Abstract
The Indus Valley has been the backdrop for several historic and prehistoric population movements between South Asia and West Eurasia. However, the genetic structure of present-day populations from Northwest India is poorly characterized. Here we report new genome-wide genotype data for 45 modern individuals from four Northwest Indian populations, including the Ror, whose long-term occupation of the region can be traced back to the early Vedic scriptures. Our results suggest that although the genetic architecture of most Northwest Indian populations fits well on the broader North-South Indian genetic cline, culturally distinct groups such as the Ror stand out by being genetically more akin to populations living west of India; such populations include prehistorical and early historical ancient individuals from the Swat Valley near the Indus Valley. We argue that this affinity is more likely a result of genetic continuity since the Bronze Age migrations from the Steppe Belt than a result of recent admixture. The observed patterns of genetic relationships both with modern and ancient West Eurasians suggest that the Ror can be used as a proxy for a population descended from the Ancestral North Indian (ANI) population. Collectively, our results show that the Indus Valley populations are characterized by considerable genetic heterogeneity that has persisted over thousands of years.
Keywords: Indus-Valley, autosomes, Ror, mtDNA, Y chromosome, ancestral North Indian, Harappan civilization, South Asia, Kurukshetra, aDNA
Introduction
The earliest evidence of farming-based economies in South Asia has been traced back to Mehrgarh, Pakistan ∼9 kya.1, 2 From there, farming and a settled way of life spread farther east, laying foundations for the later Indus Valley civilization (3300−1300 BCE). Climatic reconstruction and other studies suggest that the decline of the Indus Valley civilization in the Bronze Age was most likely driven by a long-term drought, which might have triggered a movement of its inhabitants eastward toward the Gangetic Plain in about 2300 BCE.3, 4, 5, 6, 7, 8
Contemporary populations of this region vary in their rituals and display diverse ethnic backgrounds.9, 10, 11, 12, 13, 14 The eastern Indus Basin, part of the early Vedic India (c. 2000 to c. 600 BCE), comprises the historical Kurukshetra15, 16 (now a district in the Haryana state). It adjoins Northwest (NW) India, which is the homeland of various ethnic communities whose long-term occupation of the area has been described in many Vedic and Hindu scriptures.17, 18, 19, 20, 21, 22
Previous genetic studies have revealed a higher West Eurasian affinity among Northwest Indian and Pakistani (PNWI) populations than among South and East Indians.23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 Furthermore, some recent ancient DNA (aDNA) studies have suggested that the major West Eurasian genetic contributions in South Asia derive from Neolithic Iranians and early Bronze Age steppe populations.36, 37 Other studies38, 39 suggest contributions from the Middle and Late Bronze Age steppe populations in South Asia, together with a Chalcolithic or Bronze Age Central Asian admixture scenario. Nevertheless, despite major breakthroughs in our ability to test models of the genetic history of populations with aDNA, the lack of genome-wide data from Northwest India (NWI) hinders our understanding of present-day genetic variation in the Indus Valley region.
To fill this gap, we have performed a genome-wide study of 45 samples from four NW Indian ethnic groups whose long-term presence in the Indus Valley region has been historically attested: their names—Ror, Gujjar, Jat, and Kamboj—are explicitly mentioned in ancient Vedic scriptures. In addition, we used previously published genomic data for 20 individuals from the Khatri population of Punjab.40 From the newly sampled populations, we generated mtDNA (190 individuals) and Y chromosome (248 individuals) data. We contextualized our data with 1,984 modern and 661 ancient Eurasian genomes from published sources (Tables S1 and S2). We set out to assess the extent of genetic heterogeneity among PNWI populations with regard to distinct genetic ancestries, as well as the amount of more recent ancestry (haplotype) sharing within and among neighboring regions. Furthermore, we also investigated the relationships between the PNWI groups, a set of ancient West Eurasians, and recent aDNA sources from South Asia.37, 38, 39, 41, 42, 43
Material and Methods
Sampling
Blood or saliva samples were collected from 254 individuals residing in NWI, mainly from the Haryana and Rajasthan states. Sampling of the Ror population was carried out within a 100 km radius from the historical Kurukshetra. Other sampled populations also come from an area within 100–400 km of Kurukshetra (Figure 1A). The presence of the Kamboj, Gujjar, Ror, and Jat populations in the region dates back to the early historical and prehistorical period.44, 45, 46, 47, 48, 49 They represent different occupational caste populations who practiced agriculture and pastoralism. The fifth group we studied, the Khatri, is one of the few in the area with roots as a merchant community 50 (Supplemental Material and Methods). All subjects, who voluntarily participated in the study, were healthy adults who were selected through interviews carefully designed so that unrelated individuals would be chosen. Informed consent containing a signature and a left-thumb impression was collected from each participant. The project was carried out in accordance with the guidelines approved by the Ethical Committees of the University of Tartu, Estonia and the Banaras Hindu University (BHU), India.
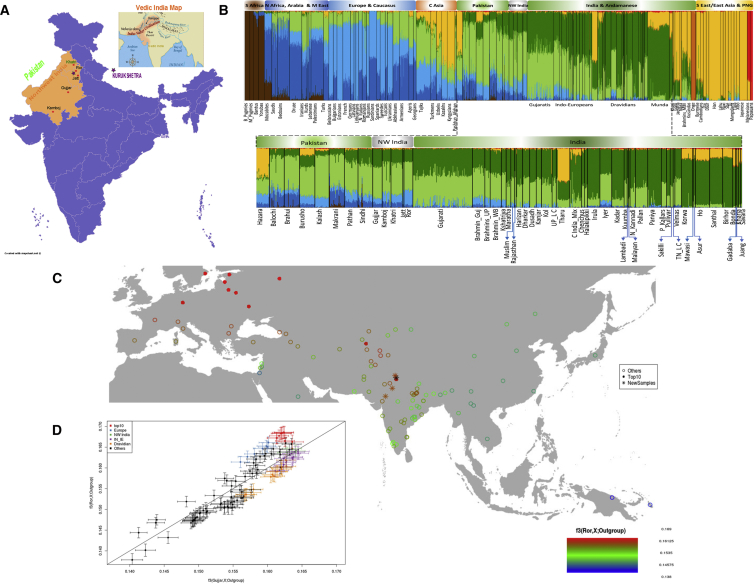
Figure 1.
Sampling Locations, ADMIXTURE, and Shared Drift in Northwest India
(A) The geographic distribution and sampling locations of newly reported modern samples from Northwest India. An inset shows a map of Vedic India. Dots denote the samples studied, and green color indicates samples from published literature.
(B) Results of ADMIXTURE analysis at K8 ancestral components with global populations. The populations are ordered geographically in a bar plot. The genetic structure of new Northwest Indians is shown with a zoom-in on South Asia. The abbreviations are as follows: S Africa, sub-Saharan Africa; N Africa, North Africa; C Asia, Central Asia; NW India, Northwest India; C India_Mix, Central India Mix (Gond individuals together with one individual each from Bhunjia and Bengali); S East Asia, Southeast Asia; PNG, Papua New Guinea; Munda, Indian Austroasiatic speakers; UP_LC, Uttar Pradesh low-caste groups; TN_LC, Tamilnadu low-caste groups; N_Kannadi, North Kannadi; P_Kallars, Piramalai Kallars.
(C) Outgroup f3 (Ror, X; Yoruba) gradient map, showing the affinity of the Ror to Eurasian populations. Red color indicates populations that have a high affinity with the Ror, green indicates groups that have a medium affinity, and blue indicates groups that have the least affinity with the Ror. The filled red circle shows the top 10 populations that share the highest amount of drift with the Ror. The star indicates the newly sampled population group, and the black star refers to the location of the Ror population.
(D) A scatterplot for outgroup f3 (Ror, X; Yoruba) versus outgroup f3 (Gujjar, X; Yoruba) plots the relative shared drift of West Eurasian populations with the Ror against the drift shared with the Gujjar. The top 10 populations sharing the most drift with the Ror are in red, and they are mostly from Europe. Other populations include Indian Austroasiatic, Central Asian, Pakistani, and Middle Eastern population groups. Abbreviations are as follows: NW India, Northwest India; IN_IE, Indian Indo-Europeans; and Dravidians, Indian Dravidians. Error bars represent jack-knife standard errors.
Genotyping and Quality Control
DNA was purified from either whole blood or saliva cells via the standard phenol and chloroform extraction procedure.51
We genotyped 45 samples, including 15 Ror and 1 Jat from Haryana, and 15 Gujjar and 14 Kamboj from Rajasthan (Figure 1A and Table S1) with the Illumina HumanOmniExpress array for 730K SNPs as per the manufacturer’s specifications. We analyzed the newly generated data together with similar data, from previously published sources, for 1,984 modern individuals across the globe29, 40, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63 (Table S1). To evaluate the genetic affinities of PNWI groups with ancient source populations, we merged the modern dataset with 661 ancient genomes, mainly from West Eurasia and South Asia, which are geographically and temporally relevant in the context of the West Eurasian contribution to modern South Asians (Table S2).37, 38, 39, 41, 42, 43 In addition, for mtDNA coding and control region polymorphisms, we genotyped 190 individuals from NWI and assigned haplogroups according to the phylotree mtDNA tree Build 17 (Table S3). For Y chromosome genotyping, we genotyped 248 NWI samples by using either sequencing or PCR-RFLP to identify 37 binary haplogroup-informative Y chromosome markers and classified them into the respective haplogroups (Table S4).
For autosomal analyses, we processed the genome-wide SNP dataset by using PLINK v1.9.64 We included only SNPs on the 22 autosomal chromosomes with minor allele frequency > 1% and removed all SNPs and samples with >3% missing data. One individual from each first- and second-degree relative pair detected with KING65 was removed at random.
mtDNA and Y Chromosome Data Analysis
To explore the relationships of population groups, we performed principal-component analysis (PCA) on the matrix of haplogroup frequencies by using prcomp in R (Figure S1). We limited the populations to the geographical range surrounding PNWI groups and removed outliers from a zoomed landscape. The sample sets include earlier-published data from literature.25, 66, 67, 68
Genome-wide SNP Data Analyses
We calculated mean pairwise FST values between PNWI groups and the regional population groups of West Eurasia (Table S5) by using the approach of Weir and Cockerham.69, 70 The Jat sample was not included in the FST analysis because there was only one sample from the population.
We carried out PC analysis by using the smartpca software (with default settings) implemented in the EIGENSOFT package71 to capture genetic variability described by the first five principal components (PCs); the two most informative are discussed in the text (Figure S2).
For PCA with merged aDNA data, we projected relevant ancient samples on the PCA space and applied default parameters (with the added options of Isqproject: YES, numoutlier: 0, and autoshrink: YES) (Figure S3). We used two population sets as projection scaffolds; the first included present-day Eurasians, and the other included present-day South Asians.
We used the model-based clustering algorithm ADMIXTURE72 to infer genomic ancestral components in PNWI in a global context. In the final settings, calculations for each of the tested ancestral clusters (K = 2 to K = 15) were repeated 25 times. The lowest cross-validation error parameter was observed at K = 12; however, we didn’t observe any significant difference of cross-validation above K = 8 (Figure S4). Because both PCA71 and structure-like analyses72 might be affected by background linkage disequilibrium (LD), we thinned the marker set by pruning out SNPs in strong LD (pairwise genotypic correlation r2 > 0.4) in a window of 200 SNPs (the window slid by 25 SNPs at a time).
We calculated f statistics by using the ADMIXTOOLS73 programs qp3Pop and qpDstat with f4mode: YES. To investigate derived allele sharing between PNWI groups and modern or ancient Eurasian populations, we computed outgroup f3 statistics73 in the form of (Pop1, X; Yoruba), where Pop1 is a PNWI group or aDNA, X is a South Asian or West Eurasian population, and Yoruba is the outgroup (Figures S5–S7). We calculated D statistics73 for various population combinations to assess gene flow among different modern populations and allele sharing between modern South Asians and published ancient sources (Figures S8 and S9 and Tables S8, S9, and S10).
We used qpWave32, 74 to test whether a set of “left” populations were related via N ancestry streams to a set of “right” populations; then we used qpAdm42 to estimate ancestry proportions in a test population (PNWI) originating from a mixture of N “reference” populations37, 38, 39 by exploiting shared genetic drift with a set of “outgroup” populations37, 38, 39 (Table S16). The populations we included in a model that compared plausible “reference” populations were Early Bronze Age Yamnaya and Middle to Late Bronze Age from the Steppe region, as well as Neolithic farmers from Iran (Iran_N) and Onge. In order to differentiate between the Early Bronze Age Yamnaya (Steppe_EMBA) and Middle to Late Bronze Age (Steppe_MLBA) groups, we used the Neolithic Anatolian farmers (Anatolia_N) in addition to other outgroups37 (Table S16) because Steppe_MLBA populations carry an Anatolian/European agriculturist component, but Steppe_EMBA populations do not.39
We applied ALDER75 to compute a weighted LD statistic and to infer the date of admixture on the basis of exponential decay of linkage disequilibrium and were thus able to approximate the time of admixture between NWI and their neighboring regional ethnic groups. We used contemporary West Eurasian and South Asian populations as putative admixing source populations (Figure S10 and Table S18).
We constructed a maximum likelihood (ML) tree for a set of global populations by using TreeMix v.1.1276 in order to place PNWI in a global context (Figure S11). We analyzed runs of homozygosity (RoHs) by using PLINK v.1.964 to investigate the parental relatedness among PNWI populations (Figure S12). RoHs were defined as a minimum of 50 consecutive SNPs in three different window sizes (1,000, 2,500, and 5,000 kb)—such that adjacent regions were fewer than 1,000 kb apart and the intraregional density of SNP coverage was no more than 1 SNP per 50 kb—allowing one heterozygous and five missing calls per window.77, 78 Because the total length and number of RoH segments varied considerably, we calculated the mean for each population.
We used the refined IBD algorithm implemented in BEAGLE v4.079, 80 to detect IBD segments that were shared by PNWI populations and a set of Eurasian populations. Refined IBD was run with default settings, and IBD segments longer than 1 cM were analyzed. IBD sharing was estimated as the average number of IBD segments per pair of individuals for each PNWI population (Figure 2A). 95% confidence intervals (CIs) for the average number of IBD segments were calculated with respect to Gujaratis and according to Kushniarevich et al..62
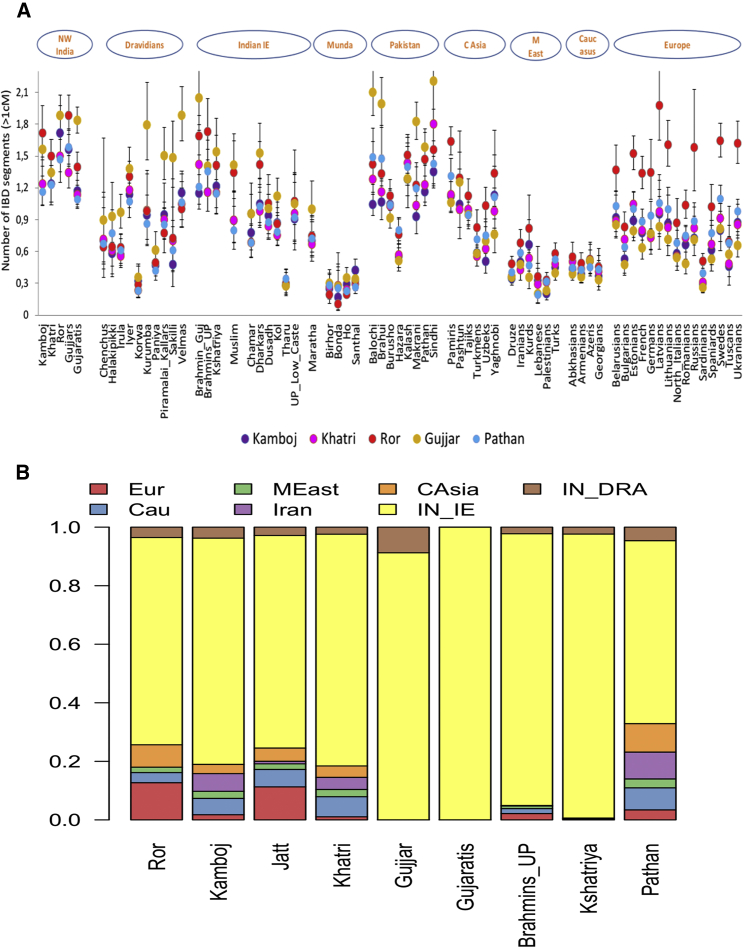
Figure 2.
IBD Sharing and Ancestry Profile of PNWI Populations
(A) Average number of IBD segments per pair of individuals for each Northwest Indian and Pathan ethnic group from Pakistan. Error bars represent 95% confidence intervals.
(B) Population-based ancestry estimates for PNWI and neighboring groups from the North Indian Gangetic Plain were inferred by CHROMOPAINTER via an NNLS-based analysis. NWI and Pakistani populations were excluded from the donor groups. Abbreviations are as follows: IN_IE, Indian Indo-Europeans; Pak, Pakistan; Eur, Europe; M East, Middle East; Cau, Caucasus; and IN_DRA, Indian Dravidians.
To perform chromosome painting, we used CHROMOPAINTER81 on data phased with SHAPEIT v2.82 We set up the -n and -M parameters after running the software’s EM option on a small subset of the populations and five randomly selected chromosomes (3, 7, 10, 17, and 22), as described in Montinaro et al.83 The estimated values for the two parameters were n = 526.701 and M = 0.00046. Recent ancestry sharing between closely related groups can hide distant relationships in CHROMOPAINTER analysis. We therefore performed two different analyses to take a more balanced approach:84 the first one excluded NWI and Pakistani groups from donors (Figure 2B), and the second one kept all samples as donors (Figures S13 and S14A). The median number of SNPs for the inferred chunks was ∼11 for both analyses (means = 13.56 and 12.79; 95% CI = 10.00–30.77 and 9.85–27.07, respectively).
We used a non-negative least-squares (NNLS)85, 86, 87 “ancestry profile” method described in previous studies83, 88 to compare the copying vectors of the PNWI and North Indian Indo-European (NI_IE) populations. We modeled each target PNWI and NI_IE population as a mixture of haplotypes that best fit the painting profile of different donor populations. This approach not only accounts for deviation in sample size across donor groups but also considers the fact that human population groups are inherently related and thus that most haplotypes are shared. However, if true donor groups are not included as a result of a sampling issue, this method is likely to choose the “closest” among available sampled groups. Thus, groups recognized via this approach should be considered as the most related populations among the available ones. We have also calculated standard errors across the NNLS results by applying the jack-knife method (Table S19).
Results
Northwest Indian Populations in the Context of Other South Asians and West Eurasians
We first visualized the genetic structure of populations from NWI in the context of other South Asian and Eurasian populations by using PCA, FST, and ADMIXTURE analyses (Figures S2 and 1B and Table S5). In PCA, the NWI populations were placed between Indo-European (IE) populations from Pakistan and North India, and they fell within the North-South gradient that differentiates Europe from South Asia in principal component 2 (PC2) (Figure S2A). A similar pattern was observed in FST (Figure S2B) and ADMIXTURE (Figure 1B) results. Among South Asian populations, we detected consistently lower FST values between NWI and Pakistani groups (compared to all groups, the Ror are closer to the Pathan, and the Khatri are closer to the Sindhi) than between NWI and their North Indian neighbors (Figure S2B and Table S5). These observations are supported by PCA (Figure S2A inset) and by the presence of a significantly higher proportion (Wilcoxon test p value < 0.05) of the European-like light-blue component for the Ror and Jat, akin to the Pathan and Kalash, in ADMIXTURE (Figure 1B). Therefore, in contrast with uniparental DNA,26, 27 our autosomal data suggest a close genetic affinity between populations currently residing east and west of the Indus.
Demographic PNWI History Based on Allele Frequency and Haplotype-Sharing Analyses
Outgroup f3 analysis in the form of (PNWI, X; Yoruba) showed that the Ror (and Jat) have distinct, high genetic similarity to modern Europeans (Figures 1C, 1D, and S5), far higher than the similarity observed in other NWI populations, such as the Gujjar (Figures 1D and S5). Among an extended set of South Asians, this pattern was repeated only in the Pathan population from Pakistan (Figure S5). This observation was further confirmed by D statistics, wherein the Pathan and Kalash share a higher proportion of alleles with the Ror than with any other group from NWI and NI_IE (Tables S7 and S8). TreeMix results (Figure S11) also indicate that the Kalash and Ror share the same branch.
Specifying various modern populations from South Asia, West Eurasia, and aDNA groups as likely sources, we used three population tests (f3 statistics) to explore putative admixture patterns73 in PNWI. We report only source combinations with a negative f3 statistics value (Z score < −3) (Table S6).
We further investigated the Ror group’s high affinity with modern Europeans at the haplotype-sharing level by performing identity by descent (IBD) (Figure 2A), CHROMOPAINTER (Figure S13), and NNLS (Figure 2B) analyses.
Refined IBD analysis highlights the general trend whereby the sharing of IBD segments declines as one moves along the cline from PNWI and NI_IE toward Dravidian and Indian Austroasiatic (IN_AA) groups (Figure 2A). Strikingly, among all PNWI groups studied, the Ror demonstrate the highest number of IBD segments shared with Europeans and Central Asians, whereas the Gujjar share a higher number of IBD segments with local Indian Indo-Europeans and Dravidians than do other PNWI groups (Figure 2A).
In CHROMOPAINTER analysis, as expected, the Ror (and Jat) exhibited a significantly higher number of chunks received from Europeans than do other NWI populations studied (t test, p value < 0.01). The excess sharing between the Ror and Europeans was made evident by the NNLS ancestry-profiling method, which we used to report the ancestry proportions of seven regional groups. Furthermore, the same analysis supports our previous observation suggesting a high degree of heterogeneity among PNWI groups. Populations such as the Ror and Jat possess more European and less Indian Indo-European (IN_IE) ancestry than other PNWI groups; however, they differ from each other in their Central Asian and Caucasus ancestry (Figure 2B). Interestingly, relative to other groups of NWI, the Khatri have a higher proportion of Caucasus ancestry, along with substantial Central Asian and Middle Eastern ancestry. Conversely, the Gujjar stand out as having the more ancestry from IN_IE groups and Dravidians than other PNWI groups do. To test whether population inbreeding could be responsible for the observed patterns, we analyzed RoHs in the genomes of PNWI groups, along with those of other neighboring Indian and West Eurasian populations. The Ror showed the smallest average number of RoHs (Figure S12), suggesting a higher effective population size (rather than inbreeding) or higher level of gene flow from other groups.
Furthermore, a putative West-South Eurasian admixture date for the Ror ∼50 generations (∼1,500 years) back, inferred by ALDER (Table S18), is corroborated by the lack of documented recent contacts between European populations and the Ror. This rules out any major impact of the recent colonial regime in India on the Ror population. Thus, the observed excess of a West Eurasian genetic component in the Ror group is most likely due to ancient migrations in the region.
Ancient Contributions to the Genome of Modern PNWI Populations
West Eurasian ancestry has been described as a composition of four main ancient components: Eastern European hunter-gatherers (EHG), Caucasus hunter-gatherers (CHG), Iran_N, and Anatolia_N.36, 41, 42, 43 However, EHG and CHG together are often associated in an ancestry from the steppe belt,37 the Steppe_EMBA, which has been suggested as a major source of ancient admixture during Bronze Age population movements in West Eurasia and South Asia.42, 43 Recent aDNA studies, through the sampling of surrounding regions and analysis of the first samples from South Asia, have contributed significantly to our understanding of ancient population dynamics in South Asia. The new sampling mainly covers Neolithic (IranTuran_N) and Bronze Age (IranTuran_BA) individuals from the eastern Iran-Turkmenistan region, hunter-gatherers from West Siberia forest zone (WestSiberia_HG), Copper Age individuals from Botai, Kazakhstan (Botai), and further Steppe_MLBA individuals. Moreover, the new sampling includes Chalcolithic Namazga (Namazga_CA) and Iron Age (Turkmenistan_IA) individuals from Turkmenistan; Bactria-Margiana Archaeological Complex (BMAC) individuals and Bronze Age outliers from BMAC and eastern Iran (Indus_Diaspora [best known as Indus_Periphery]); and the first two ancient groups from South Asia, Iron Age or prehistorical samples (SPGT) and early historical period samples (SouthAsia_H) from the Swat Valley, Pakistan (Table S2).38, 39
We first performed PCA to see the position of our PNWI group relative to these relevant ancient samples. We found that the NWI groups clustered near the Pakistani groups, close to the newly extracted proximal (temporally and geographically close) ancient sources (the Namazga_CA, Indus_Periphery, BMAC, SPGT, and SouthAsia_H individuals) and to other distal (temporally and geographically distant) ancient sources (Figure S3). We also observed a tight clustering of PNWI groups with the first ancient South Asian sources from the Swat Valley (the prehistorical SPGT and early historical SouthAsia_H individuals) and the Bronze Age outliers from BMAC region (the Indus_Periphery), who supposedly had a close connection with the ancient Indus Valley people as a result of their temporal and geographic vicinity (Figure S3).
We then used f3 and D statistics to assess ancient West Eurasian contributions to modern PNWI populations (Figures S6–S9 and Tables S9 and S10). Analysis revealed that the Ror display more genetic components related to EHG, Anatolia_N, CHG, Steppe_EMBA, and Steppe_MLBA than any other South Asian population, as well as a higher affinity with SPGT, SouthAsia_H, and BMAC than other PNWI groups. At the same time, the affinity that the Ror exhibit with Iran_N, Namazga_CA, and Indus_Periphery is identical to that exhibited by their immediate geographic neighbors (Figures S6B, S7, S8B, and S9 and Tables S9 and S10). Higher West Eurasian ancestry in the genomes of modern Ror people could be due to ancient or recent admixture with sources west or north of the Indus Valley. The excess of EHG ancestry in the Ror population, compared to modern Iranians (Figure S8D), seems to rule out admixture with Iranian sources, hence pointing to a Central Asian or Steppe-related population as the most likely West Eurasian source (Table S6) in the Ror population. The Ror are also distinguished as the only South Asian group that is significantly closer to Neolithic Anatolians than to Neolithic Iranians (Z score > +3) (Figures S6B and S8B and Table S9). However, because of a lack of support from qpAdm, we present this as a tentative result.
Furthermore, we explored the allele sharing of PNWI groups relative to a set of two ancient sources by using D statistics in the form of D (pop1, Yoruba; pop2, pop3), where Yoruba is the outgroup, pop1 is a South Asian population, pop2 is an aDNA source, and pop3 is another aDNA source or a modern Dravidian population. These D tests revealed a general trend among South Asian populations of higher affinity (Z score > +3) with Steppe_EMBA than with either Steppe_MLBA or Chalcolithic Namazga_CA (Table S9). Interestingly, we observed that PNWI groups exhibited a trend of equal allele sharing when their affinity to available ancient sources from the Copper Age to Middle-Late Bronze Age Central Asia was compared, except for a visible closeness of the Ror, Jat, Kalash, and Pathan groups to Steppe_MLBA rather than to Indus_Periphery (or Indus_Diaspora) people (Z score < −3) (Table S9). In contrast, NI_IE resembled Dravidians in that the group had a higher affinity with Indus_Periphery rather than with Steppe_MLBA, Namazga_CA, and BMAC. However, by the Iron Age or prehistorical time, the scenario changed; NI_IE caste groups, similarly to PNWI group, share an equal number of alleles with the prehistorical SPGT and Indus_Periphery groups, whereas NI_IE tribes, Dravidians, and IN_AA people are closer to the Indus_Periphery group (Z score < −3) (Table S9). Finally, we compared the affinity between South Asians and both the Iron Age SPGT or early historical SouthAsia_H and modern Dravidian (Paniya) individuals separately in respective D statistics; we observed that PNWI and NI_IE castes are closer to SPGT and SouthAsia_H than to the Paniya (Z score > +3). This is unlike the IN_AA and NI_IE tribal people, who show a clear affinity with the Paniya (Z score < −3) (Table S9).
A previous ancient-DNA study has suggested that the Iran_N and Steppe_EMBA groups are the best proxies for the ancient West Eurasian component in South Asians. The study also suggested that most South Asians can be modeled as a mixture of these two groups but also have Onge- and Han-related ancestries,37 a method sometimes referred to as distal modeling. However, other more recent aDNA studies have suggested that the Steppe_MLBA, grouped together with other ancient sources such as the Onge and either Namazga_CA or Indus_Periphery, offer a better fit in proximal modeling than do the Steppe_EMBA.38, 39
We used qpAdm to explore how the distal models (Iran_N, either Steppe_MLBA or Steppe_EMBA, and Onge) and proximal models (Namazga_CA or Indus_Periphery, Steppe_MLBA, and Onge) fit in the case of our studied PNWI groups (Tables S11–S16). We observed that a model with three source populations (Iran_N, Steppe_MLBA, and Onge) fits with the data for the majority of PNWI populations (p value > 0.05 and low standard errors of admixture proportion estimates) (Figure 3 and Table S11). The only exceptions were the Burusho, Hazara, Kalash, and ancient SPGT groups, who could not be modeled from these three sources. Similar to the early historical SouthAsia_H group the NWI and NI_IE groups have high proportions of Steppe_MLBA ancestry, but they have higher proportions than the Pakistanis (except Pathan) and the Dravidian groups do. The Ror and Jat peoples stand out for having the highest proportion of Steppe_MLBA ancestry (∼63%). The proportion of Steppe ancestry in the Ror is similar to that observed in present-day Northern Europeans.42 We also observed that when we applied the “Pearson correlation” to the Steppe ancestry inferred in Europeans by Haak et al.,42 the higher IBD sharing between the Ror people and Europeans was significantly and positively correlated with increasing Steppe ancestry in Europeans (Figure S15). Interestingly, when we used Steppe_EMBA instead of Steppe_MLBA in the distal model (Table S12), the Kalash and Iron Age Swat Valley SPGT data offered a good fit with the model, indicating a plausible Yamnaya-like impact in the Early-Middle Bronze Age; the effects of this impact might have persisted in some South Asian populations. In fact, we found that the model with Iran_N, Steppe_EMBA, and Onge works equally well for all modern and ancient South Asians. To test whether Steppe_MLBA or Steppe_EMBA fits better for modeling South Asians in the distal model, we added Neolithic Anatolians as a separate outgroup to the “right” list (Tables S13 and S16). This was motivated by the presence of a Neolithic Anatolian or European Early Neolithic component in Steppe_MLBA but not in Steppe_EMBA.39 Our qpAdm results demonstrate that although most PNWI groups have significant Steppe_MLBA ancestry along with Steppe_EMBA, the NI_IE group from the Gangetic Plain and Dravidian South Asians show a significant Steppe_EMBA component but not a Steppe_MLBA component. However, prehistorical and early historical ancient South Asian individuals have a higher proportion of Steppe_MLBA than Steppe_EMBA.
Figure 3.
Proportions of Ancient Ancestry in South Asian Populations
qpAdm plot indicating proportions of ancestry made up of ancient sources (Iran_N, Steppe_MLBA, and Onge) among different South Asian populations.
To clarify the issue of plausible biases introduced by differences in the sample size of reference ancient groups, we replicated our analyses by using an equal number of individuals for both Steppe_EMBA and Steppe_MLBA, and we performed D statistics and qpAdm tests for cross-validation (Table S17). We observed no significant differences between the results.
Because various analyses (Figures 1B, S2A, and S9 and Tables S7 and S8) had highlighted either the Ror or Kalash peoples as having the highest proportion of ancient North Indian (ANI) ancestry combined with their terminal position on the South Asian cline (Figure S11), we used D statistics in the form of D (Yoruba, Test; Ror, Kalash) to further evaluate their relative affinity with worldwide populations (test). We observed that the Onge and other Indian populations with a high proportion of the ASI component share significantly more alleles (Z score > 4) with the Ror than with the Kalash but that the opposite is true for Georgians, who share significantly more alleles with the Kalash than with the Ror, indicating a higher proportion of the ANI component in the former (Table S7). These results suggest that the Ror might be used as a plausible alternative proxy for ANI in the demographic modeling of South Asians. The modeling might benefit from the reduced genetic drift in the Ror compared to the Kalash (Figure S11), although the Ror group harbors a small fraction of additional ASI component (∼1%).
Y Chromosome and mtDNA Diversity in Northwest Indian Populations
In PC analysis of mtDNA and Y chromosomes, the NWI population fit on the broader North-South cline, consistent with the genome-wide analyses (Figures S1A and S1B). In mtDNA analysis, a substantial part (∼37%–51%) of the maternal lineages in NWI is West Eurasian (R0, R2, R2′JT, T, HV, I, J, K, U3, U7, U9, and W) (Table S3), in agreement with the results of an earlier study.25 The Y chromosome profiles of NWI revealed a high proportion (41%–76%) of South-Asian-specific lineages (C-M356, H-M69, R2-M124, and L-M11); the Gujjar stand out because they incorporate ∼76% of these lineages. Other haplogroups (J2-M172 and R1a1-M17) are also present at a substantial frequency (20%–55%) (Table S4). Markedly, we observed that, among neighboring NWI groups, the Ror carry the highest proportion (about 58%) of South-Asian-specific maternal lineages (M18, M2, M3, M4, M5, M6, R5, and U2), and they carry more West Eurasian paternal lineages (J2 and Q) than other groups of the region. A high proportion of West Eurasian lineages in both uniparental loci is thus broadly consistent with the results based on autosomal loci.
Discussion
In this study, we have investigated the genetic relationships among contemporary populations of NWI in the context of neighboring populations from South Asia and West Eurasia, also considering the influence of ancient West Eurasian sources.
Genetic Ancestry Components of Northwest Indian and Pakistani Populations
Evidence from genome-wide genotype data (Figures 1B, 2, and S5 and Table S8) and uniparental markers (Figure 1) revealed Northwest Indians (east of the Indus) to be intermediate between Pakistanis (west of the Indus) and North Indian Indo-European (NI_IE) speaking populations from the Gangetic Plain. Additionally, the genomic sharing between NWI populations and NI_IE populations from the Gangetic Plain, bolstered both by the results of analyses done by IBD (Figure 2A) and CHROMOPAINTER (Figures 2B and S13) and by their similar level of allele sharing with most ancient sources as shown by D statistics (discussed in the next section), establishes a noticeable genetic affinity between NWI and their contemporary neighbors on either side of the Indus Valley. This contrasts with earlier observations based on mtDNA and Y chromosome data.27 Broadly speaking, these results could be compatible with archaeological evidence suggesting that people had high mobility within the region during the prehistoric and historical time. This mobility could include the migration of the Indus people toward the Gangetic Plain after the demise of the Indus Valley Civilization which is suggested by archaeological evidence.3, 4, 5, 6, 7
On the other hand, our data also reveal substantial intra-region heterogeneity (Figures 1, 2, S2, S5, and S13 and Tables S5, S7–S9, and S10). For instance, the Ror and Jat peoples, together with the Pakistani Pathan, share genetic ancestry pointing to their possible connection with Central Asians. The genetic relatedness of the Khatri and Sindhi may agree with both peoples’ having been recognized as vital merchant communities of early modern India.89 Among the populations of NWI, the elevated similarity of the Gujjar to local Indian populations and their lower affinity with West Eurasians may relate to their historically documented affinity with various extant South Asian ethnic communities.90, 91, 92, 93 High genetic differentiation among NW Indian populations suggests long-term population structure within the region.
Ancient West Eurasian Components in PNWI Populations
Previous claims of the widespread distribution of an ancient West Eurasian component in the Indian sub-continent, either through distal or proximal sources,36, 37, 38, 39 are well supported by our f3, D statistics and qpAdm results (Figures 3 and S6–S9 and Tables S9, S10, and S11–S16). The observation that PNWI populations share more alleles with external sources from different time periods, including Mesolithic (EHG, CHG), Neolithic (Anatolia_N, Iran_N), Bronze (Steppe_EMBA, Steppe_MLBA), Copper (Namazga_CA, BMAC), and Iron Age (Turkmenistan_IA) groups than do other South Asian populations (Figures S7–S9 and Tables S9 and S10) can be explained by the geographic position of the Indus Valley as the gateway to the Indian sub-continent for any episode of gene flow from the west.
The higher affinity and admixture of PNWI populations with Neolithic Iranians and Anatolians (Figures S6C, S7B, and S8B and Tables S9 and S10), coupled with the substantial Middle Eastern component (dark blue, Figure 1B) and the significant influx of the Middle-East-related male lineage J2-M172 (Table S4) into the Indian sub-continent through the Northwest corridor,94 might agree with earlier archaeological work that took place at Mehargarh and that suggested the plausible influence from the Zagros or Levant region on the first evident settled way of life in South Asia.95, 96
A higher level of European ancestry in the Ror and Jat compared to other South Asians (Figures 1, 2, S2, S5, and S13 and Tables S5–S8) makes these two populations outliers within the broader Northwest South Asian landscape. This could be indicative of either a possible recent gene flow from a population related to Europe or to ancient West-Eurasian-related influx, which would agree with previous studies on adaptation, wherein the Ror and Jat have stood out for their high frequency of the lactase persistence allele (LCT-13910T) and the light-skin-color gene variant (SLC24A5).70, 97 We also report that, relative to other South Asians, the Ror group has high shared drift with the EHG and Steppe_EMBA groups, higher allele sharing with the Steppe_MLBA group, and higher affinity with the Iron Age (prehistorical) and early historical first South Asian ancient sources (Figures S6A, S6B, S7, S8A, S8D, and S9 and Tables S9 and S16). We find this indicative of multiple plausible influxes of Steppe-like ancestry into the Ror group, as well as their close connection with prehistorical to early historical South Asia.
The Ror display more affinity with Neolithic Anatolians than with Neolithic Iranians (Figures S6C and S8B and Table S8), whereas other South Asians in our dataset show almost equal allele sharing with both Neolithic aDNAs. Such an affinity might also explain the higher frequency of the light-skin-color variant (SLC24A5 allele rs1426654) in the Ror because a higher frequency of this allele has also been found both in Neolithic Anatolians and CHG.43, 98 The Ror have an affinity with Anatolian Neolithic farmers and a closeness to the Pathan and Central Asians (Figures 2, S2, S6C, and S8B and Tables S5, S8, and S10). These facts, taken together with a gradient of affinity with ancient Steppe_MLBA or Steppe_EMBA, CHG, EHG, and Anatolia_N groups that decreases from Central Asia to the Ror, point to a possible contact with Central Asian and/or Steppe peoples that took place earlier than ∼1,500 years ago, as suggested by the ALDER result (Table S18). qpAdm results consistently indicate a higher proportion of Steppe_MLBA ancestry in NWI populations than in other Indians. The Ror stand out in South Asia as the population with the highest proportion of Steppe ancestry (Figures 3 and S9 and Tables S10, and S11–S15), which could plausibly be linked to the finding that, among the South Asian groups, the Ror have the highest affinity with both the Neolithic Anatolians and northern Europeans. Interestingly, such a prominent West Eurasian link is not supported by mtDNA evidence in the Ror, perhaps hinting at a male-biased admixture scenario in the Ror from the Central Asia/Steppe region. Such a hypothesis is bolstered by the higher frequencies of the Y chromosomal haplogroups J2 and Q (Tables S3 and S4).
Among extant populations, both the Kalash and Ror groups stand out because they have the highest proportions of the ANI component, which can be modeled as a mixture of Iranian Neolithic and either Early-Middle (in case of the Kalash) or Middle-Late (in case of the Ror) Bronze Age Steppe ancestries. Although quantitatively the Kalash might have the highest ANI proportion, the Ror appear to be an important alternative to the Kalash as a proxy for ANI in demographic modeling in the absence of relevant ancient DNA data from India; this is due to diversity within the Steppe component as well as the high level of drift in the Kalash population.99
In summary, we demonstrate a higher proportion of genomic sharing between PNWI populations and ancient EHG and Steppe-related populations than we observe in other South Asians. We report that the Ror are the modern population that is closest to the first prehistorical and early historical South Asian ancient samples near the Indus Valley, and they also harbor the highest Steppe-related, EHG, and Neolithic Anatolian ancestry. However, compared to other adjoining groups, the Ror show less affinity with the Neolithic Iranians. The Ror population can plausibly be used as an alternative proxy for ANI in future demographic modeling of South Asian populations. Collectively, our results point out that the PNWI groups have high allele sharing with the region surrounding the ancient Indus Valley and that the PNWI region is an area of rich diversity in population dynamics and one where neighboring groups might harbor divergent genetic ancestries from multiple admixture events.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the Ror, Gujjar, Kamboj, and Jat communities for their support of this study and all individual volunteers for donating their samples. R.V. thanks the Swedish Collegium for Advanced Studies for support during his sabbatical stay in Uppsala. We thank Lehti Saag, Bayazit Yunusbayev, Hovhannes Sahakyan, Jose Rodrigo Flores Espinoza, and Erwan Pennarun for useful discussions and assistance. We thank Tuuli Reisberg for her assistance in genotype data curation. We also thank Mari Järve for language editing. All data analyses were performed at the High-Performance Computer Centre of the University of Tartu, Estonia (http://www.hpc.ut.ee). Support was provided by the European Union through the European Regional Development Fund Centre of Excellence for Genomics and Translational Medicine (project no. 2014–2020.4.01.15-0012) to the EBC-IG and by the Estonian Institutional Research grant IUT24-1 (to A.K.P., R.V., M. Metspalu, S.R., E.M., and T.K.). M. Metspalu was supported by Estonian Research Council grant PRG 243. M. Mondal was supported by the European Union through the European Regional Development Fund (project no. 2014-2020.4.01.16-0030). G.C. is supported by National Geographic Explorer grant HJ3-182R-18, A.K. was supported by Estonian Personal grant PUT 1339, and L.P. was supported by the European Union through the European Regional Development Fund (project no. 2014-2020.4.01.16-0024, MOBTT53). A.K.P. was supported by the European Social Fund’s Doctoral Studies and Internationalization Programme DoRa. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: December 6, 2018
Footnotes
Supplemental Data include 15 figures, 19 tables, and Supplemental Material and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.022.
Contributor Information
Ajai K. Pathak, Email: ajaipathak@gmail.com.
Gyaneshwer Chaubey, Email: gyaneshwer.chaubey@bhu.ac.in.
Accession Numbers
The data for the 45 sequences reported in this paper are available from the Gene Expression Omnibus of the National Centre for Biotechnology Information (GEO: GSE119653) and the data repository of the Estonian Biocentre (www.ebc.ee/free_data).
Web Resources
The 1000 Genomes Project, http://www.internationalgenome.org/home
HapMap3, https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html
PhyloTree, http://www.phylotree.org/
Supplemental Data
References
- 1.Jarrige J.-F. Economy and society in the Early Chalcolithic/Bronze Age of Baluchistan: New perspectives from recent excavations at Mehrgarh. South Asian Archaeol. 1981;1979:93–114. [Google Scholar]
- 2.Costantini L. The Beginning of Agriculture in the Kachi Plain: The Evidence of Mehrgarh. In: Allchin B., editor. South Asian Archaeology 1981. Cambridge University Press; 1984. pp. 29–33. [Google Scholar]
- 3.Misra V.N. Prehistoric human colonization of India. J. Biosci. 2001;26(4, Suppl):491–531. doi: 10.1007/BF02704749. [DOI] [PubMed] [Google Scholar]
- 4.Tripathi J.K., Bock B., Rajamani V., Eisenhauer A. Is Rhiver Ghaggar, Saraswati? Geochemical constraints. Curr. Sci. 2004;87:1141–1144. [Google Scholar]
- 5.Gupta A.K., Anderson D.M., Pandey D.N., Singhvi A.K. Adaptation and human migration, and evidence of agriculture coincident with changes in the Indian summer monsoon during the Holocene. Curr. Sci. 2006;90:1082–1090. [Google Scholar]
- 6.Madella M., Fuller D.Q. Palaeoecology and the Harappan civilisation of South Asia: A reconsideration. Quat. Sci. Rev. 2006;25:1283–1301. doi: 10.1016/j.quascirev.2005.10.012. [DOI] [Google Scholar]
- 7.Brooke J.L. Cambridge University Press; 2014. Climate change and the course of global history: A rough journey. [Google Scholar]
- 8.Dutt S., Gupta A.K., Wünnemann B., Yan D. A long arid interlude in the Indian summer monsoon during: 4,350 to 3,450 cal. yr BP contemporaneous to displacement of the Indus valley civilization. Quat. Int. 2018;482:83–92. [Google Scholar]
- 9.Thapar B.K. A Harappan Metropolis beyond the Indus Valley. In: Possehl G.L., editor. Ancient Cities of the Indus. Carolina Academic Press; 1979. pp. 196–202. [Google Scholar]
- 10.Possehl G.L. Aris & Phillips; 1982. Harappan civilization: A contemporary perspective. [Google Scholar]
- 11.Shaffer J.G., Lichtenstein D.A. Ethnicity and change in the Indus valley cultural tradition. Old Probl. New Perspect. Archaeol. South Asia. 1989;2:117–126. [Google Scholar]
- 12.Mughal M.R. The Decline of the Indus Civilization and the Late Harappan Period in the Indus Valley. Lahore Museum Journal. 1990;3:1–22. [Google Scholar]
- 13.Possehl G.L. Revolution in the urban revolution: The Emergence of the Indus urbanization. Annu. Rev. Anthropol. 1990;19:261–282. [Google Scholar]
- 14.Nath A. Rakhigarhi: A Harappan metropolis in the Saraswati-Drishadvati divide. Puratattva. 1998;28:39–45. [Google Scholar]
- 15.Benedetti G. The chronology of Puranic kings and Rigvedic rishis in comparison with the phases of the Sindhu-Sarasvati civilization. In: Rao N., editor. Sindu-Sarasvati Civilization: New Perspectives. D.K. Printworld (P) Ltd.; 2014. pp. 220–246. [Google Scholar]
- 16.Kenoyer J.M. Cultures and societies of the Indus tradition. In: Thapar R., editor. Historical Roots in the Making of ‘the Aryan. National Book Trust; 2006. pp. 21–49. [Google Scholar]
- 17.Chidbhavananda S. Tapovanam Publishing House; 1965. The Bhagavad Gita: Original Stanzas. [Google Scholar]
- 18.Purana B. Gita Press. Gorakhpur; 1971. Samvat 2037. [Google Scholar]
- 19.Lal P. Asia Book Corporation of America; 1980. Mahabharata of Vyasa. [Google Scholar]
- 20.O’Flaherty W.D. Penguin Books; 1981. The Rig Veda: An anthology. One hundred and eight hymns, selected, translated, and annotated. [Google Scholar]
- 21.Dange S.S. Ajanta Publications; 1984. The Bhagavata Purana: Mytho-Social Study. [Google Scholar]
- 22.Sitaramiah V., Sītārāmayya V. Sahitya Akademi; 1972. Valmiki Ramayana. [Google Scholar]
- 23.Kivisild T., Rootsi S., Metspalu M., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.-V., Stepanov V. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metspalu M., Kivisild T., Metspalu E., Parik J., Hudjashov G., Kaldma K., Serk P., Karmin M., Behar D.M., Gilbert M.T.P. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana-Murci L., Chaix R., Wells R.S., Behar D.M., Sayar H., Scozzari R., Rengo C., Al-Zahery N., Semino O., Santachiara-Benerecetti A.S. Where west meets east: The complex mtDNA landscape of the southwest and Central Asian corridor. Am. J. Hum. Genet. 2004;74:827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McElreavey K., Quintana-Murci L. A population genetics perspective of the Indus Valley through uniparentally-inherited markers. Ann. Hum. Biol. 2005;32:154–162. doi: 10.1080/03014460500076223. [DOI] [PubMed] [Google Scholar]
- 27.Thangaraj K., Naidu B.P., Crivellaro F., Tamang R., Upadhyay S., Sharma V.K., Reddy A.G., Walimbe S.R., Chaubey G., Kivisild T., Singh L. The influence of natural barriers in shaping the genetic structure of Maharashtra populations. PLoS ONE. 2010;5:e15283. doi: 10.1371/journal.pone.0015283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing J., Watkins W.S., Hu Y., Huff C.D., Sabo A., Muzny D.M., Bamshad M.J., Gibbs R.A., Jorde L.B., Yu F. Genetic diversity in India and the inference of Eurasian population expansion. Genome Biol. 2010;11:R113. doi: 10.1186/gb-2010-11-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metspalu M., Romero I.G., Yunusbayev B., Chaubey G., Mallick C.B., Hudjashov G., Nelis M., Mägi R., Metspalu E., Remm M. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am. J. Hum. Genet. 2011;89:731–744. doi: 10.1016/j.ajhg.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma G., Tamang R., Chaudhary R., Singh V.K., Shah A.M., Anugula S., Rani D.S., Reddy A.G., Eaaswarkhanth M., Chaubey G. Genetic affinities of the central Indian tribal populations. PLoS ONE. 2012;7:e32546. doi: 10.1371/journal.pone.0032546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gazi N.N., Tamang R., Singh V.K., Ferdous A., Pathak A.K., Singh M., Anugula S., Veeraiah P., Kadarkaraisamy S., Yadav B.K. Genetic structure of Tibeto-Burman populations of Bangladesh: evaluating the gene flow along the sides of Bay-of-Bengal. PLoS ONE. 2013;8:e75064. doi: 10.1371/journal.pone.0075064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moorjani P., Thangaraj K., Patterson N., Lipson M., Loh P.-R., Govindaraj P., Berger B., Reich D., Singh L. Genetic evidence for recent population mixture in India. Am. J. Hum. Genet. 2013;93:422–438. doi: 10.1016/j.ajhg.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaubey G., Kadian A., Bala S., Rao V.R. Genetic affinity of the Bhil, Kol and Gond mentioned in epic ramayana. PLoS ONE. 2015;10:e0127655. doi: 10.1371/journal.pone.0127655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaubey G., Ayub Q., Rai N., Prakash S., Mushrif-Tripathy V., Mezzavilla M., Pathak A.K., Tamang R., Firasat S., Reidla M. “Like sugar in milk”: Reconstructing the genetic history of the Parsi population. Genome Biol. 2017;18:110. doi: 10.1186/s13059-017-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reich D., Thangaraj K., Patterson N., Price A.L., Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broushaki F., Thomas M.G., Link V., López S., van Dorp L., Kirsanow K., Hofmanová Z., Diekmann Y., Cassidy L.M., Díez-Del-Molino D. Early Neolithic genomes from the eastern Fertile Crescent. Science. 2016;353:499–503. doi: 10.1126/science.aaf7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazaridis I., Nadel D., Rollefson G., Merrett D.C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damgaard P. de B., Martiniano R., Kamm J., Moreno-Mayar J.V., Kroonen G., Peyrot M., Barjamovic G., Rasmussen S., Zacho C., Baimukhanov N. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science. 2018;360:6396. doi: 10.1126/science.aar7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narasimhan V.M., Patterson N.J., Moorjani P., Lazaridis I., Mark L., Mallick S., Rohland N., Bernardos R., Kim A.M., Nakatsuka N. The genomic formation of South and Central Asia. bioRxiv. 2018 doi: 10.1101/292581. [DOI] [Google Scholar]
- 40.Basu A., Sarkar-Roy N., Majumder P.P. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc. Natl. Acad. Sci. USA. 2016;113:1594–1599. doi: 10.1073/pnas.1513197113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allentoft M.E., Sikora M., Sjögren K.-G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P.B., Schroeder H., Ahlström T., Vinner L. Population genomics of bronze age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 42.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davids T.W.R. Putnam; 1903. Buddhist India. [Google Scholar]
- 45.Elliott H.M. Lond. Trübner; 1859. Memoirs on the History, Folk-Lore, and Distribution of the Races of the North Western Provinces of India. [Google Scholar]
- 46.Smith V.A. Clarendon Press; 1924. The Early History of India, from 600 BC to the Muhammadan Conquest including the Invasion of Alexander the Great. [Google Scholar]
- 47.Law B.C. Volume I. Thacker, Spink & Co.; 1924. Ancient Mid-Indian Ksatriya Tribes. [Google Scholar]
- 48.Raychaudhuri H. Cosmo Publications; 2006. Political history of ancient India: From the accession of Parikshit to the extinction of the Gupta dynasty. [Google Scholar]
- 49.Mishra K.C. National Publishing House; 1987. Tribes in The Mahabharata: A socio-cultural study. [Google Scholar]
- 50.Levi S. The Indian merchant diaspora in early modern central Asia and Iran. Iran. Stud. 1999;32:483–512. [Google Scholar]
- 51.Mathew C.G.P. Nucleic Acids. Springer; 1984. The isolation of high molecular weight eukaryotic DNA; pp. 31–34. [DOI] [PubMed] [Google Scholar]
- 52.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 53.Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 54.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaubey G., Metspalu M., Choi Y., Mägi R., Romero I.G., Soares P., van Oven M., Behar D.M., Rootsi S., Hudjashov G. Population genetic structure in Indian Austroasiatic speakers: The role of landscape barriers and sex-specific admixture. Mol. Biol. Evol. 2011;28:1013–1024. doi: 10.1093/molbev/msq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasmussen M., Guo X., Wang Y., Lohmueller K.E., Rasmussen S., Albrechtsen A., Skotte L., Lindgreen S., Metspalu M., Jombart T. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yunusbayev B., Metspalu M., Järve M., Kutuev I., Rootsi S., Metspalu E., Behar D.M., Varendi K., Sahakyan H., Khusainova R. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol. Biol. Evol. 2012;29:359–365. doi: 10.1093/molbev/msr221. [DOI] [PubMed] [Google Scholar]
- 58.Behar D.M., Metspalu M., Baran Y., Kopelman N.M., Yunusbayev B., Gladstein A., Tzur S., Sahakyan H., Bahmanimehr A., Yepiskoposyan L. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum. Biol. 2013;85:859–900. doi: 10.3378/027.085.0604. [DOI] [PubMed] [Google Scholar]
- 59.Di Cristofaro J., Pennarun E., Mazières S., Myres N.M., Lin A.A., Temori S.A., Metspalu M., Metspalu E., Witzel M., King R.J. Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS ONE. 2013;8:e76748. doi: 10.1371/journal.pone.0076748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fedorova S.A., Reidla M., Metspalu E., Metspalu M., Rootsi S., Tambets K., Trofimova N., Zhadanov S.I., Hooshiar Kashani B., Olivieri A. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): Implications for the peopling of Northeast Eurasia. BMC Evol. Biol. 2013;13:127. doi: 10.1186/1471-2148-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raghavan M., Skoglund P., Graf K.E., Metspalu M., Albrechtsen A., Moltke I., Rasmussen S., Stafford T.W., Jr., Orlando L., Metspalu E. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kushniarevich A., Utevska O., Chuhryaeva M., Agdzhoyan A., Dibirova K., Uktveryte I., Möls M., Mulahasanovic L., Pshenichnov A., Frolova S., Genographic Consortium Genetic heritage of the Balto-Slavic speaking populations: A synthesis of autosomal, mitochondrial and Y-chromosomal data. PLoS ONE. 2015;10:e0135820. doi: 10.1371/journal.pone.0135820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yunusbayev B., Metspalu M., Metspalu E., Valeev A., Litvinov S., Valiev R., Akhmetova V., Balanovska E., Balanovsky O., Turdikulova S. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 2015;11:e1005068. doi: 10.1371/journal.pgen.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qamar R., Ayub Q., Mohyuddin A., Helgason A., Mazhar K., Mansoor A., Zerjal T., Tyler-Smith C., Mehdi S.Q. Y-chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grugni V., Battaglia V., Hooshiar Kashani B., Parolo S., Al-Zahery N., Achilli A., Olivieri A., Gandini F., Houshmand M., Sanati M.H. Ancient migratory events in the Middle East: New clues from the Y-chromosome variation of modern Iranians. PLoS ONE. 2012;7:e41252. doi: 10.1371/journal.pone.0041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derenko M., Malyarchuk B., Bahmanimehr A., Denisova G., Perkova M., Farjadian S., Yepiskoposyan L. Complete mitochondrial DNA diversity in Iranians. PLoS ONE. 2013;8:e80673. doi: 10.1371/journal.pone.0080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 70.Gallego Romero I., Basu Mallick C., Liebert A., Crivellaro F., Chaubey G., Itan Y., Metspalu M., Eaaswarkhanth M., Pitchappan R., Villems R. Herders of Indian and European cattle share their predominant allele for lactase persistence. Mol. Biol. Evol. 2012;29:249–260. doi: 10.1093/molbev/msr190. [DOI] [PubMed] [Google Scholar]
- 71.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., Ray N., Parra M.V., Rojas W., Duque C., Mesa N. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loh P.-R., Lipson M., Patterson N., Moorjani P., Pickrell J.K., Reich D., Berger B. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pickrell J.K., Pritchard J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McQuillan R., Leutenegger A.-L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joshi P.K., Esko T., Mattsson H., Eklund N., Gandin I., Nutile T., Jackson A.U., Schurmann C., Smith A.V., Zhang W. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523:459–462. doi: 10.1038/nature14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Browning B.L., Browning S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lawson D.J., Hellenthal G., Myers S., Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Delaneau O., Marchini J., Zagury J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 83.Montinaro F., Busby G.B.J., Pascali V.L., Myers S., Hellenthal G., Capelli C. Unravelling the hidden ancestry of American admixed populations. Nat. Commun. 2015;6:6596. doi: 10.1038/ncomms7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Dorp L., Balding D., Myers S., Pagani L., Tyler-Smith C., Bekele E., Tarekegn A., Thomas M.G., Bradman N., Hellenthal G. Evidence for a common origin of blacksmiths and cultivators in the Ethiopian Ari within the last 4500 Years: Lessons for clustering-based inference. PLoS Genet. 2015;11:e1005397. doi: 10.1371/journal.pgen.1005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lawson C.L., Hanson R.J. SIAM; Philadelphia: 1995. Solving least squares problems. [Google Scholar]
- 86.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.R Core Team . R Foundation for Statistical Computing; 2013. R: A language and environment for statistical computing. [Google Scholar]
- 88.Leslie S., Winney B., Hellenthal G., Davison D., Boumertit A., Day T., Hutnik K., Royrvik E.C., Cunliffe B., Lawson D.J., Wellcome Trust Case Control Consortium 2. International Multiple Sclerosis Genetics Consortium The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levi S.C. Brill; 2002. The Indian Diaspora in Central Asia and Its Trade, 1550–1900. [Google Scholar]
- 90.Bingley A.H. Ess Publications; 1978. Caste, tribes & culture of Rajputs. [Google Scholar]
- 91.Ibbetson D. Cosmo Publications; 1987. Landmarks in Indian Anthropology: Punjab Castes, Races, Castes, and Tribes of the People of Punjab. [Google Scholar]
- 92.Khatra P.S., Sharma V. Socio-economic issues in the development of nomadic Gujjars. Indian J. Agric. Econ. 1992;47:448–449. [Google Scholar]
- 93.Singh D.E. Walter de Gruyter; 2012. Islamization in Modern South Asia: Deobandi Reform and the Gujjar Response. [Google Scholar]
- 94.Singh S., Singh A., Rajkumar R., Sampath Kumar K., Kadarkarai Samy S., Nizamuddin S., Singh A., Ahmed Sheikh S., Peddada V., Khanna V. Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup. Sci. Rep. 2016;6:19157. doi: 10.1038/srep19157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jarrige C. Department of Culture and Tourism, Government of Sindh; 1995. Mehrgarh: Field reports 1974-1985, from Neolithic times to the Indus Civilization. [Google Scholar]
- 96.Gangal K., Vahia M.N., Adhikari R. Spatio-temporal analysis of the Indus urbanization. Curr. Sci. 2010;98:846–852. [Google Scholar]
- 97.Basu Mallick C., Iliescu F.M., Möls M., Hill S., Tamang R., Chaubey G., Goto R., Ho S.Y., Gallego Romero I., Crivellaro F. The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet. 2013;9:e1003912. doi: 10.1371/journal.pgen.1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones E.R., Gonzalez-Fortes G., Connell S., Siska V., Eriksson A., Martiniano R., McLaughlin R.L., Gallego Llorente M., Cassidy L.M., Gamba C. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 2015;6:8912. doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ayub Q., Mezzavilla M., Pagani L., Haber M., Mohyuddin A., Khaliq S., Mehdi S.Q., Tyler-Smith C. The Kalash genetic isolate: ancient divergence, drift, and selection. Am. J. Hum. Genet. 2015;96:775–783. doi: 10.1016/j.ajhg.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.