Biologics based on recombinant AAVs (rAAVs) are increasingly becoming attractive human gene delivery vehicles, especially after the approval of Glybera in Europe and Luxturna in the United States. However, preexisting neutralizing antibodies against the AAV capsids in a large percentage of the human population limit wide-spread utilization of these vectors. To circumvent this problem, stealth vectors must be generated that are undetectable by these antibodies. This study details the high-resolution characterization of a new antigenic region on AAV5, a vector being developed for numerous delivery applications. The structure of AAV5 complexed with HL2476, a novel antibody, was determined by cryo-EM to 3.1-Å resolution. The resolution of the density map enabled the identification of interacting residues between capsid and antibody and the determinants of neutralization. Thus, the information obtained from this study can facilitate the generation of host immune escape vectors.
KEYWORDS: AAV5, adeno-associated virus, antibody epitope, cryo-EM, electron microscopy, gene therapy vectors, monoclonal antibodies, neutralizing antibodies
ABSTRACT
Adeno-associated virus serotype 5 (AAV5) is being developed as a gene delivery vector for several diseases, including hemophilia and Huntington’s disease, and has a demonstrated efficient transduction in liver, lung, skeletal muscle, and the central nervous system. One limitation of AAV gene delivery is preexisting neutralizing antibodies, which present a significant challenge for vector effectiveness in therapeutic applications. Here, we report the cryo-electron microscopy (cryo-EM) and image-reconstructed structure of AAV5 in complex with a newly generated monoclonal antibody, HL2476, at 3.1-Å resolution. Unlike other available anti-AAV5 capsid antibodies, ADK5a and ADK5b, with epitopes surrounding the 5-fold channel of the capsid, HL2476 binds to the 3-fold protrusions. To elucidate the capsid-antibody interactions, the heavy and light chains were sequenced and their coordinates, along with the AAV5 viral protein, assigned to the density map. The high resolution of the complex enabled the identification of interacting residues at the 3-fold protrusions of the capsid, including R483, which forms two hydrogen bonds with the light chain of HL2476. A panel of AAV5 variants was generated and analyzed by native dot immunoblot and transduction assays. This identified variants with antibody escape phenotypes that maintain infectivity.
IMPORTANCE Biologics based on recombinant AAVs (rAAVs) are increasingly becoming attractive human gene delivery vehicles, especially after the approval of Glybera in Europe and Luxturna in the United States. However, preexisting neutralizing antibodies against the AAV capsids in a large percentage of the human population limit wide-spread utilization of these vectors. To circumvent this problem, stealth vectors must be generated that are undetectable by these antibodies. This study details the high-resolution characterization of a new antigenic region on AAV5, a vector being developed for numerous delivery applications. The structure of AAV5 complexed with HL2476, a novel antibody, was determined by cryo-EM to 3.1-Å resolution. The resolution of the density map enabled the identification of interacting residues between capsid and antibody and the determinants of neutralization. Thus, the information obtained from this study can facilitate the generation of host immune escape vectors.
INTRODUCTION
Adeno-associated viruses (AAVs) are nonpathogenic, single-stranded DNA viruses of the Parvoviridae utilized as vectors for gene delivery applications. These viruses are composed of nonenveloped capsids with T=1 icosahedral symmetry and a diameter of ∼260 Å (1). Currently, 13 human and nonhuman primate AAV serotypes and numerous additional isolates from different species have been described (2). The amino acid sequence of different AAV capsids can vary by up to ∼50% (3), enabling them to bind different host cell receptors, resulting in alternative cell and tissue tropisms. AAV serotype 5 (AAV5) represents one of the most divergent of the AAVs and utilizes α2,3-linked sialic acid-containing glycans as primary host cell receptors (4, 12, 47). However, the overall composition of the viral capsid is similar among all AAVs. The capsids are assembled from 60 subunits of three overlapping capsid viral proteins (VPs), VP1 (∼87 kDa), VP2 (∼73 kDa), and VP3 (∼61 kDa), in a predicted 1:1:10 ratio (6). The individual VPs are expressed within the same open reading frame and share a C terminus. VP1 and VP2 represent N-terminal extended forms of VP3. In addition, VP1 possesses ∼137 amino acids N terminal of VP2 that contain a phospholipase A2 (PLA2) domain, which is required for AAV infectivity (7). The 60 VPs create the characteristic features of the AAV capsids with cylindrical channels at the 5-fold axes, protrusions surrounding the 3-fold axes, and depressions at the 2-fold axes and surrounding the 5-fold channels that are separated by “walls,” termed 2/5-fold walls. Variable regions (VRs) I to IX (VR-I to VR-IX), defined for the AAVs based on sequence and structure differences clustered at or around these capsid features, result in phenotypic differences, such as in receptor attachment and antigenicity, between the AAV serotypes (5, 8–15).
The ability to package recombinant DNA into AAV capsids and utilize the capsids’ natural ability to deliver and express therapeutic genes in the desired target cells has made recombinant AAV vectors (rAAVs) one of the most successful tools to restore the function of a defective gene for the treatment of monogenic disorders. However, a major hurdle for the application of AAV vectors in gene therapy is preexisting neutralizing antibodies (NAbs) in a large percentage of the human population (16, 17). The prevalence of anti-AAV antibodies among healthy adults ranges from 30% to 80% depending on the AAV serotype (16, 17). In the case of AAV5, ∼40% of a selected cohort displayed antibodies against the viral capsid (16). These antibodies originate from prior exposure to natural AAVs and primarily target the capsid, which could lead to vector inactivation and treatment inefficiency (18, 19). Strategies to address this issue include the coadministration of immunosuppressive agents (reviewed in reference 20), preinjection of empty capsids to consume reactive antibodies (reviewed in reference 20), or the utilization of structurally optimized capsids to evade neutralizing antibodies (reviewed in references 21 and 22). Towards the latter approach, the antigenic regions of AAV capsids need to be identified and subsequently modified by rational capsid-surface engineering or directed evolution (reviewed in references 21 and 22). In order to identify the antigenic regions, cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction of AAV capsids in complex with antibodies has been used. This approach has led to the mapping of the binding sites of several monoclonal antibodies (MAbs) to multiple AAV serotypes, e.g., ADK1a, ADK1b, 4E4, and 5H7 to AAV1 (9, 11), A20 and C37-B to AAV2 (9, 23), ADK5a, ADK5b, and 3C5 to AAV5 (9, 11), ADK6 to AAV6 (13), and ADK8 to AAV8 (10). Most of these antibodies efficiently neutralize infectivity by the respective AAV vector. The resolutions of the reconstructed capsid-antibody complexes ranged from 6.7 to 23 Å, allowing the mapping of a “footprint” indicating a cluster of capsid surface residues that are potentially involved in the capsid-antibody interaction. In the case of AAV6 and AAV8, single-residue substitutions or loop swap mutations successfully restored infectivity in the presence of the neutralizing antibodies (10, 13).
This study characterizes a new antigenic region on the highly divergent AAV5 capsid at atomic resolution. The previously reported AAV5-antibody complexes, AAV5-ADK5a and AAV5-ADK5b, occupy similar epitopes around the 5-fold symmetry axis (11). The third reported AAV5-antibody complex, AAV5-3C5, bound at the 2/5-fold wall in a tangential orientation that covers a large fraction of the capsid surface, except the 3-fold axis (9). In order to characterize the antigenic potential of the 3-fold region, a new anti-AAV5 capsid antibody directed at the 3-fold axis, named HL2476, was generated by blocking the 5-fold region with the AVB ligand during the antibody selection process. The AAV5-HL2476 complex structure was determined by cryo-EM and 3D image reconstruction to 3.1-Å resolution. HL2476 binds on the protrusions surrounding the icosahedral 3-fold axes. Furthermore, the high resolution of the complex structure enabled direct identification of residues interacting with the HL2476 antibody. Mutational analysis confirmed the role of specific AAV5 residues as contacts for HL2476. This analysis also points to a mechanism of AAV5 neutralization by HL2476 that involves partial blockage of its’ cell surface glycan receptor attachment site. This complex structure represents the highest-resolution antibody complex structure determined by cryo-EM to date and a critical milestone in the development of cryo-EM as a structural biology tool, with new possibilities for the characterization of many functional regions of the AAV capsids via capsid-ligand complex visualization.
RESULTS
Novel anti-AAV5 capsid antibody HL2476 targets the 3-fold capsid region.
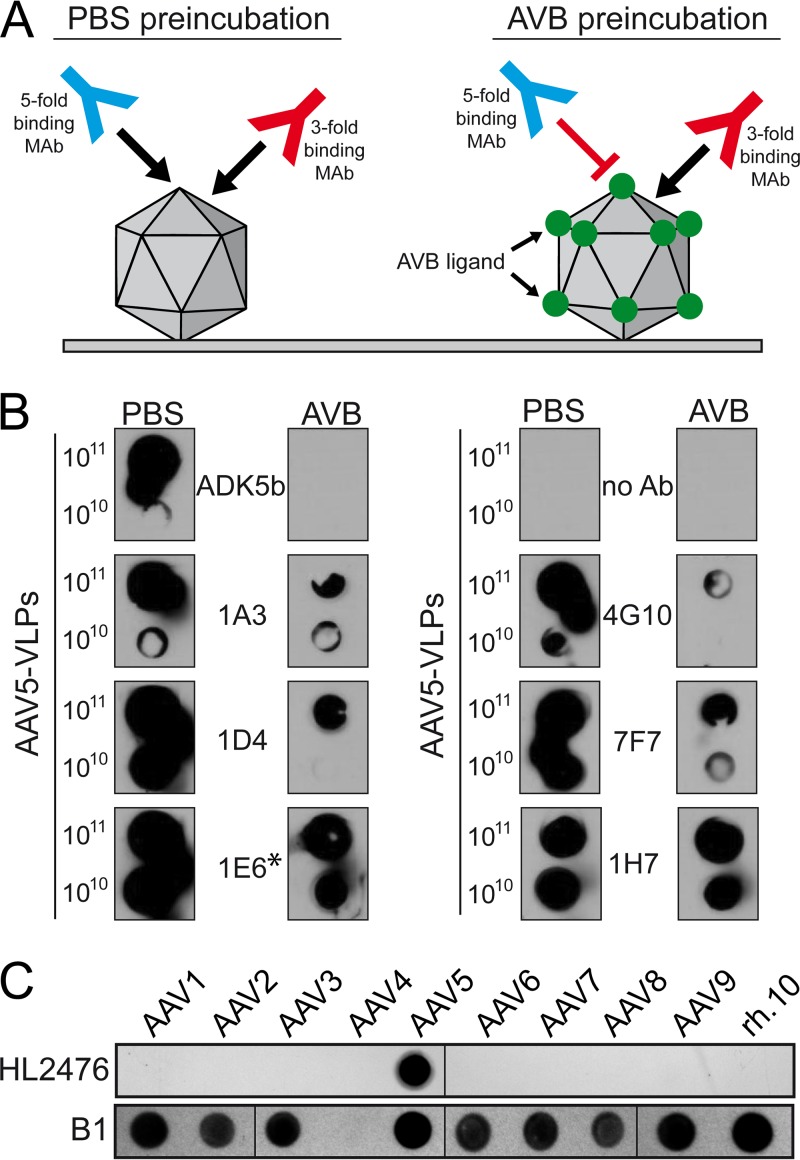
A novel MAb directed against the AAV5 3-fold capsid region was generated by immunization of mice with AAV5 VLPs and their splenocytes used to generate hybridoma cell clones as previously described for AAV8 and AAV9 (24). To select for a hybridoma cell line expressing a MAb targeting the 3-fold region of the AAV5 capsid, a native dot immunoblot analysis was performed with AAV5 capsids pretreated with either phosphate-buffered saline (PBS) or the AVB nanobody prior to addition of the hybridoma supernatant (Fig. 1A). While PBS treatment allowed the binding of all possible antibodies to the AAV5 capsid, preloading with AVB blocked access to the capsid surface area around the 5-fold region, extending partially to the 2/5-fold wall (based on a structurally mapped AVB footprint; data not shown) and, thus, should only allow binding of antibodies to the 3-fold region (Fig. 1A). Consequently, signals restricted to PBS-treated capsids indicated antibodies binding to the 5-fold region, such as for ADK5b and likely 4G10 (Fig. 1B). Similar reactivity to AAV5 capsids under both conditions indicated antibodies binding to the 3-fold region, as in 1E6 and 1H7. Hybridoma cell line 1E6 was selected and the MAb derived from it termed HL2476. HL2476 produced from the hybridoma cell line was determined to belong to the IgG2b κ subclass.
FIG 1.
Analysis of HL2476. (A) Schematic of MAb selection. (B) Native dot immunoblot assay screening of multiple monoclonal hybdridoma supernatants. The number of VLPs loaded onto the membrane is indicated on the left. AAV5 VLPs were incubated with PBS or with the AVB ligand, binding to the 5-fold region, prior to incubation with the hybridoma supernatants. AVB blocks the successive binding of antibodies binding in this region of the capsid, whereas antibodies targeting other capsid regions remain unaffected by AVB. The asterisk indicates the hybridoma sample used for monoclonization, which here is termed HL2476. (C) Native dot immunoblot assay analyzing the specificity of the HL2476 antibody. For this assay, 109 genome-containing particles of the indicated AAV serotype was loaded onto the membrane and reacted with HL2476. For the lower panel the same amounts of heat-denatured AAVs were used and analyzed with MAb B1. The AAV4 capsid proteins do not react with MAb B1.
HL2476 exhibits specificity for the AAV5 capsid.
The specificity of HL2476 for AAV5 was confirmed by probing against the capsids of serotypes AAV1-9 and AAVrh.10 by a native dot immunoblot assay. The antibody was not cross-reactive to any other AAV serotype tested (Fig. 1C). As a positive control, a native dot immunoblot assay conducted with denatured capsids and probed with the B1 antibody, which recognizes an epitope at the C terminus of denatured AAV VPs (25), reacted against all serotypes tested except AAV4 (Fig. 1C). The latter serotype does not possess the B1 epitope (8). Hence, these observations confirm the specificity of HL2476 to the AAV5 capsid.
HL2476 binds to the 3-fold region of the AAV5 capsid and orders the 5-fold channel.
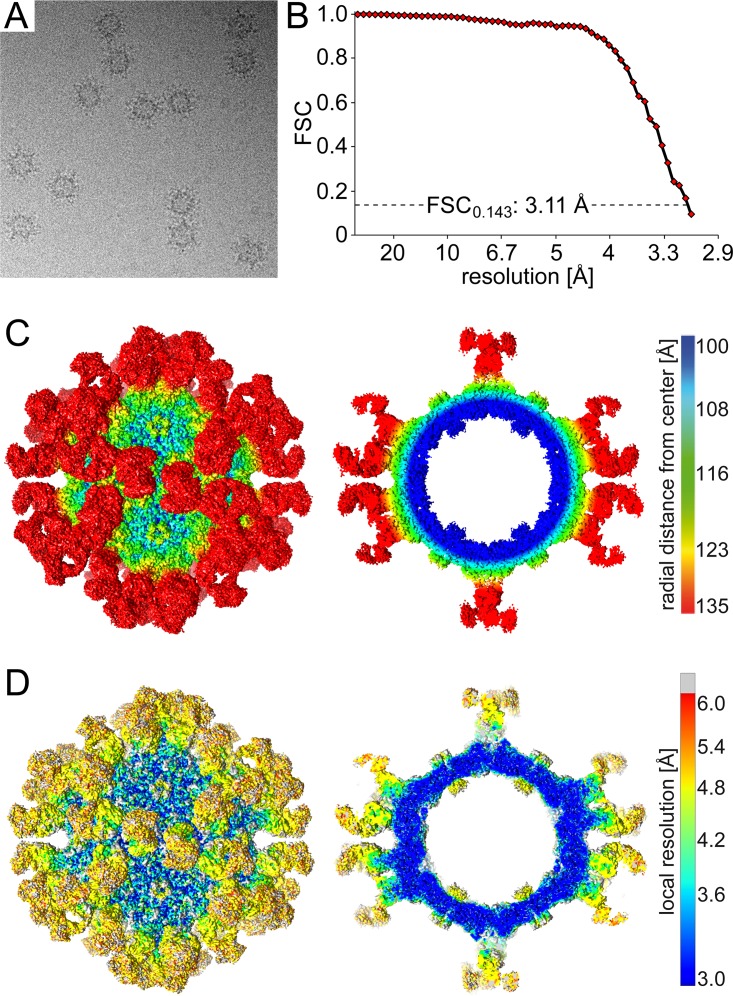
Cryo-EM and image reconstruction was used to locate the binding site of the HL2476 antibody on the AAV5 capsid. Antigen binding fragments (Fab) of HL2476 generated by papain cleavage of purified HL2476 IgGs were complexed with purified AAV5 VLPs at a molecular ratio of 120 Fabs per capsid, corresponding to two Fabs per potential VP binding site. Cryo-EM data collected on the vitrified complex sample generated 1,140 individual particles with Fab decoration (Fig. 2A). The resolution of the virus-antibody structure, estimated using a Fourier shell correlation (FSC; 0.143) plot, was 3.11 Å (Fig. 2B). Density for the bound Fabs was clearly visible and extended from the AAV5 capsid surface at protrusions surrounding the 3-fold axes (Fig. 2C and D). In contrast to previous AAV5-Fab complex structures, the 5-fold region is open on the capsid surface. However, unexpectedly, a cross-section of the map revealed additional density under the 5-fold channel, extending it toward the interior of the capsid (Fig. 2C and D). Calculation of the local resolution of the complex density map showed the core AAV5 capsid and Fab’s complementarity-determining regions (CDRs) to have the highest overall resolution, while the Fab’s constant region is less ordered (Fig. 2D). The “basket” under the 5-fold channel displayed lower local resolution than the rest of the capsid (Fig. 2D).
FIG 2.
Cryo-EM and 3D image reconstruction of the AAV5-HL2476 Fab complex. (A) Example micrograph of the AAV5-HL2476 Fab complex. (B) Fourier shell correlation (FSC) plotted against resolution for the reconstructed AAV5-HL2476 structure. The resolution of the cryo-reconstructed map was estimated to a resolution of 3.11 Å based on an FSC of 0.143. (C) Surface density map of the cryo-reconstructed AAV5-HL2476 Fab complex structure. The view is down the icosahedral 2-fold axis, and the map is contoured at a sigma (σ) threshold level of 1.0. The reconstructed map is colored according to radial distance from the particle center (blue to red), as indicated by the scale bar (right). This figure was generated using UCSF-Chimera (38). (D) Surface density map, as shown in panel C, colored by local resolution calculated using ResMap (39), as indicated by the scale bar (right).
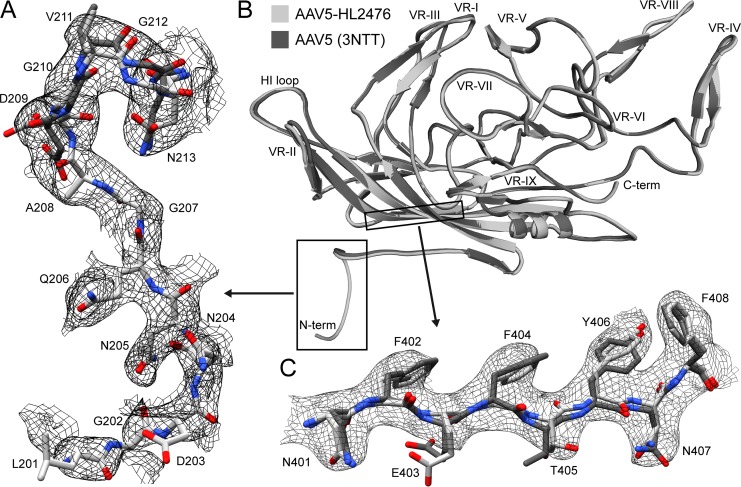
The high resolution of the AAV5-HL2476 complex structure map enabled the side-chain assignment of AAV5 VP3 amino acids starting from N-terminal residue 205 to 724, the C-terminal residue. In addition, the main chain of residues 201 to 204 was also interpretable. Residues 201 to 208 represent 8 and 6 additional ordered residues, respectively, compared to the AAV5 crystal structure (Fig. 3A) ordered from residues 209 to 724 (PDB entry 3NTT) and cryo-EM structure, determined to 2.9-Å resolution (not shown), ordered from residue 207 to 724. The VP3 N terminus of the AAVs is situated under the 5-fold channel, and these additional residues form the interior channel extension described above. As with other AAV structures, the VP1u and VP1/2 common sequences were disordered or averaged out by applying strict icosahedral symmetry (5, 8, 26). The final AAV5 VP3 60mer capsid model was refined against the cryo-reconstructed complex map, yielding a correlation coefficient (CC) of 0.89 with good geometry (Table 1). Except for the additional N terminus, the ordered VP3 structure remained largely unchanged in the complex structure compared to the original crystal structure, with an overall root mean square deviations of 0.48 Å (Fig. 3).
FIG 3.
Comparison of the AAV5 cryo-EM and X-ray structures (PBD entry 3NTT). (A) Atomic model of the N termini from both structures (residue range, amino acids 201/209 to 213) within the AAV5-HL2476 cryo-EM map at a sigma level of 1.2. (B) Superposition of a VP monomer from the cryo-EM (light gray) and X-ray structure (dark gray). The N and C termini, as well as the variable regions (VRs), are indicated. (C) Atomic models of β-strand G within the AAV5-HL2476 cryo-EM map at a sigma level of 2.5. Panels A and C are shown as zoomed-in views from panel B. This figure was generated with UCSF-Chimera (38) (A and C) and PyMol (48) (B).
TABLE 1.
Summary of data collection, image-processing parameters, and refinement statistics
| Parameter | Value for: |
|
|---|---|---|
| AAV5-HL2476 complex | AAV5 alone | |
| Total number of micrographs | 254 | |
| Defocus range (µm) | 1.16-3.48 | |
| Electron dose (e−/Å2) | 75 | |
| Frames/micrograph | 50 | |
| Pixel size (Å/pixel) | 1.05 | |
| Starting no. of particles | 1,140 | |
| No. of particles used for final map | 1,026 | |
| B-factor used for final map (Å2) | −25 | |
| Resolution of final map (Å) | 3.11 | |
| PHENIX model refinement statistics | ||
| Residue range | 205-724 (AAV5) 21-234 (LC) 20-236 (HC) | 205-724 |
| Map CC | 0.706 | 0.885 |
| RMSD (bonds) (Å) | 0.01 | 0.01 |
| RMSD (angles) (Å) | 0.91 | 0.85 |
| All-atom clash score | 13.37 | 10.76 |
| Ramachandran plot | ||
| Favored (%) | 94.1 | 96.7 |
| Allowed (%) | 5.2 | 3.3 |
| Outliers (%) | 0.7 | 0 |
| Rotamer outliers (%) | 0.12 | 0 |
| C-β deviations | 0 | 0 |
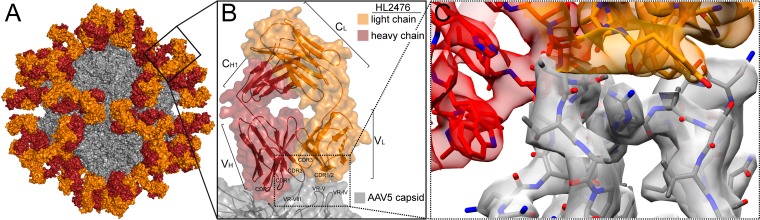
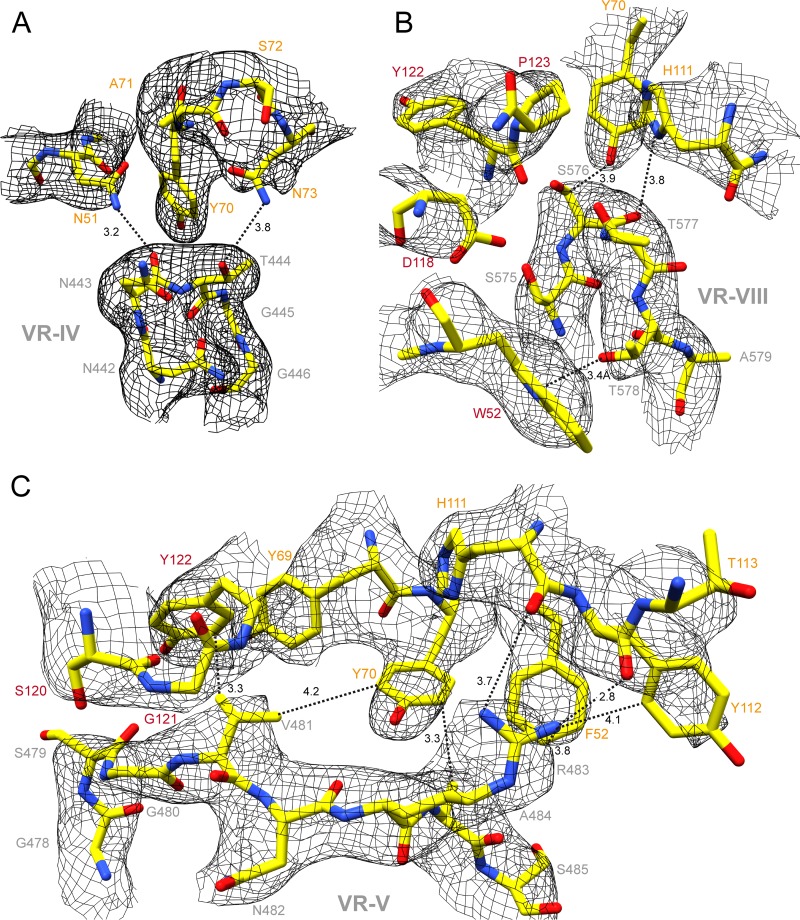
The amino acid sequence of the HL2476 heavy and light chains, obtained from the cDNA isolated from the hybridoma cell line, was used to build a 3D model for interpreting the density map. Docking of the HL2476 model showed that the light chain was bound at the apex of the AAV5 3-fold protrusions, whereas the heavy chain interacts with residues on the side of each of the protrusions (Fig. 4, Table 2). The apex of each 3-fold protrusion is assembled from two AAV5 VP3 monomers (Table 2, VP3A and VP3I in the context of a 60mer) contributing VRs to the binding footprint. The VP3A interactions occlude a surface area of 470 Å2 consisting of residues from VR-V (R471, G474, N476, S479, G480, V481, N482, R483, A484, S485), VR-VI (N519, T520), and VR-VII (T563, N564, V565, T568). In addition to the surface area coverage, of note are six stabilizing hydrogen bonds between the VP and HL2476 atoms (Table 2). VP3I also contributes a significant binding surface of 490 Å2, consisting of residues from VR-IV (N443, T444, G445) and VR-VIII (T571, N572, N573, Q574, S576, T577, T579), and eight hydrogen bonds. A third VP3 monomer contributing to the 2/5-fold wall, VP3G (in a 60mer), only makes a few surface contacts, 80 Å2, from residues in VR-1 (G257, S258, N259), VR-III (N374), and VR-V (F489, R494, E496, G499, S501), with no detectable hydrogen bond interaction. The CDR1, CDR2, and CDR3 of the light chain of HL2476 interacts with VR-IV, VR-V, and VR-VIII, while CDR1, CDR2, and CDR3 of the heavy chain interacts with VR-V, VR-VI, VR-VII, and VR-VIII (Fig. 4, Table 2). The amino acid side chain densities were well defined in the CDRs of the Fab light and heavy chains (VL and VH) and in the conserved antiparallel β-strands. The constant regions (CL and CH1) of HL2476 were interpreted using coordinates from other Fab structures, PDB entry 5EZI for the light chain and 1MRC for the heavy chain. Refinement of the AAV5-HL2476 60mer model against the cryo-reconstructed complex map resulted in a reduced overall correlation coefficient (CC) of 0.71 (Table 1). This is likely due to the lower resolution of the constant region of the HL2476 Fab (Fig. 2D).
FIG 4.
AAV5-HL2476 complex structure. (A) The complex structure model colored by molecule, with the AAV5 capsid in gray, the heavy chain in red, and the light chain in orange. (B) Close-up view of the binding interface between a Fab and a 3-fold protrusion on the AAV5 capsid surface. The localization of the complementarity-determining regions (CDRs) relative to capsid variable regions (VRs) is shown. The variable and constant regions of the light and heavy chains are indicated. (C) Further magnification of the CDR-VR binding interface. The atomic model of AAV5 and HL2476 is shown in the cryo-electron density map. This figure was generated with PyMol (48) (A and B) and UCSF-Chimera (38) (C).
TABLE 2.
Analysis and mutational outcome of the AAV5-HL2476 binding interface
| Residue VP3 | Location | HL2476 interaction residue | Distancea (Å) (interaction type) | AAV5 variant(s) | HL2476 escape |
|---|---|---|---|---|---|
| N443 VP3I | VR-IV | N51 (LC, CDR1) | 3.2 (HB) | N443Q, N443T | No |
| T444 VP3I | VR-IV | N73 (LC, CDR2) | 3.8 (HB) | T444V | No |
| R471 VP3A | VR-V | D74 (HC, CDR2) | 3.0 (HB) | R471E | No |
| V481 VP3A | VR-V | Y70 (LC, CDR2), Y122 (HC, CDR3) | 3.4 (VDW) | V481T, V481P, V481Y | No (V481T), partial (V481P), yes (V481Y) |
| R483 VP3A | VR-V | F52 (LC, CDR1), Y112 (LC, CDR3) | 3.8 (HB) | R483A, R483K, R483Q | Yes |
| A484 VP3A | VR-V | Y70 (LC, CDR2) | 3.4 (VDW) | ΔA484, A484S, A484Q | No (ΔA484, A484S), yes (A484Q) |
| T520 VP3A | VR-VI | R50 (HC, CDR1) | 2.5 (HB) | T520A, T520R | No |
| S576 VP3I | VR-VIII | P123 (HC, CDR3), Y70 (LC, CDR2) | 3.6 (HB) | S576A, S576Q | No (S576A), yes (S576Q) |
| T577 VP3I | VR-VIII | H111 (LC, CDR3) | 3.6 (HB) | T577A, T577V | No |
| T578 VP3I | VR-VIII | W52 (HC, CDR1) | 3.5 (HB) | T578A, T578Q | No |
HB, hydrogen bond; VDW, van der Waal contact.
Single-residue variants confer AAV5 escape from HL2476 neutralization.
The AAV5-HL2476 binding interface is extensive, with VR-V displaying the most interactions (Fig. 4 and 5 and Table 2). Site-directed mutagenesis of AAV5 residues, at a distance ≤4 Å from HL2476 residues with interactions, were conducted and tested by native dot immunoblot and in vitro neutralization assays to confirm their importance in binding (Fig. 5, Table 2). Mutation of numerous close contact residues had no effect, with the exception of V481, R483, and A484, located in VR-V, and S576, in VR-VIII (Fig. 5, Table 2). The VR-V residues are proximal to F52, Y69, Y70, and Y112 of the light chain, as well as G121 and Y122 of the heavy chain of HL2476 (Fig. 5C). Residue S576, located in VR-VIII, is proximal to Y70 and H111 of the light chain and P123 of the heavy chain (Fig. 5B).
FIG 5.
Binding interface of the AAV5-HL2476 complex structure. (A) Density for the apex of the AAV5 VR-IV and adjacent CDR1 and CDR2 of the light-chain (LC) residues. Atomic distances (in Å), as measured in Coot (37), of ≤4 Å between the AAV5 capsid surface and the Fab residues are indicated. The amino acid residues are shown in stick representations and are colored according to atom type: C, yellow; O, red; N, blue inside a black mesh density map. Residues are labeled with colors: AAV5 VP, gray; LC, orange; heavy chain (HC), red. (B) AAV5 VR-VIII, the LC CDR2/3, and HC CDR1/3, shown as described for panel A. (C) AAV5 VR-V, LC CDR1/2/3, and HC CDR3, shown as described for panel A. The images were generated using UCSF Chimera (38).
AAV5 variants AAV5-V481P, AAV5-V481Y, AAV5-R483K, AAV5-A484Q, and AAV5-S576Q, designed to introduce conformational changes or hydrophobic or large polar residues, were observed to successfully express VP1, VP2, and VP3, assemble capsids, and escape HL2476 recognition (Fig. 6A and B). The transduction efficiency of these variants in HEK293 cells ranged from 30% to 80% of wild-type (WT) AAV5 levels, with AAV5-V481Y having the highest expression (Fig. 6C). In an in vitro neutralization assay, WT AAV5 vectors were neutralized at low antibody concentrations corresponding to only ∼6 HL2476 IgG molecules per AAV5 capsid (Fig. 6D). The AAV5-V481P variant was observed to be neutralized at higher antibody concentration of 60 IgG molecules per capsid, while AAV5-A484Q was partially neutralized, with 6 and 60 IgG molecules displaying ∼50% transduction at these antibody concentrations (Fig. 6D). The remaining variants, AAV5-V481Y, AAV5-R483K, and AAV5-S576Q, escaped neutralization by HL2476 at all three antibody concentrations tested. Significantly, variant AAV5-S576Q showed an antibody enhancement of transduction efficiency, ∼5-fold, at an antibody-to-capsid binding site ratio of 60 IgG molecules per capsid (Fig. 6D).
FIG 6.
AAV5 variants escape HL2476. (A) SDS-PAGE of WT AAV5 and AAV5 variants. The images show the VP1, VP2, and VP3 bands and negative-stain EM images of purified samples. (B) Native dot immunoblot of AAV5 variants. ADK5b was used as a positive control. (C) HEK293 cell transduction by AAV5 variants. Relative transduction values of AAV5 variants compared to WT AAV5, as determined by luciferase reporter gene expression, are shown. (D) Neutralization assay of AAV5 variants. Preincubation was done with increasing amounts of HL2476 relative to the number of binding sites on a capsid as indicated (e.g., 1:10 IgG to binding site is equivalent to 6 IgG molecules per capsid). All experiments were performed in triplicate and are displayed as means ± standard deviations (n = 3).
DISCUSSION
Engineering AAV vectors to circumvent the detrimental effects of preexisting neutralizing antibodies (22) is key to their broad application as human gene therapy biologics. An important step in this effort is the understanding of the antigenic structure of the capsids. In this study, the footprint of a novel antibody, HL2476, directed against AAV5, was characterized at atomic resolution and used for the development of antibody escape variants. Previous structural analyses of AAV capsid-antibody complexes achieved the identification of approximate binding sites and footprints of antibodies due to insufficient resolution. These sites were then confirmed by mutagenesis. However, due to recent advances in cryo-EM technology, resulting in significant improvement in the resolution of reconstructed cryo-EM maps, the AAV5-HL2476 complex was solved to a resolution of 3.1 Å. To our knowledge, this structure currently represents the highest-resolution structure of a capsid-Fab complex for any virus. At this resolution, the AAV5-HL2476 interactions were interpretable with atomic detail (Fig. 4 and 5). A near-atomic-resolution structure, at 4.1 Å, was reported for a canine parvovirus-Fab complex (27); however, the structure with the second highest resolution reported for an AAV-Fab complex was for AAV2-A20 at 6.7 Å (23).
The structure of the AAV5 capsid within AAV5-HL2476 is very similar to that determined for the capsid alone using cryo-EM to 2.9-Å resolution (not shown), which is comparable to the 3.1-Å resolution of the complex. The most significant difference is the additional density ordered under the 5-fold channel (Fig. 2). The N terminus is ordered to residue 209 in the crystal structure (5) and 207 in the cryo-EM structure (unpublished data) of the capsid alone. In the AAV5-HL2476 structure, the N terminus is ordered to residue 201 (Fig. 3). In all previous parvovirus structures, with the exception of the human bocaviruses (28), the N terminus has been disordered up to residues equivalent to AAV5 residue 209. Flexibility of the N terminus is required for externalization of the VP1u PLA2 domain and nuclear localization signals. These functions are required for exit from the endosome/lysosomal pathway and entry into the nucleus for genome replication after receptor-mediated endocytosis (29). The improved ordering of this VP region in the presence of antibody suggests stabilization of the capsid. The effect of this stabilization on infection is unknown.
The three previously reported anti-AAV5 antibodies, ADK5a, ADK5b, and 3C5, bind either to the 5-fold region or over the 2/5-fold wall (9, 11). All of these antibodies neutralize AAV5 transduction (11, 30, and unpublished data). In order to prevent the isolation of another 5-fold binding antibody, this study adopted a novel screening procedure where the 5-fold region was blocked by the AVB affinity ligand to enable selection of a 3-fold binding HL2476 (Fig. 1A). Other 3-fold binding clones were present but were not isolated for further study (Fig. 1B). Prior to the generation and mapping of the HL2476 antibody, ADK5a, ADK5b, and 3C5 Fab footprints covered most of the capsid surface, except the 3-fold region. Like ADK5a and ADK5b, which bind around the 5-fold axes, HL2476, binding on the 3-fold protrusions, is neutralizing. These observations identify functional regions of the AAV5 capsid. An overlap of the Fab footprints, including HL2476, covers the entire surface of the AAV5 capsid (Fig. 7A and B). This image highlights the challenge that a preexisting polyclonal response poses to the engineering of escape vectors.
FIG 7.
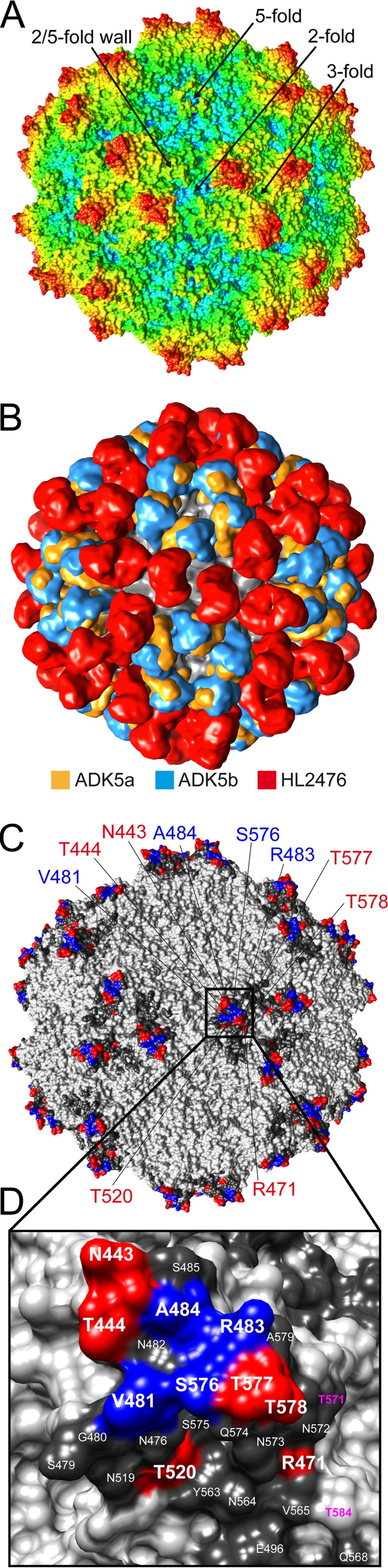
AAV5 capsid and anti-AAV5 antibody footprints. (A) Radially colored AAV5 capsid surface. The color key is the same as that in Fig. 2C. The icosahedral symmetry axes, 2-, 3-, and 5-fold axes, and the 2/5-fold wall are labeled. (B) Superposition of low-resolution density maps for the ADK5a (11 Å, orange), ADK5b (12 Å, blue), and HL2476 (9 Å, red) Fabs onto the AAV5 capsid surface. The entire surface of the AAV5 capsid is covered by these Fabs. (C) Location of the AAV5-HL2476 footprint. Capsid residues conferring HL2476 escape after mutation are colored blue, residues that do not are colored red. Additional capsid residues that are occluded by HL2476 binding are indicated in dark gray. (D) Close-up of the 3-fold protrusion from panel C with residue types and numbers indicated. Residues in magenta contribute to the sialic acid binding pocket of AAV5. This figure was generated with UCSF-Chimera (38).
Guided by mutagenesis, immunoblot experiments, and transduction studies, residues within VR-V (V481, R483, and A484) and VR-VIII (S576) of AAV5 VP3 were identified as the main determinants of HL2476 binding and neutralization (Fig. 6). These residues form a continuous surface patch on top of the 3-fold protrusion, which is flanked by the two highest points of the protrusion formed by VR-IV and VR-VIII (Fig. 7A and C). In this patch, R483 is the most critical contact residue for HL2476 binding, because all changes made to this position disrupted HL2476 binding by AAV5 (Table 2). The AAV5-R483A and -R483Q variants are defective for transduction (>90% reduction compared to WT AAV5), indicating a role for this 3-fold region in the life cycle. Interestingly, the reported sialic acid binding pocket for AAV5 is located at the 3-fold axis surrounded by the protrusions (12). The specific residues forming the AAV5 SIA binding pocket include residues M569, A570, T571, G583, T584, Y585, and L587. The heavy chain of the HL2476 antibody abuts and partially occludes this region and forms a van der Waal contact with T571 (Fig. 7D). Thus, a likely outcome of HL2476 binding is the prevention of receptor binding and cell attachment. Monoclonal antibodies binding to the 3-fold region of different AAV serotypes, e.g., AAV1-5H7 (9, 30), AAV1-ADK1a (11), AAV2-C37B (9, 25), AAV6-ADK6 (13), and AAV8-ADK8 (10), neutralize the transduction capability of the respective AAV serotype. However, the mechanism of neutralization can vary between different AAV serotypes. In the case of AAV1-5H7 (30), AAV1-ADK1a (11), AAV2-C37B (9), and AAV6-ADK6 (13), the footprint of the antibody overlaps the receptor binding site and virus binding to the cell surface is inhibited. For AAV8, the primary receptor, including its binding site, is unknown, and ADK8 neutralization occurs after cellular attachment and prenuclear entry (10). Thus, nuclear entry is inhibited.
In summary, the footprint for a novel anti-AAV5 capsid antibody has been characterized at atomic resolution by cryo-EM. At 3.1-Å resolution, the residue-level interactions analyzed provided the platform for mutations identifying the individual amino acids that determine HL2476 binding and neutralization. These residues overlap those mapped as antigenic footprints for other AAV-Fab complexes and are involved in primary receptor attachment and transduction determination. Thus, this complex structure provides information that can aid in the development of AAV5 antibody escape vectors and uniquely provides information for the engineering of the CDR of HL2476 to restrict or expand capsid recognition. The latter capability is useful for the characterization of various aspects of engineered capsid variants, especially capsid assembly.
MATERIALS AND METHODS
Animals and cell culture.
Six-week-old female CD-1 mice were used for the production of anti-AAV5 monoclonal antibodies (MAb) at the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) Hybridoma Core Laboratory, as previously reported (24). Use of animals in the UF Hybridoma Core Laboratory occurred under the guidelines of the Institutional Animal Care and Use Committee. HEK293 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and 100 U of penicillin/ml and 100 μg/ml of streptomycin at 37°C in 5% CO2.
Preparation of AAV5 VLPs.
Recombinant AAV5 virus-like particles (VLPs) were expressed using the Bac-to-Bac baculovirus-Sf9 insect cell expression system (Invitrogen, Carlsbad, CA) and purified using an AVB affinity column as described previously (31). Purified VLPs were concentrated to 3 mg/ml and buffer exchanged into 1× PBS, pH 7.4. The concentration of the sample was estimated by optical density (OD) measurements (using OD280 and E = 1.7 for calculation, in mg/ml), as well as SDS-PAGE with bovine serum albumin (BSA) concentration standards. Prior to use, the purity and integrity of the VLPs were also monitored by SDS-PAGE and negative-stain EM, respectively.
Mouse immunization and spleen hybridoma cell culture for anti-AAV5 IgG production.
Mice were immunized three times with subcutaneous injections of 50 μg of AAV5 VLPs at 21-day intervals and one intraperitoneal injection on day 120 as the last boost. The first three subcutaneous injections were accompanied by a Sigma adjuvant system (Sigma-Aldrich, St. Louis, MO), which contains 0.5 mg monophosphoryl lipid A, 0.5 mg synthetic trehalosedicorynomycolate in 44 μl squalene oil, 0.2% Tween 80, and water. Test bleeds from immunized animals were obtained 10 to 14 days after every booster injection by following animal care protocols. The collected sera were tested for highly specific antibody response at the ICBR by enzyme-linked immunosorbent assay (ELISA) and in-house using native dot immunoblot assays as described below. Four days after the final boost injection, the splenocytes of immunized mice were fused with mouse myeloma Sp2/0 cells using 50% polyethylene glycol 1500 (Roche) as the fusing agent. The fused hybrids were cultured in DMEM supplemented with HAT (hypoxanthine-aminopterin-thymidine) (Sigma-Aldrich, St. Louis, MO) to eliminate the unfused myeloma cells. To obtain the positive hybridoma clones with the highest specific anti-AAV5 capsid antibody response, the supernatants from the resulting hybridoma cells were collected and screened by native dot immunoblot assays, as described below.
Selection of IgGs by native dot immunoblot assays.
Anti-AAV5 hybridoma samples were screened before and after monoclonization by native dot immunoblot assays with AAV5 VLPs. For this assay, AAV5 VLPs were loaded onto a nitrocellulose membrane by vacuum suction using a Minifold dot-blot apparatus (GE Healthcare). Membranes were blocked in 6% milk in PBS for 1 h at room temperature, cut in multiple pieces, and then probed with the different hybridoma supernatants. Unbound antibodies were removed by three washes in PBS-T (PBS with 0.1% Tween 20) (5 min each), after which the membranes were incubated with a secondary horseradish peroxidase (HRP)-linked anti-mouse IgG antibody (GE Healthcare). Following 1 h of incubation, the membranes were washed three times in PBS-T and then imaged by ECL using Immobilon chemiluminescent substrate (Millipore). Positive hybridoma supernatants underwent a second round of screening similar to that described above. However, before blotting with the hybridoma supernatants, VLPs were incubated with a 100-fold molar excess of AVB ligand to block the 5-fold region of the capsids (32). Samples showing signal with and without AVB preincubation were selected for monoclonization.
IgG sequencing.
For sequencing, mRNA was isolated from the monoclonal hybridoma cell line expressing HL2476 antibody, and cDNA was generated. Degenerate primers to conserved IgG VH regions and kappa chain VL were used for amplification by PCR using rapid amplification of cDNA ends. Gel-purified amplicons were cloned and then purified from Escherichia coli. Plasmid DNA was sequenced and analyzed using bioinformatics tools.
IgG production and purification.
To produce sufficient quantities of MAb for purification, hybridoma cells of the selected clone were cultivated in 500 ml medium in surface roller bottles (Sigma-Aldrich, St. Louis, MO) for 12 days. Cells were centrifuged at 4,000 × g for 10 min, and the supernatant was collected. For preservation, 0.01% sodium azide was added to the supernatant. The HL2476 MAb was purified from the hybridoma supernatant by affinity chromatography using a protein G-Sepharose column (GE Healthcare). The hybridoma culture was diluted 1:1, vol/vol, in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4) and applied to the column at room temperature below the manufacturer-recommended flow rate of 1 ml/min. The column was washed with PBS and the antibody was eluted with 15 ml of 0.1 M citric acid (pH 2.0). The eluted antibody was neutralized with 1 M Tris-HCl (pH 10.0) to a final pH of 7.4. The elution fractions were concentrated using an Apollo 7-ml centrifugal concentrator (Orbital Biosciences, Topsfield, MA). The total MAb yield was determined at an OD of 280 nm, along with estimation by SDS-PAGE with BSA concentration standards.
Fab generation and purification.
For the generation of Fabs from purified HL2476 MAbs, immobilized papain was activated with l-cysteine according to the manufacturer’s instructions (Pierce, Rockford, IL) and mixed with purified sample at a suggested enzyme/substrate ratio of 1:160 (wt/wt). The slurry was incubated with moderate shaking at 37°C overnight. The reaction was stopped with sample buffer (10 mM Tris-HCl, pH 7.5) and then gently centrifuged (200 × g, 5 min) to pellet the immobilized papain-agarose beads. The aqueous mixture was carefully removed and diluted in 20 mM sodium phosphate buffer (pH 8.5) and applied to a Hi-Trap protein A column (GE Healthcare, Uppsala, Sweden) using a peristaltic pump at a rate of 1 ml/min. HL2476 Fabs were collected in the flowthrough and concentrated on Amicon-Ultra concentrators (Millipore, Billerica, MA). The purity was monitored by SDS-PAGE.
Preparation of VLP-Fab complexes.
Purified AAV5 VLPs were mixed with Fabs at a ratio of ∼2 Fabs per potential VP binding site in the 60-meric capsid, giving a final ratio of ∼1:120 (capsid to Fab). Complexes were viewed by negative-stain electron microscopy to confirm Fab decoration of the virus particles prior to sample vitrification.
Cryo-EM data collection for AAV5-Fab complexes.
AAV5-Fab complex samples (3.5 μl) were applied to glow-discharged Quantifoil copper grids with 2-nm continuous carbon support over holes (Electron Microscopy Sciences), blotted, and vitrified using a Vitrobot Mark 4 (FEI) at 95% humidity and 4°C. The particle distribution and ice quality of the grids were screened in-house using an FEI Tecnai G2 F20-TWIN microscope (FEI Co., Hillsboro, OR, USA) operated under low-dose conditions (200 kV, ∼20 e−/Å2). Images were collected on a Gatan UltraScan 4000 charge-coupled device (CCD) camera (Gatan, Inc., Pleasanton, CA, USA). Grids deemed optimal for data collection were used for collecting micrograph movie frames using the Leginon semiautomated application (33) on a Titan Krios electron microscope (FEI Co.) operated at 300 kV, with images recorded on a Gatan K2 Summit direct electron detection camera. The microscope was equipped with a Gatan postcolumn imaging filter (GIF) utilizing a slit width of 20 eV. Data collection used counting mode and an accumulated dose of 75 e−/Å2 fractionated into 50 movie frames per micrograph. Movie frame alignment used the MotionCor2 application with dose weighting (34). These data sets were collected as part of the National Institutes of Health (NIH) West/Midwest Consortium for High-Resolution Cryo-Electron Microscopy project. A nominal magnification of ×130,000 was used for data collection, resulting in a pixel size of 1.07 Å.
3D reconstruction of the AAV5-Fab complex.
Individual AAV5-HL2476 particle images were manually extracted from the micrographs using the RobEM software package (35) (http://cryoEM.ucsd.edu/programs.shtm). Preprocessing of the selected images (normalization and apodization) and estimation of the defocus level of each micrograph were performed using the AUTOPP subroutine (options F, O, and 3X) of AUTO3DEM as previously described (35, 36). The ab initio random-model computation procedure in AUTO3DEM was used to generate a starting model of the virus-Fab complex at ∼30-Å resolution from 100 particle images (35). This map was used to initiate full orientation and origin determination and refinement of the entire set of images with AUTO3DEM utilizing the gold standard protocol. Corrections to compensate for the effects of phase reversals in the microscope contrast-transfer function for each micrograph were performed as previously described, without amplitude corrections (36). An inverse B-factor (temperature factor) value of 1/25 Å2 was applied to sharpen the high-resolution features of the resulting final map, followed by visual inspection in the Coot and Chimera programs (37, 38). The final 3D map, reconstructed from 1,026 particle images, was estimated to be reliable to ∼3.1-Å resolution according to the criterion of an FSC of 0.143. For the determination of the local resolution of the complex, the map prior to B-factor correction was used as input in the ResMap (39) application.
Docking of the AAV5 capsid and HL2476 models and scaling of the cryo-EM map.
The atomic coordinates of the AAV5 capsid (60mer generated from PDB entry 3NTT [5] in the VIPERdb online server) were docked into the cryo-reconstructed density map using the fit-in-map subroutine in UCSF-Chimera (38) and to subsequently determine the absolute scale (i.e., pixel size) of the cryo-EM map by CC calculations. The pixel size of the reconstructed map was adjusted using the e2proc3d.py subroutine in EMAN2 (40), and a ccp4 format map was generated with the Mapman program (41). The heavy- and light-chain amino acid sequences of HL2476 were blasted in the NCBI BLASTP suite against the PDB database to identify the most similar antibody chain structure. 3D models of the light and heavy chain of HL2476 were generated with the SWISS Model online server (42) using PDB entries 5EZI and 1MRC as templates, respectively. The HL2476 coordinates were then docked into the ccp4 map using the fit-in-map function in Chimera by rigid-body rotations and translations monitored by CC calculation. The contact interactions for the AAV5-HL2476 complex were analyzed using the online server PDBePISA (43).
Model refinement.
The AAV5 VP monomer and the HL2476 light- and heavy-chain coordinates were refined in Coot (37) by manual building and utilizing the real-space-refinement subroutine to adjust side and main chains into the 1/25-Å2 B-factor-corrected map. From the refined model a 60mer of the complex was generated using VIPERdb (44). The capsid was further refined against the map utilizing the rigid body, real space, and B-factor refinement subroutines in Phenix (45). The CC and refinement statistics, including root mean square deviations (RMSD) from ideal bond lengths and angles (Table 1), were analyzed by Phenix (45).
Site-directed mutagenesis of HL2476 footprint residues.
The pXR5 plasmid containing AAV2 rep and AAV5 cap (46) served as the template for site-directed mutagenesis PCRs. For each mutant, complementary PCR primers were designed that contained the desired mutation, which was flanked on both sides by 10 to 15 homologous base pairs. Primers were ordered from Sigma-Aldrich (Houston, TX) and used in PCR amplification reactions using a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA) and PfuUltra high-fidelity DNA polymerase (Agilent, Santa Clara, CA). PCR products were incubated at 37°C for 1 h with DpnI restriction enzyme (NEB, Ipswich, MA) to degrade the methylated template plasmid. The reactions were then transformed into DH5α competent cells (NEB, Ipswich, MA), which were cultured on LB-ampicillin selective media and further amplified to isolate the plasmid. Clones were submitted for Sanger sequencing (Genewiz, South Plainfield, NJ) to verify that the desired mutations were generated.
rAAV5 variant production and quality control.
The plasmid pTR-UF3-Luciferase contains a recombinant AAV2 genome that expresses a luciferase reporter gene under the control of a cytomegalovirus (CMV) promoter, flanked by AAV2 inverted terminal repeats (ITRs). pTR-UF3-Luciferase, and pHelper (Stratagene), and the corresponding mutant pXR5 plasmids were cotransfected by polyethylenimine (PEI) transfection of HEK-293 cells. After 72 h of incubation at 37°C with 5% CO2, cells were harvested, washed with PBS, and centrifuged at 300 × g for 10 min, and then the resuspended cells were subjected to three freeze-thaw cycles (−80°C to 37°C) and finally incubated with 125 U/ml Benzonase for 1 h at 37°C before centrifugation at 10,000 × g for 15 min to pellet the cell debris. All variants and WT AAV5 were produced in biological triplicate, in 15-cm plates of HEK-293 cells, for use in subsequent experiments. Cell lysates were screened for expression of viral proteins VP1, VP2, and VP3 by Western blotting with B1 antibody (25). Five-μl aliquots were incubated with proteinase K at 56°C for 2 h, and the released vector genomes were purified by a PCR purification kit (Invitrogen). The genome containing titers of these preparations were determined by quantitative PCR in a Bio-Rad MyiQ2 thermocycler instrument (Bio-Rad) using SYBR green master mix (Bio-Rad).
Screening of mutants for HL2476 escape phenotypes by native dot immunoblot analysis.
AAV5 variants, including the wild type and a negative cell culture control, were adsorbed onto nitrocellulose membranes (Bio-Rad, Hercules, CA) in a dot blot manifold (Schleicher and Schuell, Dassel, Germany). Excess fluid was drawn through the membrane by vacuum filtration. The membrane was removed from the manifold and blocked with 6% milk in PBS, pH 7.4, for 1 h. Primary antibody in the form of hybridoma supernatant was applied to the membrane at a 1:100 dilution in PBS with 3% milk, 0.3% Tween 20 and incubated for 1 h. The membrane was then washed with PBS, and horseradish peroxidase (HRP)-linked secondary antibody (GE Healthcare) was applied at a dilution of 1:5,000 in PBS and incubated for 1 h. The membrane was washed with PBS, and then Immobilon chemiluminescent substrate (Millipore, Darmstadt, Germany) was applied to the membrane and the signal detected on X-ray film. The B1 and ADK5b antibodies were used as controls to confirm the presence and integrity of AAV capsid proteins using denatured and native capsids, respectively.
Measuring transduction efficiency of escape mutants by luciferase assay.
Clarified cell lysates of luciferase-packaged mutant vectors (in biological triplicate) were used to infect HEK-293 cells at a multiplicity of infection (MOI) of 105. After 48 h, cells were lysed and luciferase activity assayed using a luciferase assay kit (Promega, Madison, WI) as described in the manufacturer’s protocol. Uninfected cells of the same plate were used as a negative control, while cells infected with WT AAV5 were used as a positive control and a standard to which each mutant’s relative transduction efficiency was calculated as a percentage.
Quantifying HL2476 evasion by neutralization assay.
The ability of each mutant to evade HL2476 neutralization was measured in HEK293 cells by a luciferase assay, as described above, after incubation of purified variants with purified HL2476 MAb at various levels of binding site saturation for 1 h at 37°C.
Data availability.
The antibody is freely available to academic interests through a Materials Transfer Agreement (MTA) by contacting Mavis Agbandje-McKenna. The AAV5 capsid structure is identical to that deposited in the Protein Data Bank under entry 3NTT.
ACKNOWLEDGMENTS
We thank the Hybridoma Core of the University of Florida Interdisciplinary Center for Biotechnology Research (UF-ICBR) for the generation of the monoclonal HL2476 antibody, the UF-ICBR Gene Expression & Genotyping Core for sequencing of the HL2476 antibody, and the UF-ICBR Electron Microscopy Core for access to electron microscopes utilized for negative-stain electron microscopy and cryo-EM data collection. The Spirit and TF20 cryo-electron microscopes were provided by the UF College of Medicine (COM) and Division of Sponsored Programs (DSP). We thank Hong Zhou (University of California–Los Angeles) and the NIH West/Midwest Consortium for High-Resolution Cryo Electron Microscopy project for access to the Electron Imaging Center for Nanomachines’s Titan Krios and K2 DED, utilized for high-resolution data collection (M.P.I., H.Z., M.A.M., and others). The University of Florida COM and NIH GM082946 (to M.A.M. and R.M.) provided funds for the research efforts at the University of Florida. M.A.M. is an SAB member for Voyager Therapeutics, Inc., and AGTC has a sponsored research agreement with AGTC, Voyager Therapeutics, and Intima Biosciences, Inc., and is a consultant for Intima Biosciences, Inc. M.A.M. is a cofounder of StrideBio, Inc., a biopharmaceutical company with interest in developing AAV vectors for gene delivery application.
REFERENCES
- 1.Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol Biol 807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 2.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daya S, Berns KI. 2008. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seiler MP, Miller AD, Zabner J, Halbert CL. 2006. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum Gene Ther 17:10–19. doi: 10.1089/hum.2006.17.10. [DOI] [PubMed] [Google Scholar]
- 5.Govindasamy L, Dimattia MA, Gurda BL, Halder S, McKenna R, Chiorini JA, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. 2013. Structural insights into adeno-associated virus serotype 5. J Virol 87:11187–11199. doi: 10.1128/JVI.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becerra SP, Koczot F, Fabisch P, Rose JA. 1988. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J Virol 62:2745–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol 83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 8.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, Agbandje-McKenna M. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol 80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurda BL, DiMattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen WJ, Muzyczka N, Olson NH, Sinkovits RS, Chiorini JA, Zolotutkhin S, Kozyreva OG, Samulski RJ, Baker TS, Parrish CR, Agbandje-McKenna M. 2013. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol 87:9111–9124. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. 2012. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol 86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng YS, Gurda BL, Chipman P, McKenna R, Afione S, Chiorini JA, Muzyczka N, Olson NH, Baker TS, Kleinschmidt J, Agbandje-McKenna M. 2015. Adeno-associated virus serotype 1 (AAV1)- and AAV5-antibody complex structures reveal evolutionary commonalities in parvovirus antigenic reactivity. J Virol 89:1794–1808. doi: 10.1128/JVI.02710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afione S, DiMattia MA, Halder S, Di Pasquale G, Agbandje-McKenna M, Chiorini JA. 2015. Identification and mutagenesis of the adeno-associated virus 5 sialic acid binding region. J Virol 89:1660–1672. doi: 10.1128/JVI.02503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett AD, Wong K, Lewis J, Tseng YS, Smith JK, Chipman P, McKenna R, Samulski RJ, Kleinschmidt J, Agbandje-McKenna M. 2018. AAV6 K531 serves a dual function in selective receptor and antibody ADK6 recognition. Virology 518:369–376. doi: 10.1016/j.virol.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang LY, Patel A, Ng R, Miller EB, Halder S, McKenna R, Asokan A, Agbandje-McKenna M. 2016. Characterization of the adeno-associated virus 1 and 6 sialic acid binding site. J Virol 90:5219–5230. doi: 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell CL, Gurda BL, Van Vliet K, Agbandje-McKenna M, Wilson JM. 2012. Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J Virol 86:7326–7333. doi: 10.1128/JVI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. 2010. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 17.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. 2009. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaiss AK, Muruve DA. 2005. Immune responses to adeno-associated virus vectors. Curr Gene Ther 5:323–331. doi: 10.2174/1566523054065039. [DOI] [PubMed] [Google Scholar]
- 19.Mingozzi F, High KA. 2007. Immune responses to AAV in clinical trials. Curr Gene Ther 7:316–324. doi: 10.2174/156652307782151425. [DOI] [PubMed] [Google Scholar]
- 20.Tse LV, Moller-Tank S, Asokan A. 2015. Strategies to circumvent humoral immunity to adeno-associated viral vectors. Expert Opin Biol Ther 15:845–855. doi: 10.1517/14712598.2015.1035645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng YS, Agbandje-McKenna M. 2014. Mapping the AAV capsid host antibody response toward the development of second generation gene delivery vectors. Front Immunol 5:9. doi: 10.3389/fimmu.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JK, Agbandje-McKenna M. 2018. Creating an arsenal of adeno-associated virus (AAV) gene delivery stealth vehicles. PLoS Pathog 14:e1006929. doi: 10.1371/journal.ppat.1006929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCraw DM, O'Donnell JK, Taylor KA, Stagg SM, Chapman MS. 2012. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology 431:40–49. doi: 10.1016/j.virol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng YS, Vliet KV, Rao L, McKenna R, Byrne BJ, Asokan A, Agbandje-McKenna M. 2016. Generation and characterization of anti-adeno-associated virus serotype 8 (AAV8) and anti-AAV9 monoclonal antibodies. J Virol Methods 236:105–110. doi: 10.1016/j.jviromet.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74:9281–9293. doi: 10.1128/JVI.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A 99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Organtini LJ, Lee H, Iketani S, Huang K, Ashley RE, Makhov AM, Conway JF, Parrish CR, Hafenstein S. 2016. Near-atomic resolution structure of a highly neutralizing Fab bound to canine parvovirus. J Virol 90:9733–9742. doi: 10.1128/JVI.01112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mietzsch M, Kailasan S, Garrison J, Ilyas M, Chipman P, Kantola K, Janssen ME, Spear J, Sousa D, McKenna R, Brown K, Soderlund-Venermo M, Baker T, Agbandje-McKenna M. 2017. Structural insights into human bocaparvoviruses. J Virol 91:e00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatakrishnan B, Yarbrough J, Domsic J, Bennett A, Bothner B, Kozyreva OG, Samulski RJ, Muzyczka N, McKenna R, Agbandje-McKenna M. 2013. Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J Virol 87:4974–4984. doi: 10.1128/JVI.02524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbison CE, Weichert WS, Gurda BL, Chiorini JA, Agbandje-McKenna M, Parrish CR. 2012. Examining the cross-reactivity and neutralization mechanisms of a panel of mAbs against adeno-associated virus serotypes 1 and 5. J Gen Virol 93:347–355. doi: 10.1099/vir.0.035113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith RH, Levy JR, Kotin RM. 2009. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Lock M, Prongay AJ, Alvira MR, Petkov B, Wilson JM. 2015. Identification of an adeno-associated virus binding epitope for AVB Sepharose affinity resin. Mol Ther Methods Clin Dev 2:15040. doi: 10.1038/mtm.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM–an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilyas M, Mietzsch M, Kailasan S, Vaisanen E, Luo M, Chipman P, Smith JK, Kurian J, Sousa D, McKenna R, Soderlund-Venermo M, Agbandje-McKenna M. 2018. Atomic resolution structures of human bufaviruses determined by cryo-electron microscopy. Viruses 10:E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Kleywegt GJ, Jones TA. 1996. xdlMAPMAN and xdlDATAMAN–programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr 52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 42.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL 3rd, Reddy VS. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res 37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol 76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol 75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA: http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The antibody is freely available to academic interests through a Materials Transfer Agreement (MTA) by contacting Mavis Agbandje-McKenna. The AAV5 capsid structure is identical to that deposited in the Protein Data Bank under entry 3NTT.