Abstract
Understanding how dispersal and gene flow link geographically separated the populations over evolutionary history is challenging, particularly in migratory marine species. In southern right whales (SRWs, Eubalaena australis), patterns of genetic diversity are likely influenced by the glacial climate cycle and recent history of whaling. Here we use a dataset of mitochondrial DNA (mtDNA) sequences (n = 1327) and nuclear markers (17 microsatellite loci, n = 222) from major wintering grounds to investigate circumpolar population structure, historical demography and effective population size. Analyses of nuclear genetic variation identify two population clusters that correspond to the South Atlantic and Indo-Pacific ocean basins that have similar effective breeder estimates. In contrast, all wintering grounds show significant differentiation for mtDNA, but no sex-biased dispersal was detected using the microsatellite genotypes. An approximate Bayesian computation (ABC) approach with microsatellite markers compared the scenarios with gene flow through time, or isolation and secondary contact between ocean basins, while modelling declines in abundance linked to whaling. Secondary-contact scenarios yield the highest posterior probabilities, implying that populations in different ocean basins were largely isolated and came into secondary contact within the last 25,000 years, but the role of whaling in changes in genetic diversity and gene flow over recent generations could not be resolved. We hypothesise that these findings are driven by factors that promote isolation, such as female philopatry, and factors that could promote dispersal, such as oceanographic changes. These findings highlight the application of ABC approaches to infer the connectivity in mobile species with complex population histories and, currently, low levels of differentiation.
Subject terms: Evolutionary ecology, Population genetics
Introduction
Migratory marine species, such as sea turtles, sharks, sea birds and marine mammals, are by nature highly mobile, and many lack obvious barriers to long-distance movement (Bonfil et al. 2005; Benson et al. 2011; Mate and Best 2011). Despite this, many such species also have geographically subdivided populations. A key question when managing contemporary populations of migratory marine species is whether such geographically distinct populations are linked by current dispersal or gene flow, or are now isolated and experiencing genetic drift. Such questions are of interest in understanding the evolution of population structure, and are also of importance for management, given the disproportionate number of migratory marine species that are of conservation concern (Hoffmann et al. 2010; Croxall et al. 2012; Dulvy et al. 2014; IUCN 2017). When using genetic data to investigate such questions, it is recognised that patterns of genetic variation within and among populations result from the interplay of evolutionary forces through time. Consequently, to understand the potential genetic impacts of contemporary anthropogenic processes, it is important to ask how the current genetic structure of a species reflects its history of population dynamics and adaptive challenges (Jobling 2012).
The demographic histories of migratory marine species have been strongly influenced by oceanographic and climatic processes that shape the distribution of primary productivity and prey resources (Pastene et al. 2007, Bowen et al. 2016), and suitable habitats (Fontaine et al. 2010; Munro and Burg 2017), as well as a species’ dispersal strategy and capacity (Bowen et al. 1994; Munro and Burg 2017; Pichler et al. 2001; Veríssimo et al. 2017). Life history traits, such as philopatry (Andreotti et al. 2016; Bowen et al. 2016), social organisation (Baker et al. 1993; Palsboll et al. 1995; Whitehead 1998) and mating systems (Palsbøll et al. 2010; Hoelzel 1999), also influence the patterns of genetic diversity in natural populations of migratory marine species. Recent anthropogenic impacts, such as direct hunting and habitat degradation, have also affected the contemporary patterns of genetic diversity in such species (e.g., Pinsky and Palumbi 2014). However, interpreting the impact of recent anthropogenic activity on longer-lived migratory marine species, such as baleen whales, is controversial (Alter et al. 2012; Attard et al. 2015). The long and overlapping generation times of baleen whales (20+years: Taylor et al. 2007) may have slowed the loss of genetic diversity during the demographic bottleneck caused by whaling, which was severe for many species but relatively short-lived.
Here we focus on a migratory baleen whale species with circumpolar distribution (Fig. 1, IWC 2001; Richards 2009), the southern right whale (SRW: Eubalaena australis). During winter, SRWs typically inhabit shallow, sheltered coastal areas at mid-latitudes, where the females calve and both the sexes socialise. During spring, SRWs migrate to offshore summer feeding grounds in mid-latitudes to high-latitudes (IWC 2001). Given these habitat preferences, we suggest that the historical demography of SRWs has been influenced by the transition from last glacial maximum (LGM) to Holocene, during which the sea levels rose dramatically, primary productivity increased and the Antarctic sea ice cover decreased while becoming more seasonal (Gersonde et al. 2005; Clark et al. 2009; Denis et al. 2009; Allen et al. 2011; Scourse 2013; Bentley et al. 2014). These changes impacted the shallow, near-shore marine environment used as SRW wintering areas; many such areas would have become unsuitable due to rise in the sea level, while newer, larger potential wintering areas were created during the expansion of shallow marine habitat (Scourse 2013). These disruptions could have plausibly precipitated increased dispersal rates among SRW wintering grounds, leading to secondary contact between the previously isolated populations.
Fig. 1.
Map of a historical and b contemporary southern right whale winter habitat. The contemporary distribution is divided into large wintering aggregations (dark grey squares) and areas with sporadic sightings (coastal regions coloured white: see Supplementary Material 2 for references). Samples included in this study are from nursery grounds with bold acronyms: ARG (Argentina), SAF (South Africa), SWA (southwest Australia), and NZSA (New Zealand sub-Antarctic), but not from the BZL (Brazilian) and SCA (southcentral Australia) nursery grounds, also marked on the map. Also included are samples from the SEA nursery ground (southeast Australia, marked by open black square) and mainland New Zealand wintering habitat, which are a part of the sporadic sightings
Furthermore, whaling and the species’ subsequent recovery may have influenced the connectivity between wintering grounds (Allendorf et al. 2008). Between the eighteenth and twentieth centuries, whalers killed an estimated 150,000 or more SRWs, driving a hemispheric population size decline from ~100,000 whales to possibly fewer than 400 whales by 1920 (IWC 1986, 2001, 2012; Jackson et al. 2008). Covert Soviet whaling from 1951 to 1971, in violation of international protection from the League of Nations introduced in 1931 (IWC 1986), further slowed the species’ recovery: the 3368 SRWs killed (Tormosov et al. 1998) comprised an estimated 50% of the hemispheric population size at that time (Jackson et al. 2008). Today, the species has recovered in some parts of its former range. Large aggregations now occur in some key calving/nursery areas (e.g., Argentina, South Africa, Australia; see Fig. 1), while regular sightings of small numbers of SRWs occur in other parts of the historical range (Bannister 1990; Best 1990; Patenaude et al. 1998; Cooke et al. 2001; Groch et al. 2005; Carroll et al. 2013). The latter areas could represent the remnant populations (e.g., Chile/Peru: Reilly et al. 2008) and/or areas that are undergoing recolonisation from larger wintering aggregations (e.g., mainland New Zealand: Carroll et al. 2014). Recolonisation and asymmetric migration rates could have resulted from the differential rates of recovery shown by SRW wintering grounds (IWC 2001), promoting higher levels of connectivity in the aftermath of whaling.
Genetic studies of several extant SRW populations, defined by current calving grounds, using a short fragment of the mitochondrial (mtDNA) control region (275 bp), showed hierarchical population structure, indicating limited connectivity between the South Atlantic and Indo-Pacific ocean basins (Patenaude et al. 2007). Female philopatry was invoked as a major cause of this pattern, as long-term studies of the individually identified SRWs show long-term fidelity to natal wintering grounds (Rowntree et al. 2001; Carroll et al. 2016). Subsequent studies integrating the stable isotope and the genetic data of contemporary populations suggests some degree of maternally directed learning of both wintering and summer feeding grounds (Carroll et al. 2015; Valenzuela et al. 2009).
Here, we build on the previous work using mtDNA haplotype sequences from 1327 individuals (~10% of the current global population) and 17 nuclear DNA microsatellite genotypes for 222 individuals (~2% of the population), allowing both mtDNA and nuclear DNA diversity and population structure to be inferred for the first time on a circumpolar scale in the SRW. This extensive dataset allows us to begin to disentangle the contemporary and historical factors that account for the observed patterns of genetic variation, thereby illuminating the complex population dynamics of this widely distributed species. Specifically, we make inferences about the past and the current patterns of gene flow, while taking into account the non-equilibrium population dynamics, and provide the information for conservation and management of this species, now recovering from centuries of whaling.
We use approximate Bayesian computation (ABC) to evaluate the relative power of alternative historical scenarios to explain the phylogeographic pattern of the mtDNA haplotypes previously described (Patenaude et al. 2007), and the potential impact of whaling on connectivity. Patenaude et al. (2007) described a mtDNA phylogeographic pattern for SRW consistent with two competing hypotheses: (A) random lineage sorting in a species with continuous gene flow or (B) secondary contact between the formerly isolated populations (Avise 2000). Under hypothesis (A), we posit that there was continued gene flow, potentially male-biased, between the wintering grounds after population divergence. Under hypothesis (B), we posit that the wintering grounds in the South Atlantic and Indo-Pacific ocean basins became isolated following divergence, but came into secondary contact as a result of the environmental change following the last glacial maximum (LGM, 16–20,000 years before present: Clark et al. 2009).
We interpret our results in the context of two different conservation management frameworks. The first framework (Wade and Angliss 1997) defines subpopulations or stocks as groups for which the demographic processes operating within the group are more important for persistence than immigration from other subpopulations. The second framework (Crandall et al. 2000) views subpopulations as groups that can be defined on the basis of contemporary and historical ecology and genetic exchangeability.
Materials and methods
Genetic data generation and compilation
For both the microsatellite and mtDNA control region analyses, we used a combination of previously published data sources (Carroll et al. 2015; Valenzuela et al. 2009) and new data (Table 1, Supplementary Material 1, Supplementary Table 1). Allele calls were standardised between laboratories for the microsatellite data, and the standard quality control measures were taken, including tests for deviation from Hardy-Weinberg equilibrium and genotyping error rate estimation (Supplementary Material 1). All sampling areas were nursery grounds except for the Australian wintering habitat, which is a mixture of migratory corridors and winter nursery areas (Carroll et al. 2015). Therefore, we make some comparisons using samples from the southeast and southwest Australian nursery grounds only and others, using the entire Australian wintering habitat sample.
Table 1.
Summary of the number of samples and diversity statistics for mitochondrial DNA control region (mtDNA) and microsatellite markers (17 loci) for southern right whale nursery areas in Argentina, (ARG), South Africa (SAF), New Zealand (NZ), southwest Australia (SWA), southeast Australia (SEA) and the overall Australian wintering habitat (AUS-WH) and ocean basins (Indo-Pacific; IP and South Atlantic; SA). For mtDNA, the sample size (nm), number of haplotypes (nhap), standardised haplotype richness (nshap), haplotype (h) and nucleotide (π) diversities (with standard deviation, ± SD) are shown. For microsatellites, the sample size (2n), standardised allele richness (k) and observed and expected heterozygosities (HOBS and HEXP), and effective population size (Ne; lowest allele frequency 0.02 and jackknifed confidence intervals CI) are shown.
| mtDNA | Microsatellites | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Region | nm | nhap | nshap ± SD | h ± SD | π ± SD (%) | 2n | k ± SD | HOB ± SD | HEXP ± SD | Ne (95% CI) |
| ARG | 208 | 28 | 8.91 ± 1.24 | 0.94 ± 0.01 | 2.27 ± 1.16 | 92 | 5.37 ± 1.49 | 0.72 ± 0.03 | 0.77 ± 0.03 | 181 (120, 344) |
| SAF | 350 | 37 | 9.03 ± 1.28 | 0.94 ± 0.01 | 2.47 ± 1.26 | 94 | 5.47 ± 1.38 | 0.71 ± 0.04 | 0.76 ± 0.03 | 239 (142, 656) |
| SA | 558 | 55 | 10.04 ± 1.10 | 0.97 ± 0.01 | 2.52 ± 1.28 | 186 | 5.46 ± 1.45 | 0.72 ± 0.03 | 0.76 ± 0.03 | 365 (241, 712) |
| SWA | 16 | 5 | 4.44 ± 0.63 | 0.71 ± 0.09 | 1.79 ± 01.00 | 34 | 5.16 ± 1.25 | 0.80 ± 0.04 | 0.78 ± 0.02 | 195 (61, ∞) |
| SEA | 12 | 5 | 5.71 ± 0.92 | 0.82 ± 0.07 | 2.17 ± 1.22 | 24 | 5.39 ± 1.37 | 0.80 ± 0.04 | 0.78 ± 0.02 | 317(49, ∞) |
| AUS-WH | 77 | 10 | 5.32 ± 1.06 | 0.78 ± 0.03 | 1.90 ± 0.99 | 156 | 5.47 ± 1.35 | 0.78 ± 0.03 | 0.78 ± 0.02 | 286 (169, 792) |
| NZ | 692 | 11 | 3.88 ± 0.83 | 0.69 ± 0.01 | 1.49 ± 0.79 | 102 | 5.41 ± 1.47 | 0.72 ± 0.03 | 0.76 ± 0.03 | 412 (192, ∞) |
| IP | 769 | 13 | 4.10 ± 0.90 | 0.71 ±± 0.01 | 1.60 ± 0.84 | 258 | 5.46 ± 1.37 | 0.76 ± 0.03 | 0.78 ± 0.03 | 331(230, 556) |
| Total | 1327 | 60 | 7.91 ± 1.47 | 0.89 ± 0.01 | 2.21 ± 1.13 | 442 | 5.54 ± 1.44 | 0.74 ± 0.03 | 0.77 ± 0.03 | - |
Estimates of genetic diversity
We estimated the standardised allelic richness with FSTAT v 2.9 (Goudet 1995), and estimated the observed and the expected heterozygosity for each microsatellite locus per sample partition using GenoDive v2.0 (Meirmans and van Tienderen 2004). We estimated the haplotype and the nucleotide diversity for the mtDNA sequence data using Arlequin v3.5 (Excoffier and Lischer 2010), and tested for significant differences in these statistics between sample partitions using permutation tests (Alexander et al. 2016). To obtain comparable estimates of the number of haplotypes detected between sample partitions, we randomly selected 12 individuals from each partition, with replacement, 1000 times, to estimate the mean number of haplotypes (and its standard deviation). We estimated apparent contemporary Ne from the microsatellite genotypes using the bias-corrected version of the linkage disequilibrium method (Waples 2006), as implemented in programme NeEstimator v2.01 (Do et al. 2014).
Investigating contemporary patterns of genetic diversity
We undertook an hierarchical Analysis of Molecular Variance (AMOVA (Excoffier et al. 1992)) for both mtDNA and microsatellite data in Arlequin, with the wintering grounds grouped into ocean basins, the significance of which was assessed with a permutation test (50,000 permutations, α = 0.05). We estimated the pairwise genetic differentiation between sample partitions for the microsatellite loci by calculating the overall and pairwise FST and Jost’s D statistic (Jost 2008) using GenoDive and for the mtDNA data, by calculating the overall and pairwise FST and ΦST statistics using Arlequin. The probability of the observed level of differentiation occurring in a panmictic population was estimated using the log-likelihood G test in GenoDive (microsatellite) and the permutation test in Arlequin (mtDNA), for a total of 10,000 permutations each.
We used two complementary methods to detect the genetic clusters within the microsatellite data: discriminant analysis of principal components (DAPC) using the R package adegenet (Jombart and Ahmed 2011) and STRUCTURE v2.3.4 (Pritchard et al. 2000). DAPC is a generic multivariate method that makes few assumptions about the underlying data and seeks to maximise the between-group variation, while minimising the within-group variation. We ran the DAPC with samples grouped by wintering grounds and by nursery grounds (Australian migratory corridor samples were excluded). Data were visualised by plotting samples by linear discriminant coordinates. In contrast, STRUCTURE method attempts to group the individuals into clusters that minimise deviations from the Hardy-Weinberg equilibrium and linkage disequilibrium. The fit of the data to K populations was assessed in STRUCTURE method under the admixture and correlated allele frequency model, with and without prior information on the sampling locations of the data (location prior set as wintering ground). Ten replicates of K = 1–5 were conducted, each with burn-ins of one million iterations and runs of ten million Markov chain Monte Carlo (MCMC) iterations, and the convergence was assessed by visually inspecting the summary statistics (e.g., FST). We used CLUMPAK to summarise the modes or distinct solutions for each value of K (Kopelman et al. 2015) and assessed the most likely value of K using the mean log likelihood from across the ten runs, summarised using the STRUCTURE HARVESTER (Earl and VonHoldt 2012). The relationship between the population structure and the geographic location was quantified using ObStruct (Gayevskiy et al. 2014).
We investigated whether the phylogeographic pattern, originally documented in the mtDNA by Patenaude et al. (2007), was still evident with our larger dataset, by estimating a phylogenetic tree using MrBayes v3.2 (Ronquist et al. 2012) and the sequences from all three right whale species (E. australis, E. japonica and E. glacialis). We used MrBayes v3.2 to simultaneously select the best model of evolution and construct a phylogenetic tree, by sampling across the substitution model space in the Bayesian MCMC analysis itself. The analysis was conducted using the sequences of the unique SRW mtDNA control region haplotypes and previously published sequences from the North Atlantic (E. glacialis) and North Pacific (E. japonica) right whales (see Supplementary File 1 for accession numbers and Supplementary File 2 for the nexus file of SRW sequences used). We undertook two runs of MrBayes, each with two chains that were run for one million iterations, using the bowhead whale (Balaena mysticetus) sequence as an outgroup. We compared the standard deviation of the split frequencies (<0.01) and the potential scale reduction factor (PSRF, should be close to 1) to detect whether the convergence had been reached, and then ran the programme for additional iterations, if required. We summarized the tree and branch length information after discarding the first 25% of trees as burn-in and used MrBayes to generate the consensus tree with clade credibility (posterior probability) values. We also constructed a median joining network (Bandelt et al. 1999) for the haplotypes using POPART (Leigh and Bryant 2015) to examine the relationships and distributions of haplotypes (with ε = 0).
Estimating contemporary and long-term gene flow
We used the programme BayesAss v3.0 (Wilson and Rannala 2003) to co-estimate the recent migration rates (past two generations), individual assignment and ancestries, based on the microsatellite genotypes. Initial runs were conducted to calibrate the mixing parameters and to ensure that the acceptance rates were in the optimal range of 0.2–0.6. During this phase, we adjusted the allele-frequency mixing parameter to 0.15, but decided to keep all other parameters at their default values (0.10). We then conducted five BayesAss runs of ten million iterations with initial burn-ins of one million iterations. Parameters were sampled every 1000 iterations and the traces were visually checked for convergence in TRACER v1.6 (Rambaut et al. 2014). We reported the median migration rates with 95% HPD interval from all runs and mean assignment probabilities of individuals across the five runs.
We tested for sex-biased dispersal with the R package hierfstat (Goudet and Jombart 2015), looking for differences in FST and the variance of corrected assignment indices (vAIc) between males and females (Goudet et al. 2002). The significance of the difference was tested using null distributions generated with 1000 permutations (Goudet et al. 2002).
We attempted to estimate the long-term gene flow rates using the coalescent-based programme LAMARC v 2.0, as this programme simultaneously estimates migration rates, growth rates and θ (Kuhner 2006). In principle, this method can account for changes in abundance caused by whaling, and also avoid positively biasing diversity estimates by accounting for migration (Kuhner 2006). We used the Bayesian option for LAMARC, and ran the two replicates, each comprising one chain with 100,000 sampled genealogies, sampled every 50th genealogy, discarding the first 25,000 samples of each search. To improve search performance of the Bayesian option, we followed the suggestion from the LAMARC manual, of employing a search strategy with three heated chains (1.0, 1.1, and 1.3, respectively). We conducted one LAMARC run using all the microsatellite data, as the LAMARC manual recommends one longer run with heating to obtain the best results for the Bayesian option. Given the large size of the mtDNA dataset, we ran three replicates of LAMARC, each with a different set of 100 individuals (n = 25 each for Argentina, South Africa and New Zealand nursery areas and the Australian wintering habitat), randomly subsampled from the larger dataset without replacement. Long-term and whaling era migration rates were also estimated with ABC analyses (see below).
Inferring historical gene flow and demographic history of SRWs
We employed the approximate Bayesian computation (ABC) to test competing hypotheses regarding changes in Ne and gene flow from the observed microsatellite data. These initial scenarios were generated by previous work, characterising the mtDNA phylogeographic pattern, which has been described as either consistent with continuous gene flow after divergence or isolation followed by secondary contact (Patenuade et al. 2007). The mtDNA data were not included in the ABC analyses, as the hypotheses being tested were generated from the mtDNA phylogeny. To test the hypothesis that whaling could have impacted the connectivity between wintering grounds, we included scenarios in which the rates of gene flow rate changes in the whaling era. As a type of null model, we include a scenario with no gene flow after divergence.
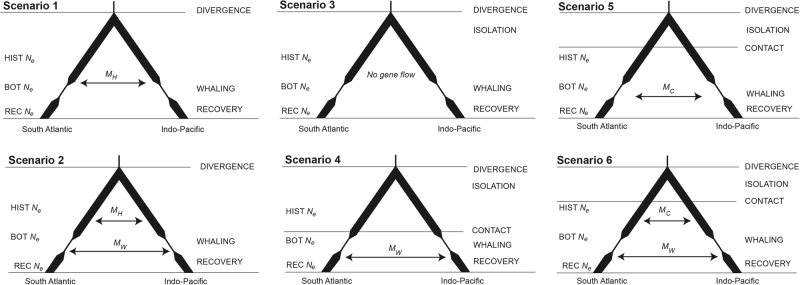
A total of six scenarios were examined (Fig. 2): continuous gene flow following population divergence at a single migration rate MH (Scenario 1) or two migration rates: one since divergence MH and one since the whaling era MW (Scenario 2); isolation following divergence, with either or no subsequent gene flow (Scenario 3); or gene flow at one migration rate since the whaling era MW (Scenario 4), or one migration rate since the secondary contact, MC (Scenario 5); or two migration rates: one since the secondary contact, MC, and one since the whaling era, MW (Scenario 6).
Fig. 2.
Scenarios compared using the approximate Bayesian computation framework. The six scenarios include continuous gene flow following population divergence with migration varying by scenario; Scenario 1: single migration rate MH; Scenario 2: two migration rates: one since divergence MH and one since the whaling era MW; or isolation following divergence, with either: Scenario 3: no subsequent gene flow; Scenario 4: gene flow at one migration rate since the whaling era MW; Scenario 5: one migration rate since secondary contact, MC; or Scenario 6: two migration rates: one since secondary contact, MC, and one since the whaling era, MW. In all scenarios, the populations diverge at time DIVERGENCE and maintain one HIST Ne until whaling, when each population declines to bottleneck population size BOT Ne during the whaling era and subsequently recover to REC Ne population size
All scenarios incorporated a reduction in Ne due to whaling, followed by a recovery (see Table 2 for prior distributions), and the timing of these events was fixed at nine and two generations before present, respectively. We assumed an average effective generation time of 25 years (Taylor et al. 2007) and a per-generation mutation rate for the microsatellite loci of 5 × 10−4 (Estoup et al. 2002). For each scenario, 100,000 coalescent simulations were run with fastsimcoal2 (Excoffier and Foll 2011), and 17 summary statistics (Supplementary Table 2) were calculated in arlsumstat v3.5.2 (Excoffier and Lischer 2010). We analysed the results in an ABC framework with the R package abc (Csilléry et al. 2012) using the neural networks algorithm (Blum and François 2010) with 1% acceptance ratio to estimate the posterior parameters, a method that is suitable for high-dimensional, correlated summary statistics (Csilléry et al. 2012). Model selection was conducted by calculating the posterior model probabilities and Bayes factors (BF) in the abc package. BF was calculated for all possible pairs of models and was interpreted following the scale of Jeffreys (1961); see Supplementary Table 3). The ability of the ABC approach to distinguish between the best-selected models was assessed using a cross-validation function in the abc package. In addition, we undertook the posterior predictive checks using nine summary statistics not employed in the initial ABC analysis. The posterior predictive checks were carried out by sampling 1000 combinations of model parameters from the posterior distributions for scenarios, and using these as the input for coalescent simulations in fastsimcoal2 with the same settings as used for the initial scenarios. For each of the 1000 simulations per scenario, we calculated the 95% confidence intervals and determined whether these encompassed the observed value.
Table 2.
Statistics for the approximate Bayesian computation (ABC) analysis of the historical demography of the southern right whale
| A. | Prior | Scenario 4 | Scenario 5 | Scenario 6 |
|---|---|---|---|---|
| Migration rate since secondary contact (MC) | ||||
| MC SA to IP | 0.001–0.50 | – | 0.0281 (0.0087, 0.2068) | 0.0114 (0.0023, 0.4259) |
| MC IP to SA | 0.001–0.50 | – | 0.0008 (0.0002, 0.0825) | 0.0128 (0.0015, 0.4349) |
| Migration rate since whaling era (MW) | ||||
| MW SA to IP | 0.001–0.50 | 0.0164 (0.0034, 0.0717) | – | 0.0025 (0.0002, 0.0830) |
| MW IP to SA | 0.001–0.50 | 0.0235 (0.0052, 0.2091) | – | 0.0025 (0.0013, 0.0718) |
| Effective population sizes (Ne) | ||||
| HIST Ne SA | 1000–100,000 | 3874 (1057, 80, 909) | 3,916 (1131, 85,781) | 3188 (920, 65,783) |
| HIST Ne IP | 1000–100,000 | 4635 (1275, 92,942) | 3,880 (1131, 82,748) | 3908 (973, 88,744) |
| BOT Ne SA | 20–2000 | 128 (34, 2447) | 90 (23, 2216) | 74 (21, 1430) |
| BOT Ne IP | 20–2000 | 88 (22, 1936) | 88 (23, 1743) | 111 (29, 1998) |
| REC Ne SA | 1000–20,000 | 1358 (466, 9433) | 888 (344, 8126) | 2494 (706, 15,728) |
| REC Ne IP | 100–20,000 | 4447 (1179, 24,463) | 2008 (814, 10,436) | 2013 (660, 12,851) |
| Timing (generations) | ||||
| CONTACT | 10–100,000 | Fixed | 38 (12, 962) | 74 (20, 2187) |
| DIVERGENCE | 2000–200,000 | 189,621 (122,048, 200,959) | 194,697 (103,040, 209,241) | 198,944 (109,871, 211,017) |
| B. | Scenario 4 | Scenario 5 | Scenario 6 | |
| Posterior model probability | 0.0048 | 0.5614 | 0.4338 | |
| Bayes factor | 117.14 | 1.00 | 1.29 | |
| Posterior predictive checks | 6/9 | 7/9 | 7/9 |
A. Prior log-uniform distributions used for ABC analysis and posterior values obtained for the South Atlantic (SA) and Indo-Pacific (IP) ocean basins under Scenarios 4–6. Posterior mode and 95% HPDs are reported for migration rates (MC: constant migration rate since secondary contact; MW: migration rate since whaling era), effective population sizes (Ne) and time in generations since secondary contact (CONTACT) and population divergence (DIVERGENCE). Effective population sizes are shown schematically in Fig. 2 and are divided into historical or pre-whaling (HIST Ne), bottleneck (BOT Ne) and recovery (REC Ne). B. Posterior support, as shown by the posterior model probability, Bayes factor (relative to Scenario 5), and the number of posterior predictive summary statistics that encompassed the observed value
Results
Microsatellite genotyping and diversity statistics
In total, 222 individuals were genotyped at an average of 16.2 of 17 microsatellite loci, with an estimated error rate of 0.7% per allele (for more information see Supplementary Material 1).
The microsatellite-based diversity statistics were broadly comparable across ocean basins and nursery areas (Table 1). In contrast, the mtDNA data (Table 1) showed generally higher levels of diversity in the South Atlantic than in the Indo-Pacific nursery grounds. Permutations confirmed that this was a statistically significant difference for both nucleotide and haplotype diversity (at α = 0.05).
As the samples comprise of overlapping generations, the estimates of Ne, based on microsatellite loci, actually reflect the number of effective breeders (Nb) that produced the sample. Estimates of Nb were broadly similar across ocean basins and Argentinean and South African nursery grounds (Table 1; Supplementary Table 4). For the two individual Australian nursery grounds and the New Zealand wintering ground, the estimates of Nb had undefined upper boundaries, and for the former, this is likely due to the small sample sizes. For the New Zealand wintering ground, we suggest it is due to the increased variance and lower precision found when using the bias-corrected linkage disequilibrium method in populations with large Ne (Waples and Do 2010). However, the lower bound has been shown to be reliable in such cases (Waples and Do 2010), so Nb = 192 is probably a reasonable lower bound for the New Zealand wintering ground.
Contemporary patterns of genetic diversity
The AMOVA analyses and fixation indices indicated greater variation between ocean basins (AMOVA: FST = 0.126 and ΦST = 0.131 for mtDNA, FST = 0.024 for microsatellites, all at p < 0.01) than among wintering grounds within the ocean basins (FST = 0.052 and ΦST = 0.082 for mtDNA, FST = 0.004 for microsatellites, all at p < 0.01; see Table 3 and Supplementary Table 5 for fixation indices). Direct comparison of the Indo-Pacific and South Atlantic ocean basins yielded the estimates of divergence for mtDNA at FST = 0.161 and ΦST = 0.189 (p < 0.001). The divergence for microsatellite loci was FST = 0.012 (95% CI: 0.007, 0.018) and Jost’s D = 0.041 (95% CI: 0.024, 0.063, all p < 0.001).
Table 3.
Genetic differentiation indices between the southern right whale nursery areas in Argentina, (ARG), South Africa (SAF), New Zealand (NZ) and southwest Australia (SWA), southeast Australia (SEA) and the overall Australian wintering habitat (AUS-WH). Pairwise FST based on mtDNA haplotype frequencies (bottom left quadrant) and microsatellite loci (top right quadrant)
| ARG-WG | SAF | SWA | SEA | AUS-WH | NZ | |
|---|---|---|---|---|---|---|
| ARG | 0.001** (0.000, 0.003) | 0.021*** (0.013, 0.031) | 0.022*** (0.007, 0.021) | 0.013*** (0.008, 0.019) | 0.012*** (0.006, 0.019) | |
| SAF | 0.048*** | 0.023*** (0.012, 0.034) | 0.017** (0.005, 0.031) | 0.014*** (0.008, 0.020) | 0.013*** (0.007, 0.019) | |
| SWA | 0.143*** | 0.136*** | 0.013* (0.000, 0.028) | – | 0.016** (0.001, 0.035) | |
| SEA | 0.104*** | 0.098** | 0.098* | – | 0.000 (0.000, 0.003) | |
| AUS-WH | 0.123*** | 0.115*** | – | – | 0.004** (0.000, 0.010) | |
| NZ | 0.190*** | 0.189*** | 0.142*** | 0.000 | 0.073*** |
The 95% confidence intervals for microsatellite-based FST, shown in parentheses, was estimated using bootstrapping over the loci (999 permutations)
*p < 0.05; **p < 0.01, ***p < 0.001
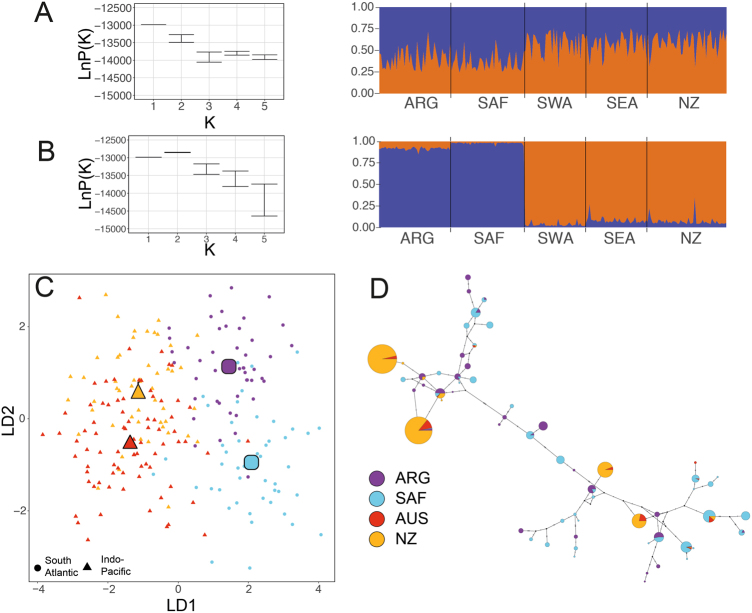
Microsatellite loci data clustered by ocean basins, based on the STRUCTURE analysis, particularly with location prior set as nursery or wintering ground (Fig. 3a, b). This was supported by log likelihood suggesting the best K = 2, with cluster corresponding to ocean basin and ObStruct analysis showing that there was a strong (R2 = 0.95) and significant (p < 0.0001) correlation between ocean basin and genetic cluster. When no prior population information was provided, the highest likelihood was for K = 1 (Fig. 3), which is not surprising, given that the STRUCTURE analysis has little power to resolve population structure FST < 0.02 (Latch et al. 2006). However, when K = 2, the next best fitting K, was analysed with ObStruct, there was a significant correlation (p < 0.0001) between ocean basin and genetic cluster (R2 = 0.45).
Fig. 3.
Inference of population structure of the southern right whale wintering habitats. Samples collected in Argentina (ARG) and South Africa (SAF) are pooled to form the South Atlantic dataset and samples from Australia (AUS) and New Zealand (NZ) are pooled to form the Indo-Pacific dataset. a, b STRUCUTRE results (left); mean log likelihood (LnP(K)), for K = 1–5, and (right) the proportion of each individual’s genome that is assigned to each cluster when K = 2 for a standard admixture setting and b location prior implemented. c Individuals plotted by linear discriminants (LD) from DAPC conducted with samples grouped by wintering ground. The large symbols show the centroid of each wintering ground. d Median joining haplotype network of mtDNA haplotypes. Haplotypes are coloured by wintering ground they were sampled in, using key shown. Inferred, unsampled haplotypes are shown by small black circles
When grouped by wintering grounds, DAPC separated samples with ocean basins along linear discriminant 1 (LD1) and by wintering grounds within ocean basins along LD2 (Fig. 3c). There was an overlap between Indo-Pacific wintering grounds, as described previously (Carroll et al. 2015), although when grouped by nursery grounds, the distinctiveness of the southwest Australian samples was emphasised (Supplementary Figure 1).
As expected, the global mitochondrial phylogenetic tree for the three right whale species shows distinct clades into which North Pacific, North Atlantic and southern hemisphere individuals sort cleanly (Supplementary Figure 2). Convergence was indicated as the average standard deviation of split frequencies was 0.005 and the PRSF was 1.00. Within SRWs, the Indo-Pacific sample contains just 13 haplotypes in a total sample of 769 individuals, while the South Atlantic sample contains 55 haplotypes (4.2 times as many) in a total sample of 558 (0.73 times as many). The 13 Indo-Pacific haplotypes are broadly distributed through the SRW genealogy, as are many other common haplotypes (Fig. 3d). This is the Type II phylogeographic pattern, described as pronounced phylogenetic gaps between some branches in a gene tree, with principal lineages showing no obvious geographic pattern (Avise 2000), as previously described (Patenaude et al. 2007) for SRWs. However, the Indo-Pacific samples could possibly be derived from expansions from a few ancestral sequences, which could imply a small founding population.
Estimates of contemporary and long-term gene flow
We pooled Argentina and South Africa to represent the ‘South Atlantic' and pooled New Zealand and Australia to represent the ‘Indo-Pacific’ for the BayesAss analysis, following the nomenclature previously used (Patenaude et al. 2007). Inspection of the traces for all five runs indicated that convergence was achieved with effective sample sizes for all parameters on each run >450. The migration rate estimates were consistent across runs (Supplementary Table 6), and the median migration rate (proportion of individuals that are migrants) from the South Atlantic to the Indo-Pacific was 0.038 (95% HPD 0.006, 0.083) and 0.028 (95% HPD < 0.001, 0.068), in the reverse direction (Supplementary Figure 2). BayesAss analysis identified two putative first-generation and four second-generation immigrants, though mostly with low confidence (Supplementary Table 7). There was no evidence of sex-biased dispersal between ocean basins, based on either the FST or vAIc metrics, using the microsatellite genotypes (p > 0.05 for all analyses).
The three runs of the coalescent sampler LAMARC with different random subsamples of the mtDNA dataset produced similar patterns and so the results were combined for parameter estimation using TRACER. While the combined effective sample sizes were sufficient (>500), the traces did not show signs of convergence for all parameters. The LAMARC analysis, using microsatellite markers, also failed to converge, so we do not present the results.
Historical demography of the SRW
The ABC analysis yielded near-zero posterior probabilities (<0.0001) for the demographic scenario with no gene flow (scenario 3) and those with continuous gene flow (scenarios 1 and 2, see Supplementary Table 8). In contrast, the posterior model probability and the BF estimates (Table 2) strongly support the scenarios, simulating secondary contact between previously isolated populations (scenarios 4–6), and we focus on the results of these scenarios in the rest of the paper (posterior distributions can be found in Supplementary File 3).
Of the three models simulating secondary contact, scenarios 5 and 6 showed the best fit to the data, based on posterior model support and BF (Table 2B). The support for scenario 5, which had one constant migration rate since secondary contact, was marginally greater than that for scenario 6, which had one post-secondary contact and one post-whaling migration rate (Table 2B, Supplementary Figure 5), with a BF = 1.29, which is barely worth mentioning on Jeffreys’ (1961) scale for interpreting BF. The cross-validation analysis showed that, while scenario 4 was distinguishable from scenarios 5 and 6, these two latter scenarios were misclassified as each other >30% of time. This implies that the ABC method was unable to distinguish between scenarios that differ by events that happened in the recent past (<10 generations), given the available dataset. In light of this, and the fact that the post-contact and post-whaling migration estimates for scenario 6 were very similar to each other and to the post-contact estimate from scenario 5 (all <0.03), we suggest that scenario 5 offers the most parsimonious scenario to explain the observed data (see Supplementary File 3 for graphs of model fit, and prior and posterior distributions).
Overall, the evidence is consistent with the hypothesis that secondary contact was stimulated by environmental changes that occurred near the end of LGM. Obvious candidates would include sea-level rise, which was dramatic and changed the spatial distribution of suitable wintering habitat (Scourse 2013), potentially triggering increased dispersal in search of better places to calve and socialise. We were unable to resolve the timing of the initial secondary contact, as the estimates span 11–960 generations ago (corresponding to 275–24,000 years, assuming a 25-year effective generation time), which includes the beginning of the present warm interglacial period.
Discussion
This first investigation of global diversity and population structure for SRW using both mtDNA and nuclear DNA markers highlights a complex interplay between the forces promoting isolation (e.g., philopatry and migratory fidelity) and the forces promoting connectivity (e.g., climate change). The ABC results robustly support a period of historical isolation that was followed by secondary contact between ocean basins. As right whales can easily swim thousands of kilometres, both the isolation and its subsequent breakdown were likely the consequences of behavioural mechanisms, presumably the same ones that continue to isolate the populations today, as reflected in the heterogeneity of gene-flow estimates across the Southern Hemisphere. The present data and analyses do not provide sufficient resolution to resolve the debates about the impact of whaling on SRW genetic diversity, but they do bear on a number of general issues in population management.
Connectivity through time: impacts of natural and anthropogenic changes
The finding of isolation and secondary contact between the Indo-Pacific and South Atlantic suggests that SRWs in the two ocean basins had limited connectivity, until less than 1000 generations ago (25,000 years, assuming a 25-year effective generation time). This implies that for thousands of generations after divergence, the populations were relatively isolated, with the most parsimonious explanation being behavioural mechanisms such as female philopatry, discussed in-depth below. The timing of secondary contact supports our hypothesis, that the transition from LGM to Holocene precipitated the secondary contact, but other, more recent factors cannot be ruled out. The impact of profound climatic change, such as the transition to Holocene after LGM, will depend on the ecology of the species. For example, both Antarctic blue (Balaenoptera musculus musculus) and bowhead whales saw decrease in effective population sizes linked to the decline of their sea ice habitat after LGM (Attard et al. 2015; Phillips et al. 2013). By contrast, the emergence of new breeding habitat and productive foraging areas coincident with deglaciation, reduction in sea ice and increasing productivity during the Holocene after LGM, led to postglacial population expansion in a number of penguin species and southern elephant seals (Mirounga leonina) in the Southern Hemisphere (Younger et al. 2016). Other species have apparently adapted to new conditions; Pyenson and Lindberg (2011) suggest that the grey whale (Eschrichtius robustus) can switch prey, enabling the species to maintain a constant population size across glacial cycles.
The impact of glaciation on connectivity within species is less well studied than its impact on effective population sizes, but its effects would be expected to depend on dispersal ability, population density and ecological necessity or opportunity (Bérubé et al. 1998; Phillips et al. 2011). In the ice-adapted and vagile bowhead whale, heavy sea ice three thousand years ago did not prove a barrier to connectivity across the Holarctic, based on a comparison of historical and contemporary mtDNA data (Alter et al. 2012). In contrast, dispersal rates increased in the Steller sealion (Eumetopias jubatus) coincident with an increase in glaciation that disrupted breeding habitat (when effective population size was high; Phillips et al. 2011). We suggest that SRWs may have experienced something similar, when their breeding areas were disrupted by sea-level rise following the LGM, prompting increased dispersal between shifting constellations of winter breeding grounds.
Our point estimates of migration from the Indo-Pacific into the South Atlantic from BayesAss are tenfold higher than the estimates of post-contact migration from ABC. Whether this difference reflects a real, very recent increase in migration, or a bias in BayesAss estimates, probably linked to the violation of continuous migration assumption of the programme, is not clear. A recent increase in migration could plausibly be linked to recovery from whaling. Allendorf et al. (2008) suggested that hunting could either increase directional gene flow into certain populations, for example, into low density areas, or it could decrease migration with concomitant reductions in population size and density. Increase in migration linked to population expansion and recolonisation has been seen in other marine mammals such as the migration from areas of high to low density implicated in the recovery of populations of grey seals (Brasseur et al. 2014). As our estimate of recent gene flow in SRW does not appear to be asymmetric, the change in migration rate could also reflect hunting’s impact on behavioural processes across populations. Hunting appears to effect the timing of migratory events in sockeye salmon (Oncorhynchus nerca) (Quinn et al. 2007), as well as characteristics such as boldness (Leclerc et al. 2017) that could influence exploratory behaviour (although this is controversial; Mueller et al. 2014; Rollins et al. 2015). Disruption to breeding behaviour or aggregations could also have precipitate increased dispersal during or subsequent to the whaling era. For example, the disruption of humpback whale breeding aggregations is thought to have influenced meta-population dynamics (Clapham and Zerbini 2015). This idea, the social aggregation hypothesis, holds that hunting-induced disruption to the humpback whale’s social system led to movement of animals from low density to high density breeding aggregations as the species recovered from whaling (Clapham and Zerbini 2015). Hunting, particularly in the early pre-industrial era, targeted coastal wintering grounds used by aggregations of SRWs as calving, socialising and breeding areas (Smith et al. 2012). Disruption of behaviours at wintering grounds by whaling could have prompted dispersal events in recent generations, just as disruption of wintering grounds by the expansion of shallow seas during the Holocene could have thousands of years prior.
An alternative explanation for a recent increase in migration rates is that the demographic history of the SRWs violates the assumptions of BayesAss, in which case the estimates of migration rate from this programme could be biased. BayesAss considers a constant population size scenario and implicitly assumes that migration is continuous through time. Linkage or gametic disequilibrium observed in a population is due to the recent migration, which increases concomitantly with levels of genetic differentiation amongst populations. However, the demographic history of SRWs, with secondary contact following isolation and the demographic decline due to whaling, likely increased the linkage disequilibrium beyond what is expected under continuous migration and constant population sizes. This would lead to an overestimate of migration rates. Simulation studies indicate that if the model assumptions are violated, BayesAss still performs well if migration rates are low (<0.01) and FST is high (>0.10) (Faubet et al. 2007), however, in the present study FST is fairly low (FST = 0.024). Additional empirical and theoretical work should be directed towards resolving this biologically interesting question, which has potentially large implications for attempts to predict how SRWs might respond to future climate change.
More generally, it is important to consider the limitations of the methods used to infer connectivity in non-equilibrium situations. The ABC framework is highly flexible and can explicitly simulate and compare scenarios involving isolation with gene flow and isolation with secondary contact scenarios. Previous suggestions that coalescent-based methods may also be able to differentiate such models based on the timing of migration estimates have proven incorrect (Gaggiotti 2011), based on both theoretical (Sousa et al. 2011) and simulation studies (Strasburg and Rieseberg 2011). Such investigations would violate the assumption, common to many coalescent based methods, that immigration rates have been constant throughout the lifespan of the coalescent tree (Kuhner 2006; Sousa et al. 2011). Violation of this assumption may have contributed to the lack of convergence of LAMARC analyses in our study, although other problems have been noted to affect the analyses using coalescent-based methods (see Putman and Carbone 2014).
Migratory fidelity and circumpolar population structure
The present analyses confirm previous findings of hierarchical global population structure in mtDNA haplotype data (Patenaude et al. 2007) using AMOVA and fixation indices, and extend these finding to microsatellite markers (Table 3, Supplementary Table 5). In particular, we document low but statistically significant differentiation between the Argentinean and South African nursery grounds at nuclear loci (microsatellite FST = 0.001, Jost’s D = 0.004, p < 0.01) (Table 2 and Fig. 3), and extend the previous findings of stronger differentiation in mtDNA haplotype data (Patenaude et al. 2007).
Reduced connectivity between ocean basins is a common characteristic in cetaceans, which has been attributed to site fidelity, social structure and resource specialisation in toothed whales, and to migratory fidelity in baleen whales (Bowen et al. 2016). Migratory fidelity, like other forms of philopatry or site fidelity, is hypothesised to be adaptive as it increased the chances of finding mates and/or suitable habitat, and may be favoured by natural selection, by enhancing the maintenance of co-adapted gene complexes (Greenwood 1980; Refsnider and Janzen 2010; Stiebens et al. 2013). In addition to many baleen whale species, such as humpback whales (Baker et al. 2013), natal philopatry is found in all seven sea-turtle species (Bowen et al. 2016), many shark (Chapman et al. 2015) and sea-bird species (e.g., Milot et al. 2008), showing its ubiquity in migratory marine species.
We did not find evidence of sex-biased dispersal in SRWs, using indirect genetic methods based on the microsatellite genotypes. It might be that in SRWs, different genetic patterns are evident on distinct habitats across a migratory network, reflective of different patterns of gene flow or dispersal. For example, both males and females show long-term fidelity to nursery grounds, based on the photo-identification and genotype recapture studies (Carroll et al. 2013; Cooke et al. 2001). Photo-identification studies, tracking individuals, and paternity analyses, showing recent patterns of gene flow, also indicate limited connectivity between the calving grounds within ocean basins, based on studies from the Indo-Pacific ocean basin (Pirzl et al. 2009; Carroll et al. 2012). In contrast, gene flow between individuals that use different calving grounds could occur on shared migratory corridors, as suggested in Australian migratory habitats based on a combination of photo-identification and genetic data (Carroll et al. 2015), or on shared feeding grounds. For example, whales from South African and Argentinean wintering grounds share summer feeding areas, based on photo-identification and stable isotope data (Best et al. 1993, Rowntree et al. 2001). This could facilitate the mating between whales from different wintering grounds on migratory corridors, or promote the temporary dispersal of whales between wintering grounds.
Sex-biased dispersal is not necessarily common in migratory marine species. In a recent review, Chapman et al. (2015) found residency and site fidelity occurred in both sexes in >50 studies in sharks, although many other studies examined such behaviours in only one sex. In turtles, which show strong natal philopatry, genetic connectivity is attributed to the occasional wanderer (Bowen et al. 2016), rather than sex-biased dispersal. Even in a species with a similar life-history pattern to SRWs, with strong female philopatry, the humpback whale, there was no evidence that dispersal was sex biased on a global scale, based on a large genetic analysis (Jackson et al. 2014).
Uncertain impact of whaling-era events in ABC analyses
The ABC analyses could not differentiate the scenarios with differences in very recent generations, including between those scenarios with changes in gene flow linked to the whaling era (e.g., between scenarios 5 and 6). This is consistent with findings that recent events are not accurately detected by the coalescent-based methods, due to data-driven and theoretical constraints (Boitard et al. 2016, Wakeley et al. 2016). In the light of this finding, we ran additional analyses and found that, with the current dataset, the method did not have the power to differentiate between scenarios with and without a whaling-related bottleneck (See R code and Supplementary File 3).
SRWs underwent a centuries-long demographic bottleneck due to whaling, and simulation studies suggest this reduced mtDNA haplotype number and diversity in the species (Jackson et al. 2008). Long-term exploitation of other cetacean species first hunted by pre-industrial whalers is also correlated with the decline in the genetic diversity (Alter et al. 2012; Waldick et al. 2002). For example, bowhead whales appear to have lost unique mitochondrial lineages in contemporary populations, compared with historical samples, and this was attributed to the habitat loss during the Little Ice Age and/or whaling (Alter et al. 2012). Any decline in diversity in nuclear genes like microsatellites is likely to be less severe than declines in mtDNA, given the larger effective population size. Indeed, high levels of nuclear diversity have been found in some great whale species, supporting the hypothesis that they have had large, long-term effective population sizes (e.g., Ruegg et al. 2013). Furthermore, low levels of genetic diversity in pygmy blue whales (Balaenoptera musculus brevicauda) were found to be related to a founder event, rather than recent whaling, in an ABC study using both mtDNA and microsatellite data (Attard et al. 2015). These examples highlight the importance of placing recent anthropogenic impacts in the long-term evolutionary context of a species.
The present study provides what appear to be the first estimates of Nb for SRWs, using the bias-corrected linkage disequilibrium method (Waples 2006), which produced similar results for the South Atlantic (Nb = 365, 95% CI 241, 712) and Indo-Pacific ocean basins (Nb = 331, 95% CI 230, 556). The estimates of Nb for the Australian, Argentinean and South African wintering grounds were broadly similar, in the low hundreds. Morin et al. (2012) estimated Nb for bowhead whale stocks using 22 microsatellite markers and also found estimates in the low hundreds: the small effective population size was attributed to whaling, but might also be due to life history traits similar across these related species. Given that the demographic bottleneck in SRWs happened in recent generations, it is likely that genomic-based methods that estimate effective population sizes through time using linkage disequilibrium methods (e.g., Hollenbeck et al. 2016) will be needed to determine the extent to which whaling impacted levels of nuclear genetic diversity.
As with all ABC studies, we could only compare a limited number of distinct hypotheses (Beaumont 2010): in the present case, these were suggested by previous analyses of mtDNA data (Patenaude et al. 2007). Future studies should investigate models that explicitly incorporate sex-biased dispersal and allow migration rates to vary in response to anthropogenic disturbance and other environmental variables such as bathymetry. Given that we could decisively reject the continuous migration scenarios (1–3) in favour of the isolation and secondary contact scenarios (5 and 6) using only 17 polymorphic nuclear markers, future studies using whole-genome data seem likely to have power to estimate many such parameters.
Conservation implications of findings
Effective management of migratory species requires consideration of both the overall migratory network and the different migratory habitats it encompasses. Long-term photographic and genetic monitoring programmes show that females return regularly to their natal calving grounds across decades (Carroll et al. 2013; Rowntree et al. 2001). As recruitment is dependent upon female reproductive success, the persistence of these calving grounds is likely reliant on these philopatric females (Avise 2000), and is unlikely to be supplemented by the recruitment from other calving grounds (Carroll et al. 2011; Clapham et al. 2008). Therefore, SRW calving grounds seem likely to be substantially and demographically independent, which would imply that they qualify as separate sub-populations under the population concepts advocated by Wade and Angliss (1997). On an evolutionary scale, female fidelity to wintering grounds will have contributed to significant differences in mtDNA haplotype frequencies between the wintering grounds, as presented here and in previous studies (Baker et al. 1999; Patenaude et al. 2007).
On a broader scale, vertical transmission of migratory preferences to both feeding and calving or breeding areas suggests an argument for ecological or demographic distinctiveness of the ocean basins, under the framework advocated by Crandall et al. (2000). While not explicitly mentioned by those authors, behavioural variability is arguably as valid as morphological variability, when deciding if individuals from different populations are exchangeable. Vertical transmission of socially learned behaviour, such as learned migratory routes, can shape adaptation, and by favouring the conservation of migratory traditions, promote isolation between populations (Brakes and Dall 2016; Whiten 2017). Recently it has been suggested that this ‘second inheritance system’ needs to be integrated into modelling and management of migratory marine species that face challenges from climate change, as migratory conservatism could limit responses to a changing environment (Keith and Bull 2017). Therefore, we suggest that SRWs in the two ocean basins should be considered distinct population segments. The ABC and genetic analyses (STRUCTURE and FST results) reject the null hypothesis of historical and contemporary genetic exchangeability, and the well-documented behavioural mechanisms are consistent with the observed levels of genetic differentiation. Our findings also suggest that non-equilibrium scenarios should be considered in future studies of population structure in migratory marine species. ABC techniques will make this feasible, even where low levels of differentiation and complex population histories pose severe challenges for other inference methods.
Electronic supplementary material
Acknowledgements
Biopsy collection in Argentina was conducted under permits from Dirección de Fauna y Flora Silvestre (DFyFS) and Secretaría de Turismo y Areas Protegidas del Organismo Provincial de Turismo (OPT), Chubut Province, using protocols approved by the University of Utah Institutional Animal Care and Use Committee under assigned protocol number 05–01003. Biopsy sampling was carried out under permits issued to PBB in terms of the South African Sea Fishery Act (no. 12 of 1988), issued 22 February 1995, 9 February 1996 and 9 July 1997. Ethics for the New Zealand and Australian datasets can be found in the original publication detailing the generation of these genetic data (Carroll et al. 2015). Claudia de Silva and Mary-Beth Rew contributed to the laboratory work on the South African SRW samples. ELC was supported while writing this paper by a EU Horizon 2020 Marie Slodowska Curie Fellowship, project BEHAVIOUR-CONNECT, and by a Newton Fellowship from the Royal Society of London. Laboratory analyses conducted by ELC were funded by a small grant from the British Ecological Society 5076 / 6118 and Bayesian analysis was supported by training from the National Science Foundation under Grant No. DEB-1145200. OEG was supported by the Marine Alliance for Science and Technology for Scotland (MASTS) funded by the Scottish Founding Council (grant reference HR09011). Genetic data from the South African right whale samples were generated by MB and PJP with the support of UC Berkeley, University of Stockholm and University of Groningen. Computational biology analyses were supported by the University of St Andrews Bioinformatics Unit, which is funded by a Wellcome Trust ISSF award. We thank the Editor and three anonymous reviewers for constructive comments that improved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0077-y) contains supplementary material, which is available to authorized users.
References
- Alexander A, Steel DJ, Hoekzema K, Mesnick S, Engelhaupt D, Kerr I, et al. What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Mol Ecol. 2016;25:2754–2772. doi: 10.1111/mec.13638. [DOI] [PubMed] [Google Scholar]
- Allen CS, Pike J, Pudsey CJ. Last glacial-interglacial sea-ice cover in the SW Atlantic and its potential role in global deglaciation. Quat Sci Rev. 2011;30:2446–2458. doi: 10.1016/j.quascirev.2011.04.002. [DOI] [Google Scholar]
- Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N. Genetic effects of harvest on wild animal populations. Trends Ecol Evol. 2008;23:327–337. doi: 10.1016/j.tree.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Alter SE, Rosenbaum HC, Postma LD, Whitridge P, Gaines C, Weber D, et al. Gene flow on ice: The role of sea ice and whaling in shaping holarctic genetic diversity and population differentiation in bowhead whales (Balaena mysticetus) Ecol Evol. 2012;2:2895–2911. doi: 10.1002/ece3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti S, Von Der Heyden S, Henriques R, Rutzen M, Meÿer M, Oosthuizen H, et al. New insights into the evolutionary history of white sharks. Carcharodon carcharias J Biogeogr. 2016;43:328–339. doi: 10.1111/jbi.12641. [DOI] [Google Scholar]
- Attard CRM, Beheregaray LB, Jenner KCS, Gill PC, Jenner MM, Morrice MG, et al. Low genetic diversity in pygmy blue whales is due to climate-induced diversification rather than anthropogenic impacts. Biol Lett. 2015;11:20141037. doi: 10.1098/rsbl.2014.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: the history and formation of species. Cambridge, MA, USA: Harvard University Press; 2000. [Google Scholar]
- Baker CS, Patenaude NJ, Bannister JL, Robins J, Kato H. Distribution and diversity of mtDNA lineages among southern right whales (Eubalaena australis) from Australia and New Zealand. Mar Biol. 1999;134:1–7. doi: 10.1007/s002270050519. [DOI] [Google Scholar]
- Baker CS, Perry A, Bannister JL, Weinrich M, Abernethy R, Calambokidis J, et al. Abundant mitochondrial DNA variation and world-wide population structure in humpback whales. Proc Natl Acad Sci. 1993;90:8239–8243. doi: 10.1073/pnas.90.17.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS, Steel DJ, Calambokidis J, Falcone E, González-Peral U, Barlow J, et al. Strong maternal fidelity and natal philopatry shape genetic structure in North Pacific humpback whales. Mar Ecol Prog Ser. 2013;494:291–306. doi: 10.3354/meps10508. [DOI] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bannister JL (1990) Southern right whales off western Australia. Rep Int Whal Comm (Special Issue 12): 279–288
- Beaumont M. Approximate Bayesian computation in evolution and ecology. Annu Rev Ecol Syst. 2010;41:379–406. doi: 10.1146/annurev-ecolsys-102209-144621. [DOI] [Google Scholar]
- Benson SR, Eguchi T, Foley DG, Forney KA, Bailey H, Hitipeuw C, et al. Large-scale movements and high-use areas of western Pacific leatherback turtles, Dermochelys coriacea. Ecosphere. 2011;2:1–27. doi: 10.1890/ES11-00053.1. [DOI] [Google Scholar]
- Bentley MJ, Ocofaigh C, Anderson JB, Conway H, Davies B, Graham AGC, et al. A community-based geological reconstruction of Antarctic Ice Sheet deglaciation since the Last Glacial Maximum. Quat Sci Rev. 2014;100:1–9. doi: 10.1016/j.quascirev.2014.06.025. [DOI] [Google Scholar]
- Bérubé M, Aguilar A, Dendanto D, Larsen F, Di Sciara GN, Sears R, et al. Population genetic structure of North Atlantic, Mediterranean Sea and Sea of Cortez fin whales, Balaenoptera physalus (Linnaeus 1758): analysis of mitochondrial and nuclear loci. Mol Ecol. 1998;7:585–599. doi: 10.1046/j.1365-294x.1998.00359.x. [DOI] [PubMed] [Google Scholar]
- Best P. Trends in the inshore right whale population off South Africa, 1969–87. Mar Mammal Sci. 1990;6:93–108. doi: 10.1111/j.1748-7692.1990.tb00232.x. [DOI] [Google Scholar]
- Best P, Payne R, Rowntree VJ, Palazzo J, Both M. Long-range movements of South Atlantic right whales Eubalaena australis. Mar Mammal Sci. 1993;9:227–234. doi: 10.1111/j.1748-7692.1993.tb00451.x. [DOI] [Google Scholar]
- Blum MGB, François O. Non-linear regression models for approximate Bayesian computation. Stat Comput. 2010;20:63–73. doi: 10.1007/s11222-009-9116-0. [DOI] [Google Scholar]
- Boitard S, Rodriguez W, Jay F, Mona S, Austerlitz F. Inferring population size history from large samples of genome-wide molecular data - an approximate Bayesian computation approach. PLoS Genet. 2016;12:e1005877. doi: 10.1371/journal.pgen.1005877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfil R, Meyer M, Scholl MC, Johnson R, O’Brien S, Oosthuizen H, et al. Transoceanic migration, spatial dynamics, and population linkages of white sharks. Science. 2005;310:100–103. doi: 10.1126/science.1114898. [DOI] [PubMed] [Google Scholar]
- Bowen BW, Gaither MR, Dibattista JD, Iacchei M, Andrews KR, Grant WS, et al. Comparative phylogeography of the ocean planet. Proc Natl Acad Sci USA. 2016;113:7962–7969. doi: 10.1073/pnas.1602404113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen BW, Kamezaki N, Limpus CJ, Hughes GR, Meylan AB, Avise JC. Global phylogeography of the loggerhead turtle (Caretta caretta) as indicated by mitochondrial DNA haplotypes. Evolution. 1994;48:1820–1828. doi: 10.1111/j.1558-5646.1994.tb02217.x. [DOI] [PubMed] [Google Scholar]
- Brakes P, Dall SRX. Marine mammal behavior: a review of conservation implications. Front Mar Sci. 2016;3:87. doi: 10.3389/fmars.2016.00087. [DOI] [Google Scholar]
- Brasseur SMJM, van Polanen Petel TD, Gerrodette T, Meesters EHWG, Reijnders PJH, Aarts G. Rapid recovery of dutch gray seal colonies fueled by immigration. Mar Mammal Sci. 2014;31:405–426. doi: 10.1111/mms.12160. [DOI] [Google Scholar]
- Carroll EL, Baker CS, Watson M, Alderman R, Bannister JL, Gaggiotti OE, et al. Cultural traditions across a migratory network shape the genetic structure of southern right whales around Australia and New Zealand. Sci Rep. 2015;5:16182. doi: 10.1038/srep16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EL, Childerhouse SJ, Christie M, Lavery S, Patenaude NJ, Alexander A, et al. Paternity assignment and demographic closure in the New Zealand southern right whale. Mol Ecol. 2012;16:3960–3973. doi: 10.1111/j.1365-294X.2012.05676.x. [DOI] [PubMed] [Google Scholar]
- Carroll EL, Childerhouse SJ, Fewster R, Patenaude N, Steel DJ, Dunshea G, et al. Accounting for female reproductive cycles in a superpopulation capture – recapture framework. Ecol Appl. 2013;23:1677–1690. doi: 10.1890/12-1657.1. [DOI] [PubMed] [Google Scholar]
- Carroll EL, Fewster R, Childerhouse SJ, Patenaude NJ, Boren L, Baker CS. First direct evidence for natal wintering ground fidelity and estimate of juvenile survival in the New Zealand southern right whale Eubalaena australis. PLoS ONE. 2016;11:e0146590. doi: 10.1371/journal.pone.0146590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EL, Patenaude NJ, Alexander A, Steel DJ, Harcourt R, Childerhouse SJ, et al. Population structure and individual movement of southern right whales around New Zealand and Australia. Mar Ecol Prog Ser. 2011;432:257–268. doi: 10.3354/meps09145. [DOI] [Google Scholar]
- Carroll EL, Rayment W, Alexander A, Baker CS, Patenaude NJ, Steel DJ, et al. Reestablishment of former wintering grounds by the New Zealand southern right whales. Mar Mammal Sci. 2014;30:206–220. doi: 10.1111/mms.12031. [DOI] [Google Scholar]
- Chapman DD, Feldheim KA, Papastamatiou YP, Hueter RE. There and back again: a review of residency and return migrations in sharks, with implications for population structure and management. Annu Rev Ecol Evol Syst. 2015;7:547–570. doi: 10.1146/annurev-marine-010814-015730. [DOI] [PubMed] [Google Scholar]
- Clapham P, Aguilar A, Hatch LT. Determining spatial and temporal scales for management of cetaceans: lessons from whaling. Mar Mammal Sci. 2008;24:183–201. doi: 10.1111/j.1748-7692.2007.00175.x. [DOI] [Google Scholar]
- Clapham P, Zerbini A. Is social aggregation driving high rates of increase in some Southern Hemisphere humpback whale populations? Mar Biol. 2015;162:625–634. doi: 10.1007/s00227-015-2610-3. [DOI] [Google Scholar]
- Clark PU, Dyke A, Shakun J, Carlson A, Clark J, Wohlfarth B, et al. The last glacial maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- Cooke J, Rowntree VJ, Payne R. Estimates of demographic parameters for southern right whales (Eubalaena australis) observed off Península Valdés, Argentina. J Cetacea Res Manag Spec Issue. 2001;2:125–132. [Google Scholar]
- Crandall K, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends Ecol Evol. 2000;15:290–295. doi: 10.1016/S0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Croxall JP, Butchart SHM, Lascelle B, Stattersfield AJ, Sullivan B, Symes A, et al. Seabird conservation status, threats and priority actions: a global assessment. Bird Conserv Int. 2012;22:1–34. doi: 10.1017/S0959270912000020. [DOI] [Google Scholar]
- Csilléry K, François O, Blum MGB. abc: an R package for approximate Bayesian computation (ABC) Methods Ecol Evol. 2012;3:475–479. doi: 10.1111/j.2041-210X.2011.00179.x. [DOI] [Google Scholar]
- Denis D, Crosta X, Schmidt S, Carson DS, Ganeshram RS, Renssen H, et al. Holocene productivity changes off Adélie Land (East Antarctica) Paleoceanography. 2009;24:PA3207. doi: 10.1029/2008PA001689. [DOI] [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR. NeEstimatorv2: re-implementation of software for the estimation of contemporary effective population size from genetic data. Mol Ecol Resour. 2014;14:209–214. doi: 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- Dulvy NK, Fowler SL, Musick JA, Cavanagh RD, Kyne M, Harrison LR, et al. Extinction risk and conservation of the world’s sharks and rays. eLife. 2014;3:e00590. doi: 10.7554/eLife.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, VonHoldt BM. Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol. 2002;11:1591–1604. doi: 10.1046/j.1365-294X.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Foll M. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics. 2011;27:1332–1334. doi: 10.1093/bioinformatics/btr124. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer H. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubet P, Waples R, Gaggiotti OE. Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol. 2007;16:11491166. doi: 10.1111/j.1365-294X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Tolley KA, Michaux JR, Birkun A, Ferreira M, Jauniaux T, et al. Genetic and historic evidence for climate-driven population fragmentation in a top cetacean predator: the harbour porpoises in European water. Proc R Soc B. 2010;277:2829–2837. doi: 10.1098/rspb.2010.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggiotti OE. Making inferences about speciation using sophisticated statistical genetics methods: Look before you leap. Mol Ecol. 2011;20:2229–2232. doi: 10.1111/j.1365-294X.2011.05104.x. [DOI] [PubMed] [Google Scholar]
- Gayevskiy V, Klaere S, Knight S, Goddard MR. ObStruct: a method to objectively analyse factors driving population structure using Bayesian ancestry profiles. PLoS ONE. 2014;9:e85196. doi: 10.1371/journal.pone.0085196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersonde R, Crosta X, Abelmann A, Armand L. Sea-surface temperature and sea ice distribution of the Southern Ocean at the EPILOG Last Glacial Maximum - A circum-Antarctic view based on siliceous microfossil records. Quat Sci Rev. 2005;24:869–896. doi: 10.1016/j.quascirev.2004.07.015. [DOI] [Google Scholar]
- Goudet J. FSTAT (version 1.2): a computer note computer program to calculate F-statistics. J Hered. 1995;86:485–486. doi: 10.1093/oxfordjournals.jhered.a111627. [DOI] [Google Scholar]
- Goudet J, Jombart T (2015) hierfstat: estimation and tests of hierarchical F-statistics. R package version 0.044-22. https://CRAN.R-project.org/package=hierfstat
- Goudet J, Perrin N, Waser PM. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol. 2002;11:1103–1114. doi: 10.1046/j.1365-294X.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- Greenwood P. Mating systems, philopatry and dispersal in birds and mammals. Anim Behav. 1980;28:1140–1162. doi: 10.1016/S0003-3472(80)80103-5. [DOI] [Google Scholar]
- Groch K, Palazzo J, Flores P, Ardler F, Fabian M. Recent rapid increase in the right whale (Eubalaena australis) population off southern Brazil. Lat Am J Aquat Mamm. 2005;4:41–47. doi: 10.5597/lajam00068. [DOI] [Google Scholar]
- Hoelzel AR. Impact of population bottlenecks on genetic variation and the importance of life history; a case study of the northern elephant seal. Biol J Linn Soc. 1999;68:23–39. doi: 10.1111/j.1095-8312.1999.tb01156.x. [DOI] [Google Scholar]
- Hoffmann M, Hilton-Taylor C, Angulo A, Bohm M, Brooks TM, Butchart SHM, et al. The impact of conservation on the status of the world’s vertebrates. Science. 2010;330:1503–1509. doi: 10.1126/science.1194442. [DOI] [PubMed] [Google Scholar]
- Hollenbeck CM, Portnoy DS, Gold JR. A method for detecting recent changes in contemporary effective population size from linkage disequilibrium at linked and unlinked loci. Hered (Edinb) 2016;117:207–216. doi: 10.1038/hdy.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN (2017) IUCN red list of threatened species. Version 2017-3. http://www.iucnredlist.org
- IWC Right whales: Past and present status. Rep Int Whal Comm Spec Issue. 1986;10:146–152. [Google Scholar]
- IWC Report of the workshop on the comprehensive assessment of right whales. J Cetacea Res Manag Spec Issue. 2001;2:1–60. [Google Scholar]
- IWC (2012). Report of the workshop on the assessment of southern right whales, Buenos Aires, Argentina 13-16 September 2011. Unpublished report (SC/64/Rep5) presented to the Scientific Committee of the International Whaling Commission, Cambridge, UK
- Jackson JA, Patenaude NJ, Carroll EL, Baker CS. How few whales were there after whaling? Inference from contemporary mtDNA diversity. Mol Ecol. 2008;17:236–251. doi: 10.1111/j.1365-294X.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- Jackson JA, Steel DJ, Beerli P, Congdon BC, Olavarría C, Leslie MS, et al. Global diversity and oceanic divergence of humpback whales (Megaptera novaeangliae) Proc R Soc B Biol Sci. 2014;281:20133222. doi: 10.1098/rspb.2013.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys H (1961). The theory of probability (3rd ed). University of Oxford Press, Oxford.
- Jobling MA. The impact of recent events on human genetic diversity. Philos Trans R Soc B. 2012;367:793–799. doi: 10.1098/rstb.2011.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. adegenet 1. 3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294X.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Keith SA, Bull JW. Animal culture impacts species’ capacity to realise climate-driven range shifts. Ecography (Cop) 2017;40:296–304. doi: 10.1111/ecog.02481. [DOI] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner MK. LAMARC 2.0: Maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Latch E, Dharmarajan G, Glaubitz J, Rhodes O. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet. 2006;7:295–302. doi: 10.1007/s10592-005-9098-1. [DOI] [Google Scholar]
- Leclerc M, Zedrosser A, Pelletier F. Harvesting as a potential selective pressure on behavioural traits. J Appl Ecol. 2017;54:1941–1945. doi: 10.1111/1365-2664.12893. [DOI] [Google Scholar]
- Leigh JW, Bryant D. Popart: Full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- Mate B, Best P. Coastal, offshore and migratory movements of South African right whales revealed by satellite telemetry. Mar Mammal Sci. 2011;27:455–476. doi: 10.1111/j.1748-7692.2010.00412.x. [DOI] [Google Scholar]
- Meirmans P, van Tienderen P. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. doi: 10.1111/j.1471-8286.2004.00770.x. [DOI] [Google Scholar]
- Milot E, Weimerskirch H, Bernatchez L. The seabird paradox: Dispersal, genetic structure and population dynamics in a highly mobile, but philopatric albatross species. Mol Ecol. 2008;17:1658–1673. doi: 10.1111/j.1365-294X.2008.03700.x. [DOI] [PubMed] [Google Scholar]
- Morin PA, Archer FI, Pease VL, Hancock-Hanser BL, Robertson KM, Huebinger RM, et al. Empirical comparison of single nucleotide polymorphisms and microsatellites for population and demographic analyses of bowhead whales. Endanger Species Res. 2012;19:129–147. doi: 10.3354/esr00459. [DOI] [Google Scholar]
- Mueller JC, Edelaar P, Carrete M, Serrano D, Potti J, Blas J, et al. Behaviour-related DRD4 polymorphisms in invasive bird populations. Mol Ecol. 2014;23:2876–2885. doi: 10.1111/mec.12763. [DOI] [PubMed] [Google Scholar]
- Munro KJ, Burg TM. A review of historical and contemporary processes affecting population genetic structure of Southern Ocean seabirds. Emu Austral Ornithol. 2017;117:4–18. [Google Scholar]
- Palsboll PJ, Clapham PJ, Mattila DK, Larsen F, Sears R, Siegismund HR, et al. Distribution of mtDNA haplotypes in North Atlantic humpback whales: The influence of behaviour on population structure. Mar Ecol Prog Ser. 1995;116:1–10. doi: 10.3354/meps116001. [DOI] [Google Scholar]
- Palsbøll PJ, Peery MZ, Bérubé M. Detecting populations in the ‘ambiguous’ zone: kinship-based estimation of population structure and genetic divergence. Mol Ecol Resour. 2010;10f:797–805. doi: 10.1111/j.1755-0998.2010.02887.x. [DOI] [PubMed] [Google Scholar]
- Pastene LA, Goto M, Kanda N, Zerbini AN, Kerem D, Watanabe K, et al. Radiation and speciation of pelagic organisms during periods of global warming: The case of the common minke whale. Balaenoptera acutorostrata Mol Ecol. 2007;16:1481–1495. doi: 10.1111/j.1365-294X.2007.03244.x. [DOI] [PubMed] [Google Scholar]
- Patenaude NJ, Baker CS, Gales N. Observations of southern right whales on New Zealand’s subantarctic wintering grounds. Mar Mammal Sci. 1998;14:350–355. doi: 10.1111/j.1748-7692.1998.tb00726.x. [DOI] [Google Scholar]
- Patenaude NJ, Portway V, Schaeff C, Bannister JL, Best P, Payne R, et al. Mitochondrial DNA diversity and population structure among southern right whales (Eubalaena australis) J Hered. 2007;98:147–157. doi: 10.1093/jhered/esm005. [DOI] [PubMed] [Google Scholar]
- Phillips CD, Gelatt TS, Patton JC, Bickham JW. Phylogeography of Steller sea lions: relationships among climate change, effective population size, and genetic diversity. J Mammal. 2011;92:1091–1104. doi: 10.1644/10-MAMM-A-305.1. [DOI] [Google Scholar]
- Phillips CD, Hoffman JI, George JC, Suydam RS, Huebinger RM, Patton JC, et al. Molecular insights into the historic demography of bowhead whales: Understanding the evolutionary basis of contemporary management practices. Ecol Evol. 2013;3:18–37. doi: 10.1002/ece3.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler FB, Robineau D, Goodall RNP, Meÿer MA, Olivarría C, Baker CS. Origin and radiation of Southern Hemisphere coastal dolphins (genus Cephalorhynchus) Mol Ecol. 2001;10:2215–2223. doi: 10.1046/j.0962-1083.2001.01360.x. [DOI] [PubMed] [Google Scholar]
- Pinsky ML, Palumbi SR. Meta-analysis reveals lower genetic diversity in overfished populations. Mol Ecol. 2014;23:29–39. doi: 10.1111/mec.12509. [DOI] [PubMed] [Google Scholar]
- Pirzl R, Patenaude NJ, Burnell S, Bannister JL. Movements of southern right whales (Eubalaena australis) between Australian and subantarctic New Zealand populations. Mar Mammal Sci. 2009;25:455–461. doi: 10.1111/j.1748-7692.2008.00276.x. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putman AI, Carbone I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol Evol. 2014;4:4399–4428. doi: 10.1002/ece3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyenson ND, Lindberg DR. What happened to gray whales during the pleistocene? the ecological impact of sea-level change on benthic feeding areas in the North Pacific ocean. PLoS ONE. 2011;6:e21295. doi: 10.1371/journal.pone.0021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TP, Hodgson S, Flynn L, Hilborn R, Rogers DE. Direction selection by fisheries and the timing of sockeye salmon (Oncorhynchus nerka) Ecol Appl. 2007;17:731–739. doi: 10.1890/06-0771. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard M, Xie D, Drummond A (2014) Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer
- Refsnider JM, Janzen FJ. Putting eggs in one basket: Ecological and evolutionary hypotheses for variation in oviposition-site choice. Annu Rev Ecol Evol Syst. 2010;41:39–57. doi: 10.1146/annurev-ecolsys-102209-144712. [DOI] [Google Scholar]
- Reilly SB, Bannister JL, Best P, Brown MW, Brownell R, Butterworth D, et al. (2008) Eubalaena australis (Chile-Peru subpopulation). In: IUCN 2009. IUCN Red List of Threatened Species. Version 2009.2. www.iucn.redlist.org
- Richards R. Past and present distributions of southern right whales (Eubalaena australis) New Zeal J Zool. 2009;36:447–459. doi: 10.1080/03014223.2009.9651477. [DOI] [Google Scholar]
- Rollins LA, Whitehead MR, Woolnough AP, Sinclair R, Sherwin WB. Is there evidence of selection in the dopamine receptor D4 gene in Australian invasive starling populations? Curr Zool. 2015;61:505–519. doi: 10.1093/czoolo/61.3.505. [DOI] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowntree VJ, Payne R, Schell D. Changing patterns of habitat use by southern right whales (Eubalaena australis) on their nursery ground at Península Valdés, Argentina, and in their long-range movements. J Cetacea Res Manag Spec Issue. 2001;2:133–143. [Google Scholar]
- Ruegg K, Rosenbaum HC, Anderson EC, Engel M, Rothschild A, Baker CS, et al. Long-term population size of the North Atlantic humpback whale within the context of worldwide population structure. Conserv Genet. 2013;14:103–114. doi: 10.1007/s10592-012-0432-0. [DOI] [Google Scholar]
- Scourse J (2013). Quaternary sea-level and palaeotidal changes: a review of impacts on, and responses of, the marine biosphere. In: Hughes RN, Huges D, Smith IP (eds) Oceanography and Marine Biology: An Annual Review, Taylor and Francis, Boca Raton (Florida), pp 1–70
- Smith TD, Reeves RR, Josephson E, Lund JN. Spatial and seasonal distribution of American whaling and whales in the age of sail. PLoS ONE. 2012;7:e34905. doi: 10.1371/journal.pone.0034905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa VC, Grelaud A, Hey J. On the nonidentifiability of migration time estimates in isolation with migration models. Mol Ecol. 2011;20:3956–3962. doi: 10.1111/j.1365-294X.2011.05247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiebens VA, Merino SE, Roder C, Lee PLM, Eizaguirre C. Living on the edge: how philopatry maintains adaptive potential. Proc R Soc B. 2013;280:20130305. doi: 10.1098/rspb.2013.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. Interpreting the estimated timing of migration events between hybridizing species. Mol Ecol. 2011;20:2353–2366. doi: 10.1111/j.1365-294X.2011.05048.x. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Chivers SJ, Larese J, Perrin WF (2007) Generation length and percent mature estimates for IUCN assessments of cetaceans. Adm Rep LJ-07-01, National Marine Fisheries Service, Southwest Fisheries Science Center. https://swfsc.noaa.gov/uploadedFiles/Divisions/PRD/Publications/Generation%20Length%20Admin%20Report.pdf
- Tormosov D, Mikhaliev Y, Best P, Zemsky V, Sekiguchi M, Brownell R. Soviet catches of Southern right whales Eubalaena australis 1951–71. Biol Conserv. 1998;86:185–197. doi: 10.1016/S0006-3207(98)00008-1. [DOI] [Google Scholar]
- Valenzuela LO, Sironi M, Rowntree VJ, Seger J. Isotopic and genetic evidence for culturally inherited site fidelity to feeding grounds in southern right whales (Eubalaena australis) Mol Ecol. 2009;18:782–791. doi: 10.1111/j.1365-294X.2008.04069.x. [DOI] [PubMed] [Google Scholar]
- Veríssimo A, Sampaio Iacute, McDowell JR, Alexandrino P, Mucientes G, Queiroz N, et al. World without borders—genetic population structure of a highly migratory marine predator, the blue shark (Prionace glauca) Ecol Evol. 2017;7:4768–4781. doi: 10.1002/ece3.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade P, Angliss R (1997) Guidelines for assessing marine mammal stocks: Report of the GAMMS workshop April 3-5, 1996, Seattle, WA. U.S.A. Department of Commerce, NOAA Technical Memoradum NMFS-OPR-12, 93 pp.
- Wakeley J, King L, Wilton PR. Effects of the population pedigree on genetic signatures of historical demographic events. Proc Natl Acad Sci USA. 2016;113:7994–8001. doi: 10.1073/pnas.1601080113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldick RC, Kraus SD, Brown MW, White BN. Evaluating the effects of historic bottleneck events: an assessment of microsatellite variability in the endangered North Atlantic right whale. Mol Ecol. 2002;11:2241–2249. doi: 10.1046/j.1365-294X.2002.01605.x. [DOI] [PubMed] [Google Scholar]
- Waples RS. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet. 2006;7:167–184. doi: 10.1007/s10592-005-9100-y. [DOI] [Google Scholar]
- Waples RS, Do C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl. 2010;3:244–262. doi: 10.1111/j.1752-4571.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead H. Cultural selection and genetic diversity in matrilineal whales. Science. 1998;282:1708–1711. doi: 10.1126/science.282.5394.1708. [DOI] [PubMed] [Google Scholar]
- Whiten A. A second inheritance system: the extension of biology through culture. Interface Focus. 2017;7:20160142. doi: 10.1098/rsfs.2016.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GA, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JL, Emmerson LM, Miller KJ. The influence of historical climate changes on Southern Ocean marine predator populations: a comparative analysis. Glob Chang Biol. 2016;22:474–493. doi: 10.1111/gcb.13104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.