Rotavirus is an important cause of childhood diarrheal disease worldwide. Two immunodominant proteins of rotavirus, VP7 and VP4, determine G and P genotypes, respectively. Recently, G12P[8] rotaviruses have become increasingly predominant. By analyzing rotavirus genome sequences from stool specimens obtained in Nashville, TN, from 2011 to 2013 and globally circulating rotaviruses, we found evidence of multiple introductions of G12 genes into the area. Based on sequence polymorphisms, VP7 proteins of these viruses are predicted to present themselves to the immune system very differently than those of vaccine strains. Many of the sick children with G12P[8] rotavirus in their diarrheal stools also were fully vaccinated. Our findings emphasize the need for continued monitoring of circulating rotaviruses and the effectiveness of the vaccines against strains with emerging G and P genotypes.
KEYWORDS: G12P[8], efficacy, genotype, phylogenetics, rotavirus, vaccine
ABSTRACT
Rotavirus is the leading global cause of diarrheal mortality for unvaccinated children under 5 years of age. The outer capsid of rotavirus virions consists of VP7 and VP4 proteins, which determine viral G and P types, respectively, and are primary targets of neutralizing antibodies. Successful vaccination depends upon generating broadly protective immune responses following exposure to rotaviruses presenting a limited number of G- and P-type antigens. Vaccine introduction resulted in decreased rotavirus disease burden but also coincided with the emergence of uncommon G and P genotypes, including G12. To gain insight into the recent predominance of G12P[8] rotaviruses in the United States, we evaluated 142 complete rotavirus genome sequences and metadata from 151 clinical specimens collected in Nashville, TN, from 2011 to 2013 through the New Vaccine Surveillance Network. Circulating G12P[8] strains were found to share many segments with other locally circulating strains but to have distinct constellations. Phylogenetic analyses of G12 sequences and their geographic sources provided evidence for multiple separate introductions of G12 segments into Nashville, TN. Antigenic epitopes of VP7 proteins of G12P[8] strains circulating in Nashville, TN, differ markedly from those of vaccine strains. Fully vaccinated children were found to be infected with G12P[8] strains more frequently than with other rotavirus genotypes. Multiple introductions and significant antigenic mismatch may in part explain the recent predominance of G12P[8] strains in the United States and emphasize the need for continued monitoring of rotavirus vaccine efficacy against emerging rotavirus genotypes.
IMPORTANCE Rotavirus is an important cause of childhood diarrheal disease worldwide. Two immunodominant proteins of rotavirus, VP7 and VP4, determine G and P genotypes, respectively. Recently, G12P[8] rotaviruses have become increasingly predominant. By analyzing rotavirus genome sequences from stool specimens obtained in Nashville, TN, from 2011 to 2013 and globally circulating rotaviruses, we found evidence of multiple introductions of G12 genes into the area. Based on sequence polymorphisms, VP7 proteins of these viruses are predicted to present themselves to the immune system very differently than those of vaccine strains. Many of the sick children with G12P[8] rotavirus in their diarrheal stools also were fully vaccinated. Our findings emphasize the need for continued monitoring of circulating rotaviruses and the effectiveness of the vaccines against strains with emerging G and P genotypes.
INTRODUCTION
Vaccines that fail to eradicate a viral pathogen can apply pressures that lead to changes in the genetic composition of a microbial population (1–4). Close human-animal interactions and increased global movement can spread new virus serotypes or variants into antigenically naive populations, providing opportunities for these viruses to gain a foothold (5, 6).
Rotavirus is the leading cause of diarrheal mortality for unvaccinated children under 5 years of age, resulting in an estimated 146,000 deaths worldwide in 2015, mostly in developing countries (7). The introduction of vaccines in the last decade has dramatically reduced the incidence of rotavirus disease and mortality in many countries. However, the effects of widespread vaccination on rotavirus evolution and the level of cross protection provided by current vaccines against antigenically distinct rotaviruses remain areas of active inquiry.
Rotavirus virions are nonenveloped particles enclosing a genome of 11 double-stranded RNA (dsRNA) segments, which encode six structural proteins (VP) and five or six nonstructural proteins (NSP) (8). The virion outer capsid consists of 260 VP7 trimers forming the glycoprotein shell and 60 VP4 trimers projecting from the surface (9). In the trypsin-activated form, cleaved VP8* fragments form dimers at the tips of VP4 spikes. VP4 and VP7 facilitate viral attachment and entry and are the primary targets of neutralizing antibodies. Human rotaviruses comprise a limited subset of G types, specified by VP7, and P types, specified by VP4 (10–12). While rotaviruses are often referred to solely by GXP[X] type, complete rotavirus nomenclature involves assignment of a genotype to each of the 11 segments (13, 14). The nine internal segments (those other than VP7 or VP4) of circulating human rotaviruses often are classified as either all genotype 1 or all genotype 2 (15).
Rotarix (RV1; GlaxoSmithKline), one of two domestically approved vaccines, is an attenuated human G1P[8] isolate introduced in the United States in 2008 (16, 17). RotaTeq (RV5; Merck) contains five human-bovine monoreassortant viruses, each containing a human VP4 (P[8]) or VP7 (G1 to G4) segment in a bovine rotavirus genetic background (18). RV5 was introduced in the United States in 2006. Rotavirus-specific IgA levels in serum and stool mostly correlate with protection from severe disease (19, 20). RV1 and RV5 have demonstrated >85% efficacy against severe rotavirus disease in developed countries and about 51% efficacy in developing countries, where there often is greater rotavirus antigenic diversity (21–23). Thus, both RV1 and RV5 can induce broadly protective responses.
Viruses with segmented RNA genomes, including rotavirus, evolve primarily through accumulation of nonsynonymous substitutions due to the error-prone viral RNA-dependent RNA polymerase (drift) and genome segment reassortment during coinfection (shift) (24). Outer-capsid gene segments (VP7 and VP4) are more diverse and reassort more readily than the remaining nine internal segments, which exhibit preferred constellations at the subgenotype level (25). Current rotavirus vaccines were derived using human strains that circulated in the 1980s, when G1, G2, G3, G4, P[4], and P[8] were the most prevalent genotypes, with G1P[8] rotaviruses predominant during most seasons (16, 26, 27). Circulating rotaviruses have subsequently diversified through genetic drift and shift. Differences in known VP7 and VP4 antigenic epitopes between vaccine strains and circulating strains have been demonstrated by studies in Belgium, where the highest variability was noted between circulating G1P[8] rotaviruses and G1 and P[8] vaccine components (28). While G1 strain prevalence declined globally from 2000 to 2007, G3 strains reemerged, and G9 and G12 strains emerged (27). These trends continued with the introduction of national immunization programs in many countries, with a slight resurgence of G1P[8] strains and transient predominance of G2P[4] (29). Rotaviruses containing G9 and G12, which are absent from RV1 and RV5, have recently gained global epidemiologic relevance, including in the United States (30–32). The impact of vaccine pressure on rotavirus evolution and selection remains unclear.
Rotaviruses with G12 genotypes were first identified in diarrheic babies in the Philippines in 1987 (G12P[4]) and then again in Thailand in 1998 (G12P[9]) (33, 34). Since then, these strains have been detected across the globe with increasing frequency, primarily as G12P[6] and G12P[8] combinations (29, 31, 35). In the United States, G12P[8] has been the predominant rotavirus type detected for the past several years (30). Human G12 VP7 segments cluster in three phylogenetic lineages, with the vast majority clustering in lineage III (31). Phylogenetic analyses suggest that human G12 strains originated from a human-porcine reassortment event (36). Within the human population, the G12 VP7 segment appears to have reassorted frequently with locally circulating strains (35).
The New Vaccine Surveillance Network (NVSN) was established to evaluate the impacts of new rotavirus vaccines through active sentinel surveillance at U.S. medical centers, including Vanderbilt University Medical Center (VUMC) in Nashville, TN. In the current study, we analyzed 142 rotavirus complete genome sequences from 151 stool samples collected between 2011 and 2013 through the VUMC NVSN. The majority were G12P[8] strains. Based on analysis of phylogenetic data and metadata, we found evidence of multiple G12 introductions into Nashville, TN, and higher probability of vaccination for G12P[8]-positive than other rotavirus-positive children, which may be related to marked antigenic variation between VP7 proteins of circulating G12P[8] and vaccine strains. These findings underscore the importance of continued surveillance and analyses aimed at determining whether responses elicited by current vaccines are fully protective against emerging rotavirus genotypes, including G12P[8].
RESULTS
Collection, sequencing, and genotyping of rotaviruses circulating in Nashville, TN, from 2011 to 2013.
The types of rotaviruses from stool specimens collected at the VUMC NVSN site and sequenced between the 2005-2006 and 2010-2011 seasons are summarized in Table 1 (37, 38). In the current study, 2,165 total stool specimens were collected from children with acute gastroenteritis (AGE) (1,447) and healthy controls (718) enrolled at the VUMC NVSN site from the 2011-2012 and 2012–2013 seasons. Of these, 151 were verified to be rotavirus positive and genotyped as described previously (30). G12P[8] rotaviruses predominated during both seasons (Table 1), and they predominated throughout the United States during this period (30). We determined complete rotavirus genome sequences from 142 VUMC NVSN specimens collected during the 2011–2012 and 2012–2013 seasons using semiautomated 11-segment reverse transcription (RT)-PCR and the Ion Torrent next-generation-sequencing platform. The terminal sequences of each segment were derived from RT-PCR primer sequences and may differ slightly from actual sequences within the 5′- and 3′-terminal 34 to 39 nucleotides. Of these sequences, 136 were derived from G12P[8] rotaviruses, with the remainder representing G1P[8] (n = 1), G2P[4] (n = 2), G3P[8] (n = 1), G9P[8] (n = 1), and mixed G1,G6P[8] (n = 1) strains, as determined by RT-PCR (Table 1). Partial genome sequences determined for the remaining rotavirus-positive specimens were excluded from this analysis.
TABLE 1.
Rotavirus G/P types collected and sequenced from children at VUMC in Nashville, TN (2005 to 2013)a
| G/P type | No. of samplesb
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005–2006c
|
2006–2007 |
2007–2008 |
2008–2009 |
2009–2010 |
2010–2011 |
2011–2012 |
2012–2013 |
|||||||||
| Coll. | Seq. | Coll. | Seq. | Coll. | Seq. | Coll. | Seq. | Coll. | Seq. | Coll. | Seq. | Coll. | Seq. | Coll. | Seq. | |
| G1P[8] | 65 | 26 | 30 | 10 | 12 | 4 | 1 | 1 | ||||||||
| G2P[4] | 5 | 4 | 5 | 12 | 12 | 5 | 2 | |||||||||
| G3P[8] | 2 | 1 | 3 | 25 | 17 | 1 | 18 | 1 | 1 | |||||||
| G9P[8] | 2 | 1 | 2 | 1 | ||||||||||||
| G12P[8] | 1 | 3 | 2 | 1 | 15 | 15 | 126 | 121 | ||||||||
| G12P[6] | 4 | 2d | 1 | |||||||||||||
| Mixed | 1 | 1 | 1 | 1 | ||||||||||||
| Total | 75 | 28 | 36 | 11 | 15 | 0 | 39 | 19 | 1 | 0 | 34 | 12 | 16 | 16 | 135 | 126 |
Numbers of samples collected (Coll.) and sequenced (Seq.) are shown by season and collection status.
For the 2005–2006 season, sample collection began on 1 January 2006.
Typed as P[6] by PCR but determined to be P[8] by sequence analysis by McDonald et al. (38).
Phylogenetic and constellation analyses of rotaviruses circulating in Nashville, TN, from 2005 to 2013.
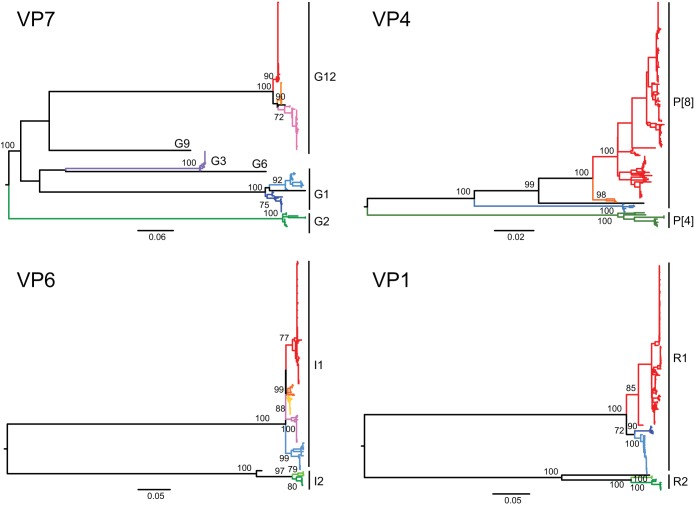
To investigate phylogenetic relationships among rotaviruses circulating in Nashville, TN, we constructed maximum-likelihood (ML) trees for each of the 11 rotavirus segments using nucleotide sequence data for all completely sequenced specimens collected between 2005 and 2013 (Fig. 1; see Fig. S1 in the supplemental material). Using the online genotyping tool RotaC (39) and phylogenetic clustering, we assigned genotypes to each of the 11 gene segments of the 142 newly sequenced viruses. Phylogenetic clustering of the outer-capsid segments was consistent with assignments made by RT-PCR (Table 1). Internal segments were classified as entirely genotype 1 (G1P[8], G3P[8], G9P[8], and G12P[8]) or entirely genotype 2 (G2P[4]) for each sequenced specimen, as in previous analyses of VUMC NVSN specimens (37, 38). The only two exceptions were the G2P[4] specimen VU12-13-26, which contained a genotype 1 VP3 segment (M1) in an otherwise genotype 2 background, and the G1,G6P[8] mixed specimen VU12-13-176. Rotaviruses containing mixtures of genotype 1 and genotype 2 are rarely detected, possibly due to requirements for coevolved gene sets to permit successful maintenance within the population (15). It will be interesting to see whether this constellation persists across seasons. The G1,G6P[8] mixed specimen VU12-13-176 has the genotype G1,G6-P[8]-I2-R2-C2-M1-A3-N2-T6-E2-H3. At the nucleotide level, VU12-13-176 G1 VP7 and P[8] VP4 segments have 100% nucleotide identity with RV5 strain WI97-9 and 99% nucleotide identity with RV5 strain WI97-4, respectively. The internal segments have 99 to 100% nucleotide identity with those from RV5 strain WI97-4 or WI97-9, suggesting that the virus is a fully vaccine-derived reassortant. Vaccine-associated AGE has been reported at multiple NVSN sites in the United States and in other countries (30, 40–44). The most commonly associated virus appears to be a vaccine-derived G1P[8] (vdG1P[8]), which results from reassortment among RV5 strains (43). The genome sequence of the G1,G6P[8] mixed specimen VU12-13-176 strongly suggests it contains vdG1P[8] rotavirus. The infant from whom the specimen was collected was just over 9 weeks old and had been admitted to the emergency department with AGE 2 days after receiving the first dose in the RV5 vaccination series. No underlying medical conditions were reported.
FIG 1.
Phylogenetic relationships among Nashville, TN, rotaviruses, 2005 to 2013. ML trees assembled from nucleotide sequences of four genome segments (VP7, VP4, VP6, and VP1) for all VUMC NVSN rotavirus specimens with completely sequenced genomes are shown. Horizontal branch lengths are drawn to scale and represent substitutions per nucleotide. Internal genotype assignments are indicated on the right, and subgenotype allele assignments for segments other than VP7 are indicated by color. Bootstrap values at nodes defining genotypes or subgenotype alleles are shown as percentages. For a detailed ML tree for each genome segment, see Fig. S1.
To investigate phylogenetic relationships at the subgenotype level, we made subgenotype “allele” assignments for phylogenetic clades with strong bootstrap support (>70%) at separating nodes (Fig. 1 and Fig. S1). For the majority of segments, two to four subgenotype alleles were assigned per genotype. Allele combinations (constellations) associated with each VP7-VP4 pair in the collection were determined by making a grid of colored boxes and revealed many shared alleles within each genotype but little overlap between genotypes (Fig. 2; see Table S2 in the supplemental material). For genotype 1 rotaviruses, some constellations persisted during multiple seasons. However, many constellations seemed to preferentially segregate with specific G-P allele combinations. The maintenance of genotype distinctions and persistence of specific constellations across seasons are consistent with previous findings (37, 38). The vast majority of G12P[8] rotaviruses (131/140) contained one of three internal segment constellations: (i) all “red” alleles, (ii) a “blue” VP1 and otherwise “red” alleles, or (iii) a “pink” VP6 and otherwise “red” alleles (Fig. 2 and Table S2). While nearly all the alleles present in these constellations have been observed in association with at least one other rotavirus type circulating in Nashville, TN, between 2005 and 2013, the three most commonly detected allele combinations are unique to the G12P[8] rotaviruses. Other than in G12P[8] constellations, the blue VP1 allele was detected in only a single G1P[8] strain collected during the 2012–2013 season, and the pink VP6 allele was detected in four G1P[8] strains collected during the 2005 to 2007 seasons. The red VP6 allele was detected only in the context of G12P[8] strains, whereas the remaining red alleles in the G12P[8] constellations were detected commonly in circulating G1P[8] and G3P[8] strains in the VUMC NVSN collection.
FIG 2.
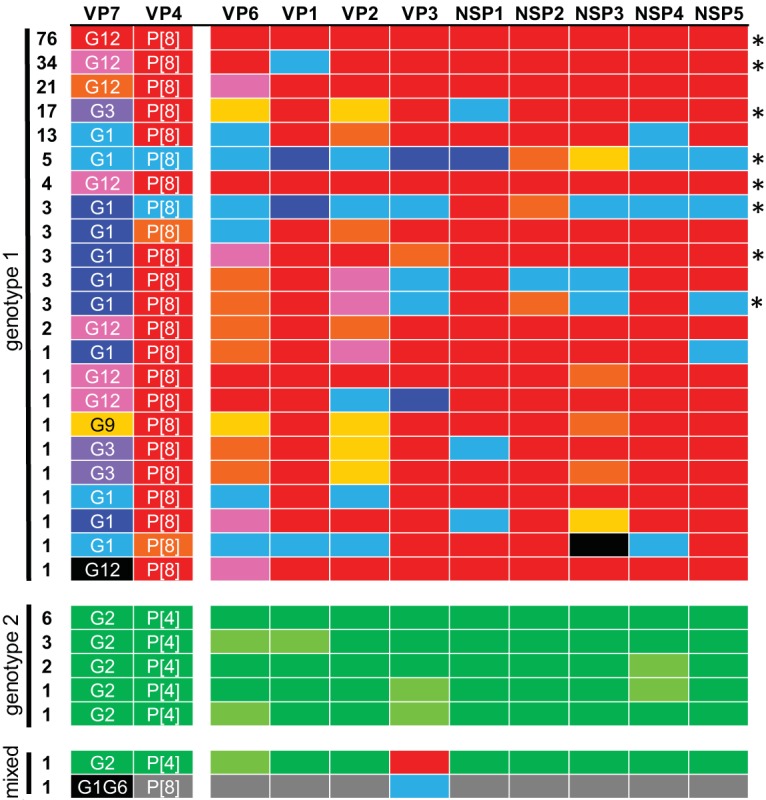
Allele-based genome constellations for rotaviruses in Nashville, TN, 2005 to 2013. The bars representing genome segments are colored based on phylogenetic analyses and subgenotype allele assignments, as shown in Fig. 1 and described in the text. Constellations are ordered based on genotype and frequency of detection, which are indicated on the left. The protein encoded by each segment is shown at the top. Allele constellations detected in multiple seasons are indicated by asterisks.
Global relationships among G12 segments.
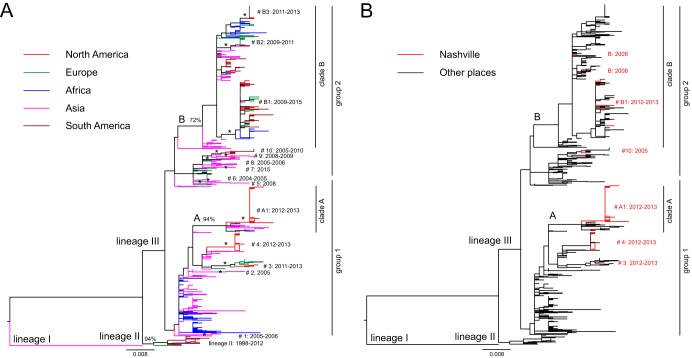
To determine the relationship of G12 segments from rotaviruses circulating in Nashville, TN, and elsewhere in the United States with those circulating globally, we generated an ML tree of 839 global complete G12 VP7 nucleotide sequences from GenBank (Fig. 3). In line with the lineages of G12 defined previously (31), lineage I comprises strain L26, isolated from the Philippines, and there are a small number of lineage II strains collected from 1998 to 2012, mainly from South America. Most global G12 strains collected from 2002 to 2015 are in lineage III (Fig. 3A). The ML phylogeny of global lineage III G12 strains suggests significant genetic diversity. Although we could not define true sublineages (groups 1 and 2) in lineage III, due to low bootstrap support values and frequent polytomies (multiple branches descending from a single node), we defined two clades (A and B) and a number of monophyletic groups or subclades (1 to 10, A1, and B1 to B3), which contain at least three sequences and are supported by ML bootstrap values of at least 70%. Nonetheless, the strains in group 2 (e.g., clade B and monophyletic groups 5 to 10) are phylogenetically distinct from those in group 1 (e.g., clade A and monophyletic groups 1 to 4) (Fig. 3A). In group 1, sequences from the same geographical origin (continent) tend to cluster together or close to each other in the phylogenetic tree. Most Asian and African sequences present polytomies, while strains from Europe (Italy and France) and North America (United States) form monophyletic groups 3, 4, and A1. The long internal branches of these monophyletic groups show the extensive genetic diversity that exists among G12 strains and suggest they evolved over many years prior to being detected or sampled (Fig. 3A). Sequences in group 2 include strains that circulated worldwide recently, and they are more geographically mixed than sequences in group 1. In particular, monophyletic group B1 contains recent strains isolated from North America, South America, Asia, Europe, and Africa from 2009 to 2015 (Fig. 3A).
FIG 3.
Phylogenetic relationships among globally circulating G12 VP7 rotavirus segments. ML trees assembled from G12 nucleotide sequences of 839 globally circulating rotaviruses. Horizontal branch lengths are drawn to scale and represent substitutions per nucleotide. Lineage (I, II, or III), group (1 or 2), clade (A or B), and monophyletic group or subclade (1 to 10, A1, and B1 to B3) assignments are indicated. Bootstrap values at nodes defining clades are shown as percentages. The same tree is shown in both panels but colored to reveal the continent of origin for each sequence (A) or a Nashville, TN, origin versus other geographic origin (B).
In Nashville, TN, G12 strains were reported sporadically before 2011 and became predominant from 2011 to 2013. In our phylogenetic analysis, two 2005 G12 sequences cluster together; however, three 2008 G12 sequences in clade B do not cluster together, suggesting at least two independent introductions of G12 strains into Nashville in 2008 (Fig. 3B). Most G12 strains from 2012 to 2013 are in group 1, monophyletic groups A1, 3, and 4, and the rest are in group 2, monophyletic group B1. Polytomies of Nashville G12 strains in A1 with very short genetic distances suggest that a single introduction of the A1 strain spread quickly and dominated activity in Nashville during the 2012–2013 season. The A1 G12 VP7 corresponds to the red subgenotype allele, 4 corresponds to the orange allele, and 3 corresponds to the black allele in Fig. 2 and Table S2 (Fig. 3B). All group 2 virus introductions into Nashville correspond to the pink subgenotype allele. A notable finding was that the majority of strains circulating in Nashville from 2012 to 2013 (A1, 3, and 4) were phylogenetically distinct from the common strains circulating worldwide (clade B) (Fig. 3B). In summary, the activity of G12 strains in Nashville is characterized by multiple introductions of G12 viruses into the area, followed by quick spread of the predominant strains, which is different from what has been observed for strains circulating in other global regions (45–48) (Fig. 3).
Antigenic-epitope comparison of circulating G12P[8] rotavirus VP4 proteins with those of vaccine strains.
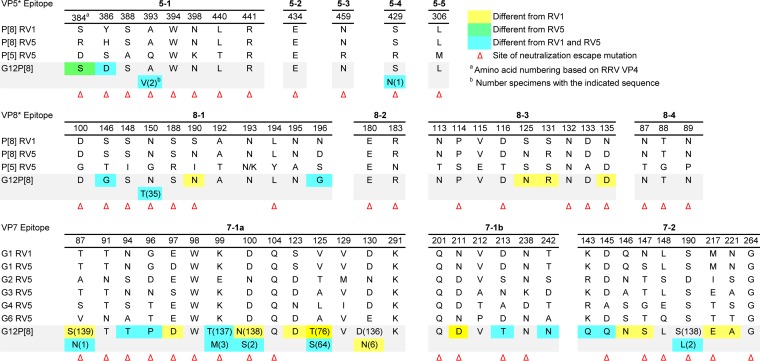
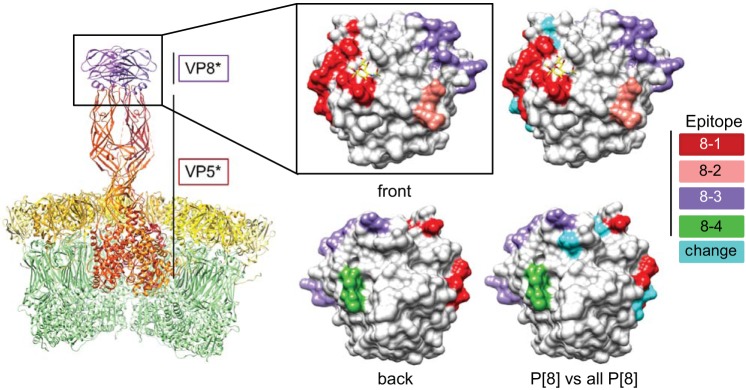
We performed amino acid sequence alignments to compare the compositions of VP4 proteins of circulating G12P[8] rotaviruses and those of vaccine strains. We identified 13 residues in the 775-amino-acid VP4 protein that differ among all the fully sequenced G12P[8] rotaviruses in the Nashville, TN, NVSN collection and the VP4 components of both RV1 and RV5. A total of 39 or 28 residues differ between all of the fully sequenced G12P[8] rotaviruses in the collection and the P[8] VP4 component of RV1 or RV5, respectively. To gain insight into the antigenic properties of G12P[8] rotaviruses, we identified VP4 amino acids that reside in predicted antigenic epitopes and differ among all G12P[8] rotaviruses in the collection and VP4 components of RV1 and RV5 (Table 2). These epitopes were previously predicted by mapping neutralization escape mutants and identifying surface-exposed amino acids in the VP4 trypsin cleavage products VP5* and VP8* that vary among prevalent human P types (25, 49, 50). We found three residues in antigenic epitopes that consistently differed from those of both the RV1 and RV5 vaccine strains and three additional residues that differed for a small number of clinical specimens (Table 2). The three changes were nonconservative; at amino acids 146 and 196, polar (Ser or Asn) or acidic (Asp) residues were replaced by flexible Gly residues. At amino acid 386, a basic His or polar Tyr was replaced by an acidic Asp residue. Four additional VP4 antigenic-epitope residues differed in all cases from those of RV1 VP4, and one differed in all cases from RV5 VP4. By mapping the locations of residues that differed between P[8] VP4 proteins of all specimens from the Nashville, TN, NVSN collection and all VP4 components of RV1 and RV5 onto the structure of rhesus rotavirus (RRV) VP4, we found that five of the residues are located in the VP8* domain (amino acids 46 to 231), and four of them are predicted to be surface exposed (Fig. 4). While they vary in sequence, VP4 proteins from different rotaviruses have highly similar structures, with the VP8* domains of different genotypes exhibiting differences in the width of a cleft between two β-sheets of some strains (51). Thus, we anticipate that the simian RRV VP4 structure approximates the locations of surface-exposed residues well.
TABLE 2.
Antigenic-epitope variation between Nashville, TN, G12P[8] VP4 and VP7 proteins and those of vaccine strains
FIG 4.
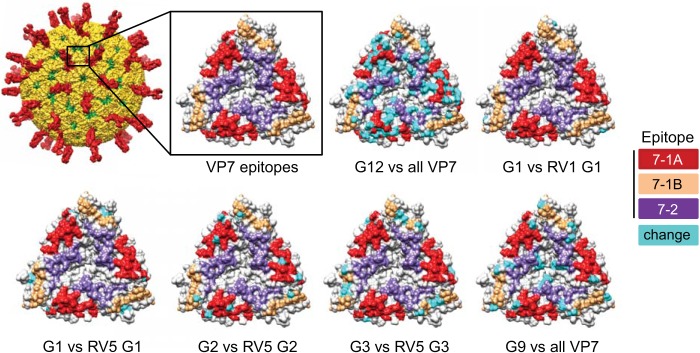
Amino acid variation between VP4 proteins of G12P[8] rotaviruses circulating in Nashville, TN, since 2005 and those of vaccine strains. (Left) Ribbon drawing showing a cutaway view of the rotavirus surface, with VP4 trypsin fragments VP5* (red) and VP8* (purple) labeled (PDB accession no. 4V7Q) (9). VP7 is colored yellow, and VP6 is colored green. (Middle) Front (as in the ribbon drawing) and back (rotated 180°) surface contour views of the enlarged VP8* domain shown in light gray, with known antigenic epitopes colored red (8-1), pink (8-2), purple (8-3), and green (8-4) (PDB no. 1KQR) (50). (Right) Additional surface representations of RRV VP8* showing positions at which all derived P[8] VP4 amino acid sequences from G12P[8] viruses in the VUMC NVSN collection differ from those of all the VP4 components of RV1 (P[8]) and RV5 (P[8] and P[5]) colored cyan.
Antigenic-epitope comparison of circulating G12P[8] rotavirus VP7 with those of vaccine strains.
Using amino acid sequence alignments, we identified 24 residues in the 326-amino-acid VP7 protein that differ among all sequenced G12P[8] rotaviruses in the VUMC NVSN collection and all VP7 components of RV1 and RV5. A total of 72 or 70 residues differ among all of the fully sequenced G12P[8] rotaviruses in the collection and the G1 VP7 components of RV1 or RV5, respectively. By mapping residues that differ among the VP7 proteins of all G12P[8] rotaviruses in the collection and all RV1 and RV5 VP7 vaccine components onto the RRV VP7 structure, we identified 16 surface-exposed residues, at least 7 of which reside in predicted antigenic epitopes (Fig. 5 and Table 2). VP7 proteins from different rotaviruses appear to have highly similar structures (9, 52–54). Furthermore, VP7 proteins from different rotavirus species are capable of forming hybrid virus-like particles and are predicted to have similar overall folds (55, 56). Based on these observations, we anticipate that the simian RRV VP7 structure approximates the locations of surface-exposed residues well. VP7 antigenic epitopes were previously predicted by mapping antibody neutralization escape mutants, using structural information from RRV rotavirus protein-antibody complexes, and identifying surface-exposed amino acids that vary among prevalent human G types (25, 52). Several of the amino acid differences in antigenic epitopes are nonconservative (e.g., G/D/N/T96P and D/A145Q) and potentially alter antigenic properties of the viruses (Table 2).
FIG 5.
Amino acid variation between VP7 proteins of rotaviruses circulating in Nashville, TN, since 2005 and those of vaccine strains. (Upper left) Surface representation of a rotavirus virion, with VP4 colored red and VP7 colored yellow (PDB accession no. 4V7Q) (9). The boxed area shows an enlarged RRV VP7 trimer colored light gray, with known antigenic epitopes colored red (7-1A), khaki (7-1b), and purple (7-2) (PDB accession no. 3FMG) (96). Positions at which all derived VP7 amino acid sequences of a particular G type from the VUMC NVSN collection (2005 to 2013) differ from those of the indicated VP7 vaccine component(s) are colored cyan. The word “all” refers to all VP7 components of RV1 (G1) and RV5 (G1, G2, G3, G4, and bovine G6).
Antigenic-epitope comparison of circulating non-G12 rotavirus VP7 with those of vaccine strains.
RV1 contains a single human G1 VP7 component, and RV5 viruses contain human G1, G2, G3, and G4 components, as well as a bovine G6 VP7. Based on amino acid alignments, there are only two amino acid differences, one of which is surface exposed, between the VP7 components of all 37 G1P[8] rotaviruses in the sequenced VUMC NVSN collection and the G1 RV1 VP7 component (Fig. 5). There are seven amino acid differences, two of which are surface exposed and one of which resides in a predicted antigenic epitope, between the VP7 of G1P[8] viruses in the collection and the G1 RV5 VP7. Between the VP7 proteins of the 14 fully sequenced G2P[4] rotaviruses in the VUMC NVSN collection and the G2 VP7 component of RV5, there are 15 differences, 3 of which are in surface-exposed residues in predicted antigenic epitopes. VP7 proteins of the 19 sequenced G3P[8] rotaviruses in the collection all differ from the RV5 G3 VP7 at seven positions, four of which are surface exposed and three of which reside in predicted antigenic epitopes. The VP7 protein of the lone sequenced G9P[8] virus from the collection differs from the six VP7 components of RV1 and RV5 at 14 positions. Six of the differing residues are surface exposed, and two reside in predicted antigenic epitopes. While the G9 VP7 is more antigenically distinct from all VP7 vaccine components than G1, G2, and G3 VP7 proteins in circulating rotaviruses are from just the homologous vaccine components, it is less divergent from all RV1 and RV5 VP7 sequences than are the G12 VP7 proteins in rotaviruses circulating in Nashville, TN (Fig. 5 and Table 2).
Vaccination status of rotavirus-positive infants and children.
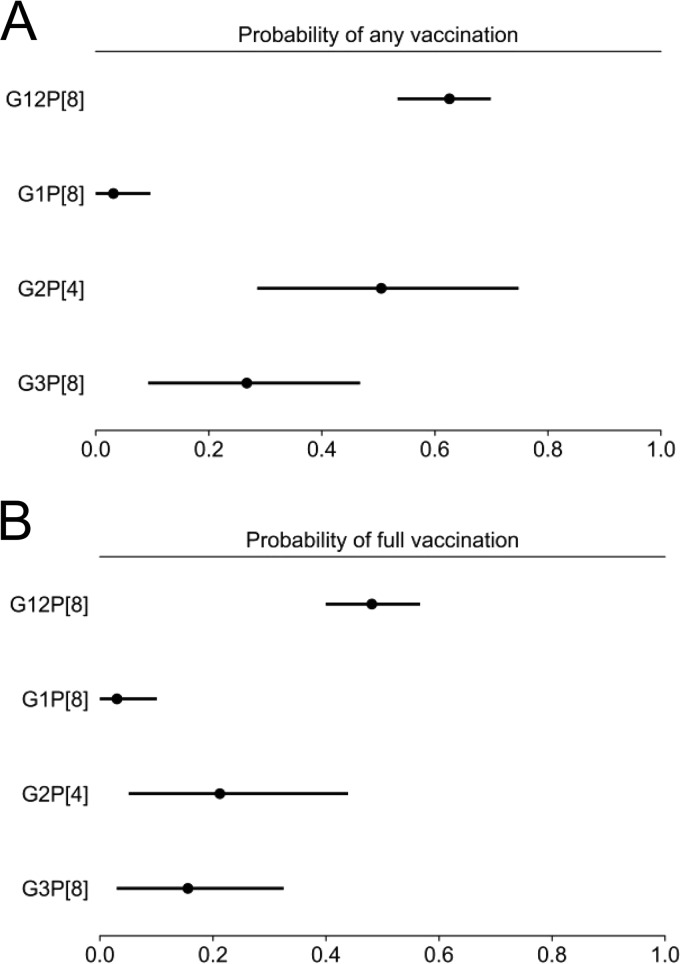
To ascertain the relationship between receipt of RV1 or RV5 vaccine and infection with specific circulating rotavirus G/P types, we determined the vaccination status of infants and children whose stools were collected between 2005 and 2013, from which rotavirus complete genomes were subsequently sequenced. Of these 212 children, 33 were unvaccinated because the vaccine was not available when they were infants or they were too young to be vaccinated, and 3 had unknown vaccination status (Table 3). Among the remaining 176 children, 1 of 10 (10%) G1P[8]-positive, 3 of 13 (∼23%) G2P[4]-positive, and 3 of 17 (∼18%) G3P[8]-positive children had received the complete series of RV5 vaccine. In contrast, 68 of 134 (∼51%) G12P[8]-positive children who were vaccine eligible had received a complete series of RV1 (two doses) or RV5 (three doses) or three mixed doses. Three of the 134 children were healthy controls. Among all the G12P[8]-positive children whose rotavirus genomes were completely sequenced, 68 of 140 (∼49%) received a complete vaccine series, and another 20 of 140 (∼14%) received at least one vaccine dose. The only child with a sequenced G9P[8]-positive specimen had received a complete series of RV5. Based on these data, we developed a model to predict the probability of full (complete) or any (complete or incomplete) vaccination associated with each rotavirus type (Fig. 6). The model predicted with 95% credibility that the probability of full vaccination was between 0.4 and 0.56 (mean = 0.48) for a child who tested positive for G12P[8] rotavirus, whereas it was less than 0.1 for G1P[8] (mean = 0.04) and between 0.03 and 0.32 for G3P[8] (mean = 0.17). A similar pattern was predicted for any vaccination, with a probability between 0.54 and 0.70 (mean = 0.62) for a child who tested positive for G12P[8] rotavirus, less than 0.09 for G1P[8] (mean = 0.04), and between 0.10 and 0.46 for G3P[8] (mean = 0.27). The highest variability was predicted for G2P[4]-positive children, with a probability between 0.05 and 0.43 (mean = 0.23) for full vaccination and between 0.29 and 0.75 (mean = 0.50) for any vaccination. The large credibility intervals and shifts in probability for full or any vaccination are likely due to the small sample size and higher variability in vaccination status for G2P[4]-positive children (Table 3).
TABLE 3.
Vaccination status of rotavirus-positive children in Nashville, TN, with sequenced viral genomes (2005 to 2013)
| G/P type | No. of children |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 dose | 1 dose |
2 doses |
3 doses |
Ud | NAe | Total | |||||||
| RV1a | RV5b | Mc | RV1 | RV5 | M | RV1 | RV5 | M | |||||
| G1P[8] | 9 | 1 | 1 | 26 | 37 | ||||||||
| G2P[4] | 6 | 1 | 2 | 1 | 3 | 1 | 14 | ||||||
| G3P[8] | 12 | 2 | 3 | 1 | 1 | 19 | |||||||
| G9P[8] | 1 | 1 | |||||||||||
| G12P[8] | 46 | 3 | 4 | 2 | 13f | 61f | 5 | 6f | 140 | ||||
| Mixed | 1 | 1 | |||||||||||
| Total | 73 | 9 | 20 | 74 | 3 | 33 | 212 | ||||||
RV1, Rotarix.
RV5, RotaTeq.
M, mixed RV1 and RV5 dosing or identity of vaccine dose unknown.
U, vaccination status unknown.
NA, vaccine not available or child <42 days old.
One sample in the category was from a healthy control.
FIG 6.
Model of vaccination probability based on rotavirus type. Analyses were of data presented in Table 3. The data points indicate the estimated posterior means, with the bars indicating the corresponding 95% HPD intervals, which represent a probabilistic statement of uncertainty about the true probabilities.
DISCUSSION
The majority of the sequenced genomes from specimens collected from 2011 to 2013 were from G12P[8] rotaviruses, which have predominated in the United States for the past several years (30). G12 strains in the VUMC NVSN collection have acquired multiple internal genes that are genetically well matched to those of other G1P[8] and G3P[8] strains that have circulated locally in recent years. However, in most cases, the G12P[8] strains have acquired new combinations of VP1 and VP6 alleles in their constellations. The VP1 segment encodes the viral RNA-dependent RNA polymerase. The VP6 segment encodes the intermediate-layer protein of the viral particle and interacts directly with the VP7 protein (9, 53). If the internal gene segment constellations associated with G12P[8] viruses confer a fitness advantage of some sort, we expect to detect them in subsequent seasons and perhaps in association with other G/P genotypes. Currently, our understanding of the selective advantages of specific gene sets in constellations is limited.
The phylogenetic analysis of global G12 strains suggests extensive genetic diversity (Fig. 3). Lineage III circulates globally and includes recent G12 strains reported from other countries. It has been proposed that G12P[8] rotaviruses in the United States originated from the introduction of the G12 VP7 segment into a globally common G1P[8] rotavirus strain (57). While our analyses support the idea that circulating G12P[8] strains have a genetic background similar to those of locally circulating strains, the 2012–2013 activity was characterized by multiple G12 introductions into the surveillance area, followed by spread of the predominant strains. Strikingly, most of the strains circulating in Nashville (in monophyletic groups A1 and 4) from 2012 to 2013 are phylogenetically distinct from the strains circulating in other places during the same period (in clade B) (Fig. 3). There were some introductions of global G12 strains (in monophyletic group B1) into the surveillance area, but they failed to spread rapidly. Nashville G12 strains in A1 and 4 are genetically closer to some Asian strains. However, the long internal branches of these monophyletic groups suggest that they evolved over a long time before being sampled and that there are gaps of unsampled regions between Asia and the United States. Thus, we cannot definitively conclude that Asia is the origin of G12 strains circulating in Nashville during 2012–2013.
The effects of widespread vaccination on rotavirus antigenic profiles and the effects of antigenic heterogeneity on vaccine-mediated protection remain unclear. We focused on amino acid differences observed between VP7 or VP4 proteins for all locally circulating strains of a particular G or P type during the past several years and those of vaccine strains. There are a relatively small number of conserved differences among VP7 proteins of circulating G1, G2, and G3 rotaviruses and the homologous vaccine components. As one would expect, G9 and G12 VP7 proteins exhibit increased numbers of surface-exposed residues that differ from those of all the vaccine components. The very small numbers of amino acid differences observed between groups of circulating rotaviruses in Nashville, TN, and the homologous G1, G2, and G4 VP7 and P[8] VP4 vaccine component proteins imply that vaccine-elicited antibodies would efficiently neutralize infection by most strains with homologous G or P types, providing some negative selective pressure on them. In contrast, the relatively high number of consistent amino acid differences on the VP7 antigenic surface, particularly in antigenic epitopes, between circulating G12P[8] strains and vaccine VP7 components could result in reduced antibody binding and, therefore, reduced selection against these viruses. The antigenic epitopes shown in Fig. 4 and 5 and listed in Table 2 were identified using animal rotaviruses and may differ from those present in the human rotaviruses under discussion (58). Nonetheless, differences in the amino acid compositions of VP7 or VP4 surfaces have the potential to alter antigenic epitopes. Reduced selection against circulating G12P[8] viruses, which is consistent with our vaccination results, may favor predominance of G12P[8] rotaviruses.
On the other hand, this model is likely oversimplified. The small numbers of conserved differences between the P[8] components of all circulating G12P[8] viruses in the VUMC NVSN collection and the P[8] components of RV1 and RV5 suggest a potential for antibodies elicited by current vaccines to efficiently neutralize G12P[8] viruses by binding VP4. Numerous studies suggest that binding of a single VP4-specific monoclonal antibody to a specific epitope is sufficient to achieve neutralization in vitro (59–64). Thus, while neutralizing antibody responses may be most efficient when they are raised against rotavirus strains with outer-capsid proteins that are antigenically matched in both VP4 and VP7 proteins of circulating strains to which a child is later exposed, sufficient protection may be induced when either outer-capsid antigen is antigenically matched with the circulating strain. Studies of antibodies generated following single or multiple natural rotavirus infections or immunizations suggest that initial exposures elicit more heterotypic VP4 and homotypic VP7 human rotavirus antibodies, whereas subsequent exposures are dominated by cross-reactive human-specific VP7 antibodies (65, 66). A recent study of human rotavirus antibodies in adults, who had presumably had multiple natural infections, suggested that homotypic anti-VP7 and nonneutralizing anti-VP8* antibodies are more common than heterotypic anti-VP7 and anti-VP4 antibodies, though both homotypic and heterotypic antibodies were detected (67). Limited antibody responses to VP4 after vaccination may result in little selective pressure, whereas highly cross-reactive VP7-specific antibodies may negatively select for rotaviruses with homologous VP7 proteins and positively select for antigenic drift and rotaviruses with the most divergent VP7 proteins. However, clear correlates of protection for rotavirus following vaccination have not been well established (19, 68–71). While protection following vaccination mostly correlates with levels of rotavirus-specific antibodies, the roles of neutralizing antibodies and other factors in mediating protection are less clear, particularly in children. In the current study, we have not sought to identify rates of genetic drift in VP4 and VP7 for each genotype. Such analyses are important and likely will reveal more complexity and subtlety in the influence of vaccination on rotavirus evolution than what is described here.
Our analyses of vaccination data for infants and children with sequenced rotavirus genomes suggest increased probability of any or complete vaccination for G12P[8]-positive infants and children (Table 3 and Fig. 6). Since nearly all of the children in the analysis had AGE, this increased vaccination probability may suggest a decreased probability of vaccine-mediated protection against G12P[8] rotavirus disease. Nonetheless, there has been a sustained decrease in the incidence of rotavirus detection in the United States in postvaccine years compared to prevaccine years (72). Thus, current rotavirus vaccines appear to provide broad cross protection. In the early years of the NVSN, when G1P[8] viruses were predominant, rotavirus vaccines were not widely available, which makes it difficult to compare the percentages of vaccinated rotavirus-positive children with AGE caused by G1P[8] and G12P[8] viruses. Still, the failure of G1P[8] viruses to regain predominance, despite remaining in circulation, as evidenced by their detection in the 2008-2009 and 2012–2013 seasons, suggests that the current vaccines protect very efficiently against AGE induced by these viruses. Future analyses of larger sample sizes are needed to determine whether these trends are supported and whether rotavirus type-specific differences in vaccine-mediated protection exist.
In Nashville, TN, and across the United States, RV5 is more commonly administered than RV1. RV1 contains only a G1 component, which differs substantially in amino acid sequence from nonhomologous VP7 proteins. While RV5 is more antigenically diverse, including four human VP7 components and one human VP4, it is comprised primarily of bovine virus segments. Despite its lack of antigenic breadth, RV1 has shown evidence of broad protection (73, 74). Considering the low neutralizing antibody responses following two doses of RV1 (75) and protection from rotavirus gastroenteritis observed for nonseropositive vaccinees (76), it is possible that the human origin of internal RV1 segments provides protection through mechanisms that involve viral proteins other than outer-capsid antigens. Such mechanisms could include antibodies directed against the intermediate-layer protein VP6, which can provide protection in mice (77), or other structural and nonstructural proteins, which could potentially contribute to T-cell recognition (78–80). When larger data sets can be assembled, it will be informative to determine the relationships between differences in vaccination probabilities and rotavirus G and P types based on the type of vaccine (RV1 or RV5) administered. Recently published data from Australia show increased G12P[8] prevalence in states using RV5 and increased G2P[4] and equine G3P[8] prevalence in states and territories using RV1, supporting the idea that the two vaccines apply different selective immunological pressures that influence circulating rotavirus population diversity (81). Modeling suggests that differences in homotypic and heterotypic protection from vaccination can influence genotype selection, particularly for RV1 (82). However, some studies have found no consistent pattern to indicate selective pressure from vaccines (29). An alternative explanation is that continuous natural variation of cocirculating rotavirus genotypes, along with other poorly understood factors, contributes significantly to fluctuations in global and regional genotype landscapes. The effects of vaccines on rotavirus populations also may be more complex than simple serotype selection, as suggested by increased detection of G2P[4] reassortants containing animal-derived segments and of G1P[8] rotaviruses containing segments genetically distinct from those of RV1 in postvaccine versus prevaccine years in Belgium (83, 84). Additional surveillance and directed studies will help to clarify the specific roles of RV1 and RV5 in rotavirus selection and evolution.
Regional differences in rotavirus genome constellations, potential variability in RV1 and RV5 efficacy against specific G- and P-type antigens, the rapid pace of segmented RNA virus evolution via genetic drift and shift, and global spread provide plausible explanations for reduced vaccine efficacy in some regions of the world and suggest future potential for current vaccines to lose efficacy against severe rotavirus disease over time. In the United States, where many circulating rotaviruses share common genetic backgrounds and a limited number of G and P types, RV1 and RV5 still seem to offer broad protection, even against rotaviruses containing emerging outer-capsid antigens. Specifically, rotavirus vaccine effectiveness (VE) in the United States in 2012–2013 for completely vaccinated children was 80% for both RV1 and RV5 (85). However, while RV5 VE was estimated at 78% for G12P[8] and 87% against G2P[4] rotaviruses, RV1 VE was estimated at 82% for G12P[8] and 53% for G2P[4]. Thus, these vaccines may apply selective pressure against homologous G and P types while promoting the success of circulating heterologous G and P types. In Europe from 2004 to 2006, RV1 VE against G2 rotaviruses was high (85.5%), though still lower than RV1 VE against G1 (96.4%) or G3 (93.7%) rotaviruses (86). Such differences may influence circulating rotavirus populations. In developing countries, emerging antigenic variation, as represented by G12 VP7, in genetic backgrounds that are antigenically mismatched with vaccine strains in both G and P types, and also potentially in internal segments, may in part explain reduced vaccine efficacy. The findings described here highlight the need for continued global surveillance to elucidate the changing rotavirus G/P type landscape and the efficacy of current vaccines against emerging rotaviruses.
MATERIALS AND METHODS
Sample collection.
Fecal specimens were collected from 2,165 children up to 18 years old who presented with AGE (1,447) at the VUMC emergency department, hospital, or outpatient clinics or who were healthy (718) and were seen at outpatient clinics in Nashville, TN, during the years 2011 to 2013. Collection was performed in accordance with an NVSN protocol approved by the Centers for Disease Control and Prevention (CDC) and the VUMC Institutional Review Board. Informed consent, including future specimen use, and permission to obtain medical records were provided by a parent or guardian at the time of enrollment. Rotavirus vaccination status was verified by reviewing medical records obtained from the child’s primary care provider and/or the Tennessee Immunization Information System. Children between the ages of 15 days and 11 years (2011-2012) or 18 years (2012–2013) were considered eligible AGE cases if they were residents of Davidson County, TN, and had diarrhea (≥3 episodes within 24 h) or vomiting (≥1 episode in 24 h) within a period of ≤10 days. Children with a noninfectious cause of diarrhea, a history of immune deficiency, or previous enrollment for the same episode of AGE or who were transferred from another hospital were excluded. Children aged 15 days through 11 years (2011-2012) or 18 years (2012–2013) from Davidson County presenting for well-child visits were eligible to serve as healthy controls unless they had acute respiratory infection symptoms within 3 days preceding the visit or AGE symptoms within 14 days preceding the visit or were immunocompromised. Fecal samples collected within 7 days of enrollment were tested for rotavirus antigen using the commercial enzyme-linked immunosorbent assay (ELISA) Premier Rotaclone (Meridian Bioscience).
RNA extraction, RT-PCR, and nucleotide sequencing.
Rotavirus-positive stool specimens were processed for RNA extraction and high-throughput RT-PCR and sequencing. Rotavirus dsRNA was extracted and sequenced from deidentified rotavirus-positive fecal specimens using TRIzol (Invitrogen) according to previously described methods (87, 88). Briefly, the extracted RNA was used in each of 11 one-step RT-PCRs (Qiagen) to amplify full-length RNA for each genome segment. The primers used to amplify all the gene segments were described previously (89, 90). The PCR products were cleaned by treatment with exonuclease I (New England BioLabs). The products were quantified using a SYBR green dsDNA detection assay (Thermo Fisher Scientific). All 11 RT-PCR products for each genome were pooled in equimolar amounts in preparation for sequencing using the Ion Torrent platform (Thermo Fisher Scientific).
Ion Torrent libraries were prepared by shearing pooled rotavirus amplicons and ligating barcoded adaptors to the sheared DNA, using the Ion Xpress Plus fragment library kit (Thermo Fisher Scientific), to create a library of approximately 200-bp fragments. Equal volumes of pooled libraries were cleaned using Ampure XP beads (Beckman Coulter, Inc.). Real-time PCR was performed on the pooled barcoded libraries to evaluate the library and adaptor quality and to determine the template dilution factor for emulsion PCR. The pool was normalized and amplified on Ion Sphere particles (ISPs) using the Ion One Touch instrument. The pool was enriched for template-positive ISPs on the Ion One Touch ES instrument. Sequencing was performed on the Ion Torrent PGM platform using an Ion 316 chip (Thermo Fisher Scientific).
The reads generated by Ion Torrent PGM were de novo assembled using CLC Bio’s clc_novo_assemble program (Qiagen) to form contigs. The resulting contigs were mapped against their closest reference sequences (CLC Bio’s clc_ref_assemble_long program). Any locus with an observed variant that was detected in the majority of sequencing reads was reflected in the assembled consensus sequence of the segment.
Nucleotide alignments and phylogenetic analyses.
For each segment of a rotavirus collected in Nashville, TN, between 2005 and 2013 with a completely sequenced genome, nucleotide sequences were aligned using the MUSCLE algorithm in MEGA 6.0 with manual adjustment (91). A general time-reversal (GTR) nucleotide substitution model with a gamma distribution of among-site rate variation (GTR + Γ) was selected as the best-fit model by Modeltest in MEGA 6.0 and used in ML phylogeny construction with 1,000 bootstrap replicates (in MEGA 6.0). ML trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Genotype assignments were made using RotaC v2.0 (39). Subgenotype allele assignments were made for phylogenetic clades with strong bootstrap support (>70%) at separating nodes.
To understand the phylogenetic relationship between G12 segments from rotaviruses collected in Nashville, TN, and in other countries, a global G12 data set with 839 complete nucleotide sequences was generated, which included 140 G12 sequences from Nashville and 699 publicly available global G12 sequences downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/; accessed 26 October 2017). The ML phylogeny of the global G12 data set was inferred using the methods described above.
Amino acid alignments and structural analyses.
VP7 and VP4 amino acid alignments were constructed with MAFFT v7.2 (92) using the E-INS-I strategy. Structural analyses of VP7 and VP4 were performed using the UCSF Chimera molecular modeling system and Protein Data Bank (PDB) files 4V7Q (9), 3FMG (52), and 1KQR (50). Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Biostatistical analysis of vaccination data.
To estimate the association between rotavirus genotype and vaccination status, we fit a Bayesian logistic model (93) predicting the probability of vaccination as a function of genotype. A generalized linear modeling approach with a logit transformation was used, with G12P[8], the most common genotype, as the baseline, thereby estimating the relative effects of the remaining genotypes. The baseline probability and genotype effects were all given vague normal prior distributions (standard deviation = 3) on the logit scale. Two alternative models were constructed, one using full vaccination as the positive outcome, with unvaccinated and partially vaccinated individuals pooled, and another using partial or full vaccination as the positive outcome versus unvaccinated individuals. The models were fit using the Markov chain Monte Carlo method (the No U-turn sampler [94]) as implemented in the PyMC3 library for Python (95). The algorithm was run for 1,000 iterations, following 2,000 tuning iterations, yielding posterior means and 95% highest probability density (HPD) intervals as a measure of posterior uncertainty (Fig. 6). The samples were checked for convergence and goodness of fit, with both checks negative.
Accession number(s).
The sequences described were submitted to GenBank under the accession numbers given in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank the children and families in Nashville, TN, who contributed to this study. We thank Susmita Shrivastava for sequence data submission to GenBank.
This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services (HHS), under award number U19AI110819. The project was supported by NIH through the Vanderbilt Institute for Clinical and Translational Research (UL1 TR000445), who provided access to REDCap, and by the CDC (1U01IP00163-01 and 3U01IP001063-01S1 to N.B.H.) and HHS (5U01IP000464-05, 3U01IP000464-05S1, and 1U01IP000464-05S2 to N.B.H.). S.R.D. is also supported by the NIH-funded Tennessee Center for AIDS Research (P30 AI110527) and U19AI095227. The Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, is supported by NIGMS grant P41-GM103311.
The content is solely our responsibility and does not represent the official views of the CDC and the NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01476-18.
REFERENCES
- 1.Chong YL, Padhi A, Hudson PJ, Poss M. 2010. The effect of vaccination on the evolution and population dynamics of avian paramyxovirus-1. PLoS Pathog 6:e1000872. doi: 10.1371/journal.ppat.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam C, Octavia S, Ricafort L, Sintchenko V, Gilbert GL, Wood N, McIntyre P, Marshall H, Guiso N, Keil AD, Lawrence A, Robson J, Hogg G, Lan R. 2014. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis 20:626–633. doi: 10.3201/eid2004.131478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Read AF, Baigent SJ, Powers C, Kgosana LB, Blackwell L, Smith LP, Kennedy DA, Walkden-Brown SW, Nair VK. 2015. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol 13:e1002198. doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pybus OG, Tatem AJ, Lemey P. 2015. Virus evolution and transmission in an ever more connected world. Proc Biol Sci 282:20142878. doi: 10.1098/rspb.2014.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.G. B. D. Child Mortality Collaborators. 2016. Global, regional, national, and selected subnational levels of stillbirths, neonatal, infant, and under-5 mortality, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1725–1774. doi: 10.1016/S0140-6736(16)31575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desselberger U. 2014. Rotaviruses. Virus Res 190:75–96. doi: 10.1016/j.virusres.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. 2011. Atomic model of an infectious rotavirus particle. EMBO J 30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desselberger U, Iturriza-Gómara M, Gray JJ. 2001. Rotavirus epidemiology and surveillance. Novartis Found Symp 238:125–147. [DOI] [PubMed] [Google Scholar]
- 12.Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis 192(Suppl 1):S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- 13.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthijnssens J, Van Ranst M. 2012. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr Opin Virol 2:426–433. doi: 10.1016/j.coviro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein DI, Smith VE, Sherwood JR, Schiff GM, Sander DS, DeFeudis D, Spriggs DR, Ward RL. 1998. Safety and immunogenicity of live, attenuated human rotavirus vaccine 89-12. Vaccine 16:381–387. doi: 10.1016/S0264-410X(97)00210-7. [DOI] [PubMed] [Google Scholar]
- 17.Anonymous. 2016. Rotarix package insert. GlaxoSmithKline, Research; Triangle Park, NC. [Google Scholar]
- 18.Anonymous. 2006. RotaTeq package insert. Merck & Co., Inc., Whitehouse Station, NJ. [Google Scholar]
- 19.Angel J, Steele AD, Franco MA. 2014. Correlates of protection for rotavirus vaccines: Possible alternative trial endpoints, opportunities, and challenges. Hum Vaccin Immunother 10:3659–3671. doi: 10.4161/hv.34361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel M, Glass RI, Jiang B, Santosham M, Lopman B, Parashar U. 2013. A systematic review of anti-rotavirus serum IgA antibody titer as a potential correlate of rotavirus vaccine efficacy. J Infect Dis 208:284–294. doi: 10.1093/infdis/jit166. [DOI] [PubMed] [Google Scholar]
- 21.Jiang V, Jiang B, Tate J, Parashar UD, Patel MM. 2010. Performance of rotavirus vaccines in developed and developing countries. Hum Vaccin 6:532–542. doi: 10.4161/hv.6.7.11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rha B, Tate JE, Payne DC, Cortese MM, Lopman BA, Curns AT, Parashar UD. 2014. Effectiveness and impact of rotavirus vaccines in the United States—2006-2012. Expert Rev Vaccines 13:365–376. doi: 10.1586/14760584.2014.877846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patton JT. 2012. Rotavirus diversity and evolution in the post-vaccine world. Discov Med 13:85–97. [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald SM, Nelson MI, Turner PE, Patton JT. 2016. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat Rev Microbiol 14:448–460. doi: 10.1038/nrmicro.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald SM, Matthijnssens J, McAllen JK, Hine E, Overton L, Wang S, Lemey P, Zeller M, Van Ranst M, Spiro DJ, Patton JT. 2009. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog 5:e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goveia MG, Ciarlet M, Owen KE, Ranucci CS. 2011. Development, clinical evaluation, and post-licensure impact of RotaTeq, a pentavalent rotavirus vaccine. Ann N Y Acad Sci 1222:14–18. doi: 10.1111/j.1749-6632.2011.05970.x. [DOI] [PubMed] [Google Scholar]
- 27.Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. 2012. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 30(Suppl 1):A122–A130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- 28.Zeller M, Patton JT, Heylen E, De Coster S, Ciarlet M, Van Ranst M, Matthijnssens J. 2012. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J Clin Microbiol 50:966–976. doi: 10.1128/JCM.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doro R, Laszlo B, Martella V, Leshem E, Gentsch J, Parashar U, Banyai K. 2014. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure?. Infect Genet Evol 28:446–461. doi: 10.1016/j.meegid.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowen MD, Mijatovic-Rustempasic S, Esona MD, Teel EN, Gautam R, Sturgeon M, Azimi PH, Baker CJ, Bernstein DI, Boom JA, Chappell J, Donauer S, Edwards KM, Englund JA, Halasa NB, Harrison CJ, Johnston SH, Klein EJ, McNeal MM, Moffatt ME, Rench MA, Sahni LC, Selvarangan R, Staat MA, Szilagyi PG, Weinberg GA, Wikswo ME, Parashar UD, Payne DC. 2016. Rotavirus strain trends during the postlicensure vaccine era: United States, 2008-2013. J Infect Dis 214:732–738. doi: 10.1093/infdis/jiw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M. 2010. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol Biol Evol 27:2431–2436. doi: 10.1093/molbev/msq137. [DOI] [PubMed] [Google Scholar]
- 32.O’Ryan M. 2009. The ever-changing landscape of rotavirus serotypes. Pediatr Infect Dis J 28:S60–S62. doi: 10.1097/INF.0b013e3181967c29. [DOI] [PubMed] [Google Scholar]
- 33.Pongsuwanna Y, Guntapong R, Chiwakul M, Tacharoenmuang R, Onvimala N, Wakuda M, Kobayashi N, Taniguchi K. 2002. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. J Clin Microbiol 40:1390–1394. doi: 10.1128/JCM.40.4.1390-1394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taniguchi K, Urasawa T, Kobayashi N, Gorziglia M, Urasawa S. 1990. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J Virol 64:5640–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, Iturriza-Gomara M, Iftekharuddin N, Azim T, Van Ranst M. 2007. Evolutionary history and global spread of the emerging G12 human rotaviruses. J Virol 81:2382–2390. doi: 10.1128/JVI.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh S, Kobayashi N, Nagashima S, Chawla-Sarkar M, Krishnan T, Ganesh B, Naik TN. 2010. Full genomic analysis and possible origin of a porcine G12 rotavirus strain RU172. Virus Genes 40:382–388. doi: 10.1007/s11262-010-0454-y. [DOI] [PubMed] [Google Scholar]
- 37.Dennis AF, McDonald SM, Payne DC, Mijatovic-Rustempasic S, Esona MD, Edwards KM, Chappell JD, Patton JT. 2014. Molecular epidemiology of contemporary G2P[4] human rotaviruses cocirculating in a single U.S. community: footprints of a globally transitioning genotype. J Virol 88:3789–3801. doi: 10.1128/JVI.03516-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald SM, McKell AO, Rippinger CM, McAllen JK, Akopov A, Kirkness EF, Payne DC, Edwards KM, Chappell JD, Patton JT. 2012. Diversity and relationships of cocirculating modern human rotaviruses revealed using large-scale comparative genomics. J Virol 86:9148–9162. doi: 10.1128/JVI.01105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maes P, Matthijnssens J, Rahman M, Van Ranst M. 2009. RotaC: a Web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol 9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donato CM, Ch'ng LS, Boniface KF, Crawford NW, Buttery JP, Lyon M, Bishop RF, Kirkwood CD. 2012. Identification of strains of RotaTeq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J Infect Dis 206:377–383. doi: 10.1093/infdis/jis361. [DOI] [PubMed] [Google Scholar]
- 41.Hemming M, Vesikari T. 2012. Vaccine-derived human-bovine double reassortant rotavirus in infants with acute gastroenteritis. Pediatr Infect Dis J 31:992–994. doi: 10.1097/INF.0b013e31825d611e. [DOI] [PubMed] [Google Scholar]
- 42.Boom JA, Sahni LC, Payne DC, Gautam R, Lyde F, Mijatovic-Rustempasic S, Bowen MD, Tate JE, Rench MA, Gentsch JR, Parashar UD, Baker CJ. 2012. Symptomatic infection and detection of vaccine and vaccine-reassortant rotavirus strains in 5 children: a case series. J Infect Dis 206:1275–1279. doi: 10.1093/infdis/jis490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Payne DC, Edwards KM, Bowen MD, Keckley E, Peters J, Esona MD, Teel EN, Kent D, Parashar UD, Gentsch JR. 2010. Sibling transmission of vaccine-derived rotavirus (RotaTeq) associated with rotavirus gastroenteritis. Pediatrics 125:e438–e441. doi: 10.1542/peds.2009-1901. [DOI] [PubMed] [Google Scholar]
- 44.Bucardo F, Rippinger CM, Svensson L, Patton JT. 2012. Vaccine-derived NSP2 segment in rotaviruses from vaccinated children with gastroenteritis in Nicaragua. Infect Genet Evol 12:1282–1294. doi: 10.1016/j.meegid.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Rougemont A, Kaplon J, Fremy C, Legrand-Guillien MC, Minoui-Tran A, Payan C, Vabret A, Mendes-Martins L, Chouchane M, Maudinas R, Huet F, Dubos F, Hober D, Lazrek M, Bouquignaud C, Decoster A, Alain S, Languepin J, Gillet Y, Lina B, Mekki Y, Morfin-Sherpa F, Guigon A, Guinard J, Foulongne V, Rodiere M, Avettand-Fenoel V, Bonacorsi S, Garbarg-Chenon A, Gendrel D, Lebon P, Lorrot M, Mariani P, Meritet JF, Schnuriger A, Agius G, Beby-Defaux A, Oriot D, Colimon R, Lagathu G, Mory O, Pillet S, Pozzetto B, Stephan JL, Aho S, Pothier P. 2016. Clinical severity and molecular characteristics of circulating and emerging rotaviruses in young children attending hospital emergency departments in France. Clin Microbiol Infect 22:737.e9–737.e15. [DOI] [PubMed] [Google Scholar]
- 46.Giammanco GM, Bonura F, F DIB, Cascio A, Ferrera G, Dones P, Saporito L, Collura A, Terranova DM, Valenzise M, Allu MT, Casuccio N, Palermo M, Banyai K, Martella V, De Grazia S. 2016. Introduction and prolonged circulation of G12 rotaviruses in Sicily. Epidemiol Infect 144:1943–1950. doi: 10.1017/S0950268815003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langa JS, Thompson R, Arnaldo P, Resque HR, Rose T, Enosse SM, Fialho A, de Assis RM, da Silva MF, Leite JP. 2016. Epidemiology of rotavirus A diarrhea in Chokwe, Southern Mozambique, from February to September, 2011. J Med Virol 88:1751–1758. doi: 10.1002/jmv.24531. [DOI] [PubMed] [Google Scholar]
- 48.Umair M, Abbasi BH, Nisar N, Alam MM, Sharif S, Shaukat S, Rana MS, Khurshid A, Mujtaba G, Aamir UB, Zaidi SSZ. 2017. Molecular analysis of group A rotaviruses detected in hospitalized children from Rawalpindi, Pakistan during 2014. Infect Genet Evol 53:160–166. doi: 10.1016/j.meegid.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Dormitzer PR, Nason EB, Prasad BV, Harrison SC. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkataram Prasad BV, Shanker S, Hu L, Choi JM, Crawford SE, Ramani S, Czako R, Atmar RL, Estes MK. 2014. Structural basis of glycan interaction in gastroenteric viral pathogens. Curr Opin Virol 7:119–127. doi: 10.1016/j.coviro.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, Dormitzer PR. 2009. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324:1444–1447. doi: 10.1126/science.1170481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. 2009. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A 106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. 2009. Rotavirus architecture at subnanometer resolution. J Virol 83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eren E, Zamuda K, Patton JT. 2016. Modeling of the rotavirus group C capsid predicts a surface topology distinct from other rotavirus species. Virology 487:150–162. doi: 10.1016/j.virol.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Chang KO, Kim WY, Saif LJ. 2002. Production of hybrid double- or triple-layered virus-like particles of group A and C rotaviruses using a baculovirus expression system. Virology 302:1–8. doi: 10.1006/viro.2002.1610. [DOI] [PubMed] [Google Scholar]
- 57.Freeman MM, Kerin T, Hull J, Teel E, Esona M, Parashar U, Glass RI, Gentsch JR. 2009. Phylogenetic analysis of novel G12 rotaviruses in the United States: a molecular search for the origin of a new strain. J Med Virol 81:736–746. doi: 10.1002/jmv.21446. [DOI] [PubMed] [Google Scholar]
- 58.Higo-Moriguchi K, Akahori Y, Iba Y, Kurosawa Y, Taniguchi K. 2004. Isolation of human monoclonal antibodies that neutralize human rotavirus. J Virol 78:3325–3332. doi: 10.1128/JVI.78.7.3325-3332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorziglia M, Larralde G, Ward RL. 1990. Neutralization epitopes on rotavirus SA11 4fM outer capsid proteins. J Virol 64:4534–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirkwood CD, Bishop RF, Coulson BS. 1996. Human rotavirus VP4 contains strain-specific, serotype-specific and cross-reactive neutralization sites. Arch Virol 141:587–600. doi: 10.1007/BF01718319. [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi N, Taniguchi K, Urasawa S. 1990. Identification of operationally overlapping and independent cross-reactive neutralization regions on human rotavirus VP4. J Gen Virol 71:2615–2623. doi: 10.1099/0022-1317-71-11-2615. [DOI] [PubMed] [Google Scholar]
- 62.Mackow ER, Shaw RD, Matsui SM, Vo PT, Dang MN, Greenberg HB. 1988. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci U S A 85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padilla-Noriega L, Dunn SJ, Lopez S, Greenberg HB, Arias CF. 1995. Identification of two independent neutralization domains on the VP4 trypsin cleavage products VP5* and VP8* of human rotavirus ST3. Virology 206:148–154. doi: 10.1016/S0042-6822(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 64.Taniguchi K, Maloy WL, Nishikawa K, Green KY, Hoshino Y, Urasawa S, Kapikian AZ, Chanock RM, Gorziglia M. 1988. Identification of cross-reactive and serotype 2-specific neutralization epitopes on VP3 of human rotavirus. J Virol 62:2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clark HF, Offit PA, Ellis RW, Eiden JJ, Krah D, Shaw AR, Pichichero M, Treanor JJ, Borian FE, Bell LM, Plotkin SA. 1996. The development of multivalent bovine rotavirus (strain WC3) reassortant vaccine for infants. J Infect Dis 174(Suppl 1):S73–S80. doi: 10.1093/infdis/174.Supplement_1.S73. [DOI] [PubMed] [Google Scholar]
- 66.Gorrell RJ, Bishop RF. 1999. Homotypic and heterotypic serum neutralizing antibody response to rotavirus proteins following natural primary infection and reinfection in children. J Med Virol 57:204–211. doi:. [DOI] [PubMed] [Google Scholar]
- 67.Nair N, Feng N, Blum LK, Sanyal M, Ding S, Jiang B, Sen A, Morton JM, He XS, Robinson WH, Greenberg HB. 2017. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci Transl Med 9:eaam5434. doi: 10.1126/scitranslmed.aam5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Angel J, Franco MA, Greenberg HB. 2012. Rotavirus immune responses and correlates of protection. Curr Opin Virol 2:419–425. doi: 10.1016/j.coviro.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clarke E, Desselberger U. 2015. Correlates of protection against human rotavirus disease and the factors influencing protection in low-income settings. Mucosal Immunol 8:1–17. doi: 10.1038/mi.2014.114. [DOI] [PubMed] [Google Scholar]
- 70.Desselberger U, Huppertz HI. 2011. Immune responses to rotavirus infection and vaccination and associated correlates of protection. J Infect Dis 203:188–195. doi: 10.1093/infdis/jiq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franco MA, Greenberg HB. 2013. Rotavirus. Microbiol Spectr 1. doi: 10.1128/microbiolspec.AID-0011-2013. [DOI] [PubMed] [Google Scholar]
- 72.Aliabadi N, Tate JE, Haynes AK, Parashar UD, Centers for Disease Control and Prevention . 2015. Sustained decrease in laboratory detection of rotavirus after implementation of routine vaccination-United States, 2000-2014. MMWR Morb Mortal Wkly Rep 64:337–342. [PMC free article] [PubMed] [Google Scholar]
- 73.Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira B, Victor JC, Gillard PH, Cheuvart BB, Han HH, Neuzil KM. 2010. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 362:289–298. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- 74.Pawinski R, Debrus S, Delem A, Smolenov I, Suryakiran PV, Han HH. 2010. Rotarix in developing countries: paving the way for inclusion in national childhood immunization programs in Africa. J Infect DIS 202:S80–S86. doi: 10.1086/653547. [DOI] [PubMed] [Google Scholar]
- 75.Ward RL, Kirkwood CD, Sander DS, Smith VE, Shao M, Bean JA, Sack DA, Bernstein DI. 2006. Reductions in cross-neutralizing antibody responses in infants after attenuation of the human rotavirus vaccine candidate 89-12. J Infect DIS 194:1729–1736. doi: 10.1086/509623. [DOI] [PubMed] [Google Scholar]
- 76.Cheuvart B, Neuzil KM, Steele AD, Cunliffe N, Madhi SA, Karkada N, Han HH, Vinals C. 2014. Association of serum anti-rotavirus immunoglobulin A antibody seropositivity and protection against severe rotavirus gastroenteritis: analysis of clinical trials of human rotavirus vaccine. Hum Vaccin Immunother 10:505–511. doi: 10.4161/hv.27097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burns JW, Siadat-Pajouh M, Krishnaney AA, Greenberg HB. 1996. Protective effect of rotavirus VP6-specific IgA monoclonal antibodies that lack neutralizing activity. Science 272:104–107. doi: 10.1126/science.272.5258.104. [DOI] [PubMed] [Google Scholar]
- 78.Franco MA, Greenberg HB. 1999. Immunity to rotavirus infection in mice. J Infect Dis 179(Suppl 3):S466–S469. doi: 10.1086/314805. [DOI] [PubMed] [Google Scholar]
- 79.Parra M, Herrera D, Calvo-Calle JM, Stern LJ, Parra-López CA, Butcher E, Franco M, Angel J. 2014. Circulating human rotavirus specific CD4 T cells identified with a class II tetramer express the intestinal homing receptors alpha4beta7 and CCR9. Virology 452-453:191–201. doi: 10.1016/j.virol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward RL. 2008. Rotavirus vaccines: how they work or don't work. Expert Rev Mol Med 10:e5. doi: 10.1017/S1462399408000574. [DOI] [PubMed] [Google Scholar]
- 81.Roczo-Farkas S, Kirkwood CD, Cowley D, Barnes GL, Bishop RF, Bogdanovic-Sakran N, Boniface K, Donato CM, Bines JE. 2018. The impact of rotavirus vaccines on genotype diversity: a comprehensive analysis of 2 decades of Australian surveillance data. J Infect Dis 218:546–554. doi: 10.1093/infdis/jiy197. [DOI] [PubMed] [Google Scholar]
- 82.Pitzer VE, Patel MM, Lopman BA, Viboud C, Parashar UD, Grenfell BT. 2011. Modeling rotavirus strain dynamics in developed countries to understand the potential impact of vaccination on genotype distributions. Proc Natl Acad Sci U S A 108:19353–19358. doi: 10.1073/pnas.1110507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeller M, Heylen E, Tamim S, McAllen JK, Kirkness EF, Akopov A, De Coster S, Van Ranst M, Matthijnssens J. 2017. Comparative analysis of the Rotarix vaccine strain and G1P[8] rotaviruses detected before and after vaccine introduction in Belgium. PeerJ 5:e2733. doi: 10.7717/peerj.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeller M, Nuyts V, Heylen E, De Coster S, Conceicao-Neto N, Van Ranst M, Matthijnssens J. 2016. Emergence of human G2P[4] rotaviruses containing animal derived gene segments in the post-vaccine era. Sci Rep 6:36841. doi: 10.1038/srep36841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Payne DC, Selvarangan R, Azimi PH, Boom JA, Englund JA, Staat MA, Halasa NB, Weinberg GA, Szilagyi PG, Chappell J, McNeal M, Klein EJ, Sahni LC, Johnston SH, Harrison CJ, Baker CJ, Bernstein DI, Moffatt ME, Tate JE, Mijatovic-Rustempasic S, Esona MD, Wikswo ME, Curns AT, Sulemana I, Bowen MD, Gentsch JR, Parashar UD. 2015. Long-term consistency in rotavirus vaccine protection: RV5 and RV1 vaccine effectiveness in US children, 2012–2013. Clin Infect Dis 61:1792–1799. doi: 10.1093/cid/civ872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vesikari T, Karvonen A, Prymula R, Schuster V, Tejedor JC, Cohen R, Meurice F, Han HH, Damaso S, Bouckenooghe A. 2007. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 370:1757–1763. doi: 10.1016/S0140-6736(07)61744-9. [DOI] [PubMed] [Google Scholar]
- 87.Nyaga MM, Jere KC, Peenze I, Mlera L, van Dijk AA, Seheri ML, Mphahlele MJ. 2013. Sequence analysis of the whole genomes of five African human G9 rotavirus strains. Infect Genet Evol 16:62–77. doi: 10.1016/j.meegid.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 88.Nyaga MM, Stucker KM, Esona MD, Jere KC, Mwinyi B, Shonhai A, Tsolenyanu E, Mulindwa A, Chibumbya JN, Adolfine H, Halpin RA, Roy S, Stockwell TB, Berejena C, Seheri ML, Mwenda JM, Steele AD, Wentworth DE, Mphahlele MJ. 2014. Whole-genome analyses of DS-1-like human G2P[4] and G8P[4] rotavirus strains from Eastern, Western and Southern Africa. Virus Genes 49:196–207. doi: 10.1007/s11262-014-1091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Magagula NB, Esona MD, Nyaga MM, Stucker KM, Halpin RA, Stockwell TB, Seheri ML, Steele AD, Wentworth DE, Mphahlele MJ. 2015. Whole genome analyses of G1P[8] rotavirus strains from vaccinated and non-vaccinated South African children presenting with diarrhea. J Med Virol 87:79–101. doi: 10.1002/jmv.23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nyaga MM, Jere KC, Esona MD, Seheri ML, Stucker KM, Halpin RA, Akopov A, Stockwell TB, Peenze I, Diop A, Ndiaye K, Boula A, Maphalala G, Berejena C, Mwenda JM, Steele AD, Wentworth DE, Mphahlele MJ. 2015. Whole genome detection of rotavirus mixed infections in human, porcine and bovine samples co-infected with various rotavirus strains collected from sub-Saharan Africa. Infect Genet Evol 31:321–334. doi: 10.1016/j.meegid.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB. 2013. Bayesian data analysis, 3rd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 94.Hoffman MD, Gelman A. 2014. The No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. J Machine Learning Res 15:1593–1623. [Google Scholar]
- 95.Salvatier J, Wiecki TV, Fonnesbeck C. 2016. Probabilistic programming in Python using PyMC3. PeerJ Computer Science 2:e55. doi: 10.7717/peerj-cs.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aoki ST, Trask SD, Coulson BS, Greenberg HB, Dormitzer PR, Harrison SC. 2011. Cross-linking of rotavirus outer capsid protein VP7 by antibodies or disulfides inhibits viral entry. J Virol 85:10509–10517. doi: 10.1128/JVI.00234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.