Abstract
In this study, Illumina Miseq sequencing of 16S rRNA gene amplicon was performed on sediments collected from Krossfjorden, Arctic for analyzing the bacterial community structure. Metagenome contained 15,936 sequences with 5,809,491 bp size and 53% G+C content. Metagenome sequence information are now available at NCBI under the Sequence Read Archive (SRA) database with accession no. SRP159159. Taxonomic hits distribution from MG-RAST analysis revealed the dominance of Alpha- and Gamma-subdivisions of Proteobacteria (88.89%) along with Bacteriodetes (8.89%) and Firmicutes (2.22%). Predominant species were Alteromonadales bacterium TW-7 (24%), Pseudoalteromonas haloplanktis (20%) and Pseudoalteromonas spp. SM9913 (18%). MG-RAST assisted analysis also detected the presence of a variety of marine taxa like Bacteriodes, Pseudovibrio, Marinobacter, Idiomarina, Teredinibacter, etc. which take part in key ecological functions and biogeochemical activities of Arctic fjord ecosystems.
Keywords: Arctic, Bacterial diversity, Metagenome, Amplicon sequencing, Illumina
Specifications table
| Subject area | Biology |
| More specific subject area | Polar metagenomics |
| Type of data | FastaQ file |
| How data was acquired | NGS sequencing on Illumina MiSeq platform |
| Data format | Raw data |
| Experimental factors | Fjord sediment samples from Krossfjorden, Arctic |
| Experimental features | Metagenomic DNA extraction from Krossfjorden sediment, NGS sequencing on Illumina MiSeq platform and MG-RAST analysis of NGS data |
| Data source location | Krossfjorden, Arctic (79°08׳60"N, 11°44׳59"E) |
| Data accessibility | The data of this metagenome is available in the NCBI BioSample Submission Portal as Bioproject ID: PRJNA488527 and SRA accession no.:SRP159159. https://www.ncbi.nlm.nih.gov/sra/SRP159159. |
Value of the data
-
•
Present study reveals the bacterial community structure in Arctic fjords, a least explored extreme environment undergoing rapid changes due to climate variability.
-
•
Metagenome based study enabled the detection of Arctic bacterial community including unculturable bacterial population.
-
•
Dominance of gram negative bacteria (97.78%) in the Krossfjorden sediments limiting the gram positive to a significantly low level 2.22% was an important observation.
-
•
Proteobacteria dominated the bacterial community followed by Bacteriodetes and Firmicutes.
1. Direct link to deposited data
Deposited data can be found here: https://www.ncbi.nlm.nih.gov/sra/?term=SRP159159.
2. Data
During recent years, metagenome based approaches has helped in unraveling the microbiome diversity in various eco-habitats overcoming the shortfalls of culture based methods, since majority of the microbes are unculturable and thereby go undetected in conventional methods. Supported with Next Generation Sequencing Technology, Metagenomics can provide vast information about the enormous uncultured microbial populations existing in any environment [1]. Microbial communities inhabiting extreme environments are adapted to extreme conditions thereby serving as a repository of novel genes and bioactive molecules. Collective operation of metagenomics, NGS platforms and annotation tools elicit the exploration of extremophiles from diverse environments [2], [3]. Present study is focused on the taxonomic composition of bacterial assemblages in Krossfjorden sediments.
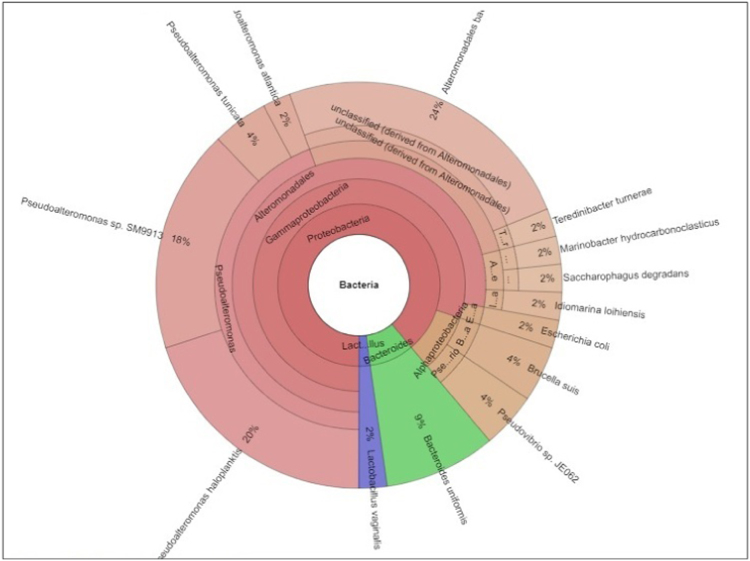
A total of 15,796 reads were analyzed and 2907 operational taxonomic units (OTU) were identified in order to reveal the diversity of bacterial flora. All the resulting fragments or OTUs were then classified into 3 phyla, 4 classes, 6 orders, 9 families and 11 genera. MG-RAST analysis for phylum level classification revealed the preponderance of Proteobacteria (88.89%) followed by Bacteriodetes (8.89%) and Firmicutes (2.22%). The classified OTUs represent distinct orders dominated by Alteromonadales, Bacteriodales, Rhizobiales and Rhodobacteriales. Family level dominance was shown by Pseudoalteromonadaceae (40.44%), Bacteriodaceae (8.89%), Rhodobacteriaceae (4.44%), Alteromonadaceae (4.44%) and Brucellaceae (4.44%). Nevertheless, an unclassified bacterial group derived from Alteromonadales (26.67%) was also found among the Krossfjorden bacterial families. The analysis identified 11 genera among which the most dominant was Pseudoalteromonas (44.44%) followed by Bacteriodes (8.89%), Pseudovibrio (4.44%) and Brucella (4.44%). Species level identification showed that Alteromonadales bacterium TW-7 (24%), Pseudoalteromonas haloplanktis (20%) and Pseudoalteromonas sp. SM9913 (18%) were the predominant ones (Fig. 1) [4].
Fig. 1.
Interactive Krona chart for visualization of bacterial communities detected from Krossfjorden sediments.
3. Experimental design, materials and methods
In this study, sediment samples were collected from Krossfjorden (79°08׳60"N, 11°44׳59"E) using a Van Veen Grab during Indian Scientific Expedition to the Arctic (2015–2016) during summer. Sediment samples were suspended in sterilized virus free seawater and subjected to low speed (320 × g) centrifugation for the removal of coarse sediment particles followed by high speed (9000 × g) centrifugation for collecting the fine sediment fraction with bacteria. The supernatant was subjected to sequential filtration using 0.8 μ, 0.45 μ and 0.22 μ filter membranes for the separation of bacterial fraction [5]. Fine sediment obtained after high speed centrifugation and the residue on filter membranes were used for metagenomic DNA extraction as per modified Zhou et al. [6] protocol. Next Generation Sequencing was done using Illumina Miseq at Interpretomics, Bangalore. Quality and quantity of extracted DNA were analysed by NanoDrop ND-2000 and Qubit. The 16S rRNA libraries were prepared from the QC passed DNA sample using 16S rRNA gene universal primers (Forward Primer: ACTCCTACGGGAGGCAGCAG and Reverse Primer: GGACTACHVGGGTWTCTAAT) with standard Illumina barcodes and adapters. The Amplicons were further purified using Ampure XP beads. The barcoded libraries were validated by Agilent DNA 1000 Bioanalyser and quantified using Qubit DNA BR reagent assay. The quantified libraries were pooled and sequenced using MiSeq.
Raw sequences from Illumina Miseq were processed and analysed using Metagenomic Rast Server (MG-RAST) version 4.0.3 (http://metagenomics.anl.gov/). Raw data were uploaded as FASTAQ files after demultiplexing of paired-end reads. Reads generated after quality processing and deduplication by MG-RAST pipeline analysis were subjected to taxonomic analysis. MG-RAST pipeline has provided an estimate of bacterial abundances present in Krossfjorden sediments and based on these, an assessment was carried out to evaluate the bacterial diversity within the sample.
3.1. Nucleotide sequence accession number
Metagenome sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under the accession number: SRP159159.
Acknowledgements
Authors are grateful to NCPOR, Goa for providing logistic and technical support for sample collection. Necessary facilities provided by Dept. of Marine biology, Microbiology and Biochemistry and National Centre for Aquatic Animal Health (NCAAH), Cochin University of Science and Technology are thankfully acknowledged.
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.11.101.
Contributor Information
Bhavya Kachiprath, Email: bhavyanirmalagiri@gmail.com.
Jayesh Puthumana, Email: jayeshp24@gmail.com.
Jayanath Gopi, Email: jayanathgopi@gmail.com.
Solly Solomon, Email: lazarsolly@hotmail.com.
K.P. Krishnan, Email: kpkrishnan@gmail.com.
Rosamma Philip, Email: rosammap@gmail.com, rose@cusat.ac.in.
Transparency document. Supplementary material
Supplementary material
References
- 1.Mandal S.D., Lalremsanga Z.H.T., Kumar N.S. Bacterial diversity of Murlen National Park located in Indo–Burman biodiversity hotspot region: a metagenomic approach. Genom. Data. 2015;5:25–26. doi: 10.1016/j.gdata.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux S., Enault F., Ravet V., Colombet J., Bettarel Y., Auguet J., Bouvier T., Lucas-Staat S., Vellet A., Prangishvili D., Forterre P., Debroas D., Sime-Ngando T. Analysis of metagenomic data reveals common features of halophilic viral communities across continents. Environ. Microbiol. 2016;18(3):889–903. doi: 10.1111/1462-2920.13084. [DOI] [PubMed] [Google Scholar]
- 3.Pramanik A., Basak P., Banerjee S., Sengupta S., Chattopadhyay D., Bhattacharyya M. Metagenomic exploration of the bacterial community structure at Paradip Port, Odisha, India. Genom. Data. 2015;7:94–96. doi: 10.1016/j.gdata.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a web browser. BMC Bioinforma. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon S., Kachiprath B., Jayanath G., Sajeevan T.P., Singh I.S.B., Philip R. High-quality metagenomic DNA from marine sediment samples for genomic studies through a preprocessing approach. 3 Biotech. 2016;6:160. doi: 10.1007/s13205-016-0482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J., Bruns M.A., Tiedje J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996;62(2):316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Deposited data can be found here: https://www.ncbi.nlm.nih.gov/sra/?term=SRP159159.