Abstract
Motile cilia move body fluids and gametes and the beating of cilia lining the airway epithelial surfaces ensures that they are kept clear and protected from inhaled pathogens and consequent respiratory infections. Dynein motor proteins provide mechanical force for cilia beating. Dynein mutations are a common cause of primary ciliary dyskinesia (PCD), an inherited condition characterized by deficient mucociliary clearance and chronic respiratory disease coupled with laterality disturbances and subfertility. Using next-generation sequencing, we detected mutations in the ciliary outer dynein arm (ODA) heavy chain gene DNAH9 in individuals from PCD clinics with situs inversus and in one case male infertility. DNAH9 and its partner heavy chain DNAH5 localize to type 2 ODAs of the distal cilium and in DNAH9-mutated nasal respiratory epithelial cilia we found a loss of DNAH9/DNAH5-containing type 2 ODAs that was restricted to the distal cilia region. This confers a reduced beating frequency with a subtle beating pattern defect affecting the motility of the distal cilia portion. 3D electron tomography ultrastructural studies confirmed regional loss of ODAs from the distal cilium, manifesting as either loss of whole ODA or partial loss of ODA volume. Paramecium DNAH9 knockdown confirms an evolutionarily conserved function for DNAH9 in cilia motility and ODA stability. We find that DNAH9 is widely expressed in the airways, despite DNAH9 mutations appearing to confer symptoms restricted to the upper respiratory tract. In summary, DNAH9 mutations reduce cilia function but some respiratory mucociliary clearance potential may be retained, widening the PCD disease spectrum.
Keywords: DNAH9, dynein, mutation, motile cilia, situs inversus, primary ciliary dyskinesia, tomography
Main Text
Cilia are highly conserved microtubular organelles found on almost every cell of the human body.1 Multiple motile cilia line the upper and lower respiratory tract, ependymal cells, Fallopian tubes, and the Eustachian tubes. Singleton motile cilia present during early development function at the embryonic left-right organizer (node). These different motile cilia beat in a coordinated fashion to transport overlying fluid. The single flagellum of sperm tails is similar in structure to motile cilia.2
Primary ciliary dyskinesia (PCD [MIM: 244400]) is an autosomal-recessive or X-linked condition in which the motile cilia are dysfunctional and fail to effectively transport secretions.3, 4 Pathogenic variants in >35 genes involved in cilia assembly, structure, and motility have been identified to cause PCD.5, 6 In keeping with the function and location of motile cilia and sperm, symptoms of PCD can include neonatal respiratory distress, chronic productive cough, recurrent infections of the upper and lower airways, middle ear symptoms such as otitis media, and male and female infertility. Due to cilia dysfunction in the embryonic node, situs inversus and other congenital cardiac and laterality disorders also manifest.3, 6 Diagnosis typically uses a combination of clinical tests including nasal nitric oxide (nNO) measurement, ciliary beat and ultrastructure analysis by high-speed video and transmission electron microscopy (HVSM, TEM), immunofluorescence staining of motile cilia proteins, and genetic analysis.6
The core structure (axoneme) of motile cilia consists of a circle of nine peripheral microtubular doublets plus or minus a central pair of microtubules, respectively termed 9+2 (multiple cilia) or 9+0 (singleton nodal cilia).1, 4 Inner (IDA) and outer (ODA) dynein arms that drive ciliary beating through ATP hydrolysis are attached to the peripheral microtubular doublets. The mammalian ODA, a complex structure attached to the microtubules by a docking complex, is composed of multiple heavy, light, and intermediate dynein chains.7, 8 In a highly evolutionarily conserved fashion, the ODA structure repeats every 24 nm along the length of the axoneme and is controlled through the nexin dynein regulatory complex and radial spokes to produce a regular ciliary waveform.9, 10 Two types of human ODAs are described.11 Type 1 ODAs reside in the proximal part of the cilium closer to the cell body in the region of the microvilli, containing the two dynein heavy chains (HC) DNAH5 (orthologous to the ODA γ-HC found in Chlamydomonas, the ancient protist that forms a key PCD model system) and DNAH11 (orthologous to the Chlamydomonas ODA β-HC). Type 2 ODAs reside in the distal cilium beyond the region of the microvilli, containing the two heavy chains DNAH5 and DNAH9 (also orthologous to Chlamydomonas ODA β-HC).11, 12
Mutations in DNAH5 (MIM: 603335) and DNAH11 (MIM: 603339) are among the commonest causes of PCD.13, 14, 15 DNAH5 mutations result in loss of the γ-HC DNAH5 and the β-HC DNAH9 and consequently a classic ODA loss visible by TEM. In contrast, pathogenic variants in DNAH11 result in apparently normal ultrastructure by TEM, with ODAs continuing to assemble DNAH5 and DNAH9 despite the absence of DNAH11.14, 16 Higher-resolution ultrastructural analysis using electron tomography reveals that DNAH11 mutations do confer a subtle structural defect, affecting only the proximal type 1 ODAs with a reduced ODA volume.16 Furthermore, in individuals with pathogenic variants in DNAH11, DNAH9 can reside in the place of DNAH11 in the proximal part of the axoneme.12
Here, we describe individuals with mutations affecting highly conserved domains of the ODA heavy chain DNAH9 (MIM: 603330). We report clinical, genetic, immunofluorescence, ciliary beat analysis, and electron tomography ultrastructural studies that increase our understanding of the arrangement of type 2 ODA heavy chains of motile cilia. These reveal that DNAH9 mutations confer a loss of ODAs in the distal portion of the ciliary axoneme, accompanied by a subtle effect on the ciliary beat pattern.
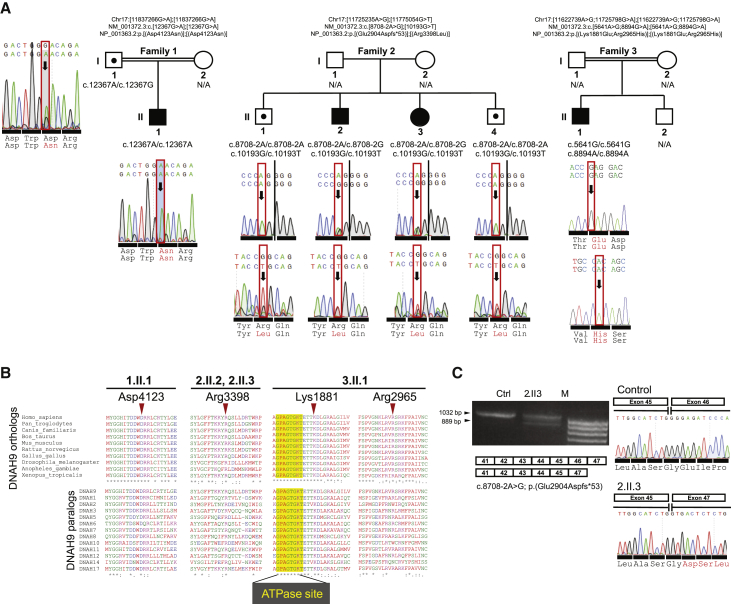
This study was approved by the ethics review boards of the Institute of Child Health/Great Ormond Street Hospital, London (UK) (08/H0713/82) and the Comité de Protection des Personnes CPP Ile-de-France III (France) (CPP07729 and CPP02748). We used a targeted next generation sequencing (NGS) gene panel (Agilent SureSelectQXT or Roche SeqCap EZ Choice) for genetic screening in a total of 536 individuals with confirmed or suggestive diagnosis of PCD. We identified loss-of-function mutations in DNAH9 (GenBank: NM_001372.3) in four individuals from three unrelated families (Figure 1A). Family 1 has one male child born in the UK to Turkish consanguineous parents with rhinosinusitis, cough, situs inversus, and complex congenital heart disease consisting of an unbalanced atrioventricular septal defect with congenitally corrected transposition of the great arteries and inferior vena cava conduit stenosis that required stenting. He also has a gastrointestinal condition (protein-losing enteropathy). He carries a homozygous missense mutation c.12367G>A (p.Asp4123Asn), confirmed by Sanger sequencing which also showed the father was a carrier (mother’s sample not available) (Figure 1A). This variant is absent from the Genome Aggregation Database (gnomAD). Phylogenetic analysis showed that Asp4123 is an amino acid highly conserved across different species and also across several dynein heavy chains (Figure 1B). This variant is predicted to be probably damaging using PolyPhen-2 with a score of 1.0 and it has a Combined Annotation Dependent Depletion (CADD) score of 33 (CADD score of ≥20 indicates a variant is among the top 1% of damaging variants in the genome).
Figure 1.
Loss-of-Function Bi-allelic DNAH9 Mutations Identified in Three Unrelated Families
(A) Pedigrees of three unrelated families with DNAH9 mutations. Family 1 has one affected child harboring homozygous c.12367G>A (p.Asp4123Asn) mutations, family 2 has two affected siblings with compound heterozygous splice acceptor and missense mutations c.8708−2A>G (p.Glu2904Aspfs∗53) and c.10193G>T (p.Arg3398Leu), and family 3 has one affected individual with two homozygous missense mutations c.5641A>G (p.Lys1881Glu) and c.8894G>A (p.Arg2965His).
(B) The four identified missense mutations all affect highly evolutionarily conserved residues which are also conserved across most axonemal dyneins. Notably in 3.II.1 carrying two homozygous missense variants, the p.Lys1881Glu substitution targets a highly conserved amino acid flanking the only ATPase site thought to retain an ATP hydrolyse activity (AAA1 domain).
(C) RT-PCR of nasal epithelial brush biopsy samples yielded one band of 1,032 bp in the healthy control subject and two bands (1,032 and 889 bp) in the sample from individual 2.II.3 carrying the DNAH9 c.8708−2A>G putative canonical splice site mutation. cDNA sequencing showed that c.8708−2A>G causes excision of DNAH9 exon 46 from the transcript and a frameshift mutation: p.Glu2904Aspfs∗53.
In family 2, a UK-based non-consanguineous Somali family with an affected brother and sister, both of whom had rhinosinusitis and situs inversus, we found compound heterozygous variants carried by both affected siblings: a missense variant c.10193G>T (p.Arg3398Leu) and an exon 46 consensus splice acceptor site variant c.8708−2A>G. Two unaffected siblings carried only the c.10193G>T (p.Arg3398Leu) allele in a heterozygous state, results confirmed by Sanger sequencing in all four siblings (Figure 1A). This variant is absent from the gnomAD, dbSNP, and EVS control databases and predicted by PolyPhen-2 to be probably damaging with a score of 0.997 and a CADD score of 34. Arg3398 is a highly conserved amino acid in different species and also across several dynein heavy chains (Figure 1B). The c.8708−2A>G splice variant is in dbSNP (rs143007518) and predicted to cause a frameshift, with a CADD score of 23.3. It was confirmed by Sanger sequencing in the two affected siblings and found to be absent in two unaffected siblings. It is present once in the gnomAD control database in a heterozygous state in a carrier from the East Asian population with a total allele frequency of 3.234e−5. To assess its functional impact on DNAH9 splicing, we isolated total RNA using Trizol Reagent (QIAGEN) from non-cultured ciliated cells obtained through nasal brushing of one of the affected siblings (2.II.3) and a healthy control subject. We used the High-Capacity RNA-to-cDNA Kit (ThermoFisher Scientific) to synthesize cDNA. Using primers in exons 41 and 47 of DNAH9, RT-PCR yielded one band of 1,032 base pairs (bp) in the healthy control subject while two bands (1,032 and 889 bp) were amplified in the affected child. We Sanger sequenced the two bands amplified in the latter sample after purification using the Monarch DNA Gel Extraction Kit (New England BioLabs). Sequence analysis revealed skipping of exon 46 leading to a predicted shifting of the expressed protein reading frame, producing a stop codon 53 residues downstream (p.Glu2904Aspfs∗53) (Figure 1C).
In family 3, the Algerian male proband born to a consanguineous union had rhinosinusitis, cough, situs inversus, and marked asthenozoospermia (sperm immotility) in fertility testing (Table S1). He was identified to carry two homozygous missense mutations, which were confirmed by Sanger sequencing in the affected individual’s DNA (Figure 1A). No DNA was available from other family members, but despite the lack of parental samples, intra-run comparative analysis of the subject’s NGS read depth ratio confirmed that two copies of all the DNAH9 coding exons were present in his genome, showing that there was no hemizygosity and that it is a complex allele with two homozygous variants present (Figure S1). The first homozygous missense variant, c.5641A>G (p.Lys1881Glu), affects a highly conserved amino acid flanking the only ATPase site thought to retain an ATP hydrolyse activity (AAA1 domain) (Figure 1B) and it is absent from dbSNP, gnomAD, and EVS control databases. The CADD score is 27.8 and PolyPhen-2 predicts it to be probably damaging with a score of 1.0. The second homozygous missense variant, c.8894G>A (p.Arg2965His), was found in 16 unaffected heterozygous carriers in gnomAD with a total allele frequency of 5.8e−5 (rs375908701). It changes a highly conserved residue Arg2965 in DNAH9 and this is predicted to be probably damaging with a score of 0.99 in PolyPhen-2 and a CADD score of 34 (Figures 1A and 1B).
Table 1 shows clinical and PCD test results from these individuals, revealing a comparatively mild phenotype. Notably, all affected individuals had situs inversus. Although showing upper respiratory tract symptoms (rhinitis), none of the affected children had recorded signs of neonatal respiratory distress, otitis media, or signs of permanent lung damage (bronchiectasis). Their nasal nitric oxide levels were within or close to the normal range (>77 nL/min). Three of the four affected probands had lung function (forced expiratory volume in one second [FEV1] % predicted) in keeping with that predicted in healthy children based on their age, height, gender, and ethnicity.6 Individual 1.II.1 had compromised lung function (49% predicted), which is likely attributed to co-morbidities and poor technique in conducting the test.
Table 1.
Clinical Features of Individuals Carrying DNAH9 Mutations
| Individual | 1.II.1 | 2.II.2 | 2.II.3 | 3.II.1 |
|---|---|---|---|---|
| DNAH9 variants (NM_001372.3) | c.12367G>A (p.Asp4123Asn) + c.12367G>A (p.Asp4123Asn) | c.8708−2A>G (p.Glu2904Aspfs∗53) + c.10193G>T (p.Arg3398Leu) | c.8708−2A>G (p.Glu2904Aspfs∗53) + c.10193G>T (p.Arg3398Leu) | c.5641A>G (p. Lys1881Glu) + c.5641A>G (p. Lys1881Glu) & c.8894G>A (p.Arg2965His) + c.8894G>A (p.Arg2965His) |
| Age at diagnosis | 8 years | 2 years | 9 years | 13 years |
| Neonatal distress | N | N | N | N |
| Rhinosinusitis | Y | Y | Y | Y |
| Wet cough | Y | Y with infection | Y with infection | Y |
| Otitis media | N | N | N | N |
| Bronchiectasis | N | N | N | N |
| Situs Inversus | Y | Y | Y | Y |
| Infertility | NA too young | NA too young | NA too young | Y |
| Lung function - forced expiratory volume in 1 s (FEV1%pred) | 49 | 108 | normal range | 92 |
| Nasal nitric oxide (normal > 77 nL/min) | 105 nL/min | 46 nL/min | 100 nL/min | 216 nL/min |
| Ciliary beat frequency (normal 8–15 Hz) | 8.4–10.1 Hz | 8–9.3 Hz | 8.4–8.6 Hz | 5.0–6.6 Hz |
| Co-morbidities | protein losing enteropathy, complex congenital heart disease | N | N | N |
Two or more nasal brush biopsies per individual were analyzed on separate occasions to assess ciliary function. Ciliary beat frequency, measured by HSVM, was at the lower end of the normal range17 in all individuals (Table 1). As summarized in Figures 2A and S2, ciliary beat pattern analysis revealed cilia with a subtle defect in the bend of the cilia in the distal portion captured in affected individuals 1.II.1, 2.II.2, and 3.II.1 (Videos S1, S2, and S3), compared to healthy control subjects (Video S4). Quantitative re-analysis of videos of ciliary beating in nasal cells from affected individuals at one center (B.L.) confirmed a lower beat frequency associated with the altered beat and potentially with a lower distance and area swept by the cilium (Table S2). As a measure of mucociliary clearance, the transport of latex microbeads across the cilia surface was also assessed on a fresh nasal sample from 3.II.1 compared to a healthy control subject (Videos S5 and S6). The ciliated edges of 3.II.1 clearly induced fluid flow, observed by displacement of the microbeads calculated as a ciliary beating efficiency index18 of 0.35 ± 0.16 mPa (evaluated on four ciliated edges), less than the control subject (2.6 ± 1.3 mPa, evaluated on 11 edges). This implies that some respiratory mucociliary clearance potential may be retained by the cilia in individuals with DNAH9 mutations. Interestingly, in contrast to their respiratory cilia motility, sperm analysis performed in individual 3.II.1 showed oligozoospermia with only 3% sperm having preserved motility (other affected individuals were too young to be assessed).
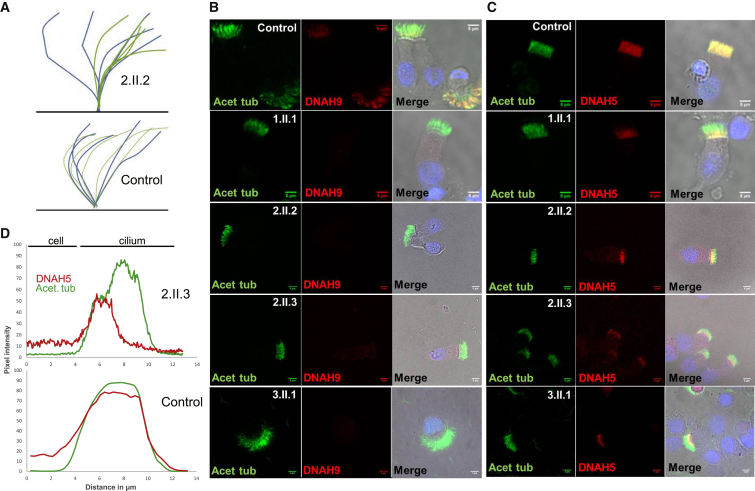
Figure 2.
Loss of Distal Ciliary ODA Type 2 Heavy Chains (DNAH9 and DNAH5) from the Respiratory Cilia of Individuals with DNAH9 Mutations
(A) Diagram of cilia beating in a control subject compared to a DNAH9 mutation case subject (2.II.2), showing the subtle defect in the bend of the distal portion of the cilia that is captured in HSVM (effective stroke shown in blue, recovery stroke in green). DNAH9-deficient cilia have reduced beating frequency accompanied by reduced amplitude of the distal cilia area.
(B and C) Respiratory epithelial cells were double stained with an antibody marker of the ciliary axoneme (acetylated alpha tubulin, green) and the ODA β-HC and γ-HC antibody markers (DNAH9 and DNAH5, red). Co-localization of acetylated tubulin and presence of dynein heavy chain appears yellow in merged images in the right column. Merged images of the red and green are overlaid with a light microscopy image to aid visualization of the cell and an image of the nucleus labeled with DAPI in blue.
(B) DNAH9 localizes distally in the cilia of control subject but is absent from cilia of affected individuals.
(C) In contrast to the control cilia where DNAH5 localizes along the whole length, it is highly reduced in the distal part of the cilia of affected DNAH9-mutation-carrying individuals but is still present proximally. All scale bars, 5 μM.
(D) A graph showing DNAH5 signal quantified by plotting the pixel intensity in images across profile plots of immunofluorescent images of antibody-labeled cilia, in individual 2.II.3. DNAH5 is present mainly in the proximal part of the cilia and reduced in the distal part (results from control are shown in Figure S4).
We next performed cilia immunofluorescence staining to investigate the effect of DNAH9 mutations on DNAH9 localization in nasal epithelial cilia. In control subjects, as expected, DNAH9 was restricted to the distal portion of the cilia (Figure 2B). We found similar results in unaffected siblings of family 2 (data not shown). In contrast, affected individuals carrying DNAH9 mutations showed heavily reduced or absent staining of DNAH9 protein in the cilia, confirming the severity of the inherited mutations (Figure 2B). Since affected individuals in all three families carried single amino acid substitution “missense” changes but showed this marked reduction in ciliary DNAH9 protein, we performed quantitative RT-PCR to investigate the abundance of DNAH9 transcript levels. For two case subjects where nasal samples were available, 2.II.3 and 3.II.1, we found that DNAH9 transcripts were still present but at variable levels presumably reflecting variable levels of ciliation in primary nasal samples as well as possible mutational effects (Figures 1C and S3). Western blot of the 3.II.1 sample also confirmed a complete lack of the DNAH9 protein (Figure S3).
We further sought to identify the location of other ODA components. In control subjects, DNAH5 stains the entire cilia length (Figure 2C) but in cilia from individuals with DNAH9 mutations, type 2 ODAs in the distal portion of the mutant cilia were lacking DNAH5 staining; this was severely reduced and usually completely absent (Figure 2C). DNAH5 staining was unaffected, being similar to control subjects, in type 1 ODAs in the proximal portion of the cilium (Figure 2C). The difference in proximal and distal DNAH5 signal along the length of the cilia from an affected individual with DNAH9 mutations could be quantified by plotting pixel intensity across ten profile plots of immunofluorescent antibody-labeled cilia (Figure 2D). These findings suggest that DNAH9 absence often results in loss or failure of assembly of DNAH5 γ-HC in the distal part of the cilia.
We analyzed the localization of other key motile ciliary components in individuals with DNAH9 mutations. We found that staining of the ODA intermediate chain DNAI18 was altered in affected individuals from all three families carrying DNAH9 mutations, being lost from the distal part of the cilia only, in parallel to the loss of DNAH9 and DNAH5 (Figures 2B, 2C, and S4). Quantification of immunofluorescent antibody pixel intensities along the cilia illustrate these results for the cilia of individual 2.II.3 compared to a control subject (Figure S5). In individual 2.II.2 with DNAH9 mutations, DNAH11, the other axonemal β-HC present in type 1 ODAs, was present in the proximal portion of the cilium according to its usual localization behavior12, 16 and in a similar distribution compared to control subjects, as judged by immunostaining (Figure S6 and quantified in Figure S5). This suggests that DNAH11 does not replace DNAH9 in the type 2 ODAs in the absence of DNAH9, but further quantitative work is required to prove this. In contrast, CCDC114, a component of the ODA docking complex, was still present as normal throughout the length of the ODA, as confirmed also by western blot, suggesting that there is no failure in docking complex assembly (Figures S3 and S7). Key components of the inner dynein arm and radial spoke head were also present as normal throughout the axoneme as measured by antibodies directed against DNALI1 and RSPH4A (Figures S7 and S8 and quantified in Figure S5).
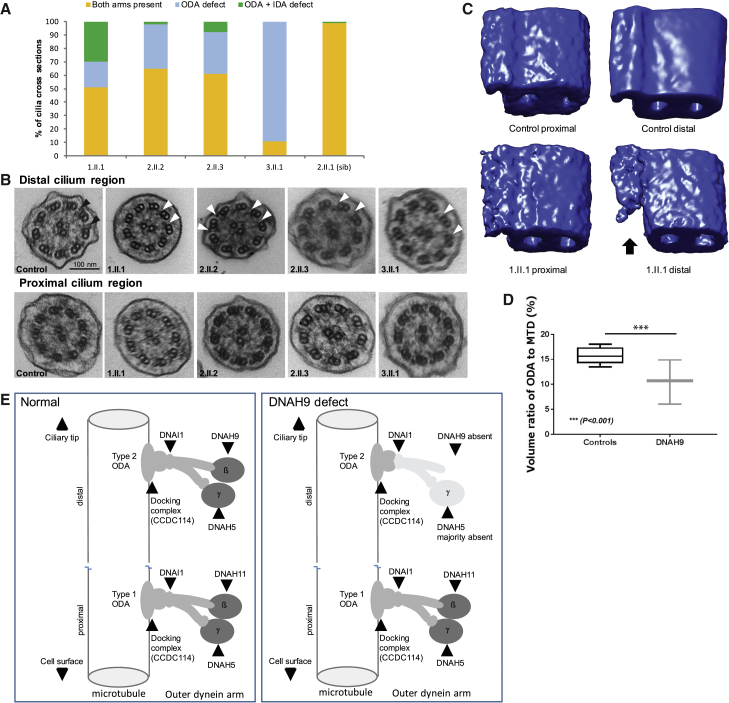
We next assessed by TEM the ultrastructural effect of DNAH9 mutations on cilia. Overall, evaluating >100 cilia cross sections without bias toward measurements in any particular cilia region, ODA defects affected 35%–89% of axonemes in individuals carrying DNAH9 mutations with significant numbers of ODAs in most cases remaining undisturbed (Figure 3A). We then divided the TEM cross sections into those taken from either the distal or the proximal cilia regions. The proximal axonemal cilia segment corresponding to the microvilli region (the location of type 1 ODAs) were of normal composition, while axonemes in the distal part of the cilium (judged by distance from the cell surface and absence of surrounding microvilli) had significant numbers of absent or truncated ODAs (Figure 3B).
Figure 3.
Ultrastructural Defects of the Distal Outer Dynein Arm in Individuals with DNAH9 Mutations
(A) Brushings of the nasal epithelium were assessed by transmission electron microscopy in four individuals with DNAH9 mutations and one unaffected sibling (2.II.1). A minimum of 100 axonemes in cross section were assessed per individual. The number of axonemes demonstrating defects of the dynein arms are expressed as a percentage. Both dynein arms were present in the majority of the axonemes assessed (11%–65%) and the remaining had absent outer dynein arms (89%–35%). Where ODAs were absent, usually the accompanying inner dynein arms remained present (orange) but in occasional axonemes the IDA was also absent (gray). The unaffected sibling has 99% normal ultrastructure in keeping with quantification in previously described healthy control subjects.28
(B) Example of transmission electron micrographs of ciliary axonemes in cross section from a healthy control subject (1st column) and four individuals with DNAH9 mutations (columns 2–5). ODAs are highlighted by black arrowheads in the control and ODA loss is highlighted by the white arrowheads. ODAs are intact in ciliary cross sections judged to be proximal to the cell surface (bottom row) whereas they are frequently absent in the distal part of the cilium (top row). Scale bar = 100 nm.
(C) Electron tomography and averaging were used to generate 3D models of the ODA, which have been falsely colored blue for visualization. Using PEET analysis, transmission electron microscope tomograms taken from an area with >6 ciliary cross sections (54 microtubular doublets) were combined into a single model. Models from tomograms from individual 1.II.1 in the proximal cilia region, judged by the presence of surrounding microvili, are similar to the models of both the proximal and the distal region of the healthy control axonemes. The model from the distal region of the axonemes of 1.II.1 is reduced in volume (indicated by the black arrow). This model was created from ODAs that appeared to remain intact in standard electron micrographs.
(D) The volume occupied by the ODA as a proportion of the entire microtubular doublet distal model was quantified for three affected individuals (1.II.1, 2.II.2, 2.II.3) and six healthy control subjects. Data are plotted as box and whisker plots, showing a difference in volume in the distal region compared to controls (p > 0.001, independent samples t test). Additional tomograms from individuals carrying DNAH9 mutations used for quantification are shown in the supplemental data (Figure S5).
(E) Information from immunofluorescence protein location studies and electron tomography ultrastructural studies were used to create a model of type 1 and type 2 ODAs in cilia from normal control subjects and individuals with DNAH9 mutations. In the distal cilium of individuals with DNAH9 mutations, we have speculatively left in a lightly shaded remnant ODA structure in the distal axoneme, since occasional outer dynein arms remain visible in TEM and electron tomography images and we cannot fully exclude their absence by immunofluorescence.
To better understand the nature of DNAH9-associated structural defects, we performed higher-resolution imaging by 3D electron tomography on axonemes from affected individuals. Tomograms were taken from ODAs that appeared complete by conventional TEM (such as those indicated by black arrows in Figure 3B). This confirmed the regionality of the DNAH9 mutational effect since in individual 1.II.1, tomography revealed a loss of volume only of the type 2 ODAs, i.e., only ODAs in the distal cilium, while in the proximal cilium the ODA volume was equivalent to that of control subjects. For type 2 ODAs of the distal cilium that were not completely lost but retained, the reduction of ODA volume was specific to their “forearm” portion, reflecting the predicted location of the DNAH9 β-HC within the ODAs (Figure 3C). The ODA forearm-specific defect was also confirmed by tomography of the ciliary distal region ODAs in individuals 2.II.2 and 2.II.3 (Figure S9). These DNAH9 data are in keeping with our previously published converse (proximal) findings in individuals with DNAH11 variants, who have a similarly reduced ODA volume for the DNAH11 β-HC but affecting only the type 1 ODAs of the proximal cilium.16 Quantification of the ODA volumes in cilia from three DNAH9-mutated individuals was compared to results from individuals without respiratory disease. This showed a significant decrease in the ODA volume in the distal portion of their cilia, while ODA volume in the proximal portion of the cilia was not affected (Figure 3D). Preliminary tomography studies in individuals with PCD carrying DNAH5 mutations show some evidence that DNAH9 mutations may confer a less reduced type 2 ODA volume than DNAH5 mutations, but further study is required (A.S., unpublished data). Figure 3E presents a model of the findings arising from the current study, showing the regional (distal cilium) effect of DNAH9 loss on the ODA structures in individuals with pathogenic variants in DNAH9.
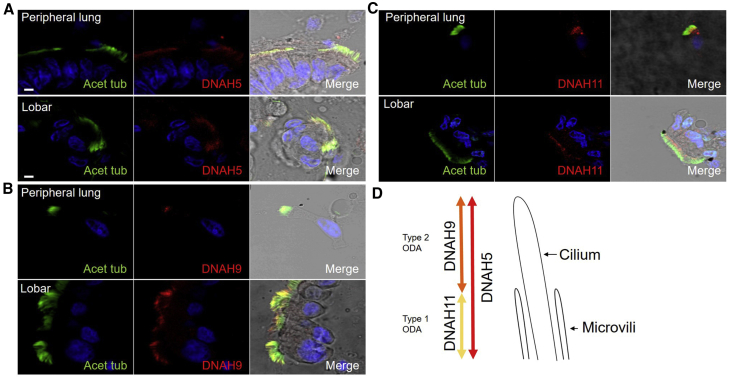
Due to the findings in individuals harboring DNAH9 mutations of upper respiratory tract symptoms but preserved lung function in three of four probands and no evidence of bronchiectasis (Table 1), we sought to confirm whether DNAH9 was present throughout the respiratory tract and not isolated to nasal tissue. Tissue obtained from an individual without PCD undergoing a lobectomy showed similar localization of DNAH9, DNAH5, and DNAH11 in the nasal passage, lobar region, and the peripheral airways of <1.5 mm. This confirms the location of both type 1 and type 2 ODAs in cilia throughout the airways (Figure 4).
Figure 4.
Consistent Location of the Outer Dynein Arm Gamma (DNAH5) and Beta Heavy Chains (DNAH9 and DNAH11) of the Ciliated Epithelial Cells at Various Nasal and Bronchopulmonary Sites
(A–C) Respiratory epithelial cells from a non-PCD donor were stained with a marker of the ciliary axoneme (acetylated alpha tubulin, green), the outer dynein arm beta and gamma heavy chain markers (DNAH9, DNAH11, or DNAH5, red), and the nucleus (DAPI, blue). DNAH5 (A) localizes along the entire length of the cilia, DNAH9 (B) localizes distally in the cilia, and DNAH11 (C) localizes proximally in the cilia (note that this antibody is associated with some non-specific background staining in the cytoplasm). The same pattern of staining localization is seen for all three heavy chain proteins in epithelial cells dissected from both the peripheral airways of less than 1.5 mm and from the lobe region, in comparison to the nasal epithelial cells shown in Figure 2. Scale bars, 5 μM.
(D) Cartoon showing the unified nasal and bronchopulmonary distribution of type 1 (DNAH5- and DNAH11-positive) and type 2 (DNAH5- and DNAH9-positive) ODAs in cilia throughout the airways.
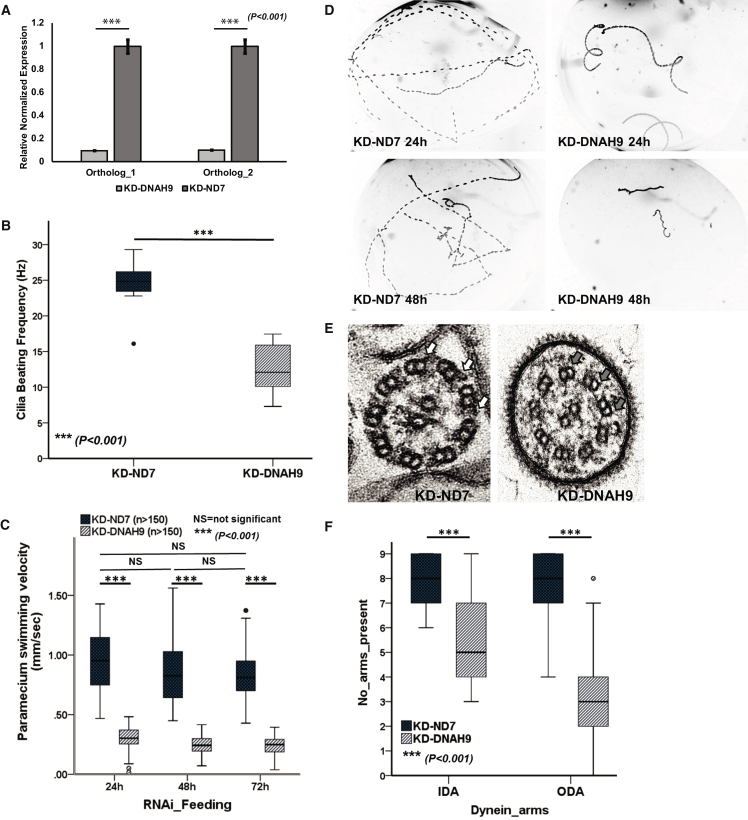
We recently reported the use of Paramecium as a model for motile ciliary disorders, due to the highly conserved nature of ciliary proteins.19 To study the role of DNAH9 in cilia motility, we employed gene knock-down in Paramecium using RNAi silencing by feeding, as described before.19 We identified two orthologs of DNAH9 due to whole genome duplication in the Paramecium genome (GenBank: XM_001443078.1; PRJEB27934) (Figure S10). Real-time quantitative PCR confirmed >80% knock down of transcript levels for both these DNAH9 orthologs by RNAi, comparing DNAH9-RNAi Paramecia (double knockdown of both orthologs) to ND7-RNAi Paramecia as a control (the ND7 gene is routinely used as a control as it has no role in cilia motility19) (Figure 5A). Paramecium DNAH9-RNAi experiments were repeated >3 times to confirm the reproducibility of the results. We showed a significant reduction of the swimming velocity and cilia beating frequency in the DNAH9 knockdown cells compared to controls over 3 days of RNAi silencing (Figures 5B–5D and Videos S7 and S8). Ultrastructural loss of ODAs from the axoneme was quantified by TEM (Figures 5E and 5F). The phenotype of the DNAH9 mutant Paramecium cilia therefore recapitulates the human phenotype.
Figure 5.
RNAi Silencing of Paramecium DNAH9 Shows Severe Defective Motile Cilia Phenotype
(A) Quantitative RT-PCR assessment of expression level of DNAH9 orthologs after RNAi knockdown in Paramecium. The expression level is reduced by >80% in the DNAH9-RNAi cells compared to its expression in the control ND7-RNAi cells (ND7 gene not associated with cilia motility); p < 0.001, independent samples t test.
(B) Reduced cilia beat frequency after DNAH9-RNAi silencing. At 48 hr of RNAi, the ciliary beat frequency of DNAH9 knockdown cells is reduced by ∼50% compared to ND7 control knockdown cells. 7–10 paramecia evaluated per condition; p < 0.001, Mann-Whitney U test.
(C) Analysis of swimming velocity of Paramecium DNAH9 and ND7 knockdown cells shows a significant reduction (∼70%) in velocity of DNAH9 knockdown cells compared to the control ND7 knockdown cells over 72 hr of RNAi silencing. More than 150 paramecia were evaluated per condition; p < 0.001, independent samples t test.
(D) Paramecium swimming pattern visualized in dark-field microscopy, shown by the Z projection of track recordings, captured using a 10× objective after 24 and 48 hr of RNAi feeding. DNAH9 knockdown cells (right) show a severe motility phenotype compared to the control ND7 silenced cells (left).
(E) Transmission electron micrographs of Paramecium cilia in cross-section showing normal 9+2 arrangement for the control ND7 knockdown (left, outer dynein arms indicated by white arrows) and absence of the outer (gray arrows) dynein arms in the DNAH9 knockdown cells (right).
(F) Quantification of TEM dynein arms counts across >40 cross-sections per knockdown experiment showed reduced ODA and IDA in DNAH9 knockdown cells compared to ND7 control knockdown cells with ODA loss more extensive than IDA loss. p > 0.001, independent samples t test. All error bars indicate SEM.
In summary, we report four individuals with PCD carrying mutations in DNAH9 that affect amino acids in highly conserved protein domains or cause a frameshift, all of which result in severely reduced or absent levels of protein. These individuals all manifest with respiratory symptoms, laterality (situs) defects, and (in one case) male infertility. Our findings are in agreement with Loges et al.,20 who also report mutations in DNAH9 that cause similar phenotypes. DNAH9-related disease is a form of primary ciliary dyskinesia, since the motile cilia of the respiratory system and embryonic node are defective (the latter since situs defects are present1, 4, 5) as well as sperm motility.
DNAH9 is the β-HC dynein of type 2 ODA, which is expressed in the airways and sperm.11, 22 We find that DNAH9 is expressed in cilia from all the human upper and lower respiratory airway epithelia we tested. DNAH9 knock-down in Paramecium mimics the ODA and cilia motility defects of DNAH9-deficient individuals, indicating the highly conserved critical role DNAH9 plays in forming the axonemal dynein arms. Recently, DNAH9 polymorphisms were associated with asthma and bronchial hyper responsiveness in response to early life tobacco smoke exposure in large GWASs, suggesting wider roles in lung pathology.23
Our protein localization and structural studies confirm the previously reported region-specific localization of DNAH9 in cilia, limited to the distal axoneme portion.11 In all the affected individuals with PCD, axonemal DNAH9 levels appear heavily reduced to undetectable in immunostaining of their ciliated nasal epithelial cells, even in affected individuals from two families carrying different single amino acid substitution (missense) changes. qRT-PCR showing that DNAH9 transcripts could still be amplified in these samples suggest that the mutant forms of the encoded protein may be expressed but not assembled to the axoneme or subject to degradation. This has been noted before for missense mutations causing PCD found in other genes.24 DNAH9 protein was also confirmed to be undetectable by western blotting of ciliated nasal epithelial cells from 3.II.1 who is homozygous for missense mutations. This suggests these affected protein residues are vital to DNAH9 axonemal localization and function, supported by their highly conserved nature across other dyneins and DNAH9 orthologs.
The absence of ciliary DNAH9 correlates to a clear loss of type 2 ODAs from the airway cilia of individuals carrying DNAH9 mutations. The heavy and intermediate ODA chains DNAH9, DNAH5, and DNAI1 are all absent or severely reduced from the distal cilia portion, contributing to the regional ODA volume reduction seen by electron tomography. Markers of the axonemal ODA docking complexes, inner dynein arms, and radial spokes seem to be unaffected and in place as normal, suggesting that the defect is restricted to affecting only the ODAs. DNAH5 mutations causing PCD have similarly been reported to disturb the ODAs but not the ODA docking complexes.25
Type 2 ODAs are located only within the distal cilium, such that DNAH9 mutations are accompanied by a subtle effect on the ciliary beat pattern. We find that individuals carrying DNAH9 mutations usually have nasal nitric oxide levels in the normal range along with other features of a sometimes mild clinical presentation. These findings challenge the current paradigm of the phenotype of PCD. DNAH9 was previously considered as a PCD candidate gene by Bartoloni et al.,26 but no pathogenic mutations were found in 31 PCD-affected families screened before the advent of next generation sequencing. We conclude that DNAH9 cases can be difficult to recognize without genetic testing. Notably, all the individuals carrying DNAH9 mutations in this study displayed laterality defects (expected to affect 50% of PCD-affected case subjects), and this may have been a lead symptom for their referral to the PCD clinic.
Interestingly, infertility tests in one available case in our study show that DNAH9 defects may have a greater impact on sperm motility than cilia motility, but further functional analysis in a larger cohort would be needed to confirm whether this is the case, including further studies of the distribution of DNAH9 in human sperm compared to cilia.11 Altogether, these clinical findings can better inform disease diagnostics, disease management, and appropriate counselling of affected families.
Previous work showed that in individuals with PCD caused by DNAH11 defects, although DNAH11 is absent from their cilia, DNAH9 shifts from a distal to pan-axonemal localization, i.e., is aberrantly present in place of the type 1 ODAs; but despite this, the shift in DNAH9 localization is not able to rescue the DNAH11 motility defects.12 In contrast to this pattern, our present study demonstrates no evidence in subjects with DNAH9 mutations for a change in their ciliary DNAH11 localization or its substituting for DNAH9, implying that DNAH11 and DNAH9 β-HCs behave differently. Interestingly, a recent study proposed a sequential assembly of DNAH5 and DNAH9, followed by DNAH11 throughout different stages of ciliogenesis toward generation of a mature cilium, so the protein distribution (and ciliary waveform) varies at different stages.27 In our study, we ensured to select only full-length cilia for quantification of signal intensity of the different ODA markers from the proximal to distal regions.
In conclusion, this study sheds new light on ODA HC biology and the effect of inherited mutations in ciliary ODA genes, broadening our understanding of the PCD clinical phenotypic spectrum.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are grateful to the families who participated in this study and thank the UK PCD Family Support Group for their continued support. We thank Ms. Dani Lee and Prof. Chris O’Callaghan (UCL Great Ormond Street Institute of Child Health) for help with assessment of Paramecium motility, Dr. Jane Hayward with genetic analysis, and Dr. Dale Moulding (GOS-ICH Confocal Microscopy Core Facility) for imaging assistance. We thank Catherine Faucon (Service d’Anatomopathologie, Hôpital intercommunal, Créteil, France) for her great technical assistance in TEM studies. Work by M. Legendre, S.A., and E.E. is supported by grants from the Fondation pour la Recherche Médicale (DEQ20120323689) and the Legs Poix from the Chancellerie des Universités and from the Investissements d'Avenir (RaDiCo program; ANR-10-COHO-0003). Other funding for this study was provided by Action Medical Research (GN2101; H.M.M.) and Great Ormond Street Children’s Charity grant (V4515; H.M.M.) and Leadership awards (V1299, V2217; H.M.M.). We acknowledge support from the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London. M.R.F. is supported by the British Council Newton-Mosharafa Fund and the Ministry of Higher Education, Egypt. Work by A.S. is independent research funded by a postdoctoral research fellowship from the National Institute of Health Research and Health Education England. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute of Health Research, or the Department of Health. The authors participate in the COST Action BEAT-PCD: Better Evidence to Advance Therapeutic options for PCD network (BM1407) and this work was supported by two BM1407 COST Action STSM Grants awarded to M.R.F. and P.L.B. The authors alone are responsible for the content and writing of the paper.
Published: November 21, 2018
Footnotes
Supplemental Data include ten figures, five tables, and eight videos and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.016.
Web Resources
dbSNP build 141, https://www.ncbi.nlm.nih.gov/projects/SNP/
gnomAD Browser, http://gnomad.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
ParameciumDB, http://paramecium-archive.i2bc.paris-saclay.fr/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RNAi off-target tool, http://paramecium.cgm.cnrs-gif.fr/cgi/tool/alignment/off_target.cgi
Supplemental Data
References
- 1.Reiter J.F., Leroux M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linck R.W., Chemes H., Albertini D.F. The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J. Assist. Reprod. Genet. 2016;33:141–156. doi: 10.1007/s10815-016-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas J.S., Burgess A., Mitchison H.M., Moya E., Williamson M., Hogg C., National PCD Service, UK Diagnosis and management of primary ciliary dyskinesia. Arch. Dis. Child. 2014;99:850–856. doi: 10.1136/archdischild-2013-304831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horani A., Ferkol T.W., Dutcher S.K., Brody S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016;18:18–24. doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchison H.M., Valente E.M. Motile and non-motile cilia in human pathology: from function to phenotypes. J. Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 6.Lucas J.S., Barbato A., Collins S.A., Goutaki M., Behan L., Caudri D., Dell S., Eber E., Escudier E., Hirst R.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2017;49:49. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King S.M. Axonemal Dynein Arms. Cold Spring Harb. Perspect. Biol. 2016;8:8. doi: 10.1101/cshperspect.a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pazour G.J., Agrin N., Walker B.L., Witman G.B. Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J. Med. Genet. 2006;43:62–73. doi: 10.1136/jmg.2005.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgoyne T., Dixon M., Luther P., Hogg C., Shoemark A. Generation of a three-dimensional ultrastructural model of human respiratory cilia. Am. J. Respir. Cell Mol. Biol. 2012;47:800–806. doi: 10.1165/rcmb.2011-0440OC. [DOI] [PubMed] [Google Scholar]
- 10.Viswanadha R., Sale W.S., Porter M.E. Ciliary Motility: Regulation of Axonemal Dynein Motors. Cold Spring Harb. Perspect. Biol. 2017;9:9. doi: 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fliegauf M., Olbrich H., Horvath J., Wildhaber J.H., Zariwala M.A., Kennedy M., Knowles M.R., Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougherty G.W., Loges N.T., Klinkenbusch J.A., Olbrich H., Pennekamp P., Menchen T., Raidt J., Wallmeier J., Werner C., Westermann C. DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes. Am. J. Respir. Cell Mol. Biol. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles M.R., Daniels L.A., Davis S.D., Zariwala M.A., Leigh M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles M.R., Leigh M.W., Carson J.L., Davis S.D., Dell S.D., Ferkol T.W., Olivier K.N., Sagel S.D., Rosenfeld M., Burns K.A., Genetic Disorders of Mucociliary Clearance Consortium Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–441. doi: 10.1136/thoraxjnl-2011-200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olbrich H., Häffner K., Kispert A., Völkel A., Volz A., Sasmaz G., Reinhardt R., Hennig S., Lehrach H., Konietzko N. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 16.Shoemark A., Burgoyne T., Kwan R., Dixon M., Patel M.P., Rogers A.V., Onoufriadis A., Scully J., Daudvohra F., Cullup T. Primary ciliary dyskinesia with normal ultrastructure: three-dimensional tomography detects absence of DNAH11. Eur. Respir. J. 2018;51:51. doi: 10.1183/13993003.01809-2017. [DOI] [PubMed] [Google Scholar]
- 17.Chilvers M.A., Rutman A., O’Callaghan C. Functional analysis of cilia and ciliated epithelial ultrastructure in healthy children and young adults. Thorax. 2003;58:333–338. doi: 10.1136/thorax.58.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottier M., Peña Fernández M., Pelle G., Isabey D., Louis B., Grotberg J.B., Filoche M. A new index for characterizing micro-bead motion in a flow induced by ciliary beating: Part II, modeling. PLoS Comput. Biol. 2017;13:e1005552. doi: 10.1371/journal.pcbi.1005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fassad M.R., Shoemark A., le Borgne P., Koll F., Patel M., Dixon M., Hayward J., Richardson C., Frost E., Jenkins L. C11orf70 Mutations Disrupting the Intraflagellar Transport-Dependent Assembly of Multiple Axonemal Dyneins Cause Primary Ciliary Dyskinesia. Am. J. Hum. Genet. 2018;102:956–972. doi: 10.1016/j.ajhg.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loges N.T., Antony D., Maver A., Deardorff M.A., Güleç E.Y., Gezdirici A., Nöthe-Menchen T., Höben I.M., Jelten L., Frank D. Recessive DNAH9 loss-of-function mutation cause laterality defects and subtle respiratory ciliary-beating defects. Am. J. Hum. Genet. 2018;103:995–1008. doi: 10.1016/j.ajhg.2018.10.020. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed W., Carson J.L., Moats-Staats B.M., Lucier T., Hu P., Brighton L., Gambling T.M., Huang C.H., Leigh M.W., Collier A.M. Characterization of an axonemal dynein heavy chain expressed early in airway epithelial ciliogenesis. Am. J. Respir. Cell Mol. Biol. 2000;23:734–741. doi: 10.1165/ajrcmb.23.6.4045. [DOI] [PubMed] [Google Scholar]
- 23.Dizier M.H., Nadif R., Margaritte-Jeannin P., Barton S.J., Sarnowski C., Gagné-Ouellet V., Brossard M., Lavielle N., Just J., Lathrop M. Interaction between the DNAH9 gene and early smoke exposure in bronchial hyperresponsiveness. Eur. Respir. J. 2016;47:1072–1081. doi: 10.1183/13993003.00849-2015. [DOI] [PubMed] [Google Scholar]
- 24.Hornef N., Olbrich H., Horvath J., Zariwala M.A., Fliegauf M., Loges N.T., Wildhaber J., Noone P.G., Kennedy M., Antonarakis S.E. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallmeier J., Shiratori H., Dougherty G.W., Edelbusch C., Hjeij R., Loges N.T., Menchen T., Olbrich H., Pennekamp P., Raidt J. TTC25 Deficiency Results in Defects of the Outer Dynein Arm Docking Machinery and Primary Ciliary Dyskinesia with Left-Right Body Asymmetry Randomization. Am. J. Hum. Genet. 2016;99:460–469. doi: 10.1016/j.ajhg.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartoloni L., Blouin J.L., Maiti A.K., Sainsbury A., Rossier C., Gehrig C., She J.X., Marron M.P., Lander E.S., Meeks M. Axonemal beta heavy chain dynein DNAH9: cDNA sequence, genomic structure, and investigation of its role in primary ciliary dyskinesia. Genomics. 2001;72:21–33. doi: 10.1006/geno.2000.6462. [DOI] [PubMed] [Google Scholar]
- 27.Oltean A., Schaffer A.J., Bayly P.V., Brody S.L. Quantifying Ciliary Dynamics during Assembly Reveals Stepwise Waveform Maturation in Airway Cells. Am. J. Respir. Cell Mol. Biol. 2018;59:511–522. doi: 10.1165/rcmb.2017-0436OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoemark A., Dixon M., Corrin B., Dewar A. Twenty-year review of quantitative transmission electron microscopy for the diagnosis of primary ciliary dyskinesia. J. Clin. Pathol. 2012;65:267–271. doi: 10.1136/jclinpath-2011-200415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.