Abstract
Neurodevelopmental disorders (NDD) are genetically and phenotypically heterogeneous conditions due to defects in genes involved in development and function of the nervous system. Individuals with NDD, in addition to their primary neurodevelopmental phenotype, may also have accompanying syndromic features that can be very helpful diagnostically especially those with recognizable facial appearance. In this study, we describe ten similarly affected individuals from six unrelated families of different ethnic origins having bi-allelic truncating variants in TMEM94, which encodes for an uncharacterized transmembrane nuclear protein that is highly conserved across mammals. The affected individuals manifested with global developmental delay/intellectual disability, and dysmorphic facial features including triangular face, deep set eyes, broad nasal root and tip and anteverted nostrils, thick arched eye brows, hypertrichosis, pointed chin, and hypertelorism. Birthweight in the upper normal range was observed in most, and all but one had congenital heart defects (CHD). Gene expression analysis in available cells from affected individuals showed reduced expression of TMEM94. Global transcriptome profiling using microarray and RNA sequencing revealed several dysregulated genes essential for cell growth, proliferation and survival that are predicted to have an impact on cardiotoxicity hematological system and neurodevelopment. Loss of Tmem94 in mouse model generated by CRISPR/Cas9 was embryonic lethal and led to craniofacial and cardiac abnormalities and abnormal neuronal migration pattern, suggesting that this gene is important in craniofacial, cardiovascular, and nervous system development. Our study suggests the genetic etiology of a recognizable dysmorphic syndrome with NDD and CHD and highlights the role of TMEM94 in early development.
Keywords: neurodevelopmental disorder, facial dysmorphism, congenital heart defects, CRISPR-Cas9, Tmem94 knockout mice
Introduction
Human brain development is a complex dynamic process that begins in the third gestational week and extends at least through early adulthood.1 Neurodevelopmental disorders (NDD) refer to impaired development and function of the central nervous system, which include developmental delay, intellectual disability (ID), learning disabilities, autism spectrum disorders, motor and tic disorders, and communication disorders.2 NDD can be accompanied by non-central nervous system defects, e.g., congenital heart defects (CHD) and facial dysmorphism, in a recognizable syndromic pattern, is clinically and genetically heterogeneous.3, 4 The genes implicated with both NDD and CHD are generally involved in early embryonic stages of nervous and neurovascular development including brain vascularization, vessel maturation and branching, development of blood brain barrier, neural proliferation and migration, and synapse formation.2, 5 For instance, neural crest cell (NCC) differentiation and migration are coordinated by a large array of genes function in a precisely orchestrated manner.6 NCC are multipotent cells derive into multiple cell types including glial cells, connective tissues, craniofacial cartilage and bone, pigmented cells of skin, and cardiac septa development. Thus, perturbation of genes associated with NCC development and function leads to multi organ abnormalities including brain, craniofacial and cardiac development.7 Owing to the increased utilization of next generation sequencing, there has been a recent surge in the identification of genetic etiologies of NDD.8, 9, 10, 11, 12 However, achieving a definitive genetic diagnosis is still incomplete because of the wide range of phenotypic and genotypic etiologies and a large number of remaining unknown genetic causes.13, 14, 15
In this study, we describe recessive truncating variants in TMEM94 in ten individuals who manifested with NDD, CHD, and similar facial gestalt identified through collaborative efforts and matchmaking platforms.16 Analysis of primary cells from affected individuals showed reduced expression of TMEM94. Further global transcription profiling showed dysregulation of several genes related to cell survival and cell growth and proliferation, among others. Deletion of Tmem94 in mice by CRISPR-Cas9 led to impaired embryonic development, especially neuro and cardiovascular development. The overlap between phenotypes observed in the affected individuals and those in homozygous mutant mice embryos emphasize the role of this gene in brain, craniofacial, and cardiac development.
Material and Methods
Human Subjects and Genetic Analysis
Affected members of family 1 were evaluated at Amrita Institute of Medical Sciences and Research Center, Kerala, India. Informed consent for research was obtained from parents under the protocol 76-HG-0238 (diagnosis and treatment of patients with inborn errors of metabolism or other genetic disorders) approved by the Institutional Review Board of the NIH, National Human Genome Research Institute (NHGRI). Affected individuals from families 2 and 6 were recruited from Qatar into an IRB-approved protocol (KFSHRC RAC#2080006). Written informed consent was obtained from the parents to participate in these studies. The proband from family 3 was enrolled in the NIH Undiagnosed Diseases Network17, 18 under the clinical protocol 15-HG-0130, “Clinical and Genetic Evaluation of Patients with Undiagnosed Disorders Through the Undiagnosed Diseases Network,” approved by the NHGRI Institutional Review Board. The proband from family 4 was recruited from Mazumdar Shaw Medical Center, Bangalore, India after written informed consent obtained by the institute from parents for the publication of clinical information and photographs. For family 5, ethical approval was obtained from the ethical commission of the Canton of Zurich and written consent was provided by the affected individuals and parents including publication of facial photographs. SNP microarray, whole-exome sequencing (WES) or whole-genome sequencing (WGS) were employed to identify the genetic etiology in each family and the detailed methodologies used are described in the Supplemental Methods.
Gene Expression Studies and Splice Analysis
Immortalized lymphoblastoid cell lines from all the affected members from family 2 and skin-derived fibroblasts from the proband of family 3 were available for gene expression studies. Lymphoblastoid cell lines and fibroblasts were maintained as previously described.19, 20 Total RNA was extracted from cultured cell lines using RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s guidelines. The extracted RNA was reverse transcribed (High Capacity RNA to cDNA synthesis kit, Applied Biosystems, Waltham, CA, USA) for cDNA sequencing and expression studies. For splice site analysis in family 3, PCR products were amplified using primers flanking the splice site and cloned into TOPO vector according to manufacturer’s protocol (Invitrogen). Recombinant colonies were selected using blue white screening and sequenced using vector specific M13 primers (Table S1). For family 5, direct sequencing was performed after PCR amplification using cDNA-specific primers flanking the intronic variant. For quantitative real-time PCR, primers specific to the longest isoform of human TMEM94 (NM_001321148.1) and POLR2A were amplified using Power SYBR Green PCR master mix, monitored, and analyzed as described.20 To understand the effect of the truncating variants in TMEM94 on global gene expression, we performed gene expression microarray of one affected (Family 2-II.3) versus four gender matched healthy controls as previously described.21 Significantly dysregulated genes were defined as those with absolute fold change (FC) > 1.5 and adjusted Welch’s t test p value < 0.05.
RNA Sequencing
RNA from lymphoblastoid cell lines of all the affected members of family 2 (2-II.1, 2-II.2, and 2-II.3) and two control cell lines were extracted using Maxwell RSC Instrument (Promega) according to manufacturer’s guidelines. After the quality control procedures, mRNA was enriched using oligo(dT) beads and then fragmented randomly in fragmentation buffer, followed by cDNA synthesis using random hexamers and reverse transcriptase. After first-strand synthesis, a custom second-strand synthesis buffer (Illumina) was added with dNTPs, RNase H, and Escherichia coli polymerase I to generate the second strand by nick-translation. The final cDNA library was used after a round of purification, terminal repair, A-tailing, ligation of sequencing adapters, size selection, and PCR enrichment. The libraries were read using Illumina HiSeq and the raw quality-filtered FASTQ files were then aligned to the human reference genome build GRCh37/hg19 using STAR version 2.5.3a in manual two-pass mapping mode.22 Low quality reads (average Phred-scaled base call quality < 20) from the RNA sequence data were removed using Trimmomatic.23 GENCODE v19 annotation was used for genome annotation on the fly while preparing STAR genome indices for the alignment.24 Gene-level raw read counts were determined using feature Counts from the Subread package with a RefSeq transcript annotation set consisting of 27,090 gene features.25, 26, 27 Differential expression analysis was performed using DESeq2 within the SARTools R package.28, 29 Differentially expressed genes with Benjamini-Hochberg (BH) adjusted false discovery rates of < 0.05 were included in an Ingenuity Pathway Analysis.30, 31
For family 3, RNA was extracted from blood and skin fibroblast using QIAGEN RNeasy Plus Micro Kit. Quantification and quality was assessed using Qubit 3.0 Fluorometer and Agilent bioanalyzer. A total of 1 μg RNA was submitted to the UCLA Neuroscience Genomics Core (UNGC) for library construction and RNA sequencing. Sequencing libraries were generated using Illumina TruSeq Stranded Total RNA RIbo-Zero Gold. Sequencing was performed to generate on average ∼65 million 69 base paired-end reads on the Illumina HiSeq 4000. FASTQ files were aligned using STAR-2.5.2b two pass Mode to the Ensembl genome release 75. Quality was assessed using RNA-SeQC v1.1.8 and gene expression was quantified with Cufflinks v.2.2.1. BAM files were loaded in IGV (Integrative Genomics Viewer; https://software.broadinstitute.org/software/igv/) to generate the sashimi plot of the splice alteration.
Generation and Characterization of Tmem94 Knock-Out Mice
All experiments in mice were approved by the NIH NHGRI Animal Care and Use Committee under the protocol G-17-5 (Mouse Models of Rare and Undiagnosed Diseases). For generating the knock out mice, we used CRISPR/Cas9 technology to introduce a frameshift in the early codons of mouse Tmem94. Two single stranded oligos targeting exon 4 were designed using online CRISPRscan software,32 with no putative off-targets. The oligos contained the T7 promoter, target sequence, and a universal reverse primer binding site (Table S1). We annealed each single stranded oligo with a universal reverse primer and extended it using a proof reading enzyme (QuickChange II XL Site directed mutagenesis kit, Agilent Technologies, CA, USA). The double strand products (∼150 bp) were in-vitro transcribed (HiScribe T7 High Yield RNA Synthesis Kit, NEB, MA, USA) and the synthesized gRNAs were purified (MEGAclear Transcription Clean-Up Kit, Ambion, MA, USA) according to the manufacturer’s instructions. The quantity of the sgRNAs was assessed on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and loaded onto 2% agarose gels to check quality. Tmem94 sgRNAs and spCas9 mRNAs (from Trilink Biotechnologies, San Diego, CA, #L-6008) were microinjected into the pronuclei of mouse zygotes as described.33 Successfully injected C57BL6/J zygotes were surgically implanted into pseudo pregnant recipient CB6F1 female mice. Tails from each embryo were genotyped by PCR and Sanger sequencing using specific primers (Table S1). For harvesting embryos, pregnant mice were euthanized using CO2 asphyxiation at 12.5, 15.5, and 18.5 days of gestation, dissected, and embryos were collected for genotyping and photography. Embryo images were taken with a stereomicroscope (Axio Zoom V16, Zeiss) attached to a light microscope. To study the expression of Tmem94 in knockout mice, we cultured mouse embryonic fibroblasts in DMEM supplemented with 10% fetal bovine serum20 and extracted RNA and synthesized cDNA as described above. The longest protein coding isoform of mouse Tmem94 (NM_028014.3) was amplified using Power SYBR Green PCR using Polr2a as a housekeeping gene to normalize the results (primers shown in Table S1).
Magnetic Resonance Imaging and Computed Tomography
Magnetic resonance images were acquired using a Bruker Biospin Avance III microimaging system using 14.1 T magnet, Paravision 5.1 software, and Micro 2.5 imaging gradients. The embryos were immersed in 0.5% (v/v) Magnetvist (aq) solution (Bayer Healthcare Pharmaceuticals, Lancaster, PA) for 10 days to improve MRI contrast and image resolution. Ex-vivo MRI was performed on a Bruker 14.1T MR imaging spectrometer (Bruker Biospin, Billerica, MA, USA) using a spoiled FLASH technique34 with TR/TE = 75/8 ms and 2 signal averages. The resulting 3D images were acquired and processed to a resolution of 49 microns isotropic or smaller. For CT, the embryos were fixed with 4% paraformaldehyde aqueous solution for 2 weeks prior to scanning. The embryos were affixed to a closed-cell Styrofoam support bed for centering in the CT field of view and covered with a Parafilm M (Bemis NA, Neenah, WI, USA) film membrane to prevent movement from excessive drying during CT scan. The embryos were scanned on a Bruker Micro-CT Skyscan 1176 in vivo Micro CT (Bruker, Inc. Billerica, MA, USA) with a 0.7-degree angular step, and 2 frame averages per step. The X-ray source was set to a current of 177 microamps, voltage of 45 kVp, and a 0.2 mm Al filter for spectral hardening. Images were reconstructed using the Bruker Micro CT Recon software with an isotropic image resolution of 18 μm over a FOV of 35 mm. The radiographs of craniofacial regions of mouse embryos were acquired at 35kV using Faxitron X-ray and Fuji computed radiography system with a digital camera attached.
Histological Staining and Microscopy
For tissue analysis, the whole embryo and dissected brains were fixed in 10% formalin and embedded in paraffin using standard methods. Slides 10 μm thick were further processed for staining either standard Nissl or Masson trichrome staining. Bright field images of stained tissues were collected using a Zeiss AxioObserver Z1 widefield microscope equipped with a 20× plan-apochromat (N.A. 0.45) objective lens, a motorized scanning stage, and an Axiocam MRc5 color CCD camera.
Results
Identification of a Recognizable Syndrome of NDD, CHD, and Facial Dysmorphism in Multiple Probands from Unrelated Families
Family 1
Proband 1 (1-II.4 in Figure 1A) is an 8-month-old male infant from Oman and the 4th child of consanguineous parents. He was diagnosed antenatally with Tetralogy of Fallot and pulmonary atresia and born at 32 weeks of gestation weighing 2.9 kg (> 97th centile). He underwent a Blalock-Taussig shunt at 1 month of age and intra cardiac repair with right ventricular and pulmonary artery conduit at 8 months of age. He was referred to the department of genetics for evaluation of dysmorphic features. On examination at 8 months of age, he presented with mild motor developmental delay; he could sit with support and reach for objects but was not able to transfer objects from one hand to another. Weight was 8.3 kg (10-25th centile), height 73 cm (75-90th centile) and head circumference 42 cm (< 3rd centile). Dysmorphic features included synophrys, thick arched eyebrows, hypertrichosis, left convergent squint, flat nasal bridge, short nose, long philtrum with thin upper vermillion, low set ears with prominent ear lobules, and widely spaced nipples (Figures 1B, 1–2, and Figure S1, 1). Cyanosis and clubbing of the fingers and toes (Figure S1, 2–3)., which were most likely due to his congenital heart disease, were noted. His array CGH was normal. Ultrasound evaluation of the abdomen was normal. On re-evaluation at 24 months of age, receptive language was delayed at 19 months and expressive language at 15 months; speech therapy was initiated. Otoacoustic emission analysis was normal. The eldest sister (1-II.1 in Figure 1A) presented with similar facial features. At 6 years of age, she had global developmental delay and poor scholastic performance. She had an inverted triangular face, thick arched eyebrows, hypertrichosis, midfacial hypoplasia, short nose with flat nasal bridge, hypertelorism, low set pinnae with prominent ear lobules, long flat philtrum with a thin upper vermillion, and pointed chin (Figures 1B, 3–4). Echocardiogram revealed an atrial septal defect. Her fingers and toes were long (Figures S1, 4–5). Karyotype analysis showed 46, XX,16qh+. MRI of the brain of both the affected siblings in family 1 were normal (Figures S1, 6–7). A 4.5-year-old elder female sibling (Figure 1A-II.2) was unaffected; another elder female sibling (Figure 1, A-II.3) died with pneumonia at 8 months of age; from the information provided by the parents, she might also be affected because she had similar facial features and an atrial septal defect.
Figure 1.
Pedigree and Clinical Features of the Families
(A) Pedigree of the six families included in the study. Filled objects indicate affected status and arrows indicate the probands.
(B) Proband of family 1 (1-II.4) is shown with arched eyebrows, synophrys, left convergent strabismus, flat nasal bridge, hypertrichosis, long philtrum, thin upper vermillion, and low set pinnae with prominent lobules (1 and 2). His elder sibling (1-II.1) has inverted triangular face, thick arched eyebrows, hypertelorism, flat nasal bridge, long philtrum, thin upper vermillion and pointed chin and low set pinnae with prominent lobules (3 and 4). Proband of family 2 (2-II.3) shown with short low anterior hairline, synophrys with thick eyebrows, long eyelashes, hypertelorism, broad flat nasal bridge, upturned nose, triangular upper lip, prominent chin, and low set ears with prominent lobules (5 and 6). His eldest sister (2-II.1) has thick eyebrows, hypertelorism, broad nasal bridge, nasal tip, posteriorly rotated prominent ears, and big ear lobules (7 and 8). His elder sister (2-II.2) has thick eyebrows, broad nasal bridge, nasal tip, and big ear lobules (9). In family 3, the proband (3-II.1) is shown with triangular face, pointed chin, thick arched eyebrows, short nose, long philtrum (10) and posteriorly rotated prominent ear lobules (11). The proband in family 4 is shown with triangular face with pointed chin, thick arched eye brows, short nose, long philtrum (12) and prominent posteriorly rotated ear lobules (13). In family 5, the elder sibling of the proband (5-II.2) (14-15) and the proband (5-II.3) (16-17) are shown with similar facial gestalt of thick arched eyebrows, low set ears with under folded helix, prominent nose, short philtrum, prominent cupid bow upper lip, and increased volume of lip vermillion. Proband of family 6 is shown with thick arched eye brows, deep set eyes, long philtrum, and pointed chin (18).
Family 2
Proband 2 (2-II.3 in Figure 1A) is a three-year-old boy born from a consanguineous Qatari couple who had two spontaneous miscarriages in the first trimester. Pregnancy was complicated by gestational diabetes managed with insulin. Birth was by normal vaginal delivery at 38 weeks of gestation with a birth weight of 4.02 kg (85th centile), head circumference of 35 cm (50th centile), and length of 55 cm (98th centile). As an infant, he developed mild respiratory distress that responded to oxygen supplementation by nasal cannula. Echocardiogram showed a small PFO with left to right shunt, multiple small VSDs with left to right shunt; all closed spontaneously on follow up. He had recurrent viral illnesses documented by PCR of respiratory secretions to be due to human metapneumo-, adeno-, rhino- and bocaviruses. He developed neutropenia, which was attributed to phenytoin therapy for episodes of generalized tonic-clonic seizures started at 2 years of age with abnormal EEG. During infancy, he initially had generalized hypotonia, but at present he has truncal hypotonia and appendicular hypertonia with brisk reflexes and ankle clonus. Dysmorphic features include short forehead with a low anterior hair line, synophrys with thick eyebrows, long eyelashes, hypertelorism (interpupillary distance at 20 months of age was 6.5 cm, 97th centile), broad flat nasal bridge, upturned nose, triangular upper lip, prominent chin, and low set ears with prominent lobules and short neck (Figures 1B, 5–6). MRI of the brain showed bilateral symmetrical restricted diffusion and high T2 signal intensity of the cerebral white matter, mainly the U fibers, subcortical white matter and external capsule (data not shown), with relative sparing of internal capsule, corona radiata, centrum semi ovale and corpus callosum. Similar changes affected the dentate nucleus bilaterally. The brainstem and basal ganglia had normal signal intensity and the ventricular system appeared normal.
The eldest sister of Proband 2 (2-II.1 in Figure 1A) is an 8-year-old girl delivered preterm at 30 weeks of gestation after an uneventful pregnancy. At birth, her weight was 1.77 kg (90th centile), head circumference was 26.5 cm (50th centile), and length was 45.5 cm (> 97th centile). She was admitted to NICU for 1 month due to respiratory distress syndrome and neonatal jaundice. Like her brother, she initially had generalized hypotonia. She had speech delay, some hyperactive behavior and learning difficulties. She presented with the same dysmorphic features as her brother, i.e., thick eyebrows, hypertelorism, broad nasal bridge and nasal tip, prominent posteriorly rotated ears and big ear lobules in addition to prominent chin (Figures 1B, 7–8). The other sister of Proband 2 (2-II.2 in in Figure 1A) is a 6-year-old girl born term after an uneventful pregnancy with a birth weight of 4.36 kg (95th centile), head circumference of 37 cm (98th centile), and length of 54 cm (98th centile). She has mild scoliosis of the dorsal spine and normal motor development after an initial delay, but had speech delay and learning difficulties. She has pectus excavatum, and the same dysmorphic features as her two affected siblings (Figure 1B, 9). MRI of the brain was normal.
Family 3
Proband 3 (3-II.1 in Figure 1A) is a 16-year-old male from a non-consanguineous couple of European American ethnicity. He was born at 36 weeks of gestation by assisted vaginal delivery; the pregnancy was complicated by preeclampsia. APGAR scores were 7 (1 minute) and 9 (5 minutes); he had severe hypotonia and was in the NICU for 4 weeks due to a VSD, ASD, and PDA. The PDA and VSD spontaneously closed without intervention. At age 2 months, the proband underwent surgery for laryngomalacia. At age 3 years he had his ASD surgically repaired. At age 6 years, Proband 3 had a fundoplication for severe GERD, pyloroplasty, inguinal hernia repair, and the removal of a Mullerian duct remnant. At age 12 years he had contralateral hernia repair and at age 15 he had laparoscopic surgery to remove an abdominal mesenteric cyst. Additionally, at age 15, he had a bony decompression and C1 laminectomy for his Chiari 1 malformation.
His early gross developmental milestones were delayed in the first 2 years of life; he sat at 8 months and walked at 19 months. He was found to have an expressive language delay. Neuropsychological testing at age 16 (WASI-II, WAIS-IV, WRAT-4, Vineland 3, Conners-3 parent form, BRIEF-2, and SCQ) found that he had a specific learning disability due to psychological processing deficits in the areas of sensory-motor and visual perception. Additional findings included deficits in cognitive abilities, word reading, reading comprehension, spelling, math computation, brief attention, working memory, number sense, and communication in the low or below average range. Socialization skills were found to be intact, but he struggles to socialize with similarly functioning peers. On physical exam, Proband 3 is above the 90th percentile for height and weight, has macrocephaly, a triangular face, hypertrichosis, a pointed chin, synophrys with thick arched eyebrows, a short nose with a flat nasal bridge, a long philtrum, and posteriorly rotated ears with prominent ear lobes (Figure 1B, 10–11). Other pertinent findings include strabismus, optic glioma, dilation of the ascending aorta, severe scoliosis (spine X-rays detected a 48-degree S-shaped lumbar scoliosis at age 14 years old), arachnodactyly with large hands, generalized hypotonia, pectus excavatum, and numerous boggy subcutaneous lesions (Figure S1, 8–11). MRI evaluations have identified a Chiari 1 malformation and a delayed myelination pattern (at age 2 years), and a T2 hyperintense lesion involving the left orbital gyrus and a stable Chiari 1 malformation (at ages 14 and 15 years old). Spinal MRI identified a hyper intense signal involving the vertebral body of T7 that could represent an intraosseous hemangioma, dextrothoracolumbar scoliosis, and a Chiari 1 malformation.
Family 4
Proband 4 (4-II.1 in Figure 1A) is a 17-month-old male born to third degree consanguineous parents who was referred with history of congenital heart disease, developmental delay and delayed speech. He was born by a lower segment caesarean section due to history of decreased fetal movements and fetal distress. His birth weight was 4.6 kg (99th centile). At 6 months of age, he was diagnosed with congenital heart disease due to complaints of cyanosis and recurrent respiratory infections. Echocardiography showed Tetralogy of Fallot with hypoplastic pulmonary annulus, large perimembranous misaligned ventricular septal defect and large secundum atrial septal defect. On examination, he has facial dysmorphism characterized by inverted triangular face, thick eyebrows with thin medial extension, down slant palpebral fissures, lateral third eversion of lower eyelids, wide nasal bridge, small nose, low set posteriorly rotated ears with large uplifted ear lobules, long philtrum, small mouth, thin vermillion of upper lip and pointed chin (Figure 1B, 12–13). Other clinical features include webbed neck, low set widely spaced nipples, fetal finger pads, shawl scrotum, distal hypospadias, bilateral undescended testes, overlapping toes, and long great toes (Figure S1, 12–15). At 17 months of age, his head circumference was 47 cm (60th centile) and his length of 88 cm (> 97th centile). At the age of 3 years 5 months, the head circumference was 50.2 cm (75th centile), height was 108 cm (> 97th centile) and weight was 17 kg (90–95th centile). The rest of the systemic examination was unremarkable. Giemsa-banded chromosome analyses and chromosome microarray were normal.
Family 5
Proband 5 (5-II.3 in Figure 1A) was a 16-year-old girl with mild developmental delay and macrocephaly born to non-consanguineous healthy Turkish parents, originating from two different small villages. She was born at 37 5/7 weeks of gestation by Caesarean section with a weight of 4.17 kg (> 97th centile), length of 52 cm (> 75th centile), and unreported head circumference. At two weeks of age, she was diagnosed with an ASD, a VSD, and a double outlet right ventricle without a pulmonic stenosis, which were surgically corrected. Her motor development was mildly delayed with unsupported sitting at 12 months and independent walking at 17 months. Her speech development was also delayed and characterized by mispronunciation of words and stuttering. Social skills were considered within normal limits. Her facial features included apparently bitemporal narrowing with normal measurements (50th centile) with a low anterior hairline, full and curly hair, hypertrichosis on the back and extremities, synophrys with thick arched eyebrows, long eyelashes, upslanted palpebral fissures, ptosis, rather wide eye distance (interpupillary distance of 6.1cm, which is +0.7SD for Turkish population35), high nasal root and bridge and broad nasal tip, anteverted nares short and deep philtrum, thick upper and lower lip vermillion, under folded helices, and large ear lobes and a triangular facial shape (Figure 1B, 14–15). Her palms were narrow (palm width/length ratio −3SD) with long slender fingers (97th centile). Her feet showed a 2–3 cutaneous toe syndactyly bilaterally. At the age of 13 years she developed focal epileptic seizures with impaired awareness and rare evolution to bilateral tonic-clonic seizures, which are partially controlled by lamotrigine treatment. At the age of 18 years she attended a special education school due to learning disability and short attention span. She showed no obvious behavioral issues and was described as very sensitive and shy. Formal testing at the age of 16 years revealed an IQ of 58. For cosmetic reasons she regularly epilates the synophrys. Hearing tests done after birth and at 8.5 years were normal. Microarray analysis using an Affymetrix Cytoscan HD array at a 20kb resolution showed no abnormalities.
Individual 5-II.2, the elder sister of individual 5-II.3, was born after three miscarriages at 37 weeks of gestation by Cesarean section due to prenatally detected omphalocele. Placentamegaly (1.1 kg) was reported but tissue for histology was unavailable. Her birthweight 3.78 kg is at 96th centile, birth length was 51 cm (50–75th centile), and head circumference (OFC) was 33.5 cm (25–50th centile). An atrioventricular septal defect (AVSD) and intestinal malrotation, which were initially suspected during the prenatal period, were confirmed at birth together with a PDA and parachute mitral valve. The congenital heart defect, omphalocele, and intestinal malrotation were successfully corrected surgically. In the neonatal period, she was noted to have large ear lobes and a hypertrichosis of the helix and of the back of the ear. She also had narrow palms (palm width/length ratio −2.75SD) and slender fingers (97th centile) as well as long toes. Neonatally, she was suspected to be affected with Beckwith-Wiedemann-syndrome and a targeted surveillance strategy aimed to detect embryonal tumors was started, but no visceral involvement was detected. Standard karyotyping, DNA methylation studies of domain 1 (H19) on chromosome 11, and paternal uniparental disomy 11 testing revealed normal results at that time. Her psychomotor development was mildly delayed and she had difficulties in reading, writing, spelling, and reasoning. She had her menarche at 18 years but remained oligomenorrheic. During puberty, she had developed hypertrichosis of the face, arms and legs. For cosmetic reasons she regularly epilates unwanted body hair including the synophrys. She had strabismus and she wore eyeglasses because of high myopia. When she was reviewed at 24 years, she was taller than her targeted height (173cm, +1.5SD; mother 25th centile, father 75–90th centile). She also had macrocephaly (OFC was 58.5cm, +2.35SD; mother was on the 75–90th centile, while her father was on the 50th centile). At this age, she was overweight (weight of 97kg, 2.89SD. Her facial appearance was similar to that of her sister: she had bitemporal narrowing with normal measurements (50th centile) with a low anterior hairline, full and curly hair, cosmetically treated hypertrichosis on extremities and face and synophrys with thick arched eyebrows, long eyelashes, upslanted palpebral fissures, rather wide eye distance (interpupillary distance of 6.3cm, which is +1.23SD for Turkish population35), high nasal root and bridge and broad nasal tip, short and deep philtrum, thick upper and lower lip vermillion, prognathism, and open bite, small teeth and wide interdental spaces, under folded helices and large ear lobes and a triangular facial shape (Figure 1B, 16–17). Her hands were long and narrow with long fingers (palm width/length ratio −2.75SD; palm length 12.6cm, > 97th centile; middle finger length 9cm, > 97th centile). She has numerous nevi and several cystic scalp formations that were removed surgically. Histopathology is available for one nevus only, which showed a melanocytic papillomatous nevus of predominantly dermal type. She attended a special education school and received training in housekeeping but was unemployed and lived with her parents.
Family 6
Proband 6 (6-II.2 in Figure 1A) is a 30-month-old girl from Egypt who presented with developmental delay, speech delay, and dysmorphic facial features. She was antenatally diagnosed to have complex congenital heart disease (double outlet right ventricle, ASD, VSD). She had a history of feeding and swallowing difficulties which is now resolved. She was born term by normal vaginal delivery. Her motor development was delayed with unsupported sitting at 16 months and currently can walk without support with wide based gait, can vocalize sounds but not words, and sometimes can understand simple orders. Her facial features include hairy forehead, thick eye brows, deep set eyes, hypertelorism (interpupillary distance of 8cm, 97th centile), depressed nasal bridge, low set ears, smooth philtrum, and thin upper lips (Figures 1B, 18). Her chromosome microarray was normal.
Detailed clinical data of all ten affected individuals from six families are summarized in Table 1.
Table 1.
Clinical Highlights of Affected Family Members with TMEM94 Variants
| Features | Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position in the pedigree | II.1 | II.4 | II.1 | II.2 | II.3 | II.1 | II.1 | II.2 | II.3 | II.2 | |
| Ethnicity | Omani | Qatari | European American | Indian | Turkish | Egypt | |||||
| Consanguinity | + | + | No | + | Not reported, but parents from two small villages 3km away | + | |||||
| Variant in cDNA∗ and protein level | c.2764C>T; p.Arg922∗ | c.840del; p.Asp280Glufs∗10 | c.2635dup; p.Met879Asnfs∗18 and c.795-1G>C | c.2000_2004dup; p.Pro669Alafs∗8 | c.4028+5G>A; p.Val1344∗ | c.3497delA; p.Asn1166Thrfs∗84 | |||||
| Age | 8 years 7 months | 8 months | 8 years | 6 years | 3 years | 17 years | 3 years 5 months | 24 years | 18 years | 30 months | |
| Gender | Female | Male | Female | Female | Male | Male | Male | Female | Female | Female | |
| Growth | Birth weight | Not available | 2.9 kg (born at 32 weeks age of gestation) (>97th centile) | 1.77 kg (born at 30 weeks age of gestation) (90th centile) | 4.36 kg (95th centile) | 4.02 kg (born at 38 weeks of gestation) (85th centile) | 4.65 kg (99th centile) | 4.6 kg (> 97th centile) | 3.7 kg (96th centile) Cesarean at 37 weeks | 4.17 kg (>97th centile; +2.35SD) Cesarean section at 37 weeks 5/7 | Not available |
| Birth length | Not available | Not available | 45.5 cm (born at 30 weeks age of gestation) (>97th centile) | 54 cm (98th centile) | 55 cm (98th centile) | Not available | Not available | 51cm (50-75th centile) | 52cm (75th centile) | Not available | |
| Birth head circumference | Not available | Not available | 26.5 cm (born at 30 weeks age of gestation) (50th centile) | 37 cm (98th centile) | 35 cm (50th centile) | Not available | Not available | 33.5cm (25-50th centile) | Not available | Not available | |
| Recent height | 120 cm (10thcentile) | 73 cm (75- 90th centile) | 129.6 cm (75.76th centile) | 124.3 cm (85th centile) | 90 cm at 90th centile | 188.0 cm (96th centile) | 108 cm (>97th centile) | 173cm (90-97th centile; +1.5SD), | 178.1cm (>97th centile; +2.3 SD) at age 16yrs | 95 cm (96th centile) | |
| Recent weight | 19 kg (<3rd centile) | 8.3 kg (10- 25th centile) | 25.7kg (61.08th centile) | 22.4kg (65.7th centile) | 17kg (88.74th centile) | 79.5kg (91st centile) | 17 kg (90-95th centile) | 97kg (>97th centile; 2.89SD) | 89.1kg (>97th centile; +2.51 SD) at age 16yrs | 14.3kg (86th centile) | |
| Recent head circumference | 50 cm (<10th centile) | 42 cm (< 3rd centile) | 53.5 cm (90th centile) | 55 cm (97th centile) | 46.5 cm (77.38th centile) (at 10 months) | 63.5 cm (>100th centile) | 50.2 cm (75th centile) | 58.5cm (>97th centile; +2.35SD) | 58.5cm (97th centile; +2.35SD) | 52cm (99th centile) | |
| Neurologic features | Motor delay (HP:0001270) | + | + | + | − | + | + | + | + | + | + |
| Learning disability (HP:0001328) | + | + | + | + | + | + | + | + | + | + | |
| Speech delay (HP:0000750) | + | + | + | + | + | Expressive language delay | + | + | + | + | |
| MRI o | Normal brain MRI | Normal brain MRI | Not available | Normal brain MRI | Bilateral symmetrical restricted diffusion and high T2 signal intensity of the cerebral white matter of the brain | Chiari type I malformation and cortical-based mass in the left inferior frontal gyrus pars orbitalis (ganglioglioma or other low grade glial tumor). Spinal MRI -T7 hyperintense signal | Not available | Not available | Enhanced signal within both hippocampi in FLAIR cuts, signs of microhemorrhages, possibly linked to hemodynamic changes during heart surgery | Not available | |
| Facial features | Triangular face (HP:0000325) | + | + | + | + | + | + | + | + | + | + |
| Hypertrichosis (HP:0004554) | + | + | + | + | + | + | + | + | + | + | |
| Pointed chin (HP:0000307) | + | + | + | + | + | + | + | + | + | + | |
| Synophrys (HP:0000664) | + | + | + | + | + | + | + | + | + | + | |
| Thick arched eyebrows (HP:0002553) | + | + | + | + | + | + | + | + | + | + | |
| Short nose (HP:0003196) | + | + | + | + | + | + | + | Prominent (HP: 0000448) | Prominent (HP: 0000448) | + | |
| Flat nasal bridge (HP:0005280) | + | + | + | + | + | + | + | − | − | + | |
| Long philtrum (HP:0000343) | + | + | + | + | + | - | + | HP:0000322 short philtrum | HP:0000322 short philtrum | + | |
| Thin upper lip vermillion (HP:0000343) | + | + | + | + | Triangular upper lip | Not noted | + | prominent cupid bow upper lip, increased volume of lip vermillion | prominent cupid bow upper lip, increased volume of lip vermillion | + | |
| Ears | Low set ears (HP:0000369) | + | + | + | + | + | Posteriorly rotated low set ears with hypoplastic helices | Large, low set posteriorly rotated ears | HP:0008544, under folded helix | HP:0008544, under folded helix | + |
| Prominent ear lobules (HP:0009748) | + | + | + | + | + | + | + | + | + | + | |
| Ophthalmological features | No gross eye abnormalities, no extraocular muscle weakness, visual acuity not tested but no corrective lenses at time of examination | No gross eye abnormalities, no extraocular muscle weakness, visual acuity not f tested but no corrective lenses at time of examination | Quiet anterior segment, Normal fundus, Visual acuity: left eye (6/12), right eye (6/18), Need glasses (+0.5-1.75) | Quiet anterior segment, Normal fundus, Visual acuity: left eye (6/9), right eye (6/9), Unaided | Bilateral nystagmus, Normal fundus, Need glasses (-1.5, -1.75) | Strabismus, deep set eye, long eye lashes, optic glioma of the right eye | Refractive error, normal fundus | Strabismus and myopia | Visual acuity not formally tested but no need for corrective lenses at time of examination; HPO:0007838 progressive Ptosis | Not available | |
| Cardiac abnormalities | Atrial septal defect | Tetralogy of Fallot and pulmonary atresia | None | Small secondum ASD, two midmuscular VSDs and mall PDA (VSDs and PDA closed spontaneously) | Small patent foramen ovale and VSD | ASD, VSD, and PDA (PDA closed spontaneously); dilation of the ascending aorta (Z +3.6), sinus tachycardia, lymphatic vessel neoplasm | Tetralogy of Fallot with hypoplastic pulmonary annulus, large peri membranous malaligned VSD and large secundum ASD | AVSD, PDA, parachute mitral valve | ASD, VSD, double outlet right ventricle without pulmonal stenosis | ASD, VSD, Double outlet double ventricle | |
| Skeletal features | Long fingers and toes | None | None | Mild scoliosis | None | Severe scoliosis, arachnodactyly, large hands, bilateral overlapping of the 3rd toe by the 2nd and 4th toes, asymmetry of the hip, osteochondroma of the proximal left tibial metaphysis | Bilateral overlapping toes, bilateral long great toes | Open bite and prognathism, long fingers, narrow hands (palm width/length- 2.75SD) | Thoracic kyphoscoliosis, long fingers, bilateral cutaneous 2,3 toe syndactyly, overlapping of the 2nd toe by the 3rd toe, narrow hands (palm width/length -3SD) | Not available | |
| Respiratory features | None | None | Bronchial asthma, recurrent infections | Wheezy chest, bronchomalacia | Neutropenia, recurrent infections | Laryngotracheomalacia, dyspnea, recurrent pneumonia | None | None | None | None | |
| Gastrointestinal features | None | None | None | None | None | GERD, mesenteric cyst, diarrhea, constipation | None | Omphalocele and malrotation | None | Not available | |
| Genitourinary features | None | None | None | None | None | Mullerian duct remnant, adrenal cyst, dysuria, urinary retention, bilateral inguinal hernia | Penile hypospadias, bilateral undescended testis | None | None | Not available | |
| Other features | None | Widely spaced nipples (HP:0006610) | Generalized hypotonia (HP:0001290) and hyper active behavior (HP:0000752); generalized tonic-clonic seizures | Pectus excavatum (HP:0000767), umbilical hernia, right ectopic kidney with no dilatation | Truncal hypotonia (HP:0008936) and appendicular hypertonia (HP:0002509) | Macrocephaly, high palate, dolichocephaly, vertigo, generalized hypotonia, pectus excavatum, numerous nevi, boggy subcutaneous lesions | Web neck, low set wide spaced nipples, shawl scrotum | Polyhydramnios, placentamegaly (1100gr), several nevi and cystic scalp formations, Macrocephaly | Polyhydramnios, large placenta (800gr), fatigable weakness of skeletal muscles, partial seizures at onset at 13 years with a sharp wave localized in the left temporal region, Macrocephaly | History of feeding and swallowing difficulties | |
Table shows the clinical features of individuals with bi-allelic variants in TMEM94. HPO (human phenotype ontology) terms were specified when available.
“+” and “−” denotes presence and absence of a clinical feature, respectively. ASD, atrial septal defect; VSD, ventricular septal defect; PDA, patent ductus arteriosus;
AVSD, atrioventricular septal defect. ∗cDNA nomenclature based on GenBank NM_001321148.1.
Identification of TMEM94 Bi-allelic Variants
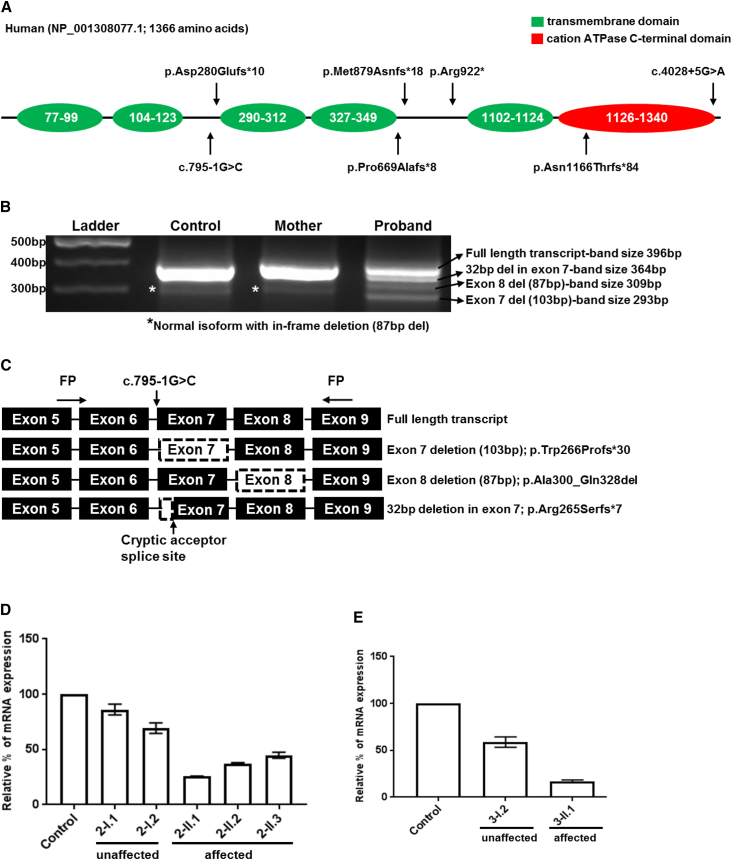
Homozygosity/autozygosity mapping using SNP array, WES and/or WGS were used in all six families to identify the genetic etiology. None of the affected individuals from six families seems to have significant pathogenic copy number variants (CNVs) and in family 3, the non-consanguineous family, did not reveal any homozygous areas > 5Mb as expected. In family 5, although the parents denied consanguinity in the family history (Table 1), nine long stretches of homozygous areas and ethnic background (Turkish origin) suggest possible consanguinity (Table S2). Final analysis of all six families revealed bi-allelic truncating variants in TMEM94 as a common genetic cause among other candidate genes (Table S3–S5). In online databases ExAC and gnomAD browser (see URL’s), there are several potential loss-of-function variants occurring in heterozygosity in TMEM94, with allele frequency less than 0.00002 (1 or 2 allele counts in each ethnic population), but no healthy homozygotes, suggesting that the gene is highly intolerant to bi-allelic truncating variants. In view of the predicted deleteriousness of the TMEM94 variant, segregation in the family, and similar phenotype in affected members of all six families, we considered this to be the best candidate gene. All the variants identified in TMEM94 were Sanger validated for confirmation and family segregation (Figure S2). All the truncating variants are predicted to abolish the highly conserved C-terminal domain including cation P-type ATPase domains (Figure 2A). Alignment of human TMEM94 (NP_001308077.1) protein sequences with different species using Clustal Omega showed > 90% identity between human, chimpanzee, cattle, hamster, mouse, and dog; chicken, frog, and zebra fish showed 82%, 71%, and 69%, respectively. The C-terminal region of the protein showed higher similarity compared to the overall protein sequence.
Figure 2.
Molecular Analysis and Gene Expression Studies
(A) Diagrammatic representation of conserved domains of TMEM94 and the position of variants in six families identified.
(B) Splice analysis of the variant NM_001321148.1: c.795-1G>C identified in the proband of family 3. For control and unaffected 20 μL of PCR products were loaded into each well, while for the proband 40 μL was loaded due to severely reduced abundance of transcripts. Normal full-length transcripts (396 bp) and an isoform with exon 8 deletion (309 bp) that predicted to lead to an in-frame deletion(87 bp) was present in control, unaffected and proband. This isoform could be benign. Proband showed two different mutant transcripts; whole exon 7 skipping (p.Trp266Profs∗30) and 32 bp deletion (p.Arg265Serfs∗7) due to the activation of a cryptic acceptor splice site in exon 7.
(C) Diagram showing the primer designing scheme for splice analysis and aberrant transcripts identified. Black boxes representing the coding region with corresponding exon numbers. Boxes with dashed outlines represents the skipped exons/areas of exons identified through TA sub cloning and sequencing (Chromatograms in Figure S3). Variants are named in reference to NM_001321148.1 and NP_001308077.1.
(D and E) Relative quantification of mRNA expression in family 2 and 3 showing markedly reduced expression of TMEM94 in the affected individuals of family 2 (D: 2-II.1, 2-II.2 and 2-II.3) and family 3 (E: 3-II.1). Error bars represent SEM.
In the four families (family 1, 2, 4, and 5), exome/genome variant filtering focused on variants located within the homozygous areas of the genome, due to possible consanguinity in the pedigree. A list of homozygous areas and variant filtering strategy are listed in Tables S2–S5. In family 1, exome variant filtering narrowed down the number of variants to four. The NEMP2 (MIM 616497, NM_001142645) missense variant is predicted to lead to a leucine to arginine change in a moderately conserved amino acid and a protein domain of unknown function. The TRIM65 (NM_173547.3) missense variant (rs61754937) changes an arginine to tryptophan at position 418, a highly conserved amino acid, but has very low constraint metrics (see ExAc). The SAMD4B (NM_018028.3) variant (rs141485885) leads to a valine to isoleucine change in a highly conserved amino acid but not in a known protein domain. By Sanger sequencing, NEMP2, TRIM65, and SAMD4B were homozygous in both affected individuals, hence segregating with the affected status in the family. Of the four variants, however, NM_001321148.1: c.2764C>T in TMEM94 gene was extremely rare (rs557746506, ExAc MAF: 1/116268), predicted to truncate the protein, and has the highest CADD score (Phred-score, 43) (Table S5). In family 2, SERINC2 (MIM614549): NM_001199037.1: c.1327T>C; p.Tyr443His and TMEM94: NM_001321148.1: c.840del; p.Asp280Glufs∗10 were identified as final candidates (Table S5). The TMEM94 variants identified in family 1 and 2 (Omani and Qatari) were absent in our in-house whole exome sequencing cohort, which is collected from 2,379 individuals from Saudi Arabia. In Family 4, the final variants included three genes, TMEM132B: c.769T>C; p.Trp257Arg, TMEM94:c.2000_2004dup; p.Pro669Alafs∗8, TRPM5 (MIM 604600): c.1934C>T;p.Thr645Met. In family 5, two final candidate genes were identified: ITGB4 (MIM 147557): c.737C>T;p.Thr246Met, and TMEM94:c.4028+5G>A;p.?. In family 6, two genes in the final list included TMEM94: c.3467delA;p.Asn1156Thrfs∗84, and GAA (MIM 606800): c.2105G>A; p.Arg702His. In family 5 and 6, the parents and unaffected siblings were carrier for the TMEM94 variants (Figure S2). The sequencing chromatogram of family 6 is not shown. In family 6, although a suspicious variant in GAA was identified (Table S5), the enzymatic studies and clinical picture are inconsistent with Pompe disease (MIM 232300); we nonetheless continue to monitor this proband clinically.
For family 5, affymetrix Cytoscan (2.65 M) Array on the proband revealed 9 regions of homozygosity greater of 3Mb (Table S2). WES in the affected individual (with an average depth of coverage of 234.5x with 98% of the targeted bases with ≥ 20 reads) identified the homozygous substitution c.4028+5G>A (NM_001321148.1) at the exon / intron boundary 31 of the TMEM94 gene as best candidate. Sanger sequencing confirmed the variant in homozygous state in both affected siblings and was heterozygous in the parents (Figure S2). The base substitution was predicted by in-silico programs to reduce the donor splice site by 66% (MaxEnt: −76%, NNSPLICE: −99.5%, HSF: −13.5%, SpliceSiteFinder-like: −76.8%), resulting in a potential abnormal splicing that possibly affects the C-terminal portion of the protein. Sequencing of RT-PCR products from blood collected in PAXgene RNA tubes confirmed abnormal splicing resulting in 36 bp intronic retention in the transcript introducing a premature termination codon, NM_001321148.1: r.4028_4029ins36; p.Val1344∗ (Figure S3A).
Of note, none of the homozygous variants of unknown significance in the affected individuals from consanguineous families, including NEMP2, TRIM65, SAMD4B, SERINC2, TRPM5, TMEM132B, and ITGB4 have been reported to cause recessively inherited neurodevelopmental phenotypes. Genome-wide association studies, however, associate low frequency variants in TRIM65 with white matter hyperintensities,36 de novo copy number variation of region involving SERINC2 with autism spectrum disorder,37 heterozygous variant in TMEM132B contributing as a risk factor to intracranial aneurysm.38
In family 3, the non-consanguineous family, the final analysis (Tables S4 and S5) revealed compound heterozygous variants in TMEM94; a splice site variant, c.795-1G>C, which is predicted to be inherited from the father (father’s DNA sample was unavailable for the analysis) and a single base pair duplication, c.2635dup; p.Met879Asnfs∗18 inherited from the mother (Figure S2 and Table S5). RNA-seq analysis of the splice site variant (NM_001321148.1:c.795-1G>C) in-silico predicted an abnormal splicing pattern (Figure S3B). For further confirmation, we PCR amplified the cDNA derived from fibroblasts using primers flanking the proposed splice defect. PCR amplification revealed normal transcript in the affected individual, control, and unaffected mother (Figure 2B), but in addition, we found multiple aberrant transcripts in 3-II.1 (Figure 2B). Sequencing of colonies revealed normal transcripts and a transcript with an in-frame deletion (p.Ala300_Gln328del), which is present in control, unaffected mother, and proband, suggesting a normal benign isoform. In addition to this, two other mutant transcripts were identified in the affected individual, which is absent in control and mother; skipping of exon 7, and 32 bp deletion in exon 7 due to the activation of a cryptic acceptor site inside exon 7 (Figure 2C). Both aberrant transcripts are predicted to shift the amino acid frame and result to premature termination codons, p.Trp266Profs∗30 and p.Arg265Serfs∗7 (Figures 2B–2C and Figure S3C). Prior to WGS at the UDN, the proband had an extensive work up and normal findings for cytogenetic testing (microarray and karyotype), molecular genetic testing for aortopathy panel (ACTA2 [MIM 102620], COL3A1 [MIM120180], COL5A1 [MIM 120215], COL5A2 [MIM 120190], FBN1 [MIM 134797], FBN2 [MIM612570], MYH11 [MIM160745], MYLK [MIM600922], SKI [MIM 164780], SLC2A10 [MIM 606145], SMAD3 [MIM 603109], TGFB2 [MIM 190220], TGFBR1 [MIM 190181], TGFBR2 [MIM 190182]) and clinical exome sequencing.
Gene Expression Profiling Is Altered in TMEM94 Mutant Cells
TMEM94 is predicted to have several isoforms, three of which are predominant coding transcripts (NM_014738.5, NM_001321149.1 and NM_001321148.1). None of the transcripts reported in GenBank39 lack the last exons of the gene, which encodes the highly conserved C-terminal domain.40 We checked the level of expression of TMEM94 transcripts in proband-derived cells available from family 2 and 3. In family 2, RNA expression was reduced by ∼60% in three affected individuals (Figure 2D). For family 3, dermal fibroblasts were available for the expression analysis and the affected individual showed 85%–90% reduced expression (Figure 2E). Global transcription profiling by microarray using proband (2-II.3) from family 2 compared to four gender matched controls using 1670 significant probes corresponding to 1257 genes identified 677 upregulated and 580 downregulated (adjusted p value < 0.05 and absolute fold change > 1.5) in the affected individual (Figure S4).
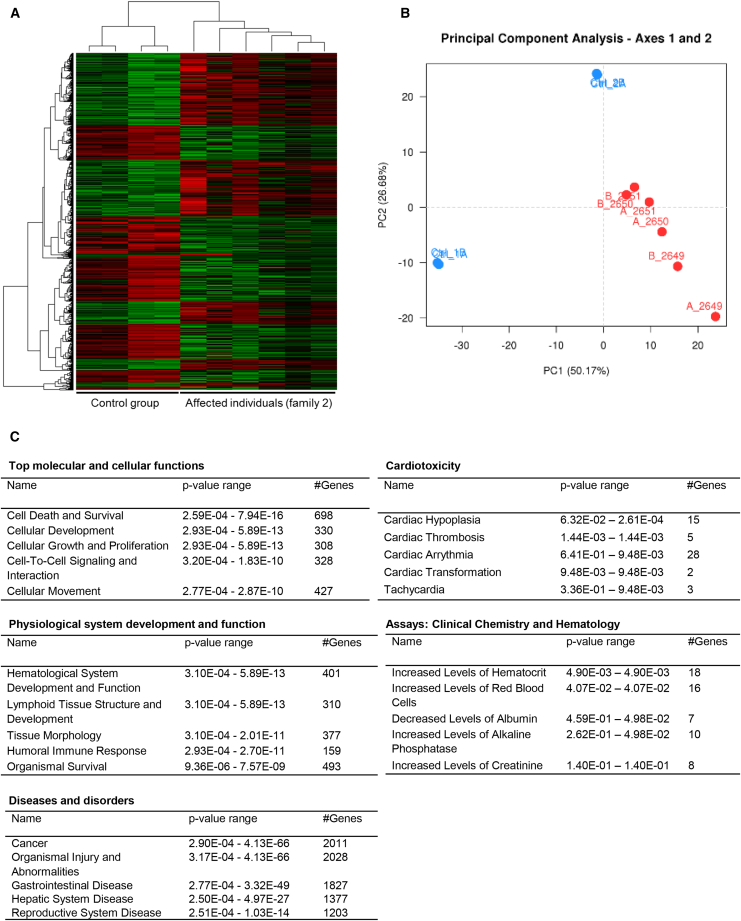
We further examined global gene-expression profile changes using all the three affected individuals from family 2 (II.1, II.2, and II.3) and two unaffected controls by RNA sequencing. Differential expression analysis comparing the affected individuals and controls revealed significant dysregulation of 2438 genes (FDR < 0.05) in the affected individuals (Figures 3A and 3B); 1219 were upregulated and 1219 were downregulated (Table S6). Ingenuity Pathway Analysis of the overall differentially expressed genes revealed the expression of various genes that regulate cellular development, cell to cell interaction, and cell growth and proliferation significantly altered in all the three affected individuals (Figure 3C). Dysregulation of these genes are predicted to have an impact on hematological system development, immune response, organismal injuries and cardiotoxicity. TMEM94 interaction network generated using dysregulated genes revealed alteration of several essential genes associated with neurological function (Table S7).
Figure 3.
Transcriptome Profiling and Ingenuity Pathway Analysis in TMEM94 Mutant Cells
(A) Heatmap of 2,438 genes that are significantly dysregulated in three (in duplicates) TMEM94 mutant individuals determined by DESeq2 (FDR < 0.05).
(B) Principal component analysis of normalized gene expression showing significant differences between the control and affected individual group. Red dots indicate individuals with disease phenotype and blue indicates controls.
(C) List of altered molecular and cellular functions, physiological functions and diseases associated with the dysregulated genes. p values and the number of dysregulated genes involved for each function are also shown.
Characterization of Tmem94 in Mice
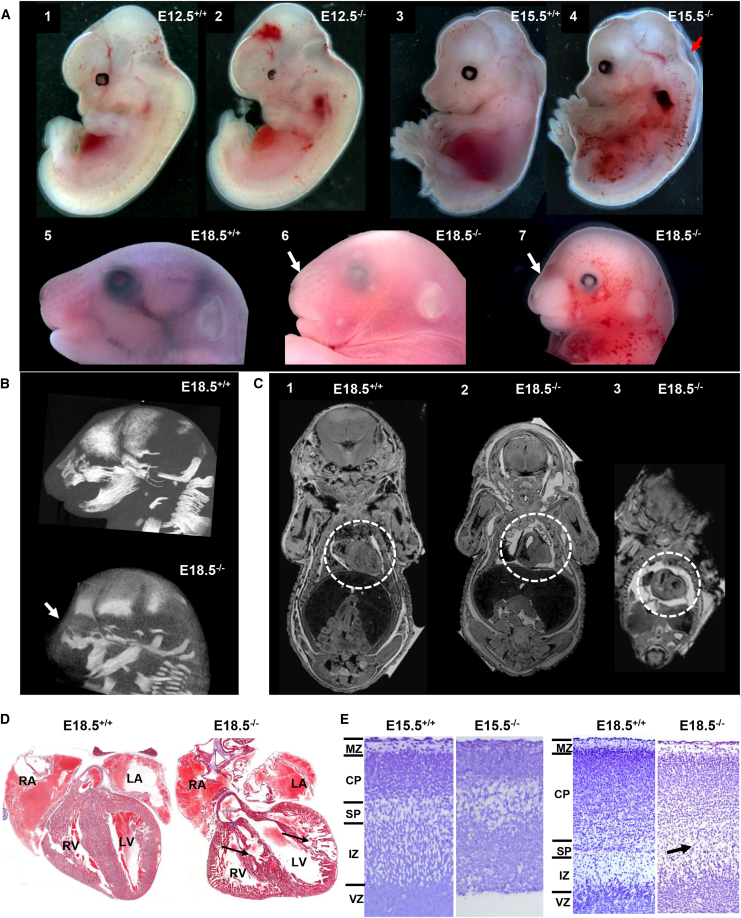
Because of the high sequence similarity and homology of human and mouse TMEM94 (∼95%), we knocked out Tmem94 in mice by using CRISPR-Cas9 technology with two different sets of gRNAs targeting exon 4 (Figure S5A). Consistent with our expectation that this gene is essential for mammalian development, the pups having either compound heterozygous or homozygous variants in Tmem94, were either embryonic lethal or born dead. The viable pups from 6 sets of injections were either wild-type or heterozygous. F1 mice with a single null allele (heterozygous; c.361_364delGC; p.Arg121Profs∗58) appeared morphologically normal; these were outbred with wild-type C57BL6/J to identify F2s, then in-crossed to identify germline mutants. About 20% homozygous mutants were identified at E18.5 and all mutants were dead by P1, suggesting an embryonic lethal phenotype. Homozygous mutants evaluated at E12.5 stage showed variable amount of hemorrhages in the frontal and hind regions of the cephalic area (Figures 4A, 1–2). Similarly, at E15.5 days, mutants showed hemorrhage and decreased body size (Figure 4A, 3–4). E18.5 days old mutants were developmentally delayed and showed dysmorphic facies, round head, decreased body size, hemorrhage and disrupted craniofacial bone on CT and X-ray (Figures 4A-B and S5B). Whole body MRI imaging at E18.5 revealed that the mutant mice showed poorly developed heart chambers (Figure 4C, 1–3). Tmem94 expression analysis of the mutants showed 80%–90% reduction of mRNA transcripts in the embryonic fibroblasts (Figure S5C).
Figure 4.
Phenotypic and Histopathological Evaluation of Tmem94 Knockout Mice
(A) Tmem94−/− mice were embryonic lethal, hemorrhagic, and showed craniofacial abnormalities. Tmem94 knock-out phenotype at different embryonic development stages showed varying degree of severity. Compared to the wild-type (1), homozygous mutant (2) at early embryonic stage (E12.5 days) showed hemorrhage at frontal and hind regions of brain. At E15.5 days, compared to wild-type (3), mutants (4) showed decreased body size, hemorrhage and fluid filled embryo sac (arrow) suggesting fetal resorption. The mutants at E18.5 showed craniofacial abnormalities (6 and 7), round head shape, and superficial hemorrhages, compared to the wild-type (5).
(B) CT images of mutant (lower panel) showing short nasal bone (arrow) and craniofacial abnormalities compared to the wild-type (upper panel).
(C) Coronal MRI scan of embryos at E18.5 days showing poorly developed heart structure in two mutants (2 and 3) compared to wild-type (1). The region of heart is highlighted in white dotted circle.
(D) In comparison to wild-type (left panel), Masson trichrome staining of coronal heart sections of mutants (right panel) at E18.5 presented small rounded heart, poorly organized auricles (RA: right auricle, LA: Left auricle), irregular inner ventricular surface (RV: right ventricle, LV: Left ventricle), disarray of heart muscle fibers at the heart wall and interventricular septum (arrows, right panel).
(E) Nissl stained sagittal sections of mouse neocortex at E15.5 and E18.5 days. Wild-type embryos show clearly recognizable marginal zone (MZ), cortical plate (CP), sub plate (SP), intermediate zone (IZ), and ventricular zone (VZ). E15.5−/− embryo has comparatively thin disorganized neocortex and lacks distinct cortical layers (left panel). E18.5−/− manifests delayed migrating neurons, dispersed CP layer and absence of distinct sub plate layer (right panel, arrow).
Histopathological Analysis of Tmem94−/− Mice
We analyzed the histology of heart and brain of mutant embryos to further understand the mutant phenotype. Tiled images of the heart sections showed that mutant mice at E18.5 presented with a small rounded heart, poorly organized auricles, irregular inner ventricular surface, and disarray of heart muscle fibers at the interventricular septum (Figure 4D), consistent with the MRI findings (Figure 4C). We examined the neuronal layering pattern of the cortical region at two different stages of gestation. The marginal zone, cortical plate, intermediate zone, and ventricular zone were clearly recognizable in wild-type embryos. In contrast, E15.5 mutants showed comparatively thin and significantly disorganized cortical layers (Figure 4E). The surviving embryos at E18.5 showed fewer neurons in the cortical plate and absence of distinct sub plate layer indicating defects in neuronal migration (Figure 4E). These results conclude that Tmem94 has a profound role in mouse embryonic development, especially neural and cardiovascular systems.
Discussion
Here we present a rare form of NDD associated with facial dysmorphism and presence of CHD in ten similarly affected individuals from six unrelated families of different ethnic background. The common clinical features include: characteristic facial features (triangular face, widely spaced eyes, thick arched eyebrows, depressed nasal bridge and broad nasal root and tip, pointed chin and posteriorly rotated low set ears) and global developmental delay. Most of them had generalized hypertrichosis; congenital overgrowth (increased birth weight, postnatal height, weight and head circumference in the upper normal range); and cardiac abnormalities (atrial septal defect, Tetralogy of Fallot, patent ductus arteriosus, and ventricular septal defect). We link this disorder to likely loss-of-function bi-allelic variants in TMEM94. Nonetheless, there are clinical features that are different among our affected individuals. Brain MRI changes seemed to be variable and one proband (2-II.1) did not manifest with a structural cardiac defect. Other variable clinical features we observed are: ophthalmological features including nystagmus (1/10), myopia (1/10) and strabismus (2/10); skeletal features of scoliosis (3/10), arachnodactyly (4/10) and overlapping toes (3/10); macrocephaly (3/10), pectus excavatum (2/10), widely spaced nipples (2/10), respiratory infections (4/10), gastrointestinal abnormalities (2/10), and seizures (2/10). These clinical features can be due to the variability in genetic background among the affected individuals. The identification of additional cases will likely help clarify the spectrum of TMEM94-associated disorder.
TMEM94, originally named KIAA019541 is located on chromosome 17q25.1, and encodes a putatively nuclear protein with predicted multi pass membrane domains and several serine residues for phosphorylation or glycosylation.42, 43, 44, 45, 46, 47 The C-terminal region of TMEM94 is evolutionary conserved in multiple mammalian species and is predicted to be perturbed in all the affected individuals due to truncating variants. None of the reported transcripts of TMEM94 lacks the C-terminal region of the protein, specifically residues that are part of the cation transporting P-type ATPase domain (NP_055553.3; residues 1116-1330),48 suggesting that this might be an important protein domain. To understand the possible role TMEM94, we studied global gene-expression pattern in the mutant cells of family 2. The altered gene list implies the role of this gene in cell-cycle progression, nervous system, and hematological system development. TMEM94 has been shown to be induced under hypoxic conditions in breast cancer cell lines.49 The transcription network is upregulated during hypoxia to promote expression of the genes involved in cell survival and to regulate high energy demanding processes such as translation, cell division, and proliferation,50 suggesting the involvement of TMEM94 in cell division. Further analysis of the cell-cycle pattern of dermal fibroblasts from the affected individual using propidium iodide staining (Supplemental Experimental Procedures) showed that at baseline, the cells from the affected and controls appeared to be predominantly in the G0/G1 phase (Figure S6). Synchronization of cell cycle with starvation did not show any marked differences between affected and controls, while nocodazole treatment increased the number of cells in G2/M phase in control cells while affected cells showed a decrease in the G2/M phase cells, suggesting defects in cell-cycle progression (Figure S6). Although, our global gene expression analysis and cell-cycle assays imply to the role of TMEM94 in cell division, further studies will be needed to confirm this hypothesis and characterize the role of this protein in the cell.
TMEM94 is known for the possible interaction with cyclin dependent kinase 5 (CDK5; MIM 123831) and acidic fibroblast growth factor intracellular-binding protein (FIBP; MIM 608296) in human embryonic kidney cells.51, 52, 53 Our expression profiling studies did not reveal any significant dysregulation of CDK5 and FIBP. CDK5 and FIBP have a proven essential role in cell division.54, 55 Bi-allelic loss-of-function variants in CDK5 and FIBP in human and mice has been shown to cause neurological manifestations; CDK5 variants in humans cause lissencephaly with cerebellar hypoplasia (MIM 616342),56 Cdk5−/− mice manifested neurodegeneration and neuronal migration defects,57 FIBP variants cause Thauvin-Robinet-Faivre syndrome (MIM 617107) with clinical highlights including facial dysmorphism, CHD, macrocephaly, learning disabilities and overgrowth.58 Interestingly, the two probands from family 2 and the probands from families 3, 4, and 5 manifested isolated overgrowth and macrocephaly, like what was reported in individuals with FIBP variants. These phenotypic similarities suggest some degree of possible functional overlap of TMEM94 with CDK5 and FIBP; further studies will be required to show the functional interaction of these three proteins within the cell.
Loss of Tmem94 in mice leads to an embryonic lethal phenotype and abnormalities in multiple tissues, including nervous and cardiovascular systems, suggesting the role of this gene in early development. In murine cerebral cortex, histogenesis and migration occurs during embryonic days E11.5 to E17.5 of gestation.59 Tmem94 mutants at E15.5 and E18.5 days showed abnormalities in the layering pattern of developing cortex suggesting delayed neurodevelopment.
The presence of loss-of-function TMEM94 variants in six unrelated families with NDD, distinct and recognizable facial dysmorphism, and variable penetrance for cardiac abnormalities highlight the importance of TMEM94 in neural and cardiovascular development. In the context of the loss-of-function variant in the affected individuals in our study and Tmem94 knockout in mice, we demonstrate a role for this gene in mammalian embryonic development. In conclusion, we report a syndrome characterized by NDD and distinct facial dysmorphism due to truncating variants in TMEM94, a gene so far not associated with a specific cellular function or human disease.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the individuals in our study and their families for participating in this study. This work was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, USA, NIH Common Fund from the Office of the Director, U01 HG007703 (SF Nelson, JA Martinez-Agosto) and the King Salman Center for Disability Research grant (FSA). We acknowledge the support of the “Saudi Human Genome Program.” The authors also would like to thank the coordinators and Olfat Al-Harazi (King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia), Valerie Maduro (NIH-UDP, NHGRI, National Institutes of Health), Ten Taro (NIH-UDP, NHGRI, National Institutes of Health), and Dr. Jan Linkenhoker (NHGRI, National Institutes of Health). We also thank Dr. Michael J. Kruhlak (NCI Experimental Immunology Branch, Microscopy Digital Imaging facility), and NIDCR Combined Technical Research Core for helping with the imaging of histology slides. The authors do not have any conflict of interest to declare. Anita Rauch was supported by radiz—Rare Disease Initiative Zürich, Clinical Research Priority Program for Rare Diseases of the University of Zurich.
Published: December 6, 2018
Footnotes
Supplemental Data include six figures, seven tables, and Supplemental Experimental Procedures and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.11.001.
Contributor Information
Fowzan S. Alkuraya, Email: falkuraya@kfshrc.edu.sa.
May Christine V. Malicdan, Email: malicdanm@mail.nih.gov.
Accession Numbers
The variants reported in this manuscript have been deposited in Leiden Open Variation Database (LOVD) and is publicly available through: https://databases.lovd.nl/shared/transcripts/00010486.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/ (last accessed January 2018)
The Human Protein Atlas, http://www.proteinatlas.org/
GeneDx, https://www.genedx.com
gnomAD Browser, http://gnomad.broadinstitute.org/ (last accessed January 2018)
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
neXtprot, https://www.nextprot.org/term/FA-00613/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RNA-SeQC, https://software.broadinstitute.org/cancer/cga/rna-seqc
STRING 9.0, http://www.string-db.org/
UniProt, http://www.uniprot.org/
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Stiles J., Jernigan T.L. The basics of brain development. Neuropsychol. Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Homberg J.R., Kyzar E.J., Nguyen M., Norton W.H., Pittman J., Poudel M.K., Gaikwad S., Nakamura S., Koshiba M., Yamanouchi H. Understanding autism and other neurodevelopmental disorders through experimental translational neurobehavioral models. Neurosci. Biobehav. Rev. 2016;65:292–312. doi: 10.1016/j.neubiorev.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Bostwick B.L., McLean S., Posey J.E., Streff H.E., Gripp K.W., Blesson A., Powell-Hamilton N., Tusi J., Stevenson D.A., Farrelly E., Members of the Undiagnosed Diseases Network Phenotypic and molecular characterisation of CDK13-related congenital heart defects, dysmorphic facial features and intellectual developmental disorders. Genome Med. 2017;9:73. doi: 10.1186/s13073-017-0463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufman L., Ayub M., Vincent J.B. The genetic basis of non-syndromic intellectual disability: a review. J. Neurodev. Disord. 2010;2:182–209. doi: 10.1007/s11689-010-9055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quaegebeur A., Lange C., Carmeliet P. The neurovascular link in health and disease: molecular mechanisms and therapeutic implications. Neuron. 2011;71:406–424. doi: 10.1016/j.neuron.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Simões-Costa M., Tan-Cabugao J., Antoshechkin I., Sauka-Spengler T., Bronner M.E. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome Res. 2014;24:281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simões-Costa M., Bronner M.E. Insights into neural crest development and evolution from genomic analysis. Genome Res. 2013;23:1069–1080. doi: 10.1101/gr.157586.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anazi S., Maddirevula S., Faqeih E., Alsedairy H., Alzahrani F., Shamseldin H.E., Patel N., Hashem M., Ibrahim N., Abdulwahab F. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry. 2017;22:615–624. doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- 9.Harripaul R., Vasli N., Mikhailov A., Rafiq M.A., Mittal K., Windpassinger C., Sheikh T.I., Noor A., Mahmood H., Downey S. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry. 2018;23:973–984. doi: 10.1038/mp.2017.60. [DOI] [PubMed] [Google Scholar]
- 10.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kochinke K., Zweier C., Nijhof B., Fenckova M., Cizek P., Honti F., Keerthikumar S., Oortveld M.A., Kleefstra T., Kramer J.M. Systematic Phenomics Analysis Deconvolutes Genes Mutated in Intellectual Disability into Biologically Coherent Modules. Am. J. Hum. Genet. 2016;98:149–164. doi: 10.1016/j.ajhg.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riazuddin S., Hussain M., Razzaq A., Iqbal Z., Shahzad M., Polla D.L., Song Y., van Beusekom E., Khan A.A., Tomas-Roca L., UK10K Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mefford H.C., Batshaw M.L., Hoffman E.P. Genomics, intellectual disability, and autism. N. Engl. J. Med. 2012;366:733–743. doi: 10.1056/NEJMra1114194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 15.Vissers L.E., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 16.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gahl W.A., Mulvihill J.J., Toro C., Markello T.C., Wise A.L., Ramoni R.B., Adams D.R., Tifft C.J., UDN The NIH Undiagnosed Diseases Program and Network: Applications to modern medicine. Mol. Genet. Metab. 2016;117:393–400. doi: 10.1016/j.ymgme.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gahl W.A., Wise A.L., Ashley E.A. The Undiagnosed Diseases Network of the National Institutes of Health: A National Extension. JAMA. 2015;314:1797–1798. doi: 10.1001/jama.2015.12249. [DOI] [PubMed] [Google Scholar]
- 19.Garland J., Stephen J., Class B., Gruber A., Ciccone C., Poliak A., Hayes C.P., Singhal V., Slota C., Perreault J. Identification of an Alu element-mediated deletion in the promoter region of GNE in siblings with GNE myopathy. Mol. Genet. Genomic Med. 2017;5:410–417. doi: 10.1002/mgg3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephen J., Vilboux T., Haberman Y., Pri-Chen H., Pode-Shakked B., Mazaheri S., Marek-Yagel D., Barel O., Di Segni A., Eyal E. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur. J. Hum. Genet. 2016;24:1268–1273. doi: 10.1038/ejhg.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaheen R., Anazi S., Ben-Omran T., Seidahmed M.Z., Caddle L.B., Palmer K., Ali R., Alshidi T., Hagos S., Goodwin L. Mutations in SMG9, Encoding an Essential Component of Nonsense-Mediated Decay Machinery, Cause a Multiple Congenital Anomaly Syndrome in Humans and Mice. Am. J. Hum. Genet. 2016;98:643–652. doi: 10.1016/j.ajhg.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varet H., Brillet-Guéguen L., Coppée J.Y., Dillies M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 32.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harms D.W., Quadros R.M., Seruggia D., Ohtsuka M., Takahashi G., Montoliu L., Gurumurthy C.B. Mouse Genome Editing Using the CRISPR/Cas System. Current protocols in human genetics. 2014;83 doi: 10.1002/0471142905.hg1507s83. 15.17.11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haase A., Frahm J., Matthaei D., Hanicke W., Merboldt K.D. Flash Imaging - Rapid Nmr Imaging Using Low Flip-Angle Pulses. J. Magn. Reson. 1986;67:258–266. doi: 10.1016/j.jmr.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 35.Yildirim Y., Sahbaz I., Kar T., Kagan G., Taner M.T., Armagan I., Cakici B. Evaluation of interpupillary distance in the Turkish population. Clin. Ophthalmol. 2015;9:1413–1416. doi: 10.2147/OPTH.S85584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fornage M., Debette S., Bis J.C., Schmidt H., Ikram M.A., Dufouil C., Sigurdsson S., Lumley T., DeStefano A.L., Fazekas F. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann. Neurol. 2011;69:928–939. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hnoonual, A., Thammachote, W., Tim-Aroon, T., Rojnueangnit, K., Hansakunachai, T., Sombuntham, T., Roongpraiwan, R., Worachotekamjorn, J., Chuthapisith, J., Fucharoen, S., et al. (2017). Chromosomal microarray analysis in a cohort of underrepresented population identifies SERINC2 as a novel candidate gene for autism spectrum disorder. Scientific reports 21; 7 (1):12096. [DOI] [PMC free article] [PubMed]
- 38.Farlow J.L., Lin H., Sauerbeck L., Lai D., Koller D.L., Pugh E., Hetrick K., Ling H., Kleinloog R., van der Vlies P. Lessons learned from whole exome sequencing in multiplex families affected by a complex genetic disorder, intracranial aneurysm. PLoS One. 2015;24:e0121104. doi: 10.1371/journal.pone.0121104. 10 (3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagase, T., Seki, N., Ishikawa, K., Tanaka, A., and Nomura, N. (1996). Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA research: an international journal for rapid publication of reports on genes and genomes 3, 17-24. [DOI] [PubMed]
- 42.Tan H., Wu Z., Wang H., Bai B., Li Y., Wang X., Zhai B., Beach T.G., Peng J. Refined phosphopeptide enrichment by phosphate additive and the analysis of human brain phosphoproteome. Proteomics. 2015;15:500–507. doi: 10.1002/pmic.201400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H., Di Palma S., Preisinger C., Peng M., Polat A.N., Heck A.J., Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 44.Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 45.Oppermann F.S., Gnad F., Olsen J.V., Hornberger R., Greff Z., Kéri G., Mann M., Daub H. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daub H., Olsen J.V., Bairlein M., Gnad F., Oppermann F.S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Bian Y., Song C., Cheng K., Dong M., Wang F., Huang J., Sun D., Wang L., Ye M., Zou H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 48.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abu-Jamous B., Buffa F.M., Harris A.L., Nandi A.K. In vitro downregulated hypoxia transcriptome is associated with poor prognosis in breast cancer. Mol. Cancer. 2017;16:105. doi: 10.1186/s12943-017-0673-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ortmann B., Druker J., Rocha S. Cell cycle progression in response to oxygen levels. Cell. Mol. Life Sci. 2014;71:3569–3582. doi: 10.1007/s00018-014-1645-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varjosalo M., Keskitalo S., Van Drogen A., Nurkkala H., Vichalkovski A., Aebersold R., Gstaiger M. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 2013;3:1306–1320. doi: 10.1016/j.celrep.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 52.Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S., Aebersold R., Bennett K.L., Colinge J., Gstaiger M., Superti-Furga G. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods. 2013;10:307–314. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 53.Xu S., Li X., Gong Z., Wang W., Li Y., Nair B.C., Piao H., Yang K., Wu G., Chen J. Proteomic analysis of the human cyclin-dependent kinase family reveals a novel CDK5 complex involved in cell growth and migration. Mol. Cell. Proteomics. 2014;13:2986–3000. doi: 10.1074/mcp.M113.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolpakova E., Wiedłocha A., Stenmark H., Klingenberg O., Falnes P.O., Olsnes S. Cloning of an intracellular protein that binds selectively to mitogenic acidic fibroblast growth factor. Biochem. J. 1998;336:213–222. doi: 10.1042/bj3360213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawauchi T. Cdk5 regulates multiple cellular events in neural development, function and disease. Dev. Growth Differ. 2014;56:335–348. doi: 10.1111/dgd.12138. [DOI] [PubMed] [Google Scholar]
- 56.Magen D., Ofir A., Berger L., Goldsher D., Eran A., Katib N., Nijem Y., Vlodavsky E., Tzur S., Behar D.M. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with a loss-of-function mutation in CDK5. Hum. Genet. 2015;134:305–314. doi: 10.1007/s00439-014-1522-5. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi S., Ohshima T., Hirasawa M., Pareek T.K., Bugge T.H., Morozov A., Fujieda K., Brady R.O., Kulkarni A.B. Conditional deletion of neuronal cyclin-dependent kinase 5 in developing forebrain results in microglial activation and neurodegeneration. Am. J. Pathol. 2010;176:320–329. doi: 10.2353/ajpath.2010.081158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thauvin-Robinet C., Duplomb-Jego L., Limoge F., Picot D., Masurel A., Terriat B., Champilou C., Minot D., St-Onge J., Kuentz P. Homozygous FIBP nonsense variant responsible of syndromic overgrowth, with overgrowth, macrocephaly, retinal coloboma and learning disabilities. Clin. Genet. 2016;89:e1–e4. doi: 10.1111/cge.12704. [DOI] [PubMed] [Google Scholar]
- 59.Kwan K.Y., Sestan N., Anton E.S. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.