to be affected. Neurons from FXS PSCs do have functional deficits,312, 328, 329 but it is still unclear how these phenotypes link to disease characteristics. FXS-affected individuals are hypothesized to have synaptic and plasticity defects, as has been demonstrated in FXS mouse models, but assessment of synaptic function and plasticity in human FXS neurons has not been carried out primarily because of the limitations of the system, in which the human neurons in culture are immature. These discrepancies and limitations suggest that phenotypic modeling of FXS disease phenotypes with hiPSCs remains challenging. One way to overcome the immaturity of hPSC neurons in culture is to transplant FMRP-deficient human NPCs into immune-deficient mouse brains to study the development, maturation, and functions of human neurons without functional FMRP. As discussed below, these chimeras could enable the detection of synaptic differences in human FXS neurons.330
On the other hand, FXS hPSCs do provide a unique and much-needed model for studying FMR1 inactivation and reactivation. Because the FMRP coding sequence in most FXS-affected individuals is intact, efforts have been made to reactivate the silenced FMR1.331 Early attempts have involved FXS lymphocytes, which have limited expansion ability and non-neural lineage. With the development of gene editing, we created a FXS reporter line by inserting nanoluciferase into the 3′ region of endogenous FMR1 of FXS iPSCs.312, 332 The FXS reporter hPSC-derived neural cells are suitable for large-scale drug or genetic screening and will potentially lead to insight into the mechanisms of FMR1 silencing and identify ways to reactivate the gene.
Furthermore, a number of ongoing projects in various laboratories are using hPSCs to study FMRP targets and validate molecular mechanisms of FMRP regulation of neuronal development. FMRP has been shown to target a large number of mRNAs in the mouse brain.333, 334, 335 However, few of these targets have been validated in humans, and the identity of FMRP-bound mRNAs in humans remain unknown. FMRP targets most likely differ between humans and rodents, leading to different phenotypes when the protein expression is lost. As described above, an example is the dysregulation of NOS1 in developing human FXS brains, but not in FMRP-deficient mice.95
Neuronal functions, specifically synaptic activities, have not been well studied because of the immaturity of hPSC-derived neurons. Yet, analysis of this functional readout is critical for understanding underlying cortical dysfunction in FXS and other neurodevelopmental disorders. For example, about one-third of FXS-affected individuals exhibit cortical hyperexcitability.336 If such hyperexcitability can be modeled in FXS iPSC-differentiated neurons, it will provide an excellent platform for understanding the molecular mechanism of the presence and absence of hyperexcitability and provide an opportunity to prescreen individuals for clinical trials. Therefore, hPSCs are essential for validating the molecular, cellular, and functional FXS deficits that we have known for years on the basis of animal studies. More importantly, hPSCs will help us to discover the hidden differences and molecular targets that we have failed to discover with animal models.
Down Syndrome. DS hESCs337, 338, 339, 340 and hiPSCs320, 321, 322, 323, 324 have been established. The development of neurons has been primarily studied with iPSC-derived neurons, and these studies have revealed a common phenotype of oxidative stress and mitochondrial dysfunction that are consistent with many cell types from DS-affected individuals.320, 323, 324, 341 These results validate reports that neurons and progenitor cells derived from DS post-mortem fetal tissue have increased susceptibility to oxidative stress.286, 342, 343 Recent bioinformatics analyses of human and mouse DS fetal-brain transcriptomes have indicated that specific common pathway abnormalities leading to oxidative stress, neuroinflammation, apoptosis, and cell-cycle delays are present in both as early as mid-gestation.344
In addition, studies using DS iPSCs have validated initial neurodevelopmental deficits, including those in neurogenesis and synaptogenesis, found in DS-affected individuals.323, 341, 345, 346, 347 But, the inability thus far to analyze neuronal function and plasticity in human neurons with trisomy 21 has hindered the validation of hypotheses of cortical dysfunction described in mouse models. The use of DS hPSCs, especially from those DS-affected individuals with different genetic backgrounds and/or well-documented clinical history, will significantly help us to understand the genes that contribute to DS symptoms and identify targeted therapies for DS.
Altogether, the use of hPSCs is helping us to discover the hidden differences and human-specific molecular targets that we have failed to discover with animal models. Yet, there exist several challenges that currently constrain our ability to gain critical knowledge about foundational mechanisms of neurodevelopmental disorders.
Challenges and Perspectives
Clinical Variability
Despite their incredible utility, hPSC studies encounter challenges that significantly impede research progress in the field. Variability in disease characteristics can introduce variability in modeling of disease-related phenotypes in cells. Both FXS and DS have highly variable clinical presentations.132, 158 The considerable clinical variability could complicate the investigation of underlying mechanisms that lead to intellectual disability, the identification of therapeutic targets, and the application of therapeutic strategies. For example, recent work has indicated that variability in behavior and cognition in FXS could be linked to variability in electrical activity in the brain of FXS-affected individuals, as measured by EEG.223, 336 One solution is the establishment of high-quality iPSCs from individuals with well-characterized and stratified clinical features. Stratification of affected individuals and cells could enable more effective linkage of disease characteristics with cellular phenotypes and better design of clinical trials.
Genomic Variability
Unlike model organisms that tend to be inbred, genetic variation between hPSCs is a challenge that largely results from inherent genomic variability among human beings. Simply increasing the sample size does not readily compensate for this challenge because of the enormous time and cost associated with research involving hPSCs. To make disease modeling using hiPSCs even more difficult, cellular and molecular changes in human cells are often not as robust as those seen in transgenic animals, thus amplifying the effects of variability. Challenges in distinguishing biologically relevant changes leading to complex traits from those resulting from variability between hPSCs (as a result of differences in the contributing individuals’ total genomic makeups) can be addressed with isogenic “control” cells. For monogenic traits, isogenic controls can now be generated through CRISPR-Cas9 gene-editing technology, which enables facile gene editing in hPSCs. Isogenic controls can be generated through correction of the gene mutation in disease cells or introduction of the gene mutation in control cells. Use of isogenic controls enables validation of disease-related phenotypes such as FXS.348 Given that the phenotypic effects of some disorders, including trisomy 21, involve multiple genes, isogenic iPSCs cannot be developed for these conditions through current gene-editing technology. A strategy for silencing the extra chromosome has been developed but cannot yet be easily applied on a large scale.349 An alternative method for teasing apart the biologically relevant changes attributable to a trisomic imbalance is to derive PSCs from people with mosaicism for trisomy 21 (mDs).324
Pure and Enriched Populations of Cells
Pure populations of specific neural cell types can be derived from hiPSCs. These enriched, homogeneous cultures are particularly useful for the rapid screening of pharmaceutical compounds to accelerate drug discovery and advance treatments. They are also potentially useful for defining cell-intrinsic consequences of a given mutation. However, they do not recapitulate the makeup of the human brain and so might be less useful in mechanistic studies, especially those that rely on cell-cell interactions. For example, only about 20% of neurons in the cortex are GABAergic and 80% are glutamatergic according to analysis of rodents. This proportion could be different in humans. Yet, many hPSC-derived cultures utilize pure cultures of each subtype and do not always include the critical presence of glia. Advancement in tissue engineering could allow for obtaining pure population of cells grown in the presence of tissue-like cues.
Neuronal Maturation
A serious limitation of hPSC-derived neuron cultures for modeling both normal human brain development and function and maldevelopment and dysfunction is the fact that our current culture systems model immature, fetal neurons. Although these cells provide information about early developmental processes, they are less valuable for investigating neuronal function and plasticity, which is the basis of intellectual disability. This major limitation is being addressed in culture through the use of three-dimensional cultures, mixed neuron and astrocyte cultures, and the development of new media.350, 351, 352 Chimeras, in which hPSC-derived neurons are implanted into the mouse central nervous system, address the challenge by providing an environment that stimulates the maturation of human neurons.
Self-Organization of Neural Tissue
Some of the limitations of two-dimensional culture of hPSCs for modeling the development of organs, including the nervous system, have been addressed with self-organizing three-dimensional cultures, commonly termed organoids or spheroids. As was initially shown in the nervous system by Sasai’s group, a simple cell-culture system can undergo self-organization to generate a complex structure without external cues.353 Within the last 5 years, several groups have developed these methods to model the mixed cell types and increased complexity of the human cortex by beginning with the initial formation of dorsal forebrain from hPSCs.354, 355, 356, 357, 358 Recent reviews, over 70 in the last 4 years, have praised the use of organoids for modeling some aspects of human brain development but have also elucidated considerable limitations of the technology.359, 360, 361 Organoids do appear to mimic two critical aspects of human cortical development: timing and the presence of the oSVZ, which enables cortical expansion in primates.356, 362, 363 Yet, the heterogeneity that is inherent in self-organization presents a considerable challenge for identifying significant, reproducible, and disease-relevant phenotypes in these systems. For example, the assembly of individual organoids under seemingly identical conditions results in variation between organoids.359, 364 Further, the maturation of neural cells within organoids is still limited because it is in two-dimensional culture, and neurons are similar to fetal rather than adult neurons, thus limiting the assessment of function. Finally, the diverse cell types that contribute to the structure and function of the brain, including astrocytes, vascular cells, and microglia, are only beginning to be included in organoid models.330, 365, 366, 367, 368 Despite these limitations, the ability to model human-specific early development advocates for the use of organoid modeling in neurodevelopmental disorders where there is an expectation that early events in cortical development are affected, such as DS. It is likely that as organoid technology matures, so will the ability to use these cultures to model aspects of human cortical development beyond early developmental processes.
Chimeras
Implantation of PSC-derived neural progenitors into the developing or adult mouse brain enables the analysis of human cells in an in vivo setting, where complex external cues are present.369, 370, 371, 372 This strategy of “humanized rodents” shows that human neurons can influence and be influenced by the mouse neural tissue. Recently, Mansour et al. demonstrated that hPSC-differentiated forebrain organoids can survive in rodent brains and form connections with rodent neurons.330 In this paradigm, human cells can mature and form connections in a manner that is not observed in culture. Combining transplantation with methods such as optogenetics (which uses light-sensitive channels and pumps) or chemogenetics (which uses chemically engineered receptors and exogenous molecules specific to those receptors) to manipulate the activity of neurons enables control over the transplanted human neurons.373, 374, 375, 376, 377 These technical developments could lead to the ability to manipulate and analyze complex functions of human neurons, such as plasticity, in an in vivo setting and could provide better understanding of cognitive function and intellectual disability. Further, in an effort to move potential therapeutic strategies that target human-specific aspects of cells from in vitro proof of principle to in vivo testing, human cells can be transplanted into the mouse brain and mice can be treated with potential therapeutics. The current focus on reactivation of FMR1, for example, cannot be carried out in mouse knockout models and necessitates chimeras for testing reactivation strategies in vivo. It is therefore likely that chimeras will better inform human clinical trials.
Looking Forward
Fundamental differences in the development and function of the neocortex between humans and the animal models outlined above include expansion of the neocortex, neuronal subtype complexity, and human-specific aspects of gene expression and regulation. These critical differences limit the ability of animal models to recapitulate human brain development and, more importantly, to identify underlying cellular and molecular mechanisms of disorders that lead to intellectual disability. The failure of animal-study-based clinical trials for intellectual disorders further underscores the limitations of animal-model-focused human disease research. Human stem cell models will better inform clinical trials in neurological disorders generally by identifying disease-associated human neural cell phenotypes that have not been identified in mouse models. For example, hPSC models of amyotrophic lateral sclerosis exhibit phenotypes that are more consistent with affected individuals than several mouse models.378
Human-stem-cell-based models conceptually provide a valid alternative to rodent models of human brain development. It is important to remember that the use of human stem cells to model neurodevelopmental disorders must be achieved through their ability to recapitulate phenotypes that are found in affected individuals, including molecular, morphological, and genetic deficits. The phenotypes that are currently modeled in human-stem-cell-derived neurons are limited to those that are developmentally early, such as neural progenitor proliferation and differentiation. We anticipate that continual development of biotechnology and innovative bioengineering methods will allow for long-term and developmentally appropriate differentiation of neurons and other brain cell types to mimic neural circuit formation and human brain development in three dimensions. In addition, emerging imaging technology (e.g., fluorescent, optical, and voltage imaging) with high time and space resolution, as well as live imaging, will be applied to in vitro hPSC-derived human models. Furthermore, high-sensitivity and single-cell genetics, as well as sophisticated bioinformatic network analysis and computation models, will allow us to understand the dynamic changes in gene expression in different cell types during human neuronal development. These exciting new methods will allow us to analyze the phenotypes that manifest in human developmental disorders related to neuron maturation, functional integration, and plasticity. Realizing the need for human models is the first step toward this future.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01MH078972, 1R01MH116582, and R56MH113146 to X.Z.; R21HD085288 and R03HD083538 to A.B.; R21NS105339 to A.B. and X.Z.; and U54HD090256 to the Waisman Center), the University of Wisconsin-Madison and Wisconsin Alumni Research Foundation (to X.Z.), a Jenni and Kyle Professorship (to X.Z.), and the John Merck Fund (to X.Z. and A.B.).
Contributor Information
Xinyu Zhao, Email: xinyu.zhao@wisc.edu.
Anita Bhattacharyya, Email: bhattacharyy@waisman.wisc.edu.
References
- 1.Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sidman R.L., Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- 3.Bystron I., Blakemore C., Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 4.Clancy B., Darlington R.B., Finlay B.L. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- 5.Freund T.F., Buzsáki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 6.Klausberger T., Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaacson J.S., Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapray D., Lasztoczi B., Lagler M., Viney T.J., Katona L., Valenti O., Hartwich K., Borhegyi Z., Somogyi P., Klausberger T. Behavior-dependent specialization of identified hippocampal interneurons. Nat. Neurosci. 2012;15:1265–1271. doi: 10.1038/nn.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parnavelas J.G., Anderson S.A., Lavdas A.A., Grigoriou M., Pachnis V., Rubenstein J.L. The contribution of the ganglionic eminence to the neuronal cell types of the cerebral cortex. Novartis Found. Symp. 2000;228:129–139, discussion 139–147. doi: 10.1002/0470846631.ch10. [DOI] [PubMed] [Google Scholar]
- 10.Anderson S.A., Kaznowski C.E., Horn C., Rubenstein J.L., McConnell S.K. Distinct origins of neocortical projection neurons and interneurons in vivo. Cereb. Cortex. 2002;12:702–709. doi: 10.1093/cercor/12.7.702. [DOI] [PubMed] [Google Scholar]
- 11.Kriegstein A., Parnavelas J.G. Progress in corticogenesis. Cereb. Cortex. 2006;16(Suppl 1):i1–i2. doi: 10.1093/cercor/bhk041. [DOI] [PubMed] [Google Scholar]
- 12.Noctor S.C., Martinez-Cerdeño V., Kriegstein A.R. Neural stem and progenitor cells in cortical development. Novartis Found. Symp. 2007;288:59–73, discussion 73–78, 96–98. [PubMed] [Google Scholar]
- 13.Kepecs A., Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu J., Anderson S.A. Development of cortical interneurons. Neuropsychopharmacology. 2015;40:16–23. doi: 10.1038/npp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arber C., Li M. Cortical interneurons from human pluripotent stem cells: prospects for neurological and psychiatric disease. Front. Cell. Neurosci. 2013;7:10. doi: 10.3389/fncel.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelman D.M., Marin O., Rubenstein J.L.R. The generation of cortical interneurons. In: Noebels J.L., Avoli M., Rogawski M.A., Olsen R.W., Delgado-Escueta A.V., editors. Fourth Edition. National Center for Biotechnology Information; 2012. (Jasper's Basic Mechanisms of the Epilepsies). [PubMed] [Google Scholar]
- 17.Batista-Brito R., Fishell G. The developmental integration of cortical interneurons into a functional network. Curr. Top. Dev. Biol. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wonders C.P., Anderson S.A. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 19.Flames N., Marín O. Developmental mechanisms underlying the generation of cortical interneuron diversity. Neuron. 2005;46:377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 20.Kessaris N., Magno L., Rubin A.N., Oliveira M.G. Genetic programs controlling cortical interneuron fate. Curr. Opin. Neurobiol. 2014;26:79–87. doi: 10.1016/j.conb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q., Liu J., Fang A., Li R., Bai Y., Kriegstein A.R., Wang X. The dynamics of neuronal migration. Adv. Exp. Med. Biol. 2014;800:25–36. doi: 10.1007/978-94-007-7687-6_2. [DOI] [PubMed] [Google Scholar]
- 22.Marín O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur. J. Neurosci. 2013;38:2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- 23.Kriegstein A.R., Noctor S.C. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Anderson S.A., Marín O., Horn C., Jennings K., Rubenstein J.L. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 25.Anderson S.A., Eisenstat D.D., Shi L., Rubenstein J.L. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 26.Fish J.L., Dehay C., Kennedy H., Huttner W.B. Making bigger brains-the evolution of neural-progenitor-cell division. J. Cell Sci. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- 27.Lui J.H., Hansen D.V., Kriegstein A.R. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim T.G., Yao R., Monnell T., Cho J.H., Vasudevan A., Koh A., Peeyush K.T., Moon M., Datta D., Bolshakov V.Y. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem Cells. 2014;32:1789–1804. doi: 10.1002/stem.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Liu H., Sauvey C., Yao L., Zarnowska E.D., Zhang S.C. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat. Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maroof A.M., Keros S., Tyson J.A., Ying S.W., Ganat Y.M., Merkle F.T., Liu B., Goulburn A., Stanley E.G., Elefanty A.G. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholas C.R., Chen J., Tang Y., Southwell D.G., Chalmers N., Vogt D., Arnold C.M., Chen Y.J., Stanley E.G., Elefanty A.G. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen D.V., Lui J.H., Flandin P., Yoshikawa K., Rubenstein J.L., Alvarez-Buylla A., Kriegstein A.R. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czepiel M., Balasubramaniyan V., Schaafsma W., Stancic M., Mikkers H., Huisman C., Boddeke E., Copray S. Differentiation of induced pluripotent stem cells into functional oligodendrocytes. Glia. 2011;59:882–892. doi: 10.1002/glia.21159. [DOI] [PubMed] [Google Scholar]
- 34.Gaspard N., Bouschet T., Hourez R., Dimidschstein J., Naeije G., van den Ameele J., Espuny-Camacho I., Herpoel A., Passante L., Schiffmann S.N. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 35.Hansen D.V., Rubenstein J.L., Kriegstein A.R. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. doi: 10.1016/j.neuron.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X.J., Du Z.W., Zarnowska E.D., Pankratz M., Hansen L.O., Pearce R.A., Zhang S.C. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 37.Shaltouki A., Peng J., Liu Q., Rao M.S., Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31:941–952. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- 38.Sharp J., Hatch M., Nistor G., Keirstead H. Derivation of oligodendrocyte progenitor cells from human embryonic stem cells. Methods Mol. Biol. 2011;767:399–409. doi: 10.1007/978-1-61779-201-4_29. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe K., Kamiya D., Nishiyama A., Katayama T., Nozaki S., Kawasaki H., Watanabe Y., Mizuseki K., Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 41.Clowry G., Molnár Z., Rakic P. Renewed focus on the developing human neocortex. J. Anat. 2010;217:276–288. doi: 10.1111/j.1469-7580.2010.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molnár Z., Clowry G. Cerebral cortical development in rodents and primates. In: Hofman M.A., Falk D., editors. Progress in Brain Research. Elsevier; 2012. pp. 45–70. [DOI] [PubMed] [Google Scholar]
- 43.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson M.B., Kawasawa Y.I., Mason C.E., Krsnik Z., Coppola G., Bogdanović D., Geschwind D.H., Mane S.M., State M.W., Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy H., Dehay C. Self-organization and interareal networks in the primate cortex. Prog. Brain Res. 2012;195:341–360. doi: 10.1016/B978-0-444-53860-4.00016-7. [DOI] [PubMed] [Google Scholar]
- 46.Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 47.Hansen D.V., Lui J.H., Parker P.R., Kriegstein A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 48.Smart I.H., Dehay C., Giroud P., Berland M., Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dehay C., Kennedy H., Kosik K.S. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 50.Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., Haffner C., Sykes A., Wong F.K., Peters J. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 51.Florio M., Heide M., Pinson A., Brandl H., Albert M., Winkler S., Wimberger P., Huttner W.B., Hiller M. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife. 2018;7:e32332. doi: 10.7554/eLife.32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosik K.S. MicroRNAs tell an evo-devo story. Nat. Rev. Neurosci. 2009;10:754–759. doi: 10.1038/nrn2713. [DOI] [PubMed] [Google Scholar]
- 53.Marques A.C., Ponting C.P. Intergenic lncRNAs and the evolution of gene expression. Curr. Opin. Genet. Dev. 2014;27:48–53. doi: 10.1016/j.gde.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Somel M., Liu X., Tang L., Yan Z., Hu H., Guo S., Jiang X., Zhang X., Xu G., Xie G. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silbereis J.C., Pochareddy S., Zhu Y., Li M., Sestan N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron. 2016;89:248–268. doi: 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lavdas A.A., Grigoriou M., Pachnis V., Parnavelas J.G. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Rourke N.A., Chenn A., McConnell S.K. Postmitotic neurons migrate tangentially in the cortical ventricular zone. Development. 1997;124:997–1005. doi: 10.1242/dev.124.5.997. [DOI] [PubMed] [Google Scholar]
- 59.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 60.Angevine J.B., Jr., Sidman R.L. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 61.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 62.Kempermann G., Gage F.H., Aigner L., Song H., Curtis M.A., Thuret S., Kuhn H.G., Jessberger S., Frankland P.W., Cameron H.A. Human Adult Neurogenesis: Evidence and Remaining Questions. Cell Stem Cell. 2018;23:25–30. doi: 10.1016/j.stem.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakken T.E., Miller J.A., Luo R., Bernard A., Bennett J.L., Lee C.K., Bertagnolli D., Parikshak N.N., Smith K.A., Sunkin S.M. Spatiotemporal dynamics of the postnatal developing primate brain transcriptome. Hum. Mol. Genet. 2015;24:4327–4339. doi: 10.1093/hmg/ddv166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olmos-Serrano J.L., Kang H.J., Tyler W.A., Silbereis J.C., Cheng F., Zhu Y., Pletikos M., Jankovic-Rapan L., Cramer N.P., Galdzicki Z. Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination. Neuron. 2016;89:1208–1222. doi: 10.1016/j.neuron.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones E.G. The origins of cortical interneurons: mouse versus monkey and human. Cereb. Cortex. 2009;19:1953–1956. doi: 10.1093/cercor/bhp088. [DOI] [PubMed] [Google Scholar]
- 66.Al-Jaberi N., Lindsay S., Sarma S., Bayatti N., Clowry G.J. The early fetal development of human neocortical GABAergic interneurons. Cereb. Cortex. 2015;25:631–645. doi: 10.1093/cercor/bht254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jakovcevski I., Mayer N., Zecevic N. Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cereb. Cortex. 2011;21:1771–1782. doi: 10.1093/cercor/bhq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Letinic K., Zoncu R., Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 69.Radonjić N.V., Ayoub A.E., Memi F., Yu X., Maroof A., Jakovcevski I., Anderson S.A., Rakic P., Zecevic N. Diversity of cortical interneurons in primates: the role of the dorsal proliferative niche. Cell Rep. 2014;9:2139–2151. doi: 10.1016/j.celrep.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reinchisi G., Ijichi K., Glidden N., Jakovcevski I., Zecevic N. COUP-TFII expressing interneurons in human fetal forebrain. Cereb. Cortex. 2012;22:2820–2830. doi: 10.1093/cercor/bhr359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu X., Zecevic N. Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J. Neurosci. 2011;31:2413–2420. doi: 10.1523/JNEUROSCI.5249-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma T., Wang C., Wang L., Zhou X., Tian M., Zhang Q., Zhang Y., Li J., Liu Z., Cai Y. Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 73.Hladnik A., Džaja D., Darmopil S., Jovanov-Milošević N., Petanjek Z. Spatio-temporal extension in site of origin for cortical calretinin neurons in primates. Front. Neuroanat. 2014;8:50. doi: 10.3389/fnana.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arshad A., Vose L.R., Vinukonda G., Hu F., Yoshikawa K., Csiszar A., Brumberg J.C., Ballabh P. Extended Production of Cortical Interneurons into the Third Trimester of Human Gestation. Cereb. Cortex. 2016;26:2242–2256. doi: 10.1093/cercor/bhv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paredes M.F., James D., Gil-Perotin S., Kim H., Cotter J.A., Ng C., Sandoval K., Rowitch D.H., Xu D., McQuillen P.S. Extensive migration of young neurons into the infant human frontal lobe. Science. 2016;354:aaf7073. doi: 10.1126/science.aaf7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.LaMonica B.E., Lui J.H., Wang X., Kriegstein A.R. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr. Opin. Neurobiol. 2012;22:747–753. doi: 10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tyson J.A., Anderson S.A. The protracted maturation of human ESC-derived interneurons. Cell Cycle. 2013;12:3129–3130. doi: 10.4161/cc.26351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marín O. Human cortical interneurons take their time. Cell Stem Cell. 2013;12:497–499. doi: 10.1016/j.stem.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 79.Džaja D., Hladnik A., Bičanić I., Baković M., Petanjek Z. Neocortical calretinin neurons in primates: increase in proportion and microcircuitry structure. Front. Neuroanat. 2014;8:103. doi: 10.3389/fnana.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boldog E., Bakken T.E., Hodge R.D., Novotny M., Aevermann B.D., Baka J., Bordé S., Close J.L., Diez-Fuertes F., Ding S.L. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 2018;21:1185–1195. doi: 10.1038/s41593-018-0205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bakken T.E., Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Dalley R.A., Royall J.J., Lemon T. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–375. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng H., Shen E.H., Hohmann J.G., Oh S.W., Bernard A., Royall J.J., Glattfelder K.J., Sunkin S.M., Morris J.A., Guillozet-Bongaarts A.L. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fougerousse F., Bullen P., Herasse M., Lindsay S., Richard I., Wilson D., Suel L., Durand M., Robson S., Abitbol M. Human-mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum. Mol. Genet. 2000;9:165–173. doi: 10.1093/hmg/9.2.165. [DOI] [PubMed] [Google Scholar]
- 84.Dehay C., Kennedy H. Transcriptional regulation and alternative splicing make for better brains. Neuron. 2009;62:455–457. doi: 10.1016/j.neuron.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 85.Sousa A.M.M., Meyer K.A., Santpere G., Gulden F.O., Sestan N. Evolution of the Human Nervous System Function, Structure, and Development. Cell. 2017;170:226–247. doi: 10.1016/j.cell.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carver E.A., Stubbs L. Zooming in on the human-mouse comparative map: genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- 87.Bae B.-I., Jayaraman D., Walsh C.A. Genetic changes shaping the human brain. Dev. Cell. 2015;32:423–434. doi: 10.1016/j.devcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qiu J., McQueen J., Bilican B., Dando O., Magnani D., Punovuori K., Selvaraj B.T., Livesey M., Haghi G., Heron S. Evidence for evolutionary divergence of activity-dependent gene expression in developing neurons. eLife. 2016;5:e20337. doi: 10.7554/eLife.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Franchini L.F., Pollard K.S. Human evolution: the non-coding revolution. BMC Biol. 2017;15:89. doi: 10.1186/s12915-017-0428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gittelman R.M., Hun E., Ay F., Madeoy J., Pennacchio L., Noble W.S., Hawkins R.D., Akey J.M. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 2015;25:1245–1255. doi: 10.1101/gr.192591.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prabhakar S., Noonan J.P., Pääbo S., Rubin E.M. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 92.Prabhakar S., Visel A., Akiyama J.A., Shoukry M., Lewis K.D., Holt A., Plajzer-Frick I., Morrison H., Fitzpatrick D.R., Afzal V. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de la Torre-Ubieta L., Stein J.L., Won H., Opland C.K., Liang D., Lu D., Geschwind D.H. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell. 2018;172:289–304.e18. doi: 10.1016/j.cell.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwan K.Y., Lam M.M., Johnson M.B., Dube U., Shim S., Rašin M.R., Sousa A.M., Fertuzinhos S., Chen J.G., Arellano J.I. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu Z.X., Peng J., Su B. A human-specific mutation leads to the origin of a novel splice form of neuropsin (KLK8), a gene involved in learning and memory. Hum. Mutat. 2007;28:978–984. doi: 10.1002/humu.20547. [DOI] [PubMed] [Google Scholar]
- 97.Charrier C., Joshi K., Coutinho-Budd J., Kim J.E., Lambert N., de Marchena J., Jin W.L., Vanderhaeghen P., Ghosh A., Sassa T., Polleux F. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chou H.H., Hayakawa T., Diaz S., Krings M., Indriati E., Leakey M., Paabo S., Satta Y., Takahata N., Varki A. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc. Natl. Acad. Sci. USA. 2002;99:11736–11741. doi: 10.1073/pnas.182257399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dankwa S., Lim C., Bei A.K., Jiang R.H., Abshire J.R., Patel S.D., Goldberg J.M., Moreno Y., Kono M., Niles J.C., Duraisingh M.T. Ancient human sialic acid variant restricts an emerging zoonotic malaria parasite. Nat. Commun. 2016;7:11187. doi: 10.1038/ncomms11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stone I. The genetic disease, Hypoascorbemia. A fresh approach to an ancient disease and some of its medical implications. Acta Genet. Med. Gemellol. (Roma) 1967;16:52–62. doi: 10.1017/s1120962300013287. [DOI] [PubMed] [Google Scholar]
- 101.Wu X., Wakamiya M., Vaishnav S., Geske R., Montgomery C., Jr., Jones P., Bradley A., Caskey C.T. Hyperuricemia and urate nephropathy in urate oxidase-deficient mice. Proc. Natl. Acad. Sci. USA. 1994;91:742–746. doi: 10.1073/pnas.91.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.MacDermot K.D., Bonora E., Sykes N., Coupe A.M., Lai C.S., Vernes S.C., Vargha-Khadem F., McKenzie F., Smith R.L., Monaco A.P., Fisher S.E. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am. J. Hum. Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lai C.S., Gerrelli D., Monaco A.P., Fisher S.E., Copp A.J. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–2462. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- 104.Enard W., Przeworski M., Fisher S.E., Lai C.S., Wiebe V., Kitano T., Monaco A.P., Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 105.Shu W., Lu M.M., Zhang Y., Tucker P.W., Zhou D., Morrisey E.E. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- 106.Shu W., Cho J.Y., Jiang Y., Zhang M., Weisz D., Elder G.A., Schmeidler J., De Gasperi R., Sosa M.A., Rabidou D. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc. Natl. Acad. Sci. USA. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Groszer M., Keays D.A., Deacon R.M., de Bono J.P., Prasad-Mulcare S., Gaub S., Baum M.G., French C.A., Nicod J., Coventry J.A. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr. Biol. 2008;18:354–362. doi: 10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujita E., Tanabe Y., Shiota A., Ueda M., Suwa K., Momoi M.Y., Momoi T. Ultrasonic vocalization impairment of Foxp2 (R552H) knockin mice related to speech-language disorder and abnormality of Purkinje cells. Proc. Natl. Acad. Sci. USA. 2008;105:3117–3122. doi: 10.1073/pnas.0712298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hammerschmidt K., Schreiweis C., Minge C., Pääbo S., Fischer J., Enard W. A humanized version of Foxp2 does not affect ultrasonic vocalization in adult mice. Genes Brain Behav. 2015;14:583–590. doi: 10.1111/gbb.12237. [DOI] [PubMed] [Google Scholar]
- 110.Vernes S.C., Spiteri E., Nicod J., Groszer M., Taylor J.M., Davies K.E., Geschwind D.H., Fisher S.E. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am. J. Hum. Genet. 2007;81:1232–1250. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Batzoglou S., Pachter L., Mesirov J.P., Berger B., Lander E.S. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andersen R.E., Lim D.A. Forging our understanding of lncRNAs in the brain. Cell Tissue Res. 2018;371:55–71. doi: 10.1007/s00441-017-2711-z. [DOI] [PubMed] [Google Scholar]
- 113.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., Attenello F.J., Villalta J.E., Cho M.Y., Chen Y. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:aah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mitchell C., Silver D.L. Enhancing our brains: Genomic mechanisms underlying cortical evolution. Semin. Cell Dev. Biol. 2018;76:23–32. doi: 10.1016/j.semcdb.2017.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jobe E.M., McQuate A.L., Zhao X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Front. Neurosci. 2012;6:59. doi: 10.3389/fnins.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu H.Y., He L., Fominykh K., Yan Z., Guo S., Zhang X., Taylor M.S., Tang L., Li J., Liu J. Evolution of the human-specific microRNA miR-941. Nat. Commun. 2012;3:1145. doi: 10.1038/ncomms2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Davisson M.T., Schmidt C., Reeves R.H., Irving N.G., Akeson E.C., Harris B.S., Bronson R.T. Segmental trisomy as a mouse model for Down syndrome. Prog. Clin. Biol. Res. 1993;384:117–133. [PubMed] [Google Scholar]
- 119.Yu T., Li Z., Jia Z., Clapcote S.J., Liu C., Li S., Asrar S., Pao A., Chen R., Fan N. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum. Mol. Genet. 2010;19:2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Doherty A., Ruf S., Mulligan C., Hildreth V., Errington M.L., Cooke S., Sesay A., Modino S., Vanes L., Hernandez D. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ashley C.T., Sutcliffe J.S., Kunst C.B., Leiner H.A., Eichler E.E., Nelson D.L., Warren S.T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat. Genet. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 122.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 123.Fu Y.H., Kuhl D.P., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J., Fenwick R.G., Jr., Warren S.T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 124.Chen L.S., Tassone F., Sahota P., Hagerman P.J. The (CGG)n repeat element within the 5′ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 2003;12:3067–3074. doi: 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- 125.Hagerman R.J., Ono M.Y., Hagerman P.J. Recent advances in fragile X: a model for autism and neurodegeneration. Curr. Opin. Psychiatry. 2005;18:490–496. doi: 10.1097/01.yco.0000179485.39520.b0. [DOI] [PubMed] [Google Scholar]
- 126.Lozano R., Rosero C.A., Hagerman R.J. Fragile X spectrum disorders. Intractable Rare Dis. Res. 2014;3:134–146. doi: 10.5582/irdr.2014.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pastori C., Peschansky V.J., Barbouth D., Mehta A., Silva J.P., Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum. Genet. 2014;133:59–67. doi: 10.1007/s00439-013-1356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peschansky V.J., Pastori C., Zeier Z., Wentzel K., Velmeshev D., Magistri M., Silva J.P., Wahlestedt C. The long non-coding RNA FMR4 promotes proliferation of human neural precursor cells and epigenetic regulation of gene expression in trans. Mol. Cell. Neurosci. 2016;74:49–57. doi: 10.1016/j.mcn.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 129.Lejeune J.T.R.G.M. Le mogolisme, premier exemple d’aberration autosomique humaine. Ann. Genet. 1959;1:41–49. [Google Scholar]
- 130.Antonarakis S.E. Chromosome 21: from sequence to applications. Curr. Opin. Genet. Dev. 2001;11:241–246. doi: 10.1016/s0959-437x(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 131.Reeves R.H., Baxter L.L., Richtsmeier J.T. Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet. 2001;17:83–88. doi: 10.1016/s0168-9525(00)02172-7. [DOI] [PubMed] [Google Scholar]
- 132.Roper R.J., Reeves R.H. Understanding the basis for Down syndrome phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hattori M., Fujiyama A., Taylor T.D., Watanabe H., Yada T., Park H.S., Toyoda A., Ishii K., Totoki Y., Choi D.K., Chromosome 21 mapping and sequencing consortium The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 134.Sturgeon X., Gardiner K.J. Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm. Genome. 2011;22:261–271. doi: 10.1007/s00335-011-9321-y. [DOI] [PubMed] [Google Scholar]
- 135.Pelleri M.C., Cicchini E., Locatelli C., Vitale L., Caracausi M., Piovesan A., Rocca A., Poletti G., Seri M., Strippoli P., Cocchi G. Systematic reanalysis of partial trisomy 21 cases with or without Down syndrome suggests a small region on 21q22.13 as critical to the phenotype. Hum. Mol. Genet. 2016;25:2525–2538. doi: 10.1093/hmg/ddw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Delabar J.M., Theophile D., Rahmani Z., Chettouh Z., Blouin J.L., Prieur M., Noel B., Sinet P.M. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 137.Korenberg J.R. Toward a molecular understanding of Down syndrome. Prog. Clin. Biol. Res. 1993;384:87–115. [PubMed] [Google Scholar]
- 138.Korenberg J.R., Kawashima H., Pulst S.M., Ikeuchi T., Ogasawara N., Yamamoto K., Schonberg S.A., West R., Allen L., Magenis E. Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am. J. Hum. Genet. 1990;47:236–246. [PMC free article] [PubMed] [Google Scholar]
- 139.McCormick M.K., Schinzel A., Petersen M.B., Stetten G., Driscoll D.J., Cantu E.S., Tranebjaerg L., Mikkelsen M., Watkins P.C., Antonarakis S.E. Molecular genetic approach to the characterization of the “Down syndrome region” of chromosome 21. Genomics. 1989;5:325–331. doi: 10.1016/0888-7543(89)90065-7. [DOI] [PubMed] [Google Scholar]
- 140.Rahmani Z., Blouin J.L., Creau-Goldberg N., Watkins P.C., Mattei J.F., Poissonnier M., Prieur M., Chettouh Z., Nicole A., Aurias A. Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc. Natl. Acad. Sci. USA. 1989;86:5958–5962. doi: 10.1073/pnas.86.15.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gupta M., Dhanasekaran A.R., Gardiner K.J. Mouse models of Down syndrome: gene content and consequences. Mamm. Genome. 2016;27:538–555. doi: 10.1007/s00335-016-9661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Herault Y., Delabar J.M., Fisher E.M.C., Tybulewicz V.L.J., Yu E., Brault V. Rodent models in Down syndrome research: impact and future opportunities. Dis. Model. Mech. 2017;10:1165–1186. doi: 10.1242/dmm.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sago H., Carlson E.J., Smith D.J., Kilbridge J., Rubin E.M., Mobley W.C., Epstein C.J., Huang T.T. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc. Natl. Acad. Sci. USA. 1998;95:6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Villar A.J., Belichenko P.V., Gillespie A.M., Kozy H.M., Mobley W.C., Epstein C.J. Identification and characterization of a new Down syndrome model, Ts[Rb(12.1716)]2Cje, resulting from a spontaneous Robertsonian fusion between T(171)65Dn and mouse chromosome 12. Mamm. Genome. 2005;16:79–90. doi: 10.1007/s00335-004-2428-7. [DOI] [PubMed] [Google Scholar]
- 145.Xing Z., Li Y., Pao A., Bennett A.S., Tycko B., Mobley W.C., Yu Y.E. Mouse-based genetic modeling and analysis of Down syndrome. Br. Med. Bull. 2016;120:111–122. doi: 10.1093/bmb/ldw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rueda N., Mostany R., Pazos A., Flórez J., Martínez-Cué C. Cell proliferation is reduced in the dentate gyrus of aged but not young Ts65Dn mice, a model of Down syndrome. Neurosci. Lett. 2005;380:197–201. doi: 10.1016/j.neulet.2005.01.039. [DOI] [PubMed] [Google Scholar]
- 147.Coyle J.T., Oster-Granite M.L., Reeves R.H., Gearhart J.D. Down syndrome, Alzheimer’s disease and the trisomy 16 mouse. Trends Neurosci. 1988;11:390–394. doi: 10.1016/0166-2236(88)90075-6. [DOI] [PubMed] [Google Scholar]
- 148.Reeves R.H., Robakis N.K., Oster-Granite M.L., Wisniewski H.M., Coyle J.T., Gearhart J.D. Genetic linkage in the mouse of genes involved in Down syndrome and Alzheimer’s disease in man. Brain Res. 1987;388:215–221. doi: 10.1016/0169-328x(87)90028-3. [DOI] [PubMed] [Google Scholar]
- 149.Epstein C.J., Cox D.R., Epstein L.B. Mouse trisomy 16: an animal model of human trisomy 21 (Down syndrome) Ann. N Y Acad. Sci. 1985;450:157–168. doi: 10.1111/j.1749-6632.1985.tb21490.x. [DOI] [PubMed] [Google Scholar]
- 150.Davisson M.T., Schmidt C., Akeson E.C. Segmental trisomy of murine chromosome 16: a new model system for studying Down syndrome. Prog. Clin. Biol. Res. 1990;360:263–280. [PubMed] [Google Scholar]
- 151.Gilman S.R., Iossifov I., Levy D., Ronemus M., Wigler M., Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Auranen M., Vanhala R., Varilo T., Ayers K., Kempas E., Ylisaukko-Oja T., Sinsheimer J.S., Peltonen L., Järvelä I. A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am. J. Hum. Genet. 2002;71:777–790. doi: 10.1086/342720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Crawley J.N. Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol. 2007;17:448–459. doi: 10.1111/j.1750-3639.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ellegood J., Crawley J.N. Behavioral and Neuroanatomical Phenotypes in Mouse Models of Autism. Neurotherapeutics. 2015;12:521–533. doi: 10.1007/s13311-015-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kazdoba T.M., Leach P.T., Crawley J.N. Behavioral phenotypes of genetic mouse models of autism. Genes Brain Behav. 2016;15:7–26. doi: 10.1111/gbb.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kazdoba T.M., Leach P.T., Silverman J.L., Crawley J.N. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis. Res. 2014;3:118–133. doi: 10.5582/irdr.2014.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Berry-Kravis E.M., Lindemann L., Jonch A.E., Apostol G., Bear M.F., Carpenter R.L., Crawley J.N., Curie A., Des Portes V., Hossain F. Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat. Rev. Drug Discov. 2018;17:280–299.. doi: 10.1038/nrd.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.The Dutch-Belgian Fragile X Consortium Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 160.Mientjes E.J., Nieuwenhuizen I., Kirkpatrick L., Zu T., Hoogeveen-Westerveld M., Severijnen L., Rifé M., Willemsen R., Nelson D.L., Oostra B.A. The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol. Dis. 2006;21:549–555. doi: 10.1016/j.nbd.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 161.Berry-Kravis E. Mechanism-based treatments in neurodevelopmental disorders: fragile X syndrome. Pediatr. Neurol. 2014;50:297–302. doi: 10.1016/j.pediatrneurol.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 162.Heulens I., Suttie M., Postnov A., De Clerck N., Perrotta C.S., Mattina T., Faravelli F., Forzano F., Kooy R.F., Hammond P. Craniofacial characteristics of fragile X syndrome in mouse and man. Eur. J. Hum. Genet. 2013;21:816–823. doi: 10.1038/ejhg.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bagni C., Tassone F., Neri G., Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lokanga R.A., Entezam A., Kumari D., Yudkin D., Qin M., Smith C.B., Usdin K. Somatic expansion in mouse and human carriers of fragile X premutation alleles. Hum. Mutat. 2013;34:157–166. doi: 10.1002/humu.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao X.N., Usdin K. Ups and Downs: Mechanisms of Repeat Instability in the Fragile X-Related Disorders. Genes (Basel) 2016;7:E70. doi: 10.3390/genes7090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Veeraragavan S., Graham D., Bui N., Yuva-Paylor L.A., Wess J., Paylor R. Genetic reduction of muscarinic M4 receptor modulates analgesic response and acoustic startle response in a mouse model of fragile X syndrome (FXS) Behav. Brain Res. 2012;228:1–8. doi: 10.1016/j.bbr.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sidhu H., Dansie L.E., Hickmott P.W., Ethell D.W., Ethell I.M. Genetic removal of matrix metalloproteinase 9 rescues the symptoms of fragile X syndrome in a mouse model. J. Neurosci. 2014;34:9867–9879. doi: 10.1523/JNEUROSCI.1162-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gauducheau M., Lemaire-Mayo V., D’Amato F.R., Oddi D., Crusio W.E., Pietropaolo S. Age-specific autistic-like behaviors in heterozygous Fmr1-KO female mice. Autism Res. 2017;10:1067–1078. doi: 10.1002/aur.1743. [DOI] [PubMed] [Google Scholar]
- 169.Richtsmeier J.T., Zumwalt A., Carlson E.J., Epstein C.J., Reeves R.H. Craniofacial phenotypes in segmentally trisomic mouse models for Down syndrome. Am. J. Med. Genet. 2002;107:317–324. doi: 10.1002/ajmg.10175. [DOI] [PubMed] [Google Scholar]
- 170.Starbuck J.M., Dutka T., Ratliff T.S., Reeves R.H., Richtsmeier J.T. Overlapping trisomies for human chromosome 21 orthologs produce similar effects on skull and brain morphology of Dp(16)1Yey and Ts65Dn mice. Am. J. Med. Genet. A. 2014;164A:1981–1990. doi: 10.1002/ajmg.a.36594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Birger Y., Goldberg L., Chlon T.M., Goldenson B., Muler I., Schiby G., Jacob-Hirsch J., Rechavi G., Crispino J.D., Izraeli S. Perturbation of fetal hematopoiesis in a mouse model of Down syndrome’s transient myeloproliferative disorder. Blood. 2013;122:988–998. doi: 10.1182/blood-2012-10-460998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kirsammer G., Jilani S., Liu H., Davis E., Gurbuxani S., Le Beau M.M., Crispino J.D. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lana-Elola E., Watson-Scales S., Slender A., Gibbins D., Martineau A., Douglas C., Mohun T., Fisher E.M., Tybulewicz V.Lj. Genetic dissection of Down syndrome-associated congenital heart defects using a new mouse mapping panel. eLife. 2016;5:e11614. doi: 10.7554/eLife.11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li H., Edie S., Klinedinst D., Jeong J.S., Blackshaw S., Maslen C.L., Reeves R.H. Penetrance of Congenital Heart Disease in a Mouse Model of Down Syndrome Depends on a Trisomic Potentiator of a Disomic Modifier. Genetics. 2016;203:763–770. doi: 10.1534/genetics.116.188045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Daunhauer L.A., Fidler D.J. The down syndrome behavioral phenotype: implications for practice and research in occupational therapy. Occup. Ther. Health Care. 2011;25:7–25. doi: 10.3109/07380577.2010.535601. [DOI] [PubMed] [Google Scholar]
- 176.Fidler D.J., Nadel L. Education and children with Down syndrome: neuroscience, development, and intervention. Ment. Retard. Dev. Disabil. Res. Rev. 2007;13:262–271. doi: 10.1002/mrdd.20166. [DOI] [PubMed] [Google Scholar]
- 177.Tomaszewski B., Fidler D., Talapatra D., Riley K. Adaptive behaviour, executive function and employment in adults with Down syndrome. J. Intellect. Disabil. Res. 2018;62:41–52. doi: 10.1111/jir.12450. [DOI] [PubMed] [Google Scholar]
- 178.Lott I.T. Neurological phenotypes for Down syndrome across the life span. Prog. Brain Res. 2012;197:101–121. doi: 10.1016/B978-0-444-54299-1.00006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lott I.T., Dierssen M. Cognitive deficits and associated neurological complications in individuals with Down’s syndrome. Lancet Neurol. 2010;9:623–633. doi: 10.1016/S1474-4422(10)70112-5. [DOI] [PubMed] [Google Scholar]
- 180.Chapman R.S., Hesketh L.J. Language, cognition, and short-term memory in individuals with Down syndrome. Downs Syndr. Res. Pract. 2001;7:1–7. doi: 10.3104/reviews.108. [DOI] [PubMed] [Google Scholar]
- 181.Silverman W. Down syndrome: cognitive phenotype. Ment. Retard. Dev. Disabil. Res. Rev. 2007;13:228–236. doi: 10.1002/mrdd.20156. [DOI] [PubMed] [Google Scholar]
- 182.Dierssen M. Down syndrome: the brain in trisomic mode. Nat. Rev. Neurosci. 2012;13:844–858. doi: 10.1038/nrn3314. [DOI] [PubMed] [Google Scholar]
- 183.Davis W.E., Kelso J.A. Analysis of “invariant characteristics” in the motor control of down’s syndrome and normal subjects. J. Mot. Behav. 1982;14:194–212. doi: 10.1080/00222895.1982.10735273. [DOI] [PubMed] [Google Scholar]
- 184.Davis W.E., Sinning W.E. Muscle stiffness in down syndrome and other mentally handicapped subjects. J. Mot. Behav. 1987;19:130–144. doi: 10.1080/00222895.1987.10735404. [DOI] [PubMed] [Google Scholar]
- 185.Latash M.L. Learning motor synergies by persons with Down syndrome. J. Intellect. Disabil. Res. 2007;51:962–971. doi: 10.1111/j.1365-2788.2007.01008.x. [DOI] [PubMed] [Google Scholar]
- 186.Head E., Silverman W., Patterson D., Lott I.T. Aging and down syndrome. Curr. Gerontol. Geriatr. Res. 2012;2012:412536. doi: 10.1155/2012/412536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Prasher V.P., Kapadia H.M., Haque M.S. Season of birth: dementia in Alzheimer’s disease in adults with Down Syndrome. Int. J. Geriatr. Psychiatry. 2008;23:441–442. doi: 10.1002/gps.1883. [DOI] [PubMed] [Google Scholar]
- 188.Schupf N., Patel B., Pang D., Zigman W.B., Silverman W., Mehta P.D., Mayeux R. Elevated plasma beta-amyloid peptide Abeta(42) levels, incident dementia, and mortality in Down syndrome. Arch. Neurol. 2007;64:1007–1013. doi: 10.1001/archneur.64.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schupf N., Zigman W.B., Tang M.X., Pang D., Mayeux R., Mehta P., Silverman W. Change in plasma Aß peptides and onset of dementia in adults with Down syndrome. Neurology. 2010;75:1639–1644. doi: 10.1212/WNL.0b013e3181fb448b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Castro P., Zaman S., Holland A. Alzheimer’s disease in people with Down’s syndrome: the prospects for and the challenges of developing preventative treatments. J. Neurol. 2017;264:804–813. doi: 10.1007/s00415-016-8308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Neale N., Padilla C., Fonseca L.M., Holland T., Zaman S. Neuroimaging and other modalities to assess Alzheimer’s disease in Down syndrome. Neuroimage Clin. 2017;17:263–271. doi: 10.1016/j.nicl.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zis P., Strydom A. Clinical aspects and biomarkers of Alzheimer’s disease in Down syndrome. Free Radic. Biol. Med. 2018;114:3–9. doi: 10.1016/j.freeradbiomed.2017.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Belichenko P.V., Kleschevnikov A.M., Becker A., Wagner G.E., Lysenko L.V., Yu Y.E., Mobley W.C. Down Syndrome Cognitive Phenotypes Modeled in Mice Trisomic for All HSA 21 Homologues. PLoS ONE. 2015;10:e0134861. doi: 10.1371/journal.pone.0134861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hall J.H., Wiseman F.K., Fisher E.M., Tybulewicz V.L., Harwood J.L., Good M.A. Tc1 mouse model of trisomy-21 dissociates properties of short- and long-term recognition memory. Neurobiol. Learn. Mem. 2016;130:118–128. doi: 10.1016/j.nlm.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Martínez-Cué C., Martínez P., Rueda N., Vidal R., García S., Vidal V., Corrales A., Montero J.A., Pazos Á., Flórez J. Reducing GABAA α5 receptor-mediated inhibition rescues functional and neuromorphological deficits in a mouse model of down syndrome. J. Neurosci. 2013;33:3953–3966. doi: 10.1523/JNEUROSCI.1203-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Roubertoux P.L., Baril N., Cau P., Scajola C., Ghata A., Bartoli C., Bourgeois P., Christofaro J.D., Tordjman S., Carlier M. Differential Brain, Cognitive and Motor Profiles Associated with Partial Trisomy. Modeling Down Syndrome in Mice. Behav. Genet. 2017;47:305–322. doi: 10.1007/s10519-017-9835-5. [DOI] [PubMed] [Google Scholar]
- 197.Smith-Hicks C.L., Cai P., Savonenko A.V., Reeves R.H., Worley P.F. Increased Sparsity of Hippocampal CA1 Neuronal Ensembles in a Mouse Model of Down Syndrome Assayed by Arc Expression. Front. Neural Circuits. 2017;11:6. doi: 10.3389/fncir.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Whitney K.N., Wenger G.R. Impulsivity and motor activity in aged, male Ts65Dn mice. Exp. Clin. Psychopharmacol. 2013;21:345–354. doi: 10.1037/a0033965. [DOI] [PubMed] [Google Scholar]
- 199.Witton J., Padmashri R., Zinyuk L.E., Popov V.I., Kraev I., Line S.J., Jensen T.P., Tedoldi A., Cummings D.M., Tybulewicz V.L.J. Hippocampal circuit dysfunction in the Tc1 mouse model of Down syndrome. Nat. Neurosci. 2015;18:1291–1298. doi: 10.1038/nn.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zampieri B.L., Fernandez F., Pearson J.N., Stasko M.R., Costa A.C. Ultrasonic vocalizations during male-female interaction in the mouse model of Down syndrome Ts65Dn. Physiol. Behav. 2014;128:119–125. doi: 10.1016/j.physbeh.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 201.Belichenko N.P., Belichenko P.V., Kleschevnikov A.M., Salehi A., Reeves R.H., Mobley W.C. The “Down syndrome critical region” is sufficient in the mouse model to confer behavioral, neurophysiological, and synaptic phenotypes characteristic of Down syndrome. J. Neurosci. 2009;29:5938–5948. doi: 10.1523/JNEUROSCI.1547-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hyde L.A., Frisone D.F., Crnic L.S. Ts65Dn mice, a model for Down syndrome, have deficits in context discrimination learning suggesting impaired hippocampal function. Behav. Brain Res. 2001;118:53–60. doi: 10.1016/s0166-4328(00)00313-2. [DOI] [PubMed] [Google Scholar]
- 203.Kleschevnikov A.M., Belichenko P.V., Gall J., George L., Nosheny R., Maloney M.T., Salehi A., Mobley W.C. Increased efficiency of the GABAA and GABAB receptor-mediated neurotransmission in the Ts65Dn mouse model of Down syndrome. Neurobiol. Dis. 2012;45:683–691. doi: 10.1016/j.nbd.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Morice E., Andreae L.C., Cooke S.F., Vanes L., Fisher E.M., Tybulewicz V.L., Bliss T.V. Preservation of long-term memory and synaptic plasticity despite short-term impairments in the Tc1 mouse model of Down syndrome. Learn. Mem. 2008;15:492–500. doi: 10.1101/lm.969608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu C., Belichenko P.V., Zhang L., Fu D., Kleschevnikov A.M., Baldini A., Antonarakis S.E., Mobley W.C., Yu Y.E. Mouse models for Down syndrome-associated developmental cognitive disabilities. Dev. Neurosci. 2011;33:404–413. doi: 10.1159/000329422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cramer N., Galdzicki Z. From abnormal hippocampal synaptic plasticity in down syndrome mouse models to cognitive disability in down syndrome. Neural Plast. 2012;2012:101542. doi: 10.1155/2012/101542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Garner C.C., Wetmore D.Z. Synaptic pathology of Down syndrome. Adv. Exp. Med. Biol. 2012;970:451–468. doi: 10.1007/978-3-7091-0932-8_20. [DOI] [PubMed] [Google Scholar]
- 208.Holtzman D.M., Santucci D., Kilbridge J., Chua-Couzens J., Fontana D.J., Daniels S.E., Johnson R.M., Chen K., Sun Y., Carlson E. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc. Natl. Acad. Sci. USA. 1996;93:13333–13338. doi: 10.1073/pnas.93.23.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rueda N., Flórez J., Martínez-Cué C. Mouse models of Down syndrome as a tool to unravel the causes of mental disabilities. Neural Plast. 2012;2012:584071. doi: 10.1155/2012/584071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Goodliffe J.W., Olmos-Serrano J.L., Aziz N.M., Pennings J.L., Guedj F., Bianchi D.W., Haydar T.F. Absence of Prenatal Forebrain Defects in the Dp(16)1Yey/+ Mouse Model of Down Syndrome. J. Neurosci. 2016;36:2926–2944. doi: 10.1523/JNEUROSCI.2513-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sabaratnam M. Pathological and neuropathological findings in two males with fragile-X syndrome. J. Intellect. Disabil. Res. 2000;44:81–85. doi: 10.1046/j.1365-2788.2000.00261.x. [DOI] [PubMed] [Google Scholar]
- 212.Jäkälä P., Hänninen T., Ryynänen M., Laakso M., Partanen K., Mannermaa A., Soininen H. Fragile-X: neuropsychological test performance, CGG triplet repeat lengths, and hippocampal volumes. J. Clin. Invest. 1997;100:331–338. doi: 10.1172/JCI119538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Greco C.M., Navarro C.S., Hunsaker M.R., Maezawa I., Shuler J.F., Tassone F., Delany M., Au J.W., Berman R.F., Jin L.W. Neuropathologic features in the hippocampus and cerebellum of three older men with fragile X syndrome. Mol. Autism. 2011;2:2. doi: 10.1186/2040-2392-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kooy R.F., Reyniers E., Verhoye M., Sijbers J., Bakker C.E., Oostra B.A., Willems P.J., Van Der Linden A. Neuroanatomy of the fragile X knockout mouse brain studied using in vivo high resolution magnetic resonance imaging. Eur. J. Hum. Genet. 1999;7:526–532. doi: 10.1038/sj.ejhg.5200348. [DOI] [PubMed] [Google Scholar]
- 215.Ellegood J., Pacey L.K., Hampson D.R., Lerch J.P., Henkelman R.M. Anatomical phenotyping in a mouse model of fragile X syndrome with magnetic resonance imaging. Neuroimage. 2010;53:1023–1029. doi: 10.1016/j.neuroimage.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 216.Lai J.K., Lerch J.P., Doering L.C., Foster J.A., Ellegood J. Regional brain volumes changes in adult male FMR1-KO mouse on the FVB strain. Neuroscience. 2016;318:12–21. doi: 10.1016/j.neuroscience.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 217.Hinton V.J., Brown W.T., Wisniewski K., Rudelli R.D. Analysis of neocortex in three males with the fragile X syndrome. Am. J. Med. Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- 218.Irwin S.A., Patel B., Idupulapati M., Harris J.B., Crisostomo R.A., Larsen B.P., Kooy F., Willems P.J., Cras P., Kozlowski P.B. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 219.Rudelli R.D., Brown W.T., Wisniewski K., Jenkins E.C., Laure-Kamionowska M., Connell F., Wisniewski H.M. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol. 1985;67:289–295. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- 220.Irwin S.A., Idupulapati M., Gilbert M.E., Harris J.B., Chakravarti A.B., Rogers E.J., Crisostomo R.A., Larsen B.P., Mehta A., Alcantara C.J. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am. J. Med. Genet. 2002;111:140–146. doi: 10.1002/ajmg.10500. [DOI] [PubMed] [Google Scholar]
- 221.Comery T.A., Harris J.B., Willems P.J., Oostra B.A., Irwin S.A., Weiler I.J., Greenough W.T. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl. Acad. Sci. USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Grossman A.W., Elisseou N.M., McKinney B.C., Greenough W.T. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res. 2006;1084:158–164. doi: 10.1016/j.brainres.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 223.Wang J., Ethridge L.E., Mosconi M.W., White S.P., Binder D.K., Pedapati E.V., Erickson C.A., Byerly M.J., Sweeney J.A. A resting EEG study of neocortical hyperexcitability and altered functional connectivity in fragile X syndrome. J. Neurodev. Disord. 2017;9:11. doi: 10.1186/s11689-017-9191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Heard T.T., Ramgopal S., Picker J., Lincoln S.A., Rotenberg A., Kothare S.V. EEG abnormalities and seizures in genetically diagnosed Fragile X syndrome. Int. J. Dev. Neurosci. 2014;38:155–160. doi: 10.1016/j.ijdevneu.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 225.van der Molen M.J., Stam C.J., van der Molen M.W. Resting-state EEG oscillatory dynamics in fragile X syndrome: abnormal functional connectivity and brain network organization. PLoS ONE. 2014;9:e88451. doi: 10.1371/journal.pone.0088451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Van der Molen M.J., Van der Molen M.W. Reduced alpha and exaggerated theta power during the resting-state EEG in fragile X syndrome. Biol. Psychol. 2013;92:216–219. doi: 10.1016/j.biopsycho.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 227.Sinclair D., Featherstone R., Naschek M., Nam J., Du A., Wright S., Pance K., Melnychenko O., Weger R., Akuzawa S. GABA-B Agonist Baclofen Normalizes Auditory-Evoked Neural Oscillations and Behavioral Deficits in the Fmr1 Knockout Mouse Model of Fragile X Syndrome. eNeuro. 2017;4 doi: 10.1523/ENEURO.0380-16.2017. ENEURO.0380-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Contractor A., Klyachko V.A., Portera-Cailliau C. Altered Neuronal and Circuit Excitability in Fragile X Syndrome. Neuron. 2015;87:699–715. doi: 10.1016/j.neuron.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wisniewski K.E. Down syndrome children often have brain with maturation delay, retardation of growth, and cortical dysgenesis. Am. J. Med. Genet. Suppl. 1990;7:274–281. doi: 10.1002/ajmg.1320370755. [DOI] [PubMed] [Google Scholar]
- 230.Schmidt-Sidor B., Wisniewski K.E., Shepard T.H., Sersen E.A. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin. Neuropathol. 1990;9:181–190. [PubMed] [Google Scholar]
- 231.Kesslak J.P., Nagata S.F., Lott I., Nalcioglu O. Magnetic resonance imaging analysis of age-related changes in the brains of individuals with Down’s syndrome. Neurology. 1994;44:1039–1045. doi: 10.1212/wnl.44.6.1039. [DOI] [PubMed] [Google Scholar]
- 232.Colon E.J. The structure of the cerebral cortex in Down’s syndrome: a quantitative analysis. Neuropediatrics. 1972;3:362–376. [Google Scholar]
- 233.Becker L., Mito T., Takashima S., Onodera K. Growth and development of the brain in Down syndrome. Prog. Clin. Biol. Res. 1991;373:133–152. [PubMed] [Google Scholar]
- 234.Coyle J.T., Oster-Granite M.L., Gearhart J.D. The neurobiologic consequences of Down syndrome. Brain Res. Bull. 1986;16:773–787. doi: 10.1016/0361-9230(86)90074-2. [DOI] [PubMed] [Google Scholar]
- 235.Benda C.E. Grune and Stratton; 1947. Mongolism and cretinism. [Google Scholar]
- 236.Crome L.S.J. J&A Churchill; 1967. Pathology of mental retardation. [Google Scholar]
- 237.Davidoff L.M. The brain in mongolian idiocy: a report of ten cases. Arch. Neurol. Psychiatry. 1928;20:1229–1257. [Google Scholar]
- 238.Golden J.A., Hyman B.T. Development of the superior temporal neocortex is anomalous in trisomy 21. J. Neuropathol. Exp. Neurol. 1994;53:513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- 239.Ross M.H., Galaburda A.M., Kemper T.L. Down’s syndrome: is there a decreased population of neurons? Neurology. 1984;34:909–916. doi: 10.1212/wnl.34.7.909. [DOI] [PubMed] [Google Scholar]
- 240.Weitzdoerfer R., Dierssen M., Fountoulakis M., Lubec G. Fetal life in Down syndrome starts with normal neuronal density but impaired dendritic spines and synaptosomal structure. In: Lubec G., editor. Protein Expression in Down Syndrome Brain. Springer; 2001. pp. 59–70. [DOI] [PubMed] [Google Scholar]
- 241.Contestabile A., Fila T., Ceccarelli C., Bonasoni P., Bonapace L., Santini D., Bartesaghi R., Ciani E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus. 2007;17:665–678. doi: 10.1002/hipo.20308. [DOI] [PubMed] [Google Scholar]
- 242.Guidi S., Bonasoni P., Ceccarelli C., Santini D., Gualtieri F., Ciani E., Bartesaghi R. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008;18:180–197. doi: 10.1111/j.1750-3639.2007.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Guidi S., Giacomini A., Stagni F., Emili M., Uguagliati B., Bonasoni M.P., Bartesaghi R. Abnormal development of the inferior temporal region in fetuses with Down syndrome. Brain Pathol. 2018 doi: 10.1111/bpa.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Stagni F., Giacomini A., Emili M., Guidi S., Bartesaghi R. Neurogenesis impairment: An early developmental defect in Down syndrome. Free Radic. Biol. Med. 2018;114:15–32. doi: 10.1016/j.freeradbiomed.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 245.Chakrabarti L., Galdzicki Z., Haydar T.F. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2007;27:11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Haydar T.F., Blue M.E., Molliver M.E., Krueger B.K., Yarowsky P.J. Consequences of trisomy 16 for mouse brain development: corticogenesis in a model of Down syndrome. J. Neurosci. 1996;16:6175–6182. doi: 10.1523/JNEUROSCI.16-19-06175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sweeney J.E., Höhmann C.F., Oster-Granite M.L., Coyle J.T. Neurogenesis of the basal forebrain in euploid and trisomy 16 mice: an animal model for developmental disorders in Down syndrome. Neuroscience. 1989;31:413–425. doi: 10.1016/0306-4522(89)90384-9. [DOI] [PubMed] [Google Scholar]
- 248.Chakrabarti L., Best T.K., Cramer N.P., Carney R.S., Isaac J.T., Galdzicki Z., Haydar T.F. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat. Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Petit T.L., LeBoutillier J.C., Alfano D.P., Becker L.E. Synaptic development in the human fetus: a morphometric analysis of normal and Down’s syndrome neocortex. Exp. Neurol. 1984;83:13–23. doi: 10.1016/0014-4886(84)90041-4. [DOI] [PubMed] [Google Scholar]
- 250.Takashima S., Iida K., Mito T., Arima M. Dendritic and histochemical development and ageing in patients with Down’s syndrome. J. Intellect. Disabil. Res. 1994;38:265–273. doi: 10.1111/j.1365-2788.1994.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 251.Wisniewski K.E., Laure-Kamionowska M., Wisniewski H.M. Evidence of arrest of neurogenesis and synaptogenesis in brains of patients with Down’s syndrome. N. Engl. J. Med. 1984;311:1187–1188. doi: 10.1056/NEJM198411013111818. [DOI] [PubMed] [Google Scholar]
- 252.Kaufmann W.E., Moser H.W. Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 253.Marin-Padilla M. Structural abnormalities of the cerebral cortex in human chromosomal aberrations: a Golgi study. Brain Res. 1972;44:625–629. doi: 10.1016/0006-8993(72)90324-1. [DOI] [PubMed] [Google Scholar]
- 254.Marin-Padilla M. Pyramidal cell abnormalities in the motor cortex of a child with Down’s syndrome. A Golgi study. J. Comp. Neurol. 1976;167:63–81. doi: 10.1002/cne.901670105. [DOI] [PubMed] [Google Scholar]
- 255.Purpura D.P. Normal and aberrant neuronal development in the cerebral cortex of human fetus and young infant. UCLA Forum Med. Sci. 1975;1975:141–169. doi: 10.1016/b978-0-12-139050-1.50014-8. [DOI] [PubMed] [Google Scholar]
- 256.Belichenko P.V., Kleschevnikov A.M., Salehi A., Epstein C.J., Mobley W.C. Synaptic and cognitive abnormalities in mouse models of Down syndrome: exploring genotype-phenotype relationships. J. Comp. Neurol. 2007;504:329–345. doi: 10.1002/cne.21433. [DOI] [PubMed] [Google Scholar]
- 257.Haas M.A., Bell D., Slender A., Lana-Elola E., Watson-Scales S., Fisher E.M.C., Tybulewicz V.L.J., Guillemot F. Alterations to dendritic spine morphology, but not dendrite patterning, of cortical projection neurons in Tc1 and Ts1Rhr mouse models of Down syndrome. PLoS ONE. 2013;8:e78561. doi: 10.1371/journal.pone.0078561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dierssen M., Benavides-Piccione R., Martínez-Cué C., Estivill X., Flórez J., Elston G.N., DeFelipe J. Alterations of neocortical pyramidal cell phenotype in the Ts65Dn mouse model of Down syndrome: effects of environmental enrichment. Cereb. Cortex. 2003;13:758–764. doi: 10.1093/cercor/13.7.758. [DOI] [PubMed] [Google Scholar]
- 259.Bear M.F., Huber K.M., Warren S.T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 260.Braat S., Kooy R.F. Insights into GABAAergic system deficits in fragile X syndrome lead to clinical trials. Neuropharmacology. 2015;88:48–54. doi: 10.1016/j.neuropharm.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 261.Lohith T.G., Osterweil E.K., Fujita M., Jenko K.J., Bear M.F., Innis R.B. Is metabotropic glutamate receptor 5 upregulated in prefrontal cortex in fragile X syndrome? Mol. Autism. 2013;4:15. doi: 10.1186/2040-2392-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Borrie S.C., Brems H., Legius E., Bagni C. Cognitive Dysfunctions in Intellectual Disabilities: The Contributions of the Ras-MAPK and PI3K-AKT-mTOR Pathways. Annu. Rev. Genomics Hum. Genet. 2017;18:115–142. doi: 10.1146/annurev-genom-091416-035332. [DOI] [PubMed] [Google Scholar]
- 263.Wang T., Bray S.M., Warren S.T. New perspectives on the biology of fragile X syndrome. Curr. Opin. Genet. Dev. 2012;22:256–263. doi: 10.1016/j.gde.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Becker W., Soppa U., Tejedor F.J. DYRK1A: a potential drug target for multiple Down syndrome neuropathologies. CNS Neurol. Disord. Drug Targets. 2014;13:26–33. doi: 10.2174/18715273113126660186. [DOI] [PubMed] [Google Scholar]
- 265.Duchon A., Herault Y. DYRK1A, a Dosage-Sensitive Gene Involved in Neurodevelopmental Disorders, Is a Target for Drug Development in Down Syndrome. Front. Behav. Neurosci. 2016;10:104. doi: 10.3389/fnbeh.2016.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Stringer M., Goodlett C.R., Roper R.J. Targeting trisomic treatments: optimizing Dyrk1a inhibition to improve Down syndrome deficits. Mol. Genet. Genomic Med. 2017;5:451–465. doi: 10.1002/mgg3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Bellmaine S.F., Ovchinnikov D.A., Manallack D.T., Cuddy C.E., Elefanty A.G., Stanley E.G., Wolvetang E.J., Williams S.J., Pera M. Inhibition of DYRK1A disrupts neural lineage specificationin human pluripotent stem cells. eLife. 2017;6:e24502. doi: 10.7554/eLife.24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ermak G., Davies K.J.A. Chronic high levels of the RCAN1-1 protein may promote neurodegeneration and Alzheimer disease. Free Radic. Biol. Med. 2013;62:47–51. doi: 10.1016/j.freeradbiomed.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Li Y., Wang J., Zhou Y., Li D., Xiong Z.Q. Rcan1 deficiency impairs neuronal migration and causes periventricular heterotopia. J. Neurosci. 2015;35:610–620. doi: 10.1523/JNEUROSCI.1003-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Patel A., Yamashita N., Ascaño M., Bodmer D., Boehm E., Bodkin-Clarke C., Ryu Y.K., Kuruvilla R. RCAN1 links impaired neurotrophin trafficking to aberrant development of the sympathetic nervous system in Down syndrome. Nat. Commun. 2015;6:10119. doi: 10.1038/ncomms10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sun X., Wu Y., Herculano B., Song W. RCAN1 overexpression exacerbates calcium overloading-induced neuronal apoptosis. PLoS ONE. 2014;9:e95471. doi: 10.1371/journal.pone.0095471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Adorno M., Sikandar S., Mitra S.S., Kuo A., Nicolis Di Robilant B., Haro-Acosta V., Ouadah Y., Quarta M., Rodriguez J., Qian D. Usp16 contributes to somatic stem-cell defects in Down’s syndrome. Nature. 2013;501:380–384. doi: 10.1038/nature12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zorrilla de San Martin J., Delabar J.M., Bacci A., Potier M.C. GABAergic over-inhibition, a promising hypothesis for cognitive deficits in Down syndrome. Free Radic. Biol. Med. 2018;114:33–39. doi: 10.1016/j.freeradbiomed.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 274.Martínez-Cué C., Delatour B., Potier M.C. Treating enhanced GABAergic inhibition in Down syndrome: use of GABA α5-selective inverse agonists. Neurosci. Biobehav. Rev. 2014;46:218–227. doi: 10.1016/j.neubiorev.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 275.Potier M.C., Braudeau J., Dauphinot L., Delatour B. Reducing GABAergic inhibition restores cognitive functions in a mouse model of Down syndrome. CNS Neurol. Disord. Drug Targets. 2014;13:8–15. doi: 10.2174/18715273113126660185. [DOI] [PubMed] [Google Scholar]
- 276.Gardiner K.J. Molecular basis of pharmacotherapies for cognition in Down syndrome. Trends Pharmacol. Sci. 2010;31:66–73. doi: 10.1016/j.tips.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Deidda G., Parrini M., Naskar S., Bozarth I.F., Contestabile A., Cancedda L. Reversing excitatory GABAAR signaling restores synaptic plasticity and memory in a mouse model of Down syndrome. Nat. Med. 2015;21:318–326. doi: 10.1038/nm.3827. [DOI] [PubMed] [Google Scholar]
- 278.Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 279.Carpenter M.K., Cui X., Hu Z.Y., Jackson J., Sherman S., Seiger A., Wahlberg L.U. In vitro expansion of a multipotent population of human neural progenitor cells. Exp. Neurol. 1999;158:265–278. doi: 10.1006/exnr.1999.7098. [DOI] [PubMed] [Google Scholar]
- 280.Svendsen C.N., ter Borg M.G., Armstrong R.J., Rosser A.E., Chandran S., Ostenfeld T., Caldwell M.A. A new method for the rapid and long term growth of human neural precursor cells. J. Neurosci. Methods. 1998;85:141–152. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- 281.Vescovi A.L., Parati E.A., Gritti A., Poulin P., Ferrario M., Wanke E., Frölichsthal-Schoeller P., Cova L., Arcellana-Panlilio M., Colombo A., Galli R. Isolation and cloning of multipotential stem cells from the embryonic human CNS and establishment of transplantable human neural stem cell lines by epigenetic stimulation. Exp. Neurol. 1999;156:71–83. doi: 10.1006/exnr.1998.6998. [DOI] [PubMed] [Google Scholar]
- 282.Caldwell M.A., He X., Wilkie N., Pollack S., Marshall G., Wafford K.A., Svendsen C.N. Growth factors regulate the survival and fate of cells derived from human neurospheres. Nat. Biotechnol. 2001;19:475–479. doi: 10.1038/88158. [DOI] [PubMed] [Google Scholar]
- 283.Bhattacharyya A., McMillan E., Chen S.I., Wallace K., Svendsen C.N. A critical period in cortical interneuron neurogenesis in down syndrome revealed by human neural progenitor cells. Dev. Neurosci. 2009;31:497–510. doi: 10.1159/000236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Bhattacharyya A., Svendsen C.N. Human neural stem cells: a new tool for studying cortical development in Down’s syndrome. Genes Brain Behav. 2003;2:179–186. doi: 10.1034/j.1601-183x.2003.00025.x. [DOI] [PubMed] [Google Scholar]
- 285.Bahn S., Mimmack M., Ryan M., Caldwell M.A., Jauniaux E., Starkey M., Svendsen C.N., Emson P. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down’s syndrome: a gene expression study. Lancet. 2002;359:310–315. doi: 10.1016/S0140-6736(02)07497-4. [DOI] [PubMed] [Google Scholar]
- 286.Esposito G., Imitola J., Lu J., De Filippis D., Scuderi C., Ganesh V.S., Folkerth R., Hecht J., Shin S., Iuvone T. Genomic and functional profiling of human Down syndrome neural progenitors implicates S100B and aquaporin 4 in cell injury. Hum. Mol. Genet. 2008;17:440–457. doi: 10.1093/hmg/ddm322. [DOI] [PubMed] [Google Scholar]
- 287.Lu J., Esposito G., Scuderi C., Steardo L., Delli-Bovi L.C., Hecht J.L., Dickinson B.C., Chang C.J., Mori T., Sheen V. S100B and APP promote a gliocentric shift and impaired neurogenesis in Down syndrome neural progenitors. PLoS ONE. 2011;6:e22126. doi: 10.1371/journal.pone.0022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Gearhart J. New potential for human embryonic stem cells. Science. 1998;282:1061–1062. doi: 10.1126/science.282.5391.1061. [DOI] [PubMed] [Google Scholar]
- 289.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 290.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 291.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 292.Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang S.C. Neural subtype specification from embryonic stem cells. Brain Pathol. 2006;16:132–142. doi: 10.1111/j.1750-3639.2006.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zhang S.C., Wernig M., Duncan I.D., Brüstle O., Thomson J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 295.Yan Y., Yang D., Zarnowska E.D., Du Z., Werbel B., Valliere C., Pearce R.A., Thomson J.A., Zhang S.C. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Li X.J., Zhang S.C. In vitro differentiation of neural precursors from human embryonic stem cells. Methods Mol. Biol. 2006;331:169–177. doi: 10.1385/1-59745-046-4:168. [DOI] [PubMed] [Google Scholar]
- 297.Li X.J., Hu B.Y., Jones S.A., Zhang Y.S., Lavaute T., Du Z.W., Zhang S.C. Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells. 2008;26:886–893. doi: 10.1634/stemcells.2007-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hu B.Y., Du Z.W., Zhang S.C. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat. Protoc. 2009;4:1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Krencik R., Weick J.P., Liu Y., Zhang Z.J., Zhang S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Liu Y., Weick J.P., Liu H., Krencik R., Zhang X., Ma L., Zhou G.M., Ayala M., Zhang S.C. Medial ganglionic eminence-like cells derived from human embryonic stem cells correct learning and memory deficits. Nat. Biotechnol. 2013;31:440–447. doi: 10.1038/nbt.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Yuan F., Fang K.H., Cao S.Y., Qu Z.Y., Li Q., Krencik R., Xu M., Bhattacharyya A., Su Y.W., Zhu D.Y., Liu Y. Efficient generation of region-specific forebrain neurons from human pluripotent stem cells under highly defined condition. Sci. Rep. 2015;5:18550. doi: 10.1038/srep18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Pickering S.J., Braude P.R., Patel M., Burns C.J., Trussler J., Bolton V., Minger S. Preimplantation genetic diagnosis as a novel source of embryos for stem cell research. Reprod. Biomed. Online. 2003;7:353–364. doi: 10.1016/s1472-6483(10)61877-9. [DOI] [PubMed] [Google Scholar]
- 304.Ananiev G., Williams E.C., Li H., Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS ONE. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Cheung A.Y., Horvath L.M., Grafodatskaya D., Pasceri P., Weksberg R., Hotta A., Carrel L., Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum. Mol. Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Séguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 307.Kim K.Y., Hysolli E., Park I.H. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Armstrong L.C., Westlake G., Snow J.P., Cawthon B., Armour E., Bowman A.B., Ess K.C. Heterozygous loss of TSC2 alters p53 signaling and human stem cell reprogramming. Hum. Mol. Genet. 2017;26:4629–4641. doi: 10.1093/hmg/ddx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Julian L.M., Delaney S.P., Wang Y., Goldberg A.A., Doré C., Yockell-Lelièvre J., Tam R.Y., Giannikou K., McMurray F., Shoichet M.S. Human Pluripotent Stem Cell-Derived TSC2-Haploinsufficient Smooth Muscle Cells Recapitulate Features of Lymphangioleiomyomatosis. Cancer Res. 2017;77:5491–5502. doi: 10.1158/0008-5472.CAN-17-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Li Y., Cao J., Chen M., Li J., Sun Y., Zhang Y., Zhu Y., Wang L., Zhang C. Abnormal Neural Progenitor Cells Differentiated from Induced Pluripotent Stem Cells Partially Mimicked Development of TSC2 Neurological Abnormalities. Stem Cell Reports. 2017;8:883–893. doi: 10.1016/j.stemcr.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Doers M.E., Musser M.T., Nichol R., Berndt E.R., Baker M., Gomez T.M., Zhang S.C., Abbeduto L., Bhattacharyya A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Eiges R., Urbach A., Malcov M., Frumkin T., Schwartz T., Amit A., Yaron Y., Eden A., Yanuka O., Benvenisty N., Ben-Yosef D. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 314.Sheridan S.D., Theriault K.M., Reis S.A., Zhou F., Madison J.M., Daheron L., Loring J.F., Haggarty S.J. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Urbach A., Bar-Nur O., Daley G.Q., Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Boland M.J., Nazor K.L., Tran H.T., Szücs A., Lynch C.L., Paredes R., Tassone F., Sanna P.P., Hagerman R.J., Loring J.F. Molecular analyses of neurogenic defects in a human pluripotent stem cell model of fragile X syndrome. Brain. 2017;140:582–598. doi: 10.1093/brain/aww357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Yoon K.J., Nguyen H.N., Ursini G., Zhang F., Kim N.S., Wen Z., Makri G., Nauen D., Shin J.H., Park Y. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Martins-Taylor K., Hsiao J.S., Chen P.F., Glatt-Deeley H., De Smith A.J., Blakemore A.I., Lalande M., Chamberlain S.J. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum. Mol. Genet. 2014;23:2364–2373. doi: 10.1093/hmg/ddt628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Urraca N., Hope K., Victor A.K., Belgard T.G., Memon R., Goorha S., Valdez C., Tran Q.T., Sanchez S., Ramirez J. Significant transcriptional changes in 15q duplication but not Angelman syndrome deletion stem cell-derived neurons. Mol. Autism. 2018;9:6. doi: 10.1186/s13229-018-0191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Briggs J.A., Sun J., Shepherd J., Ovchinnikov D.A., Chung T.L., Nayler S.P., Kao L.P., Morrow C.A., Thakar N.Y., Soo S.Y. Integration-Free Induced Pluripotent Stem Cells Model Genetic and Neural Developmental Features of Down Syndrome Etiology. Stem Cells. 2013;31:467–478. doi: 10.1002/stem.1297. [DOI] [PubMed] [Google Scholar]
- 321.Hibaoui Y., Feki A. Concise Review: Methods and Cell Types Used to Generate Down Syndrome Induced Pluripotent Stem Cells. J. Clin. Med. 2015;4:696–714. doi: 10.3390/jcm4040696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Weick J.P., Held D.L., Bonadurer G.F., 3rd, Doers M.E., Liu Y., Maguire C., Clark A., Knackert J.A., Molinarolo K., Musser M. Deficits in human trisomy 21 iPSCs and neurons. Proc. Natl. Acad. Sci. USA. 2013;110:9962–9967. doi: 10.1073/pnas.1216575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Murray A., Letourneau A., Canzonetta C., Stathaki E., Gimelli S., Sloan-Bena F., Abrehart R., Goh P., Lim S., Baldo C. Brief report: isogenic induced pluripotent stem cell lines from an adult with mosaic down syndrome model accelerated neuronal ageing and neurodegeneration. Stem Cells. 2015;33:2077–2084. doi: 10.1002/stem.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Verlinsky Y., Strelchenko N., Kukharenko V., Rechitsky S., Verlinsky O., Galat V., Kuliev A. Human embryonic stem cell lines with genetic disorders. Reprod. Biomed. Online. 2005;10:105–110. doi: 10.1016/s1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- 326.Ben-Yosef D., Malcov M., Eiges R. PGD-derived human embryonic stem cell lines as a powerful tool for the study of human genetic disorders. Mol. Cell. Endocrinol. 2008;282:153–158. doi: 10.1016/j.mce.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 327.Telias M., Segal M., Ben-Yosef D. Neural differentiation of Fragile X human Embryonic Stem Cells reveals abnormal patterns of development despite successful neurogenesis. Dev. Biol. 2013;374:32–45. doi: 10.1016/j.ydbio.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 328.Telias M., Kuznitsov-Yanovsky L., Segal M., Ben-Yosef D. Functional Deficiencies in Fragile X Neurons Derived from Human Embryonic Stem Cells. J. Neurosci. 2015;35:15295–15306. doi: 10.1523/JNEUROSCI.0317-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Telias M., Segal M., Ben-Yosef D. Immature Responses to GABA in Fragile X Neurons Derived from Human Embryonic Stem Cells. Front. Cell. Neurosci. 2016;10:121. doi: 10.3389/fncel.2016.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Mansour A.A., Goncalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Bhattacharyya A., Zhao X. Human pluripotent stem cell models of Fragile X syndrome. Mol. Cell. Neurosci. 2016;73:43–51. doi: 10.1016/j.mcn.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Li M., Zhao H., Ananiev G.E., Musser M., Ness K.H., Maglaque D.L., Saha K., Bhattacharyya A., Zhao X. Establishment of reporter lines for detecting fragile X mental retardation (FMR1) gene reactivation in human neural cells. Stem Cells. 2017;35:158–169. doi: 10.1002/stem.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Brown V., Jin P., Ceman S., Darnell J.C., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 334.Darnell J.C., Mostovetsky O., Darnell R.B. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 335.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ethridge L.E., White S.P., Mosconi M.W., Wang J., Pedapati E.V., Erickson C.A., Byerly M.J., Sweeney J.A. Neural synchronization deficits linked to cortical hyper-excitability and auditory hypersensitivity in fragile X syndrome. Mol. Autism. 2017;8:22. doi: 10.1186/s13229-017-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Biancotti J.C., Narwani K., Buehler N., Mandefro B., Golan-Lev T., Yanuka O., Clark A., Hill D., Benvenisty N., Lavon N. Human embryonic stem cells as models for aneuploid chromosomal syndromes. Stem Cells. 2010;28:1530–1540. doi: 10.1002/stem.483. [DOI] [PubMed] [Google Scholar]
- 338.Bosman A., Letourneau A., Sartiani L., Del Lungo M., Ronzoni F., Kuziakiv R., Tohonen V., Zucchelli M., Santoni F., Guipponi M. Perturbations of heart development and function in cardiomyocytes from human embryonic stem cells with trisomy 21. Stem Cells. 2015;33:1434–1446. doi: 10.1002/stem.1961. [DOI] [PubMed] [Google Scholar]
- 339.Dumevska B., Bosman A., McKernan R., Main H., Schmidt U., Peura T. Derivation of Trisomy 21 affected human embryonic stem cell line Genea021. Stem Cell Res. (Amst.) 2016;16:401–404. doi: 10.1016/j.scr.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 340.Dumevska B., McKernan R., Goel D., Schmidt U. Derivation of Trisomy 21 affected human embryonic stem cell line Genea053. Stem Cell Res. (Amst.) 2016;16:500–502. doi: 10.1016/j.scr.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 341.Shi Y., Kirwan P., Smith J., MacLean G., Orkin S.H., Livesey F.J. A human stem cell model of early Alzheimer’s disease pathology in Down syndrome. Sci. Transl. Med. 2012;4:124ra29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Busciglio J., Yankner B.A. Apoptosis and increased generation of reactive oxygen species in Down’s syndrome neurons in vitro. Nature. 1995;378:776–779. doi: 10.1038/378776a0. [DOI] [PubMed] [Google Scholar]
- 343.Lu J., Delli-Bovi L.C., Hecht J., Folkerth R., Sheen V.L. Generation of neural stem cells from discarded human fetal cortical tissue. J. Vis. Exp. 2011;(51):2681. doi: 10.3791/2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Guedj F., Pennings J.L., Massingham L.J., Wick H.C., Siegel A.E., Tantravahi U., Bianchi D.W. An Integrated Human/Murine Transcriptome and Pathway Approach To Identify Prenatal Treatments For Down Syndrome. Sci. Rep. 2016;6:32353. doi: 10.1038/srep32353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Hibaoui Y., Grad I., Letourneau A., Sailani M.R., Dahoun S., Santoni F.A., Gimelli S., Guipponi M., Pelte M.F., Béna F. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol. Med. 2014;6:259–277. doi: 10.1002/emmm.201302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Lu H.E., Yang Y.C., Chen S.M., Su H.L., Huang P.C., Tsai M.S., Wang T.H., Tseng C.P., Hwang S.M. Modeling neurogenesis impairment in down syndrome with induced pluripotent stem cells from Trisomy 21 amniotic fluid cells. Exp. Cell Res. 2013;319:498–505. doi: 10.1016/j.yexcr.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 347.Huo H.Q., Qu Z.Y., Yuan F., Ma L., Yao L., Xu M., Hu Y., Ji J., Bhattacharyya A., Zhang S.C., Liu Y. Modeling Down Syndrome with Patient iPSCs Reveals Cellular and Migration Deficits of GABAergic Neurons. Stem Cell Reports. 2018;10:1251–1266. doi: 10.1016/j.stemcr.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Park C.Y., Halevy T., Lee D.R., Sung J.J., Lee J.S., Yanuka O., Benvenisty N., Kim D.W. Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep. 2015;13:234–241. doi: 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]
- 349.Jiang J., Jing Y., Cost G.J., Chiang J.C., Kolpa H.J., Cotton A.M., Carone D.M., Carone B.R., Shivak D.A., Guschin D.Y. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Bardy C., van den Hurk M., Kakaradov B., Erwin J.A., Jaeger B.N., Hernandez R.V., Eames T., Paucar A.A., Gorris M., Marchand C. Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Mol. Psychiatry. 2016;21:1573–1588. doi: 10.1038/mp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Kemp P.J., Rushton D.J., Yarova P.L., Schnell C., Geater C., Hancock J.M., Wieland A., Hughes A., Badder L., Cope E. Improving and accelerating the differentiation and functional maturation of human stem cell-derived neurons: role of extracellular calcium and GABA. J. Physiol. 2016;594:6583–6594. doi: 10.1113/JP270655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Lam R.S., Töpfer F.M., Wood P.G., Busskamp V., Bamberg E. Functional Maturation of Human Stem Cell-Derived Neurons in Long-Term Cultures. PLoS ONE. 2017;12:e0169506. doi: 10.1371/journal.pone.0169506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 354.Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O’Rourke N.A., Nguyen K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Di Lullo E., Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Pașca S.P. The rise of three-dimensional human brain cultures. Nature. 2018;553:437–445. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]
- 361.Quadrato G., Arlotta P. Present and future of modeling human brain development in 3D organoids. Curr. Opin. Cell Biol. 2017;49:47–52. doi: 10.1016/j.ceb.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 362.Otani T., Marchetto M.C., Gage F.H., Simons B.D., Livesey F.J. 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell. 2016;18:467–480. doi: 10.1016/j.stem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Bershteyn M., Nowakowski T.J., Pollen A.A., Di Lullo E., Nene A., Wynshaw-Boris A., Kriegstein A.R. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell. 2017;20:435–449.e4. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Quadrato G., Brown J., Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat. Med. 2016;22:1220–1228. doi: 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- 365.Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Krencik R., Seo K., van Asperen J.V., Basu N., Cvetkovic C., Barlas S., Chen R., Ludwig C., Wang C., Ward M.E. Systematic Three-Dimensional Coculture Rapidly Recapitulates Interactions between Human Neurons and Astrocytes. Stem Cell Reports. 2017;9:1745–1753. doi: 10.1016/j.stemcr.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Paşca S.P. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Pham M.T., Pollock K.M., Rose M.D., Cary W.A., Stewart H.R., Zhou P., Nolta J.A., Waldau B. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Hargus G., Cooper O., Deleidi M., Levy A., Lee K., Marlow E., Yow A., Soldner F., Hockemeyer D., Hallett P.J. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc. Natl. Acad. Sci. USA. 2010;107:15921–15926. doi: 10.1073/pnas.1010209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Muotri A.R., Nakashima K., Toni N., Sandler V.M., Gage F.H. Development of functional human embryonic stem cell-derived neurons in mouse brain. Proc. Natl. Acad. Sci. USA. 2005;102:18644–18648. doi: 10.1073/pnas.0509315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Espuny-Camacho I., Michelsen K.A., Gall D., Linaro D., Hasche A., Bonnefont J., Bali C., Orduz D., Bilheu A., Herpoel A. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 372.Guillaume D.J., Johnson M.A., Li X.J., Zhang S.C. Human embryonic stem cell-derived neural precursors develop into neurons and integrate into the host brain. J. Neurosci. Res. 2006;84:1165–1176. doi: 10.1002/jnr.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Korecka J.A., Levy S., Isacson O. In vivo modeling of neuronal function, axonal impairment and connectivity in neurodegenerative and neuropsychiatric disorders using induced pluripotent stem cells. Mol. Cell. Neurosci. 2016;73:3–12. doi: 10.1016/j.mcn.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 374.Chen Y., Xiong M., Dong Y., Haberman A., Cao J., Liu H., Zhou W., Zhang S.C. Chemical Control of Grafted Human PSC-Derived Neurons in a Mouse Model of Parkinson’s Disease. Cell Stem Cell. 2016;18:817–826. doi: 10.1016/j.stem.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Steinbeck J.A., Choi S.J., Mrejeru A., Ganat Y., Deisseroth K., Sulzer D., Mosharov E.V., Studer L. Optogenetics enables functional analysis of human embryonic stem cell-derived grafts in a Parkinson’s disease model. Nat. Biotechnol. 2015;33:204–209. doi: 10.1038/nbt.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Weick J.P., Johnson M.A., Skroch S.P., Williams J.C., Deisseroth K., Zhang S.C. Functional control of transplantable human ESC-derived neurons via optogenetic targeting. Stem Cells. 2010;28:2008–2016. doi: 10.1002/stem.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Weick J.P., Liu Y., Zhang S.C. Human embryonic stem cell-derived neurons adopt and regulate the activity of an established neural network. Proc. Natl. Acad. Sci. USA. 2011;108:20189–20194. doi: 10.1073/pnas.1108487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Sances S., Bruijn L.I., Chandran S., Eggan K., Ho R., Klim J.R., Livesey M.R., Lowry E., Macklis J.D., Rushton D. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 2016;19:542–553. doi: 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]