Abstract
FUK encodes fucokinase, the only enzyme capable of converting L-fucose to fucose-1-phosphate, which will ultimately be used for synthesizing GDP-fucose, the donor substrate for all fucosyltransferases. Although it is essential for fucose salvage, this pathway is thought to make only a minor contribution to the total amount of GDP-fucose. A second pathway, the major de novo pathway, involves conversion of GDP-mannose to GDP-fucose. Here we describe two unrelated individuals who have pathogenic variants in FUK and who presented with severe developmental delays, encephalopathy, intractable seizures, and hypotonia. The first individual was compound heterozygous for c.667T>C (p.Ser223Pro) and c.2047C>T (p.Arg683Cys), and the second individual was homozygous for c.2980A>C (p.Lys994Gln). Skin fibroblasts from the first individual confirmed the variants as loss of function and showed significant decreases in total GDP-[3H] fucose and [3H] fucose-1-phosphate. There was also a decrease in the incorporation of [5,6-3H]-fucose into fucosylated glycoproteins. Lys994 has previously been shown to be an important site for ubiquitin conjugation. Here, we show that loss-of-function variants in FUK cause a congenital glycosylation disorder characterized by a defective fucose-salvage pathway.
Keywords: congenital disorder of glycosylation; glycan, fucose; fucosylation; intellectual disability; encephalopathy; salvage
Main Text
Congenital disorders of glycosylation (CDG) are a group of clinically heterogenous disorders resulting from abnormal protein or lipid glycosylation.1, 2, 3 Genetic defects altering N-linked glycosylation account for the majority of the nearly 130 distinct glycosylation disorders; however, exome sequencing has uncovered many disorders involving the glycosylphosphatidylinositol (GPI) anchor and the glycosaminoglycan (GAG), O-linked, and glycolipid pathways.3, 4
A common form of glycosylation termed fucosylation involves incorporating the monosaccharide L-fucose (also known as 6-deoxy-L-galactose) into many glycans.5 It requires both the generation of the donor substrate guanosine diphosphate L-fucose (GDP-fucose) and the presence of a set of fucosyltransferases that transfer L-fucose to the specific glycan.5 In mammals, GDP-fucose is generated via two distinct mechanisms, a de novo pathway and a salvage pathway5 (Figure 1A). The de novo pathway utilizes a multistep reaction that converts GDP-mannose to GDP-fucose and requires both GDP-mannose 4,6-dehydratase (GMDS) and GDP-keto-6-deoxymannose 3,5 epimerase (TSTA3) (aka FX protein). The de novo pathway relies heavily on the cell’s ability to convert glucose and mannose to GDP-mannose and ultimately GDP-fucose (Figure 1A).5, 6, 7 Loss of either GMDS (MIM: 602884) or TSTA3 (MIM: 137020) by genetic manipulation or chemical inhibitors results in a near-complete loss of all cellular fucosylation.8, 9, 10
Figure 1.
Identification of FUK Pathogenic Variants in Two Unrelated Families
(A) Schematic depiction showing the fucose de novo (left) and salvage (right) pathways.
(B) Pedigrees showing FUK pathogenic variants and segregation in families for individual 1 (CDG-0440) and individual 2 (CDG-9286647).
(C) A schematic depiction of human FUK highlights each pathogenic variant on mRNA (GenBank: NM_145059.2) and protein amount (Uniprot: Q8N0W3).
(D) Alignment of several organisms and the conservation of the residue of each identified variant.
The salvage pathway uses β-L-fucose provided by two routes. The first route is through the transport of exogenous β-L-fucose into the cell via an as-yet-unknown transporter.11 The second is by the recycling of salvaged fucose inside the cell. Fucokinase (FUK) phosphorylates β-L-fucose to generate β-L-fucose-1-phosphate (β-L-Fuc1p), which subsequently undergoes a fucose-1-phosphate guanylyltransferase (FPGT)-dependent condensation reaction with guanosine triphosphate (GTP) to form GDP-β-L-fucose (Figure 1A).5, 12
It is important to note that only the β-anomers of L-fucose can be utilized by human cells, and when a fucosyltransferase (such as FUT8) utilizes GDP-β-L-fucose to create a glycosidic linkage, an inversion that switches β-L-fucose anomers to α-L-fucose occurs.5
This anomeric shift is critical because fucosylated-glycan hydrolysis, which occurs exclusively within the lysosome, is performed by the lysosomal α-fucosidases (FUCA1) and liberates free α-L-fucose to be transported out of the lysosome and into the cytoplasm (Figure 1A).13, 14 The final step of the process is catalyzed by the fucose mutarotase (FUOM), which carries out another anomeric reaction converting α-L-fucose back to the biologically active β-L-fucose.15 From now on, all reference to L-fucose will be to β-L-fucose, unless otherwise specified.
Although the roles for fucosylation, particularly FUT8-dependent core fucosylation, have been well documented in immune regulation, cancer metastasis, and inflammation, the precise contributions of either the de novo or the salvage pathways have not been extensively studied.16, 17, 18, 19, 20, 21 However, recent studies have shed light on the importance of the salvage pathway and the roles of GMDS, TSTA3, and FUK in modulating the invasive and metastatic properties of several forms of cancer, including melanoma, colorectal, and hepatocellular carcinomas.22, 23, 24, 25
To date, no genetic disorders have been specifically identified within either the de novo or salvage pathways, although pathogenic mutations have been identified in several downstream components of the fucosylation pathway. For example, recessive mutations in SLC35C1 (MIM: 605881) cause leukocyte adhesion deficiency type II (LADII), also known as SLC35C1-CDG (MIM: 266265), and are due to an inability to transport GDP-fucose into the Golgi.26, 27 Recessive mutations in the fucosyltransferase 8 (FUT8) (MIM: 602589) cause FUT8-CDG (MIM: 618005), characterized as a severe, multisystem disorder involving a defect in the most prevalent type of fucosylation, core fucosylation.28 Finally, recessive mutations in α-fucosidase 1 (FUCA1) (MIM: 612280) result in an inability to hydrolyze fucose from fucosylated glycoproteins and glycolipids, and they cause the rare lysosomal-storage disorder fucosidosis (MIM: 230000).29
We detail two unrelated individuals who had profound neurological impairments and who were found to have variants of uncertain significance (VUS) in FUK (MIM: 608675); here, we show evidence for the variants’ pathogenicity. Prior to the research studies, informed consent was obtained as part of the Undiagnosed Diseases Network, according to the institutional review board of the National Human Genome Research Institute.
Individual 1 (CDG-0440) is a male born without complications to healthy, non-consanguineous parents of Hispanic origin. He has a history of feeding difficulties secondary to hypotonia, dysphagia, and GI dysmotility. He has reflux and is noted to have poor gastric emptying. Consequently, he does not take food by mouth and has a G-J tube placement for feedings (Table 1). He presents with severe developmental delays and multiple neurological complications, including severe intellectual disabilities, hypotonia, and intractable seizures (Table 1). An electroencephalogram (EEG) showed extremely abnormal background activity consistent with severe diffuse disturbance in cerebral function: multifocal epileptiform activity involving the hemispheres bilaterally and brief multifocal seizures. This activity was interpreted as generalized tonic-clonic-type seizures in the setting of epileptic encephalopathy (Table 1). MRI findings showed a dysmorphic corpus callosum, a supratentorial ventricular system, and borderline macrocephaly in the presence of generous extra-axial CSF spaces and delays in deep-white-matter myelination (Table 1). He has had recurrent respiratory infections and episodes of apnea, one of which required lifesaving intervention. He is also noted to have symmetric maculopathy with severe visual impairment. Extensive laboratory studies have shown an elevated CSF:plasma glycine ratio and a mild, yet persistent elevation of homocysteine (18.2–22.3 μmol/L [normal range 4–14 μmol/L]) and methylmalonic acid (MMA) (0.94–1.2 nmol/L [normal range 0–0.5 nmol/L]) concentrations in the blood; these elevated amounts decreased slightly in response to hydroxocobalamin supplementation. Importantly, at three months of age and prior to hydroxocobalamin treatment, the B12 measurement was 1,353 pg/mL (normal is 211–911 pg/mL). The concentrations of homocysteine and MMA did not normalize when the individual was given B12, and thus it is not a B12 deficiency that is causing the elevated methylmalonic acid concentrations, but rather a problem in the transport or activation of B12.
Table 1.
Molecular and Clinical Characterization of Two Individuals with Pathogenic Variants in FUK
| Individual 1 (CDG0440) | Individual 2 (CDG9286647) | |
|---|---|---|
| Sex | male | female |
| Current age | 6 years | 7 years |
| Ancestry | Hispanic | Middle Eastern (Qatari) |
| Age at onset of symptoms | 3 years | 4 years |
| FUK pathogenic variants | c.667T>C (p.Ser223Pro); c.2047C>T (p.Arg683Cys) | c.2980A>C (p.Lys994Gln) |
| Pregnancy complications | No | born premature at 25 weeks (810 g) |
| Feeding problems | Feeding difficulties were secondary to hypotonia, dysphagia, and GI dysmotility. Positive for reflux and is noted to have poor gastric emptying. Does not take food by mouth and has a G-J tube placement for feedings. | Feeding difficulties were categorized by aspiration with oral feeds, necessitating G-tube feeding. Noted for chronic malabsorption with diarrhea. |
| Facial features or dysmorphism | no | no |
| Developmental delay | yes | yes |
| Stature | normal for age | normal for age |
| Intellectual disability | yes, severe | yes, severe |
| Seizures/epilepsy | multifocal epileptiform activity involving the hemispheres bilaterally and brief multifocal seizures, interpreted as generalized tonic-clonic type seizures in the setting of epileptic encephalopathy. | Seizures were initially consistent with infantile spasms (flexor spasms), and EEG findings showed hypsarrhythmia. |
| Ataxia or gait problems | nonambulatory | nonambulatory |
| Hypotonia | central hypotonia | central hypotonia with limb spasticity |
| Brain anomalies | yes, dysplastic corpus callosum, delayed myelination in deep white matter | yes, cerebellar atrophy, agenesis of corpus callosum, white matter abnormalities that were characterized as severe periventricular leukomalacia with paucity of white matter |
| Ocular | symmetric maculopathy with severe visual impairment | Strabismus, nystagmus, cortical blindness, optic nerve atrophy |
| Skeletal | contractures | contractures |
| Cardiac | none | none |
| Respiratory | recurrent respiratory infections | respiratory difficulties |
| Hepatopathy | yes, elevated gamma glutamyl transferase (GGT) | no |
| Gastrointestinal | GI problems. G-J-tube, motility issues | GI Problems. G-tube, history of neonatal necrotizing enterocolitis with bowel perforation |
| Immunological | recurrent infections, enlarged platelets | no |
| Biochemical testing abnormalities | normal carbohydrate-deficient transferrin result | none reported |
Whole-exome-sequencing was performed on trio genomic DNA and did not identify any candidates that could explain the abnormal biochemical results. A de novo VUS was identified in KMT2b (MIM: 606834) c.644C>A (p.Thr215Asn); however, the variant was predicated to be benign or tolerated, and the clinical phenotype associated with KMT2B, autosomal-dominant Dystonia 28, childhood-onset (MIM: 617284), did not fit the presentation of individual 1. WES did identify two VUSs in FUK (GenBank: NM_145059.2), c.667T>C (p.Ser223Pro) and c.2047C>T (p.Arg683Cys) (Figure 1B, Table 1).
Individual 2 (CDG-9286647) is a female born prematurely at 25 weeks to healthy, consanguineous parents of Qatari origin. Like individual 1, she has a history of feeding problems, respiratory difficulties, developmental delays, severe intellectual disabilities, intractable seizures, and hypotonia (Table 1). Her feeding difficulties were characterized by aspiration with oral feeds, necessitating G-tube feeding. She had chronic malabsorption with diarrhea, although this was at least partly due to the fact that she had a history of necrotizing enterocolitis and had had a partial small-bowel resection (Table 1). She did present with failure to thrive, but this was well managed early on and has since improved. An MRI showed cerebellar atrophy, agenesis of the corpus callosum, and white-matter abnormalities; these symptoms were all characterized as severe periventricular leukomalacia with paucity of white matter and were more consistent with a destructive process rather than delayed myelination or demyelination. Also present were cystic encephalomalacia, as well as a small pons and cerebellum that were initially read as consistent with pontocerebellar hypoplasia (Table 1). Her seizures were initially consistent with infantile spasms (flexor spasms), and the EEG findings showed hypsarrhythmia. After treatment for infantile spasms, seizures evolved and were primarily characterized by eye blinking and brief arrest in activity; EEGs continued to show variant hypsarrhythmia (continued disorganization typical of hypsarrhythmia but low voltage). Spasms were responsive to treatment with prednisone, but eye-blinking seizures were refractory to multiple medications (Table 1).
She also presented with ocular problems, including strabismus, optic-nerve atrophy, and cortical blindness (Table 1). Furthermore, she did not show the same abnormal metabolic profile as individual 1. WES was performed on trio genomic DNA and identified a homozygous FUK VUS, c.2980A>C (p.Lys994Gln) (Figure 1B, Table 1). Both individuals have had their WES data reanalyzed and did not show any additional variants of interest to prioritize.
In silico modeling using the Combined Annotation Dependent Depletion (CADD) scoring method30 showed that all three FUK variants, c.667T>C (p.Ser223Pro) (CADD = 24.8), c.2047C>T (p.Arg683Cys) ( CADD = 32), and c.2980A>C (p.Lys994Gln) (CADD = 28.1), were predicted to be in the top 0.5% of deleterious variants.
We used the Genome Aggregation Database (gnomAD) of 123,136 exomes and 15,496 genomes (Ver2: accessed October 12, 2018) to highlight the rarity of these three variants; we found the c.667T>C (p.Ser223Pro) variant in 17 carriers from 246,034 alleles, the c.2047C>T (p.Arg683Cys) variant in 27 carriers from 168,584 alleles, and the c.2980A>C (p.Lys994Gln) variant in 65 carriers from 276,772 alleles. Importantly, we did not identify any homozygotes for any of the three variants. Given the rarity of these three variants and the fact that they are predicted to be deleterious, we focused on FUK as a plausible candidate for biochemical investigation.
Fucokinase shares protein homology with other kinases within the highly conserved GHMP kinase superfamily, which includes galactokinase (GALK) and the plant-specific L-arabinokinase (ARA1). Like FUK, GALK and ARA1 are both sugar-1 phosphate-generating kinases.31 The p.Ser223Pro variant occurs in the L-fucokinase domain, but the p.Arg683Cys variant occurs between the L-fucokinase domain and the GHMP kinase N-terminal domain, where the ATP binding motif resides (Figure 1C). The p.Lys994Gln variant occurs within the GHMP kinase C-terminal domain (Figure 1C). All three amino acids are highly conserved within higher eukaryotes (Figure 1D), which is reflected in the previously mentioned CADD scores.
For individual 1, both primary fibroblast and transformed lymphoblast cell lines were available for biochemical analysis. Unfortunately, neither was available for individual 2.
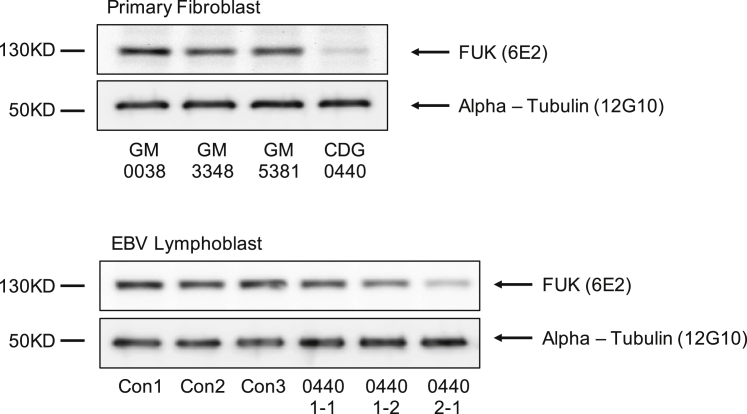
Immunoblot analysis with a monoclonal FUK antibody (Thermo Fisher MA5-15847) showed that fibroblasts from individual 1 had an 80% reduction in the amount of FUK antigen when they were compared to three commercially available control fibroblast cultures (Figure 2). Furthermore, lymphoblasts derived from this individual also showed a clear reduction (60%) in the amount of FUK antigens, although to a lesser extent than in the fibroblasts (Figure 2). All FUK immunoblots were normalized to alpha tubulin levels (Figure 2).
Figure 2.
Immunoblot Analysis of FUK Protein in Control and Individual 1 (CDG-0440) Fibroblasts and Lymphoblasts
Upper panel: immunoblot analysis with a monoclonal FUK antibody was used for comparison of three controls and individual 1 and shows an 80% reduction in FUK protein amount. Lower panel: immunoblot analysis of FUK in EBV-transformed lymphoblasts from three controls, the parents of individual 1 (1-1, father; 1-2, mother), and individual 1 (2-1, proband). The mAb to FUK (6E2) (Thermo Fisher MA5-15847) was used at a 1:1000 dilution.
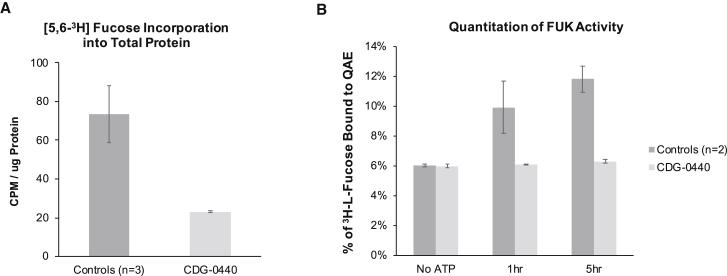
We used two independent methods to directly assay FUK activity in fibroblasts from individual 1. The first method measured how much [5,6-3H]-fucose had been incorporated into cell-associated glycoproteins (i.e., fucosylated glycoproteins). Normally, cultured cells are not grown with exogenous L-fucose, except for that present in the supplemented bovine/calf serum. Any exogenously provided L-fucose will be utilized by the salvage pathway and incorporated into fucosylated glycoproteins via the previously mentioned salvage pathway. Therefore, providing exogenous [5,6-3H]-fucose and observing its incorporation should be a direct measure of FUK activity. When compared to the average of three controls, the amount of [5,6-3H]-fucose incorporated into cell-associated fucosylated glycoproteins in individual 1 was 70% lower (Figure 3A). Furthermore, we determined a 60% reduction in both GDP-[3H] fucose and [3H]-Fuc1p (Figure S1A). Both of these results are consistent with a loss of FUK activity. Importantly, lentiviral transduction of wild-type FUK into fibroblasts from individual 1 was able to restore [5,6-3H]-fucose incorporation (Figure S1B and S1C).
Figure 3.
Determination of FUK Activity in Control and Individual 1 (CDG-0440) Fibroblasts
(A) Metabolic labeling of fibroblasts (3 controls and individual 1 all in triplicate) with 2 μCi/mL [5,6-3H]-fucose for 24 hr and determination of [3H]-fucose incorporation into cell-associated protein. After the cells were normalized to protein content, individual 1 had a 70% reduction in fucose incorporation when this individual was compared to the three controls.
(B) In vitro FUK assay performed with crude cytoplasmic extracts and [5,6-3H]-fucose as the substrate. After the kinase reaction, the product [5,6-3H] fucose-1-phosphate was purified via ion-exchange chromatography and counted with a scintillation counter.
The assay was performed in three biological replicates with error bars representing standard deviations.
The second method is an in vitro assay of FUK kinase activity. Unlike the previous method, which involved radiolabeling live cells, the in vitro method required growing substantial amounts of cells to prepare crude cytoplasmic cell extracts in which to assay FUK activity.32, 33 After lysis, FUK activity decreases rapidly due to enzyme instability.32, 33 After optimizing assay conditions for primary control fibroblasts, we found that individual 1 had nearly no FUK activity, in contrast to the higher activity in two control cell lines (Figure 3B). Although these results support individual 1’s having no FUK activity, we know there is residual FUK activity from the cell-based [5,6-3H]-fucose labeling experiments. These results can be explained in part by the inherent instability of wild-type FUK once the cells are lysed and by the further instability that individual 1 has as a result of FUK variants.
Because there is no other known fucose kinase, both the cell-based and in vitro assays can be seen as direct measurements of FUK activity. To confirm this assumption, we created a CRISPR/Cas9 FUK knockout (KO) HAP1 cell line and showed that when we provided exogenous [5,6-3H]-fucose, there was no incorporation into either cell-associated fucosylated glycoproteins or purified soluble fractions where GDP-fucose or fucose-1p would be present; thus, we proved there is no other kinase capable of substituting for FUK (Figure S2A).
Because the vast majority of GDP-fucose is generated via the de novo pathway, we hypothesized that the loss of the salvage pathway would have minimal effects on total cellular fucosylation. Flow-cytometric analysis of both fibroblasts from individual 1 and FUK KO cells supported this hypothesis and showed no detectable changes in fucose-dependent lectin binding (data not shown). Furthermore, mass-spectrometry analysis of total serum N- and O-glycans from individual 1 showed no obvious changes in either total or individual N-glycan composition (data not shown). These results support the notion that the salvage pathway might be more critical in some cell types but that others can function without it.
Unfortunately, no cell lines from individual 2 were available to assay. Attempts to use lentiviral complementation of our FUK knockout (KO) HAP1 cells highlighted that FUK protein level is highly regulated and likely cell type-dependent. For example, in fibroblasts from individual 1, we consistently saw an 80% reduction in FUK protein amount with the corresponding reduction in [5,6-3H]-fucose incorporation into cell-associated material (Figures 2 and 3A). However, when we used lentivirus to infect the FUK KO cells with either FUK variant from individual 1, individually or in combination, the protein amounts of either variant were comparable to those of wild-type cells (data not shown). This suggests either that FUK protein amount is highly regulated and varies among different cell types or that perhaps the lentiviral promoter used was producing too much FUK protein. We addressed the latter possibility by infecting the FUK KO cells with lentiviral constructs containing both a weak EF1 and strong CMV promoter and obtained similar outcomes (data not shown).
Despite being unable to create a usable cell-based model to assay the p.Lys994Gln variant from individual 2, we identified published proteomic studies highlighting Lys994 as an important site of ubiquitin conjugation in both mouse and human tissue samples.34, 35 Because we cannot perform cell-based assays to confirm the pathogenicity of the p.Lys994Gln found in individual 2, we are classifying it as a likely pathogenic variant.
The primary function of FUK as the only known fucose-1-phosphate kinase across nearly all species is well established. However, the biological consequence of losing FUK or the fucose salvage pathway is not well understood. To date, no Fuk- or Fpgt-knockout (KO) mouse models are available, but studies using Tsta3 KO mice have shown how critical the de novo pathway is to both animal survival and total cellular fucosylation.9 In fact, HAP1 cells null for FUK do not show a clear fucosylation defect as determined by lectin staining (data not shown). Clearly, the de novo pathway provides adequate levels of GDP-fucose needed to carry out fucosylation in this cell line, and this most likely holds true for other cell types as well. Cancer cells lacking GMDS, CHO cells treated with specific GMDS inhibitors, or even Tsta3-KO mice have a near-complete deficiency of cellular fucosylation.9, 10, 36, 37 However, these defects in de novo-pathway-dependent fucosylation can be completely reversed if cells are provided with exogenous L-fucose.9 Some positive results were seen in several LAD-II cases, and thus it is quite likely that L-fucose could be tested as a therapeutic option for this disorder.
Surprisingly, the commonly-utilized model organism Drosophila melanogaster lacks not only a protein ortholog to human FUK, but also an ortholog to FPGT, suggesting that the salvage pathway might not be critical for fly development.38 Studies in zebrafish show that salvage-pathway enzymes are provided maternally; detectable transcripts for FUK and FPGT are not detected until the 16–32 cell stage, prior to the beginning of zygotic transcription.39 In contrast, enzymes encoding for the de novo pathway are present throughout development.39
Although next-generation sequencing will certainly identify more potential FUK-CDG cases, confirming their diagnosis might be difficult unless reliable biomarkers can be discovered. Until then, fibroblast cell lines will be needed to directly assay FUK activity.
Here we show that FUK, a core component of the fucose-salvage pathway, causes a congenital glycosylation disorder characterized by defective fucose salvage.
Consortia
Members of the Undiagnosed Diseases Network include David R. Adams, Aaron Aday, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Eva Baker, Ashok Balasubramanyam, Hayk Barseghyan, Gabriel F. Batzli, Alan H. Beggs, Babak Behnam, Hugo J. Bellen, Jonathan A. Bernstein, Gerard, T. Berry, Anna Bican, David P. Bick, Camille L. Birch, Devon Bonner, Braden E. Boone, Bret L. Bostwick, Lauren C. Briere, Elly Brokamp, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Shan Chen, Gary D. Clark, Terra R. Coakley, Joy D. Cogan, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Precilla D’Souza, Mariska Davids, Jean M. Davidson, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, Emilie D. Douine, David D. Draper, Annika M. Dries, Laura Duncan, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Gregory M. Enns, Ascia Eskin, Cecilia Esteves, Tyra Estwick, Liliana Fernandez, Carlos Ferreira, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Noah D. Friedman, William A. Gahl, Rena A. Godfrey, Alica M. Goldman, David B. Goldstein, Sarah E. Gould, Jean-Philippe F. Gourdine, Catherine A. Groden, Andrea L. Gropman, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Frances High, Ingrid A. Holm, Jason Hom, Ellen M. Howerton, Yong Huang, Fariha Jamal, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Lefkothea Karaviti, Emily G. Kelley, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Susan Korrick, Mary Koziura, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, C. Christopher Lau, Jozef Lazar, Kimberly LeBlanc, Brendan H. Lee, Hane Lee, Shawn E. Levy, Richard A. Lewis, Sharyn A. Lincoln, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Ronit Marom, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Thomas May, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Jason D. Merker, Thomas O. Metz, Matthew Might, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, David R. Murdock, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, James P. Orengo, J. Carl Pallais, Christina GS. Palmer, Jeanette C. Papp, Neil H. Parker, Loren DM. Pena, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Jacinda B. Sampson, Susan L. Samson, Kelly Schoch, Daryl A. Scott, Lisa Shakachite, Prashant Sharma, Vandana Shashi, Rebecca Signer, Edwin K. Silverman, Janet S. Sinsheimer, Kevin S. Smith, Rebecca C. Spillmann, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, David A. Sweetser, Queenie K.-G. Tan, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Eric Vilain, Tiphanie P. Vogel, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Melissa Walker, Jijun Wan, Michael F. Wangler, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, John Yang, Yaping Yang, Amanda J. Yoon, Guoyun Yu, Diane B. Zastrow, Chunli Zhao, and Allison Zheng.
Declaration of Interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing offered by Baylor Genetics Laboratories.
Acknowledgments
The Rocket Fund, National Institutes of Health (NIH) grant R01DK099551, and Diana & Gabriel Wisdom supported this work. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination in the Office of the NIH Director under award number U01HG007709. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding was also provided by the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (U54 HD083092). L.C.B. is supported by NIH grant K08DK106453 and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. The authors would like to thank the SBP Medical Discovery Institute FACS Core and Colin W. McSweeney for their help.
Published: November 29, 2018
Footnotes
Supplemental Data include two figures and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.021.
Web Resources
Combined Annotation Dependent Depletion (CADD), https://cadd.gs.washington.edu/
Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org/
Supplemental Data
References
- 1.Freeze H.H., Eklund E.A., Ng B.G., Patterson M.C. Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci. 2015;38:105–125. doi: 10.1146/annurev-neuro-071714-034019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng B.G., Freeze H.H. Perspectives on glycosylation and its congenital disorders. Trends Genet. 2018;34:466–476. doi: 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira C.R., Altassan R., Marques-Da-Silva D., Francisco R., Jaeken J., Morava E. Recognizable phenotypes in CDG. J. Inherit. Metab. Dis. 2018;41:541–553. doi: 10.1007/s10545-018-0156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeze H.H., Schachter H., Kinoshita T. Genetic Disorders of Glycosylation. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., editors. Essentials of Glycobiology, rd. Cold Spring Harbor, NY; 2015. pp. 569–582. [Google Scholar]
- 5.Schneider M., Al-Shareffi E., Haltiwanger R.S. Biological functions of fucose in mammals. Glycobiology. 2017;27:601–618. doi: 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yurchenco P.D., Atkinson P.H. Equilibration of fucosyl glycoprotein pools in HeLa cells. Biochemistry. 1977;16:944–953. doi: 10.1021/bi00624a021. [DOI] [PubMed] [Google Scholar]
- 7.Yurchenco P.D., Atkinson P.H. Fucosyl-glycoprotein and precursor polls in HeLa cells. Biochemistry. 1975;14:3107–3114. doi: 10.1021/bi00685a011. [DOI] [PubMed] [Google Scholar]
- 8.Kanda Y., Imai-Nishiya H., Kuni-Kamochi R., Mori K., Inoue M., Kitajima-Miyama K., Okazaki A., Iida S., Shitara K., Satoh M. Establishment of a GDP-mannose 4,6-dehydratase (GMD) knockout host cell line: a new strategy for generating completely non-fucosylated recombinant therapeutics. J. Biotechnol. 2007;130:300–310. doi: 10.1016/j.jbiotec.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 9.Smith P.L., Myers J.T., Rogers C.E., Zhou L., Petryniak B., Becker D.J., Homeister J.W., Lowe J.B. Conditional control of selectin ligand expression and global fucosylation events in mice with a targeted mutation at the FX locus. J. Cell Biol. 2002;158:801–815. doi: 10.1083/jcb.200203125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen J.G., Mujacic M., Frohn M.J., Pickrell A.J., Kodama P., Bagal D., San Miguel T., Sickmier E.A., Osgood S., Swietlow A. Facile modulation of antibody fucosylation with small molecule fucostatin inhibitors and cocrystal structure with GDP-mannose 4,6-dehydratase. ACS Chem. Biol. 2016;11:2734–2743. doi: 10.1021/acschembio.6b00460. [DOI] [PubMed] [Google Scholar]
- 11.Leck J.R., Wiese T.J. Purification and characterization of the L-fucose transporter. Protein Expr. Purif. 2004;37:288–293. doi: 10.1016/j.pep.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Quirk S., Seley K.L. Identification of catalytic amino acids in the human GTP fucose pyrophosphorylase active site. Biochemistry. 2005;44:13172–13178. doi: 10.1021/bi051288d. [DOI] [PubMed] [Google Scholar]
- 13.Opheim D.J., Touster O. The purification and characterization of rat liver lysosomal alpha-L-fucosidase. J. Biol. Chem. 1977;252:739–743. [PubMed] [Google Scholar]
- 14.Dawson G., Tsay G. Substrate specificity of human alpha-L-fucosidase. Arch. Biochem. Biophys. 1977;184:12–23. doi: 10.1016/0003-9861(77)90321-6. [DOI] [PubMed] [Google Scholar]
- 15.Park D., Ryu K.S., Choi D., Kwak J., Park C. Characterization and role of fucose mutarotase in mammalian cells. Glycobiology. 2007;17:955–962. doi: 10.1093/glycob/cwm066. [DOI] [PubMed] [Google Scholar]
- 16.Ito Y., Miyauchi A., Yoshida H., Uruno T., Nakano K., Takamura Y., Miya A., Kobayashi K., Yokozawa T., Matsuzuka F. Expression of alpha1,6-fucosyltransferase (FUT8) in papillary carcinoma of the thyroid: Its linkage to biological aggressiveness and anaplastic transformation. Cancer Lett. 2003;200:167–172. doi: 10.1016/s0304-3835(03)00383-5. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal P., Fontanals-Cirera B., Sokolova E., Jacob S., Vaiana C.A., Argibay D., Davalos V., McDermott M., Nayak S., Darvishian F. A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell. 2017;31:804–819e7. doi: 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rombouts Y., Jónasdóttir H.S., Hipgrave Ederveen A.L., Reiding K.R., Jansen B.C., Freysdottir J., Hardardottir I., Ioan-Facsinay A., Giera M., Wuhrer M. Acute phase inflammation is characterized by rapid changes in plasma/peritoneal fluid N-glycosylation in mice. Glycoconj. J. 2016;33:457–470. doi: 10.1007/s10719-015-9648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda M., Kamada Y., Takamatsu S., Shimomura M., Maekawa T., Sobajima T., Fujii H., Nakayama K., Nishino K., Yamada M. Specific increase in serum core-fucosylated haptoglobin in patients with chronic pancreatitis. Pancreatology. 2016;16:238–243. doi: 10.1016/j.pan.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Yu R., Ma B., Yang Y., Jiao X., Liu Y., Cao H., Dong W., Liu L., Ma K. Core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production. J. Immunol. 2015;194:2596–2606. doi: 10.4049/jimmunol.1402678. [DOI] [PubMed] [Google Scholar]
- 21.Collins E.S., Galligan M.C., Saldova R., Adamczyk B., Abrahams J.L., Campbell M.P., Ng C.T., Veale D.J., Murphy T.B., Rudd P.M., Fitzgerald O. Glycosylation status of serum in inflammatory arthritis in response to anti-TNF treatment. Rheumatology (Oxford) 2013;52:1572–1582. doi: 10.1093/rheumatology/ket189. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Huang D., Chen K.Y., Cui M., Wang W., Huang X., Awadellah A., Li Q., Friedman A., Xin W.W. Fucosylation deficiency in mice leads to colitis and adenocarcinoma. Gastroenterology. 2017;152:193–205e10. doi: 10.1053/j.gastro.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama K., Moriwaki K., Imai T., Shinzaki S., Kamada Y., Murata K., Miyoshi E. Mutation of GDP-mannose-4,6-dehydratase in colorectal cancer metastasis. PLoS ONE. 2013;8:e70298. doi: 10.1371/journal.pone.0070298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau E., Feng Y., Claps G., Fukuda M.N., Perlina A., Donn D., Jilaveanu L., Kluger H., Freeze H.H., Ronai Z.A. The transcription factor ATF2 promotes melanoma metastasis by suppressing protein fucosylation. Sci. Signal. 2015;8:ra124. doi: 10.1126/scisignal.aac6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeley T., Lin S., Lester D.K., Lau E.K., Yang S. The fucose salvage pathway inhibits invadopodia formation and extracellular matrix degradation in melanoma cells. PLoS ONE. 2018;13:e0199128. doi: 10.1371/journal.pone.0199128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lühn K., Wild M.K., Eckhardt M., Gerardy-Schahn R., Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- 27.Lübke T., Marquardt T., Etzioni A., Hartmann E., von Figura K., Körner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- 28.Ng B.G., Xu G., Chandy N., Steyermark J., Shinde D.N., Radtke K., Raymond K., Lebrilla C.B., AlAsmari A., Suchy S.F. Biallelic mutations in FUT8 cause a congenital disorder of glycosylation with defective fucosylation. Am. J. Hum. Genet. 2018;102:188–195. doi: 10.1016/j.ajhg.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zielke K., Veath M.L., O’Brien J.S. Fucosidosis: Deficiency of alpha-L-fucosidase in cultured skin fibroblasts. J. Exp. Med. 1972;136:197–199. doi: 10.1084/jem.136.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange B.M., Croteau R. Isopentenyl diphosphate biosynthesis via a mevalonate-independent pathway: Isopentenyl monophosphate kinase catalyzes the terminal enzymatic step. Proc. Natl. Acad. Sci. USA. 1999;96:13714–13719. doi: 10.1073/pnas.96.24.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S.H., Pastuszak I., Drake R., Elbein A.D. Purification to apparent homogeneity and properties of pig kidney L-fucose kinase. J. Biol. Chem. 1998;273:5685–5691. doi: 10.1074/jbc.273.10.5685. [DOI] [PubMed] [Google Scholar]
- 33.Ishihara H., Massaro D.J., Heath E.C. The metabolism of L-fucose. 3. The enzymatic synthesis of beta-L-fucose 1-phosphate. J. Biol. Chem. 1968;243:1103–1109. [PubMed] [Google Scholar]
- 34.Wagner S.A., Beli P., Weinert B.T., Schölz C., Kelstrup C.D., Young C., Nielsen M.L., Olsen J.V., Brakebusch C., Choudhary C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics. 2012;11:1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Udeshi N.D., Svinkina T., Mertins P., Kuhn E., Mani D.R., Qiao J.W., Carr S.A. Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics. 2013;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriwaki K., Noda K., Furukawa Y., Ohshima K., Uchiyama A., Nakagawa T., Taniguchi N., Daigo Y., Nakamura Y., Hayashi N. Deficiency of GMDS leads to escape from NK cell-mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology. 2009;137:188–198. doi: 10.1053/j.gastro.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Moriwaki K., Noda K., Nakagawa T., Asahi M., Yoshihara H., Taniguchi N., Hayashi N., Miyoshi E. A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology. 2007;17:1311–1320. doi: 10.1093/glycob/cwm094. [DOI] [PubMed] [Google Scholar]
- 38.Roos C., Kolmer M., Mattila P., Renkonen R. Composition of Drosophila melanogaster proteome involved in fucosylated glycan metabolism. J. Biol. Chem. 2002;277:3168–3175. doi: 10.1074/jbc.M107927200. [DOI] [PubMed] [Google Scholar]
- 39.Dehnert K.W., Beahm B.J., Huynh T.T., Baskin J.M., Laughlin S.T., Wang W., Wu P., Amacher S.L., Bertozzi C.R. Metabolic labeling of fucosylated glycans in developing zebrafish. ACS Chem. Biol. 2011;6:547–552. doi: 10.1021/cb100284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.