ABSTRACT
Congenital heart disease (CHD) is a major cause of infant mortality and morbidity, yet the genetic causes and mechanisms remain opaque. In a patient with CHD and heterotaxy, a disorder of left-right (LR) patterning, a de novo mutation was identified in the chromatin modifier gene WDR5. WDR5 acts as a scaffolding protein in the H3K4 methyltransferase complex, but a role in LR patterning is unknown. Here, we show that Wdr5 depletion leads to LR patterning defects in Xenopus via its role in ciliogenesis. Unexpectedly, we find a dual role for WDR5 in LR patterning. First, WDR5 is expressed in the nuclei of monociliated cells of the LR organizer (LRO) and regulates foxj1 expression. LR defects in wdr5 morphants can be partially rescued with the addition of foxj1. Second, WDR5 localizes to the bases of cilia. Using a mutant form of WDR5, we demonstrate that WDR5 also has an H3K4-independent role in LR patterning. Guided by the patient phenotype, we identify multiple roles for WDR5 in LR patterning, providing plausible mechanisms for its role in ciliopathies like heterotaxy and CHD.
KEY WORDS: Congenital heart disease, Heterotaxy, Xenopus, H3K4 methylation, Cilia
Highlighted Article: The candidate congenital heart disease gene WDR5 is essential for left-right patterning via its role in ciliogenesis. WDR5, a chromatin modifier, regulates transcription of foxj1 but also has a nonchromatin role at the ciliary base.
INTRODUCTION
Congenital heart disease (CHD) is the most common developmental defect (Van der Linde et al., 2011). However, our understanding of the genetic etiologies of CHD remains poor. A recent genetic analysis of CHD patients identified a marked enrichment in genes involved in chromatin modification, specifically genes involved in H3K4/H3K27 methylation or H2BK120 ubiquitination (Zaidi et al., 2013; Homsy et al., 2015). However, how global regulators of chromatin states could lead to specific phenotypes, such as CHD, remains unanswered. To address this question, we began our studies with WDR5, a recent candidate gene for CHD and a critical member of the histone (H3K4) methylation pathway (Zaidi et al., 2013; Wysocka et al., 2005).
The patient with a de novo missense mutation, K7Q, in WDR5 had a conotruncal defect with a right aortic arch (normally the aortic arch is on the left), a mild heterotaxy (Htx) phenotype (Zaidi et al., 2013). Htx results from aberrant left-right (LR) patterning of internal organs and can be associated with a severe form of CHD (Sutherland and Ware, 2009; Amack and Yost, 2010; Hamada et al., 2002). Cilia are well known to be essential for LR patterning (Li et al., 2015; Blum et al., 2014; Basu and Brueckner, 2008; Capdevila et al., 2000; Hamada et al., 2002). However, a mechanism connecting cilia, chromatin modifiers, and CHD/Htx has not been established. Here, we show that WDR5, a histone modifier, is necessary for both LR patterning and ciliogenesis.
LR patterning is established at a conserved ciliated structure, the LR organizer (LRO) [the node in mouse, Kupffer's vesicle in fish and gastrocoel roof plate (GRP) in frogs] (Burdine and Schier, 2000; Capdevila et al., 2000; Hamada et al., 2002; Blum et al., 2014). Two types of monociliated cells populate the LRO: cells in the middle are enriched in motile cilia that create leftward extracellular fluid flow, which is detected by peripheral cells that have a preponderance of immotile sensory cilia (Doerks et al., 2002; Basu and Brueckner, 2008; Schweickert et al., 2007; Boskovski et al., 2013). Once flow is sensed at the LRO left margin, dand5 (alternatively referred to as Cerl2 in mouse and coco in Xenopus; a Nodal antagonist) is downregulated, leading to pitx2 upregulation in the left lateral plate mesoderm (Burdine and Schier, 2000; Capdevila et al., 2000; Hamada et al., 2002; Blum et al., 2014; Vonica and Brivanlou, 2007). The activation of pitx2 in the lateral plate mesoderm is associated with organ lateralization (Lin et al., 1999).
WDR5 is a core subunit of the human MLL and SET1 histone H3 Lys4 methyltransferase (H3K4MT) complexes that are essential for chromatin modification and transcriptional regulation (Wysocka et al., 2005; Couture et al., 2006; Dou et al., 2006; Trievel and Shilatifard, 2009; Couture and Skiniotis, 2013; Patel et al., 2008b). In particular, H3K4MTs function as a complex consisting of one catalytic subunit [SET1A/B (SETD1A/B) or MLL1-4 (KMT2A-D) proteins] and four core regulatory subunits (WDR5, RbBP5, ASH2L and DPY-30) (Trievel and Shilatifard, 2009; Couture and Skiniotis, 2013; Takahashi et al., 2011). These regulatory subunits form a subcomplex that binds the catalytic subunit and dramatically enhances its H3K4MT activity (Patel et al., 2009). Depletion of any of the regulatory subunits impairs H3K4 methylation (Steward et al., 2006; Dou et al., 2006; Wysocka et al., 2005; Cao et al., 2010). WDR5, a highly conserved scaffolding protein, is essential for the association of RbBP5, ASH2L and DPY-30 with MLL1 via its β-propeller structure (Odho et al., 2010; Trievel and Shlatifard, 2009; Patel et al., 2008b). The central pocket made by the sevenfold propeller is crucial for binding H3 and MLL (Song and Kingston, 2008; Patel et al., 2008a; Schuetz et al., 2006; Ruthenburg et al., 2006; Couture et al., 2006). Point mutations, for example, in the arginine-binding cavity of WDR5 (S91K or F133A), can disrupt the H3K4MT complex (Patel et al., 2008b). Interestingly, the patient mutation (K7Q) lies outside the β-propeller, in the N-terminal tail (first ∼30 amino acids) that is not required for H3K4MT function (Schuetz et al., 2006), making the disease relevance of this mutation uncertain.
RESULTS AND DISCUSSION
Wdr5 depletion alters LR patterning in Xenopus tropicalis
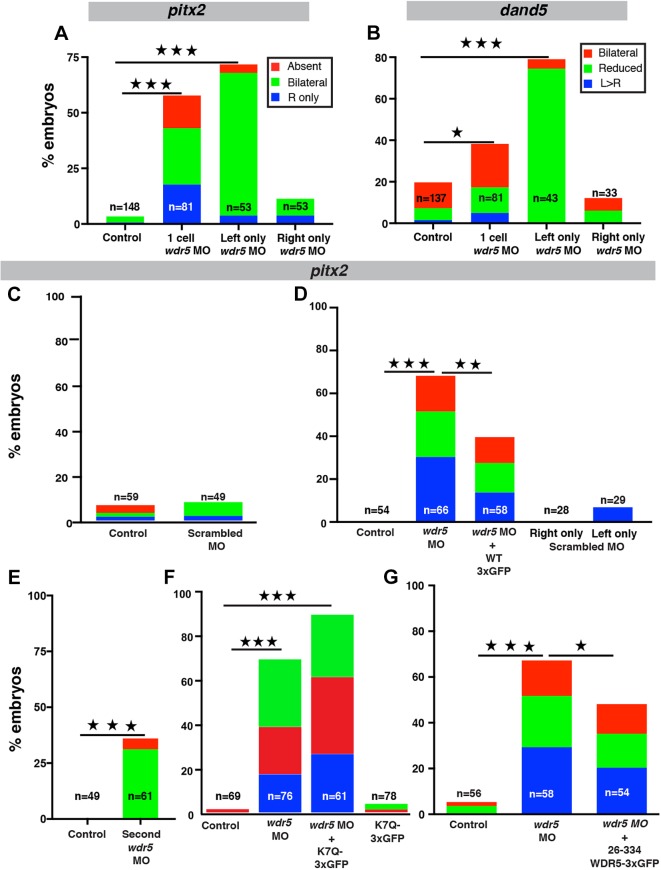
Previously, in a CHD patient with a right (rather than the normally left) aortic arch, a de novo mutation (K7Q) implicated WDR5 as a candidate disease gene (Zaidi et al., 2013). Therefore, we investigated whether WDR5 is essential for LR patterning in Xenopus. Depletion of Wdr5 in Xenopus leads to substantial pericardial edema that alters the structure of the heart, precluding accurate determination of its cardiac looping. Therefore, we examined global markers of LR patterning: pitx2c and dand5 (Fig. S1). Using a wdr5 morpholino oligonucleotide (MO), Wdr5 knockdown led to abnormal patterns of both pitx2c (∼55%) and dand5 (∼40%) (Fig. 1A,B). To examine the specificity and efficiency of our Wdr5 depletion, we employed multiple tests. First by western blotting, we previously showed that Wdr5 protein is reduced in morphants, which is rescued by injecting human wild-type (WT) WDR5 mRNA (Kulkarni et al., 2018). In addition, we demonstrated that the anti-WDR5 antibody was specific using a serial dilution of a blocking peptide that reduced the WDR5 signal on the western blot (Kulkarni et al., 2018). Second, injection of a scrambled MO does not alter pitx2c expression compared with that of uninjected controls (Fig. 1C). Third, we can partially rescue alterations in pitx2c expression in Wdr5-depleted embryos by co-injecting WT human WDR5 mRNA tagged with 3xGFP (Fig. 1D). Fourth, the depletion of Wdr5 using a second ATG MO also led to abnormal patterns of pitx2c (Fig. 1E). Together, we conclude that the Wdr5 depletion by MO is specific and is essential for global LR patterning.
Fig. 1.
Wdr5 depletion alters LR patterning in Xenopus tropicalis. (A,B) Percentage of embryos that have abnormal pitx2c (A) or dand5 (B) expression. Embryos were injected at the one-cell stage, or in one cell at the two-cell stage, with wdr5 MO. Absent, signal absent on both sides; bilateral, signal present on both sides; L, left; R, right; Reduced: signal reduced on both sides. (C-G) Percentage of embryos that have abnormal pitx2c expression. Embryos were injected at the one-cell stage with scrambled MO (C); wdr5 MO, wdr5 MO+human WDR5-3xGFP, scrambled MO injected on the right or left side of embryos (D); second wdr5 MO (E); wdr5 MO, wdr5 MO+K7Q-WDR5-3xGFP or K7Q-WDR5-3xGFP (F); or wdr5 MO and wdr5 MO+human 26-334-3xGFP (G). Experiments were repeated three times. ‘n’=number of embryos. ★P<0.05, ★★P<0.005 and ★★★P<0.0005.
Disruptions in both dand5 and pitx2c point to a defect in the LRO, possibly due to cilia-mediated signaling (Hamada et al., 2002; Doerks et al., 2002; Schweickert et al., 2007). Previous studies have shown that cilia motility on the left but not on the right side of the LRO is crucial for proper LR patterning (Vick et al., 2009). Indeed, left-sided wdr5 knockdown led to a higher percentage of abnormal expression patterns for both LR markers (dand5 ∼80% and pitx2c ∼70%) compared with knockdown on the right (dand5 ∼12% and pitx2c ∼12%), an effect not seen with scrambled MO injection (Fig. 1A,B,D). These results suggest a ciliary role for Wdr5 in LR development.
WDR5 (K7Q) is a loss-of-function allele
The WDR5 patient had a de novo K7Q missense mutation; however, whether this variant is detrimental to function was unclear. To test this hypothesis, we attempted to rescue the pitx2c phenotype in wdr5 morphants. In contrast to the WT protein, the K7Q-WDR5 variant failed to rescue pitx2c patterning in wdr5 morphants, supporting the hypothesis that it is a loss-of-function allele (Fig. 1F) (Zhu et al., 2017). Interestingly, the first 30 amino acids are dispensable for forming the β-propeller structure of WDR5 and assembling a functional H3K4MT complex (Schuetz et al., 2006). Therefore, we wondered whether this allele might suggest a role independent of chromatin modification.
Wdr5 is essential for cilia in the LRO
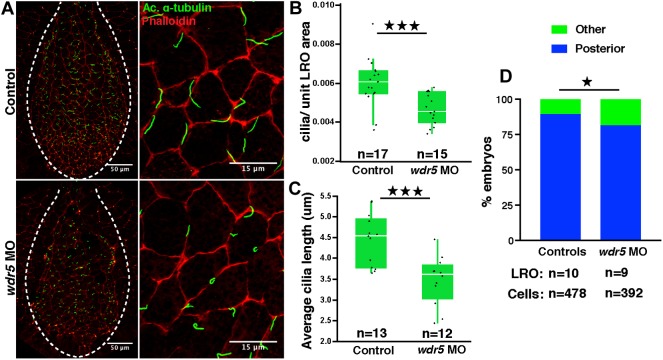
LR patterning is established at the LRO. Because we see defects in dand5 expression, an immediate downstream target of cilia signaling, we decided to examine cilia morphology in the LRO of wdr5 morphants. Using immunofluorescence, we observed that cilia in morphants were abnormal compared with those in WT embryos (Fig. 2A). Specifically, in wdr5 morphants, the cilia were shorter and fewer in number (Fig. 2B,C). We confirmed our results with scanning electron microscopy (Fig. S2). In addition, we measured cilia polarity in the LRO according to established methods (Walentek et al., 2013). Normally, the LRO monocilia are posteriorly polarized, which is essential for establishing leftward flow. Depletion of Wdr5 mildly affected cilia polarity (Fig. 2D). These results suggest that Wdr5 is essential for cilia in the LRO.
Fig. 2.
Wdr5 depletion affects cilia morphology in the LRO of X.tropicalis. (A) Cilia in control and wdr5 morphant Xenopus LROs marked by anti-acetylated (Ac.) α-tubulin (green) antibody and F-actin marked by phalloidin (red). (B,C) Number of cilia normalized to the LRO area (B) and length of cilia (C) in uninjected controls and wdr5 morphants. Data are presented as box plot with 95% confidence interval. ‘n’=number of embryos. (D) Number of cilia posteriorly localized in the LRO cells in uninjected controls and wdr5 morphants. Experiments were repeated two times. ★P<0.05 and ★★★P<0.0005.
We investigated multiple potential reasons for cilia defects. First, LRO patterning is dependent on proper dorsoventral (DV) patterning, so we began by examining DV markers in gastrula (stages 10-11) or postgastrula (stage 14) embryos (Fig. S3A,B) (Khokha et al., 2005; Heasman, 2006; Niehrs et al., 2001). Interestingly, following Wdr5 depletion, we detected an increase in expression in dkk1, a known WNT protein antagonist (Glinka et al., 1998). Previous studies have shown that the transcriptional regulation of some WNT regulators and target genes is dependent on WDR5; thus, expansion of dkk1 in our wdr5 morphants supports these studies (Zhu et al., 2008; Gori et al., 2006). However, we did not observe any change in the expression of other key DV patterning genes, including foxj1, at gastrula stage. Second, although DV patterning appears generally normal at the gastrula stages, we considered the possibility that the LRO itself was mispatterned. Prior to the onset of cilia-mediated flow (stage 16), dand5 expression is symmetric at the lateral margins, which is unchanged in control or Wdr5-depleted embryos (Fig. S3C,F). Further, we checked the expression of xnr1, which is also symmetric at the lateral margins at these stages. It was also similar between controls and morphants, demonstrating that LRO cell fate specification was not drastically affected in morphants (Fig. S3C,F). Mispatterning of the LR axis could also arise from defects in establishing the midline barrier. Therefore, we checked the expression of sonic hedgehog (shh) and lefty at the midline and found that expression was unaltered between controls and morphants (Fig. S3D,F). Based on these results, we conclude that the LRO in wdr5 morphants is normally patterned overall and sought other explanations for the defects in cilia.
Wdr5 regulates LR patterning via transcriptional regulation of foxj1
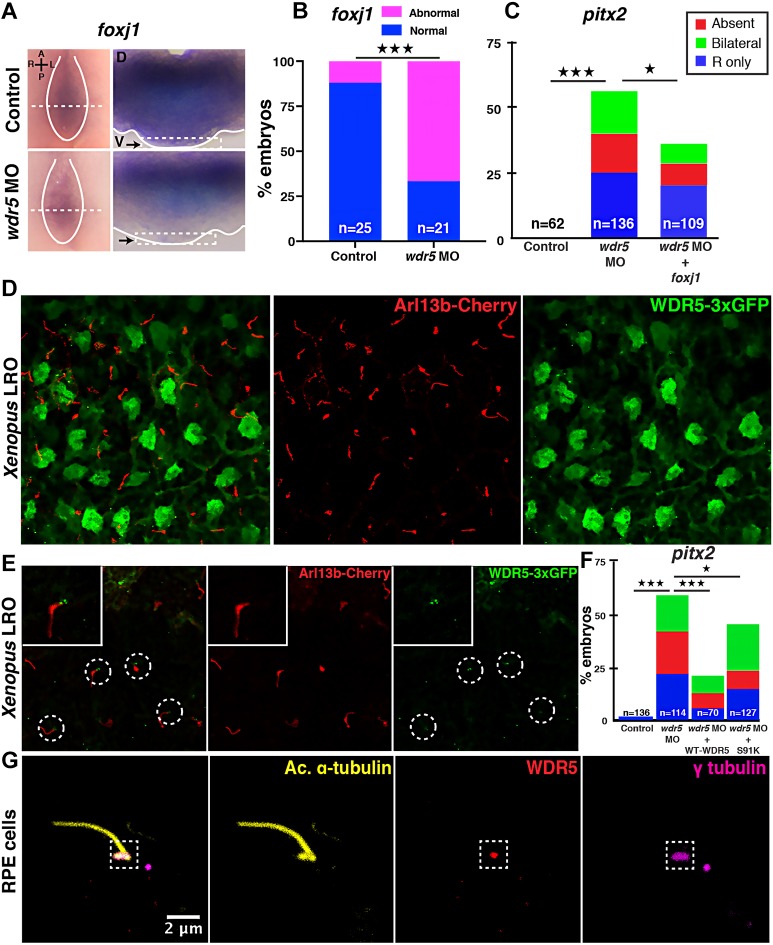
Given the established role for WDR5 as a chromatin modifier, we next examined the mRNA expression of three key cilia-specific genes in the LRO: dnah9, rfx2 and foxj1 (Chung et al., 2012; Yu et al., 2008; Stubbs et al., 2008; Vick et al., 2009). We did not detect a difference in dnah9 and rfx2 expression (Fig. S3E,F); however, foxj1 expression appeared reduced in the LRO of morphants compared with controls (Fig. 3A,B, black arrows). Therefore, we tested the possibility that Wdr5 regulates LRO cilia via foxj1 transcription using a rescue experiment. We injected foxj1 mRNA into Wdr5-depleted embryos, which partially rescued the pitx2c defects (Fig. 3C), supporting our hypothesis. Our results are consistent with multiple different findings: (1) the Foxj1 locus is methylated (Mikkelsen et al., 2007), (2) global chromatin immunoprecipitation sequencing identifies the Foxj1 locus as a target of WDR5, RbBP5 and H3K4m3 (Ang et al., 2011), and (3) Foxj1 is an established regulator of monocilia (Yu et al., 2008; Stubbs et al., 2008).
Fig. 3.
WDR5 has chromatin-dependent and -independent roles in ciliogenesis in the LRO. (A) In situ hybridization for foxj1 in the Xenopus LRO in uninjected controls and wdr5 morphants at stage 16. Embryos were bisected to reveal expression of foxj1 in the entire GRP (left column, dorsal to the top, ventral view so right margin of the LRO is to the left; the margin of the GRPs is marked by white lines). GRPs were again bisected within the LRO (dashed white lines) to reveal foxj1 expression within the tissue [right column, dorsal to the top; arrows mark the ventral surface (indicated by the dashed white line boxes at the bottom) where the LRO resides, showing loss of foxj1 in the ventral tissue]. A, anterior; D, dorsal; L, left; P, posterior; R, right; V, ventral. (B) Percentage of embryos that have abnormal foxj1 expression in control and wdr5 morphant Xenopus LROs. (C) Percentage of embryos that have abnormal pitx2 expression. Embryos were injected at one-cell stage with wdr5 MO or wdr5 MO+Xenopus foxj1 mRNA. (D) A Z-stack confocal image of Xenopus LRO, visualizing WDR5-3xGFP (green) and cilia (red). The green channel is taken at the plane of the nuclei; the red channel is taken at the apical plane to reveal cilia. Cilia are detected using Arl13b-mCherry expression. For WDR5, the look-up table (LUT) is green; for Arl13b-mCherry, the LUT is red. (E) A Z-stack confocal image of the Xenopus LRO visualizing WDR5-3xGFP (green) and cilia (red). Both the green and red channels are taken at the apical surface of the Xenopus LRO cells, showing WDR5-3xGFP localizing at the ciliary bases. Cilia are marked with Arl13b-mCherry. For WDR5-3xGFP, the LUT is green; for Arl13b-mCherry, the LUT is red. (F) Percentage of embryos that have abnormal pitx2 expression. Embryos were injected at one-cell stage with wdr5 MO+WT WDR5 mRNA or wdr5 MO+S91K-WDR5 mRNA. (G) WDR5 localizes to the ciliary base in hRPE cells. Cilia and basal bodies are marked with anti-acetylated (Ac.) α-tubulin and γ-tubulin antibodies, respectively. For Ac. α-tubulin, the LUT is yellow; for WDR5, the LUT is red; for γ-tubulin, the LUT is magenta. Experiments were repeated three to four times. ‘n’=number of embryos. ★P<0.05 and ★★★P<0.0005.
Wdr5 is expressed in the nuclei and at the bases of LRO cilia
Because Wdr5 regulates foxj1 transcription in the LRO, we decided to confirm that Wdr5 is localized to the nuclei of the LRO. Unfortunately, although our anti-WDR5 antibody detects Xenopus Wdr5 protein by western blotting, we could not detect Wdr5 in the Xenopus LRO using immunofluorescence. Therefore, we used a WDR5-3xGFP construct to examine its localization. We note that WDR5-3xGFP can partially rescue the wdr5 depletion phenotype (Fig. 1D), suggesting that it is properly localized and functioning. We injected WDR5-3xGFP in Xenopus embryos at the one-cell stage and, as expected, WDR5-3xGFP was localized to the nuclei of the LRO cells (Fig. 3D, GFP channel imaged at the level of the nucleus). However, to our surprise, we found that WDR5-3xGFP was also localized to the bases of cilia (Fig. 3E, GFP channel imaged at the apical surface). To see whether this finding was generalized to mammalian monociliated cells, we tested WDR5 localization in human retinal pigmented epithelium (hRPE) cells using immunofluorescence. Indeed, WDR5 localized to γ-tubulin, a component of the basal body in hRPE cells (Fig. 3G), the specificity of which we confirmed using a blocking peptide (Fig. S4).
WDR5 has an H3K4-independent role in LR patterning
Our unexpected discovery of WDR5 at the bases of monocilia suggested a chromatin-independent role, which might also be consistent with a deleterious effect of the K7Q allele. To test this hypothesis, we decided to employ the S91K WDR5 mutant, which disrupts binding to MLL and leads to abrogation of H3K4MT activity (Patel et al., 2008b). Consistent with a chromatin-independent role, the S91K WDR5 allele partially rescued pitx2c expression, possibly via its localization at the bases of cilia in the LRO (Fig. 3F).
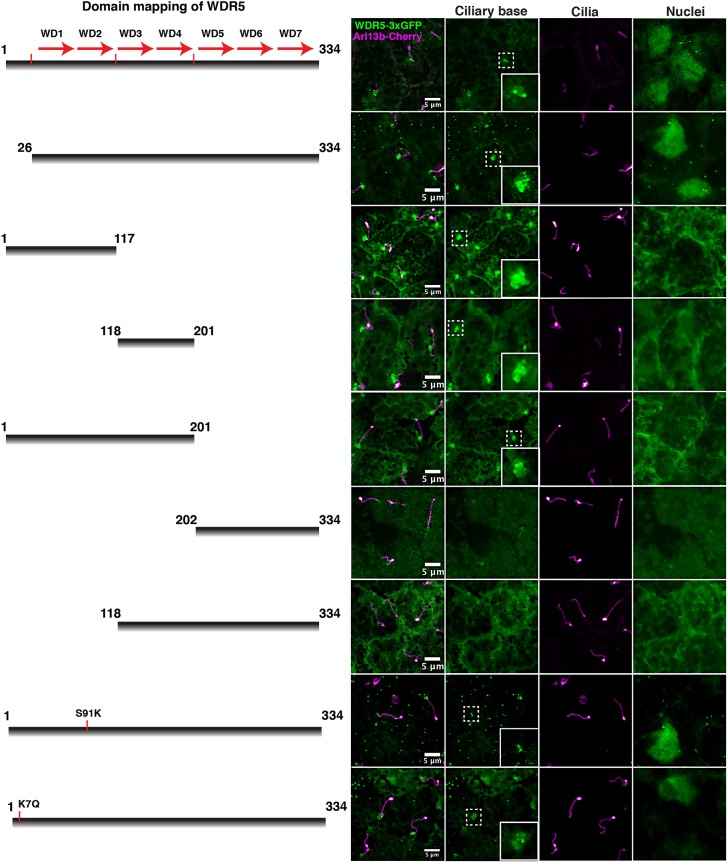
The β-propeller structure of WDR5 is sufficient for nuclear but not ciliary localization
In ciliogenesis and LR patterning, we found a chromatin-dependent and a chromatin-independent role for WDR5 that correlates with protein localization. Therefore, we sought to identify any protein domains within WDR5 that might define these two different localizations. To study protein localization, we made different deletion constructs tagged with 3xGFP and expressed them in the Xenopus LRO (Fig. 4). First, we deleted amino acids 1-26, which contains the patient variant but is dispensable for H3K4MT activity (construct 26-334-3xGFP). As expected, the 26-334-3xGFP construct partially rescues the pitx2c defect when wdr5 is depleted, indicating that it is functional via the H3K4MT pathway (Fig. 1G) (Schuetz et al., 2006). We also divided WDR5 into five more constructs: 1-117 (first and second WD repeats), 118-201 (third and fourth WD repeats), 1-201 (one to four WD repeats), 118-334 (three to seven WD repeats) and 202-334 (five to seven WD repeats) (Fig. 4). To begin, we examined nuclear localization. Of the different constructs, only the WT-3xGFP and the 26-334-3xGFP localized to the nucleus, suggesting that the whole β-propeller is essential for nuclear localization and confirming that the first 26 N-terminal amino acids are not (Schuetz et al., 2006). At the bases of cilia, the WT-3xGFP localized as sharp puncta, whereas the 26-334-3xGFP had a more diffuse localization. The GFP signal appeared more dispersed, suggesting that the β-propeller is not sufficient for precise localization at the bases of cilia. This is specifically interesting because the patient's missense mutation K7Q lies in the N-terminal tail (first 25 base pairs) of WDR5, which is dispensable for the assembly of H3K4MT complex but required for precise ciliary localization (see below). Therefore, it is possible that that patient phenotype arises from the function of WDR5 at the bases of cilia, rather than its role in the assembly of a H3K4MT complex.
Fig. 4.
Domain analysis of WDR5. Localization of different deletion constructs for WDR5-3xGFP at the ciliary base and nuclei of the Xenopus LRO. For WDR5-3xGFP, the LUT is green; for Arl13b-Cherry, the LUT is hot magenta.
In general, N-terminal constructs that include the first 25 amino acids localize to the bases of cilia (including 1-117 and 1-201, Fig. 4). However, the localizations of these constructs appear more diffuse than the full-length constructs, suggesting that an intact β-propeller is required for precise localization of WDR5 to the bases of cilia. On the other hand, the C-terminal constructs (118-334 and 202-334) fail to localize to the bases of cilia or the nucleus, producing a diffuse pattern across much of the cell, suggesting that the N-terminal part of the protein is crucial for localization to the cilia bases. Of note, the construct 118-201 also localizes to the bases of cilia, albeit in a more diffuse pattern than that of the full-length construct. However, 118-334 does not localize to the cilium base, suggesting that the C-terminal end might contain a repressive motif for cilia localization. From these data, we conclude that the N-terminal 26 amino acids (that include the patient variant) are required for precise cilium localization, although the relationship between ciliary localization and protein domains appears complex. Interestingly, different parts of WDR5 can localize to the cilium base independently of nuclear localization.
We also examined the localization of the S91K mutant, which cannot participate in H3K4MT activity, and the patient's mutation (K7Q) in the ciliated cells of the LRO (Fig. 4). As predicted, localization of the S91K variant to the bases of cilia was similar to that shown by WT-WDR5 (Fig. 4). In addition, the S91K variant also localized to the nucleus, confirming that its effect on H3K4MT activity is via assembly of the H3K4MT complex rather than nuclear localization. Interestingly, the K7Q variant did not precisely localize to the bases of cilia. The K7Q localization was even more diffuse than that of the 26-334 construct, suggesting that the K7Q missense mutation might interfere with the ability of WDR5 to localize to the bases of LRO cilia.
In conclusion, our results emphasize the importance of patient-driven gene discovery tightly coupled with developmental biology, which together can be a powerful strategy to elucidate disease mechanisms. Recent studies in CHD and autism clearly point to a role for chromatin modifiers in disease pathogenesis, but the molecular mechanisms remain unclear (De Rubeis et al., 2014; Zaidi et al., 2013; Carneiro et al., 2011). In our search for the underlying molecular mechanism for WDR5 in CHD using Xenopus, we uncovered an H3K4-dependent, and an unexpected H3K4-independent, cilia role in the LRO. Importantly, this cilia phenotype could have important clinical implications for patient management.
MATERIALS AND METHODS
Animal husbandry
X. tropicalis were housed and cared for in our aquatics facility according to established protocols that were approved by the Yale Institutional Animal Care and Use Committee.
Cell culture
hRPE cells were obtained from the Brueckner laboratory of Yale School of Medicine and were contamination free. Cells were cultured in Dulbecco's modified eagle medium/Ham's F12 medium supplemented with 10% fetal bovine serum.
Microinjection of MOs and mRNA in Xenopus
Embryos were produced by in vitro fertilization and raised to appropriate stages in 1/9×MR+gentamycin according to established protocols (Khokha et al., 2002; del Viso and Khokha, 2012). Staging of Xenopus tadpoles was as previously described (Nieuwkoop, 1994). MOs or mRNA were injected into one-cell or two-cell embryos as described previously (Khokha et al., 2002). The following MOs were injected: wdr5 translation blocking (2.5-4 ng 5′-CGGGTTTCTTTTCTTCTGTTGCCAT-3′), second wdr5 translation blocking (15 ng/embryo 5′-TGCAAGAACAACTTGTGGCCGGATA-3′) and scrambled control MO (4 ng 5′-CCTCTTACCTCAGTTACAATTTATA-3′). Alexa Fluor 488 (Invitrogen), mini-ruby (Invitrogen) or GFP (100 pg) were injected as tracers. We generated in vitro capped mRNA using a mMessage mMachine Kit (Ambion) and followed the manufacturer's instructions. Full-length human WDR5 was obtained from the IMAGE consortium collection (Thermo Fisher Scientific IMAGE clone 3538255) and subcloned into the pCSDest vector using Gateway recombination techniques. We generated the K7Q mutation using PCR amplification and cloning into PCS2 vector. We injected 400 pg human WT-WDR5, WDR5-GFP, WDR5-S91K, WDR5-3xGFP, WDR5-K7Q-3xGFP, WDR5-26-334-3xGFP and 150 pg Xenopus foxj1 RNA for rescue of wdr5 morphants. Mutant constructs for domain analysis were generated using PCR amplification and cloning into PCS2 vector. Human WT and the mutant constructs of WDR5 were tagged with 3xGFP and were injected (200 pg) into one-cell embryos to mark WDR5 in the LRO. We injected 200 pg Arl13b-Cherry to mark LRO cilia.
RNA in situ hybridization
X. tropicalis embryos were collected at various stages and in situ hybridization was performed according to standard protocols (Khokha et al., 2002). We collected stage 10 embryos for foxj1, wnt8, dkk1, xnr3, otx2 and myf5 expression, and stage 14 embryos for sox2 expression. For GRPs, we collected stage 16 embryos for dand5, foxj1, shh, dnah9, xnr1 and rfx2, stage 19-21 for dand5 and lefty, and stage 28 for pitx2. GRPs were dissected as described previously (Schweickert et al., 2007). Additional probe information is available upon request.
Image analysis
Images were captured using a Zeiss 710 Live confocal microscope. Images were processed in Fiji (National Institutes of Health), ImageJ (National Institutes of Health) or Adobe Photoshop. Quantification of GRP cilia length was performed using Volocity (Quorum Technologies) software on 3D image stacks. The number of GRP cilia was measured using the ‘Analyze particle’ function in Fiji. Final figures were made in Adobe Illustrator. Cilia polarity was measured using previously described methods (Walentek et al., 2013).
Statistical analysis
Sample size (n) is defined as the number of embryos. Statistical analysis was performed using GraphPad Prism, JMP (SAS) and VassarStats (VassarStats.net) software. pitx2c, dand5, rfx2, dnah9, foxj1, xnr1, lefty and shh were compared using Chi-square analysis. All other comparisons were made using Student's t-tests after confirming the normal distribution of the data. We randomly picked one-cell X. tropicalis embryos from fertilization as uninjected controls or for MO or RNA injections.
Antibodies
Mouse monoclonal anti-acetylated α-tubulin (Sigma-Aldrich, T-6793; 1:1000) and Alexa Fluor 488 phalloidin (1:40) were used to mark the ciliary axoneme and F-actin, respectively. Rabbit polyclonal anti-WDR5 (Bethyl, A302-430A; 1:150), mouse monoclonal anti-Arl13b (NeuroMab, N295B/66; 1:100) and mouse monoclonal anti-γ tubulin (Sigma-Aldrich, T6557; 1:100) were used for immunofluorescence.
Supplementary Material
Acknowledgements
We thank the patients and their families who were the inspiration for this study; Sarah Kubek and Michael Slocum for animal husbandry; the Electron Microscopy core and Center for Cellular and Molecular Imaging at Yale for confocal imaging; Dr Ann Miller for the C-3xGFP construct; and Dr Michael Cosgrove for the human WT and S91K-WDR5 constructs.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S.K., M.K.K.; Methodology: S.S.K.; Validation: S.S.K.; Formal analysis: S.S.K.; Investigation: S.S.K.; Data curation: S.S.K.; Writing - original draft: S.S.K.; Writing - review & editing: S.S.K., M.K.K.; Visualization: S.S.K.; Supervision: M.K.K.; Project administration: M.K.K.; Funding acquisition: S.S.K., M.K.K.
Funding
This work was supported by the National Heart, Lung, and Blood Institute (NHLBI) [Pilot Project as part of 5U01HL098188; K99/R00-5K99HL133606-02 to S.S.K.; R33HL120783 to M.K.K.] and the National Institute of Child Health and Human Development [R01HD081379 to M.K.K.]. M.K.K. is an Edward Mallinckrodt, Jr. Foundation Scholar. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NHLBI. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.159889.supplemental
References
- Amack J. D. and Yost H. J. (2010). Cardiac left–right asymmetry. In Heart Development and Regeneration (ed. Rosenthal N. and Harvey R. P.), pp. 281-293. San Diego: Academic Press. [Google Scholar]
- Ang Y.-S., Tsai S.-Y., Lee D.-F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B. et al. (2011). Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183-197. 10.1016/j.cell.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu B. and Brueckner M. (2008). Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr. Top. Dev. Biol. 85, 151-174. 10.1016/S0070-2153(08)00806-5 [DOI] [PubMed] [Google Scholar]
- Blum M., Schweickert A., Vick P., Wright C. V. and Danilchik M. V. (2014). Symmetry breakage in the vertebrate embryo: when does it happen and how does it work? Dev. Biol. 393, 109-123. 10.1016/j.ydbio.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovski M. T., Yuan S., Pedersen N. B., Goth C. K., Makova S., Clausen H., Brueckner M. and Khokha M. K. (2013). The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504, 456-459. 10.1038/nature12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine R. D. and Schier A. F. (2000). Conserved and divergent mechanisms in left-right axis formation. Genes Dev. 14, 763-776. [PubMed] [Google Scholar]
- Cao F., Chen Y., Cierpicki T., Liu Y., Basrur V., Lei M. and Dou Y. (2010). An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS ONE 5, e14102 10.1371/journal.pone.0014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J., Vogan K. J., Tabin C. J. and Izpisúa Belmonte J. C. (2000). Mechanisms of left–right determination in vertebrates. Cell 101, 9-21. 10.1016/S0092-8674(00)80619-4 [DOI] [PubMed] [Google Scholar]
- Carneiro K., Donnet C., Rejtar T., Karger B. L., Barisone G. A., Díaz E., Kortagere S., Lemire J. M. and Levin M. (2011). Histone deacetylase activity is necessary for left-right patterning during vertebrate development. BMC Dev. Biol. 11, 29 10.1186/1471-213X-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.-I., Peyrot S. M., Leboeuf S., Park T. J., Mcgary K. L., Marcotte E. M. and Wallingford J. B. (2012). RFX2 is broadly required for ciliogenesis during vertebrate development. Dev. Biol. 363, 155-165. 10.1016/j.ydbio.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J.-F. and Skiniotis G. (2013). Assembling a COMPASS. Epigenetics 8, 349-354. 10.4161/epi.24177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J.-F., Collazo E. and Trievel R. C. (2006). Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 13, 698-703. 10.1038/nsmb1116 [DOI] [PubMed] [Google Scholar]
- Del Viso F. and Khokha M. (2012). Generating diploid embryos from Xenopus tropicalis. Methods Mol. Biol. 917, 33-41. 10.1007/978-1-61779-992-1_3 [DOI] [PubMed] [Google Scholar]
- De Rubeis S., He X., Goldberg A. P., Poultney C. S., Samocha K., Cicek A. E., Kou Y., Liu L., Fromer M., Walker S. et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209-215. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Copley R. R., Schultz J., Ponting C. P. and Bork P. (2002). A two-cilia model for vertebrate left-right axis specification. Genome Res. 12, 47-56. 10.1101/gr.203201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D. and Roeder R. G. (2006). Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713-719. 10.1038/nsmb1128 [DOI] [PubMed] [Google Scholar]
- Glinka A., Wu W., Delius H., Monaghan A. P., Blumenstock C. and Niehrs C. (1998). Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357-362. 10.1038/34848 [DOI] [PubMed] [Google Scholar]
- Gori F., Friedman L. G. and Demay M. B. (2006). Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev. Biol. 295, 498-506. 10.1016/j.ydbio.2006.02.031 [DOI] [PubMed] [Google Scholar]
- Hamada H., Meno C., Watanabe D. and Saijoh Y. (2002). Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 3, 103-113. 10.1038/nrg732 [DOI] [PubMed] [Google Scholar]
- Heasman J. (2006). Patterning the early Xenopus embryo. Development 133, 1205-1217. 10.1242/dev.02304 [DOI] [PubMed] [Google Scholar]
- Homsy J., Zaidi S., Shen Y., Ware J. S., Samocha K. E., Karczewski K. J., Depalma S. R., Mckean D., Wakimoto H., Gorham J. et al. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262-1266. 10.1126/science.aac9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha M. K., Chung C., Bustamante E. L., Gaw L. W., Trott K. A., Yeh J., Lim N., Lin J. C., Taverner N., Amaya E. et al. (2002). Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 225, 499-510. 10.1002/dvdy.10184 [DOI] [PubMed] [Google Scholar]
- Khokha M. K., Yeh J., Grammer T. C. and Harland R. M. (2005). Depletion of three BMP antagonists from Spemann's organizer leads to a catastrophic loss of dorsal structures. Dev. Cell 8, 401-411. 10.1016/j.devcel.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Kulkarni S. S., Griffin J. N., Date P. P., Liem K. F. Jr. and Khokha M. K. (2018). WDR5 stabilizes actin architecture to promote multiciliated cell formation. Dev. Cell 46, 595-610.e3. 10.1016/j.devcel.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Klena N. T., Gabriel G. C., Liu X., Kim A. J., Lemke K., Chen Y., Chatterjee B., Devine W., Damerla R. R. et al. (2015). Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521, 520-524. 10.1038/nature14269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. R., Kioussi C., O'connell S., Briata P., Szeto D., Liu F., Izpisua-Belmonte J. C. and Rosenfeld M. G. (1999). Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279-282. 10.1038/45803 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R. P. et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553-560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C., Kazanskaya O., Wu W. and Glinka A. (2001). Dickkopf1 and the Spemann-Mangold head organizer. Int. J. Dev. Biol. 45, 237-240. [PubMed] [Google Scholar]
- Nieuwkoop P. D. F. (1994). Normal Table of Xenopus Laevis (Daudin). Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- Odho Z., Southall S. M. and Wilson J. R. (2010). Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J. Biol. Chem. 285, 32967-32976. 10.1074/jbc.M110.159921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Dharmarajan V. and Cosgrove M. S. (2008a). Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 283, 32158-32161. 10.1074/jbc.C800164200 [DOI] [PubMed] [Google Scholar]
- Patel A., Vought V. E., Dharmarajan V. and Cosgrove M. S. (2008b). A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 283, 32162-32175. 10.1074/jbc.M806317200 [DOI] [PubMed] [Google Scholar]
- Patel A., Dharmarajan V., Vought V. E. and Cosgrove M. S. (2009). On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 284, 24242-24256. 10.1074/jbc.M109.014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A. J., Wang W., Graybosch D. M., Li H., Allis C. D., Patel D. J. and Verdine G. L. (2006). Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13, 704-712. 10.1038/nsmb1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A., Allali-Hassani A., Martin F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A. N., Arrowsmith C. H. and Min J. (2006). Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 25, 4245-4252. 10.1038/sj.emboj.7601316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweickert A., Weber T., Beyer T., Vick P., Bogusch S., Feistel K. and Blum M. (2007). Cilia-driven leftward flow determines laterality in Xenopus. Curr. Biol. 17, 60-66. 10.1016/j.cub.2006.10.067 [DOI] [PubMed] [Google Scholar]
- Song J.-J. and Kingston R. E. (2008). WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 283, 35258-35264. 10.1074/jbc.M806900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward M. M., Lee J.-S., O'donovan A., Wyatt M., Bernstein B. E. and Shilatifard A. (2006). Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 13, 852-854. 10.1038/nsmb1131 [DOI] [PubMed] [Google Scholar]
- Stubbs J. L., Oishi I., Izpisua Belmonte J. C. and Kintner C. (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 40, 1454-1460. 10.1038/ng.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland M. J. and Ware S. M. (2009). Disorders of left-right asymmetry: heterotaxy and situs inversus. Am. J. Med. Genet. C Semin. Med. Genet. 151C, 307-317. 10.1002/ajmg.c.30228 [DOI] [PubMed] [Google Scholar]
- Takahashi Y.-H., Westfield G. H., Oleskie A. N., Trievel R. C., Shilatifard A. and Skiniotis G. (2011). Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl. Acad. Sci. USA 108, 20526-20531. 10.1073/pnas.1109360108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel R. C. and Shilatifard A. (2009). WDR5, a complexed protein. Nat. Struct. Mol. Biol. 16, 678-680. 10.1038/nsmb0709-678 [DOI] [PubMed] [Google Scholar]
- Van Der Linde D., Konings E. E. M., Slager M. A., Witsenburg M., Helbing W. A., Takkenberg J. J. and Roos-Hesselink J. W. (2011). Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 58, 2241-2247. 10.1016/j.jacc.2011.08.025 [DOI] [PubMed] [Google Scholar]
- Vick P., Schweickert A., Weber T., Eberhardt M., Mencl S., Shcherbakov D., Beyer T. and Blum M. (2009). Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog Xenopus laevis. Dev. Biol. 331, 281-291. 10.1016/j.ydbio.2009.05.547 [DOI] [PubMed] [Google Scholar]
- Vonica A. and Brivanlou A. H. (2007). The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Dev. Biol. 303, 281-294. 10.1016/j.ydbio.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Walentek P., Schneider I., Schweickert A. and Blum M. (2013). Wnt11b is involved in cilia-mediated symmetry breakage during Xenopus left-right development. PLoS ONE 8, e73646 10.1371/journal.pone.0073646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H. and Allis C. D. (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859-872. 10.1016/j.cell.2005.03.036 [DOI] [PubMed] [Google Scholar]
- Yu X., Ng C. P., Habacher H. and Roy S. (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40, 1445-1453. 10.1038/ng.263 [DOI] [PubMed] [Google Scholar]
- Zaidi S., Choi M., Wakimoto H., Ma L. J., Jiang J. M., Overton J. D., Romano-Adesman A., Bjornson R. D., Breitbart R. E., Brown K. K. et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220-223. 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E. D., Demay M. B. and Gori F. (2008). Wdr5 is essential for osteoblast differentiation. J. Biol. Chem. 283, 7361-7367. 10.1074/jbc.M703304200 [DOI] [PubMed] [Google Scholar]
- Zhu J. Y., Fu Y., Nettleton M., Richman A. and Han Z. (2017). High throughput in vivo functional validation of candidate congenital heart disease genes in Drosophila. Elife 6, e22617 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.