A fast and efficient whole-genome sequencing method identifies insertional mutations and reveals complex DNA rearrangements accompanied insertional mutagenesis in Chlamydomonas reinhardtii.
Abstract
Insertional mutagenesis, in which a piece of exogenous DNA is integrated randomly into the genomic DNA of the recipient cell, is a useful method to generate new mutants with phenotypes of interest. The unicellular green alga Chlamydomonas reinhardtii is an outstanding model for studying many biological processes. We developed a new computational algorithm, MAPINS (mapping insertions), to efficiently identify insertion sites created by the integration of an APHVIII (aminoglycoside 3′-phosphotransferase VIII) cassette that confers paromomycin resistance. Using whole-genome sequencing data, this method eliminates the need for genomic DNA manipulation and retains all the sequencing information provided by paired-end sequencing. We experimentally verified 38 insertion sites out of 41 sites (93%) identified by MAPINS from 20 paromomycin-resistant strains. Using meiotic analysis of 18 of these strains, we identified insertion sites that completely cosegregate with paromomycin resistance. In six of the seven strains with a mutant phenotype, we demonstrated complete cosegregation of the mutant phenotype and the verified insertion site. In addition, we provide direct evidence of complex rearrangements of genomic DNA in five strains, three of which involve the APHVIII insertion site. We suggest that strains obtained by insertional mutagenesis are more complicated than expected from previous analyses in Chlamydomonas. To map the locations of some complex insertions, we designed 49 molecular markers based on differences identified via whole-genome sequencing between wild-type strains CC-124 and CC-125. Overall, MAPINS provides a low-cost, efficient method to characterize insertional mutants in Chlamydomonas.
Unbiased forward genetics screens provide a powerful tool to generate mutants with phenotypes of interest to study biological mechanisms (Krysan et al., 1999; Alonso et al., 2003). Next-generation sequencing is a rapid and efficient method to identify causative mutations from forward genetics screens. Direct sequencing of the whole genome (or whole exome), followed by comparison of the sequenced reads with the reference or parental genome sequence, allows the rapid identification of single-nucleotide variants and some copy number/structure variants (Schneeberger et al., 2009; Nordström et al., 2013). The identification of causative events in insertional mutant strains, on the other hand, usually requires enrichment of the targeted insert regions via adaptor ligation and PCR before whole-genome sequencing (WGS). Even through insertion sites have been identified, the efficiency is often reduced due to failure in adaptor ligation or PCR (Lepage et al., 2013; Hsia et al., 2017).
The unicellular biciliate green alga Chlamydomonas reinhardtii is a premier model in which to study biological processes that include photosynthesis, lipid metabolism, cilia assembly and motility, and cell-cell recognition (Jinkerson and Jonikas, 2015). Single-nucleotide polymorphisms (SNPs) or small insertions/deletions generated by radiation/chemical means can be identified directly by WGS (Dutcher et al., 2012; Lin et al., 2013; Tulin and Cross, 2014). However, methods to determine gene disruption/deletion caused by insertional mutagenesis are mostly PCR based and can be technically challenging and labor intensive (Dent et al., 2005; González-Ballester et al., 2005; Meslet-Cladière and Vallon, 2012; Cheng et al., 2017). Recently, several large insertional mutant collections were constructed (Zhang et al., 2014; Li et al., 2016; Cheng et al., 2017). However, smaller forward genetic screens and suppressor/enhancer screens remain attractive to collect multiple insertional mutants with a desired phenotype. Therefore, we developed a low-cost and efficient approach to identify causative insertions obtained from insertional mutagenesis.
In this study, we introduced an APHVIII (aminoglycoside 3′-phosphotransferase VIII) cassette that confers paromomycin resistance (paroR) into Chlamydomonas and pooled 20 insertional strains for WGS. We developed a new computational method named MAPINS (mapping insertions) to identify 41 insertions caused by the APHVIII cassette with a PCR validation rate of 93% in the 20 strains. Based on information provided by paired-end sequencing reads, we observed complex DNA rearrangements in five strains that may interfere with data interpretation in the absence of this type of analysis. To map the exact locations of some complex insertions, we developed a set of 49 new molecular markers for proximity mapping.
RESULTS
Isolation of Chlamydomonas Insertional Mutants
To obtain insertional mutants in Chlamydomonas, we introduced an ∼1.8-kb exogenous DNA fragment into mitotically grown cells by electroporation. This DNA fragment (cassette) includes the APHVIII gene under the regulation of a constitutive Chlamydomonas HSP70/RBCS2 promoter (Sizova et al., 2001). Successful integration of this DNA fragment into the Chlamydomonas genome confers paroR. In total, 864 paroR transformants were collected. We selected six strains that display abnormal ciliary phenotypes, one that grows slowly, and 13 random strains that have no obvious phenotype other than paroR. To save the cost of library construction, we paired the 20 strains into 10 indexed genomic DNA libraries and subjected them to WGS.
Identification and Verification of Cassette Insertion Sites
For each pair of strains, we obtained 34 to 41 million pair-ended 101-bp reads, and ∼93% of the reads were mapped to the Chlamydomonas Phytozome genome version 5.5. The average coverage for each strain was ∼15× (Supplemental Table S1). The 101-bp reads that did not completely align to the Chlamydomonas genomic DNA sequence were collected (Fig. 1). These reads were mapped to the cassette sequence in two forms: they were either full length (101 bp) or truncated (truncated from the 3′ end of each read until the reads are 45 bp long). Some DNA reads with both genomic and cassette DNA, which carried 45 to 100 bp of the cassette sequence at the 5′ end, were completely aligned to the cassette sequence in their truncated forms but not in their full-length forms. In the full-length form of these reads, the junctions between the cassette DNA and the flanking genomic DNA defined the cassette insertion sites. Therefore, these reads were collected and aligned back to the Chlamydomonas genome. We identified coordinates in these alignments, and they represent the cassette insertion sites. Through this method, which we named MAPINS, we identified 41 insertions in the 20 strains, with read coverage varying from 1 to 21 (Table 1).
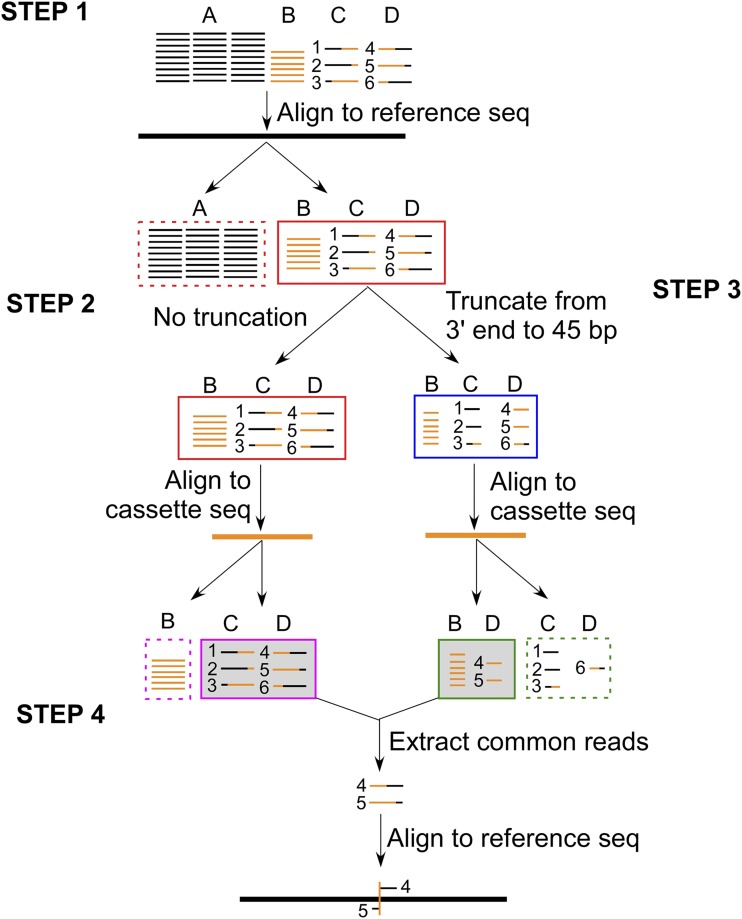
Figure 1.
MAPINS identifies flanking genomic DNA sequences around insertion sites. Paired-end 101-bp reads are composed of four different groups. They are reads completely aligned to the Chlamydomonas genome (A, black thin lines); reads completely aligned to the cassette sequence (B, orange thin lines); and reads that are chimeric between Chlamydomonas genomic sequences and the cassette sequence (C and D, mixed black-orange lines). All reads in C have the Chlamydomonas sequence at the 5′ end, and all reads in D have the cassette sequence at the 5′ end. Reads #1 to #6 represent different patterns of chimeric DNA from the 5′ to 3′ end. Step 1, All reads are first aligned to the Chlamydomonas reference genome (black thick line), and all completely aligned reads (group A) are discarded (red dashed box). Two parallel analyses are performed for the remaining reads (groups B, C, and D; red solid box) in steps 2 and 3. Step 2, They are aligned to the cassette sequence (orange thick lines), and all completely aligned reads (group B) are discarded (magenta dashed box). Reads in groups C and D are retained (magenta solid shaded box). Step 3, They are truncated from the 3′ end to 45 bp long in length. These 45-bp reads are aligned to the cassette DNA sequence. Reads that aligned completely (group B and some reads [#4 and #5] in group D) are retained (green solid shaded box). Reads that are not aligned completely are discarded (green dashed box). Step 4, Reads retained from steps 2 (magenta solid box) and 3 (green solid box) are compared. Common retained reads (#4 and #5 in group D) are extracted, and the full-length reads are realigned to the Chlamydomonas reference genome. Breakpoints in these reads define the cassette insertion site (orange vertical line) in the genome.
Table 1. Breakpoint identification and PCR verification.
| Pairs of Strains | Chromosome | Breakpoint Range | No. of Reads | Insertion Gap | Gene Identifier | Gene Name | Mutant in Strain 1 | Mutant in Strain 2 |
|---|---|---|---|---|---|---|---|---|
| bp | ||||||||
| 1A4-1D4 | 2 | Unknown–6,876,017 | 1 | Open ended | Cre02.g119651 | Cre02.g119651 | Yes | No |
| 3 | 4,539,389–4,539,406 | 6 | 18 | Cre03.g176651 | MYSM1 | No | Yes | |
| 17 | 4,322,754–4,322,816 | 6 | 63 | Cre17.g730950 | FLA10 | Yes | No | |
| 2F1-4C2 | 1 | 6,737,064–6,737,071 | 11 | 8 | Cre01.g048400 | DZIP1L | Yes | No |
| 3 | 7,296,662–7,296,698 | 11 | 37 | Cre03.g206950 | Cre03.g206950 | Yes | No | |
| 5 | 1,027,021–1,027,042 | 4 | 22 | Cre05.g246553 | Cre05.g246553 | Yes | No | |
| 17 | 3,105,419–3,105,428 | 11 | 10 | Cre17.g721250 | FAP22 | No | Yes | |
| 3F8-3D1 | 1 | 4,336,001–4,336,135 | 9 | 135 | Cre01.g029400 | TRPC3 | No | Yes |
| 2 | 7,226,958–7,227,798 | 9 | 841 | Cre02.g145950 | Cre02.g145950 | Yes | No | |
| 2 | 7,290,053–unknown | 2 | Open ended | Cre02.g145500 | PTK24 | Yes | No | |
| 4 | 1,568,203–1,568,528 | 8 | 326 | Cre04.g213400 | SUB7 | Yes | No | |
| 10 | 4,519,930–4,520,053 | 4 | 124 | No predicted gene | No predicted gene | Yes | No | |
| 12 | 7,660,573–unknown | 8 | Open ended | No predicted gene | No predicted gene | Yes | No | |
| 17 | Unknown–768,566 | 3 | Open ended | No predicted gene | No predicted gene | No | No | |
| 17 | 768,925–unknown | 7 | Open ended | No predicted gene | No predicted gene | No | No | |
| 5E3-5F11 | 6 | 5,109,304–5,109,339 | 6 | 36 | Cre06.g281050 | VPS5A | No | Yes |
| 9 | 2,129,462–2,129,485 | 3 | 24 | Cre09.g393551 | Cre09.g393551 | Yes | No | |
| 16 | 6,582,926–6,582,930 | 8 | 5 | Cre16.g672200 | BLD11 | Yes | No | |
| 8D6-9H4 | 14 | 1,860,469–1,860,478 | 14 | 10 | Cre14.g620500 | Cre14.g620500 | No | Yes |
| 16 | 6,583,673–6,583,708 | 12 | 36 | Cre16.g672200 | BLD11 | Yes | No | |
| 16 | 5,635,438–5,635,438 | 15 | 0 | Cre16.g679550 | FAP277 | No | Yes | |
| 8C12-9D5 | 2 | 1,736,814–unknown | 7 | Open ended | Cre02.g086300 | Cre02.g086300 | No | Yes |
| 6 | 5,398,524–5,398,526 | 13 | 3 | Cre06.g283900 | TMEM45B | Yes | No | |
| 9 | 6,286,926–6,286,995 | 9 | 70 | Cre09.g406500 | Cre09.g406500 | No | Yes | |
| 16 | Unknown–6,571,444 | 1 | Open ended | Cre16.g672250 | MPA13 | No | No | |
| 6C2-6A12 | 5 | 1,676,418–unknown | 2 | Open ended | Cre05.g242850 | Cre05.g242850 | No | Yes |
| 6 | Unknown–3,512,412 | 1 | Open ended | Cre06.g278107 | Cre06.g278107 | No | Yes | |
| 6 | 4,339,867–4,339,917 | 13 | 51 | Cre06.g278262 | Cre06.g278262 | Yes | No | |
| 12 | 762,534–unknown | 2 | Open ended | Cre12.g495500 | Cre12.g495500 | No | Yes | |
| 6F2-6B10 | 16 | 1,141,900–unknown | 2 | Open ended | Cre16.g650200 | MITC17 | Yes | No |
| 16 | 1,266,125–1,266,130 | 15 | 6 | Cre16.g651350 | ATG11 | No | Yes | |
| 7F3-9A9 | 6 | 2,254,537–2,254,612 | 19 | 76 | Cre06.g266450 | Cre06.g266450 | Yes | No |
| 6 | 1,938,823–1,938,849 | 10 | 27 | Cre06.g263650 | Cre06.g263650 | Yes | No | |
| 9 | Unknown–497,660 | 3 | Open ended | Cre09.g404201 | PHC10 | No | Yes | |
| 13 | 3,741,836–unknown | 3 | Open ended | Cre13.g589450 | Cre13.g589450 | No | Yes | |
| 17 | 3,468,906–3,468,911 | 9 | 6 | No predicted gene | No predicted gene | No | Yes | |
| 1D11-6D11 | 11 | 1,722,733–1,722,733 | 21 | 0 | Cre11.g467784 | SOUL | Yes | No |
| 16 | 1,549,854–1,549,855 | 11 | 2 | Cre16.g653450 | BAR1 | Yes | No | |
| 17 | Unknown–5,178,817 | 3 | Open ended | Cre17.g735550 | Cre17.g735550 | No | Yes | |
| 17 | 5,244,723–unknown | 1 | Open ended | Cre17.g736000 | Cre17.g736000 | No | Yes | |
| 17 | Unknown–5,430,365 | 4 | Open ended | Cre17.g736800 | Cre17.g736800 | Yes | No |
We designed PCR primers that amplify 100- to 600-bp fragments in the wild type (Supplemental Table S2). The same set of primers amplify bigger fragments in the insertion strains than in the wild type (Fig. 2, A and B) or do not amplify in the insertion strains due to the large insertion of the cassette (Fig. 2, C and D). Most insertions behaved as expected. In total, we experimentally verified 38 insertions (92.7%). On average, each insertional mutant generated by electroporation has 1.9 insertions, which is higher than the ∼1.4 insertions per mutant generated by the glass bead method (Dent et al., 2005).
Figure 2.
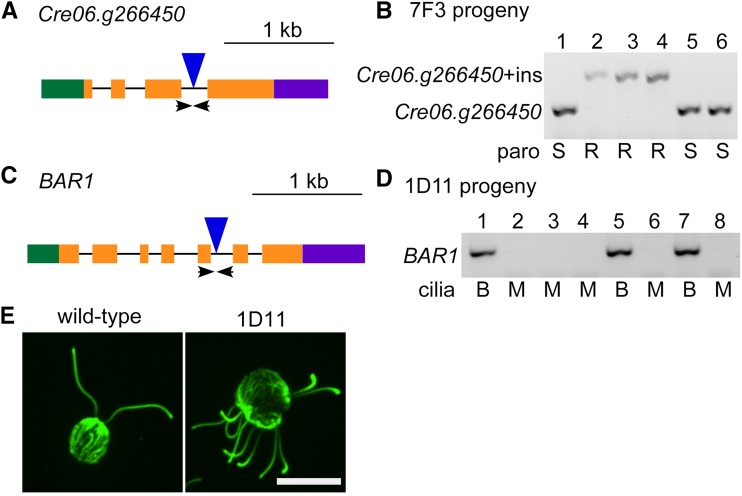
Cosegregation of PCR-validated insertion sites, paroR, and the mutant phenotype. A, Gene structure of the Cre06.g266450 gene. Green box, 5′ Untranslated region (UTR); orange boxes, exons; black lines, introns; purple box, 3′ UTR; blue triangle, insertion site of the cassette; arrows, positions of the primers used in B. The lengths of the blue triangle and arrows are not drawn to scale. B, Insertion of a truncated cassette allows PCR amplification in both mutant and wild-type cells. In six progeny from an octad between 7F3 and CC-125, the primers flanking the Cre06.g266450 insertion sites amplify two different PCR products. In this octad, resistance (R) to paromomycin cosegregates with the larger band (Cre06.g266450+ins [insert]) and sensitivity (S) to paromomycin cosegregates with the smaller band (Cre06.g266450). C, Gene structure of the BAR1 gene. Arrows indicate the positions of the primers used in D. The lengths of the blue triangle and arrows are not drawn to scale. D, The multiciliary phenotype in 1D11 always cosegregates with the insertion in BAR1, as shown in eight random progeny. The absence of a PCR product cosegregates with multiciliary cells (M) and the presence of a PCR product is always found in biciliary cells (B). E, The 1D11 mutant assembles multiple cilia. An antibody to α-tubulin (green) stains cilia protruding outside the round cell body. Bar = 10 µm.
Cosegregation of the Cassette Insertion, paroR, and the Mutant Phenotype
Our study indicates that nine out of 20 strains contain single-cassette insertions, six contain double-cassette insertions, and five strains contain three or more cassette insertions (Table 1). The fact that more than 50% of mutants contain more than one insert makes it important to test the cosegregation of cassette insertion, paroR, and the desired mutant phenotype through meiotic mapping to help with identifying the causative insertions. We backcrossed 18 of our insertional strains, which included the seven mutants that have trackable mutant phenotypes. PCR-based assays were used to analyze backcrossed progeny, and the cosegregation results are summarized in Table 2.
Table 2. Experimentally verified breakpoints by meiotic mapping.
n/a, Not available; ND, not determined.
| No. of Inserts | Strain | Phenotype | Chromosome | Gene Name | Location in Gene | Cosegregation with paroRa | Cosegregation with Phenotypea | Linkage with paroR |
|---|---|---|---|---|---|---|---|---|
| 1 | 1D4 | n/a | 3 | MYSM1 | Intron 9 | 5/5 | n/a | Yes |
| 1 | 4C2 | n/a | 17 | FAP22 | 3′ UTR | 8/8 | n/a | Yes |
| 1 | 5F11 | n/a | 6 | VPS5A | Exon 3 | 11/11 | n/a | Yes |
| 1 | 6C2 | n/a | 6 | Cre06.g278262 | Exon 4 | 9/9 | n/a | Yes |
| 1 | 6F2 | Slow growth | 16 | MITC17 | Intron 1 | 9/9 | 9/9 | Yes |
| 1 | 6B10 | n/a | 16 | ATG11 | 3′ UTR | 12/12 | n/a | Yes |
| 1 | 8C12 | n/a | 6 | TMEM45B | 5′ UTR | 16/16 | n/a | Yes |
| 1 | 8D6 | No cilia | 16 | BLD11 | Intron 14 + exon 15 | 24/24 | 24/24 | Yes |
| 2 | 1A4 | No cilia | 17 | FLA10 | Exon 18 | 11/11 | 11/11 | Yes |
| 2 | 5E3 | No cilia | 16 | BLD11 | Exon 13 | 3/3 | 3/3 | Yes |
| 2 | 6D11 | n/a | 17 | Cre17.g735550 | 5′ UTR | 10/10 | n/a | Yes |
| 17 | Cre17.g736000 | Exon 5 | 10/10 | n/a | Yes | |||
| 2 | 7F3 | n/a | 6 | Cre06.g266450 | Intron 3 | 11/14 | n/a | No |
| 6 | Cre06.g263650 | Exon 13 | 14/14 | n/a | Yes | |||
| 2 | 9D5 | n/a | 2 | Cre02.g086300 | 5′ UTR | 9/9 | n/a | Yes |
| 3 | 1D11 | Multicilia | 11 | SOUL | 3′ UTR | 3/3 | n/a | Yes |
| 16 | BAR1 | Intron 5 | 96/96b | 96/96b | Yes | |||
| 17 | Chr17-4.678M | n/a | 1/3 | n/a | No | |||
| 3 | 2F1 | Paralyzed cilia | 1 | DZIP1L | Exon 18 | 12/12 | 12/12 | Yes |
| 3 | 6A12 | n/a | 5 | Cre05.g242850 | 3′ UTR | 10/10 | n/a | Yes |
| 6 | Cre06.g278107 | 5′ UTR | 10/10 | n/a | Yes | |||
| 3 | 9A9 | n/a | 13 | Cre13.g589450 | Intron 1 | 1/6 | n/a | No |
| 17 | No predicted gene | n/a | 6/6 | n/a | Yes | |||
| 5 | 3F8 | No cilia | 2 | Cre02.g145950 | Exons 5 to 8 + introns 5 to 7 | 8/8 | 8/8 | Yes |
| 2 | PTK24 | 3′ UTR | 8/8 | 8/8 | Yes | |||
| 4 | SUB7 | Introns 1 and 2 + exon 2 | 3/8 | 3/8 | No | |||
| 12 | No predicted gene | n/a | 8/8 | 8/8 | Yes |
Number of complete tetrads.
Number of random progeny. n/a not available
In single-insertion strains, all eight strains tested showed complete cosegregation of paroR and insertion sites (Table 2). We also observed complete cosegregation of the mutant phenotype in the two strains with a trackable phenotype (6F2 and 8D6).
Two double-insertion strains, 1A4 and 5E3, lack cilia. In 1A4, one of the insertions falls into exon 18 of FLA10 (flagellar assembly10), which encodes one of the kinesin-2 motor subunits involved in intraflagellar transport (Vashishtha et al., 1996). We observed complete cosegregation of the mutant phenotype, paroR, and the FLA10 insertion in 11 tetrads of a 1A4 backcross. In 5E3, one insertion falls into exon 13 of Cre16.g672200 (gene identifier from Chlamydomonas Phytozome genome version 5.5). Given that 8D6 has a single insertion that deletes part of intron 14 and exon 15 in the same gene and has the same mutant phenotype, we proposed that the mutation in Cre16.g672200 is the causative mutation. Consistently, we observed complete cosegregation of the Cre16.g672200 insertion, paroR, and lack of cilia in 5E3 progeny.
Two double-insertion strains, 6D11 and 7F3, both contain two insertions on the same chromosome. 6D11 has no obvious mutant phenotype, and the two insertions are ∼66 kb apart on chromosome 17. As expected for a short distance, we found complete cosegregation of both insertions and paroR in 10 tetrads (Table 2). In 7F3, the insertions in Cre06.g266450 and Cre06.g263650 are ∼300 kb apart on chromosome 6. Cre06.g266450 contains a protein kinase domain and Cre06.g263650 has a conserved domain that is usually found in the C terminus of plant phosphoribosyltransferases. PCR results indicate that the insertion in Cre06.g266450 is ∼600 bp (Fig. 2, A and B), which is shorter than the coding region of APHVIII (819 bp). This insertion is unlikely to confer resistance and it does not show complete cosegregation in 14 tetrads (11 of 14) with paroR. The linked insertion in Cre06.g263650 cosegregates in all 14 tetrads with paroR (Table 2).
The 1D11 mutant assembles multiple cilia (Fig. 2E). MAPINS predicted three insertions, consistent with varying ratios of paroR and paromomycin-sensitive progeny in tetrads. In three tetrads that show 2:2 segregation, the insertions in chromosomes 11 and 16 cosegregate with paroR (Table 2). A third insertion on chromosome 17 falls into a region that has highly repetitive sequences and could not be analyzed by PCR directly. Instead, we used a polymorphism marker ∼750 kb away (chromosome 17; 4.678 Mb) and found that it is not tightly linked to paroR. In 96 random progeny of 1D11, the multiciliate phenotype always cosegregates with the insertion in the BAR1 (Bin/Amphiphysin/Rvs domain1; Cre16.g653450) gene on chromosome 16 (Fig. 2, C and D). Therefore, this insertion is likely to be the causative mutation of the mutant phenotype.
Validation of the Insertional Genes as the Causes of Mutant Phenotypes
In order to verify that we identified the correct causative insertions, rescue of the mutants with wild-type genes or identification of additional alleles that have the same mutant phenotype is needed (Lin et al., 2015; Cross et al., 2017). We verified the genes causing mutant ciliary phenotypes based on the availability of multiple alleles.
The lack-of-cilia mutant 1A4 carries an insertion in FLA10. A null fla10 mutant, generated by insertional mutagenesis that deletes at least part of the N terminus of the gene, results in no assembly of cilia (Matsuura et al., 2002). In addition, there are multiple temperature-sensitive alleles that result in a lack of cilia at the restrictive temperature (Vashishtha et al., 1996; Miller et al., 2005). Our identification of 1A4 provides an additional mutant allele of FLA10.
The 2F1 strain carries an insertion in exon 18 of DZIP1L (DAZ interacting zinc finger protein1 like; Cre01.g048400). Chlamydomonas DZIP1L is a zinc finger protein that shares 38% sequence identity (e−19) with the zebrafish (Danio rerio) Iguana protein and 32% identity (e−14) with human (Homo sapiens) DZIP1L. A nonsense mutation in the zebrafish iguana/dzip1 gene results in a reduced number of motile cilia. RNA interference of human dZIP1L in human telomerase reverse transcriptase-immortalized retinal pigment epithelial cell line 1 cells leads to defective ciliogenesis of primary cilia (Glazer et al., 2010). These results suggest that DZIP1L is involved in ciliary assembly or function. To verify that insertional mutations in the Chlamydomonas DZIP1L have ciliary defects, we obtained a second insertional mutant of DZIP1L (LMJ.RY0402.253233) from the CLiP library (Li et al., 2016). We verified that the strain has an insertion in exon 14 and shows a paralyzed cilia phenotype similar to the 2F1 strain. We obtained 12 tetrads from a backcross of the CLiP strain. All 12 tetrads showed cosegregation of the DZIP1L insertion and the paralyzed cilia phenotype. Therefore, it indicates that two different insertions in DZIP1L cause paralyzed cilia rather than a ciliary assembly defect in Chlamydomonas.
Both the 5E3 and 8D6 mutants contain insertions at different positions in Cre16.g672200, which we now name BLD11 (bald11), given that they both lack cilia. A sequence similarity search indicated that homologs of this Chlamydomonas protein are found only in the multicellular green algae Gonium pectorale and Volvox carteri. Our results from two independent insertional mutants suggest that it is involved in ciliogenesis.
Dissection of Complex Genomic DNA Rearrangements
Similar to previous studies (Zhang et al., 2014; Li et al., 2016), we identified only one side of the flanking genomic DNA sequences (open-ended insertions) in ∼40% (16 of 41) of the identified insertions (Table 1). Li et al. (2016) suggested that the insertion sometimes is accompanied by an insertion of short genomic DNA from elsewhere in the genome (junk DNA). Since we generated our reads by directly sequencing the whole genome without enzymatic digestion (Zhang et al., 2014) or single-stranded DNA extension (Li et al., 2016), we asked if we could observe complex genomic DNA rearrangements, either adjacent to insertions of the cassette or elsewhere randomly in the genome, in our mutants.
We took advantage of our paired-end sequencing by looking for reads that mapped to a different chromosome from their corresponding pair (uncoordinated reads; see “Materials and Methods”). Some of these reads may represent DNA rearrangements in the cells. We will refer to these reads as chimeric DNA. These reads were extracted from each pair of mutants and the recipient strain used for transformation (CC-124; Lin et al., 2013), and uncoordinated reads that are found in CC-124 were eliminated (Supplemental Table S3). After the elimination of CC-124 reads, the 3F8-3D1 pair contains 806 uncoordinated reads, which is significantly more than those found in other pairs (372–584). If we assume that the formation of chimeric DNA is random in each strain, then the common uncoordinated reads among different pairs of strains will be background noise. These shared chimeric reads were eliminated. The uncoordinated reads found in the 3F8-3D1 pair were eliminated from the other nine pairs of strains, and we eliminated uncoordinated reads found in 1A4-1D4 from the 3F8-3D1 pair. After this step, 21 to 77 reads remained in each pair of strains. We performed BLASTN on these reads to the Chlamydomonas genome. The majority of these reads contain repetitive sequences, aligned to multiple sites in the genome, and were eliminated from consideration.
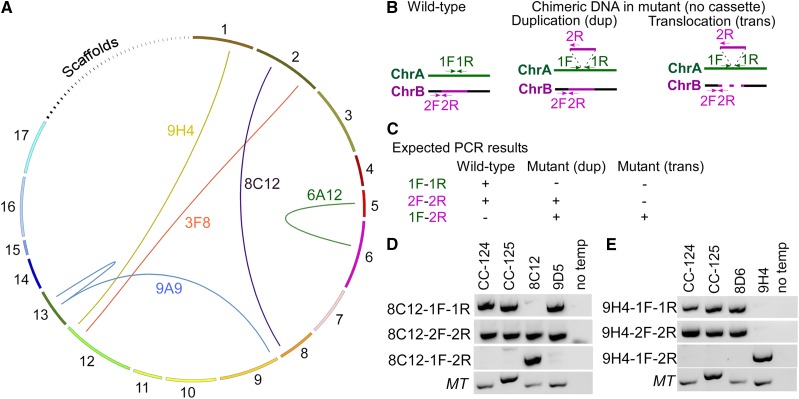
We identified five events of chimeric DNA in five pairs of strains, and the arcs in Figure 3A indicate the chromosomes involved (Supplemental Table S3). Two chimeric DNA events (in strains 8C12 and 9H4) are not associated with the insertion of the cassette, and the other three events (in strains 3F8, 6A12, and 9A9) are.
Figure 3.
Complex DNA rearrangements in multiple insertional strains. A, Diagram of chimeric DNA formed across different chromosomes in five strains (yellow, 9H4; red, 3F8; purple, 8C12; green, 6A12; blue, 9A9). The ends of the arcs indicate the junction sites. Chromosomes 1 to 17 and scaffolds 18 to 54 are drawn to scale in a clockwise direction. B, Positions of the primers in the wild type and in the mutant that form chimeric DNA due to DNA duplication or DNA translocation. Expectations for events on two different chromosomes (A, green; B, magenta) are shown. We assume that the insertion happens on chromosome A and the inserted DNA originates from chromosome B. C, Expected outcome of PCR products from the wild type and mutants with different combinations of primers. D, PCR of wild-type fragments (1F-1R on chromosome 2, 2F-2R on chromosome 8) and a chimeric DNA fragment (1F-2R) in the wild type (CC-124 and CC-125), 8C12, and 9D5. Primers for the mating-type loci (MT, including the MTA1 and MTD1 genes) are included as loading controls. No temp indicates that no DNA template was added to the PCR. E, PCR of wild-type fragments (1F-1R on chromosome 1, 2F-2R on chromosome 12) and a chimeric DNA fragment (1F-2R) in the wild type (CC-124 and CC-125), 8D6, and 9H4.
We first analyzed the events that have no association with the cassette. To simplify the analysis (Fig. 3B), we assume that a piece of genomic DNA from chromosome B (insert; magenta) integrates into chromosome A (green). We designed the 1F-1R primer set to span the insertion site on chromosome A and the 2F-2R primer set to span the junction between the insert and its flanking region on chromosome B. Once the insert integrates into chromosome A, the primer set 1F-2R amplifies the chimeric DNA in the mutant but not in wild-type cells (Fig. 3C). We expect that primer set 1F-1R does not amplify in the mutant due to the integration of the insert. If the insert is generated by DNA duplication and a copy is present at its original position, we expect the 2F-2R set to amplify in the mutant. If the insert translocates from its original position on chromosome B to chromosome A, the 2F-2R primer set should not amplify. We tested these scenarios in 8C12 and 9H4.
In the 8C12-9D5 pair of strains (Fig. 3D), chimeric DNA between chromosomes 2 and 8 is supported by four reads. The 2F-2R primer set on chromosome 8 amplified in all strains tested. This suggests that the formation of chimeric DNA is due to the duplication of genomic DNA that originated from chromosome 8. The chimeric DNA formed between chromosomes 1 and 12 found in the 8D6-9H4 pair is supported by 28 reads (Fig. 3E). Primer sets 1F-1R (on chromosome 1) and 2F-2R (on chromosome 12) amplified products in the wild type and 8D6 but not in 9H4. This suggests a translocation event.
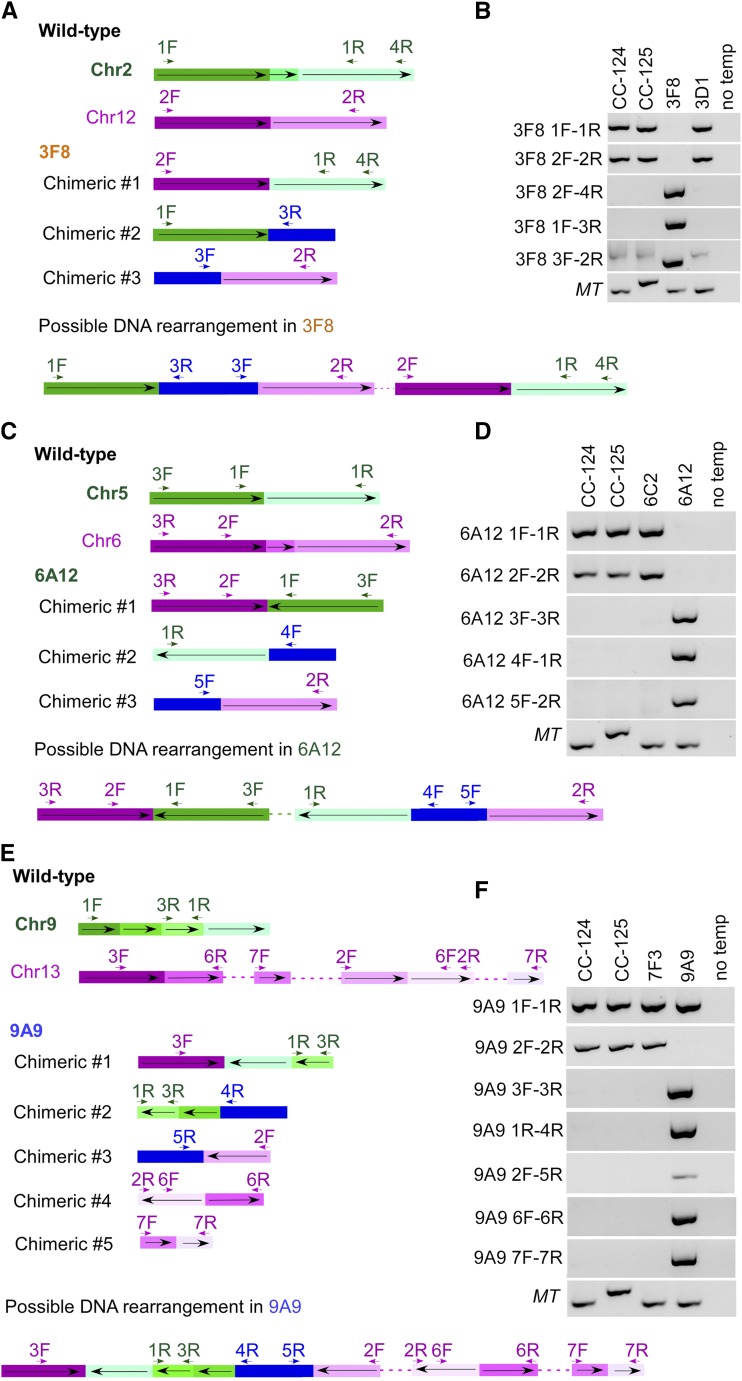
In the strains that show complex DNA rearrangement in association with the APHVIII cassette, the analyses are more complicated (Fig. 4). In the 3F8-3D1 pair (Fig. 4, A and B), chimeric DNA between chromosomes 2 (green) and 12 (magenta) is supported by 10 reads. Primers 1F-1R and 2F-2R amplified products in the wild type and 3D1 but failed in the 3F8 strain. Three different pieces of chimeric DNA were detected by sequencing reads and verified by PCR. The chimeric DNA #1 formed between chromosomes 2 and 12 was verified by primer set 2F-4R, the chimeric DNA #2 formed between part of chromosome 2 and the APHVIII cassette (blue) was verified by primer set 1F-3R, and the chimeric DNA #3 between part of chromosome 12 and the APHVIII cassette was verified by primer set 3F-2R. Based on the positions of each chimeric DNA and results from proximal mapping (see below), we attempted to assemble the DNA rearrangement event in 3F8. We suggest that fragments originating from chromosome 12 were translocated, rearranged, and inserted into chromosome 2, along with the APHVIII cassette(s). Even though the cassette is drawn as a single blue block, we cannot determine how many partial or full-length cassettes are involved in this block. Through a similar strategy, we verified the chromosome 5-6 chimeric DNA, the chromosome 5-cassette chimeric DNA, and the chromosome 6-cassette chimeric DNA in 6A12 (Fig. 4, C and D). Fragments originated from chromosome 5 were translocated, rearranged, and inserted to chromosome 6 along with the APHVIII cassette(s).
Figure 4.
Complex DNA rearrangement adjacent to the APHVIII cassette. A, Diagram of events in 3F8 involving chromosome 2 (green) and chromosome 12 (magenta) in the wild type, chimeric DNA between chromosomes 2 and 12 (#1), between chromosome 2 and the APHVIII cassette (blue; #2), and between chromosome 12 and the APHVIII cassette (#3). The positions of the primers used in B along the chromosomes and the cassette are indicated by short arrows. The orientation of the DNA fragments along the wild-type chromosome (from small to large coordinates) is indicated by long arrows. The diagram is not drawn to scale. A possible DNA rearrangement event in 3F8 is indicated at the bottom. The dashed line indicates the undetermined composition of DNA. B, PCR amplification of multiple DNA fragments in the wild type (CC-124 and CC-125), 3F8, and 3D1. No temp indicates that no DNA template was added to the PCR. C, Diagram of events in 6A12 involving chromosome 5 (green) and chromosome 6 (magenta) in the wild type, chimeric DNA between chromosomes 5 and 6 (#1), between chromosome 5 and the APHVIII cassette (#2), and between chromosome 6 and the APHVIII cassette (#3). The positions of the primers used in D along the chromosomes and the cassette are indicated by short arrows. The orientation of the DNA fragments along the wild-type chromosome (from small to large coordinates) is indicated by long arrows. The diagram is not drawn to scale. A possible DNA rearrangement event in 6A12 is indicated at the bottom. The dashed line indicates the undetermined composition of DNA. D, PCR amplification of multiple DNA fragments in the wild type (CC-124 and CC-125), 6C2, and 6A12. No temp indicates that no DNA template was added to the PCR. E, Diagram of events in 9A9 involving chromosome 9 (green) and chromosome 13 (magenta) in the wild type, chimeric DNA between chromosomes 9 and 13 (#1), between chromosome 9 and the APHVIII cassette (#2), between chromosome 13 and the APHVIII cassette (#3), and between regions of chromosome 13 (#4 and #5). The positions of the primers used in F along the chromosomes and the cassette are indicated by short arrows. The orientation of the DNA fragments along the wild-type chromosome (from small to large coordinates) is indicated by long arrows. The diagram is not drawn to scale. A possible DNA rearrangement event in 9A9 is indicated at the bottom. The dashed line indicates the undetermined composition of DNA. F, PCR amplification of multiple DNA fragments in the wild type (CC-124 and CC-125), 7F3, and 9A9. No temp indicates that no DNA template was added to the PCR.
In the 7F3-9A9 pair (Fig. 4, E and F), chromosome 9-chromosome 13-2.468M chimeric DNA (#1) was supported by six reads. The primer set 1F-1R amplified a wild-type chromosome 9 product in all strains tested. This suggests a duplication of this region in the mutant. When we manually analyzed the reads around 2.468M on chromosome 13, we identified and PCR verified a piece of chimeric DNA (#4) between 3.741760 to 3.741907 Mb and 2.468729 to 2.468877 Mb and a second piece (#5) between 2.469244 to 2.469281 Mb and 3.802295 to 3.802364 Mb. Additional pieces of chimeric DNA (#2 and #3) between the chromosomes and the APHVIII cassette also were verified by PCR. In summary, for 9A9, we observed duplication of part of chromosome 9 and translocation of part of chromosome 13, adjacent to the APHVIII cassette(s), and additional translocation events on chromosome 13 that are independent of the inserted cassette.
Deconvolution of Multiple Insertions and Proximity Mapping
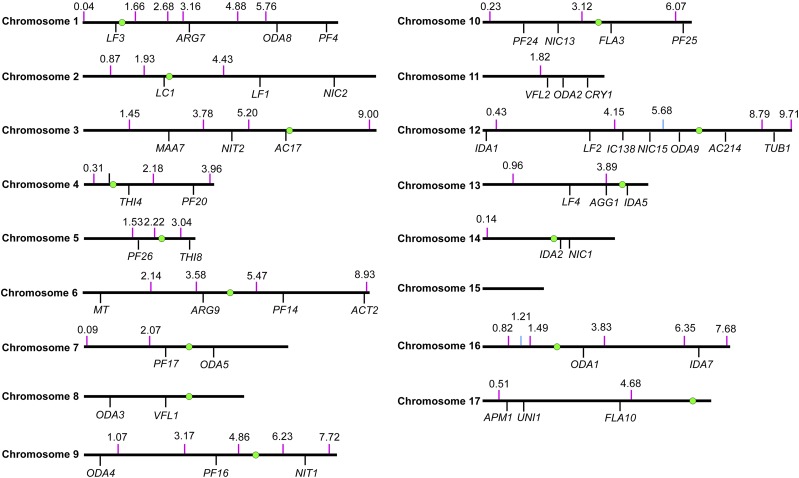
Although we detected chimeric DNA as described, we could not determine the chromosomal location of the chimera. To pinpoint the location, we relied on meiotic mapping. Previously identified molecular markers in Chlamydomonas use polymorphisms between 137c and S1D2 (Gross et al., 1988; Bowers et al., 2003; Kathir et al., 2003). To facilitate rapid mapping, we designed 49 pairs of primers across 15 of the 17 chromosomes (Fig. 5; Supplemental Table S4) based on polymorphisms between CC-124 and CC-125 identified via WGS (Lin et al., 2013). The distance between pairs of primers on each chromosome ranges from 483 kb to 4.165 Mb.
Figure 5.
Distribution of 49 new molecular markers for meiotic mapping. Newly designed molecular markers and their positions along different chromosomes are indicated by magenta vertical lines. Their positions along the chromosomes in version 5.5 are indicated above the line. One marker, which maps to both chromosome 12: 5.68 Mb and chromosome 16: 1.21 Mb, is indicated as light blue vertical lines. A subset of previously defined genes used in meiotic mapping is indicated by black vertical lines. Centromeres on each chromosome are indicated as green circles as mapped in Supplemental Table S5. We did not identify centromeres on chromosome 11 or 15.
In addition, we mapped the centromeres on 15 of the 17 chromosomes. The AC17 (acetate-requiring17), α-tubulin2, and paralyzed flagella27 loci are tightly linked to their respective centromeres (Huang et al., 1982; James et al., 1989; Kathir et al., 2003). Examination of the sequence in the region around these genes showed that there are multiple copies of a reverse transcriptase-like gene. Using this sequence to BLAST the genome, we found that 15 of 17 chromosomes have a repetitive stretch of this sequence and that it spans 200 to 800 kb. We reasoned that these regions are likely to surround or to be the centromeres in Chlamydomonas, based on their proximity to several genes that are known to be tightly linked to their centromeres. To test this idea, DNA from 25 tetrads from a cross of ac17 by CC-1952 (S1C5), a highly polymorphic strain, was isolated. Primers to genes within several hundred kilobases of the repeats were used to determine if these regions behaved as centromeres using PCR. We expect that a tightly centromere-linked probe will produce parental ditypes and nonparental ditypes and no tetratype progeny. Twenty primer sets produced easily discernible differences between the ac17 strain and CC-1952. These probes show little recombination between the probe and the centromere, as judged by behavior with respect to ac17 and the other probes. Zero to three tetratypes were found for the primers (Supplemental Tables S5 and S6). The centromeric regions are shown in Figure 5. Many of the scaffold assemblies that have not been placed on a chromosome have reverse transcription genes on them; several of them may reside on chromosomes 11 and 15 that lack a block of reverse transcriptase-like genes.
We used the polymorphism markers to identify the chromosomal locations of the chimera we identified in two insertional strains. In 6A12, the flanking DNA from chromosomes 5 and 6 cosegregate with paroR in 10 tetrads (Table 2). This is consistent with our PCR analysis result that pieces of chromosome 5, chromosome 6, and the cassette form chimeric DNA (Fig. 4C). To determine whether this chimeric DNA maps to chromosome 5 or 6, we used a polymorphism marker ∼545 kb away from the chromosome 5 sequence (2.22 Mb) and a second one ∼1.37 Mb away from the chromosome 6 insertion sequence (2.14 Mb). For parental ditype:nonparental ditype:tetratype ratios, we observed 7:1:1 for the chromosome 5 marker and 8:0:1 for the chromosome 6 marker. Therefore, part of chromosome 5 genomic DNA was inserted into chromosome 6.
In 3F8, both the aflagellate phenotype and paroR show complete cosegregation with each other and with insertions on chromosomes 2 and 12 (8 of 8). This is consistent with the chimeric DNA formation we observed (Fig. 4A). A polymorphism marker ∼1.13 Mb from the chimeric DNA on chromosome 12 (chromosome 12; 8.79 Mb) is unlinked (5 of 8). Therefore, the chimeric DNA inserted into chromosome 2. There is no predicted gene from the junk DNA that originated on chromosome 12. Since the two insertions on chromosome 2 are ∼60 kb apart, we cannot determine the causative mutation for the aflagellate phenotype with the number of tetrads we analyzed. Other approaches (plasmid rescue or isolation of a different mutant allele) would be required to identify the causative gene.
DISCUSSION
Highly Efficient Identification of Insertion Sites
Insertional mutagenesis has been used widely in Chlamydomonas. Both PCR-based and WGS-based approaches have been used to identify causative mutations. The roadblocks in PCR-based methods include tandem repeats of the insert DNA, poor annealing between degenerate primers and templates, and deletion or rearrangement of genomic DNA around the insertion sites (Dent et al., 2005; González-Ballester et al., 2005; Pollock et al., 2017). WGS-based approaches, ChlaMmeSeq and LEAP-Seq (Zhang et al., 2014; Li et al., 2016), are advantageous for identifying flanking sequences systematically in large-scale mutagenesis experiments. The ChlaMmeSeq method relies on enzymatic digestion to analyze 20- to 21-bp genomic DNA flanking the insertion sites. The low validation rate by this method (∼70%) is likely due to short flanking DNA sequences and not accounting for complex DNA rearrangement around the insertion sites. The LEAP-Seq method is an improvement that includes up to 1.5-kb flanking DNA around the insert cassette. These reads may help to identify complex DNA rearrangement. However, it is unclear why the paired-end reads were trimmed to 21 to 30 bp during LEAP-Seq analysis. The short sequence reads, along with multiple steps of initial DNA manipulation (single-stranded DNA extension and biotin-streptavidin capture), may contribute to the low validation rate (∼75%).
In this study, we present a fast and highly efficient method, MAPINS, to identify insertion sites with a PCR validation rate of 93%. A similar approach was used to map transposons in bacteria and worms (Smith, 2011). The significant difference in our method is that we retain all the information provided by 101-bp paired-end reads, which provides longer continuous flanking sequences and easily identifies chimeric DNA sequences. It does not require additional manipulation of genomic DNA, as does enzyme digestion or affinity purification. The same sequencing data can be used to analyze both SNP/short insertions/deletions and insertion sites, if needed (Lin and Dutcher, 2015). This method is highly sensitive and requires low coverage. In fact, we verified three out of four insertions that had only one read (Table 1).
Our method also is cost effective. Based on our study, 15× coverage of each strain is sufficient to detect insertion sites. Therefore, an Illumina HiSeq 2500 sequencing lane (pair-ended 101 bp), which provides ∼300 million reads, is sufficient to detect insertion sites in 18 mutants. To reduce the cost, we combined genomic DNA from two mutants before library preparation and WGS. With current sequencing technology and price, the cost to identify the causative mutation in an insertion strain is comparable to the cost to obtain a CLiP strain (Li et al., 2016) from the Chlamydomonas Source Center. Thus, this method makes it feasible and affordable to perform suppressor/enhancer screens as well as small-scale mutant phenotype screens and to identify causative insertional mutations.
The Dark Side of Insertional Mutagenesis
Both glass bead-mediated transformation and electroporation have been used widely in the Chlamydomonas community to generate insertional mutants. It has been reported that glass bead transformation usually leads to large deletions and/or DNA rearrangement (Dent et al., 2015). In our study as well as in other electroporation-based studies (Li et al., 2016; Pollock et al., 2017), deletions associated with insertions are relatively small. While the distribution of insertions is largely random, we and others (Li et al., 2016) did observe linked insertions (less than 100 kb apart) in ∼10% of the insertional strains. This may be due to changes in chromatin dynamics (Nyswaner et al., 2008) caused by the first insertion.
In this study, we provided direct evidence that five insertional strains (25%) have chimeric DNA. Three of these cases are adjacent to the insertion sites, while two are not. It is unclear how or when these chimeric DNAs formed. Previous studies proposed concatenation of insert cassettes and genomic DNA fragments due to cell lysis prior to electroporation (Zhang et al., 2014; Li et al., 2016). If this were the cause, we would expect duplication of the junk DNA and the junk DNA would always be adjacent to the inserted cassette. However, we observed both duplication and translocation events of the inserted genomic DNA in our strains. They do not necessarily associate with the inserted APHVIII cassette. Our study suggests that even electroporation leads to smaller deletions associated with the insertion of the cassette; DNA rearrangement is prevalent and sometimes complicated. Therefore, it is extremely important to perform additional verification of insertional mutants that go beyond PCR validation and phenotype cosegregation assays. This can be performed using methods such as plasmid rescue or the identification of a second mutant allele with a similar mutant phenotype.
Power of WGS
Using WGS, we and others have successfully identified causative mutations in over 50 UV/chemical-induced mutants (Lin et al., 2013; Tulin and Cross, 2014). We demonstrate here that WGS allows rapid and sensitive (93% successful rate) identification of insertional sites as well. In addition, WGS of CC-124 and CC-125 from our previous study (Lin et al., 2013) provided a new set of molecular markers (Fig. 5; Supplemental Table S4). This eliminates the need for an outcross to the highly polymorphic S1D2/S1C5 strains and facilitates faster meiotic mapping.
During the analysis of uncoordinated reads found in the insertion strains, we identified over 134,000 uncoordinated reads in the wild-type CC-124 strain (Supplemental Table S3), and they account for ∼0.26% of total reads (Lin et al., 2013). There are multiple possibilities for why we observed these uncoordinated reads. First, reads were mapped to more than one chromosome due to sequence similarity. Second, reads were mapped to repetitive sequences. Third, the formation of chimeric DNA during library preparation prior to WGS occurred. Fourth, errors and gaps occur in genome assembly, some of which were described elsewhere (Lin et al., 2013; Tulin and Cross, 2016). BLAST searches against the genome and experimental validation rule out the first three possibilities. In the last scenario, we can use the uncoordinated reads to correct some mistakes in the genome assembly.
MATERIALS AND METHODS
Chlamydomonas Strains, Media, and Growth Conditions
Chlamydomonas reinhardtii strains CC-124 and CC-125 were maintained on solid rich growth medium. For electroporation, cells were grown in Tris-acetate phosphate medium at 25°C under constant illumination. Transformants were selected on solid Tris-acetate phosphate medium supplied with 10 µg mL−1 paromomycin (Lin et al., 2013). Meiotic crosses and tetrad analysis were performed as described previously (Lin and Dutcher, 2015).
Manipulation of DNA and Electroporation
The pSI103 plasmid, which contains APHVIII (Sizova et al., 2001), was digested with BamHI, and an ∼1.8-kb fragment was gel purified. One microgram of DNA was used in electroporation. Electroporation of Chlamydomonas was performed using a NEPA21 square-pulse electroporator (Onishi and Pringle, 2016). Extraction of Chlamydomonas genomic DNA was performed as described previously (Lin and Dutcher, 2015). Library preparation, genomic DNA sequencing, demutiplexing of reads, and alignment to the Chlamydomonas reference genome were performed by the Genome Technology Access Core (Department of Genetics, Washington University).
Identification of Breakpoints by MAPINS
The workflow used in MAPINS is included in Supplemental File S1. Reads that were not mapped to the Chlamydomonas reference genome were extracted by SAMTools (Li et al., 2009; samtools view -u -f 4 -F 264 for unmapped read, samtools view -u -f 8 -F 260 for unmapped mate, and samtools view -u -f 12 -F 256 for unmapped read and mate) and aligned to the 1,853-bp insert DNA by Novoalign (Novocraft) with either full-length reads or reads clipped to 45 bp long. A customized Perl script (Supplemental File S2) was used to collect sequence identifiers of reads that aligned at 45 bp long but not at 101 bp long. A second Perl script (Supplemental File S3) was used to extract the paired-end reads with these sequence identifiers from original FASTQ files. These sequences were then aligned to the Chlamydomonas genome using Novoalign. The positions of these alignments were sorted by chromosome and analyzed for breakpoints.
Identification of Uncoordinated Reads
Reads that did not map to the reference genome coordinately were extracted by SAMTools (samtools view -F 14). This command extracts reads whose corresponding paired reads map either to a different chromosome or to positions on the same chromosome that span longer or shorter regions than expected. To simplify the study, we focus on paired-end reads spanning different chromosomes. The coordinates of the beginning of each read were collapsed into 100-bp windows before analysis. We required at least two reads in each 100-bp window to be considered for further analysis. Reads (or its corresponding pair) found in CC-124 were eliminated. The remaining reads were further filtered by comparing to the 3F8-3D1 pair and eliminating common reads. In the case of 3F8-3D1, comparison with 1A4-1D4 and elimination of common reads were performed. Reads that mapped to repetitive sequences identified by BLASTN were eliminated. We used Circos (Krzywinski et al., 2009) to draw the diagram of chimeric DNA formed across different chromosomes.
Design of Primers for Proximity Mapping
SNP differences between CC-124 and CC-125 were analyzed as described previously (Lin et al., 2013). The detection of breakpoints in sequencing reads was performed by SoftSearch (Hart et al., 2013). Primers were designed to amplify fragments across the breakpoints or SNPs.
Accession Numbers
All original WGS reads have been deposited into the National Center for Biotechnology Information Sequencing Read Archive with accession number SRP155877. The insertional mutants have been deposited at the Chlamydomonas Resource Center. All gene identifiers can be found in Chlamydomonas Phytozome genome version 5.5 (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=KEYWORD&method=Org_Creinhardtii).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Summary of WGS reads.
Supplemental Table S2. Primers used to verify insertions identified by MAPINS.
Supplemental Table S3. Identification of chimeric DNA in five strains.
Supplemental Table S4. The 49 new molecular markers used in proximity mapping.
Supplemental Table S5. Predicted centromere position and segregation in tetrads from crosses of ac17 × S1C5.
Supplemental Table S6. Primers for centromere mapping.
Supplemental File S1. Workflow used in MAPINS.
Supplemental File S2. Customized Perl script used to collect sequence identifiers of reads that aligned.
Supplemental File S3. Customized Perl script to extract paired-end reads from the original FASTQ files.
Acknowledgments
We thank Mihaela Stoyanova and John Garza for technical support and Dr. Mathieu Bottier, Gervette M. Penny, and Manishi Pandey for comments on the article. Special thanks to Dr. Gary Stormo for the name of MAPINS. We thank one of the reviewers who suggested that we study the frequency of chimeric reads that are not linked to the selectable marker.
Footnotes
This work was supported by grants from the Children’s Discovery Institute (PD-II-2014-379) and the National Institute of General Medical Sciences (R01 GM32843) to S.K.D. The Genome Technology Access Center (GTAC) in the Department of Genetics at Washington University School of Medicine performed whole sequencing, and it is partially supported by National Cancer Institute Cancer Center Support grant #P30 CA91842 to the Siteman Cancer Center and by the Institute for Clinical and Translational Sciences (ICTS)/Clinical and Translational Science Award (CTSA) grant UL1RR024992 from the National Center for Research Resources (NCRR), a component of the NIH and NIH Roadmap for Medical Research.
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bowers AK, Keller JA, Dutcher SK (2003) Molecular markers for rapidly identifying candidate genes in Chlamydomonas reinhardtii: ery1 and ery2 encode chloroplast ribosomal proteins. Genetics 164: 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Liu G, Ke W, Zhao L, Lv B, Ma X, Xu N, Xia X, Deng X, Zheng C, et al. (2017) Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Breker M, Lieberman K (2017) Validated Bayesian differentiation of causative and passenger mutations. G3 (Bethesda) 7: 2081–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Sharifi MN, Malnoë A, Haglund C, Calderon RH, Wakao S, Niyogi KK (2015) Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J 82: 337–351 [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Li L, Lin H, Meyer L, Giddings TH, Kwan AL, Lewis BL (2012) Whole-genome sequencing to identify mutants and polymorphisms in Chlamydomonas reinhardtii. G3 (Bethesda) 2: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AM, Wilkinson AW, Backer CB, Lapan SW, Gutzman JH, Cheeseman IM, Reddien PW (2010) The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev Biol 337: 148–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, de Montaigu A, Galván A, Fernández E (2005) Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal Biochem 340: 330–335 [DOI] [PubMed] [Google Scholar]
- Gross CH, Ranum LPW, Lefebvre PA (1988) Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr Genet 13: 503–508 [DOI] [PubMed] [Google Scholar]
- Hart SN, Sarangi V, Moore R, Baheti S, Bhavsar JD, Couch FJ, Kocher JPA (2013) SoftSearch: integration of multiple sequence features to identify breakpoints of structural variations. PLoS ONE 8: e83356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia MM, O’Malley R, Cartwright A, Nieu R, Gordon SP, Kelly S, Williams TG, Wood DF, Zhao Y, Bragg J, et al. (2017) Sequencing and functional validation of the JGI Brachypodium distachyon T-DNA collection. Plant J 91: 361–370 [DOI] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ (1982) Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28: 115–124 [DOI] [PubMed] [Google Scholar]
- James SW, Silflow CD, Thompson MD, Ranum LP, Lefebvre PA (1989) Extragenic suppression and synthetic lethality among Chlamydomonas reinhardtii mutants resistant to anti-microtubule drugs. Genetics 122: 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinkerson RE, Jonikas MC (2015) Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J 82: 393–412 [DOI] [PubMed] [Google Scholar]
- Kathir P, LaVoie M, Brazelton WJ, Haas NA, Lefebvre PA, Silflow CD (2003) Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot Cell 2: 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage É, Zampini É, Boyle B, Brisson N (2013) Time- and cost-efficient identification of T-DNA insertion sites through targeted genomic sequencing. PLoS ONE 8: e70912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al. (2016) An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Dutcher SK (2015) Genetic and genomic approaches to identify genes involved in flagellar assembly in Chlamydomonas reinhardtii. Methods Cell Biol 127: 349–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Miller ML, Granas DM, Dutcher SK (2013) Whole genome sequencing identifies a deletion in protein phosphatase 2A that affects its stability and localization in Chlamydomonas reinhardtii. PLoS Genet 9: e1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhang Z, Guo S, Chen F, Kessler JM, Wang YM, Dutcher SK (2015) A NIMA-related kinase suppresses the flagellar instability associated with the loss of multiple axonemal structures. PLoS Genet 11: e1005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K, Lefebvre PA, Kamiya R, Hirono M (2002) Kinesin-II is not essential for mitosis and cell growth in Chlamydomonas. Cell Motil Cytoskeleton 52: 195–201 [DOI] [PubMed] [Google Scholar]
- Meslet-Cladière L, Vallon O (2012) A new method to identify flanking sequence tags in Chlamydomonas using 3′-RACE. Plant Methods 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MS, Esparza JM, Lippa AM, Lux FG III, Cole DG, Dutcher SK (2005) Mutant kinesin-2 motor subunits increase chromosome loss. Mol Biol Cell 16: 3810–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJV, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G, Schneeberger K (2013) Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat Biotechnol 31: 325–330 [DOI] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ (2008) Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Pringle JR (2016) Robust transgene expression from bicistronic mRNA in the green alga Chlamydomonas reinhardtii. G3 (Bethesda) 6: 4115–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Mukherjee B, Bajsa-Hirschel J, Machingura MC, Mukherjee A, Grossman AR, Moroney JV (2017) A robust protocol for efficient generation, and genomic characterization of insertional mutants of Chlamydomonas reinhardtii. Plant Methods 13: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Smith HE. (2011) Identifying insertion mutations by whole-genome sequencing. Biotechniques 50: 96–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F, Cross FR (2014) A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26: 4019–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F, Cross FR (2016) Patching holes in the Chlamydomonas genome. G3 (Bethesda) 6: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha M, Walther Z, Hall JL (1996) The kinesin-homologous protein encoded by the Chlamydomonas FLA10 gene is associated with basal bodies and centrioles. J Cell Sci 109: 541–549 [DOI] [PubMed] [Google Scholar]
- Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, Jonikas MC (2014) High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26: 1398–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]