Abstract
Papain-like cysteine proteases (PLCP) are prominent peptidases found in most living organisms. In plants, PLCPs was divided into nine subgroups based on functional and structural characterization. They are key enzymes in protein proteolysis and involved in numerous physiological processes. In this paper, we reviewed the updated achievements of physiological roles of plant PLCPs in germination, development, senescence, immunity, and stress responses.
Keywords: papain-like cysteine proteases, programmed cell death, immunity, stress responses, senescence
Introduction
Proteases include diverse families (e.g., cysteine-, serine-, aspartic-, metallo-, and threonine-proteases) and play crucial roles in protein proteolysis (van der Hoorn, 2008). Based on the evolutionary relationships, they have been subdivided into 61 clans of 253 families (Rawlings et al., 2016). Among them, papain-like cysteine proteases (PLCPs), featuring a nucleophilic cysteine thiol at the active site (i.e., Cys, His, and Asn), are one of the most abundant groups of cysteine proteases (Rawlings et al., 2010).
Papain-like cysteine proteases are found in most organisms, including virus (Rawlings et al., 1992), bacteria (Kantyka et al., 2011), yeast (Enenkel and Wolf, 1993), protozoa, plants, and animals (Rawlings et al., 2010; Novinec and Lenarcic, 2013). These enzymes are produced as inactive precursors with a signal peptide for protein secretion and an auto-inhibitory prodomain to prevent unwanted protein degradation (Figure 1; Coulombe et al., 1996). The active protease domain contains the catalytic triad Cys-His-Asn (Figure 1).
FIGURE 1.
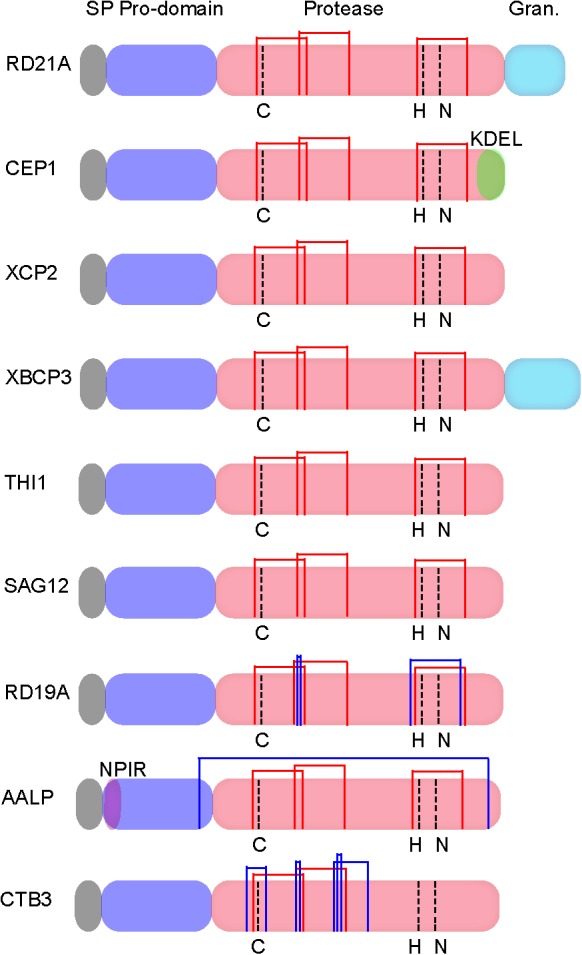
Protein structures of Arabidopsis papain-like cysteine proteases (PLCPs) [modified from Richau et al. (2012) and Misas-Villamil et al. (2016)]. SP, N-terminal signal peptide; Pro-domain, an auto-inhibitory domain; Protease, the catalytic domain contains the catalytic triad Cys-His-Asn; Granulin, C-terminal granulin domain; NPIR, a vacuolar targeting signal; KDEL, a C-terminal retrieval signal for ER localization. Common disulphide bridges and subfamily specific disulphide bridges are indicated with red thin lines and blue thin lines, respectively.
Papain-like cysteine proteases genes belong to a large multigenic family with 31, 43, 40, 26, 40, and 24 PLCP family members were identified in Arabidopsis, rubber, cassava, castor, poplar, and grapevine, respectively, and they were divided into 9 subfamilies based on structural characteristics (Figures 1, 2; Martinez and Diaz, 2008; Zou et al., 2017b). In animals, PLCPs are well known lysosomal proteases that perform significant functions in the terminal degradation of proteins within autolysosomes (Kroemer and Jaattela, 2005; Man and Kanneganti, 2016). In recent years, vast majority of plant PLCPs have been characterized during many plant processes. In this review, we summarized what is known of plant PLCPs in growth, seed germination, anther development, senescence, immunity and stress responses (Figure 3 and Table 1).
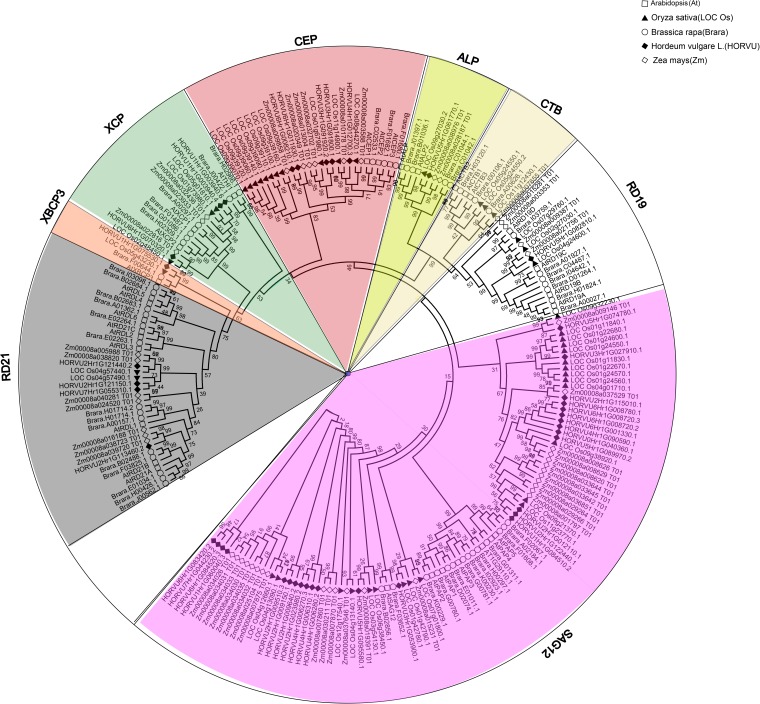
FIGURE 2.
Phylogenetic analysis of PLCPs in Arabidopsis, rice, maize, barley, and Brassica rapa. Sequence alignment was performed using MUSCLE and the phylogenetic tree was constructed using bootstrap maximum likelihood tree (1000 replicates) method of MEGA6.
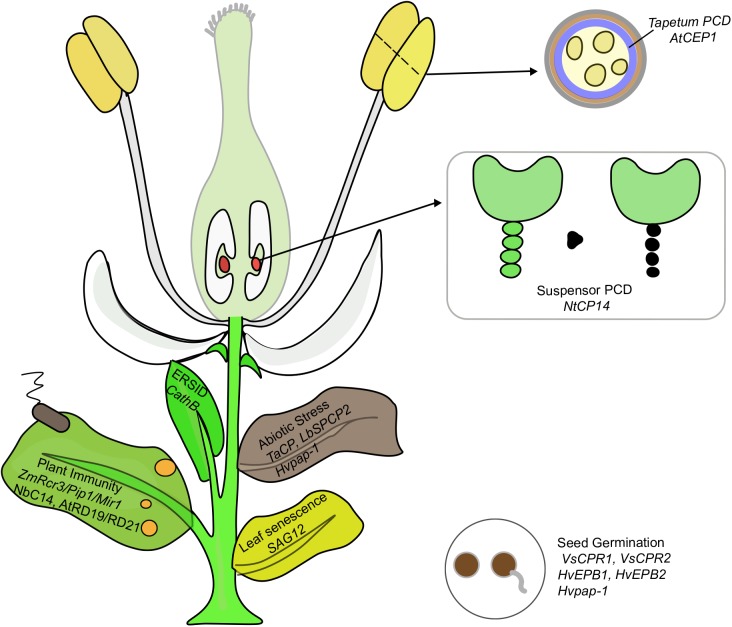
FIGURE 3.
PLCPs play important functions in multiple processes of plant growth including seed germination, PCD, abiotic stress and immunity. The genes implicated in each process are discussed in this review.
Table 1.
Catalog of plant papain-like cysteine proteases (PLCPs) involved in germination, development, senescence, immunity, and stress responses.
| PLCP | Species | Function | Reference |
|---|---|---|---|
| EP-B | Barley | Induced expression in the germinating seeds; | Koehler and Ho, 1990; |
| Degrade the endosperm storage proteins to provide | Mikkonen et al., 1996. | ||
| nitrogenous nutrients for young seedlings. | |||
| Pap-1 | Barley | Involved in grain protein mobilization during germination; | Cambra et al., 2012; |
| Silencing decreased germination rate and delayed | Diaz-Mendoza et al., 2016; | ||
| senescence process. | Velasco-Arroyo et al., 2016. | ||
| CathB | Arabidopsis | Mutants displayed reduced PCD during abiotic stress and | Ge et al., 2016; |
| endoplasmic reticulum stress. | Cai et al., 2018. | ||
| Pap-1/6/9 | Barley | Inhibition of cathepsins increased the stress-induced | Barany et al., 2018. |
| microspore embryogenesis. | |||
| CP14 | Tobacco | Silencing delayed PCD of suspensor. | Zhao et al., 2013. |
| CEP1 | Arabidopsis | Mutants delayed tapetal PCD and decreased pollen | Zhang D. et al., 2014 |
| production. | |||
| SAG12 | Arabidopsis | Induced during senescence; | Lohman et al., 1994; |
| Mutants decreased yield under low nitrogen (LN) conditions. | James et al., 2018. | ||
| SAG12-H1RD21 | Rubber tree | Highly expressed only in senescent leaves. | Zou et al., 2017a. |
| 1A | Arabidopsis | Involved in dehydration stress; Mutants enhanced susceptibility to Botrytis cinerea. | Koizumi et al., 1993; Shindo et al., 2012. |
| RD19A | Arabidopsis | Involved in dehydration stress; Mutants enhanced susceptibility to Ralstonia solanacearum. | Koizumi et al., 1993; Bernoux et al., 2008. |
| CP | Wheat | Increased expression under abiotic stress and played a role in water deficit; Silencing enhanced tolerance to salt and osmotic stress. | Zang et al., 2010 |
| CP20.1 | Pepper Brassica napus | Ectopic expression leads to premature degradation of tapetum, involvement in tapetum degradation and pollen wall synthesis. | Xiao et al., 2014.Song et al., 2016; |
| SPCP2 | Sweet potato | Enhanced resistance to drought and salt stress when overexpressing;Increased sensitivity to drought stress when overexpressing. | Chen et al., 2010;Chen et al., 2013; |
| Mir1 | Maize | Induced expression at wounding site;Enhanced resistance to caterpillar;Acts as ethylene signal conferring resistance to corn leaf aphid; | Chang et al., 2000;Pechan et al., 2002;Louis et al., 2015. |
| AALP | Arabidopsis Tomato | Increased protein activity in senescent leaves, mutants delay leaf senescence.Up-regulated upon pathogen attack and inhibited by pathogen-derived inhibitors; | Pruzinska et al., 2017.Kruger et al., 2002;Tian et al., 2007; |
| Rcr3 | Tomato | Resistance to Phytophthora infestans, Cladosporium fulvum, and Globodera rostochiensis. | Dixon et al., 2000;Song et al., 2009;Lozano-Torres et al., 2012. |
| Pip1 C14 |
Nicotiana benthamiana | Silencing plants susceptible to C. fulvum, Pseudomonas syringae, and P. infestans. | Ilyas et al., 2015.Kaschani et al., 2010. |
| XCP2 | Arabidopsis | Silencing plants susceptible to P. infestans. Mutants decreased susceptibility to R. solanacearum. | Zhang B. et al., 2014. |
PLCPs Function in Seed Germination
Storage, structural, metabolic, and protective proteins are stored in seeds. During germination, these seed proteins were mobilized or degraded to nourish growing seedlings. These processes were
mainly triggered by PLCPs (Grudkowska and Zagdanska, 2004). In germinating maize and wheat, cysteine proteinases amount up to 90% of total proteolytic activity of prolamins (Grudkowska and Zagdanska, 2004). During Vicia sativa seed germination, four PLCPs (CPR1, CPR2, proteinase A, and CPR4) were identified with CPR1 and CPR2 were involved in the mobilization of vicilin and 7S storage globulin (Fischer et al., 2000; Schlereth et al., 2000, 2001). 27 PLCPs were identified during barley grain germination (Zhang and Jones, 1995), and two PLCPs (EP-A and EP-B) were purified (Poulle and Jones, 1988). Two isoforms of EP-B (EPB1 and EPB2) were identified in the germinating seeds with its expression was induced by gibberellin (GA) and suppressed by abscisic acid (ABA) (Koehler and Ho, 1990; Mikkonen et al., 1996). The homolog cysteine endopeptidase EP8 of barley EP-A (HvEPA) in triticale was responsible for mobilizing stored proteins during seed germination, and its activity can be inhibited by endogenous cystatin TrcC-4 (Prabucka et al., 2013). During seed germination, these proteases were secreted from the scutellar epithelium and aleurone layer to the endosperm which degrade the endosperm storage proteins to provide nitrogenous nutrients for young seedlings (Mikkonen et al., 1996). In addition, HvPap-1, a GA induced PLCP, was reported to play essential functions in protein mobilization during barley grain germination (Cambra et al., 2012). HvPap-1 was localized to the protein bodies and vesicles in the embryo, and it could degrade barley endosperm proteins (hordeins, albumins, and globulins). The overexpression of HvPap-1 decreased starch amount in seeds and increased germination rate, while silencing HvPap-1 displayed an opposite phenotype with increased starch amount in seeds and decreased germination rate (Diaz-Mendoza et al., 2016). These results indicate that PLCPs are important factors in mobilizing storage proteins to promote seed germination, and their expression and/or activity is regulated by GA, ABA, and cystatins. Arabidopsis PLCPs genes display differential expression in different organs, with PAP2, PAP3, and RD19A display high expression in seed, while PAP4 and PAP5 display high expression in leaf. These researches highlight divergent functions of different PLCP proteins. In addition, a lot of PLCPs display high expression in various tissues and organs, such as RD21A, RD21B, RD19A, indicating a housekeeping role in plant growth and development (Richau et al., 2012).
In view of the important roles of PLCP proteins in seed germination, the regulation of PLCPs activity should play essential function in seed germination and seedling development. Phytocystatins (PhyCys) are a group of small proteins and can directly inhibit PLCPs activity (Arai et al., 2002). Many PhyCys have been identified, such as TrcC-6 (Siminska et al., 2015) and TrcC-8 protein that exert an inhibitory effect on PLCP through the interaction with PLCPs (Prabucka et al., 2017). The activities of PhyCys and PLCPs need to maintain a relatively balanced level to ensure the normal seed germination (Szewinska et al., 2016).
The nutrients provided by plant seeds are the basis for the growth and development of offspring. PLCPs are one of the key factors to initiate and complete this process (Szewinska et al., 2016). It is well known that seed germination may be destroyed under adversity stresses, resulting in the inability to form seedlings. Under stress conditions, the activity of PLCPs and their regulatory factors may be destroyed, but the specific mechanism of “destruction” and the resulting consequences need further analysis in various plants.
PLCPs Associated With Programmed Cell Death
Programmed cell death (PCD) is a highly ordered and genetically controlled process that removes unwanted or damaged cells in both eukaryotic and prokaryotic organisms, playing important roles in protecting against environmental stresses and pathogen invasions. DNA fragmentation, reactive oxygen species (ROS) accumulation and organelle “degradation” were general features of PCD process. PCD played essential functions throughout the plant’s life cycle from embryogenesis to plant death (Staal and Dixelius, 2007; Lord and Gunawardena, 2012).
Papain-like cysteine proteases are essential regulators of plant PCD. In Arabidopsis, a lot of PLCPs were reported in the PCD of tracheary element (TE), tapetum, suspensor, and ER-stress-induced cell death, respectively (Zhao et al., 2000; Cai et al., 2018). AtXCP1 and AtXCP2, two xylem-specific PLCPs, were expressed at a high level in xylem during the PCD process. However, no developmental differences were observed in the single xcp1, xcp2 mutants and the double mutant xcp1/xcp2, suggesting that they act redundantly with other regulators to regulate TE-PCD (Zhao et al., 2000). Cathepsins are the cysteine protease of papain-like C1A and are important regulators involved in numerous plant biological processes, including leaf senescence and PCD. Three Arabidopsis cathepsin B proteins (AtCathB1-3) were identified with tandem mass spectrometry, and its triple mutant displayed a strong reduction in the PCD induced by abiotic stress (e.g., ultraviolet, oxidative stress) and endoplasmic reticulum stress (Ge et al., 2016). Further research showed that the silencing of cathepsin B reduced ROS accumulation and ER-stress-induced PCD (ERSID), while the downregulation of PBA1 increased ERSID, demonstrating that ERSID was positively and negatively regulated by cathepsin B and PBA1, respectively (Cai et al., 2018). High catalytic activity of tobacco PLCP protein NtCP14 was restricted only to the suspensor at the 8- and 32-celled embryo stages, which correlating with the onset of PCD (Zhao et al., 2013). The overexpression of NtCP14 induces premature cell death in the basal cell lineage and leads to embryonic arrest and seed abortion, whereas the silencing of NtCP14 leads to profound delay of suspensor PCD (Zhao et al., 2013). These results indicate that PLCPs play key regulatory roles in seed, TE, tapetum, suspension, and ERSID.
Tapetum plays a crucial role in pollen development by secreting numerous nutritive proteins, enzymes, and sporopollenin precursors for pollen maturation (Plackett et al., 2011). Tapetum undergoes PCD during the late stages of pollen development and disruption PCD of tapetal cells will result in male sterility (Ku et al., 2003; Kawanabe et al., 2006; Li et al., 2017). AtCEP1, one KDEL-tailed PLCP, is expressed in tapetum and localized in endoplasmic reticulum (Zhang D. et al., 2014). In cep1 mutant, tapetal PCD was delayed and pollen production was reduced. Further transcriptomic analysis showed that the expression of genes involved in tapetum degradation and pollen development were changed in cep1 mutant (Zhang D. et al., 2014). These results showed that CEP1 plays an essential function in tapetal cell PCD and pollen development (Zhang D. et al., 2014). Moreover, a papain-like cysteine protease from Brassica napus, BnaC.CP20.1, is significant to tapetal degeneration and pollen-wall formation (Song et al., 2016). The ectopic expression of BnaC.CP20.1 prompt tapetum PCD and lead to male sterile (Song et al., 2016). In summary, the tapetum PCD is extremely important for pollen development, and PLCPs play important roles in this process. Although some PLCPs have been studied, the PLCPs which play key roles in the tapetum PCD process have not been identified or studied in depth.
PLCPs Involved in Leaf Senescence
Leaf senescence is a physiological process that recycling the endogenous nutrients from the senesencing leaves to support the growth of younger leaves and reproductive organs. Protein breakdown is one of the most fundamentally important reactions during leaf senescent and PLCPs play important functions in protein proteolysis during leaf senescence (Bhalerao et al., 2003; Roberts et al., 2012; Diaz-Mendoza et al., 2014). In Arabidopsis, many members of PLCPs, including SAG12, RD21A, AtRD19A, RD19C, ALP/SAG2/AALP/ALEU, CTB1, and CTB3, have been described as participants in leaf senescence (Lohman et al., 1994; Yamada et al., 2001; Gepstein et al., 2003; Guo and Cai, 2004; Guo and Gan, 2014).
SAG12 exhibits a strictly senescence-associated expression pattern in leaves and thus has been widely used as a senescence marker gene (Lohman et al., 1994). The sag12 mutant did not show any discernible variation in phenotype under normal conditions, whereas, under low nitrogen (LN) conditions, the yield was decreased in sag12 mutant, suggesting that SAG12 participate in the N remobilization that sustains seed production. In addition, the B. napus homolog of SAG12 and RD21A proteases were also reported in protein degradation and response to N limitation in senescent leaves (Poret et al., 2016).
Based on the senescence-specific characterization of SAG12, an autoregulatory senescence inhibition system (PSAG12-IPT) has been explored by fusing the SAG12 promoter (PSAG12) to a cytokinin-biosynthesizing enzyme-isopentenyl transferase (IPT) (Gan and Amasino, 1996). This fusion activates the expression of IPT at the onset of senescence and subsequently increases the cytokinin levels which, in turn, delays the decay of plant senescence (Gan and Amasino, 1996). This technology has been successfully applied in practical applications in many plant species and is approaching commercialization (Guo and Gan, 2014). Various SAG12 orthologs genes with similar functions have beed identified in a variety of plant species including rice, B. napus, sweet potato and tobacco (Noh and Amasino, 1999; Chen et al., 2002; Beyene et al., 2006; Gombert et al., 2006; Liu et al., 2010). Similar to Arabidopsis SAG12, rice PSAG39-IPT transgenic plants displayed a delayed leaf senescence phenotype and greater number of emerged panicles, suggesting the homologs function as Arabidopsis SAG12 (Liu et al., 2010). Similarly, in rubber tree (Hevea brasiliensis), there are 17 Arabidopsis SAG12 orthologs (HbSAG12H1–17) with HbSAG12H1 displayed the same expression pattern as senescence-associated genes, indicating that HbSAG12H1 can also act as a molecular marker to study the leaf senescence mechanism of Hevea (Zou et al., 2017a). Subsequently, Zou et al. (2017b) used HbSAG12H1 as the indicator to successfully identify six new PLCP genes, i.e., HbRD21B, HbRD21E, HbRD21F, HbCEP1, HbXBCP3L, and HbRD19B by deep sequencing of the senescence rubber leaf transcriptome, suggesting that these PLCPs may play important roles in leaf senescence (Zou et al., 2017b). In addition, the individual dark treatment of Arabidopsis leaves (8 weeks old) showed that the leaf senescence program was induced to start, and the activities of many PLCPs were increased, among which RD21A and AALP displayed the highest induction in senescing leaves (Pruzinska et al., 2017). By phenotypic analysis of the aalp-1 and rd21A-1/aalp-1 mutants, fewer senescent leaves were identified and senescence was delayed than wild type, indicating that AALP may be helpful to the senescence process of plants (Pruzinska et al., 2017).
In summary, leaf senescence is a finely regulated process involving the degradation of many substances. The enzymatic reactions catalyzed by PLCPs encoded by senescence-associated genes (SAGs) are an important pathway for protein degradation. At present, SAG12 is the most intensively studied senescence-associated PLCPs, whose function has been characterized in many species. In addition to SAG12 and other PLCPs that have been studied, there are many more PLCPs participating to leaf senescence remain largely unknown.
PLCPs Mediate Plant Abiotic Stress Response
Plants are constantly challenged by environmental abiotic stresses (e.g., heat, drought, cold, or salinity). Plants have evolved delicate mechanisms to cope with abiotic stresses by reprogramming the expression of gene subsets and inducing an adaptive response. The recycling of proteins by plant proteolysis is a primary defense line for plant survival. Among protease families, PLCPs are the predominantly up-regulated plant proteases, and exhibit increased expression in response to multiple environmental stresses (Rabbani et al., 2003; Kempema et al., 2007; Roberts et al., 2012; Diaz-Mendoza et al., 2014).
AtRD21A and AtRD19A, two important protein markers for dehydration stress adaptation, were highly induced by drought and salt stresses (Koizumi et al., 1993). Under PEG, salt and cold stresses, the expression of wheat PLCP gene (TaCP) was upregulated (Zang et al., 2010). In addition, transgenic Arabidopsis overexpressing TaCP showed stronger drought tolerance under water-stressed conditions than wild-type, indicating that TaCP plays a role in mediating dehydration tolerance (Zang et al., 2010). Sweet potato SPCP2 gene showed enhanced expression during senescence and was also regulated in response to dark, ABA, JA and ethephon treatment (Chen et al., 2010). The overexpression of SPCP2 in Arabidopsis enhanced resistance to drought and salt stress (Chen et al., 2010). Whereas, the overexpression of sweet potato SPCP3 in Arabidopsis conferred sensitivity to drought stress (Chen et al., 2013). The expression of pepper (Capsicum annuum) PLCP gene (CaCP) was induced during leaf senescence and was also significantly unregulated by abiotic and biotic stress treatments (Xiao et al., 2014). The suppression of CaCP in pepper enhanced tolerance to salt and osmotic stress (Xiao et al., 2014). HvPap-1 was induced in response to dark and nitrogen starvation (Velasco-Arroyo et al., 2016). The overexpression of HvPap-1 in barley accelerated leaf senescence, while silencing HvPap-1 with amiRNA delayed senescence process (Velasco-Arroyo et al., 2016). In addition, under stress condition, the expression of three barley cathepsin-like proteins (HvPap-1, HvPap-6, and HvPap-19) were increased and autophagy was activated in barley micropores, and the inhibition of cathepsins by caspase-3 inhibitors reduced apoptosis and increased the stress-induced microspore embryogenesis (Barany et al., 2018). In water deficient barley leaves, cystatin HvCPI-2 and HvCPI-4 delay the natural senescence process and increase tolerance to drought by regulating the expression and activity of HvPap-1, HvPap-12, and HvPap-16 C1A proteases, which may be due to the tight control of protease activity to avoid sudden degradation of proteins (Velasco-Arroyo et al., 2018).
Protein hydrolysis is very important for plants to response adversity stresses. In addition to enhancing plant resistance to stress, many PLCP proteases also accelerate plant leaf senescence or enhance plant sensitivity to abiotic stress. In this case, plants often regulate the gene expression or protein activity of PLCPs through some regulators to promote plant growth and increase crop yield. The currently deep-study regulatory factors of PLCP are phytocystatins, and many researches have revealed that the overexpression of phytocystatins significantly delays plant leaf senescence and increases stress tolerance, and the direct inhibition of protease activity may be the main reason (Kunert et al., 2015; Subburaj et al., 2017; Tan et al., 2017a,b). In conclusion, the differential effects of PLCPs in different species are determined by various factors such as the spatiotemporal pattern of protease expression and action, action substrate, and specific growth period and growth state in which the plant is located. Therefore, in different specific environments, the regulation of PLCPs activity is extremely important to enhance plant stress tolerance.
PLCPs Play a Key Role in Plant Immunity
In natural environment, plants are also attacked by a diverse array of pathogens and pests, including bacteria, fungi, oomycetes, nematodes, insects, and microbes. Many studies have highlighted the importance of PLCP in defense against plant pathogen. In most cases, a lack of PLCP expression leads to alterations of pathogen resistance because PLCP mutations are more susceptible to pathogen infection (Misas-Villamil et al., 2016).
Maize inbred resistance 1 (Mir1), a secreted maize PLCP that localized in vesicle, showed high accumulation at the wounding site after larval feeding (Chang et al., 2000). Tobacco budworm larvae feeding with Mir1-overexpressing plants caused severe damage to caterpillar for Mir1 degraded the peritrophic matrix of the insect gut (Pechan et al., 2000, 2002). Recent research showed that ethylene (ET) was required for accumulation of Mir1 and contributed to heighten resistance to corn leaf aphid (CLA) in maize (Louis et al., 2015). In addition, PLCPs are also key regulators of salicylic acid (SA)-dependent defense signaling. Previous studies have shown that SA treatment can strongly induce PLCP protein activity in maize leaves, and the activated apoplastic PLCPs can in turn induce the expression of SA-related immune genes (van der Linde et al., 2012). The latest research reveals that PLCPs need to release Z. mays immune signaling peptide 1 (Zip1) through their protein precursors to induce SA accumulation and activate SA defense signaling in leaves (Ziemann et al., 2018). These results indicate that the interaction of SA with PLCPs plays a key role in the expression regulation of downstream defense genes under bioticstress.
Tomato Rcr3 (Required for Cladosporium resistance-3) and Pip1 (Phytophthora inhibited protease-1) were up-regulated upon pathogen challenge and their activities were inhibited by pathogen-derived inhibitors (Kruger et al., 2002; Tian et al., 2007). Deletion of Rcr3 enhanced plants susceptibility to the pathogen Phytophthora infestans (Song et al., 2009), Cladosporium fulvum (Dixon et al., 2000), and nematode Globodera rostochiensis (Lozano-Torres et al., 2012). Similar with Rcr3, the Pip1 mutant plants were also more susceptible to C. fulvum, Pseudomonas syringae, and P. infestans (Ilyas et al., 2015). Silencing of C14 in Nicotiana benthamiana increased plants susceptibility to P. infestans (Kaschani et al., 2010). In addition, Arabidopsis rd19 and rd21 mutants are more susceptible to bacterial pathogen Ralstonia solanacearum and fungal pathogen Botrytis cinerea, respectively (Bernoux et al., 2008; Shindo et al., 2012). Whereas, xcp2 mutant displayed decreased susceptibility to R. solanacearum (Zhang B. et al., 2014).
Taken together, these data demonstrate that PLCPs play a determinative role in regulating pathogen defense. However, as can be seen from the above studies, previous studies have focused on the reduction of plant immunity after mutation of PLCP genes, but there is relatively little understanding of how PLCPs participate in plant immune defenses; furthermore, PLCPs play a key role in the plant defense hormone signaling pathways such as ET- and SA-pathway. We have known that the coordinated interaction among various defensive hormones of plant is crucial for the plant immunity. Therefore, studying the role of PLCPs in various plant hormone pathways and the mechanism of action will be an effective way to understand how PLCPs maintain or enhance plant immunity. On the other hand, synthetically revealing the role of PLCPs in plant immune processes is another important research content for the comprehensive exploration of plant immune mechanisms.
Concluding Remarks
Over the past few years, the study of plant PLCPs has widened considerably, and deciphering the molecular function of these proteases is advanced. PLCPs have a broad substrate specificity, and their protein location, activation and inactivation are tightly regulated in a number of ways. However, the direct link between PLCPs activation and perception in plant signaling has not been fully explored. Functional redundancy of PLCPs has hampered defining their biological functions. Therefore, multiple experimental approaches including double or even triple mutants are needed to address its biological functions. Clearly, further investigation is required to understand how PLCPs perceive stresses and signals and what is the downstream players in PLCP pathways. Sensitive and novel techniques, such as quantitative proteomics and labeling probes, were used to uncover protease substrates and function (Demir et al., 2018; van der Hoorn and Rivas, 2018), thereby may facilitate to advance our mechanical understanding to the function of plant PLCPs.
Author Contributions
HL and ZZ conceived and wrote the review. MH, QW, and LC critically reviewed the manuscript. All authors listed approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The authors are financially supported by the Natural Science Foundation of Henan Provincial Science and Technology (No. 182300410063), Key Scientific Research Projects of Henan Higher Education Institutions (No. 18A180031), the National Natural Science Foundation of China (No. U1604112), the Funding Scheme for Young Core Teachers of Henan Province (2017), and Nanhu Scholars Program for Young Scholars of Xinyang Normal University.
References
- Arai S., Matsumoto I., Emori Y., Abe K. (2002). Plant seed cystatins and their target enzymes of endogenous and exogenous origin. J. Agric. Food Chem. 50 6612–6617. 10.1021/jf0201935 [DOI] [PubMed] [Google Scholar]
- Barany I., Berenguer E., Solis M. T., Perez-Perez Y., Santamaria M. E., Crespo J. L., et al. (2018). Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 69 1387–1402. 10.1093/jxb/erx455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Timmers T., Jauneau A., Briere C., De Wit P. J., Marco Y., et al. (2008). RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20 2252–2264. 10.1105/tpc.108.058685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene G., Foyer C. H., Kunert K. J. (2006). Two new cysteine proteinases with specific expression patterns in mature and senescent tobacco (Nicotiana tabacum L.) leaves. J. Exp. Bot. 57 1431–1443. 10.1093/jxb/erj123 [DOI] [PubMed] [Google Scholar]
- Bhalerao R., Keskitalo J., Sterky F., Erlandsson R., Bjorkbacka H., Birve S. J., et al. (2003). Gene expression in autumn leaves. Plant Physiol. 131 430–442. 10.1104/pp.012732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. M., Yu J., Ge Y., Mironov A., Gallois P. (2018). Two proteases with caspase-3-like activity, cathepsin B and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis. New Phytol. 218 1143–1155. 10.1111/nph.14676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambra I., Martinez M., Dader B., Gonzalez-Melendi P., Gandullo J., Santamaria M. E., et al. (2012). A cathepsin F-like peptidase involved in barley grain protein mobilization, HvPap-1, is modulated by its own propeptide and by cystatins. J. Exp. Bot. 63 4615–4629. 10.1093/jxb/ers137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. M., Luthe D. S., Davis F. M., Williams W. P. (2000). Influence of whorl region from resistant and susceptible corn genotypes on fall armyworm (Lepidoptera: Noctuidae) growth and development. J. Econ. Entomol. 93 477–483. 10.1603/0022-0493-93.2.477 [DOI] [PubMed] [Google Scholar]
- Chen G. H., Huang L. T., Yap M. N., Lee R. H., Huang Y. J., Cheng M. C., et al. (2002). Molecular characterization of a senescence-associated gene encoding cysteine proteinase and its gene expression during leaf senescence in sweet potato. Plant Cell Physiol. 43 984–991. 10.1093/pcp/pcf125 [DOI] [PubMed] [Google Scholar]
- Chen H. J., Su C. T., Lin C. H., Huang G. J., Lin Y. H. (2010). Expression of sweet potato cysteine protease SPCP2 altered developmental characteristics and stress responses in transgenic Arabidopsis plants. J. Plant Physiol. 167 838–847. 10.1016/j.jplph.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Chen H. J., Tsai Y. J., Shen C. Y., Tsai T. N., Huang G. J., Lin Y. H. (2013). Ectopic expression of sweet potato cysteine protease SPCP3 alters phenotypic traits and enhances drought stress sensitivity in transgenic Arabidopsis plants. J. Plant Growth Regul. 32 108–121. 10.1007/s00344-012-9281-9 [DOI] [Google Scholar]
- Coulombe R., Grochulski P., Sivaraman J., Menard R., Mort J. S., Cygler M. (1996). Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 15 5492–5503. 10.1002/j.1460-2075.1996.tb00934.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F., Niedermaier S., Villamor J. G., Huesgen P. F. (2018). Quantitative proteomics in plant protease substrate identification. New Phytol. 218 936–943. 10.1111/nph.14587 [DOI] [PubMed] [Google Scholar]
- Diaz-Mendoza M., Dominguez-Figueroa J. D., Velasco-Arroyo B., Cambra I., Gonzalez-Melendi P., Lopez-Gonzalvez A., et al. (2016). HvPap-1 C1A protease and HvCPI-2 cystatin contribute to barley grain filling and germination. Plant Physiol. 170 2511–2524. 10.1104/pp.15.01944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Mendoza M., Velasco-Arroyo B., Gonzalez-Melendi P., Martinez M., Diaz I. (2014). C1A cysteine protease-cystatin interactions in leaf senescence. J. Exp. Bot. 65 3825–3833. 10.1093/jxb/eru043 [DOI] [PubMed] [Google Scholar]
- Dixon M. S., Golstein C., Thomas C. M., Van Der Biezen E. A., Jones J. D. (2000). Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. U.S.A. 97 8807–8814. 10.1073/pnas.97.16.8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C., Wolf D. H. (1993). BLH1 codes for a yeast thiol aminopeptidase, the equivalent of mammalian bleomycin hydrolase. J. Biol. Chem. 268 7036–7043. [PubMed] [Google Scholar]
- Fischer J., Becker C., Hillmer S., Horstmann C., Neubohn B., Schlereth A., et al. (2000). The families of papain- and legumain-like cysteine proteinases from embryonic axes and cotyledons of Vicia seeds: developmental patterns, intracellular localization and functions in globulin proteolysis. Plant Mol. Biol. 43 83–101. 10.1023/A:1006456615373 [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R. M. (1996). Cytokinins in plant senescence: from spray and pray to clone and play. Bioessays 18 557–565. 10.1002/bies.950180707 [DOI] [Google Scholar]
- Ge Y., Cai Y. M., Bonneau L., Rotari V., Danon A., Mckenzie E. A., et al. (2016). Inhibition of cathepsin B by caspase-3 inhibitors blocks programmed cell death in Arabidopsis. Cell Death. Differ. 23 1493–1501. 10.1038/cdd.2016.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S., Sabehi G., Carp M. J., Hajouj T., Nesher M. F., Yariv I., et al. (2003). Large-scale identification of leaf senescence-associated genes. Plant J. 36 629–642. 10.1046/j.1365-313X.2003.01908.x [DOI] [PubMed] [Google Scholar]
- Gombert J., Etienne P., Ourry A., Le Dily F. (2006). The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J. Exp. Bot. 57 1949–1956. 10.1093/jxb/erj142 [DOI] [PubMed] [Google Scholar]
- Grudkowska M., Zagdanska B. (2004). Multifunctional role of plant cysteine proteinases. Acta Biochim. Pol. 51 609–624. [PubMed] [Google Scholar]
- Guo Y., Cai Z. S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27 521–549. 10.1111/j.1365-3040.2003.01158.x [DOI] [Google Scholar]
- Guo Y., Gan S. S. (2014). Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 65 3901–3913. 10.1093/jxb/eru248 [DOI] [PubMed] [Google Scholar]
- Ilyas M., Horger A. C., Bozkurt T. O., Van Den Burg H. A., Kaschani F., Kaiser M., et al. (2015). Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 25 2300–2306. 10.1016/j.cub.2015.07.030 [DOI] [PubMed] [Google Scholar]
- James M., Poret M., Masclaux-Daubresse C., Marmagne A., Coquet L., Jouenne T., et al. (2018). SAG12, a major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol. 59 2052–2063. 10.1093/pcp/pcy125 [DOI] [PubMed] [Google Scholar]
- Kantyka T., Shaw L. N., Potempa J. (2011). Papain-like proteases of Staphylococcus aureus. Adv. Exp. Med. Biol. 712 1–14. 10.1007/978-1-4419-8414-2_1 [DOI] [PubMed] [Google Scholar]
- Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., et al. (2010). An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154 1794–1804. 10.1104/pp.110.158030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe T., Ariizumi T., Kawai-Yamada M., Uchimiya H., Toriyama K. (2006). Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 47 784–787. 10.1093/pcp/pcj039 [DOI] [PubMed] [Google Scholar]
- Kempema L. A., Cui X., Holzer F. M., Walling L. L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs, similarities and distinctions in responses to aphids. Plant Physiol. 143 849–865. 10.1104/pp.106.090662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler S. M., Ho T. H. (1990). Hormonal regulation, processing, and secretion of cysteine proteinases in barley aleurone layers. Plant Cell 2 769–783. 10.1105/tpc.2.8.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Yamaguchi-Shinozaki K., Tsuji H., Shinozaki K. (1993). Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129 175–182. 10.1016/0378-1119(93)90266-6 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Jaattela M. (2005). Lysosomes and autophagy in cell death control. Nat. Rev. Cancer 5 886–897. 10.1038/nrc1738 [DOI] [PubMed] [Google Scholar]
- Kruger J., Thomas C. M., Golstein C., Dixon M. S., Smoker M., Tang S., et al. (2002). A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744–747. 10.1126/science.1069288 [DOI] [PubMed] [Google Scholar]
- Ku S., Yoon H., Suh H. S., Chung Y. Y. (2003). Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217 559–565. 10.1007/s00425-003-1030-7 [DOI] [PubMed] [Google Scholar]
- Kunert K. J., Van Wyk S. G., Cullis C. A., Vorster B. J., Foyer C. H. (2015). Potential use of phytocystatins in crop improvement, with a particular focus on legumes. J. Exp. Bot. 66 3559–3570. 10.1093/jxb/erv211 [DOI] [PubMed] [Google Scholar]
- Li D. D., Xue J. S., Zhu J., Yang Z. N. (2017). Gene regulatory network for tapetum development in Arabidopsis thaliana. Front. Plant. Sci. 8:1559. 10.3389/fpls.2017.01559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhou Y., Szczerba M. W., Li X., Lin Y. (2010). Identification and application of a rice senescence-associated promoter. Plant Physiol. 153 1239–1249. 10.1104/pp.110.157123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman K. N., Gan S., John M. C., Amasino R. M. (1994). Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 92 322–328. 10.1111/j.1399-3054.1994.tb05343.x [DOI] [Google Scholar]
- Lord C. E., Gunawardena A. H. (2012). Programmed cell death in C. elegans, mammals and plants. Eur. J. Cell Biol. 91 603–613. 10.1016/j.ejcb.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Louis J., Basu S., Varsani S., Castano-Duque L., Jiang V., Williams W. P., et al. (2015). Ethylene contributes to maize insect resistance1-mediated maize defense against the phloem sap-sucking corn leaf aphid. Plant Physiol. 169 313–324. 10.1104/pp.15.00958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Torres J. L., Wilbers R. H., Gawronski P., Boshoven J. C., Finkers-Tomczak A., Cordewener J. H., et al. (2012). Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A. 109 10119–10124. 10.1073/pnas.1202867109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S. M., Kanneganti T. D. (2016). Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy 12 2504–2505. 10.1080/15548627.2016.1239679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Diaz I. (2008). The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol. Biol. 8:198. 10.1186/1471-2148-8-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkonen A., Porali I., Cercos M., Ho T. H. (1996). A major cysteine proteinase, EPB, in germinating barley seeds: structure of two intronless genes and regulation of expression. Plant Mol. Biol. 31 239–254. 10.1007/BF00021787 [DOI] [PubMed] [Google Scholar]
- Misas-Villamil J. C., Van Der Hoorn R. A., Doehlemann G. (2016). Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 212 902–907. 10.1111/nph.14117 [DOI] [PubMed] [Google Scholar]
- Noh Y. S., Amasino R. M. (1999). Regulation of developmental senescence is conserved between Arabidopsis and Brassica napus. Plant Mol. Biol. 41 195–206. 10.1023/A:1006389803990 [DOI] [PubMed] [Google Scholar]
- Novinec M., Lenarcic B. (2013). Papain-like peptidases: structure, function, and evolution. Biomol. Concepts 4 287–308. 10.1515/bmc-2012-0054 [DOI] [PubMed] [Google Scholar]
- Pechan T., Cohen A., Williams W. P., Luthe D. S. (2002). Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc. Natl. Acad. Sci. U.S.A. 99 13319–13323. 10.1073/pnas.202224899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechan T., Ye L., Chang Y., Mitra A., Lin L., Davis F. M., et al. (2000). A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12 1031–1040. 10.1105/tpc.12.7.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett A. R., Thomas S. G., Wilson Z. A., Hedden P. (2011). Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 16 568–578. 10.1016/j.tplants.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Poret M., Chandrasekar B., Van Der Hoorn R. A. L., Avice J. C. (2016). Characterization of senescence-associated protease activities involved in the efficient protein remobilization during leaf senescence of winter oilseed rape. Plant Sci. 246 139–153. 10.1016/j.plantsci.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Poulle M., Jones B. L. (1988). A Proteinase from germinating barley: I. Purification and some physical properties of a 30 kD cysteine endoproteinase from green malt. Plant Physiol. 88 1454–1460. 10.1104/pp.88.4.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabucka B., Drzymała A., Grabowska A. (2013). Molecular cloning and expression analysis of the main gliadin-degrading cysteine endopeptidase EP8 from triticale. J. Cereal Sci. 58 284–289. 10.1016/j.jcs.2013.06.004 [DOI] [Google Scholar]
- Prabucka B., Mielecki M., Chojnacka M., Bielawski W., Czarnocki-Cieciura M., Orzechowski S. (2017). Structural and functional characterization of the triticale (x Triticosecale Wittm.) phytocystatin TrcC-8 and its dimerization-dependent inhibitory activity. Phytochemistry 142 1–10. 10.1016/j.phytochem.2017.06.008 [DOI] [PubMed] [Google Scholar]
- Pruzinska A., Shindo T., Niessen S., Kaschani F., Toth R., Millar A. H., et al. (2017). Major Cys protease activities are not essential for senescence in individually darkened Arabidopsis leaves. BMC Plant Biol. 17:4. 10.1186/s12870-016-0955-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani M. A., Maruyama K., Abe H., Khan M. A., Katsura K., Ito Y., et al. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133 1755–1767. 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J., Bateman A. (2010). MEROPS: the peptidase database. Nucleic Acids Res. 38 D227–D233. 10.1093/nar/gkp971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J., Finn R. (2016). Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44 D343–D350. 10.1093/nar/gkv1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Pearl L. H., Buttle D. J. (1992). The baculovirus Autographa californica nuclear polyhedrosis virus genome includes a papain-like sequence. Biol. Chem. Hoppe Seyler 373 1211–1215. 10.1515/bchm3.1992.373.2.1211 [DOI] [PubMed] [Google Scholar]
- Richau K. H., Kaschani F., Verdoes M., Pansuriya T. C., Niessen S., Stuber K., et al. (2012). Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 158 1583–1599. 10.1104/pp.112.194001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I. N., Caputo C., Criado M. V., Funk C. (2012). Senescence-associated proteases in plants. Physiol. Plant. 145 130–139. 10.1111/j.1399-3054.2012.01574.x [DOI] [PubMed] [Google Scholar]
- Schlereth A., Becker C., Horstmann C., Tiedemann J., Muntz K. (2000). Comparison of globulin mobilization and cysteine proteinases in embryonic axes and cotyledons during germination and seedling growth of vetch (Vicia sativa L.). J. Exp. Bot. 51 1423–1433. [PubMed] [Google Scholar]
- Schlereth A., Standhardt D., Mock H. P., Muntz K. (2001). Stored cysteine proteinases start globulin mobilization in protein bodies of embryonic axes and cotyledons during vetch (Vicia sativa L.) seed germination. Planta 212 718–727. 10.1007/s004250000436 [DOI] [PubMed] [Google Scholar]
- Shindo T., Misas-Villamil J. C., Horger A. C., Song J., Van Der Hoorn R. A. (2012). A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS One 7:e29317. 10.1371/journal.pone.0029317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminska J., Orzechowski S., Bielawski W. (2015). Analysis of expression and inhibitory activity of a TrcC-6 phytocystatin present in developing and germinating seeds of triticale (×Triticosecale Wittm.). Plant Physiol. Biochem. 96 209–216. 10.1016/j.plaphy.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Song J., Win J., Tian M., Schornack S., Kaschani F., Ilyas M., et al. (2009). Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. U.S.A. 106 1654–1659. 10.1073/pnas.0809201106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Zhou Z., Tang S., Zhang Z., Xia S., Qin M., et al. (2016). Ectopic expression of BnaC.CP20.1 results in premature tapetal programmed cell death in Arabidopsis. Plant Cell Physiol. 57 1972–1984. 10.1093/pcp/pcw119 [DOI] [PubMed] [Google Scholar]
- Staal J., Dixelius C. (2007). Tracing the ancient origins of plant innate immunity. Trends Plant Sci. 12 334–342. 10.1016/j.tplants.2007.06.014 [DOI] [PubMed] [Google Scholar]
- Subburaj S., Zhu D., Li X., Hu Y., Yan Y. (2017). Molecular characterization and expression profiling of Brachypodium distachyon L. Cystatin genes reveal high evolutionary conservation and functional divergence in response to abiotic stress. Front. Plant Sci. 8:743. 10.3389/fpls.2017.00743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewinska J., Siminska J., Bielawski W. (2016). The roles of cysteine proteases and phytocystatins in development and germination of cereal seeds. J. Plant Physiol. 207 10–21. 10.1016/j.jplph.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Tan Y., Li M., Yang Y., Sun X., Wang N., Liang B., et al. (2017a). Overexpression of MpCYS4, A Phytocystatin Gene from Malus prunifolia (Willd.) Borkh, enhances stomatal closure to confer drought tolerance in transgenic Arabidopsis and apple. Front. Plant Sci. 8:33. 10.3389/fpls.2017.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Yang Y., Li C., Liang B., Li M., Ma F. (2017b). Overexpression of MpCYS4, a phytocystatin gene from Malus prunifolia (Willd.) Borkh., delays natural and stress-induced leaf senescence in apple. Plant Physiol. Biochem. 115 219–228. 10.1016/j.plaphy.2017.03.025 [DOI] [PubMed] [Google Scholar]
- Tian M., Win J., Song J., Van Der Hoorn R., Van Der Knaap E., Kamoun S. (2007). A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143 364–377. 10.1104/pp.106.090050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R. A. (2008). Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59 191–223. 10.1146/annurev.arplant.59.032607.092835 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R. A. L., Rivas S. (2018). Unravelling the mode of action of plant proteases. New Phytol. 218 879–881. 10.1111/nph.15156 [DOI] [PubMed] [Google Scholar]
- van der Linde K., Hemetsberger C., Kastner C., Kaschani F., Van Der Hoorn R. A., Kumlehn J., et al. (2012). A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24 1285–1300. 10.1105/tpc.111.093732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Arroyo B., Diaz-Mendoza M., Gandullo J., Gonzalez-Melendi P., Santamaria M. E., Dominguez-Figueroa J. D., et al. (2016). HvPap-1 C1A protease actively participates in barley proteolysis mediated by abiotic stresses. J. Exp. Bot. 67 4297–4310. 10.1093/jxb/erw212 [DOI] [PubMed] [Google Scholar]
- Velasco-Arroyo B., Diaz-Mendoza M., Gomez-Sanchez A., Moreno-Garcia B., Santamaria M. E., Torija-Bonilla M., et al. (2018). Silencing barley cystatins HvCPI-2 and HvCPI-4 specifically modifies leaf responses to drought stress. Plant Cell Environ. 41 1776–1790. 10.1111/pce.13178 [DOI] [PubMed] [Google Scholar]
- Xiao H. J., Yin Y. X., Chai W. G., Gong Z. H. (2014). Silencing of the CaCP gene delays salt- and osmotic-induced leaf senescence in Capsicum annuum L. Int. J. Mol. Sci. 15 8316–8334. 10.3390/ijms15058316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Matsushima R., Nishimura M., Hara-Nishimura I. (2001). A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 127 1626–1634. 10.1104/pp.010551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Q. W., Wang C. X., Li X. Y., Guo Z. A., Jing R. L., Zhao J., et al. (2010). Isolation and characterization of a gene encoding a polyethylene glycol-induced cysteine protease in common wheat. J. Biosci. 35 379–388. 10.1007/s12038-010-0043-1 [DOI] [PubMed] [Google Scholar]
- Zhang B., Tremousaygue D., Denance N., Van Esse H. P., Horger A. C., Dabos P., et al. (2014). PIRIN2 stabilizes cysteine protease XCP2 and increases susceptibility to the vascular pathogen Ralstonia solanacearum in Arabidopsis. Plant J. 79 1009–1019. 10.1111/tpj.12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Liu D., Lv X., Wang Y., Xun Z., Liu Z., et al. (2014). The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26 2939–2961. 10.1105/tpc.114.127282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. Y., Jones B. L. (1995). Characterization of germinated barley endoproteolytic enzymes by two-dimensional gel electrophoresis. J. Cereal Sci. 21 4499–4503. 10.1016/0733-5210(95)90030-6 [DOI] [Google Scholar]
- Zhao C., Johnson B. J., Kositsup B., Beers E. P. (2000). Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 123 1185–1196. 10.1104/pp.123.3.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhou X. M., Zhang L. Y., Wang W., Ma L. G., Yang L. B., et al. (2013). A bipartite molecular module controls cell death activation in the Basal cell lineage of plant embryos. PLoS Biol. 11:e1001655. 10.1371/journal.pbio.1001655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann S., Van Der Linde K., Lahrmann U., Acar B., Kaschani F., Colby T., et al. (2018). An apoplastic peptide activates salicylic acid signalling in maize. Nat Plants 4 172–180. 10.1038/s41477-018-0116-y [DOI] [PubMed] [Google Scholar]
- Zou Z., Liu J., Yang L., Xie G. (2017a). Survey of the rubber tree genome reveals a high number of cysteine protease-encoding genes homologous to Arabidopsis SAG12. PLoS One 12:e0171725. 10.1371/journal.pone.0171725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Xie G., Yang L. (2017b). Papain-like cysteine protease encoding genes in rubber (Hevea brasiliensis): comparative genomics, phylogenetic, and transcriptional profiling analysis. Planta 246 999–1018. 10.1007/s00425-017-2739-z [DOI] [PubMed] [Google Scholar]