Human parvovirus B19 (B19V) infection causes hematological disorders and is the leading cause of nonimmunological fetal hydrops during pregnancy. During infection, B19V expresses two structural proteins, VP1 and VP2, and three nonstructural proteins, NS1, 11-kDa, and 7.5-kDa. While NS1 is essential, 11-kDa plays an enhancing role in viral DNA replication. Here, we elucidated a mechanism underlying 11-kDa protein-regulated B19V DNA replication. 11-kDa is tightly associated with cellular growth factor receptor-bound protein 2 (Grb2) during infection. In vitro, 11-kDa interacts with Grb2 with high affinity through three proline-rich motifs, of which at least one is indispensable for the regulation of viral DNA replication. 11-kDa and Grb2 interaction disrupts extracellular signal-regulated kinase (ERK) signaling, which mediates upregulation of B19V replication. Thus, our study reveals a novel mechanism of how a parvoviral small nonstructural protein regulates viral DNA replication by interacting with a host protein that is predominately expressed in the cytoplasm.
KEYWORDS: B19 virus, DNA replication, parvovirus
ABSTRACT
Lytic infection of human parvovirus B19 (B19V) takes place exclusively in human erythroid progenitor cells of bone marrow and fetal liver, which disrupts erythropoiesis. During infection, B19V expresses three nonstructural proteins (NS1, 11-kDa, and 7.5-kDa) and two structural proteins (VP1 and VP2). While NS1 is essential for B19V DNA replication, 11-kDa enhances viral DNA replication significantly. In this study, we confirmed the enhancement role of 11-kDa in viral DNA replication and elucidated the underlying mechanism. We found that 11-kDa specially interacts with cellular growth factor receptor-bound protein 2 (Grb2) during virus infection and in vitro. We determined a high affinity interaction between 11-kDa and Grb2 that has an equilibrium dissociation constant (KD) value of 18.13 nM. In vitro, one proline-rich motif was sufficient for 11-kDa to sustain a strong interaction with Grb2. In consistence, in vivo during infection, one proline-rich motif was enough for 11-kDa to significantly reduce phosphorylation of extracellular signal-regulated kinase (ERK). Mutations of all three proline-rich motifs of 11-kDa abolished its capability to reduce ERK activity and, accordingly, decreased viral DNA replication. Transduction of a lentiviral vector encoding a short hairpin RNA (shRNA) targeting Grb2 decreased the expression of Grb2 as well as the level of ERK phosphorylation, which resulted in an increase of B19V replication. These results, in concert, indicate that the B19V 11-kDa protein interacts with cellular Grb2 to downregulate ERK activity, which upregulates viral DNA replication.
IMPORTANCE Human parvovirus B19 (B19V) infection causes hematological disorders and is the leading cause of nonimmunological fetal hydrops during pregnancy. During infection, B19V expresses two structural proteins, VP1 and VP2, and three nonstructural proteins, NS1, 11-kDa, and 7.5-kDa. While NS1 is essential, 11-kDa plays an enhancing role in viral DNA replication. Here, we elucidated a mechanism underlying 11-kDa protein-regulated B19V DNA replication. 11-kDa is tightly associated with cellular growth factor receptor-bound protein 2 (Grb2) during infection. In vitro, 11-kDa interacts with Grb2 with high affinity through three proline-rich motifs, of which at least one is indispensable for the regulation of viral DNA replication. 11-kDa and Grb2 interaction disrupts extracellular signal-regulated kinase (ERK) signaling, which mediates upregulation of B19V replication. Thus, our study reveals a novel mechanism of how a parvoviral small nonstructural protein regulates viral DNA replication by interacting with a host protein that is predominately expressed in the cytoplasm.
INTRODUCTION
Human parvovirus B19 (B19V) is one of only two parvoviruses that have been confirmed to be pathogenic to humans (1, 2). While B19V infection does not constitute a severe outcome in healthy people, it can result in serious, and occasionally fatal, hematologic diseases in susceptible patients. Acute B19V infection can cause transient aplastic crisis in sickle-cell disease patients. Pure red cell aplasia and chronic anemia often happen in immunocompromised patients infected with B19V (2, 3). More importantly, B19V infection is the leading cause of nonimmunological fetal hydrops during pregnancy (4) and is one of the key etiological agents of congenital viral infections which traverse the uterine-placental interface (5). It can cause aplastic crisis in the fetus and hydrops fetalis, which occurs as a result of infection-induced anemia in pregnant women (6–9). Infection of B19V in the first trimester of pregnancy has an approximate 10% risk of fetal loss.
B19V belongs to species Primate erythroparvovirus 1 in the genus Erythroparvovirus of the family Parvoviridae (10). Productive infection of B19V takes place exclusively in CD36+ human erythroid progenitor cells (EPCs) at the stage from the late burst-forming unit-erythroid (BFU-E) to CFU-erythroid (CFU-E) in human bone marrow and fetal liver (11–15). From the 5.6-kb linear single-stranded DNA (ssDNA) genome, which is flanked by two identical terminal repeats (ITRs) (16), a total of five viral proteins are encoded, including three nonstructural proteins (NS1, 11-kDa, and 7.5-kDa) and two capsid proteins (VP1 and VP2) (17). As the most studied large nonstructural protein, NS1 has been shown to serve multiple functions in viral DNA replication, transcription transactivation, cytotoxicity, and cell cycle regulation (18–22). The functions of the other two nonstructural proteins, 7.5-kDa and 11-kDa, are poorly understood. The 11-kDa protein is highly expressed during infection (23), suggesting its potential importance in the B19V life cycle. It has been reported to interact with human growth factor receptor binding protein 2 (Grb2) in vitro (24), to induce apoptosis (23), and to play a role in virus egress (18). The function of the 7.5-kDa protein is currently unknown.
Recently, we identified the function of the 11-kDa protein, in the context of a B19V reverse genetics system, as having an important role in viral DNA replication (25). Grb2 is an adaptor protein that couples activated receptor tyrosine kinases to downstream effectors and regulators. Grb2 protein can be illustrated based on its functional Src homology 3 (SH3) domains as nSH3-SH2-cSH3 (where nSH3 and cSH3 are N-terminal SH3 and C-terminal SH3, respectively) (26). It binds to receptor tyrosine kinases via its SH2 domain (27, 28). Upon activation, the SH3 domains of Grb2 serve as docking sites for a variety of proteins containing proline-rich (PR) motifs. Examples of the best-characterized binding partners of Grb2 SH3 domains are the SOS guanine nucleotide exchange factor 1 (SOS1) (29, 30) and the adaptor protein Grb2-associated binding protein 1 (Gab1) (31, 32). The engagement of these partners further mediates the downstream cellular signaling cascades, including the classic SOS–Ras–mitogen-activated protein kinase (MEK)–extracellular signal-regulated kinase 1 and 2 (ERK1/2) activation (33–36). Knockdown of Grb2 expression usually results in significantly reduced phosphorylation of ERK1/2 in various cell types (37–39), including CD34+ hematopoietic stem cells (HSCs) (40) that are the precursors of the CD36+ EPCs used in the current study.
During B19V infection of CD36+ EPCs, MEK/ERK signaling was found to be a negative regulator of B19V replication (41). While overactivation of ERK, shown by an increased level of phosphorylated ERK (pERK), significantly inhibits B19V infection, blockage of ERK activation boosts viral replication dramatically. Interestingly, ERK is a well-known classic effector downstream of Grb2 (42). Therefore, we hypothesize that during infection, B19V expresses 11-kDa abundantly, which interacts with and Grb2 and blocks activation of the downstream ERK, which enhances B19V DNA replication.
In this study, we examined this hypothesis by generating an 11-kDa knockout and proline-rich motif mutants in a B19V infectious clone (M20), which were then used to determine the efficiency in viral DNA replication of transfected UT7/Epo-S1 cells, and by short hairpin RNA (shRNA) knockdown of Grb2 during B19V infection of CD36+ EPCs. We showed the enhancement role of 11-kDa in B19V DNA replication. The interaction between 11-kDa and Grb2 was confirmed both in vitro and in vivo. By performing site-directed mutagenesis, we identified that 11-kDa processes three functionally redundant proline-rich motifs, of which at least one is required for Grb2 binding to 11-kDa. Finally, the 11-kDa and Grb2 interaction significantly reduced cellular ERK phosphorylation in both transfected UT7/Epo-S1 cells and B19V-infected CD36+ EPCs.
RESULTS
Knockout of 11-kDa reduces viral DNA replication of the B19V infectious clone independent of expression of other viral proteins.
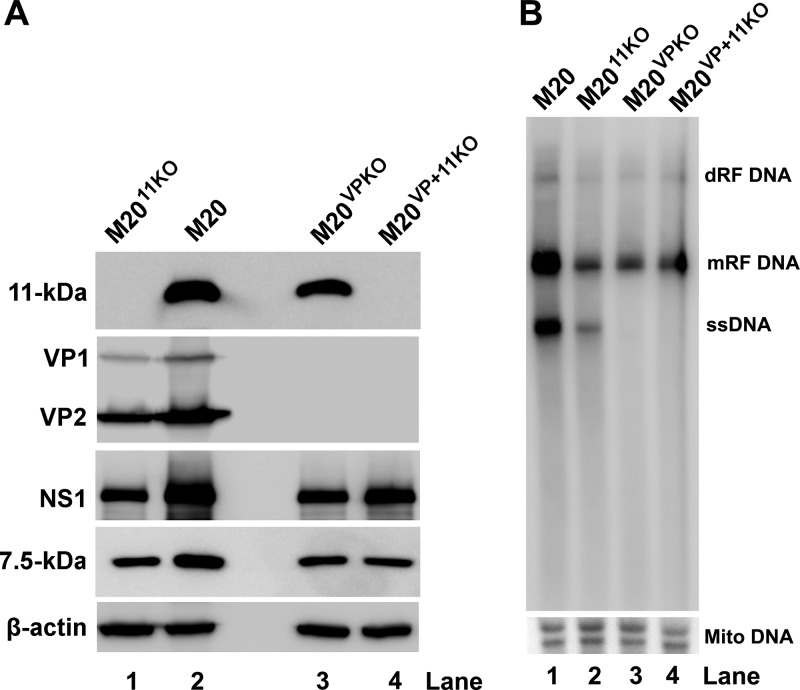
To systematically examine the role of 11-kDa expression in viral DNA replication, wild-type (WT) B19V infectious clone pM20 and its mutants with knockout (KO) of 11-kDa, VP1/2, or both 11-kDa and VP1/2 (pM2011KO, pM20VPKO, and pM20VP+11KO, respectively) were digested with SalI to obtain the linear replicative-form (RF) B19V genomes, which were then delivered into hypoxia-primed UT7/Epo-S1 cells via nucleofection. At 2 days postnucleofection, Western blotting results showed a complete knockout of 11-kDa in M2011KO (Fig. 1A, lane 1). M2011KO still expressed nonstructural proteins NS1 and 7.5-kDa at levels similar to those from the M20VPKO and M20VP+11KO but at levels lower than those from the WT M20 (Fig. 1A, NS1 and 7.5-kDa). As expected, M2011KO expressed VP1/2, but M20VPKO and M20VP+11KO did not (Fig. 1A, VP1/2).
FIG 1.
Knockout of 11-kDa significantly reduces the level of B19V replication. UT7/Epo-S1 cells were primed in hypoxia for 2 days prior to nucleofection. Wild-type (WT) and mutants of pM20 were digested with SalI before being nucleofected. (A) At 48 h postnucleofection, cells were harvested for Western blotting using antibodies, as indicated. β-Actin was probed as a loading control. (B) At 48 h postnucleofection, Hirt DNA was extracted from the cells and analyzed with Southern blotting using an M20 (B19V nt 1 to 5596) probe. The blot was reprobed with a mitochondrial DNA (Mito DNA) probe. dRF, double replicative form; mRF, monomer replicative form.
We then tested the effect of the 11-kDa null mutant on viral DNA replication by transfecting the RF B19V genomes into UT7/Epo-S1 cells and analyzing viral DNA replication using Southern blotting. We found that 11-kDa knockout drastically reduced the level of both monomer RF (mRF) DNA and ssDNA (Fig. 1B, lane 1 versus lane 2). In comparison to the VP1/2 knockout-only mutant, M20VPKO, the double (both VP1/2 and 11-kDa)-knockout mutant, M20VP+11KO, showed no appreciable difference in terms of the levels of mRF DNA (Fig. 1B, lanes 3 and 4). Notably, no obvious ssDNA was produced in either of the VP knockout mutants (Fig. 1B, lanes 3 and 4), suggesting that ssDNA production relies on the presence of the assembled viral capsids. In other words, the ssDNA is likely not presented as the free-DNA form but, rather, only as encapsidated into virions.
Taken together, our results suggest that it is unlikely that the 11-kDa-enhanced B19V DNA replication is mediated by regulating expression of other viral proteins.
The 11-kDa protein interacts with Grb2 in vitro with a high affinity.
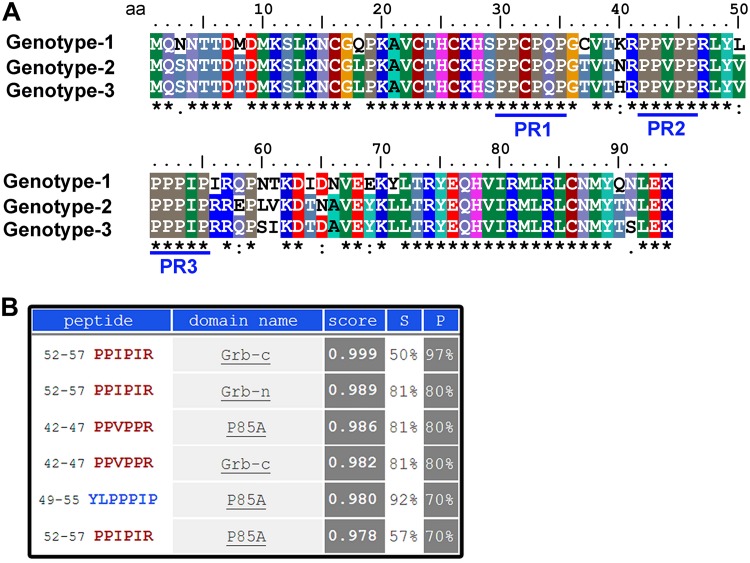
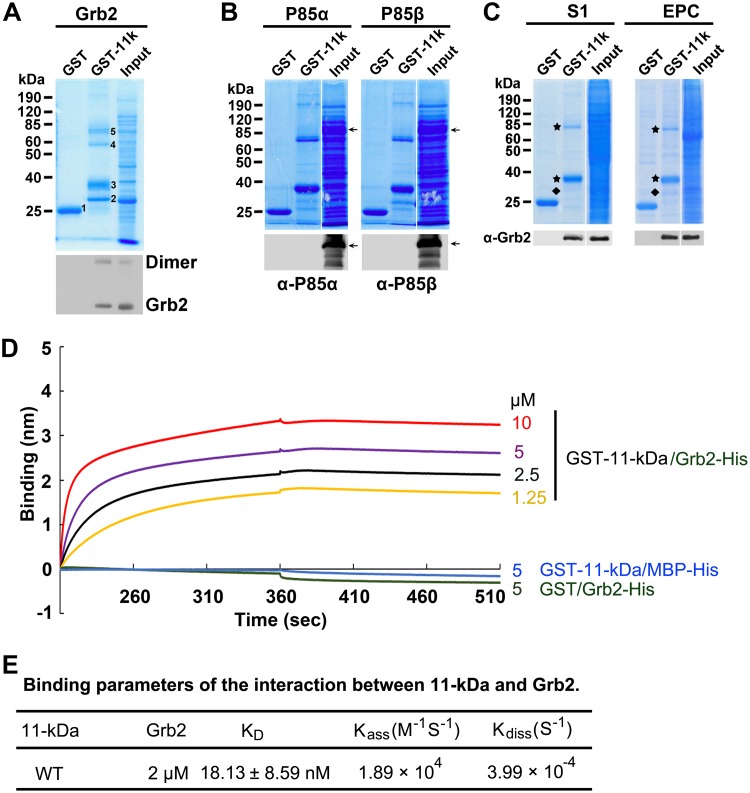
11-kDa contains three well-conserved putative Src homology 3 (SH3) binding domains (proline-rich motifs) at amino acids (aa) 30 to 35, 42 to 46, and 51 to 55, respectively, which are conserved in three genotypes of B19V (Fig. 2A). The candidates that potentially bind to the 11-kDa protein were identified by in silico analysis at http://cbm.bio.uniroma2.it/SH3-Hunter. The candidates with scores higher than threshold levels were chosen from the program output and are presented in Fig. 2B. Two of the three SH3 binding domains on 11-kDa showed a nearly perfect score for binding to human Grb2 protein at the C terminus (Grb-c), while one of the two also showed good computational affinity to the N terminus (Grb-n). In addition to human Grb2, the p85α subunit of the phosphoinositide 3-kinase (PI3K) also had good scores for binding to the proline-rich motif at aa 42 to 46. To test the computational predictions, we first employed an in vitro pulldown assay to determine the binding between glutathione S-transferase (GST)–11-kDa (N-terminal GST) and the two candidates (Grb2 and PI3K p85α) plus the p85β subunit of PI3K as a control. While human Grb2 showed an interaction with GST–11-kDa (Fig. 3A), both p85α and p85β failed to be pulled down by GST–11-kDa (Fig. 3B). We further used purified GST–11-kDa to pull down the native form of Grb2 out of the whole-cell lysate prepared from M20-transfected UT7/Epo-S1 cells or B19V-infected CD36+ EPCs. The result further confirmed the interaction between Grb2 and 11-kDa (Fig. 3C).
FIG 2.
11-kDa protein contains three conserved proline-rich motifs. (A) Alignment of amino acid sequence of the 11-kDa proteins. The 11-kDa amino acid sequences of the B19V representative genotype 1 (J35 isolate; GenBank accession no. AY386330), genotype 2 (A6 isolate; GenBank accession no. AY064475), and genotype 3 (V9 isolate; GenBank accession no. AJ249437) were chosen for alignment using ClustalW2 (65). Identical amino acids are indicated with a star symbol, while semiconserved and conserved amino acids are indicated with periods and colons, respectively, under the amino acid sequences of the 11-kDa proteins. The three conserved proline-rich motifs (PR1 to PR3) are underlined. (B) In silico analysis. The potential 11-kDa-interacting proteins were analyzed by the SH3 hunter program (http://cbm.bio.uniroma2.it/SH3-Hunter/).
FIG 3.
11-kDa specifically interacts with Grb2 but not P85α in vitro. (A to C) GST pulldown assay. Purified GST and GST–11-kDa were used as bait to pull down Grb2 from bacteria lysate. Numbered bands are as follows: 1, GST control; 2, Grb2; 3, GST–11-kDa; 4, putative Grb2 dimer; 5, putative GST–11-kDa dimer (A). Purified GST and GST–11-kDa were used as bait to pull down PI3K p85α and p85β subunits from bacterial lysate. Arrow shows the expressed p85α and p85β at ∼85 kDa (B). Purified GST and GST–11-kDa were used as bait to pull down Grb2 from lysate of M20-transfected UT7/Epo-S1 (S1) cells or B19V-infected EPCs. Both Coomassie blue staining and Western blotting were performed to examine the pulldown results (C). Diamond, Grb2; star, GST-11kDa monomers and dimers. (D and E) Biolayer interferometry (BLI) analysis of the interaction between 11-kDa and Grb2. BLI sensograms show the association and dissociation of Grb2 protein (at 2 μM) with WT 11-kDa at different concentrations, as indicated. Concentrations of 5 μM GST–11-kDa with 2 μM MBP-His and 5 μM GST with 2 μM MBP-Grb2-His were set up as negative controls (D). Binding parameters and Kdiss and Kass were used to calculate KD values (ratio of dissociation to association rate constant). Experiments were repeated at least three times for calculating the means and standard deviations of the KD.
We next used biolayer interferometry (BLI) to determine the kinetics of the 11-kDa and Grb2 protein interaction in vitro. We purified GST–11-kDa and maltose binding protein (MBP)-Grb2-His for this purpose. We first dipped the Ni-nitrilotriacetic acid (NTA) biosensor tips into 2 µM MBP-Grb2-His and then into GST–11-kDa at various concentrations. We observed a strong association of GST–11-kDa with the sensor at 10, 5, 2.5, or 1.25 µM, and almost no dissociations in the binding buffer containing 100 mM NaCl were observed (Fig. 3D). As controls, neither an interaction between MBP and GST–11-kDa nor between Grb2 and GST was observed (Fig. 3D). The equilibrium dissociation constant (KD) between 11-kDa and Grb2 was determined as 18.13 ± 8.59 nM by dividing the dissociation rate constant (Kdiss) by the association rate constant (Kass) (Fig. 3E), suggesting a high-affinity protein-protein interaction.
The 11-kDa protein associates with Grb2 during infection.
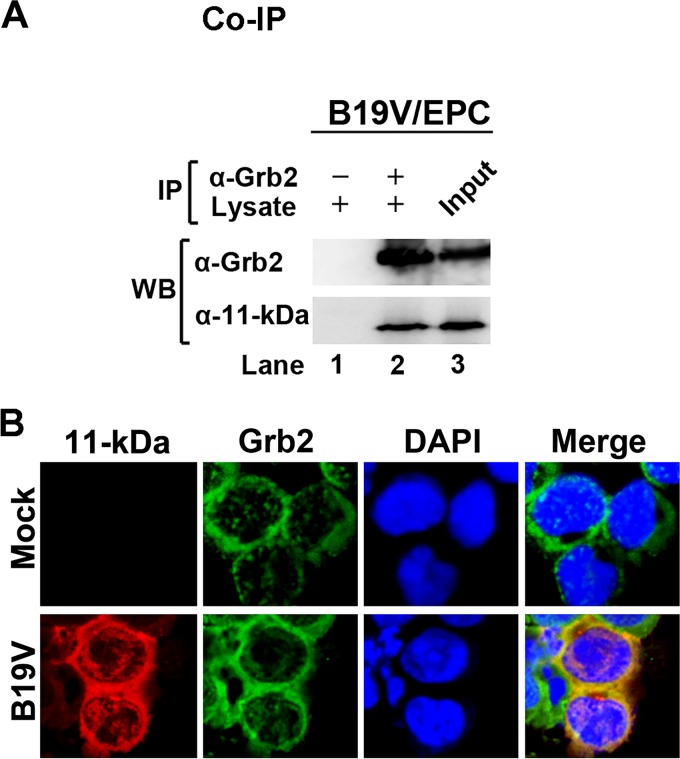
The binding of 11-kDa and the native Grb2 was further observed in a coimmunoprecipitation (co-IP) assay using B19V-infected CD36+ EPCs. Both cellular Grb2 and 11-kDa were coimmunoprecipitated when anti-Grb2 was added to the infected cell lysate (Fig. 4A). Furthermore, the immunofluorescence assay showed colocalization of 11-kDa with Grb2 predominately in the cytoplasm of B19V-infected CD36+ EPCs (Fig. 4B). There were minor associations of 11-kDa with Grb2 in the nucleus, which is consistent with a previous report that partial Grb2 was also located in the nucleus (43). These results demonstrated an association between viral 11-kDa and cellular Grb2 in B19V-infected CD36+ EPCs.
FIG 4.
11-kDa specifically interacts with cellular Grb2 in vivo during infection. (A) Coimmunoprecipitation (co-IP) assay. An anti-Grb2 antibody was used to bind with and cross-link to beads, followed by pulldown of 11-kDa from lysate of B19V-infected EPCs at 48 h postinfection. Western blotting (WB) was used to identify the pulled down proteins, 11-kDa and Grb2, using anti-11-kDa and anti-Grb2, respectively. (B) Immunofluorescence assay. At 48 h postinfection, B19V-infected EPCs were harvested and coimmunostained for 11-kDa (red), Grb2 (green), and nucleus (blue) simultaneously. The cells were then visualized by confocal microscopy. DAPI, 4′,6′-diamidino-2-phenylindole.
Knockdown of cellular Grb2 facilitates B19V infection.
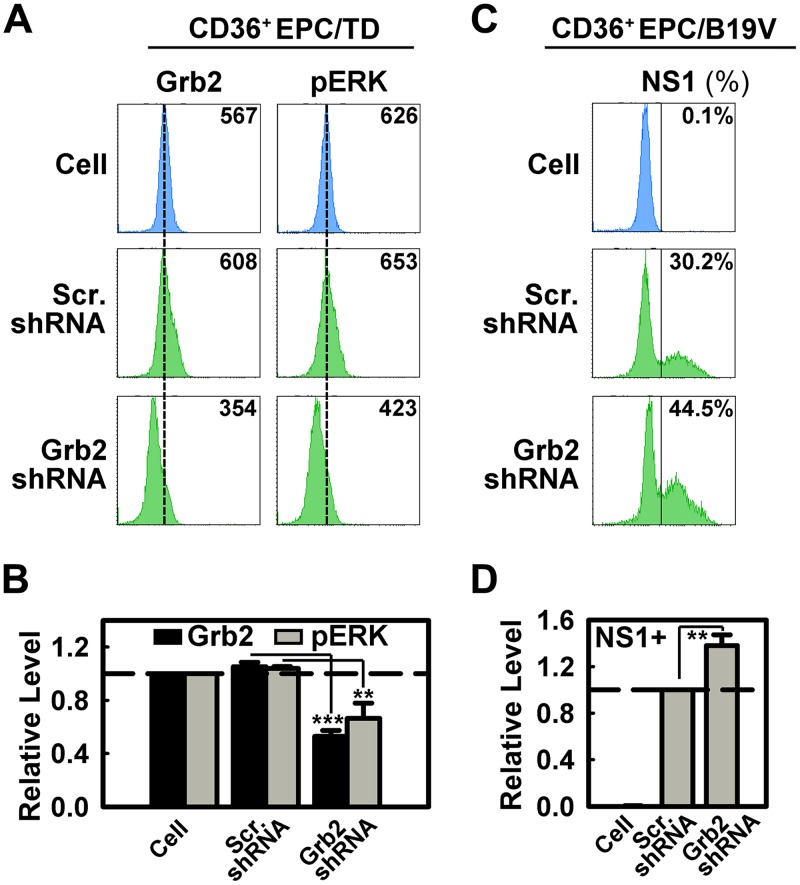
Given the specific 11-kDa–Grb2 interaction, the importance of 11-kDa in B19V replication, and the high abundance of the 11-kDa protein during infection (23), we hypothesized that cellular Grb2 plays an important role in B19V infection. To test this hypothesis, we produced lentivirus encoding an shRNA specifically targeting the human Grb2 gene and applied it to CD36+ EPCs. In a representative experiment shown in Fig. 5A, the pERK peak matched the GrB2 peak. The Grb2-specific shRNA decreased the level of cellular Grb2 expression by ∼50% and pERK expression by ∼40%, as determined as mean fluorescence intensities (MFI) by flow cytometry (Fig. 5B). This result is consistent with previous reports that downregulation of cellular Grb2 resulted in a decrease of ERK activation (phosphorylation of ERK) (37–40). In agreement with our previous finding that ERK activation negatively regulates B19V infection (41), downregulation of cellular Grb2 using a Grb2-specific shRNA consistently increased the level of NS1-positive cells by ∼40% in B19V-infected CD36+ EPCs (Fig. 5C and D).
FIG 5.
Grb2 knockdown facilitates B19V infection. (A and B) CD36+ EPCs on day 7 were transduced with lentiviruses. Cells were harvested at 48 h postransduction. The cells were analyzed by flow cytometry for cellular Grb2 and phosphorylated ERK (pERK) expression as values of mean fluorescence intensity (MFI). A representative experiment is shown in panel A. Statistical analyses of three independent experiments are plotted with levels relative to those of control cells. (C and D) CD36+ EPCs on day 7 were transduced with lentivirus expressing a scrambled shRNA (Scr.shRNA) or shRNA targeting Grb2 (Grb2shRNA), followed by B19V infection on day 9. Cells were harvested at 48 h postinfection and analyzed for NS1 expression as a percentage of NS1-expressing cells. A representative experiment is shown in panel C. Statistical analyses of NS1 expression levels from three independent experiments are plotted with levels relative to those of the Scr.shRNA-treated group. **, P < 0.01; ***, P < 0.001.
Thus, the above results support the idea that Grb2 regulates the ERK activity of CD36+ EPCs, which controls B19V infection.
ERK activity is reduced in cells expressing 11-kDa or infected with B19V.
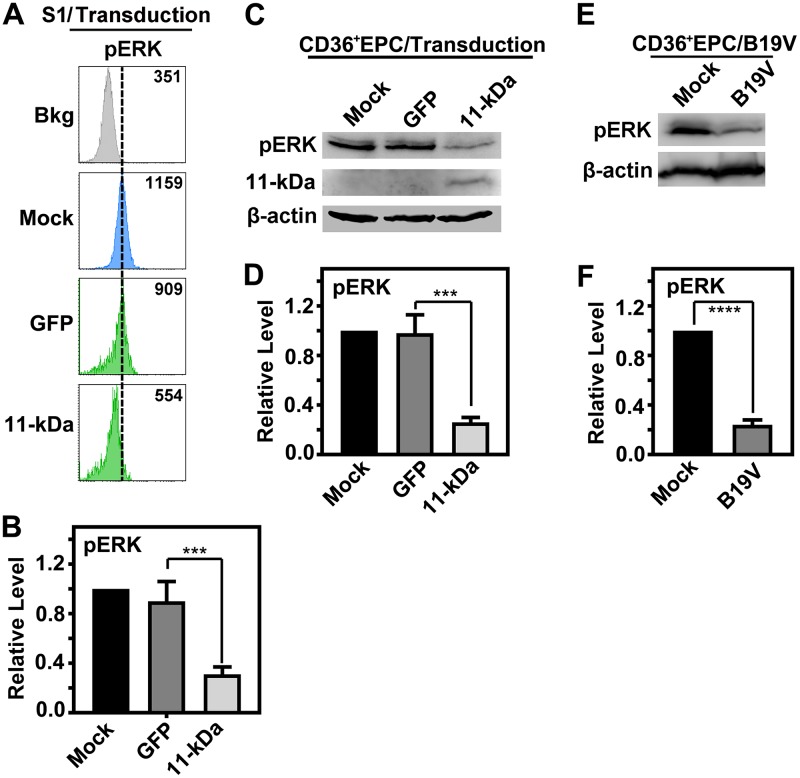
To further interrogate the 11-kDa–Grb2–ERK pathway, we examined the level of pERK in UT7/Epo-S1 cells which expressed green fluorescent protein (GFP) or both GFP and 11-kDa. By selection of the GFP-positive population, the reduction of ERK activity by 11-kDa expression was significantly detected by flow cytometry analysis (Fig. 6A and B). Consistently, in CD36+ EPCs transduced with 11-kDa-encoding retrovirus, the level of pERK was significantly decreased as well (Fig. 6C and D). As a control, during B19V infection, the level of ERK activity was also consistently decreased (41) (Fig. 6E and F).
FIG 6.
B19V infection and 11-kDa expression only reduce the level of cellular phosphorylated ERK (pERK). (A and B) UT7/Epo-S1 cells were primed in hypoxia for 2 days prior to treatment. Cells were transduced with either a retrovirus made from pMSCV-IRES-GFP (GFP) or from pMSCV-OPT11k-IRES-GFP (11-kDa), as indicated. At 48 h postransduction, cells were harvested and stained with anti-pERK, followed by flow cytometry analysis. (A) Mean fluorescence intensity (MFI) values are shown. (B) Statistical analyses of the relative levels of ERK phosphorylation are shown. (C and D) CD36+ EPCs were transduced with respective retroviruses as described above, expressing either GFP or 11-kDa. At 48 h postransduction, cells were harvested and analyzed by Western blotting. Statistical analyses of the relative levels of ERK phosphorylation are shown. (E and F) CD36+ EPCs were infected with B19V. At 48 h postinfection, cells were collected for Western blot analysis. A representative blot is shown. Statistical analyses of the relative levels of ERK phosphorylation are shown. ***, P < 0.001; ****, P < 0.0001.
Collectively, these results confirmed that 11-kDa expression decreases cellular ERK (phosphorylation) activity.
The 11-kDa protein interacts with Grb2 via three redundant proline-rich motifs.
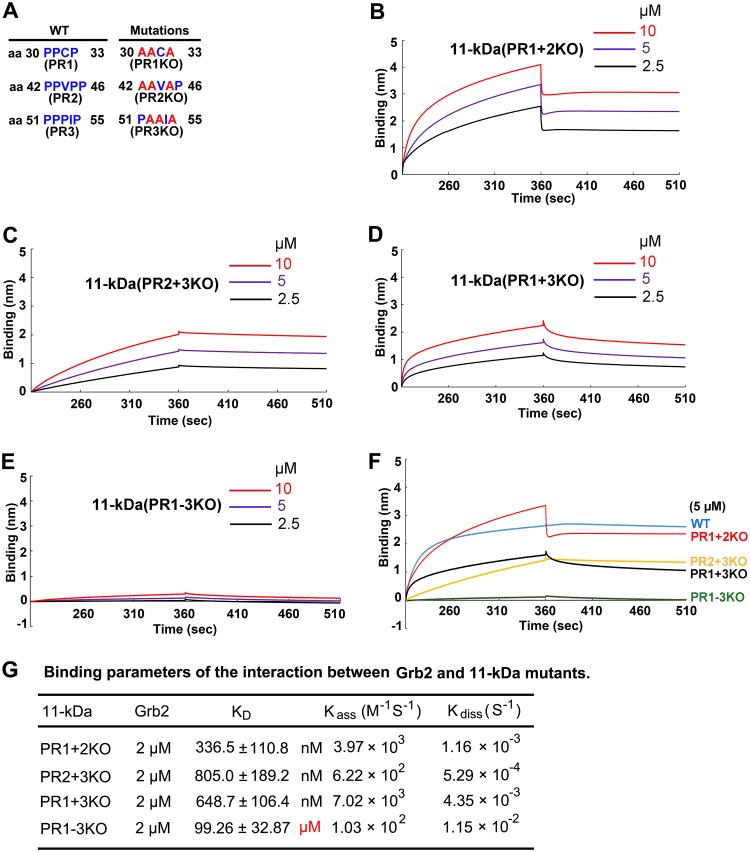
B19V 11-kDa contains three proline-rich motifs (Fig. 7A), and all three motifs have the potential to bind cellular Grb2 (24). To investigate whether all of these SH3 binding motifs are important in facilitating B19V replication, we mutated the proline-rich motifs in various combinations. Knockout of two out of three proline-rich motifs decreased the binding affinity of the mutant 11-kDa with Grb2 compared with that of the WT 11-kDa but did not disrupt the interaction between 11-kDa and Grb2 in vitro, as shown in the biolayer interferometry (Fig. 7B to D). Only knockout of all three proline-rich motifs (PR1 to PR3) completely abolished the 11-kDa and Grb2 interaction (Fig. 7E). A comparison of the interaction kinetics of Grb2 with the WT and four mutants is shown in Fig. 7F, and their KD values were calculated and are shown in Fig. 7G. These results indicate that these three proline-rich motifs are functionally redundant in binding with cellular Grb2 and that at least one is required for a functional binding.
FIG 7.
One proline-rich motif is sufficient for 11-kDa to interact with Grb2 in vitro. (A) Mutations of proline-rich motifs. The proline (blue) residues which were mutated to alanine (red) in each mutant are shown. The numbers indicate the position of the amino acid residue in the 11-kDa protein. (B to E) Biolayer interferometry (BLI) BLI sensograms show association and dissociation of the MBP-Grb2-His protein (2 μM) with GST–11-kDa proline-rich (PR) motif mutants at different concentrations, as indicated. (F) Comparison of binding kinetics of all four PR mutants with that of WT 11-kDa. (G) Binding parameters. The KD value is the ratio of the dissociation to the association rate constant. Experiments were repeated at least three times for calculating the means and standard deviations of the KD.
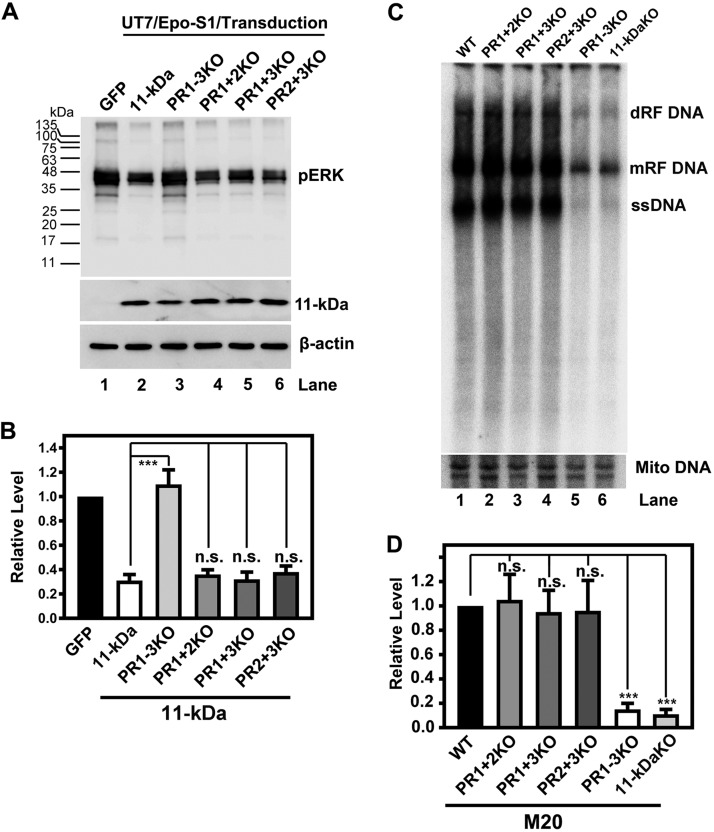
To examine the function of the proline-rich motifs of 11-kDa in cells, the mutant 11-kDa proteins were expressed in UT7/Epo-S1 cells to determine the level of ERK activity. While knockout of all three WT proline-rich motifs (PR1 to PR3)successfully rescued the decrease of pERK (Fig. 8A and B, PR1–3KO), knockout of any two of them still significantly reduced the level of cellular pERK as did the WT (Fig. 8A and B, lanes 4 to 6). This result, in line with the in vitro interaction assay (Fig. 7), indicates the redundancy of these three proline-rich motifs in mediating 11-kDa and Grb2 interaction.
FIG 8.
One proline-rich motif is sufficient for 11-kDa to decrease ERK phosphorylation and to increase B19V DNA replication. (A and B) Mutations of two proline-rich (PR) motifs in the 11-kDa protein are not enough to abolish the inhibition of ERK phosphorylation. Hypoxia-primed UT7/Epo-S1 cells were transduced with the indicated retroviruses made from pMSCV-IRES-GFP (GFP) and pMSCV-OPT11k-IRES-GFP (11-kDa) and 11-kDa mutant retroviruses (PR1–3KO, PR1 + 2KO, PR1 + 3KO, and PR2 + 3KO). (A) At 48 h postransduction, cells were harvested and analyzed by Western blotting using anti-pERK. The blot was reprobed with anti-11-kDa and anti-β-actin. (B) Statistical analyses of the relative levels of ERK phosphorylation from three independent experiments are shown. (C and D) Mutations of two proline-rich motifs in the 11-kDa protein are not enough to decrease B19V DNA replication. UT7/Epo-S1 cells were nucleofected with the indicated plasmids, including WT pM20 (WT), M20 11-kDa KO. and proline-rich-domain knockouts of 11-kDa (PR1–3KO, PR1 + 2KO, PR1 + 3KO, and PR2 + 3KO). At 48 h postnucleofection, cells were harvested for Hirt DNA extraction. The Hirt DNA samples were then digested with DpnI, followed by Southern blot analysis using the M20 probe. The blot was reprobed using a mitochondrial (Mito) DNA probe. (D) Statistical analyses of the relative levels of the mRF DNA from three independent experiments are shown. ***, P < 0.001; ns, not significant (P > 0.05).
In order to examine the function of the proline-rich motifs in the context of viral DNA replication, the same mutations were made to the infectious clone, pM20. After nucleofecting these proline-rich domain M20 mutants into UT7/Epo-S1 cells, Southern blotting was employed to determine the effect of these mutations on viral DNA replication. Knockout of all three proline-rich motifs showed an effect on viral DNA replication similar to that of the complete knockout of 11-kDa (Fig. 8C, compare lanes 5 and 6). Importantly, knockout of any two of the three proline-rich motifs did not significantly decrease the level of viral DNA replication (Fig. 8C and D, lanes 2 to 4). As controls, knockout of any two of the three proline-rich motifs still significantly reduced the level of pERK but not that of the protein that has all the three proline-rich motifs mutated (PR1–3KO) (data not shown), which is consistent with what is shown in Fig. 8A and B.
Taken together, all of these results support the conclusion that these three proline-rich motifs are functionally redundant in interacting with cellular Grb2 and in facilitating B19V DNA replication.
DISCUSSION
The interaction between 11-kDa and Grb2 was first demonstrated in an in vitro binding assay (24). Fortunately, with the B19V reverse genetics system, we were able to explore the significance of this interaction in B19V-permissive cells. By engineering 11-kDa and VP knockouts in the infectious clone, we showed that the 11-kDa protein plays an enhancement role in B19V DNA replication while it does not affect the basal level of B19V DNA replication driven by NS1. Furthermore, we demonstrated the specific interaction between 11-kDa and Grb2 during B19V infection, which is mediated by three functionally redundant proline-rich motifs in vitro. Importantly, even one proline-rich motif is sufficient to convey the interaction between 11-kDa and Grb2, which significantly reduces cellular ERK activity, and, as a result, facilitates B19V replication.
Upregulation of ERK activity has been observed during infection by various viruses and implicated as a positive factor for both virus entry and intracellular trafficking during infection (44). For some DNA viruses, ERK also facilitates virus replication through cell cycle regulation (45). Interestingly, in consistency with our previous findings (41), ERK signaling is a negative regulator in B19V replication. The MEK/ERK pathway is indispensable in downstream signaling of Epo and Epo receptor (EpoR)-dependent proliferation and survival of EPCs. Phosphorylation of EpoR at the tyrosine 489 (Y489) position mediates the interaction of EpoR and the SH2 domain of Grb2, and the N-terminal SH3 domain of the Grb2 binds to the Ras-guanine exchange factor SOS, which transduces the signal through the Grb2-SOS-Ras-Raf1-MEK-ERK pathway (Fig. 9) (46). We propose that the interaction of 11-kDa with Grb2 disrupts the physiological binding of Grb2 with SOS, which therefore decreases ERK activity. We previously demonstrated that the hypoxic culture condition provided a fine-tuned balance between proliferation (ERK activity) and differentiation (STAT5 phosphorylation) that facilitates B19V replication (41). It is intriguing to see that B19V expresses 11-kDa, one of the only three nonstructural proteins expressed at high abundance (23), to specifically target cellular Grb2 and block the activation of the downstream ERK (Fig. 9). This finding underscores the remarkable negative regulation of B19V replication by ERK signaling. These results encourage future studies to further explore the significance of ERK-regulated downstream molecules and their potential effect on B19V replication. Any strategies to increase ERK activity in EPCs would be beneficial to limit B19V infection.
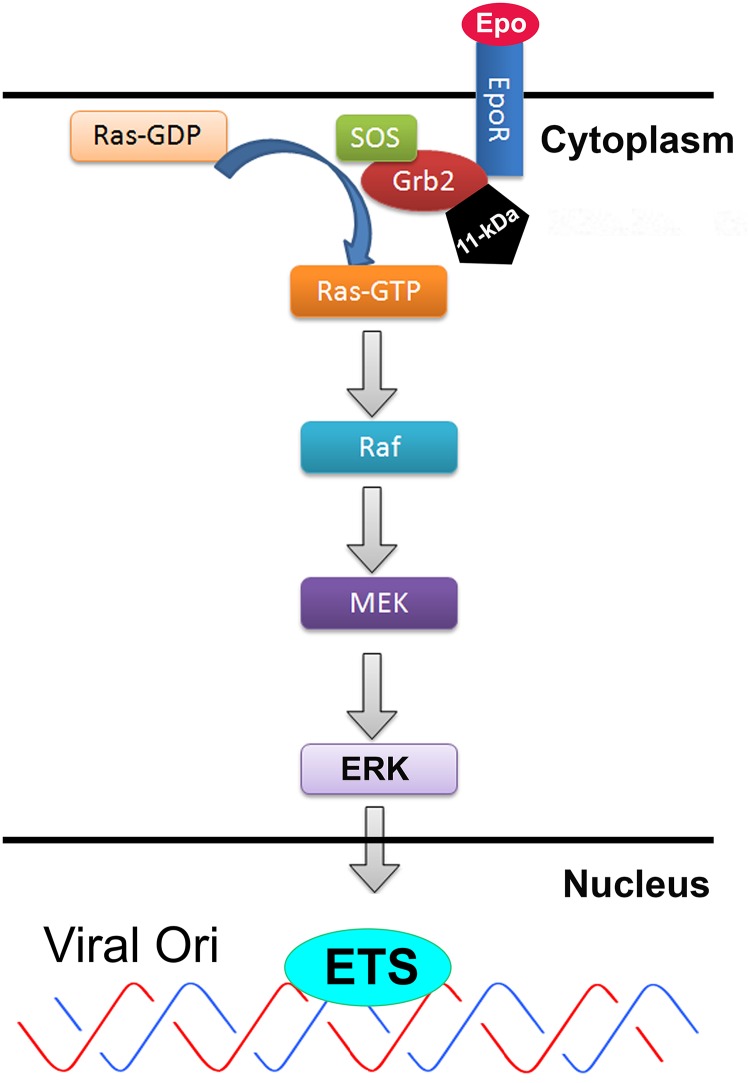
FIG 9.
Proposed model of the 11-kDa disruption of the EpoR-Grb2-SOS-Ras-MEK-ERK pathway. Upon Epo receptor (EpoR) activation through binding of Epo, Grb2 binds to tyrosine-phosphorylated EpoR either directly, through its SH2 domain, or indirectly, by binding to EpoR-associated tyrosine-phosphorylated SHC. Through the SH3 domain, Grb2 is constitutively associated with the guanine nucleotide-releasing factor (SOS) (29, 66, 67). Binding of Grb2 to EpoR leads to translocation of SOS to the membrane, where it triggers the exchange of GDP for GTP on Ras (68, 69); Ras-GTP then activates the Raf/MEK/ERK/ETS cascade (42). 11-kDa binding to Grb2 disrupts the interaction of Grb2 with SOS, which would block the exchange of GDP for GTP on Ras, and thereafter inhibits the Raf/MEK/ERK/ETS pathway. ERK activates ETS transcription factors, i.e., Elk-1 and c-Ets, which have been predicted to bind to the B19V replication origin (viral Ori) (20, 53).
ERK signaling plays an important role in cell cycle progression (47). Particularly, activation of ERK facilitates G2/M transition by phosphorylation (activation) of CDC25 (48, 49). B19V replication requires S-phase factors to replicate the viral genome (50). B19V infection induces a late S-phase arrest, represented by a prolonged S phase (BrdU+/DNA4N) at an early stage followed by a complete G2-phase (BrdU−/DNA4N) arrest at a late stage of infection (51, 52). The late S-phase arrest is induced by the B19V replication-induced DNA damage response, while the G2-phase arrest is largely induced by the NS1 protein (22). Only 11-kDa expression in cells induced apoptosis (23), but it did not induce G2-phase arrest (data not shown), suggesting that the positive role of ERK inhibition in B19V replication is not likely to be mediated by the cell cycle regulation. On the other hand, importantly, it has been predicted that there are consensus binding sites of two ERK-driven ETS transcription factors, Elk-1 and c-Ets, in the replication origin of B19V (20, 53). Of note, the binding sites of Elk-1 and c-Ets overlap the putative host factor binding sites in the minimal viral DNA replication origin (54). Thus, we hypothesize that ERK activation may phosphorylate ETS transcription factors, which bind to the B19V replication origin in competition with other host factors that enhance B19V DNA replication (Fig. 9). Overall, the mechanism underlying ERK-regulated B19V replication warrants further investigation.
In conclusion, the present study, in concert with our previous report (41), demonstrated the negative regulatory effect of ERK signaling on B19V DNA replication. In order to ensure the inhibition of ERK activity that optimizes viral DNA replication, the viral genome encodes the small nonstructural protein 11-kDa, which is abundantly expressed during infection, to interrupt signaling transduction by directly binding to Grb2 at proline-rich motifs in the cytoplasm. Thus, our study provides a unique model of viral DNA replication regulation by a small viral nonstructural protein which is expressed predominantly in the cytoplasm.
MATERIALS AND METHODS
Ethics statement.
CD34+ hematopoietic stem cells (HSCs) were isolated from bone marrow of a healthy human donor. We purchased them from the National Disease Research Interchange (NDRI) (Philadelphia, PA) without any identification information on the cells, and, therefore, an institutional review board (IRB) review was waived.
Primary and cell line cells.
(i) Ex vivo expansion of CD36+ EPCs. Upon arrival at the laboratory (defined as day 0), CD34+ HSCs were cultured in erythroid progenitor cell (EPC) expansion Wong medium (23, 55) under normoxia (5% CO2 and 21% O2) at 37°C until day 4 and were then frozen in liquid nitrogen at 0.5 × 106 cells/vial. These cells were defined as day 4 HSCs (23, 55). The day 4 HSCs were thawed and cultured in the Wong medium under hypoxia (5% CO2 and 1% O2) at 37°C until the time of treatment and B19V infection, as indicated in each figure legend. The hypoxia condition was achieved using the three-door chamber of a Heracell 150 Tri-Gas incubator (ThermoFisher).
(ii) UT7/Epo-S1 cells. UT7/Epo-S1 cells were obtained from Kevin Brown with permission from Kazuo Sugamura. They are megakaryoblastoid cell line cells with erythroid characteristics supporting B19V replication and were grown as described previously (56). While UT7/Epo-S1 cells were maintained under normoxia, they were primed under hypoxia for 2 days prior to nucleofection and kept under hypoxia until the time of analysis (41).
Virus and infection.
B19V viremic plasma (no. 13) was obtained from Viracor Eurofins Laboratories (Lee’s Summit, MO), and the numbers of B19V genome copies (vgc) per milliliter (1012 vgc/ml) were quantified as previously described (41). B19V infection was carried out by adding the B19V-containing plasma directly to the culture at a multiplicity of infection (MOI) of 2,000 vgc/cell.
Construction of plasmids.
(i) Mutants of the B19V infectious clone. pM2011KO, the 11-kDa knockout M20 mutant, was generated by mutating all the three ATGs (nucleotides [nt] 4890 to 4892, nt 4911 to 4913, and nt 4917 to 4919) (GenBank accession no. AY386330) at the 5ʹ end of the coding region to ACGs and has been described previously (25). pM20VPKO, the VP1/2 knockout mutant, was generated by mutating nt 3413 to 3424 to 5ʹ-TAA GCT AGC-3′, which introduced not only an early termination codon to capsid genes but also an XbaI site for cloning confirmation. The double-knockout mutant pM20VP+11KO was generated by combining the above two mutants. Based on pM20, the 11-kDa proline-rich domain knockout mutants were generated by mutating the proline residues in the 11-kDa coding region to alanines, as indicated in Fig. 7A. Mutations in the three proline-rich motifs did not alter the VP1/2 open reading frame (ORF) or NS1, 7.5-kDa, and VP1/2 expression (data not shown).
(ii) Bacterial expression plasmids. The wild-type (WT) 11-kDa open reading frame (ORF) was inserted in the EcoRI and XhoI sites of vector pGEX-4T3 (GE Life Sciences) to generate pGEX4T–11-kDa. The respective proline-rich motif mutants of 11-kDa were generated by mutating prolines to alanines, as indicated in Fig. 7A. ORFs of human Grb2 and the PI3K p85α and p85β subunits were cloned into EcoRI (BamHI) and XhoI sites of pET-26b(+) (MilliporeSigma).
pMBP-Grb2-His plasmid was constructed by cloning a Grb2-6×Histidine ORF in the pMAL-c5X vector (New England Biolabs) through NdeI and BamHI sites. pMBP-His was constructed by fusing 6×His to the MBP ORF at the C terminus in pMAL-c5X through the SacI site.
(iii) Lentiviral vectors expressing shRNAs. Plasmids pLKO-GFP and pLKO-GFP-Scramble-shRNA have been described previously (57). The validated shRNA sequence (5ʹ-CCG GTC CTC TCT GTC AAG TTT GGA AAC TCG AGT TTC CAA ACT TGA CAG AGA GGT TTT TC-3ʹ) (MilliporeSigma, St. Louis, MO) was cloned into pLKO-GFP using the AgeI and EcoRI sites to generate the pLKO-GFP-Grb2shRNA plasmid.
(iv) Retroviral vectors expressing codon-optimized 11-kDa and its mutants.The 11-kDa coding sequence was codon optimized for optimal expression in human cells at Integrated DNA Technologies, Inc. (Coralville, Iowa). The optimized 11-kDa coding sequence was inserted in the EcoRI and XhoI sites of the pMSCV-IRES-GFP plasmid (where MSCV is mouse stem cell virus and IRES is internal ribosome entry site), to generate pMSCV-OPT11k-IRES-GFP, according to a method described previously (41). Respective proline-rich motif mutants of 11-kDa were constructed by mutating the same prolines as indicated in Fig. 7A to alanines.
Viral vector production and transduction.
Lentivirus was produced and concentrated according to instructions from Addgene (http://www.addgene.org/plko). The retroviruses Retro-OPT11k-IRES-G and Retro-IRES-G (control) were produced by transfecting GP293 cells (TaKaRa Clontech) with pMSCV-OPT11k-IRES-GFP-WRE and pMSCV-MCS-IRES-GFP-WRE (57), respectively, together with pCMV-VSVG (where CMV is cytomegalovirus and VSVG is vesicular stomatitis virus G protein). The retroviral vectors were concentrated according to the manufacturer’s instructions (catalog no. PT3132-1; Clontech).
Concentrated lentiviral or retroviral vectors were inoculated to CD36+ EPCs (day 7 culture) at an MOI of ∼4 fluorescent focus units/cell as described previously (57).
Protein purification.
Glutathione S-transferase (GST) and GST-fused proteins were expressed in BL21(DE3) Escherichia coli cells and purified according to the manufacturer’s protocols by using GSTrap FF columns (GE Life Sciences) attached to a BioLogic LP low-pressure chromatography system (Bio-Rad). His-tagged proteins Grb2 and PI3K p85α and p85β were expressed in BL21(DE3) E. coli and used as crude lysates.
MBP-Grb2-His and MBP-His proteins were expressed in BL21(DE3) E. coli. Crude extracts were incubated with amylose resin according to the manufacturer’s instructions (New England Biolabs). After cells were washed with the washing buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]), proteins were then eluted with 10 mM maltose.
All proteins for biolayer interferometry analysis were dialyzed with Tris binding buffer (25 mM Tris-HCl, pH 7.4, and 100 mM NaCl), and the concentrations were measured using a Coomassie (Bradford) protein assay kit according to the manufacturer’s instructions (catalog no. 23200; Thermo Scientific).
Biolayer interferometry analysis.
A BLItz biolayer interferometry system (Pall ForteBio) was used to determine the binding kinetics of the GST-fused 11-kDa (GST–11-kDa) and 11-kDa proline rich (PR) motif mutants, GST–11-kDa (PR1 and PR2 KO, PR1 + 2KO; PR1 and PR3 KO, PR1 + 3KO; PR2 and PR3 KO, PR2 + 3KO; and PR1–3KO), with MBP-Grb2-His as described previously (25). Briefly, Ni-NTA biosensors were hydrated in Tris binding buffer (25 mM Tris-HCl, pH 7.4, and 100 mM NaCl) for 10 min, and MBP-Grb2-His or MBP-His (at 2 μM) was mounted on Ni-NTA biosensors and equilibrated with the binding buffer. Then the Ni-NTA biosensors were dipped into the binding buffer containing GST–11-kDa and its mutant proteins or GST alone (control) at different concentrations to determine the binding parameters, Kass (association rate constant) and Kdiss (dissociation rate constant), using BLItz Pro software.
Western and far-Western blot analysis.
Western blot analysis was carried out as previously described (58, 59). Signals were developed using SuperSignal West Pico chemiluminescent substrate (ThermoFisher) under the Fujifilm imager LAS 4000 (Fujifilm Life Sciences). Images were processed for quantification with Multi Gauge, version 2.3, software (Fujifilm Life Sciences). Antibodies used for Western blotting were as follows: mouse anti-11-kDa, anti-B19V VP1/2, anti-β-actin, rabbit anti-Grb2, and anti-pERK1/2. Secondary antibodies were horseradish peroxidase (HRP)-conjugated anti-rat and HRP-conjugated anti-mouse or anti-rabbit. β-Actin was used as a loading control.
For far-Western blotting, the procedure was essentially identical to that of Western blotting except that an additional 60-min incubation step was included in which lysates of protein-expressing bacteria in 1 ml of culture were added in 10 ml of blocking buffer. Then the membrane was further incubated with anti-Grb2, followed by incubation with a secondary antibody.
Nucleofection (transfection).
The B19V infectious clone (M20) (60) or various M20 mutants, as indicated in each figure legend, were digested with SalI. Two million hypoxia-primed UT7/Epo-S1 cells were electroporated with 3 µg of the digested DNA in Nucleofector Solution V using an Amaxa Nucleofector (Lonza, Basel, Switzerland), as described previously (61). After transfection, cells were cultured under hypoxia conditions (1% O2).
Southern blot analysis of viral DNA replication.
Low-molecular-weight (Hirt) DNA was extracted from the cells. The Southern bot was probed with 32P-labeled B19V M20 DNA as described previously (62) and was reprobed with a 32P-labeled probe for mitochondrial DNA (63). The blot was exposed to a phosphorimaging screen, scanned on a PhosphorImager (Typhoon FLA9400), and analyzed using Image Quant TL software (GE Life Sciences).
Pulldown assays.
(i) GST pulldown assay. A GST protein pulldown assay was performed using a MagneGST Pull-Down System (TM249; Promega) essentially according to the manufacturer’s instructions.
(ii) Co-IP assay. A coimmunoprecipitation (co-IP) assay was performed using a Pierce Crosslink IP kit (catalog no. 26147; ThermoFisher) according to the manufacturer’s instructions.
Immunofluorescence staining.
Cells were collected and poured on slides by using cytospin (Shandon Cytospin III). After cells were fixed and permeabilized in 100% ice-cold acetone, the slides were then blocked and probed with rabbit anti-Grb2 and mouse anti-11-kDa antibodies. The slides were visualized with a Nikon confocal microscope, and images were taken at a magnification of ×100 .
Flow cytometry analysis.
Intracellular staining was performed at room temperature, essentially as described previously (64). The following antibodies were used: rabbit anti-pERK1/2 and rat anti-NS1 (23). For flow cytometry by GFP selection, the secondary antibody used was Cy5-conjugated anti-rat or anti-rabbit antibody.
Antibodies used.
Mouse anti-B19V 11-kDa and rat anti-NS1 sera were obtained as described previously (23). The following antibodies were purchased from the indicated vendors: anti-VP1/2 (MAB8293, clone R92-F6) from Millipore, rabbit anti-Grb2 (NB110-57013) from Novus, anti-pERK1/2 (Thr202/Tyr204) monoclonal antibody (4377) from Cell Signaling, and anti-β-actin (A5441) from Sigma. Cy5-conjugated anti-rat (112-176-143) or anti-rabbit (111-176-144) from Jackson ImmunoResearch, HRP-conjugated anti-rat (112-035-003) from Jackson Laboratory, and HRP-conjugated anti-mouse (A4416) or anti-rabbit (A0545) from Sigma were also used.
Statistical analysis.
We used GraphPad Prism, version 7.0, to perform statistical analysis. The P values were determined by using one-way analysis of variance (ANOVA) for comparison of three or more groups and by Student’s t test for comparison of two groups. Error bars show mean and standard deviations (SD), and P values are defined in the figure legends.
ACKNOWLEDGMENTS
We thank members in the Qiu lab for technical support and valuable discussions. We acknowledge the Flow Cytometry Core Laboratory, The University of Kansas Medical Center, for help in flow cytometry analysis. We are indebted to the Lutkenhaus lab for the use of BLItz system.
This study was supported in full by PHS grant R01 AI070723 from the National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Brown KE. 2010. The expanding range of parvoviruses which infect humans. Rev Med Virol 20:231–244. doi: 10.1002/rmv.648. [DOI] [PubMed] [Google Scholar]
- 2.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young NS, Brown KE. 2004. Parvovirus B19. N Engl J Med 350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 4.Jelliffe-Pawlowski L, Baer R, Moon-Grady AJ, Currier RJ. 2011. Second trimester serum predictors of congenital heart defects in pregnancies without chromosomal or neural tube defects. Prenat Diagn 31:466–472. doi: 10.1002/pd.2720. [DOI] [PubMed] [Google Scholar]
- 5.Pereira L. 2018. Congenital viral infection: traversing the uterine-placental interface. Annu Rev Virol 5:273–299. doi: 10.1146/annurev-virology-092917-043236. [DOI] [PubMed] [Google Scholar]
- 6.Lamont R, Sobel J, Vaisbuch E, Kusanovic J, Mazaki-Tovi S, Kim S, Uldbjerg N, Romero R. 2011. Parvovirus B19 infection in human pregnancy. BJOG 118:175–186. doi: 10.1111/j.1471-0528.2010.02749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong EP, de Haan TR, Kroes AC, Beersma MF, Oepkes D, Walther FJ. 2006. Parvovirus B19 infection in pregnancy. J Clin Virol 36:1–7. doi: 10.1016/j.jcv.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Al-Khan A, Caligiuri A, Apuzzio J. 2003. Parvovirus B-19 infection during pregnancy. Infect Dis Obstet Gynecol 11:175–179. doi: 10.1080/10647440300025518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonvicini F, Bua G, Gallinella G. 2017. Parvovirus B19 infection in pregnancy-awareness and opportunities. Curr Opin Virol 27:8–14. doi: 10.1016/j.coviro.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Söderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozawa K, Kurtzman G, Young N. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233:883–886. doi: 10.1126/science.3738514. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava A, Lu L. 1988. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J Virol 62:3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young NS, Mortimer PP, Moore JG, Humphries RK. 1984. Characterization of a virus that causes transient aplastic crisis. J Clin Invest 73:224–230. doi: 10.1172/JCI111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young N, Harrison M, Moore J, Mortimer P, Humphries RK. 1984. Direct demonstration of the human parvovirus in erythroid progenitor cells infected in vitro. J Clin Invest 74:2024–2032. doi: 10.1172/JCI111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi T, Ozawa K, Takahashi K, Asano S, Takaku F. 1990. Susceptibility of human erythropoietic cells to B19 parvovirus in vitro increases with differentiation. Blood 75:603–610. [PubMed] [Google Scholar]
- 16.Deiss V, Tratschin JD, Weitz M, Siegl G. 1990. Cloning of the human parvovirus B19 genome and structural analysis of its palindromic termini. Virology 175:247–254. doi: 10.1016/0042-6822(90)90205-6. [DOI] [PubMed] [Google Scholar]
- 17.Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol 61:2395–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol 80:5941–5950. doi: 10.1128/JVI.02430-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffatt S, Yaegashi N, Tada K, Tanaka N, Sugamura K. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J Virol 72:3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gareus R, Gigler A, Hemauer A, Leruez-Ville M, Morinet F, Wolf H, Modrow S. 1998. Characterization of cis-acting and NS1 protein-responsive elements in the p6 promoter of parvovirus B19. J Virol 72:609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D, Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S, Young NS. 2010. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J Clin Invest 120:3530–3544. doi: 10.1172/JCI41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu P, Zhou Z, Xiong M, Zou W, Deng X, Ganaie SS, Kleiboeker S, Peng J, Liu K, Wang S, Ye SQ, Qiu J. 2017. Parvovirus B19 NS1 protein induces cell cycle arrest at G2-phase by activating the ATR-CDC25C-CDK1 pathway. PLoS Pathog 13:e1006266. doi: 10.1371/journal.ppat.1006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen AY, Zhang EY, Guan W, Cheng F, Kleiboeker S, Yankee TM, Qiu J. 2010. The small 11 kDa non-structural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood 115:1070–1080. doi: 10.1182/blood-2009-04-215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan MM, Tamburic L, Shippam-Brett C, Zagrodney DB, Astell CR. 2001. The small 11-kDa protein from B19 parvovirus binds growth factor receptor-binding protein 2 in vitro in a Src homology 3 domain/ligand-dependent manner. Virology 291:285–291. doi: 10.1006/viro.2001.1217. [DOI] [PubMed] [Google Scholar]
- 25.Ganaie SS, Chen AY, Huang C, Xu P, Du A, Kleiboeker S, Qiu J. 2018. RNA binding protein RBM38 regulates expression of the 11-kDa protein of parvovirus B19 which facilitates viral DNA replication. J Virol 92:e02050-17. doi: 10.1128/JVI.02050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chardin P, Cussac D, Maignan S, Ducruix A. 1995. The Grb2 adaptor. FEBS Lett 369:47–51. doi: 10.1016/0014-5793(95)00578-W. [DOI] [PubMed] [Google Scholar]
- 27.Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, Schlessinger J, Pawson T. 1992. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360:689–692. doi: 10.1038/360689a0. [DOI] [PubMed] [Google Scholar]
- 28.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. 1992. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 70:431–442. doi: 10.1016/0092-8674(92)90167-B. [DOI] [PubMed] [Google Scholar]
- 29.Chardin P, Camonis JH, Gale NW, van AL, Schlessinger J, Wigler MH, Bar-Sagi D. 1993. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science 260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. 1993. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 31.Schaeper U, Gehring NH, Fuchs KP, Sachs M, Kempkes B, Birchmeier W. 2000. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J Cell Physiol 149:1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewitzky M, Kardinal C, Gehring NH, Schmidt EK, Konkol B, Eulitz M, Birchmeier W, Schaeper U, Feller SM. 2001. The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P-x-x-P core motif. Oncogene 20:1052–1062. doi: 10.1038/sj.onc.1204202. [DOI] [PubMed] [Google Scholar]
- 33.Ling Y, Maile LA, Lieskovska J, Badley-Clarke J, Clemmons DR. 2005. Role of SHPS-1 in the regulation of insulin-like growth factor I-stimulated Shc and mitogen-activated protein kinase activation in vascular smooth muscle cells. Mol Biol Cell 16:3353–3364. doi: 10.1091/mbc.e04-10-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald CB, Seldeen KL, Deegan BJ, Lewis MS, Farooq A. 2008. Grb2 adaptor undergoes conformational change upon dimerization. Arch Biochem Biophys 475:25–35. doi: 10.1016/j.abb.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Reuther GW, Der CJ. 2000. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol 12:157–165. doi: 10.1016/S0955-0674(99)00071-X. [DOI] [PubMed] [Google Scholar]
- 36.Robinson MJ, Cobb MH. 1997. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol 9:180–186. doi: 10.1016/S0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 37.Xu TR, Vyshemirsky V, Gormand A, von KA, Girolami M, Baillie GS, Ketley D, Dunlop AJ, Milligan G, Houslay MD, Kolch W. 2010. Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species. Sci Signal 3:ra20. doi: 10.1126/scisignal.2000517. [DOI] [PubMed] [Google Scholar]
- 38.Riera L, Lasorsa E, Ambrogio C, Surrenti N, Voena C, Chiarle R. 2010. Involvement of Grb2 adaptor protein in nucleophosmin-anaplastic lymphoma kinase (NPM-ALK)-mediated signaling and anaplastic large cell lymphoma growth. J Biol Chem 285:26441–26450. doi: 10.1074/jbc.M110.116327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinoda T, Taya S, Tsuboi D, Hikita T, Matsuzawa R, Kuroda S, Iwamatsu A, Kaibuchi K. 2007. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci 27:4–14. doi: 10.1523/JNEUROSCI.3825-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modi H, Li L, Chu S, Rossi J, Yee JK, Bhatia R. 2011. Inhibition of Grb2 expression demonstrates an important role in BCR-ABL-mediated MAPK activation and transformation of primary human hematopoietic cells. Leukemia 25:305–312. doi: 10.1038/leu.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIF alpha. PLoS Pathog 7:e1002088. doi: 10.1371/annotation/59703f7f-9506-49d1-b339-09ee31510e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dance M, Montagner A, Salles JP, Yart A, Raynal P. 2008. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell Signal 20:453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Romero F, Ramos-Morales F, Domínguez A, Rios RM, Schweighoffer F, Tocqué B, Pintor-Toro JA, Fischer S, Tortolero M. 1998. Grb2 and its apoptotic isoform Grb3-3 associate with heterogeneous nuclear ribonucleoprotein C, and these interactions are modulated by poly(U) RNA. J Biol Chem 273:7776–7781. doi: 10.1074/jbc.273.13.7776. [DOI] [PubMed] [Google Scholar]
- 44.Pleschka S. 2008. RNA viruses and the mitogenic Raf/MEK/ERK signal transduction cascade. Biol Chem 389:1273–1282. doi: 10.1515/BC.2008.145. [DOI] [PubMed] [Google Scholar]
- 45.Moffat JF, Greenblatt RJ. 2010. Effects of varicella-zoster virus on cell cycle regulatory pathways. Curr Top Microbiol Immunol 342:67–77. doi: 10.1007/82_2010_28. [DOI] [PubMed] [Google Scholar]
- 46.Lodish HF, Ghaffari S, Socolovsky M, Tong W, Zhang J. 2009. Intracellular signaling by the erythropoietin receptor, p 155–174. In Elliott SG, Foote MA, Molineux G (ed), Erythropoietins, erythropoietic factors and erythropoiesis. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 47.Zhang W, Liu HT. 2002. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res 12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 48.Wang R, He G, Nelman-Gonzalez M, Ashorn CL, Gallick GE, Stukenberg PT, Kirschner MW, Kuang J. 2007. Regulation of Cdc25C by ERK-MAP kinases during the G2/M transition. Cell 128:1119–1132. doi: 10.1016/j.cell.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 49.Hayne C, Tzivion G, Luo Z. 2000. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J Biol Chem 275:31876–31882. doi: 10.1074/jbc.M002766200. [DOI] [PubMed] [Google Scholar]
- 50.Zou W, Wang Z, Xiong M, Chen AY, Xu P, Ganaie SS, Badawi Y, Kleiboeker S, Nishimune H, Ye SQ, Qiu J. 2018. Human parvovirus B19 utilizes cellular DNA replication machinery for viral DNA replication. J Virol 92:e01881-17. doi: 10.1128/JVI.01881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol 85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo Y, Kleiboeker S, Deng X, Qiu J. 2013. Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol 87:12766–12775. doi: 10.1128/JVI.02333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganaie SS, Qiu J. 2018. Recent advances in replication and infection of human parvovirus B19. Front Cell Infect Microbiol 8:166. doi: 10.3389/fcimb.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tewary SK, Zhao H, Deng X, Qiu J, Tang L. 2014. The human parvovirus B19 non-structural protein 1 N-terminal domain specifically binds to the origin of replication in the viral DNA. Virology 449:297–303. doi: 10.1016/j.virol.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong S, Zhi N, Filippone C, Keyvanfar K, Kajigaya S, Brown KE, Young NS. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol 82:2470–2476. doi: 10.1128/JVI.02247-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morita E, Tada K, Chisaka H, Asao H, Sato H, Yaegashi N, Sugamura K. 2001. Human parvovirus B19 induces cell cycle arrest at G2 phase with accumulation of mitotic cyclins. J Virol 75:7555–7563. doi: 10.1128/JVI.75.16.7555-7563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen AY, Guan W, Lou S, Liu Z, Kleiboeker S, Qiu J. 2010. Role of erythropoietin receptor signaling in parvovirus B19 replication in human erythroid progenitor cells. J Virol 84:12385–12396. doi: 10.1128/JVI.01229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen W, Deng X, Zou W, Cheng F, Engelhardt JF, Yan Z, Qiu J. 2015. Identification and functional analysis of novel non-structural proteins of human bocavirus 1. J Virol 89:10097–10109. doi: 10.1128/JVI.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, Qiu J, Cheng F, Chu Y, Yoto Y, O'Sullivan MG, Brown KE, Pintel DJ. 2004. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J Virol 78:12929–12939. doi: 10.1128/JVI.78.23.12929-12939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhi N, Zadori Z, Brown KE, Tijssen P. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142–152. doi: 10.1016/j.virol.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 61.Ganaie SS, Zou W, Xu P, Deng X, Kleiboeker S, Qiu J. 2017. Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLoS Pathog 13:e1006370. doi: 10.1371/journal.ppat.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guan W, Cheng F, Yoto Y, Kleiboeker S, Wong S, Zhi N, Pintel DJ, Qiu J. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J Virol 82:9951–9963. doi: 10.1128/JVI.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng X, Yan Z, Cheng F, Engelhardt JF, Qiu J. 2016. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog 12:e1005399. doi: 10.1371/journal.ppat.1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen AY, Luo Y, Cheng F, Sun Y, Qiu J. 2010. Bocavirus infection induces a mitochondrion-mediated apoptosis and cell cycle arrest at G2/M-phase. J Virol 84:5615–5626. doi: 10.1128/JVI.02094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 66.Barber DL, Corless CN, Xia K, Roberts TM, D'Andrea AD. 1997. Erythropoietin activates Raf1 by an Shc-independent pathway in CTLL-EPO-R cells. Blood 89:55–64. [PubMed] [Google Scholar]
- 67.Skolnik EY, Batzer A, Li N, Lee CH, Lowenstein E, Mohammadi M, Margolis B, Schlessinger J. 1993. The function of GRB2 in linking the insulin receptor to Ras signaling pathways. Science 260:1953–1955. doi: 10.1126/science.8316835. [DOI] [PubMed] [Google Scholar]
- 68.Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. 1993. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature 363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 69.Gout I, Dhand R, Hiles ID, Fry MJ, Panayotou G, Das P, Truong O, Totty NF, Hsuan J, Booker GW, Campbell ID, Waterfield MD. 1993. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 75:25–36. doi: 10.1016/S0092-8674(05)80081-9. [DOI] [PubMed] [Google Scholar]