Norovirus is one of the leading causes of food-borne illness worldwide. Despite its prevalence, our understanding of norovirus biology is limited due to the difficulty in growing human norovirus in vitro and a lack of an animal model. Murine norovirus (MNoV) is a model norovirus system because MNoV replicates robustly in cell culture and in mice. To identify host genes that can restrict norovirus replication when overexpressed, we performed genome-wide CRISPR activation screens to induce gene overexpression at the native locus through recruitment of transcriptional activators to individual gene promoters. We found 49 genes that could block murine norovirus replication in human cells. Several of these genes are associated with classical immune signaling pathways, while many of the molecules we identified have not been previously associated with antiviral activity. Our data are a resource for those studying noroviruses, and we provide a robust approach to identify novel antiviral genes.
KEYWORDS: CRISPR, noroviruses, virology
ABSTRACT
Noroviruses (NoVs) are a leading cause of gastroenteritis worldwide, yet host factors that restrict NoV replication are not well understood. Here, we use a CRISPR activation genome-wide screening to identify host genes that can inhibit murine norovirus (MNoV) replication in human cells. Our screens identified with high confidence 49 genes that can inhibit MNoV infection when overexpressed. A significant number of these genes are in interferon and immune regulation signaling networks, but surprisingly, the majority of the genes identified are neither associated with innate or adaptive immunity nor associated with any antiviral activity. Confirmatory studies of eight of the genes validate the initial screening data. Mechanistic studies on TRIM7 demonstrated a conserved role of the molecule in mouse and human cells in restricting MNoV in a step of infection after viral entry. Furthermore, we demonstrate that two isoforms of TRIM7 have differential antiviral activity. Taken together, these data provide a resource for understanding norovirus biology and demonstrate a robust methodology for identifying new antiviral molecules.
IMPORTANCE Norovirus is one of the leading causes of food-borne illness worldwide. Despite its prevalence, our understanding of norovirus biology is limited due to the difficulty in growing human norovirus in vitro and a lack of an animal model. Murine norovirus (MNoV) is a model norovirus system because MNoV replicates robustly in cell culture and in mice. To identify host genes that can restrict norovirus replication when overexpressed, we performed genome-wide CRISPR activation screens to induce gene overexpression at the native locus through recruitment of transcriptional activators to individual gene promoters. We found 49 genes that could block murine norovirus replication in human cells. Several of these genes are associated with classical immune signaling pathways, while many of the molecules we identified have not been previously associated with antiviral activity. Our data are a resource for those studying noroviruses, and we provide a robust approach to identify novel antiviral genes.
INTRODUCTION
Noroviruses are nonenveloped viruses with positive-sense RNA genomes that infect the gastrointestinal tract of mammals (1). Human norovirus (HNoV) is the leading cause of viral gastroenteritis worldwide and is estimated to have an economic burden of $1 billion annually (2). Despite the high infectivity of HNoV between individuals, it has proven difficult to culture in vitro with significant advances made only in the past few years (3, 4). Because of the challenges in growing HNoV in cell culture and lack of an animal model, murine norovirus (MNoV) has emerged as a system for understanding norovirus biology. MNoV grows robustly in cell culture, is amenable to reverse genetics, and is a natural pathogen of laboratory mice. MNoV is similar to HNoV in that both viruses are spread fecal-orally, infect intestinal tissues, have similar genomic organization and protein function, and can establish persistent infection in individuals (1).
Host factors that limit or modulate norovirus replication are still not well understood. We and others previously used a whole-genome CRISPR/Cas9 loss-of-function screen to identify essential host factors required for MNoV growth in macrophage-like cell lines, leading to the identification of CD300lf as a proteinaceous receptor for MNoV (5, 6). Importantly, expression of mouse CD300lf in cells is sufficient for replication in all cell types tested, including human cells, thus expanding the repertoire of cell lines that can be utilized to study norovirus infection with MNoV (5, 6). These studies also demonstrate that the cellular machinery for replication of MNoV is highly conserved with species specificity conferred by the receptor. Although CRISPR knockout studies for MNoV and other viruses have identified host factors necessary for viral infection, they have yet to identify restriction factors that antagonize viral replication (5–11). Recent modifications of the CRISPR/Cas9 system have led to the ability to transactivate gene expression through specific recruitment of transcriptional activators to individual promoters and has been broadly called CRISPR activation (CRISPRa [12–14]). cDNA overexpression systems have been used to identify viral restriction factors, but this approach has several technical hurdles, most notably the cloning, delivery, and expression of large transcripts and the need to select individual protein isoforms rather than screening for the effects of all transcripts that emanate from a given cellular gene promoter. Because CRISPRa induces gene expression from the native locus and relies only on single guide RNA (sgRNA) homology for specificity, it overcomes many of these obstacles. We therefore utilized CRISPRa to identify genes whose overexpression block MNoV replication.
We use recently developed CRISPRa genome-wide libraries to screen for host genes that antagonize MNoV replication in human cells expressing the MNoV receptor CD300lf. Our screens and subsequent validation identify numerous anti-MNoV genes, including canonical interferon-stimulated genes (ISGs) and several genes with novel antiviral activity. We demonstrate that the antiviral activity of one such gene, TRIM7, is highly specific to only one of the two major TRIM7 isoforms, highlighting the utility and power of the CRISPRa screening approach. Further mechanistic studies demonstrate that TRIM7 inhibits MNoV replication after viral entry. These data provide a resource for understanding norovirus biology and a methodology for identifying unappreciated viral restriction factors.
(This article was submitted to an online preprint archive [15].)
RESULTS
Generation of CRISPRa libraries in HeLa-CD300lf cells.
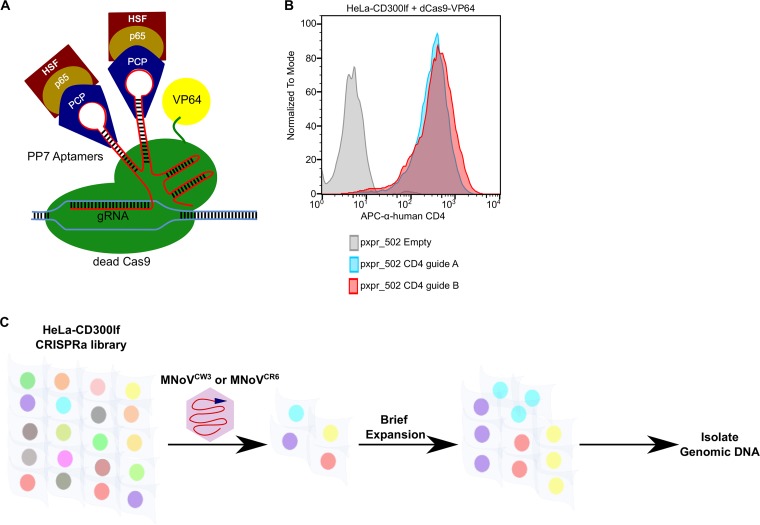
We set out to identify genes in human cells that have antinorovirus activity when overexpressed. Since noroviruses exhibit strict species tropism and at this time HNoV replication is not robust or scalable for whole-genomic pooled screens, we selected HeLa cells to use as a model human cell line and generated HeLa-CD300lf stable cells that can be infected by MNoV. We introduced the catalytically dead Cas9-VP64 (dCas9-VP64) fusion construct into HeLa-CD300lf cells (HeLa-CD300lf-dCas9-VP64). Before proceeding with a genome-wide screen, we developed a simple assay to test transcriptional activation activity in HeLa-CD300lf-dCas9-VP64 cells relying on the low level of expression of CD4 in these cells. sgRNAs targeting the CD4 transcriptional start site robustly increased the surface levels of CD4 in HeLa-CD300lf-dCas9-VP64 cells compared to an empty vector control (Fig. 1A and B).
FIG 1.
CRISPRa screen for antinorovirus genes. (A) Schematic of the CRISPRa system used in this study. Dead Cas9 (dCas9) fused to VP64 interacts with sgRNAs at the transcriptional start site of a gene. sgRNA have PP7 hairpins that recruit a chimeric protein containing heat shock factor (HSF), p65 transcription factor, and phage coat protein (PCP) fused together. In combination, this leads to the transcriptional activation of the targeted gene. (B) HeLa-CD300lf cells expressing dead Cas9-VP64 fusion were transduced with the indicated sgRNA plasmids and assayed for CD4 expression 1 week after antibiotic selection. At baseline CD4 levels are low, but they dramatically increase upon expression of the CRISPRa machinery. (C) Cartoon overview of the cell survival CRISPRa screen performed with HeLa-CD300lf cells. sgRNAs from cells surviving MNoV challenge were analyzed and compared to relative abundance of a mock-infected sample.
We next introduced the Calabrese CRISPRa library into HeLa-CD300lf-dCas9-VP64 cells. Importantly, this library uses an updated CRISPRa sgRNA vector system (37). This system (pXPR_502) is bicistronic with one promoter expressing the sgRNAs with PP7 stem loops, while the second promoter drives expression of the synergistic transcriptional activators (Fig. 1A). This new system is advantageous over the previous synergistic activation mediator (SAM) vectors as it consolidates the transcription activators and sgRNA into one lentivirus (14). The Calabrese library contains 113,240 total sgRNAs with each gene targeted by three to six sgRNAs split between two pools, SetA and SetB. After introduction of these pools into HeLa-CD300lf-dCas9-VP64 cells, we challenged the resulting cellular pools with either MNoVCW3 and MNoVCR6 and isolated resistant cells (Fig. 1C). MNoVCW3 causes acute, systemic infection in mice, while MNoVCR6 establishes a persistent infection in intestinal tuft cells, a specialized intestinal epithelial cell (16–18). MNoVCW3 and MNoVCR6 have been used to model human noroviruses, transkingdom interactions, and innate immunity restriction of viral tropism (4, 19–24). In summary, we performed two independent CRISPRa screens using two MNoV strains (MNoVCW3 and MNoVCR6) in an effort to provide a broad resource for understanding norovirus biology.
CRISPRa screen for anti-MNoV genes in HeLa-CD300lf cells.
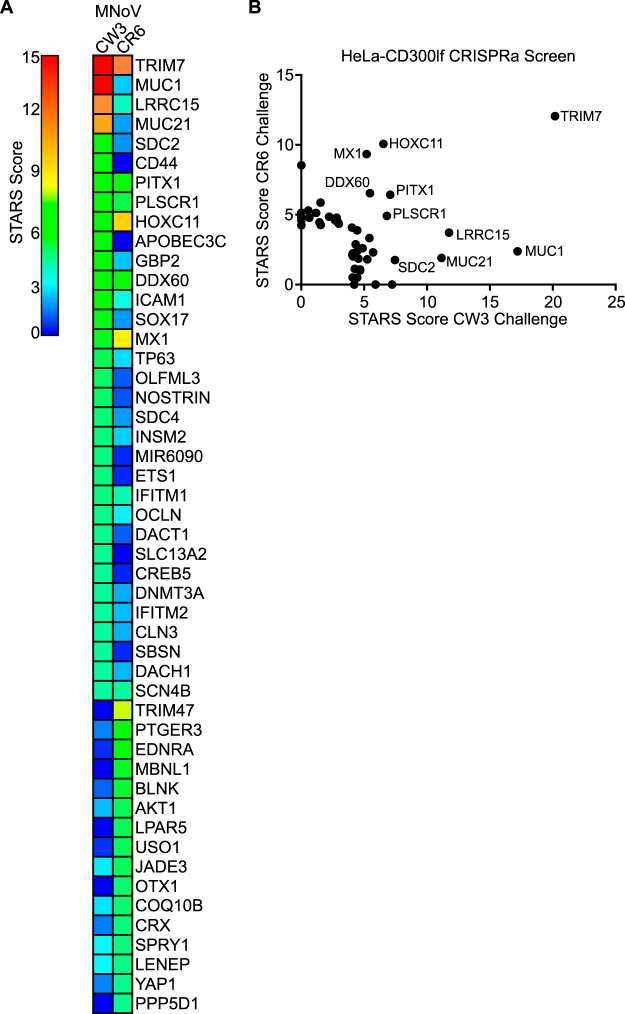
HeLa-CD300lf-dCas9-VP64 cells cellular pools containing the Calabrese library were infected with MNoV at a multiplicity of infection (MOI) of 50. In addition, a second challenge of MNoVCR6 was preformed to exert a high selective pressure for this strain of virus (see Materials and Methods for more details). Surviving cells were harvested after a brief expansion period (Fig. 1C). sgRNAs from resistant cells were compared to mock-infected controls using STARS (25). The STARS algorithm calculates a score and a false discovery rate (FDR) for a gene with at least two sgRNAs enriched in the top 10% of a ranked ordered list of total sequenced sgRNAs. We consider a gene a hit in an individual screen if the FDR was <0.25. We found that 33 and 22 genes in MNoVCW3- and MNoVCR6-infected HeLa-CD300lf, respectively, scored as hits (see Tables S1 and S2 and Fig. 2A and B). Six genes (TRIM7, PITX1, HOXC11, DDX60, MX1, and PLSCR1) were enriched after challenge with both viral strains.
FIG 2.
Identification of anti-MNoV genes in human cells. (A) Heat map showing the enrichment of genes in the two indicated conditions. Genes are color coded based upon their STARS score. (B) Comparison of STARS scores from HeLa-CD300lf cells challenged with MNoVCW3 (x axis) and MNoVCR6 (y axis). Genes that did not meet the criteria to receive a STARS score are assigned a value of 0.
To explore the possibility that individual hits function in a similar pathway, we took the 49 genes that scored for either viral strains and analyzed them using gene set enrichment analysis (GSEA) (26, 27). Comparison of the hallmark genes with our data set indicated a significant enrichment in genes related to interferon alpha responses, including GBP2, which has been recently shown to block MNoV replication complex formation (28). When analyzing the function of the genes by GO terms, we also found an enrichment for interferon and cytokine responses, along with genes involved in embryogenesis and RNA polymerase gene expression (Table 1). These data demonstrate the ability to discover expected candidate antiviral genes (e.g., those regulated by inflammatory cytokines) and candidate genes involved in other pathways (e.g., embryogenesis).
TABLE 1.
GSEA of CRISPRa screening hits
| Gene set name | No. of genes in overlap |
P | FDR q-value |
|---|---|---|---|
| GO negative regulation of response to stimulus | 16 | 3.19E–12 | 1.90E–08 |
| GO positive regulation of biosynthetic process | 17 | 1.93E–11 | 5.76E–08 |
| GO transcription from RNA polymerase II promoter | 12 | 5.05E–11 | 1.00E–07 |
| GO positive regulation of gene expression | 16 | 1.15E–10 | 1.72E–07 |
| GO RNA polymerase II transcription factor activity sequence specific DNA binding | 11 | 2.01E–10 | 2.40E–07 |
| GO embryo development | 12 | 5.56E–10 | 5.53E–07 |
| GO positive regulation of multicellular organismal process | 14 | 7.19E–10 | 5.66E–07 |
| GO response to cytokine | 11 | 7.59E–10 | 5.66E–07 |
| GO negative regulation of cell communication | 13 | 1.19E–09 | 7.90E–07 |
| GO regulation of transcription from RNA polymerase II promoter | 15 | 1.78E–09 | 9.76E–07 |
| Hallmark interferon alpha response | 6 | 2.02E–09 | 9.76E–07 |
| GO positive regulation of transcription from RNA polymerase II promoter | 12 | 2.05E–09 | 9.76E–07 |
| GO tissue development | 14 | 2.13E–09 | 9.76E–07 |
| GO sequence-specific DNA binding | 12 | 2.94E–09 | 1.25E–06 |
| GO negative regulation of viral genome replication | 5 | 3.73E–09 | 1.48E–06 |
| GO transcriptional activator activity RNA polymerase II transcription regulatory region sequence-specific binding |
8 | 4.10E–09 | 1.53E–06 |
| GO transcription factor activity RNA polymerase II core promoter-proximal region sequence-specific binding |
8 | 5.62E–09 | 1.97E–06 |
| GO transcriptional activator activity RNA polymerase II core promoter-proximal region sequence-specific binding |
7 | 1.05E–08 | 3.49E–06 |
| GO epithelium development | 11 | 1.38E–08 | 4.18E–06 |
| GO nucleic acid binding transcription factor activity | 12 | 1.47E–08 | 4.18E–06 |
Validation of CRISPRa screening results in HeLa cells.
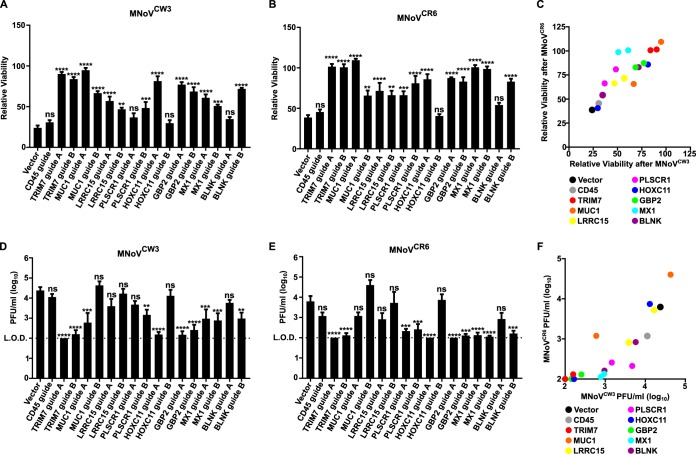
To validate our screening data, we individually introduced two sgRNAs per gene into HeLa CD300lf-dCas9-VP64 cells targeting TRIM7, MUC1, LRRC15, PLSCR1, HOXC11, GBP2, MX1, and BLNK and compared them to cells transduced with an empty pXPR_502 vector or pXPR_502 with a sgRNA targeting CD45. These genes encompass 7 of the top 15 enriched genes after MNoVCW3 challenge. In addition, three of the eight genes showed enrichment in an MNoV-strain-dependent manner (MUC1 and GBP2 for MNoVCW3 and BLNK for MNoVCR6) and thus were ideal for testing strain specificity for potentially antiviral molecules.
Since the screen was designed to enrich for genes that protected cells from virus-induced cytopathic effect (CPE), we first tested the ability of cells to survive MNoV infection. All genes tested had at least one sgRNA show statistical growth advantage over control cells when challenged with either MNoVCW3 or MNoVCR6 (Fig. 3A and B). Furthermore, five and six of the eight genes, respectively, had both sgRNAs provide a survival phenotype when challenged with MNoVCW3 and MNoVCR6 (Fig. 3A and B). We were unable to validate an MNoV strain-specific protective effect for any gene tested (Fig. 3C). These false negatives in our data sets likely arise from multiple factors, including the different selective pressures exerted by the two strains of MNoV. More specifically, MNoVCW3 only required a single challenge of virus, while MNoVCR6 required two successive challenges to elicit enough CPE for our screening efforts. Taken together, these data validate the reproducibility of the CRISPRa screening platform and indicate the potential for false negatives.
FIG 3.
Validation of anti-MNoV genes in human cells. (A and B) HeLa-CD300lf cells expressing dCas9-VP64 and the indicated sgRNAs were challenged with MNoVCW3 (A) or MNoVCR6 (B) at an MOI of 50. Cellular viability was assessed 72 h postinfection (hpi). Values are normalized for each cell line to uninfected control wells to determine the percent viability. The data are shown as means ± the standard errors of the mean (SEM) from three independent experiments, and data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. (C) Graph depicting the correlation between the relative viabilities of cell lines after 72 h of MNoVCW3 (x axis) and MNoVCR6 (y axis) infection. Each dot represents the average relative viability over three independent experiments that the indicated sgRNA-expressing cell line exhibited. (D and E) HeLa-CD300lf cells expressing dCas9-VP64 and the indicated sgRNAs were infected with MNoVCW3 (D) or MNoVCR6 (E) at an MOI of 0.05. Viral production was assessed at 24 hpi by plaque assay. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. (F) Graph depicting the correlation between the viral production 24 h after MNoVCW3 (x axis) and MNoVCR6 (y axis) infection. Each dot represents the average PFU/ml value across over three independent experiments that the indicated sgRNA-expressing cell line exhibited.
We next tested whether overexpression of these genes inhibited production of infectious virions by analyzing the amount of MNoV replication 24 h postinfection (hpi) (Fig. 3D and E). Seven of the eight (MNoVCW3) genes tested and six of the eight (MNoVCR6) genes tested had at least one sgRNA significantly reduce the amount of MNoV produced (Fig. 3D to F). In addition, both sgRNAs for three genes (TRIM7, GBP2, and MX1) blocked the production of MNoVCW3 and MNoVCR6. In summary, these data provide the first systematic overview of the molecules that can inhibit MNoV infection in human cells.
Isoform specificity and evolutionary conservation of the antiviral activity of TRIM7.
Given that the top gene in both screens was TRIM7, we decided to focus our efforts on characterizing the anti-MNoV activity of TRIM7. TRIM7 (also GNIP) is an E3-ligase previously shown to interact with glycogenin and ubiquitinate the AP-1 coactivator RACO1; however, the role of TRIM7 during viral infection has not been explored (29, 30).
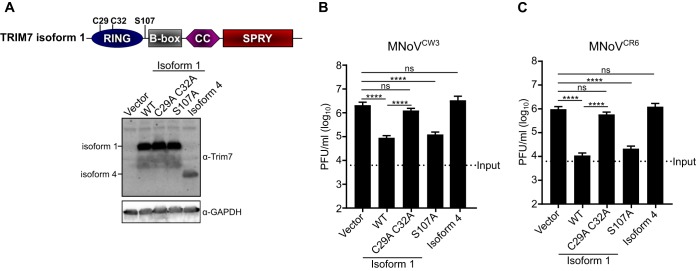
TRIM7 has at least four isoforms in humans, with isoform 1 and isoform 4 containing a functional RING domain, while isoforms 2 and 3 do not. TRIM7 isoforms 1 and 4 are the most commonly studied and display different expression pattern in muscular tissues and cells (31). We generated stable HeLa-CD300lf cells overexpressing TRIM7 isoform 1 or isoform 4 (Fig. 4A). Although the expression of TRIM7 isoform 1 protected HeLa-CD300lf from CPE from both MNoVCW3 and MNoVCR6, TRIM7 isoform 4 provided no protection (Fig. 4B and C). In addition, TRIM7 isoform 1, but not isoform 4, reduced the growth of MNoVCW3 and MNoVCR6 in HeLa-CD300lf cells (Fig. 4D and E). Taken together, these data demonstrate an isoform specificity for the anti-MNoV activity.
FIG 4.
A specific TRIM7 isoform inhibits MNoV replication in HeLa cells. (A, top) Cartoon diagram of TRIM7 isoform 1 and isoform 4. RING, B-box, coiled-coil (CC), and SPRY domains are displayed. Isoform 4 has an alternative, shorter coiled-coil domain and does not encode a SPRY domain. (A, bottom) Representative Western blot for the indicated protein expression in HeLa-CD300lf cells expressing either an empty vector, TRIM7 isoform 1, or TRIM7 isoform 4. (B and C) HeLa-CD300lf cells expressing the indicated constructs infected at an MOI of 50 with MNoVCW3 (B) or MNoVCR6 (C). Cellular viability was assessed at 72 hpi. Values are normalized for each cell line to uninfected control wells to determine the percent viability. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. **, P < 0.01; ***, P < 0.001; ns, not significant. (D and E) HeLa-CD300lf cells expressing the indicated constructs infected at an MOI of 0.05 with MNoVCW3 (D) or MNoVCR6 (E). Viral production was assessed at 24 hpi by plaque assay. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. ****, P < 0.0001; ns, not significant.
We next tested whether the previously described activities of TRIM7 were responsible for its antiviral function. We generated mutants of Trim7 that blocked ubiquitination (C29A and C32A) and phosphorylation by mitogen- and stress-activated protein kinase 1, MSK1 (S107A) (Fig. 5A) (30). HeLa-CD300lf cells expressing TRIM7 isoform 1 or the S107A mutant of isoform 1 inhibited the production of MNoVCW3 and MNoVCR6, while mutations in the RING domain (C29 and C32A) led to a loss of antiviral function (Fig. 5B and C). Therefore, while the phosphorylation of S107 on TRIM7 is dispensable for antiviral activity, the ability to ubiquitinate substrates is essential to inhibit MNoV replication.
FIG 5.
TRIM7 E3-ligase activity but not MSK1 phosphorylation is required to inhibit MNoV replication. (A, top) Cartoon diagram of TRIM7 isoform 1 with the indicated point mutations highlighted. C29 and C32 are required to catalyze ubiquitination, while S107 is phosphorylated by MSK1. (A, bottom) Representative Western blots of the indicated protein expressions in HeLa-CD300lf cells. (B and C) HeLa-CD300lf cells expressing indicated constructs infected at an MOI of 0.05 with MNoVCW3 (D) or MNoVCR6 (E). Viral production was assessed at 24 hpi as measured by plaque assay. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. ****, P < 0.0001; ns, not significant.
Given that Trim proteins have been implicated in species-specific restriction of viruses, we sought to determine whether human Trim7 would function in mouse cells (32). We transduced BV2 cells, a mouse microglial cell line that is susceptible to MNoV infection, with Trim7 expression constructs and generated stable cell lines. Similar to our findings in HeLa-CD300lf cells, the expression of Trim7 isoform 1, but not isoform 4, blocked the ability of MNoV to replicate and induce CPE (Fig. 6A to E). These data demonstrate an isoform specificity for the anti-MNoV mechanism that is conserved across species.
FIG 6.
TRIM7 blocks MNoV replication in mouse BV2 cells postentry. (A) Representative Western blot of human TRIM7 expression in BV2 cells transduced with the indicated constructs. (B and C) BV2 cells expressing the indicated constructs infected at an MOI of 5 with MNoVCW3 (B) or MNoVCR6 (C). Cellular viability was assessed at 24 hpi. Values are normalized for each cell line to the uninfected control wells to determine the percent viability. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. ***, P < 0.001; ****, P < 0.0001; ns, not significant. (D and E) BV2 cells expressing indicated constructs infected at an MOI of 0.05 with MNoVCW3 (D) or MNoVCR6 (E). Viral production was assessed at 12 hpi by plaque assay. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. ****, P < 0.0001; ns, not significant. (F) BV2 expressing the indicated constructs were transfected with viral RNA from MNoVCW3 and harvested at 12 h posttransfection. Viral production was measured by plaque assay. The data are shown as means ± the SEM from three independent experiments, and data were analyzed by one-way ANOVA with Tukey’s multiple-comparison test. ****, P < 0.0001; ns, not significant.
Finally, we determined which step of the viral life cycle was targeted by TRIM7. Since ubiquitination can lead to proteasome-mediated degradation of proteins, we first tested whether the MNoV receptor, CD300lf, was reduced upon TRIM7 overexpression. In HeLa-CD300lf we found no effect of TRIM7 on CD300lf protein levels (Fig. 4A). We next tested the hypothesis that TRIM7 targets a critical step of MNoV entry. Transfection of MNoVCW3 RNA was unable to rescue MNoV production in human TRIM7 isoform 1 expressing BV2 cells, indicating that TRIM7 can target a postentry step of the MNoV life cycle to inhibit viral replication (Fig. 6F).
DISCUSSION
We present here a genome-wide screen to identify genes that can inhibit murine norovirus replication upon overexpression in human cells. Our results provide a resource for those studying norovirus biology and validate CRISPRa screening as an approach to uncover novel antiviral molecules. Since human norovirus studies are limited due to the difficulty in culturing the virus in cell culture, we speculate that our screens with MNoV in may identify evolutionary conserved genes that may broadly inhibit NoV replication. Such genes might be targeted through genetic or chemical perturbagens to enhance HNoV replication in cell culture systems or to generate therapeutic effects.
With the discovery that Cas9 binding to DNA does not require nuclease activity, several new technologies have emerged to synthetically modulate mammalian genomes with unprecedented precision and scale (33). By leveraging the cytotoxicity of lytic viruses, these pooled screening approaches are ideal to identify genes that alter viral infection. While most investigations using CRISPR/Cas9 genome-wide screens have focused on determining host factors that facilitate viral infection, our results highlight the utility of CRISPRa to identify antiviral molecules.
Genes identified in these screens exhibited enrichment in viral and immune related genes not previously appreciated to have anti-MNoV activity (Fig. 2A). Given that the tropism of both acute and persistent strains of MNoV are dictated by interferons, these data provide the first insight into potential ISGs that mediate the viral restriction of MNoV (19, 20, 22). It will be important to determine whether these ISGs contribute to determining norovirus tropism in vivo.
Surprisingly, in addition to the ISGs, a number of genes not appreciated to have immune functions were identified as possessing anti-MNoV activity (Fig. 2A). These represent unique opportunities to identify novel norovirus vulnerabilities and to determine whether these genes possess antiviral activity against other viruses that may explain tropism or be exploited for therapeutic potential. One of these genes, TRIM7, inhibited MNoV replication after viral entry (Fig. 6F). Since ubiquitination can have diverse effects on protein stability and function, it will be essential to determine the substrate and type of lysine linkage in order to understand the mechanism of viral restriction (34). In addition, we mapped the antiviral function of TRIM7 to a specific isoform, while a closely related isoform had no detectable anti-MNoV activity (Fig. 4A to E). The only differences between these isoforms are within the protein-protein interaction coiled-coil domain SPRY domains. Because these molecules can be differentially and coexpressed in different cellular contexts and TRIM proteins are capable of homodimerization, it will be important to determine whether isoform 4 can regulate the antiviral activity or the normal physiological functions of isoform1 (35). Furthermore, since traditional cDNA-based screens require the scientist to select a specific isoform, CRISPRa approaches enable complex transcriptional variance to be reduced and encompassed by sgRNAs, which is highlighted by the specificity of anti-MNoV by TRIM7 isoforms (14).
In conclusion, our data provide the first systematic insight into genes that can restrict norovirus replication when overexpressed and provide a methodological approach to identify novel antiviral molecules using CRISPRa technology that may be broadly applicable to other viral systems.
MATERIALS AND METHODS
Cell culture.
BV2 cells (BV-2 cells were provided by Yuanan Lu, University of Hawaii at Manoa), 293T (American Type Culture Collection [ATCC]) cells, and HeLa cells (ATCC) were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum and 1% HEPES. HeLa cells were transduced with pCDH-MSCV-CD300lf-T2A-hygromycin and selected with hygromycin for 2 weeks prior to sorting CD300lf high-expressing cells to generate HeLa-CD300lf-hygromycin cells. For BV2 cells, 4 µg/ml of puromycin (Sigma-Aldrich) was added where appropriate. For HeLa cells, 1 µg/ml puromycin, 5 µg/ml blasticidin, and 350 µg/ml hygromycin (Invitrogen) were added as appropriate.
Lentivirus was generated by transfecting lentiviral vectors with packaging vector (psPAX2) and pseudotyping vector (pCMV-VSV-G) into 293Ts using TransIT-LT1 (Mirus). At 48 h posttransfection, supernatants were collected, filtered through a 0.45-μm-pore size filter (Millipore), and added to the indicated cells. After 48 h, the cells were selected with the appropriate antibiotic.
Plasmids.
Guide constructs were cloned into pxpr_502 (Addgene, catalog no. 96923). A complete list of guide sequences used in this study can be found in the supplemental material. Human TRIM7 isoform 1 (NP_976038.1) and isoform 4 (NP_203128.1) were codon optimized and cloned into pCDH-MCS-T2A-Puro-MSCV (Systems Biosciences, CD522A-1). Codon-optimized cDNA sequences for the constructs can be found in the supplemental material. All plasmids were sequenced verified prior to use.
Transcriptional activation reporter assays.
HeLa-CD300lf cells were transduced with pLenti-dCas9-VP64-BLAST (14). Transcriptional activity was assessed in these cells by transducing either parental or dCas9-VP64 cells with pxpr_502 lentiviruses targeting CD4. Cells were transduced for 2 days and subsequently selected for 7 days with puromycin, and the frequency of CD4 expression was assessed by flow cytometry.
HeLa CRISPRa screen.
HeLa-CD300lf cells expressing dCas9-VP64 were transduced with SetA and SetB of the Caprano CRISPRa library at an MOI of 0.3. Each Calabrese pool was delivered by lentiviral transduction of 1.2 × 108 HeLa cells at an MOI of ∼0.3. This equates to 3.6 × 107 transduced cells, which is sufficient for the integration of each sgRNA at least 500 independent times. At 2 days postransduction, puromycin was added to the media, and transduced cells were selected for a total of 7 days. A total of 5 × 106 HeLa CRISPRa cells were seeded in a T-175 tissue culture flask, and each experimental condition was evalauted in duplicate with six independent flasks per replicate (3 × 107 cells per replicate). Cells were infected with either MNoVCW3 or MNoVCR6 at an MOI of 50 at the time of seeding. At 2 days after infection, mock-infected cells were harvested for genomic DNA extraction. At this time point, roughly 85% of the MNoVCW3-infected cells displayed a CPE, whereas 35 to 50% MNoVCR6-infected displayed a CPE, and for both sets of conditions the cells were washed twice with phosphate-buffered saline (PBS), and fresh medium was added. After an additional 2 days of viral challenge, MNoVCR6-challenged cells had roughly 25% viable cells and thus were washed, trypsinized, collected, counted, replated at 5 × 106 cells/flask, and rechallenged. The MNoVCW3-challenged cells had <10% viable at this time point, and thus only a single challenge was performed. Cells were harvested after the second challenge with MNoVCR6 and after the single challenge with MNoVCW3 when they started to form discernible, stable colonies, i.e., between 7 and 14 days after the initial challenge. All flasks from a replicate were harvested together. Genomic DNA was isolated from surviving cells using a QIAamp DNA minikit according to the manufacturer’s instructions (Qiagen).
CRISPR screen sequencing and analysis.
Illumina sequencing and STARS analysis were performed as described previously (5). Briefly, genomic DNA was aliquoted into multiple wells of a 96-well plate with up to 10 µg of DNA in 50-µl total volume. A PCR Mastermix consisting of ExTaq DNA polymerase (Clontech), ExTaq buffer, deoxynucleoside triphosphate, P5 stagger primer, and water was generated. PCR master mix (40 µl) and 10 µl of a barcoded primer were added to each well containing 50 µl of DNA. Samples were PCR amplified as follows: 95°C for 1 min; followed by 28 cycles of 94°C for 30 s, 52.5°C for 30 s, and 72°C for 30 s; followed in turn by a final 10 min at 72°C. PCR product was purified with Agencourt AMPure XP SPRI beads according to the manufacturer’s protocol (Beckman Coulter). Samples were sequenced on an Illumina HiSeq 2000. Barcodes in the P7 primer were deconvoluted, and the sgRNA sequence was mapped to a reference file of sgRNAs in the Caprano or Calabrese library. To normalize for different numbers of reads per condition, read counts per sgRNA were normalized to 107 total reads per sample. This normalized value was then log2 transformed. sgRNAs that were not sequenced were arbitrarily assigned a read count of 1. Replicates were then averaged together, and sgRNA frequencies were then analyzed using STARS software (http://www.broadinstitute.org/rnai/public/software/index) (25). STARS computes a score for each gene of rank-ordered sgRNA hits that was above 10% of total sequenced sgRNAs. A STAR score was assigned to genes that had a sgRNA score above this threshold in either of the two independent pools.
For GSEA, screen hits were queried at the BROAD GSEA portal (https://software.broadinstitute.org/gsea/index.jsp). Only the top 20 gene set names with FDRs of <0.05 are shown in the tables.
MNoV assays.
MNoVCW3 (GenBank accession no. EF014462.1) and MNoVCR6 (GenBank accession no. JQ237823) were generated by transfecting MNoV cDNA clones into 293T cells as described previously (17). For cell survival assays, 104 of the indicated HeLa cells were seeded in wells of a white-walled 96-well plate (Corning) with indicated MNoV strains at an MOI of 50. At 72 h postinfection, viability was measured using CellTiter-Glo (Promega) according to the manufacturer’s protocol. The viability for each cell line was normalized to a mock-infected sample. Each condition was assessed at least in triplicate in three independent experiments. A similar experimental setup was utilized for BV2 cells, except that an MOI of 5.0 was used and the viability was assessed after 24 h of infection.
For MNoV growth curves, 5 × 104 cells were infected in suspension with MNoVCW3 or MNoVCR6 at an MOI of 0.05 in the wells of a 96-well plate. Plates were frozen at 12 or 24 h postinfection at −80°C for BV2 and HeLa cells, respectively. Total cell lysate was used in subsequent plaque assays, as previously described (36). All infections were performed in triplicate in each of at least three independent experiments.
Viral RNA (vRNA) from MNoVCW3 was extracted from cell-free viral preparations using TRIzol (Invitrogen) according to the manufacturer’s instructions. Purified vRNA was tested via plaque assay to ensure complete inactivation of MNoV. Then, 4 µg of vRNA was transfected into BV2 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Transfected cells were frozen 12 h later. Each condition was assayed by plaque assay in duplicate in three independent experiments.
Antibodies and Western blotting.
The following antibodies were used for flow cytometry or Western blotting: rabbit α-TRIM7 (Sigma), mouse α-GAPDH-HRP (Sigma), Armenian hamster α-CD300lf clone 3F6 (Genentech), anti-human CD4 (Invitrogen, clone S3.5), and anti-mouse CD4 (BioLegend, clone RM4-5).
Cells were placed on ice, washed once in PBS, prior to lysis in cold radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% IGEPAL, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) with HALT protease and phosphatase inhibitor cocktail (Sigma). Lysates were clarified by centrifugation prior to resolution on SDS-PAGE gels (Bio-Rad) and transfer to polyvinylidene difluoride membranes.
Data availability.
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
ACKNOWLEDGMENTS
We thank Megan Baldridge and Craig Wilen for helpful conversations and review of the manuscript.
Washington University School of Medicine holds patents on several aspects of murine norovirus. These patents have been licensed, generating income for the university and the inventors, including H.W.V.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01324-18.
REFERENCES
- 1.Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. 2014. Advances in norovirus biology. Cell Host Microbe 15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartsch SM, Lopman BA, Hall AJ, Parashar UD, Lee BY. 2012. The potential economic value of a human norovirus vaccine for the United States. Vaccine 30:7097–7104. doi: 10.1016/j.vaccine.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353(6306):1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orchard RC, Wilen CB, Doench JG, Baldridge MT, McCune BT, Lee YC, Lee S, Pruett-Miller SM, Nelson CA, Fremont DH, Virgin HW. 2016. Discovery of a proteinaceous cellular receptor for a norovirus. Science 353:933–936. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haga K, Fujimoto A, Takai-Todaka R, Miki M, Doan YH, Murakami K, Yokoyama M, Murata K, Nakanishi A, Katayama K. 2016. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc Natl Acad Sci U S A 113:E6248–E6255. doi: 10.1073/pnas.1605575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, Schumann K, Yu H, Krupzcak KM, Garcia-Beltran W, Piechocka-Trocha A, Krogan NJ, Marson A, Sabatini DM, Lander ES, Hacohen N, Walker BD. 2017. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet 49:193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, Diamond MS. 2016. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding S, Diep J, Feng N, Ren L, Li B, Ooi YS, Wang X, Brulois KF, Yasukawa LL, Li X, Kuo CJ, Solomon DA, Carette JE, Greenberg HB. 2018. STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat Commun 9:1485. doi: 10.1038/s41467-018-03782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, Lee Y, Andrade J, tenOever B, Manicassamy B. 2018. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep 23:596–607. doi: 10.1016/j.celrep.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orchard RC, Sullender ME, Dunlap BF, Balce DR, Doench JG, Virgin HW. 2018. Identification of antinorovirus genes in human cells using genome-wide CRISPR activation screening. bioRxiv 10.1101/350090. [DOI] [PMC free article] [PubMed]
- 16.Nice TJ, Strong DW, McCune BT, Pohl CS, Virgin HW. 2013. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J Virol 87:327–334. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strong DW, Thackray LB, Smith TJ, Virgin HW. 2012. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J Virol 86:2950–2958. doi: 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, Hykes BL, McAllaster MR, Balce DR, Feehley T, Brestoff JR, Hickey CA, Yokoyama CC, Wang YT, MacDuff DA, Kreamalmayer D, Howitt MR, Neil JA, Cadwell K, Allen PM, Handley SA, van Lookeren Campagne M, Baldridge MT, Virgin HW. 2018. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360:204–208. doi: 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 2015. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nice TJ, Osborne LC, Tomov VT, Artis D, Wherry EJ, Virgin HW. 2016. Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog 12:e1005684. doi: 10.1371/journal.ppat.1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. 2015. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 23.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kernbauer E, Ding Y, Cadwell K. 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature 516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biering SB, Choi J, Halstrom RA, Brown HM, Beatty WL, Lee S, McCune BT, Dominici E, Williams LE, Orchard RC, Wilen CB, Yamamoto M, Coers J, Taylor GA, Hwang S. 2017. Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe 22:74–85. doi: 10.1016/j.chom.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skurat AV, Dietrich AD, Zhai L, Roach PJ. 2002. GNIP, a novel protein that binds and activates glycogenin, the self-glucosylating initiator of glycogen biosynthesis. J Biol Chem 277:19331–19338. doi: 10.1074/jbc.M201190200. [DOI] [PubMed] [Google Scholar]
- 30.Chakraborty A, Diefenbacher ME, Mylona A, Kassel O, Behrens A. 2015. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signaling. Nat Commun 6:6782. doi: 10.1038/ncomms7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montori-Grau M, Pedreira-Casahuga R, Boyer-Díaz Z, Lassot I, García-Martínez C, Orozco A, Cebrià J, Osorio-Conles O, Chacón MR, Vendrell J, Vázquez-Carrera M, Desagher S, Jiménez-Chillarón JC, Gómez-Foix AM. 2018. GNIP1 E3 ubiquitin ligase is a novel player in regulating glycogen metabolism in skeletal muscle. Metabolism 83:177–187. [DOI] [PubMed] [Google Scholar]
- 32.van Gent M, Sparrer KMJ, Gack MU. 2018. TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol 5:385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doench JG. 2018. Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet 19:67–80. doi: 10.1038/nrg.2017.97. [DOI] [PubMed] [Google Scholar]
- 34.Yau R, Rape M. 2016. The increasing complexity of the ubiquitin code. Nat Cell Biol 18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 35.Esposito D, Koliopoulos MG, Rittinger K. 2017. Structural determinants of TRIM protein function. Biochem Soc Trans 45:183–191. doi: 10.1042/BST20160325. [DOI] [PubMed] [Google Scholar]
- 36.Hwang S, Alhatlani B, Arias A, Caddy SL, Christodoulou C, Cunha JB, Emmott E, Gonzalez-Hernandez M, Kolawole A, Lu J, Rippinger C, Sorgeloos F, Thorne L, Vashist S, Goodfellow I, Wobus CE. 2014. Murine norovirus: propagation, quantification, and genetic manipulation. Curr Protoc Microbiol 33:1–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JD. 2018. Up, down, and out: optimized libraries for CRISPRa, CRISPRi, and CRISPR-knockout genetic screens. bioRxiv doi: 10.1101/356626. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.