Abstract
Interactions between phytoplankton and bacteria play a central role in mediating biogeochemical cycling and food web structure in the ocean. The cosmopolitan diatoms Thalassiosira and Chaetoceros often dominate phytoplankton communities in marine systems. Past studies of diatom-bacterial associations have employed community-level methods and culture-based or natural diatom populations. Although bacterial assemblages attached to individual diatoms represents tight associations little is known on their makeup or interactions. Here, we examined the epibiotic bacteria of 436 Thalassiosira and 329 Chaetoceros single cells isolated from natural samples and collection cultures, regarded here as short- and long-term associations, respectively. Epibiotic microbiota of single diatom hosts was analyzed by cultivation and by cloning-sequencing of 16S rRNA genes obtained from whole-genome amplification products. The prevalence of epibiotic bacteria was higher in cultures and dependent of the host species. Culture approaches demonstrated that both diatoms carry distinct bacterial communities in short- and long-term associations. Bacterial epibonts, commonly associated with phytoplankton, were repeatedly isolated from cells of diatom collection cultures but were not recovered from environmental cells. Our results suggest that in controlled laboratory culture conditions bacterial–diatom and bacterial–bacterial interactions select for a simplified, but specific, epibiotic microbiota shaped and adapted for long-term associations.
Keywords: diversity, heterotrophic bacteria, interactions, diatoms, Thalassiosira, Chaetoceros, microbiome, Western English Channel
Introduction
Bacteria eukaryotic microalgae are the major components of the plankton in the upper and ocean layers and their metabolism largely controls pelagic energy flow and nutrient cycling (Falkowski et al., 2008). Determining how they interact is therefore essential to strengthen the understanding of these groups and how they impact marine biogeochemical cycles.
While heterotrophic prokaryotes and phytoplankton are known to interact through complex mechanisms (Azam and Malfatti, 2007), it is expected that they are very closely related in the planktonic environment. The immediate environment of marine phytoplankton cells or phycosphere (Bell and Mitchell, 1972) is considered as physically and chemically distinct from the surrounding seawater, which promote the growth of specific microbial taxa, thus creating a dynamic of interactions which can help to explain the complexity of marine food webs (Seymour et al., 2017 and references therein). The use of rRNA gene sequencing and barcoding approaches allowed establishing links between phytoplankton and bacterial community dynamics in natural communities (Rooney-Varga et al., 2005; Teeling et al., 2012) and culture collections (Schäfer et al., 2002; Jasti et al., 2005; Sapp et al., 2007). These partners often co-occur which lead to beneficial, neutral or parasitic interactions (Amin et al., 2012; Cooper and Smith, 2015; Seymour et al., 2017).
Diatoms are a large component of marine biomass and produce about 25% of the total C fixed on Earth (Nelson et al., 1995; Field et al., 1998). These key ecological players of the modern ocean have been described as the most diverse group of phytoplankton (Armbrust, 2009). Their ecological success is mainly due to their numerous metabolic properties and to their silicified cell wall (Raven and Waite, 2004). Most of their evolutionary adaptations are due to the acquisition of genes from their endosymbiotic ancestors, and by indisputable horizontal gene transfers from marine bacteria, which are rarely documented in other eukaryotic organisms (Armbrust et al., 2004; Bowler et al., 2008). The co-occurrence of bacteria and diatoms in common habitats for more than 200 million years and their intimate associations likely played a major role in the ecological success and species diversification of diatoms (Amin et al., 2012).
Although few reports of diatom–bacterial interactions have used natural diatom populations (Kaczmarska et al., 2005; Rooney-Varga et al., 2005; Amin et al., 2012), most studies were performed using cultures (Grossart, 1999; Schäfer et al., 2002; Grossart et al., 2005; Kaczmarska et al., 2005; Grossart and Simon, 2007; Sapp et al., 2007; Behringer et al., 2018). Consistent associations between specific bacterial and diatom taxa have been found (Schäfer et al., 2002; Amin et al., 2012; Behringer et al., 2018), although other work suggests that the composition of diatom-associated bacterial assemblages shifts over weeks to months in culture (Sapp et al., 2007). Today, however, it remains unclear whether bacteria associated with diatom cells are species-specific (Grossart et al., 2005; Jasti et al., 2005; Rooney-Varga et al., 2005) or determined by bacterial source communities (Kaczmarska et al., 2005; Sapp et al., 2007).
Previous studies on the associations between bacteria and diatoms have mostly considered the bacteria at the population and community levels. The attachment of bacteria to algal cells represents, however, tight associations (Cole, 1982; Grossart et al., 2005). Indeed, Gärdes et al. (2011) demonstrated that attachment of specific bacteria to diatoms Thalassiosira weissflogii was required for transparent exopolymer particle formation and aggregation. Surprisingly, little is known regarding the interactions of bacterial assemblages attached to single host cells. The sole exceptions are a microscopy study documenting the abundance and mode of attachment of bacteria attached to individual diatoms (Kaczmarska et al., 2005) and a report evaluating the composition and variability of bacterial assemblages attached to individual diatoms (Baker and Kemp, 2014). More recently, Baker et al. (2016) examined the effect of abiotic and biotic factors on the composition of the attached bacteria associated to a Chaetoceros spp. culture.
In this study, our aim was to study tight associations between attached bacteria and diatom partners. For this, we focused on bacteria attached to single cells of the environmentally relevant diatom genera Thalassiosira and Chaetoceros in natural communities and in culture that were regarded as short- and long-term associations, respectively. These diatoms are ubiquitous and often numerically abundant phytoplankton species in marine systems (Leblanc et al., 2012) and they display the highest species diversity in the pelagic temperate phytoplankton community (Round et al., 1990; Hasle and Syvertsen, 1996). Based on a recent characterization of diatom diversity patterns on a global scale (Malviya et al., 2016), Chaetoceros and Thalassiosira represented the first and third most abundant ribotypes and were among the three most diverse genera. Although specific Thalassiosira–bacteria interactions have been studied (Gärdes et al., 2011; Durham et al., 2015; van Tol et al., 2017), the diversity of heterotrophic bacteria associated with both these globally significant phytoplankton genera is not well known. Furthermore, most past studies of the associations between phytoplankton and bacteria have used population or community-level approaches that may obscure cell-to-cell interactions. In this study, our major goal was to evaluate diatom-bacteria associations at an appropriate scale in focusing on the epibiotic microflora associated to Chaetoceros and Thalassiosira species. We evaluated the prevalence of attached bacteria to diatom cells and compared bacterial assemblages in both situations. We hypothesized that in situ bacterial associations differ from those in cultures and that the attachment of bacteria specific to each diatom was also favored in laboratory culture conditions.
Materials and Methods
Diatom Cultures and Natural Samples
Clonal strains of T. delicatula RCC 2560 and Chaetoceros danicus RCC 2565 were obtained from the Roscoff Culture Collection (RCC). Both diatoms have been isolated from the same sample of surface seawater (1 m depth) collected in January 2011 offshore Roscoff at the Astan observatory site (60 m depth, 48°46′40′′ N, 3°56′15′′ W) using the RV Neomysis. The long-term maintenance of both strains in the RCC since their isolation is performed by regular subculturing at intervals of 2 weeks. Both strains are grown at 19°C in K medium for diatoms (Keller et al., 1987) with a 14:10 h light:dark cycle at 80 μE.m-2.s-1.
Natural surface seawater samples (1 m depth) were collected at the Astan and Estacade (48°43′56′′ N, 3°58′58′′ W) sites in March/April 2014 (molecular approach) and in July/August 2014 (culture approach) to isolate Thalassiosira and Chaetoceros cells. A recent analysis of the microphytoplankton abundance and diversity at Astan demonstrated that Thalassiosira and Chaetoceros are abundant (above 500 cells.L-1 on average in all the 157 samples analyzed from 2000 to 2010) and able to become dominating at times (Guilloux et al., 2013). Diatom populations in this system vary throughout the year in both species diversity and abundance of individual species.
Scanning Electron Microscopy
To visualize the epibionts attached on diatom cells, we roughly followed the protocol described by Kaczmarska et al. (2005). Diatom cultures (about 2 ml) were fixed for 2 h in 3% glutaraldehyde, filtered by gravity on 5 μm polycarbonate membranes (Isopore, Millipore) and washed three times with 10 ml of 0.22 μm filter-sterilized seawater. Samples were then dehydrated in a graded series of ethanol (two successive ethanol baths at 30, 50, 70, 90% and three at 100%) for at least 10 min at each grade. Ethanol solutions (3 ml) in the filtration tower were exchanged with gentle vacuum. After critical point drying (Bal-Ted CDP 030, Balzers, Liechtenstein), the samples were sputtered with gold and examined using a Phenom G2 Pro desktop scanning electron microscope (Phenom world). Only intact cells allowing visualization of one fully exposed side of the cingulum and valve or spine (according the diatom) at a time were analyzed. Only the bacteria that demonstrated clear evidence of attachment were counted (Kaczmarska et al., 2005).
Single Diatom Cell Isolation
Single diatom cells were isolated under sterile conditions in a laminar flow hood. To lower the number of free-living bacteria in the algal cultures and to concentrate microalgae from natural seawater samples, algal cells were first gently separated by gravity using a 47 mm diameter, 11 μm pore-size nylon filter (Millipore) and washed three times with 50 mL of autoclaved seawater. Single cells were picked with a sterile glass capillary micropipette and washed 3–4 times with filter-sterilized seawater for further bacterial epibiont culture or with sterile phosphate-buffered saline (PBS; 37 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5) for further direct molecular identification of epibionts. We previously observed that replacement of sterile seawater by PBS as washing solution improved the PCR amplification success. Single cells were directly transferred in culture medium or kept on ice until further DNA extraction. For both approaches, controls were performed for each diatom cell isolated by checking the absence of bacteria in the last seawater or PBS drop used in the washing series (see below). Since these isolation steps were time-consuming, several independent late exponential cultures of both diatoms and seawater samples were needed to isolate a sufficient number of epibionts using both approaches.
Culture of Bacterial Epibionts
For cultivation of diatom epibionts, single isolated algal cells and controls were directly transferred in 48-well plates containing low-nutrient heterotrophic medium (LNHM) (Rappé et al., 2002) prepared by dissolving 35 g/L of commercial sea salts (Red Sea Europe) instead of using natural seawater. Bacterial cultures were incubated at 19°C for up to 6 weeks and growth was examined by flow cytometry using a BD Accuri C6 cytometer (BD Biosciences). For flow cytometry, 100 μl cultures were fixed with glutaraldehyde (0.25%, final concentration) and stained with SYBR Green (Life Technologies) as described by Marie et al. (1997). Cultures that contained bacteria were streaked on LNHM agar and selected colonies were purified by subculturing. Some of the isolates obtained in this study are available at the RCC.
Dereplication of Bacterial Isolates
In order to eliminate duplicates, bacterial isolates were dereplicated by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Ghyselinck et al., 2011). Colonies were obtained by growing the isolates on marine agar medium (1:10; 0.5 g peptone, 0.1 g yeast extract, 35 g sea salts dissolved in 1 L of Milli-Q water) at room temperature for 4–7 days according to their growth rate. A small amount of colony was directly applied onto a polished steel MSP 96 target plate (Bruker Daltonics). After drying, the deposited bacteria were overlaid with 1 μl of HCCA matrix (a saturated α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid; Bruker Daltonics) and air dried at room temperature. Mass spectra were acquired on a microflex LT MALDI-TOF mass spectrometer (Bruker Daltonics) configured with Bruker flexControl software using the default settings. Mass spectra were obtained in t2d format and were converted to txt files using the Data Explorer 4.9 software (AB Sciex). The txt files were imported in BioNumerics 5.1 software (Applied Maths) and converted to fingerprints for further analyses. To obtain reliable data analysis, the spectra with extensive noise and/or insufficient signal intensities were excluded. The similarity between the spectra was expressed using Pearson’s product moment correlation coefficient and the spectra were clustered using the UPGMA clustering algorithm.
Molecular Analysis of Epibiotic Microflora
DNA Extraction From Single Cells and Whole-Genome Amplification (WGA)
DNA from single diatom cells and their attached bacteria was extracted following chemical treatment and thermal shock. Cells were lysed using lysis and neutralization buffers prepared as described in Humily et al. (2014). After addition of 0.5 μL of lysis buffer, the mixture was incubated at 4°C for 10 min in a thermocycler. The lysate was further incubated at 95°C for 1 min, cooled at 4°C before adding 0.5 μL of neutralization buffer, and kept 3 min on ice until WGA.
Whole-Genome Amplification reactions were carried out under a HEPA/UV3 PCR Workstation (UVP) using the Genomiphi v2 kit (GE Healthcare). WGA reactions were carried out in 12 μL final volume by adding sample buffer (3.5 μL), reaction buffer (4.5 μL), and phi29 enzyme (0.5 μL) and then incubated for 4 h at 30°C before inactivating the enzyme for 5 min at 65°C. Positive controls consisting of 1–9 bacterial cells in 3 μL of PBS were performed to check the efficiency of WGA reaction. Blank controls with sterile PBS were also performed for each experiment. WGA products were stored at -20°C until processing.
PCR Amplification and Sequencing of rRNA Genes
Whole-Genome Amplification products were tested for the presence of bacteria with primer 1492R (Turner et al., 1999) in combination with primer 799F (Chelius and Triplett, 2001; Ghyselinck et al., 2013) that strongly discriminates against chloroplast 16S rDNA. Reaction mixtures (12.5 μL) contained 0.75 U of GoTaq G2 Flexi DNA polymerase (Promega), 1X polymerase buffer, 2.0 mM MgCl2, 0.1 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer, and 1 μL of WGA product. The program consisted of an initial denaturation step of 3 min at 95°C, followed by 35 cycles (20 s at 95°C, 40 s at 53°C, and 40 s at 72°C), and a final extension step of 10 min at 72°C. 16S rRNA genes from cultured epibionts were amplified with primers 8F (Turner et al., 1999) and 1492R using the conditions above. The program consisted of an initial denaturation step of 10 min at 94°C, followed by 35 cycles (30 s at 95°C, 1 min at 55°C, and 1 min at 72°C), and a final extension step of 10 min at 72°C.
In order to identify the diatom single cells isolated from the natural environment, the genes encoding their 18S rRNA gene and the large sub-unit (LSU) D1–D3 region were amplified using primers 63F and 1818R and D1R and D3Ca, respectively (Orsini et al., 2002). Reaction mixtures (15 μL) contained 0.75 U of GoTaq G2 Flexi DNA polymerase, 1X polymerase buffer, 2.0 mM MgCl2, 0.2 μM of each primer, 0.1 mM of each deoxynucleoside triphosphate, and 1 μL of extracted DNA. The program consisted of an initial denaturation step of 5 min at 95°C, followed by 40 cycles (30 s at 95°C, 30 s at 50°C (for 18S rRNA amplification) or 30 s at 55°C (for LSU amplification), and 1 min at 72°C) and a final extension step of 10 min at 72°C.
Whole-Genome Amplification products that proved positive for 16S rRNA but negative for 18S rRNA were removed from further analysis. PCR products of 16S rRNA genes were cloned using TOPO TA Cloning Kit® (Invitrogen) as recommended by the manufacturer. Insert-containing clones were identified by agarose gel electrophoresis of PCR products amplified using M13F and M13R primers. Clones and PCR products were sequenced by Macrogen Europe (Amsterdam, Netherlands) or by the Biogenouest sequencing platform at the Station Biologique (Roscoff, France). Bacterial taxon for each sequence was identified and named by the homologous 16S sequence in Genbank using BLAST (Altschul et al., 1990). Phylogenetic analyses of 16S and 18S rRNA gene sequences were performed using the neighbor joining tree method implemented in MEGA6 software (Tamura et al., 2013).
Statistical Analyses of the Bacterial Communities
Each single algal cell was considered as an environment to which 16S rDNA sequences were assigned. We used the unweighted UniFrac distance measure (Lozupone and Knight, 2005; Hamady et al., 2010) to compare the presence or absence of taxa with the bacterial communities. To determine whether the cultured and natural bacterial communities in both diatoms were significantly different than random, we used the Unifrac significance test along with the principal coordinates analysis (PCoA) both run in Mothur (Schloss et al., 2009).
Nucleotide Sequence Accession Numbers
All nucleotide sequences obtained in this study are available in GenBank database under the accession numbers KX197296 to KX197383, and KU926270 (16S rRNA of cultured bacteria), KX197247 to KX197295 (16S rRNA of uncultured bacteria), and KX226392 to KX226398 (16S rRNA and LSU region of microalgae).
Results and Discussion
Bacterial Colonization of Diatom Cells
In this study, we first compare the prevalence of bacterial cell attachment in both diatom species. It is well known that bacterial colonization may be influenced by the algal growth state in cultures and the bloom stage in natural samples (Grossart et al., 2005; Kaczmarska et al., 2005). To circumvent this issue, algal cultures used for single cell isolation were all in early stationary growth phase and the same natural samples were used to isolate Chaetoceros and Thalassiosira cells. A total of 296 and 469 cells were manually isolated from independent cultures and natural samples, respectively (Table 1). Both culture-based and molecular approaches yielded generally nearly identical proportions of Chaetoceros cells colonized by bacteria. However, both methods yielded different epibiont proportions in Thalassiosira. This was somewhat surprising because (i) the observed epibiont proportions were remarkably stable in the different Thalassiosira RCC 2560 cultures used for epibiont isolation whatever the approach used (see the section “Materials and Methods”) and (ii) the high epibiont prevalences observed using the culture-based approach were consistent with that measured by scanning electron microsopy (Supplementary Table 1). From these results, a reasonable explanation is that the molecular approach is most prone to yield variable results. Epibiont proportions in diatom cells isolated from the environment were generally lower than that from the cultures as measured using the culture-based approach. This result makes sense because we observed a majority of diatom cells free of epibiotic bacteria (Table 1), with proportions exceeding 90% for Chaetoceros in natural waters. These observations are in agreement with early studies that led to the conclusion that living pelagic diatoms are not colonized by bacteria (Droop and Elson, 1966) and with more recent reports that showed high proportions of bacteria-free diatom cells in old cultures (Kaczmarska et al., 2005). They also support the hypothesis that the extent of algal colonization by bacteria are positively related to the densities of both free bacteria and microalgae (Vaqué et al., 1989). This may also explain the differential proportions of diatom cells colonized by bacteria we found in cultures and in natural samples. However, we can also assume that cells in culture are confined to uniform laboratory conditions favoring stable interactions between partners and colonization while a range of environmental factors can positively or negatively influence temporary algal–bacterial relationships in natural conditions.
Table 1.
Proportions of algal cells with bacterial epibionts in cultures and natural samples as evaluated by culture-based and molecular approaches and numbers of isolates and bacterial sequences obtained in this study.
| Cultures |
Natural samples |
|||
|---|---|---|---|---|
|
Chaetoceros RCC 2565 |
Thalassiosira RCC 2560 |
Chaetoceros spp. |
Thalassiosira spp. |
|
| Culture approach | ||||
| Algal cells isolateda | 75 | 88 | 206 | 209 |
| Algal cells with epibiontsb (%) | 27 (36) | 69 (78) | 19 (9) | 68 (33) |
| Epibiont isolatesc | 38 | 63 | 12 | 64 |
| Bacterial speciesd | 5 | 14 | 7 | 38 |
| Molecular approach | ||||
| Algal cells isolatede | 20 | 113 | 28 | 26 |
| Algal cells with epibiontsf (%) | 6 (30) | 38 (34) | 2 (7) | 14 (54) |
| PCR libraries from positive WGAs | 6 | 18 | 2 | 14 |
| Bacterial phylotypes | 9 | 19 | 3 | 14 |
| Bacterial speciesd | 7 | 4 | 3 | 10 |
aCells for which no bacterial growth was obtained from controls. bPositive enrichments in LNHM as measured by flow cytometry. cEpibionts isolated on LNHM agar plates. dDefined at the 98% similarity level. eCells for which no amplification products were obtained from controls. fProportions of positive WGAs with negative controls.
Only limited quantitative information exists on the interactions between attached bacteria and phytoplankton in pelagic aquatic environments. However, the actual view is that bacterial colonization of planktonic algae may vary with respect to algal species and physiological state (Grossart et al., 2005, 2006). Indeed, we found that Thalassiosira cells harbored significantly higher proportions of epibionts than Chaetoceros cells whatever the approach used (Table 1) and this was confirmed by SEM data (Supplementary Table 1). Two possibilities can bring out the difference observed. Since associations between bacteria and microalgae are known to change over the course of algal bloom cycles (Grossart et al., 2005, 2006; Mayali et al., 2011), one possibility is that the effect of temporal variation in algal hosts collections. However, it is not the case in our samples, because Thalassiosira and Chaetoceros cells were isolated from the same natural samples although there was a 4–5 month time difference between cell collections for both approaches. Since the epibiont proportions obtained by both approaches for each diatom showed the same trends, it is unlikely that the differences observed are linked to different physiological states. A likely possibility is that diatom hosts are quite distinct in their size and structure characteristics, their release of exopolymers and their production of inhibitory substances (Vaqué et al., 1989; Myklestad, 1995; Amin et al., 2012).
Using the cultivation and molecular data, we calculated the average number of species or OTUs attached to single diatoms cells. Interestingly, whatever the approach used, we found that each host cell harbored between 1 to 2.3 epibiotic species or OTUs, suggesting a rather low attachment. This average number is a minimum estimate of the number of bacterial cells that occurred on the host cell, but with the methods used it was not possible to evaluate how many bacterial cells were attached to the host cells examined. However, these numbers are close to that we obtained using SEM on late exponential cultures (0.8–3.8 bacterial epibionts on cingulum or spines/valve; Supplementary Table 1). These low numbers were generally in agreement with that reported by Baker and Kemp (2014) although these authors found also higher numbers of phylotypes (up to 11) per algal cell for some species or strains.
Microbial Community Comparisons Between Hosts in Short- and Long-Term Associations
Using the molecular method, a total of 45 bacterial phylotypes were obtained after WGA of 40 single diatom cells isolated from collection cultures and natural samples (Tables 1, 2). When clustered at the 98% similarity level, 24 OTUs representing diverse lineages of bacterial phyla were identified and their distribution did not overlap among host species and between collection cultures and environmental samples. The identified OTUs were classified into the classes Alpha-, Beta- and Gammaproteobacteria, Actinobacteria, Flavobacteriia, Bacilli, Cytophagia, and Sphingobacteria. Bacterial cultivation using a low-nutrient organic medium and extended incubation periods of up to 6 weeks were used to increase the overall assessment of the species richness. Cultivation identified a total of 177 unique isolates (Tables 1, 3, 4). The isolated strains fell generally into the same bacterial classes than environmental clones but none belonged to the classes Betaproteobacteria and Bacilli.
Table 2.
Epibiotic bacterial phylotypes recovered from diatom cells in culture and from natural waters and their closest cultivated relative identified by Blastn.
| Diatom host | na | Phylum/class | Order | Family | Species | 16S rRNA similarity (%) |
|---|---|---|---|---|---|---|
| Cultures | ||||||
| C. danicus RCC 2565 | 3 | Alphaproteobacteria | Rhodobacterales | Hyphomonadaceae | Algimonas sp. | 99.6 |
| 1 | Betaproteobacteria | Burkholderiales | Burkholderiaceae | Limnobacter thiooxidans | 100 | |
| 1 | Comamonadaceae | Pelomonas sp. | 99.9 | |||
| 1 | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | Halomonas sp | 99.9 | |
| 1 | Flavobacteriia | Flavobacteriales | Cryomorphaceae | Brumimicrobium mesophilum | 93.5 | |
| 1 | Actinobacteria | Micrococcales | Micrococcaceae | Nesterenkonia flava | 99.4 | |
| 1 | Bacilli | Bacillales | Bacillaceae | Aeribacillus pallidus | 99.9 | |
| T. delicatula RCC 2560 | 1 | Alphaproteobacteria | Sphingomonadales | Erythrobacteraceae | Erythrobacter litoralis | 98.8 |
| 1 | Rhodobacterales | Rhodobacteraceae | Maribius pelagius | 94.7 | ||
| 1 | Bacilli | Bacillales | Bacillaceae | Aeribacillus pallidus | 100 | |
| 16 | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Tenacibaculum skagerrakense | 97.8 | |
| Environmental cellsb | ||||||
| Chaetoceros debilis | 1 | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Marivita cryptomonadis | 98.5 |
| 1 | Pseudoruegeria lutimaris | 97 | ||||
| Chaetoceros protuberans | 1 | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Saccharophagus degradans | 95.1 |
| Thalassiosira punctigera | 1 | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Roseovarius nubinhibens | 97.9 |
| 1 | Gammaproteobacteria | Alteromonadales | Colwelliaceae | Colwellia psychroerythraea | 99.3 | |
| 1 | Chromatiales | Ectothiorhodospiraceae | Ectothiorhodospira mobilis | 94.3 | ||
| 1 | Cellvibrionales | Halieaceae | Chromatocurvus halotolerans | 95.7 | ||
| 1 | Spongiibacteraceae | Dasania marina | 93 | |||
| 1 | Oceanicoccus sagamiensis | 96.8 | ||||
| 1 | Pseudomonadales | Pseudomonadaceae | Pseudomonas trivialis | 90.7 | ||
| 4 | Cytophagia | Cytophagales | Amoebophilaceae | Candidatus Amoebophilus asiaticus | 93 | |
| 2 | Flavobacteriia | Flavobacteriales | Cryomorphaceae | Salinirepens amamiensis | 94.4 | |
| 1 | Sphingobacteriia | Sphingobacteriales | Saprospiraceae | Lewinella cohaerens | 89.1 |
aNumber of individual diatom cell carrying the corresponding bacterial phylotype; bas identified by 18S rRNA sequencing.
Table 3.
Epibiotic bacterial strains isolated from diatom cells in natural waters and their closest cultivated relative identified by Blastn.
| Diatom host | Strain (n) | Phylum/class | Order | Family | Species | 16S rRNA similarity (%) |
|---|---|---|---|---|---|---|
| Chaetoceros spp. | KC58 (2) | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | Hoeflea phototrophica | 97.9 |
| KC43 (1) | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Glaciecola punicea | 96 | |
| KC55B (4) | Pseudoalteromonadaceae | Pseudoalteromonas undina/marina | 99 | |||
| KC57 (1) | Pseudoalteromonas carrageenovora | 100 | ||||
| KC61 (1) | Vibrionales | Vibrionaceae | Photobacterium aquimaris | 99.9 | ||
| KC46A (1) | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Tenacibaculum gallaicum | 97.7 | |
| KC79A (2) | Actinobacteria | Actinomycetales | Micrococcaceae | Arthrobacter subterraneus | 99.8 | |
| Thalassiosira spp. | KC76 (1) | Alphaproteobacteria | Kiloniellales | Kiloniellaceae | Kiloniella laminariae | 96.3 |
| KC56 (1) | Rhizobiales | Hyphomicrobiaceae | Hyphomicrobium nitrativorans | 95.3 | ||
| KC41B (1) | Phyllobacteriaceae | Nitratireductor indicus | 95.2 | |||
| KC41A (1) | Nitratireductor pacificus | 95.2 | ||||
| KC45 (1) | Rhodobacterales | Rhodobacteraceae | Jannaschia sp. | 98 | ||
| KC59 (1) | Litoreibacter meonggei | 97.7 | ||||
| KC77A (2) | Loktanella maricola | 99.6 | ||||
| KC68 (1) | Loktanella vestfoldensis | 96 | ||||
| KC75 (1) | Nereida ignava | 97.3 | ||||
| KC63A (3) | Octadecabacter antarcticus | 98.1 | ||||
| KC47 (1) | Roseobacter litoralis | 98.8 | ||||
| KC52A (2) | Shimia marina | 98.2 | ||||
| KC42A (2) | Sphingomonadales | Erythrobacteraceae | Altererythrobacter luteolus | 97 | ||
| KC74 (1) | Croceicoccus marinus | 97.3 | ||||
| KC49 (1) | Erythrobacter aquimaris | 99.6 | ||||
| KC67 (1) | Sphingomonadaceae | Sphingopyxis flavimaris | 98.8 | |||
| KC60 (1) | Cytophagia | Cytophagales | Flammeovirgaceae | Roseivirga ehrenbergii | 93.2 | |
| KC65A (2) | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Croceitalea eckloniae | 97.3 | |
| KC50A (2) | Dokdonia genica | 99.7 | ||||
| KC62 (1) | Lacinutrix sp. | 97.3 | ||||
| KC44 (1) | Lutimonas vermicola | 100 | ||||
| KC51B (2) | Maribacter aquivivus | 97.7 | ||||
| KC51A (1) | Maribacter forsetii | 96 | ||||
| KC81 (1) | Maribacter ulvicola | 99.3 | ||||
| KC73A (2) | Olleya namhaensis | 97.3 | ||||
| KC46B (2) | Tenacibaculum gallaicum | 97.7 | ||||
| KC48 (1) | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Aestuariibacter litoralis | 97.8 | |
| KC78 (1) | Alteromonas tagae | 99 | ||||
| KC72 (1) | Pseudohaliea rubra | 98 | ||||
| KC53A (5) | Pseudoalteromonadaceae | Pseudoalteromonas undina/marina | 99.9 | |||
| KC66 (1) | Shewanellaceae | Shewanella japonica | 99.3 | |||
| KC70 (1) | Colwelliaceae | Thalassomonas agariperforans | 96.9 | |||
| KC64 (1) | Chromatiales | Granulosicoccaceae | Granulosicoccus coccoides | 96.9 | ||
| KC80 (1) | Vibrionales | Vibrionaceae | Vibrio breoganii | 100 | ||
| KC54 (1) | Vibrio splendidus | 100 | ||||
| KC71 (1) | Unclassified | Chromatocurvus halotolerans | 97.3 | |||
| Gammaproteobacteria | ||||||
| KC69 (1) | Sphingobacteriia | Sphingobacteriales | Saprospiraceae | Portibacter lacus | 91.5 | |
| KC79C (1) | Actinobacteria | Actinomycetales | Micrococcaceae | Arthrobacter subterraneus | 99.8 |
aNumber of individual diatom cell carrying the corresponding bacterial isolate; bbased on partial sequences >663 bp.
Table 4.
Epibiotic bacterial strains isolated from diatom cells in culture and their closest cultivated relative identified by Blastn.
| Diatom host | Strain (n) | Phylum/class | Order | Family | Species | 16S rRNA similarity (%) |
|---|---|---|---|---|---|---|
| C. danicus RCC 2565 | KC05A (17) | Alphaproteobacteria | Caulobacterales | Hyphomonadaceae | Algimonas ampicilliniresistens | 98 |
| KC39 (2) | Rhodobacterales | Rhodobacteraceae | Sulfitobacter dubius | 99.9 | ||
| KC25 (11) | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Marinobacter sediminum | 99.8 | |
| KC36 (1) | Marinobacter lipolyticus | 99.8 | ||||
| KC86 (1) | Silicimonas algicola | 100 | ||||
| T. delicatula RCC 2560 | KC04 (1) | Alphaproteobacteria | Sphingomonadales | Erythrobacteraceae | Altererythrobacter ishigakiensis | 97.9 |
| KC12 (26) | Erythrobacter gaetbuli | 98 | ||||
| KC18 (1) | Erythrobacter citreus | 98.3 | ||||
| KC90 (15) | Rhodobacterales | Rhodobacteraceae | Silicimonas algicola | 100 | ||
| KC10 (10) | Paracoccus aminophilus | 96.7 | ||||
| KC17B (1) | Paracoccus stylophorae | 97.8 | ||||
| KC38C (1)KC16 (3) | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Sulfitobacter dubiusMarinobacter salarius | 99.999.9 | |
| KC31 (3) | Marinobacter sediminum | 100 | ||||
| KC14 (3) | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Tenacibaculum sp | 95 | |
| KC02 (1) | Actinobacteria | Actinomycetales | Micrococcaceae | Kocuria rosea | 99.7 | |
| KC23 (1) | Micrococcus yunnanensis | 99.4 | ||||
| KC03 (1) | Nocardioidaceae | Nocardioides furvisabuli | 99.6 | |||
| KC24 (1) | Dermacoccaceae | Dermacoccus nishinomiyaensis | 98.8 |
aNumber of individual diatom cell carrying the corresponding bacterial isolate; bbased on partial sequences >663 bp.
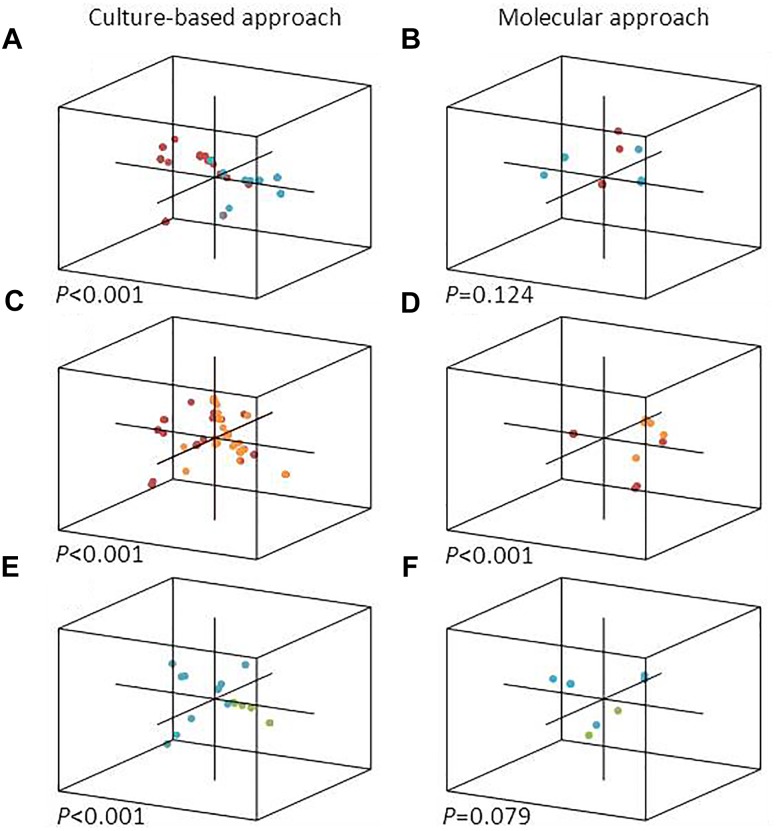
In this study, we asked if microalgal species could influence the epibiont community structure in cultures and in natural communities. Unweighted UniFrac analyses, which takes into account only presence/absence data for OTUs/species, showed that the epibiotic bacterial communities associated with T. delicatula RCC 2560 and C. danicus RCC 2565 were significantly distinct (P < 0.001) as determined by the culture-based approach. Principal Coordinate Analysis (PCoA) also separated the corresponding libraries (Figure 1A). No significant dissimilarities were shown between epibiont communities of the cultures as determined by the molecular approach (P = 0.124) (Figure 1B). Our results complement, however, earlier observations suggesting that microalgal cultures harbor specific bacterial communities (Schäfer et al., 2002; Grossart et al., 2005; Sapp et al., 2007; Guannel et al., 2011; Behringer et al., 2018). We also compared the culture-based and molecular bacterial diversity attached to Thalassiosira and Chaetoceros single cells isolated from natural samples and collection cultures, considered here as short- and long-term associations, respectively. Both methods indicated that epibiotic bacterial communities associated with cultures and environmental cells of Thalassiosira showed a highly significant difference (P < 0.001) (Figures 1C,D). Epibiont assemblages in cultured and environmental Chaetoceros cells also differed significantly as determined by culture-based approach (P < 0.001) while molecular libraries showed only marginally significant difference (P = 0.079) (Figures 1E,F). Since single cells isolated from natural waters belonged to two Chateoceros species, we enlarged the specificity of in situ associations to the genus level. Nevertheless, distinctness between in situ epibiotic communities of both diatom genera was observed in the same samples collected at the same place. This result is in line with previous findings showing that bacterial assemblages associated with phytoplankton cultures can be very different from the natural bacterial assemblages during blooms of the same species (Garcés et al., 2007). We acknowledge, however, that the diatom hosts isolated from natural waters differed at the species level from their cultured relatives.
FIGURE 1.
PCoA representation of each single diatom cell analyzed in this study, with a 3D position depending on the composition of its epibiotic bacterial community. Culture-based (left) and molecular (right) bacterial diversity attached to diatoms isolated from collection cultures (A,B), natural samples (E,F) and both (C,D) are shown. Diatom cells correspond to T. delicatula RCC 2560 (red), C. danicus RCC 2565 (blue), environmental Thalassiosira spp. (orange), and environmental Chaetoceros spp. (green).
Diversity of Diatom Epibionts
Although the molecular approach was designed and performed using necessary precautions (decontamination and cleaning procedures) to prevent contamination, the successive steps that include single cell isolation, whole genome and PCR amplification presented opportunities for the inclusion of non-host derived bacterial sequences. We also removed samples with any negative control amplification from further analysis. We didn’t find sequences recognized as being commonly associated to human skin but a few phylotypes and strains were not typical of bacterial taxa found previously with algal cultures (Tables 2–4) Among them, we identified sequences of Aeribacillus pallidus (Firmicutes) that were abundantly found in skin-associated bacterial communities of marine fishes (Larsen et al., 2013). Like other Gram-positive bacteria frequently found in marine microalgal laboratory cultures, we suspect their presence is due to a contamination from handling (Nicolas et al., 2004). Although Actinobacteria are known to be rare in pelagic marine environments (Pommier et al., 2007), we obtained several actinobacterial strains and sequences attached to diatom cells in cultures and natural samples (Tables 2–4). Interestingly, the isolates belong to genera with species obtained from diatom cultures (Le Chevanton et al., 2013) and from marine environments, most of them in association with invertebrates, algae and microalgae (Green et al., 2004; Pathom-aree et al., 2006; Menezes et al., 2010; Palomo et al., 2013). We therefore cannot exclude that these bacteria represent rare members in the diatom phycosphere.
Epibiotic bacteria isolated from natural diatom populations were highly diverse (Table 3) and only a few species overlapped in Chaetoceros and Thalassiosira cells. Interestingly, the overlapping bacterial species were also the most frequently isolated from both genera. They consisted in species of the genera Pseudoalteromonas and Tenacibaculum, the majority of which appear to be associated with eukaryotic hosts, including various marine algae (Akagawa-Matsushita et al., 1992; Egan et al., 2000, 2001; Suzuki et al., 2001). Strains and phylotypes of Tenacibaculum were also repeatedly isolated and amplified from cells of T. delicatula RCC 2560, suggesting a possible physiological adaptation to an epiphytic lifestyle. This hypothesis is in line with the ecology of Tenacibaculum species known to be often associated with surfaces of marine invertebrates, fishes and microalgae and responsible of skin lesions and algicidal activity (Piñeiro-Vidal et al., 2012; Li et al., 2013).
Alpha- and Gammaproteobacteria were the two main classes identified in the cultivable epibiotic communities associated with cultures (Table 4 and Supplementary Figure 1). Interestingly, some epibionts were regularly isolated from both diatom cultures. Among them, strains belonging to the gammaproteobacterial genus Marinobacter are common inhabitants of phytoplankton cultures (Amin et al., 2009, 2015; Le Chevanton et al., 2013; Sison-Mangus et al., 2014; Green et al., 2015; Lupette et al., 2016). While investigating the bacterial community associated with dinoflagellates and coccolithophores, Amin et al. (2009) observed that members of the genus Marinobacter were present in over 80% of cultures. Further studies demonstrated that Marinobacter isolates formed specific beneficial associations with diverse phytoplankton that could require cell-to-cell adhesion (Amin et al., 2009; Bolch et al., 2011; Gärdes et al., 2011). Other frequently isolated epibiotic bacteria were affiliated with the Rhodobacterales and they differed with the algal species. Rhodobacterales appear also to be particularly well-adapted to close association with phytoplankton in general and have been shown to increase during phytoplankton blooms (Mayali et al., 2011; Teeling et al., 2012). Laboratory experiments involving specific co-culture experiments also indicate that widespread interactions may occur between phytoplankton and Roseobacters (Durham et al., 2015; Segev et al., 2016; Seymour et al., 2017). Cultivated attached microflora of C. danicus RCC 2565 were related to the genera Algimonas while Erythrobacter, Paracoccus and a new genus within the family Rhodobacteraceae, recently described as Silicimonas algicola (Crenn et al., 2016), prevailed in T. delicatula RCC 2560. Algimonas, Erythrobacter, and Paracoccus are also reported for their common occurrence in algal cultures or as epibionts of macroalgae (Alavi et al., 2001; Schäfer et al., 2002; Jasti et al., 2005; Kaczmarska et al., 2005; Le Chevanton et al., 2013; Cruz-López and Maske, 2016).
Phylotypes recovered from diatom cells in culture and from natural waters were diverse and only a few of them were repeatedly obtained (Table 2). Similarly to Tenacibaculum species that were regularly amplified and isolated from cells of T. delicatula RCC 2560, sequences and strains of Algimonas were often obtained from cells of C. danicus RCC 2565, confirming the presence of members of this genus in Chaetoceros cultures (Le Chevanton et al., 2013). A phylotype distantly related to the obligate amoeba endosymbiont “Candidatus Amoebophilus asiaticus” was retrieved from environmental cells of T. punctigera. Although it was repeatedly amplified from single cells, it is impossible to draw any hypothesis on the possible relationships between these partners in natural waters.
A total of 56 unique isolates were obtained in this study (Tables 3, 4). We used the 16S rRNA gene identity cut-off value of 98.7% to define isolates that may represent novel species (Stackebrandt and Ebers, 2006). Overall, 35 unique isolates (>60% of the total isolates) showed 16S rRNA gene identity below this threshold and therefore may be potential candidates of new taxa. Of the new taxa, 5 isolates may represent novel genera at the conservative 95–96% identity cut-off (Yarza et al., 2014). Although 16S rRNA gene identities are based on partial sequences and our study does not prove that these isolates represent novel species, it provides a framework for isolating large numbers of epibiotic bacteria for possible novel taxa that may be of ecological importance. Indeed, repeated isolation of particular bacterial species from microalgae could be indicative of niche specificity or even established mutualistic relationships (Amin et al., 2009, 2012).
Our data demonstrated that diversity of the cultured epibiotic microflora was lower in diatom culture than in natural waters. Furthermore, the most frequently isolated taxa from diatom cultures were unusually isolated or amplified from environmental cells (Tables 2, 3). This is exemplified by Marinobacter species that we regularly grown from diatoms originally isolated from Roscoff coastal waters but were not found as diatom epibionts in natural waters off Roscoff, confirming the typically low annual relative abundance of these bacteria in the Western English Channel (Green et al., 2015). Together, our results provide some support to the hypothesis that, in uniform laboratory culture conditions, bacteria–bacteria and bacteria–diatom interactions selected for a simplified, but specific, epibiotic microbiota shaped and adapted for long-term associations. Since recent observations showed that cultivation in the laboratory for longer than 1 year resulted in only small changes in the bacteria composition suggesting robust associations between diatoms and their associated bacterial communities (Behringer et al., 2018), further work is needed to how these interactions can differ across algal species. Our findings reinforce also previous reports that showed that the amount and composition of the organic matter released by phytoplankton like polysaccharides, small amino acids, sugars, proteoglycans or glycoproteins, may act as selective agents for bacterial types (Myklestad, 1995; Sapp et al., 2007; Sarmento et al., 2013). We assume that bacterial groups able to develop an algal-attached lifestyle are probably more affected by this selection process.
Methodological Considerations
This study addressed the examination of interactions between attached bacteria and two environmentally relevant diatom genera. Most studies that tried to tackle specifically this question first separated free-living bacteria from diatom cells by filtration on membranes and examined the bacterial assemblages associated to the diatom cell fraction (Kaczmarska et al., 2005; Mayali et al., 2011). We assume that the issue of specific associations cannot be answered using this method because non-attached bacteria may also be retained by membranes, and remain on them even after extensive washing steps. The methods described in our study and that employed by Baker and Kemp (2014) ensure the epibiotic status of the bacterial communities analyzed. The main limitation of our approach is that the manual isolation of single cells is time-consuming and limits the number of analyzed cells. Future studies might apply different strategies to pursue this question further. Although they require an expensive and sophisticated equipment and special infrastructure such as a clean room (Baker and Kemp, 2014), flow cytometry cell sorting systems may represent powerful tools to facilitate the rapid and efficient isolation of microalgae. They could greatly improve the analysis of attached bacterial assemblages in multiple cultures and environmental cells.
Conclusion
Our observations complement previous studies which addressed the existence of algal-specific bacterial communities. The present analysis of the microflora attached to ubiquitous marine diatoms demonstrated conclusively that abundance and community composition of epibiotic bacteria may vary significantly with algal species. The dominance of certain epibiotic bacteria, either common or specific to algal species, together with the simplification of bacterial communities along regular algal subculturing indicate selection of bacteria highly adapted to long-term interactions with hosts.
In the context of finding bacteria that could have symbiotic interactions with diatoms, the bacterial strains we repeatedly isolated from cultures and environmental cells represent good candidates. Further co-cultures experiments with axenic cultures of T. delicatula RCC 2560 and C. danicus RCC 2565 evaluating the effect of bacteria on both microalgae could help us to determine the functional role of specific isolates.
Author Contributions
KC designed and performed the experiments (diversity analysis) and wrote the manuscript. DD performed the experiments (diversity analysis). CJ designed the experiments and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly acknowledge Raffaele Siano and Diana Sarno for their help in the identification of both diatom cultures used in this study. We also thank Geneviève Héry-Arnaud and colleagues (Unité de Bactériologie, CHRU de la Cavale Blanche, Brest, France) for the use of their Microflex LT MALDI-TOF mass spectrometer (Bruker Daltonics). Many thanks to Biogenouest sequencing platform at the Station Biologique de Roscoff for help and advices. Camille Poirier, Mathilde Miossec, and Sophie Le Panse are warmly acknowledged for their help in sequencing and scanning electron microscopy.
Footnotes
Funding. This work was supported by the French national programme EC2CO-Microbien (project MICROMAR) and the MaCuMBA project funded by the European Union’s Seventh Framework Programme (Grant agreement no. 311975). KC received a doctoral grant funded by Région Bretagne and CNRS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02879/full#supplementary-material
Maximum-likelihood tree based on 16S rRNA gene sequences showing the position of bacterial epibionts isolated from C. danicus RCC 2565 and T. delicatula RCC 2560 (Table 4). Only bootstrap values (expressed as percentages of 1000 replications) of >80% are shown. Filled circles indicate that the corresponding nodes were also recovered using the neighbor-joining algorithm. Thermococcus litoralis JCM8560T was used as outgroup. Bar, 0.05 substitutions per nucleotide position.
Attached bacteria on diatom cultures during late exponential (5 days) and late stationary (19 days) growth phases.
References
- Akagawa-Matsushita M., Matsuo M., Koga Y., Yamasato K. (1992). Alteromonas atlantica sp. nov. and Alteromonas carrageenovora sp. nov., bacteria that decompose algal polysaccharides. Int. J. Syst. Bacteriol. 42 621–627. 10.1099/00207713-42-4-621 [DOI] [Google Scholar]
- Alavi M., Miller T., Erlandson K., Schneider R., Belas R. (2001). Bacterial community associated with Pfesteria-like dinoflagellate cultures. Environ. Microbiol. 3 380–396. 10.1046/j.1462-2920.2001.00207.x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amin S. A., Green D. H., Hart M. C., Küpper F. C., Sunda W. G., Carrano C. J. (2009). Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc. Natl. Acad. Sci. U.S.A. 106 17071–17076. 10.1073/pnas.0905512106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A., Hmelo L. R., van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522 98–101. 10.1038/nature14488 [DOI] [PubMed] [Google Scholar]
- Amin S. A., Parker M. S., Armbrust E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76 667–684. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust E. V. (2009). The life of diatoms in the world’s oceans. Nature 459 185–192. 10.1038/nature08057 [DOI] [PubMed] [Google Scholar]
- Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306 79–86. 10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- Azam F., Malfatti F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5 782–791. 10.1038/nrmicro1798 [DOI] [PubMed] [Google Scholar]
- Baker L., Alegado R. A., Kemp P. (2016). Response of diatom-associated bacteria to host growth stage, nutrient concentrations, and viral host infection in a model system. Environ. Microbiol. Rep. 8 917–927. 10.1111/1758-2229.12456 [DOI] [PubMed] [Google Scholar]
- Baker L., Kemp P. (2014). Exploring bacteria–diatom associations using single-cell whole genome amplification. Aquat. Microb. Ecol. 72 73–88. 10.3354/ame01686 [DOI] [Google Scholar]
- Behringer G., Ochsenkühn M. A., Fei C., Fanning J., Koester J. A., Amin S. A. (2018). Bacterial communities of diatoms display strong conservation across strains and time. Front. Microbiol. 9:659. 10.3389/fmicb.2018.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell W., Mitchell R. (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143 265–277. 10.2307/1540052 [DOI] [Google Scholar]
- Bolch C. J. S., Subramanian T. A., Green D. H. (2011). The toxic dinoflagellate gymnodinium catenatum (Dinophyceae) requires marine bacteria for growth. J. Phycol. 47 1009–1022. 10.1111/j.1529-8817.2011.01043.x [DOI] [PubMed] [Google Scholar]
- Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456 239–244. 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- Chelius M. K., Triplett E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41 252–263. 10.1007/s002480000087 [DOI] [PubMed] [Google Scholar]
- Cole J. J. (1982). Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13 291–314. 10.1146/annurev.es.13.110182.001451 [DOI] [Google Scholar]
- Cooper M. B., Smith A. G. (2015). Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr. Opin. Plant Biol. 26 147–153. 10.1016/j.pbi.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Crenn K., Serpin D., Lepleux C., Overmann J., Jeanthon C. (2016). Silicimonas algicola gen. nov., sp. nov., a member of the Roseobacter clade isolated from the cell surface of the marine diatom Thalassiosira delicatula. Int. J. Syst. Evol. Microbiol. 66 4580–4588. 10.1099/ijsem.0.001394 [DOI] [PubMed] [Google Scholar]
- Cruz-López R., Maske H. (2016). The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front. Microbiol. 7:560. 10.3389/fmicb.2016.00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droop M. R., Elson K. G. R. (1966). Are pelagic diatoms free from bacteria? Nature 211 1096–1097. 10.1038/2111096a0 [DOI] [Google Scholar]
- Durham B. P., Sharma S., Luo H., Smith C. B., Amin S. A., Bender S. J., et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112 453–457. 10.1073/pnas.1413137112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S., James S., Holmstro C., Holmström C., Kjelleberg S. (2001). Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiol. Ecol. 35 67–73. 10.1111/j.1574-6941.2001.tb00789.x [DOI] [PubMed] [Google Scholar]
- Egan S., Thomas T., Holmström C., Kjelleberg S. (2000). Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ. Microbiol. 2 343–347. 10.1046/j.1462-2920.2000.00107.x [DOI] [PubMed] [Google Scholar]
- Falkowski P. G., Fenchel T., Delong E. F. (2008). The microbial engines that drive Earth ’s biogeochemical cycles. Science 320 1034–1039. 10.1126/science.1153213 [DOI] [PubMed] [Google Scholar]
- Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281 237–240. 10.1126/science.281.5374.237 [DOI] [PubMed] [Google Scholar]
- Garcés E., Vila M., Reñé A., Alonso-Sáez L., Anglès S., Lugliè A., et al. (2007). Natural bacterioplankton assemblage composition during blooms of Alexandrium spp. (Dinophyceae) in NW Mediterranean coastal waters. Aquat. Microb. Ecol. 46 55–70. 10.3354/ame046055 [DOI] [Google Scholar]
- Gärdes A., Iversen M. H., Grossart H. P., Passow U., Ullrich M. S. (2011). Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5 436–445. 10.1038/ismej.2010.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck J., Pfeiffer S., Heylen K., Sessitsch A., De Vos P. (2013). The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. PLoS One 8:e71360. 10.1371/journal.pone.0071360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck J., Van Hoorde K., Hoste B., Heylen K., De Vos P. (2011). Evaluation of MALDI-TOF MS as a tool for high-throughput dereplication. J. Microbiol. Methods 86 327–336. 10.1016/j.mimet.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Green D. H., Echavarri-bravo V., Brennan D., Hart M. C. (2015). Bacterial diversity associated with the coccolithophorid algae Emiliania huxleyi and Coccolithus pelagicus f. braarudii. Biomed. Res. Int. 2015:194540. 10.1155/2015/194540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D. H., Llewellyn L. E., Negri A. P., Blackburn S. I., Bolch C. J. S. (2004). Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 47 345–357. 10.1016/S0168-6496(03)00298-8 [DOI] [PubMed] [Google Scholar]
- Grossart H. P. (1999). Interactions between marine bacteria and axenic diatoms (Cylindrotheca fusiformis, Nitzschia laevis, and Thalassiosira weissflogii) incubated under various conditions in the lab. Aquat. Microb. Ecol. 19 1–11. 10.3354/ame019001 [DOI] [Google Scholar]
- Grossart H. P., Czub G., Simon M. (2006). Algae-bacteria interactions and their effects on aggregation and organic matter flux in the sea. Environ. Microbiol. 8 1074–1084. 10.1111/j.1462-2920.2006.00999.x [DOI] [PubMed] [Google Scholar]
- Grossart H. P., Levold F., Allgaier M., Simon M., Brinkhoff T. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7 860–873. 10.1111/j.1462-2920.2005.00759.x [DOI] [PubMed] [Google Scholar]
- Grossart H. P., Simon M. (2007). Interactions of planktonic algae and bacteria: effects on algal growth and organic matter dynamics. Aquat. Microb. Ecol. 47 163–176. 10.3354/ame047163 [DOI] [Google Scholar]
- Guannel M. L., Horner-Devine M. C., Rocap G. (2011). Bacterial community composition differs with species and toxigenicity of the diatom Pseudo-nitzschia. Aquat. Microb. Ecol. 64 117–133. 10.3354/ame01513 [DOI] [Google Scholar]
- Guilloux L., Rigaut-Jalabert F., Jouenne F., Ristori S., Viprey M., Not F., et al. (2013). An annotated checklist of marine phytoplankton taxa at the SOMLIT-Astan time series off Roscoff (Western English Channel, France): data collected from 2000 to 2010. Cah. Biol. Mar. 54 247–256. [Google Scholar]
- Hamady M., Lozupone C., Knight R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4 17–27. 10.1038/ismej.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle G. R., Syvertsen E. (1996). “Marine diatoms,” in Identifying Marine Diatoms and Dinoflagellates, ed. Tomas C. R. (San Diego, CA: Academic Press, Inc; ), 5–385. [Google Scholar]
- Humily F., Farrant G. K., Marie D., Partensky F., Mazard S., Perennou M., et al. (2014). Development of a targeted metagenomic approach to study a genomic region involved in light harvesting in marine Synechococcus. FEMS Microbiol. Ecol. 88 231–249. 10.1111/1574-6941.12285 [DOI] [PubMed] [Google Scholar]
- Jasti S., Sieracki M. E., Poulton N. J., Giewat M. W., Rooney-Varga J. N. (2005). Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71 3483–3494. 10.1128/AEM.71.7.3483-3494.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarska I., Ehrman J. M., Bates S. S., Green D. H., Léger C., Harris J. (2005). Diversity and distribution of epibiotic bacteria on Pseudo-nitzschia multiseries (Bacillariophyceae) in culture, and comparison with those on diatoms in native seawater. Harmful Algae 4 725–741. 10.1016/j.hal.2004.10.001 [DOI] [Google Scholar]
- Keller M. D., Seluin R. C., Claus W., Guillard R. R. L. (1987). Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23 633–638. 10.1016/0198-0254(88)92621-0 [DOI] [Google Scholar]
- Larsen A., Tao Z., Bullard S. A., Arias C. R. (2013). Diversity of the skin microbiota of fishes: evidence for host species specificity. FEMS Microbiol. Ecol. 85 483–494. 10.1111/1574-6941.12136 [DOI] [PubMed] [Google Scholar]
- Le Chevanton M., Garnier M., Bougaran G., Schreibera N., Lukomska E., Beìrard J.-B., et al. (2013). Screening and selection of growth-promoting bacteria for Dunaliella cultures. Algal Res. 2 212–222. 10.1016/j.algal.2013.05.003 [DOI] [Google Scholar]
- Leblanc K., Arístegui J., Armand L., Assmy P., Beker B., Bode A., et al. (2012). A global diatom database – abundance, biovolume and biomass in the world ocean. Earth Syst. Sci. Data 4 149–165. 10.5194/essdd-5-147-2012 [DOI] [Google Scholar]
- Li Y., Wei J., Yang C., Lai Q., Chen Z., Li D., et al. (2013). Tenacibaculum xiamensense sp. nov., an algicidal bacterium isolated from coastal seawater. Int. J. Syst. Evol. Microbiol. 63 3481–3486. 10.1099/ijs.0.050765-0 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupette J., Lami R., Krasovec M., Grimsley N., Moreau H., Piganeau G., et al. (2016). Marinobacter dominates the bacterial community of the Ostreococcus tauri phycosphere in culture. Front. Microbiol. 7:1414. 10.3389/fmicb.2016.01414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Bittner L., et al. (2016). Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U.S.A. 113 E1516–E1525. 10.1073/pnas.1509523113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D., Partensky F., Jacquet S., Vaulot D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63 186–193. 10.1111/j.1365-294X.2009.04480.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayali X., Franks P. J. S., Burton R. S. (2011). Temporal attachment dynamics by distinct bacterial taxa during a dinoflagellate bloom. Aquat. Microb. Ecol. 63 111–122. 10.3354/ame01483 [DOI] [Google Scholar]
- Menezes C. B. A., Bonugli-Santos R. C., Miqueletto P. B., Passarini M. R. Z., Silva C. H. D., Justo M. R., et al. (2010). Microbial diversity associated with algae, ascidians and sponges from the north coast of São Paulo state, Brazil. Microbiol. Res. 165 466–482. 10.1016/j.micres.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Myklestad S. M. (1995). Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci. Total Environ. 165 155–164. 10.1016/0048-9697(95)04549-G [DOI] [Google Scholar]
- Nelson D. M., Tréguer P., Brzezinski M. A., Leynaert A., Quéguiner B. (1995). Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycles 9 359–372. 10.1029/95GB0107059 [DOI] [Google Scholar]
- Nicolas J.-L., Corre S., Cochard J.-C. (2004). Bacterial population association with phytoplankton cultured in a bivalve hatchery. Microb. Ecol. 48 400–413. 10.1007/s00248-003-2031-6 [DOI] [PubMed] [Google Scholar]
- Orsini L., Sarno D., Procaccini G., Poletti R., Dahlmann J., Montresor M. (2002). Toxic Pseudo-nitzschia multistriata (Bacillariophyceae) from the Gulf of Naples: morphology, toxin analysis and phylogenetic relationships with other Pseudo-nitzschia species. Eur. J. Phycol. 37 247–257. 10.1017/S0967026202003608 [DOI] [Google Scholar]
- Palomo S., Gonzalez I., De La Cruz M., Martin J., Tormo J. R., Anderson M., et al. (2013). Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 11 1071–1086. 10.3390/md11041071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathom-aree W., Stach J. E. M., Ward A. C., Horikoshi K., Bull A. T., Goodfellow M. (2006). Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 10 181–189. 10.1007/s00792-005-0482-z [DOI] [PubMed] [Google Scholar]
- Piñeiro-Vidal M., Gijón D., Zarza C., Santos Y. (2012). Tenacibaculum dicentrarchi sp. nov., a marine bacterium of the familiy Flavobacteriaceae isolated from European sea bass. Int. J. Syst. Evol. Microbiol. 62(Pt 2), 425–429. 10.1099/ijs.0.025122-0 [DOI] [PubMed] [Google Scholar]
- Pommier T., Canbäck B., Riemann L., Boström K. H., Simu K., Lundberg P., et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16 867–880. 10.1111/j.1365-294X.2006.03189.x [DOI] [PubMed] [Google Scholar]
- Rappé M. S., Connon S. A., Vergin K. L., Giovannoni S. J. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418 630–633. 10.1038/nature00917 [DOI] [PubMed] [Google Scholar]
- Raven J. A., Waite A. M. (2004). The evolution of silicification in diatoms: inescapable sinking and sinking as escape? New Phytol. 162 45–61. 10.1111/j.1469-8137.2004.01022.x [DOI] [Google Scholar]
- Rooney-Varga J. N., Giewat M. W., Savin M. C., Sood S., Legresley M., Martin J. L. (2005). Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49 163–175. 10.1007/s00248-003-1057-0 [DOI] [PubMed] [Google Scholar]
- Round F. E., Crawford R. M., Mann D. G. (1990). The Diatoms: Biology and Morphology of the Genera. Cambridge: Cambridge University Press. [Google Scholar]
- Sapp M., Schwaderer A. S., Wiltshire K. H., Hoppe H. G., Gerdts G., Wichels A. (2007). Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53 683–699. 10.1007/s00248-006-9162-5 [DOI] [PubMed] [Google Scholar]
- Sarmento H., Romera-Castillo C., Lindh M., Pinhassi J., Sala M. M., Gasol J. M., et al. (2013). Phytoplankton speciesspecific release of dissolved free amino acids and their selective consumption by bacteria. Limnol. Oceanogr. 58 1123–1135. 10.4319/lo.2013.58.3.1123 [DOI] [Google Scholar]
- Schäfer H., Abbas B., Witte H., Muyzer G. (2002). Genetic diversity of “satellite” bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42 25–35. 10.1016/S0168-6496(02)00298-2 [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E., Wyche T. P., Kim K. H., Petersen J., Ellebrandt C., Vlamakis H., et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 18:e17473. 10.7554/eLife.17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour J. R., Amin S. A., Raina J.-B., Stocker R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. 10.1038/nmicrobiol.2017.65 [DOI] [PubMed] [Google Scholar]
- Sison-Mangus M. P., Jiang S., Tran K. N., Kudela R. M. (2014). Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J. 8 63–76. 10.1038/ismej.2013.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Ebers J. (2006). Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33 152–155. [Google Scholar]
- Suzuki M., Nakagawa Y., Harayama S., Yamamoto S. (2001). Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. Int. J. Syst. Evol. Microbiol. 51 1639–1652. 10.1099/00207713-51-5-1639 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336 608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Turner S., Pryer K. M., Miao V. P., Palmer J. D. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46 327–338. 10.1111/j.1550-7408.1999.tb04612.x [DOI] [PubMed] [Google Scholar]
- van Tol H. M., Amin S. A., Armbrust E. V. (2017). Ubiquitous bacterium inhibits diatom celle division. ISME J. 11 31–42. 10.1038/ismej.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué D., Duarte C. M., Marrasé C. (1989). Phytoplankton colonization by bacteria: encounter probability as a limiting factor. Mar. Ecol. Prog. Ser. 54 137–140. 10.3354/meps054137 [DOI] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12 635–645. 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum-likelihood tree based on 16S rRNA gene sequences showing the position of bacterial epibionts isolated from C. danicus RCC 2565 and T. delicatula RCC 2560 (Table 4). Only bootstrap values (expressed as percentages of 1000 replications) of >80% are shown. Filled circles indicate that the corresponding nodes were also recovered using the neighbor-joining algorithm. Thermococcus litoralis JCM8560T was used as outgroup. Bar, 0.05 substitutions per nucleotide position.
Attached bacteria on diatom cultures during late exponential (5 days) and late stationary (19 days) growth phases.