Arabidopsis thaliana KEA4, KEA5, and KEA6 are endosomal K+ transporters that function in maintaining pH and ion homeostasis in the endomembrane network.
Abstract
KEA4, KEA5, and KEA6 are members of the Arabidopsis (Arabidopsis thaliana) K+ efflux antiporter (KEA) family that share high sequence similarity but whose function remains unknown. Here, we show their gene expression pattern, subcellular localization, and physiological function in Arabidopsis. KEA4, KEA5, and KEA6 had similar tissue expression patterns, and the three KEA proteins localized to the Golgi, the trans-Golgi network, and the prevacuolar compartment/multivesicular bodies, suggesting overlapping roles of these proteins in the endomembrane system. Phenotypic analyses of single, double, and triple mutants confirmed functional redundancy. The triple mutant kea4 kea5 kea6 had small rosettes, short seedlings, and was sensitive to low K+ availability and to the sodicity imposed by high salinity. Also, the kea4 kea5 kea6 mutant plants had a reduced luminal pH in the Golgi, trans-Golgi network, prevacuolar compartment, and vacuole, in accordance with the K/H exchange activity of KEA proteins. Genetic analysis indicated that KEA4, KEA5, and KEA6 as well as endosomal Na+/H+exchanger5 (NHX5) and NHX6 acted coordinately to facilitate endosomal pH homeostasis and salt tolerance. Neither cancelling nor overexpressing the vacuolar antiporters NHX1 and NHX2 in the kea4 kea5 kea6 mutant background altered the salt-sensitive phenotype. The NHX1 and NHX2 proteins in the kea4 kea5 kea6 mutant background could not suppress the acidity of the endomembrane system but brought the vacuolar pH close to wild-type values. Together, these data signify that KEA4, KEA5, and KEA6 are endosomal K+ transporters functioning in maintaining pH and ion homeostasis in the endomembrane network.
The endomembrane system, which sorts proteins and membranes through the secretory and endocytic pathways, is critical for cellular functions in eukaryotes (Otegui and Spitzer, 2008; Richter et al., 2009; Contento and Bassham, 2012). The plant endomembrane system comprises the endoplasmic reticulum (ER), Golgi complex, trans-Golgi network (TGN), prevacuolar compartment (PVC)/multivesicular bodies (MVB), and vacuoles (Contento and Bassham, 2012). The endomembrane system, by enabling protein modification and sorting, plays important roles in cell polarity, cytokinesis, cell wall formation, signaling, and stress tolerance (Jurgens, 2004; Bar and Avni, 2009; Kang et al., 2010; Rose and Lee, 2010; Contento and Bassham, 2012).
Maintaining a specific luminal pH is critical for the operation of the endomembrane system (Marshansky and Futai, 2008; Casey et al., 2010; Shen et al., 2013; Schumacher, 2014). Studies have shown that the lumen of the endomembrane system becomes more acidic along the progress through the secretory or endocytic pathways in plant and animal cells (Marshansky and Futai, 2008; Casey et al., 2010; Martinière et al., 2013). Cellular and organelle pH homeostasis is maintained by the concerted activity of both H+ pumps and other anion or cation transporters permeating protons (Demaurex, 2002; Casey et al., 2010; Schumacher, 2014). The acidity of the organelles is generated by the vacuolar H+-ATPases in yeast and animals (Paroutis et al., 2004; Marshansky and Futai, 2008) and by the vacuolar H+-ATPases and H+-pyrophosphatase in plants (Martinière et al., 2013; Schumacher, 2014). The anion or cation transporters work as either shunt conductance to prevent the buildup of an electrical membrane potential or as proton leaks in order to counteract the cellular or organelle acidification created by the H+ pumps. Studies have shown that animal Na+/H+ antiporters of the Na+/H+ exchanger family counter organelle acidification through H+ leakage to maintain proper pH (Demaurex, 2002; Orlowski and Grinstein, 2007). In plants, the endosomal Na+,K+/H+ antiporters NHX5 and NHX6, which are phylogenetically related to animal Na+/H+ exchangers, help regulate cellular pH and ion homeostasis in Arabidopsis (Arabidopsis thaliana; Bassil et al., 2011a; Martinière et al., 2013; Reguera et al., 2015; Wang et al., 2015; Qiu, 2016a). NHX5 and NHX6 proteins share high sequence similarity, are genetically redundant, and both localize in the endosomal compartments, including the Golgi, TGN, and PVC/MVB (Yokoi et al., 2002; Bassil et al., 2011a; Reguera et al., 2015; Wang et al., 2015). NHX5 and NHX6 are required for the transport of seed storage proteins into the protein storage vacuole (Reguera et al., 2015; Qiu, 2016b; Wu et al., 2016). Using organelle specific pHluorin-based pH sensors, Reguera et al. (2015) found that the nhx5 nhx6 mutant had reduced pH in Golgi, TGN, and the late PVC. Wang et al. (2015) also showed that the nhx5 nhx6 mutant had reduced pH in the mature roots and the cell sap extracted from rosette leaves. A recent report showed that NHX5 and NHX6 might regulate auxin transport across the ER via the pH gradient created by their transport activity (Fan et al., 2018). These results suggest that NHX5 and NHX6 facilitate Na+,K+/H+ exchange and may act as an H+ leak system to regulate cellular pH homeostasis (Qiu, 2016a, 2016b).
The K+ efflux antiporter (KEA) proteins of plants are phylogenetically related to the K+ efflux transporters EcKefB and EcKefC of Escherichia coli. The KEA gene family is a subgroup of the large Cation Proton Antiporter2 (CPA2) family of cation/proton antiporters of Arabidopsis (Mäser et al., 2001; Chanroj et al., 2012). Phylogenetic analysis divided the KEA clade into two subgroups: KEA1 to KEA3 and KEA4 to KEA6 (Chanroj et al., 2012; Zheng et al., 2013). In Arabidopsis, KEA4 to KEA6 are 75% to 83.4% similar to each other, whereas similarities within the KEA1 to KEA3 isoforms are low (21.9%–30%; Zheng et al., 2013). KEA gene expression is responsive to environmental cues. The expression of KEA1, KEA3, and KEA4 was enhanced under low-K+ stress, whereas KEA2 and KEA5 were induced by sorbitol and abscisic acid treatments, suggesting that KEAs may play a role in K+ homeostasis and osmotic adjustment in Arabidopsis (Zheng et al., 2013). Reverse genetic and proteomic studies have shown that the KEA1, KEA2, and KEA3 group is important for chloroplast physiology and function. KEA1 and KEA2 localize to inner envelope microdomains near the two caps of the organelle separated by the fission plane, where they serve important roles for plastid division and thylakoid membrane biogenesis (Aranda-Sicilia et al., 2016). The kea1 kea2 mutant had swollen chloroplasts with disrupted envelope membranes and reduced thylakoid membrane density (Kunz et al., 2014). KEA3 localizes to the thylakoid membrane to accelerate the down-regulation of pH-dependent energy dissipation under fluctuating light, a process necessary for recovering high photosynthetic efficiency and CO2 assimilation. As a result, KEA3 increases photosynthetic efficiency under fluctuating light (Armbruster et al., 2014; Kunz et al., 2014). The pH component of the proton motive force across the thylakoid membrane was decreased in the kea1 kea2 mutant but increased in the kea3 mutant, indicating a critical function for the KEA1 to KEA3 proteins in chloroplast pH homeostasis (Kunz et al., 2014). These data have been interpreted as evidence that KEA proteins catalyze K+/H+ exchange, but experimental confirmation has been produced only for KEA2 (Aranda-Sicilia et al., 2012). Notably, chloroplastic KEA1 to KEA3 proteins have been involved in rapid Ca2+ responses to hyperosmotic stimuli preceding secondary Ca2+ signaling events that propagate along the plant axis from roots to shoots (Stephan et al., 2016). Single kea3 and double kea1 kea2 mutants displayed reduced amplitude of the hyperosmosis-induced Ca2+ response. Thus, KEA1 to KEA3 proteins may contribute to the important role of Ca2+-storing plastids in stress-induced Ca2+ signaling.
Contrary to the KEA1 to KEA3 group, the subcellular localization and physiological role of KEA4, KEA5, and KEA6 remain unknown. In this report, we characterized the gene expression, subcellular localization, and physiological roles of KEA4 to KEA6 proteins from Arabidopsis. We found that these three KEA proteins were localized to the Golgi, TGN, and PVC/MVB. We further found that the kea4 kea5 kea6 triple mutant had reduced growth and was sensitive to the treatments of low K+, high Na+, or high Li+. Notably, the triple mutant kea4 kea5 kea6 had a more acidic pH in the Golgi, TGN, PVC, and vacuole. Mutant analyses showed that KEA4, KEA5, KEA6, NHX5, and NHX6 act coordinately to facilitate endosomal pH homeostasis, whereas genetic interaction with the vacuolar antiporters NHX1 and NHX2 indicated that the endomembrane compartments and the vacuole acted independently in salt tolerance. We demonstrate that KEA4, KEA5, and KEA6 are endosomal antiporters that mediate the pH and ion homeostasis in endomembrane compartments.
RESULTS
KEA4, KEA5, and KEA6 Are Important for Growth in Arabidopsis
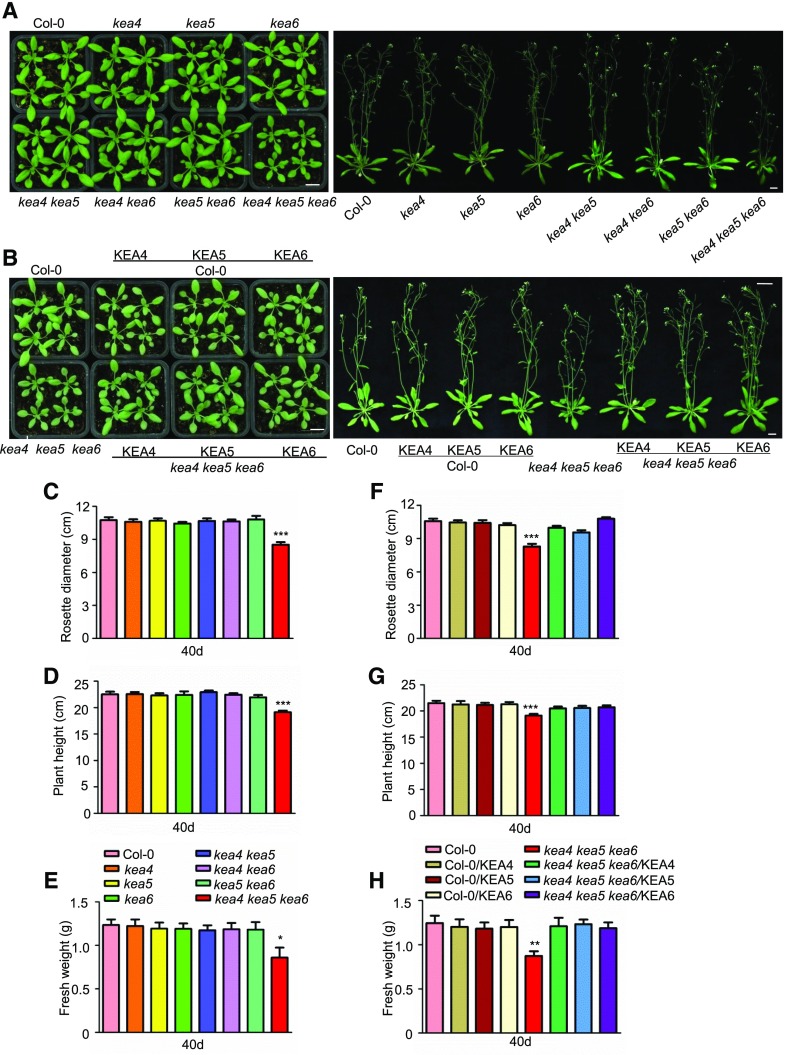
To examine the role of KEA4, KEA5, and KEA6 in seedling growth with a reverse genetics approach using Arabidopsis T-DNA mutants, we first obtained two separate T-DNA insertion lines for each of the genes: KEA4 (kea4-1 and kea4-3), KEA5 (kea5-1 and kea5-2), and KEA6 (kea6-1 and kea6-3). The absence of the KEA4, KEA5, and KEA6 transcripts in these single knockout lines was confirmed by quantitative reverse transcription PCR (RT-qPCR; Supplemental Fig. S1). We then generated two sets of the double and triple mutant lines by genetic crossing: kea4-1 kea5-1, kea4-1 kea6-1, kea5-1 kea6-1, and kea4-1 kea5-1 kea6-1 on the one hand and kea4-3 kea5-2, kea4-3 kea6-3, kea5-2 kea6-3, and kea4-3 kea5-2 kea6-3 on the other. The absence of KEA4, KEA5, and KEA6 transcripts in the double and triple knockout lines was confirmed by RT-qPCR (Supplemental Fig. S1). These two sets of the double and triple knockout lines had identical growth phenotypes (Fig. 1; Supplemental Fig. S1). Hence, only the double mutant lines kea4-1 kea5-1, kea4-1 kea6-1, and kea5-1 kea6-1 as well as the triple mutant line kea4-1 kea5-1 kea6-1 were used in subsequent experiments.
Figure 1.
The kea4 kea5 kea6 mutant is defective in growth. A and B, Seedling growth. A, Mutants. B, Overexpression lines. Photographs were taken at 21 d (left) and 40 d (right). Bars = 1 cm. C to E, Rosette diameters (C), plant heights (D), and fresh weight (E) of mutants measured at 40 d (mean ± se; n = 15 plants). F to H, Rosette diameters (F), plant heights (G), and fresh weight (H) of the overexpression lines measured at 40 d (mean ± se; n = 15 plants). In C to F, statistics by Student’s t test are shown: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
The single and double mutants resembled the wild-type plants in growth and development, and only the kea4 kea5 kea6 triple mutant showed reduced growth and smaller rosettes (Fig. 1). The growth delay of the kea4 kea5 kea6 mutant became visible after 20 d of growth in soil (Supplemental Fig. S2). At the mature stage (40 d), kea4 kea5 kea6 plants had 21%, 15%, and 26% reductions in rosette diameter, plant height, and fresh weight, respectively (Fig. 1, C–E), while rosette leaf numbers were unchanged (Supplemental Fig. S3A) compared with the wild-type plants. The kea4 kea5 kea6 mutant flowered early; the average flowering time for the wild-type plants was 26 d, whereas for the kea4 kea5 kea6 mutant it was 24 d (Supplemental Fig. S3C). Complementation with KEA4, KEA5, or KEA6 (with C-terminal fusions to RFP) restored normal development of the triple mutant, whereas overexpression had no effect on the wild type (Fig. 1; Supplemental Figs. S2 and S3). These results indicate that KEA4, KEA5, and KEA6 have similar biochemical functions important for growth and development in Arabidopsis.
KEA4, KEA5, and KEA6 Have Similar Expression Patterns
We examined the expression of KEA4, KEA5, and KEA6 in different organs by RT-qPCR (Supplemental Fig. S4). They were expressed in roots, stems, rosette leaves, flowers, and siliques. The expression of KEA4 was higher than that of KEA5 and KEA6 except in rosette leaves (Supplemental Fig. S4).
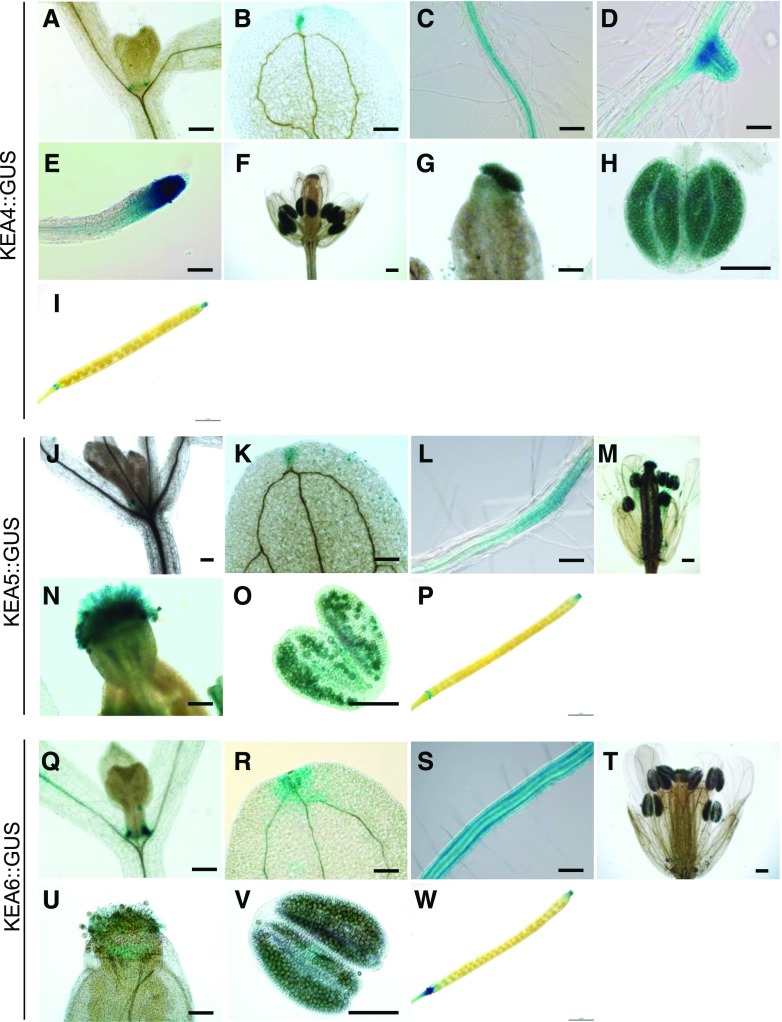
We then examined the tissue expression patterns of KEA4, KEA5, and KEA6 genes by GUS staining. The promoters of KEA4, KEA5, and KEA6 genes, 2,040, 1,545, and 1,500 bp upstream of their open reading frames, respectively, were amplified by PCR with the gene-specific primers. The promoter regions of KEA4, KEA5, and KEA6 genes were fused to the GUS reporter gene, and the resulting constructs were transformed into wild-type Arabidopsis plants. T3 seeds were collected from three independent transgenic lines, and all three lines, KEA4::GUS, KEA5::GUS, and KEA6::GUS, produced similar expression patterns. GUS staining showed largely overlapping expression patterns for KEA4, KEA5, and KEA6 genes, with a few exceptions (Fig. 2). In roots, the expression of KEA4 and KEA5 promoters was restricted to the vasculature and the tips of primary and secondary roots, whereas KEA6 was expressed in all cell types, including root hairs. All three promoters drove GUS expression in the vascular bundles and tips of cotyledons, the base of true leaves, the ovarian stigma and pollen grains within the anthers, and the tip and base of the siliques. These results suggest that KEA4, KEA5, and KEA6 may have largely overlapping functions in Arabidopsis development and physiology but also that differences may exist in roots.
Figure 2.
GUS staining assay of KEA4, KEA5, and KEA6 expression in Arabidopsis. A to I, GUS staining of ProKEA4::GUS plants. A, True leaf. B, Cotyledon. C, Root vasculature. D, Secondary root. E, Root tip. F, Flower. G, Stigma. H, Anther. I, Silique. Bars = 0.2 mm (A, B, and F), 100 μm (C–E and G), 150 μm (H), and 2 mm (I). J to P, GUS staining of ProKEA5::GUS plants. J, True leaf. K, Cotyledon. L, Root vasculature. M, Flower. N, Stigma. O, Anther. P, Silique. Bars = 0.2 mm (J, K, and M), 100 μm (L and N), 150 μm (O), and 2 mm (P). Q to W, GUS staining of ProKEA6::GUS plants. Q, True leaf. R, Cotyledon. S, Root vasculature. T, Flower. U, Stigma. V, Anther. W, Silique. Bars = 0.2 mm (Q, R, and T), 100 μm (S and U), 150 μm (V), and 2 mm (W).
KEA Proteins Mediate H+-Linked K+ Transport
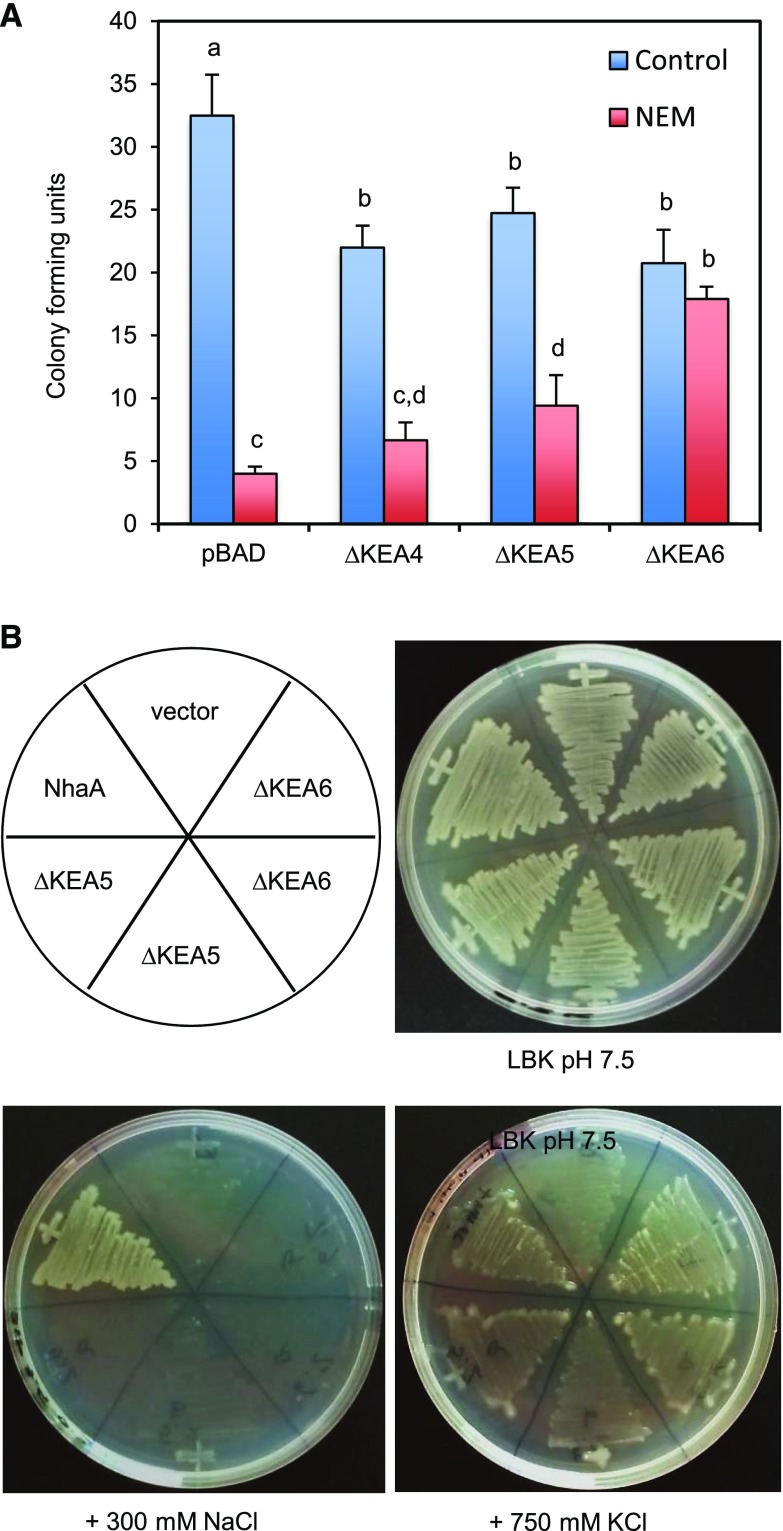
The KEA proteins of plants are thought to be K+ transporters (Chanroj et al., 2012), but only KEA2 has been confirmed to mediate electroneutral K+/H+ exchange in reconstituted proteoliposomes (Aranda-Sicilia et al., 2012). The KEA1 to KEA3 family members are critical for the pH homeostasis of chloroplasts, presumably by facilitating K+/H+ exchange (Kunz et al., 2014). To deduce the transport activity of KEA4 to KEA6 proteins, we used the E. coli strain MJF276 lacking the phylogenetically related K+ transporters KefB and KefC, which are important for protecting cells against the toxic effects of the electrophile N-ethylmaleimide (NEM). Survival of NEM is correlated with fast K+ release and cytoplasmic acidification upon the activation of KefB and KefC (Ferguson et al., 1993, 1997). The expression of full-length KEA4, KEA5, and KEA6 proteins in E. coli MJF276 failed to suppress the sensitivity to NEM. KEA proteins present long and variable N-terminal regions with no detectable conserved domains (Supplemental Fig. S5) that presumably are used to traffic the protein to the target membrane and to tether the proteins to specific microdomains; removal of these N-terminal domains is required to achieve transport activity in heterologous systems (Aranda-Sicilia et al., 2012, 2016). Therefore, N-terminal truncations of KEA4, KEA5, and KEA6 (∆KEA4, ∆KEA5, and ∆KEA6) were produced to express in E. coli only the protein domains with sequence similarity to bacterial KefB/C-like transporters, which lack such N-terminal domains (Supplemental Fig. S5). Bacterial cultures were exposed to 0.1 mm NEM for 45 min and then plated to count colony-forming units. Inducing the expression of truncated KEA proteins in control samples reduced the bacterial cell count by 23% to 36% relative to the empty vector, indicating moderate toxicity (Fig. 3). In spite of this, ∆KEA6, and to a lesser extent ∆KEA5, protected the kefB kefC mutant against NEM. ∆KEA4 expression had no statistically significant effect in this test. To determine the substrate selectivity of KEA4, KEA5, and KEA6, we also expressed the plant proteins in the E. coli strain EP432 lacking the Na+/H+ exchangers NhaA and NhaB. In the absence of these Na+ extrusion proteins, bacteria are sensitive to NaCl. Moreover, high external concentrations of K+ at alkaline pH also are toxic to EP432 cells, which require robust K+ efflux systems coupled to cytoplasmic acidification (proton influx) to withstand these conditions (Radchenko et al., 2006). Thus, EP432 cells enabled testing whether KEA proteins transport K+ and/or Na+. Again, the expression of full-length KEA4, KEA5, and KEA6 proteins in E. coli EP432 failed to modify the sensitivity to NaCl or KCl, whereas truncated ∆KEA5 and ∆KEA6 proteins imparted resistance to high KCl, but not NaCl, at pH 7.5 (Fig. 3). Complementation with the control protein NhaA permitted growth in both salts, whereas ∆KEA4 could not be assayed due to severe toxicity to EP432 cells. Together, these results strongly suggest that KEA5 and KEA6 are K+/H+ exchangers with no apparent ability to transport Na+.
Figure 3.
KEA5 and KEA6 mediate H+-linked K+ transport in bacteria. N-terminal truncations of KEA4, KEA5, and KEA6 were expressed in E. coli strains lacking K+ and Na+ transporters. A, Survival of MJF276 cells with mutated K+/H+ exchangers KefB and KefC after treatment with NEM. Shown are the colony-forming units with and without NEM treatment after appropriate serial dilutions. Data represent means and se of two biological and four technical replicates. Lettering indicates significantly different means (P < 0.05) by the one-way ANOVA test. B, EP432 cells lacking the Na+/H+ exchangers NhaA and NhaB were streaked on agar plates with LKB medium at pH 7.5 with and without supplemental NaCl and KCl as indicated. Two independent ∆KEA5 and ∆KEA6 transformants are shown. The E. coli NhaA and empty vector were used as positive and negative controls.
KEA4, KEA5, and KEA6 Are Essential to K+ Homeostasis in Arabidopsis
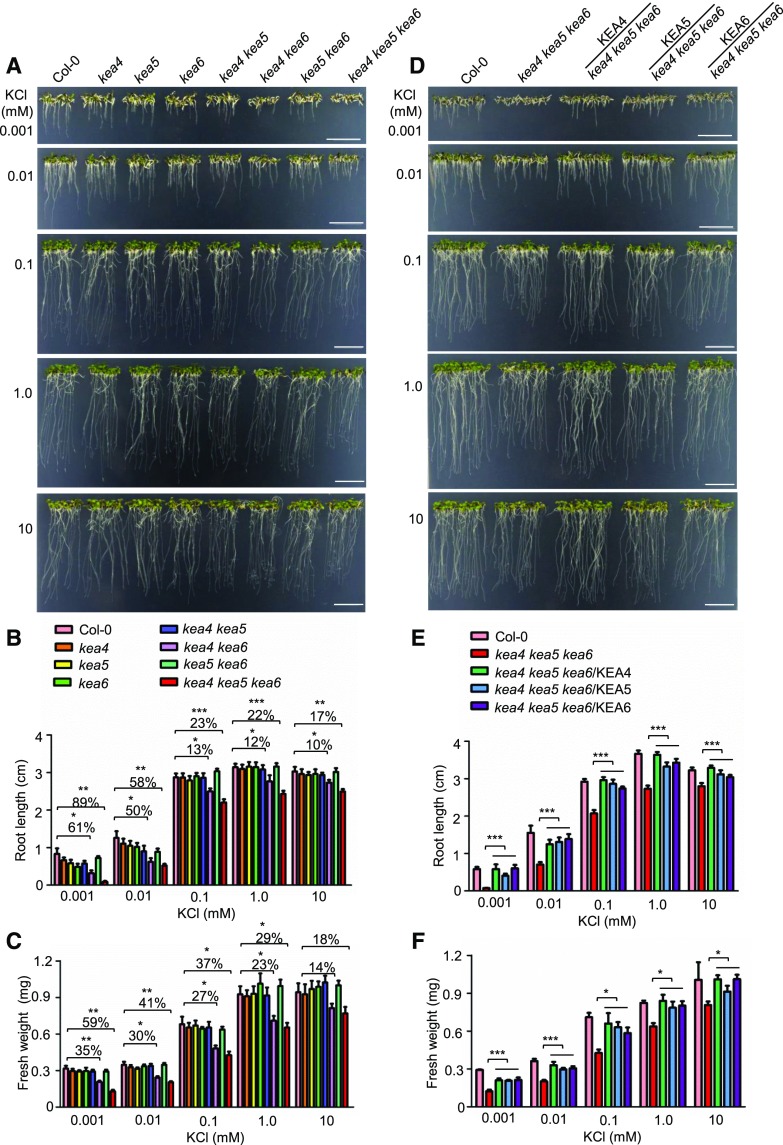
To test the functions of KEA4, KEA5, and KEA6 in the K+ homeostasis of plants, we examined the growth of kea4 kea5 kea6 seedlings on medium containing various levels of K+. The low-K+ media were made by adding different amounts of KCl (0.001–10 mm) to the modified K+-free medium (Pandey et al., 2007). As shown in Figure 4, A to C, at 10 mm KCl, the root length and seedling fresh weight of kea4 kea6 and kea4 kea5 kea6 plants were reduced slightly, while no significant difference was observed for other single and double mutants compared with ecotype Columbia (Col-0). The root growth and seedling fresh weight of kea4 kea6 and kea4 kea5 kea6 plants were reduced further when K+ was decreased (Fig. 4, A–C). At 10 and 1 µm K+, both kea4 kea6 and kea4 kea5 kea6 plants had dramatically reduced root growth and seedling fresh weight (Fig. 4, A–C). The roots were barely growing in the triple mutants at 1 µm K+ (Fig. 4). Overexpression of KEA4, KEA5, or KEA6 in the kea4 kea5 kea6 mutant recovered root growth and seedling fresh weight (Fig. 4, D–F). These results suggest that KEA4, KEA5, and KEA6 are critical to K+ homeostasis in Arabidopsis and that these proteins are biochemically exchangeable.
Figure 4.
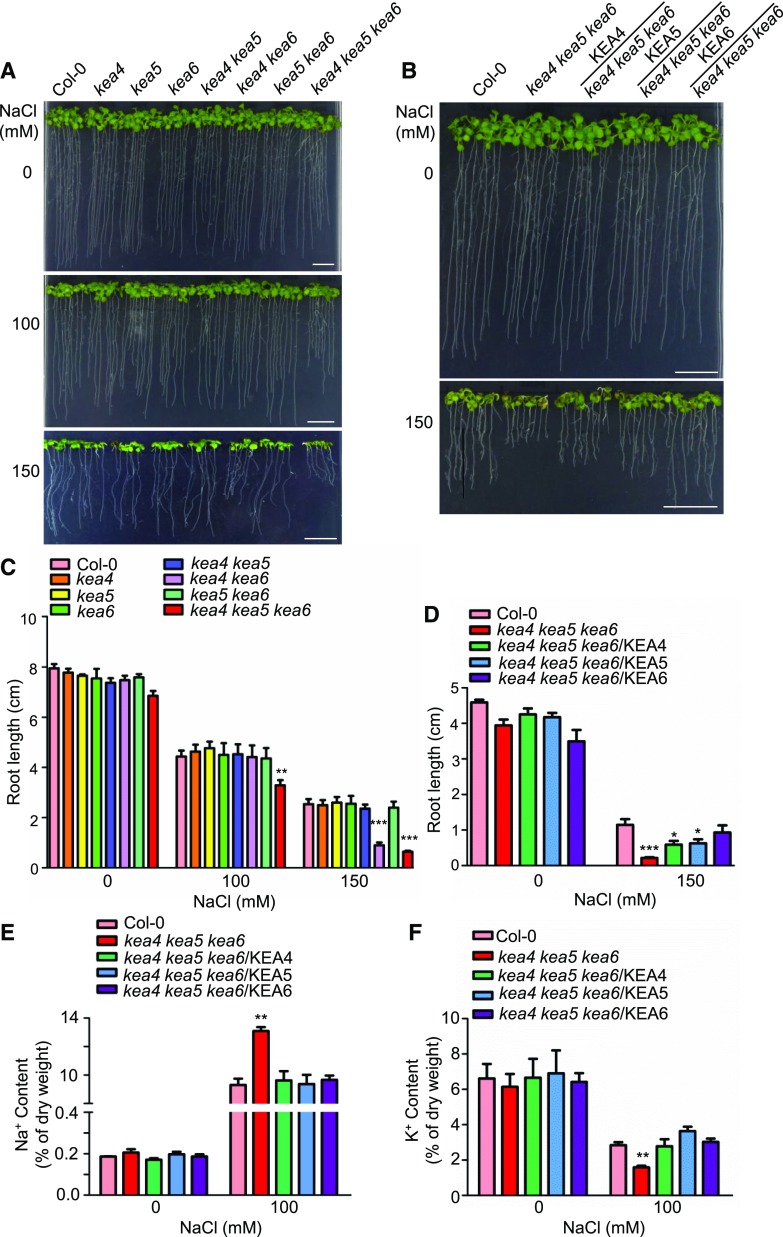
Mutant kea4 kea5 kea6 is sensitive to low-K+ treatment. A and D, Growth of the mutants and overexpression lines at low K+. A, The mutants. D, The overexpression lines. Seedlings were grown on modified MS medium containing different levels of K+ (0.001–10 mm KCl). For the solidification of MS medium, 1% ultra-pure agarose was used (Pandey et al., 2007). Photographs were taken at 7 d under low-K+ treatment. Bars = 1 cm. B and E, Root length for the seedlings from A and D, respectively (mean ± se; n = 10 seedlings). C and F, Fresh weight for the seedlings from A and D, respectively (mean ± se; n = 30 seedlings). In B, C, E, and F, statistics by Student’s t test are shown: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
The kea4 kea5 kea6 Mutant Is Sensitive to Salt Stress
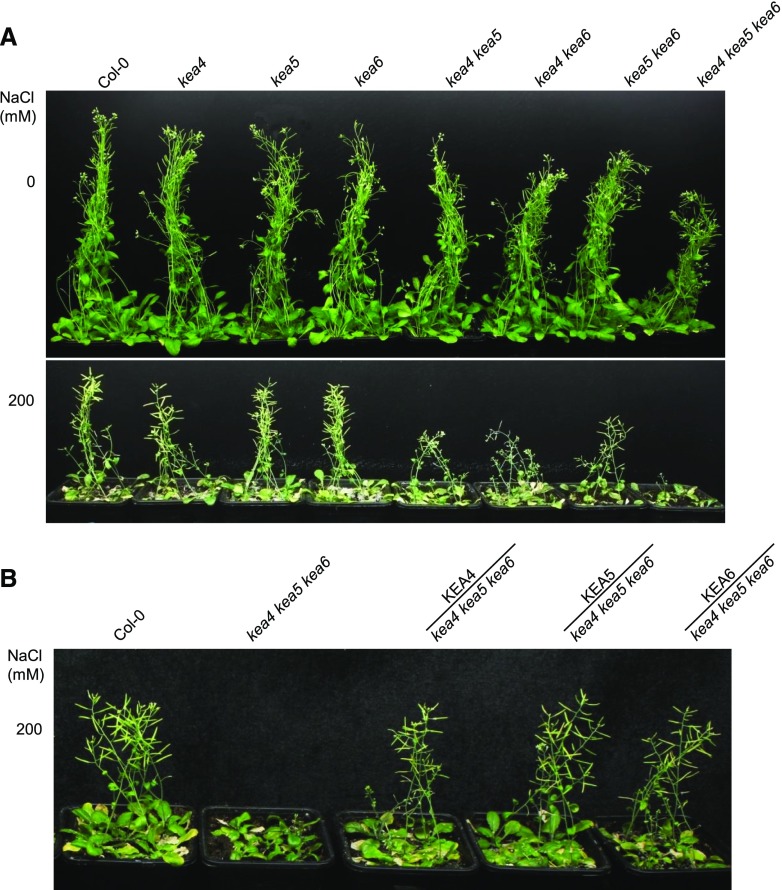
Since the K+/Na+ content ratio is considered a key predictor of the salt tolerance of plants, we also examined whether KEA4, KEA5, and KEA6 are involved in Na+ tolerance in Arabidopsis. Seed germination was tested on one-half-strength Murashige and Skoog (1/2 MS) medium containing NaCl (Supplemental Fig. S6). At 100 mm NaCl, no significant difference was observed between mutants and Col-0. At 150 mm NaCl, the wild-type seeds germinated well (90.6% germination rate), while the kea4 kea5 kea6 mutant had a very low germination rate of 9.5% (Supplemental Fig. S6). Again, the double mutant kea4 kea6 departed from other double mutants and germination was partially inhibited (75.6% germination rate) at 150 mm NaCl, while the rest of the single and double mutants were unaffected (Supplemental Fig. S6). In addition, the root growth of the kea4 kea5 kea6 mutant was inhibited under salt by 26% and 74.8% at 100 and 150 mm NaCl, respectively, compared with the wild type (Fig. 5, A and C). The root growth of the kea4 kea6 mutant also was inhibited by 64.5% in 100 mm NaCl relative to the wild type. Overexpression of KEA4, KEA5, or KEA6 in the kea4 kea5 kea6 mutant cancelled the sensitivity to salt stress (Fig. 5, B and D). The development of soil-grown plants irrigated with 200 mm NaCl was delayed in double mutants of either combination and severely impaired in the triple mutant (Fig. 6A). Again, overexpression of KEA4, KEA5, or KEA6 in the kea4 kea5 kea6 mutant significantly improved plant growth under salt stress (Fig. 6B).
Figure 5.
The kea4 kea5 kea6 mutant is sensitive to salt stress. A and B, Growth of the mutants and overexpression lines under NaCl treatment. A, The mutants. B, The overexpression lines. Four-day-old seedlings grown on 1/2 MS medium were transferred to 1/2 MS medium containing 0, 100, and 150 mm NaCl. Photographs were taken at 10 d under salt stress. Bars = 1 cm. C, Root length for seedlings from A (mean ± se; n = 8 seedlings). D, Root length for seedlings from B (mean ± se; n = 8 seedlings). E, Na+ content assay (mean ± se; n = 3 times). F, K+ content assay (mean ± se; n = 3 times). In C to F, statistics by Student’s t test are shown: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Figure 6.
Seedling growth is suppressed in the kea4 kea5 kea6 mutant under salt stress. Growth of the mutants and overexpression lines in soil under salt stress is shown. A, The mutants. B, The overexpression lines. After growing for 2 weeks, the seedlings were treated with 0 or 200 mm NaCl every 3 d. Photographs were taken after the seedlings were treated with NaCl for 4 weeks.
We also determined Na+ and K+ contents of the kea4 kea5 kea6 seedlings and the overexpression lines by atomic absorption spectrophotometry (Fig. 5E). When seedlings grew on 1/2 MS medium, there was no obvious difference in the Na+ concentration between Col-0 and kea4 kea5 kea6 plants. However, at 100 mm NaCl, the kea4 kea5 kea6 mutant accumulated a higher amount of Na+ (1.4-fold that of the wild type; Fig. 5E). Overexpression of KEA4, KEA5, or KEA6 in the kea4 kea5 kea6 mutant reduced Na+ concentrations to wild-type levels (Fig. 5E). In addition, without salt stress, the K+ levels were unchanged in the kea4 kea5 kea6 mutant relative to Col-0 (Fig. 5F). However, at 100 mm NaCl, the kea4 kea5 kea6 mutant had 44.5% less K+ compared with Col-0 (Fig. 5F), and overexpression of KEA4, KEA5, or KEA6 in the kea4 kea5 kea6 mutant restored K+ levels to the wild-type level (Fig. 5F). Together, these results suggest that KEA4, KEA5, and KEA6 are critical to Na+ and K+ homeostasis under salt stress in Arabidopsis.
High concentrations of NaCl impose both an ionic stress and osmotic challenges. Therefore, we also examined the roles of KEA4, KEA5, and KEA6 in Li+ tolerance, since Li+ often is used as an analog of Na+ that becomes toxic to cells at lower concentrations that do not represent a significant osmotic stress. The sensitivity of the kea4 kea5 kea6 triple mutant and the kea4 kea6 double mutant to LiCl recapitulated the sensitivity to NaCl, and overexpression of the KEA4, KEA5, or KEA6 gene in the kea4 kea5 kea6 mutant recovered the growth to wild-type levels (Supplemental Fig. S7). Collectively, these results show that KEA4, KEA5, and KEA6 are critically important for tolerance to the sodic component of salinity stress, presumably as the result of faulty K+ homeostasis.
KEA4, KEA5, and KEA6 Are Localized to the Endomembrane Compartments
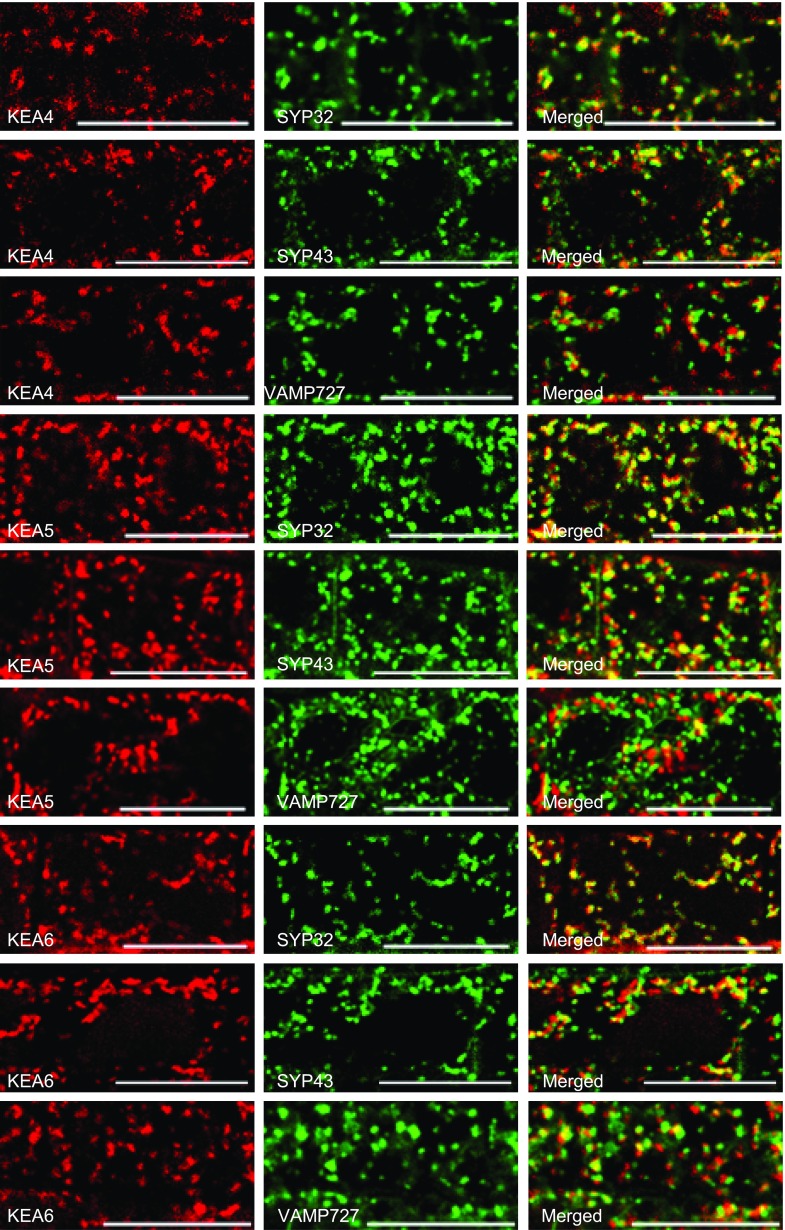
The subcellular localizations of KEA4, KEA5, and KEA6 were first visualized in Nicotiana benthamiana epidermal cells by transient coexpression of 35S:KEA-RFP constructs with the endosomal markers Man49-GFP (Golgi marker), SYP61-GFP (TGN marker), and ARA6-GFP (PVC/MVB marker). KEA4-RFP, KEA5-RFP, and KEA6-RFP fluorescent signals appeared as punctate structures in the cytosol that colocalized extensively with the endosomal markers, suggesting that KEA4, KEA5, and KEA6 are distributed along the Golgi, TGN, and PVC/MVB compartments (Supplemental Fig. S8). The subcellular localizations of KEA4, KEA5, and KEA6 were verified in seedlings of stably transformed Arabidopsis that were crossed to reporter lines expressing the Golgi marker SYP32-YFP, the TGN marker GFP-SYP43, or the PVC/MVB marker GFP-VAMP727 (Ebine et al., 2008). Consistent with the transient expression assay, the fluorescent signals of tagged KEA proteins were visualized as punctate structures within the cells that colocalized extensively with the endosomal markers (Fig. 7). The intensity correlation quotient and the Pearson’s correlation coefficient (Li et al., 2004; Reguera et al., 2015) are given in Table 1. As shown in Supplemental Movies S1 to S3, the punctate structures were moving rapidly in root cells, reflecting the highly dynamic trafficking of the KEA4-, KEA5-, and KEA6-positive bodies. Together, the results from both the transient expression in N. benthamiana cells and stably transformed Arabidopsis lines indicate that KEA4, KEA5, and KEA6 are distributed along the Golgi, TGN, and PVC/MVB compartments.
Figure 7.
Subcellular localizations of KEA4, KEA5, and KEA6 in Arabidopsis. Coexpression of KEA4-RFP, KEA5-RFP, and KEA6-RFP with the endosomal markers in root cells is shown. Markers used were SYP32 (Golgi marker), SYP43 (TGN marker), and VAMP727 (PVC marker). Bars = 20 μm.
Table 1. Subcellular localization analysis.
ICQ, Intensity correlation quotient; PSC, Pearson’s correlation coefficient. For ICQ, 0 stands for no colocalization and 0.5 stands for high colocalization; for PSC, 1 is total positive correlation and 0 is no correlation (Li et al., 2004; Reguera et al., 2015). Values are means ± se; 14 to 16 fluorescence images taken from seven different seedlings were collected for each statistical analysis.
| Seedling | SYP32-YFP | GFP-SYP43 | GFP-VAMP727 | |||
|---|---|---|---|---|---|---|
| PSC | ICQ | PSC | ICQ | PSC | ICQ | |
| KEA4-RFP | 0.76 ± 0.06 | 0.28 ± 0.01 | 0.47 ± 0.03 | 0.25 ± 0.02 | 0.43 ± 0.02 | 0.23 ± 0.01 |
| KEA5-RFP | 0.61 ± 0.02 | 0.28 ± 0.02 | 0.58 ± 0.03 | 0.26 ± 0.01 | 0.29 ± 0.03 | 0.17 ± 0.01 |
| KEA6-RFP | 0.71 ± 0.02 | 0.33 ± 0.01 | 0.41 ± 0.04 | 0.22 ± 0.01 | 0.35 ± 0.02 | 0.18 ± 0.01 |
KEA4, KEA5, and KEA6 Regulate the pH in Endomembrane Compartments
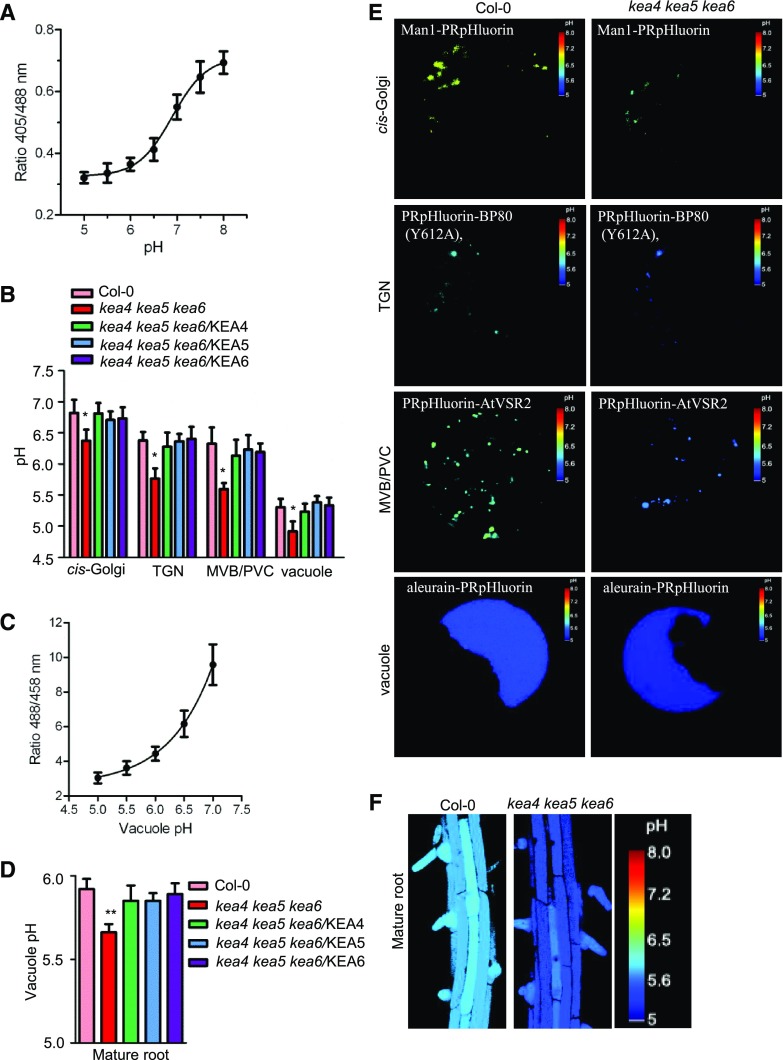
The KEA3 protein has been suggested to behave as a proton shunt from the thylakoid lumen by K+/H+ antiport (Armbruster et al., 2014). To examine whether KEA4, KEA5, and KEA6 regulate the pH in endomembrane compartments, we used the pHluorin-based ratiometric pH sensor and quantitative live-cell imaging to measure the luminal pH of the endomembrane compartments (Martinière et al., 2013; Shen et al., 2013). The pHluorin-based pH sensors targeted to the endomembrane compartments, including the cis-Golgi-targeted pH sensor Man1-PRpHluorin, the TGN-targeted PRpHluorin-BP80 (mutant Y612A), the PVC/MVB-targeted PRpHluorin-AtVSR2, and the vacuole-targeted pH sensor aleurain-PRpHluorin, were transiently expressed in the Arabidopsis protoplasts. The protoplasts were prepared from the leaves of 3-week-old seedlings of genotypes Col-0, kea4 kea5 kea6, and the triple mutant overexpressing KEA4 (kea4 kea5 kea6/KEA4), KEA5 (kea4 kea5 kea6/KEA5), or KEA6 (kea4 kea5 kea6/KEA6). The calibration curve was created with wild-type protoplasts (Fig. 8A). Representative pseudo-colored images of Man1-PRpHluorin, PRpHluorin-BP80 (Y612A), PRpHluorin-AtVSR2, and aleurain-PRpHluorin are shown in Figure 8E. As shown in Figure 8B, for the wild-type plants, the organelles were increasingly acidic along the endocytic pathways: Golgi (pH 6.8), TGN (pH 6.4), PVC/MVB (pH 6.3), and vacuole (pH 5.3). Notably, the kea4 kea5 kea6 mutant had a significantly reduced pH in the endomembrane compartments compared with the wild type: Golgi (pH 6.4), TGN (pH 5.7), PVC/MVB (pH 5.6), and vacuole (pH 4.9).
Figure 8.
The kea4 kea5 kea6 mutant has a more acidic pH in the endomembrane compartments. A, In vivo calibration curve of the pH of the cytoplasm. pH was measured using Arabidopsis protoplasts transiently expressing the Golgi-specific Man1-PRpHluorin, TGN-specific PRpHluorin-BP80 (Y612A), PVC/MVB-specific PRpHluorin-AtVSR2, and vacuole-specific aleurain-PRpHluorin. The images were taken with a Leica TCS SP5 laser scanning confocal microscope. The calibration curve was achieved by equilibrating intracellular pH with 25 μm nigericin, 60 mm KCl, and 10 mm MES/HEPES Bis-Tris-propane, pH 5 to 8 (mean ± se; n ≥ 20 protoplasts). B, pH of the Golgi, TGN, PVC/MVB, and vacuole (mean ± se; n ≥ 20 protoplasts). Statistics by Student’s t test are shown: *, P < 0.05. C, In situ calibration curve of the pH of BCECF-AM dye-loaded roots. The calibration was performed by plotting the ratio of emission fluorescence (505–550 nm), obtained when dye-loaded roots were excited with 458 and 488 nm, against the pH of equilibration buffers as described in “Materials and Methods.” Fluorescence images were collected 15 min after roots were incubated in equilibration buffers (mean ± se; n = 10 roots). D, Vacuolar pH of the mature root measured by BCECF-AM (mean ± se; n = 10 roots). Statistics by Student’s t test are shown: **, P < 0.01. E, Representative pseudocolored images for the organellar pH assays. The organelle-specific pHlurions were described in A. F, Ratio images of epidermal cells of the mature root zone in Col-0 and the kea4 kea5 kea6 mutant.
Moreover, we also determined the vacuolar pH using the fluorescein-based ratiometric pH indicator 2′,7′-bis-(2-carboxyethyl)-5(6)-carboxy fluorescein (BCECF), which was loaded into vacuoles of intact roots using its membrane-permeant acetoxymethyl (AM) ester. pH values were calculated from fluorescence ratios of confocal images using an in situ calibration curve (Fig. 8C). Figure 8F shows the representative ratio images of the BCECF-stained vacuolar fluorescence in mature roots of wild-type and kea4 kea5 kea6 seedlings. The results showed that, for the root mature region cells, the vacuolar pH was reduced in the kea4 kea5 kea6 mutant (pH 5.66) compared with the wild type (pH 5.92; Fig. 8D). Therefore, both pHluorin and BCECF assays showed the same trends of luminal pH reduction in the triple mutant. It should be noted that the vacuolar pH measured with BCECF was slightly higher than with pHluorin. This difference may be due to either different methods (pHluorin versus BCECF) or different materials (leaf protoplasts versus root cells) or both. In addition, the pH was restored to the wild-type level in kea4 kea5 kea6/KEA4, kea4 kea5 kea6/KEA5, and kea4 kea5 kea6/KEA6 complementation lines. These results suggest that KEA4, KEA5, and KEA6 function to release protons, presumably by K+/H+ exchange, to regulate the pH in the Golgi, TGN, PVC/MVB, and vacuole.
Distinct and Overlapping Roles of KEA and NHX Transporters
KEA proteins belong to the CPA2 family. Evolutionarily distant K+,Na+/H+ antiporters of the NHX group within the CPA1 family also are localized to intracellular membranes. In Arabidopsis, proteins NHX1 to NHX4 localize to the tonoplast (Barragán et al., 2012; McCubbin et al., 2014), whereas NHX5 and NHX6 are retained in Golgi and TGN (Bassil et al., 2011a). Similar to the kea4 kea5 kea6 mutant, the nhx5 nhx6 double mutant had more acidic TGN and PVC compartments compared with the wild type and was salt sensitive (Reguera et al., 2015). However, it remains unclear whether sensitivity results from defective sorting of ion transporters, perturbed recycling of membranes and embedded proteins (Bassil et al., 2011a), or even the vesicular transport of salts as cargo by a process similar to pinocytosis, since salinity-induced endocytosis correlated with the development of larger vacuoles in Arabidopsis roots and cell cultures (Hamaji et al., 2009; Baral et al., 2015a, 2015b). To address these questions and to better understand the specific roles of endosomal KEA proteins in salt tolerance, we examined the interactions between KEA4, KEA5, and KEA6 with the endosomal antiporters NHX5 and NHX6 and with the vacuolar antiporters NHX1 and NHX2 that have been shown to impart salt tolerance through the accumulation of alkali cations inside the vacuole (Leidi et al., 2010; Barragán et al., 2012).
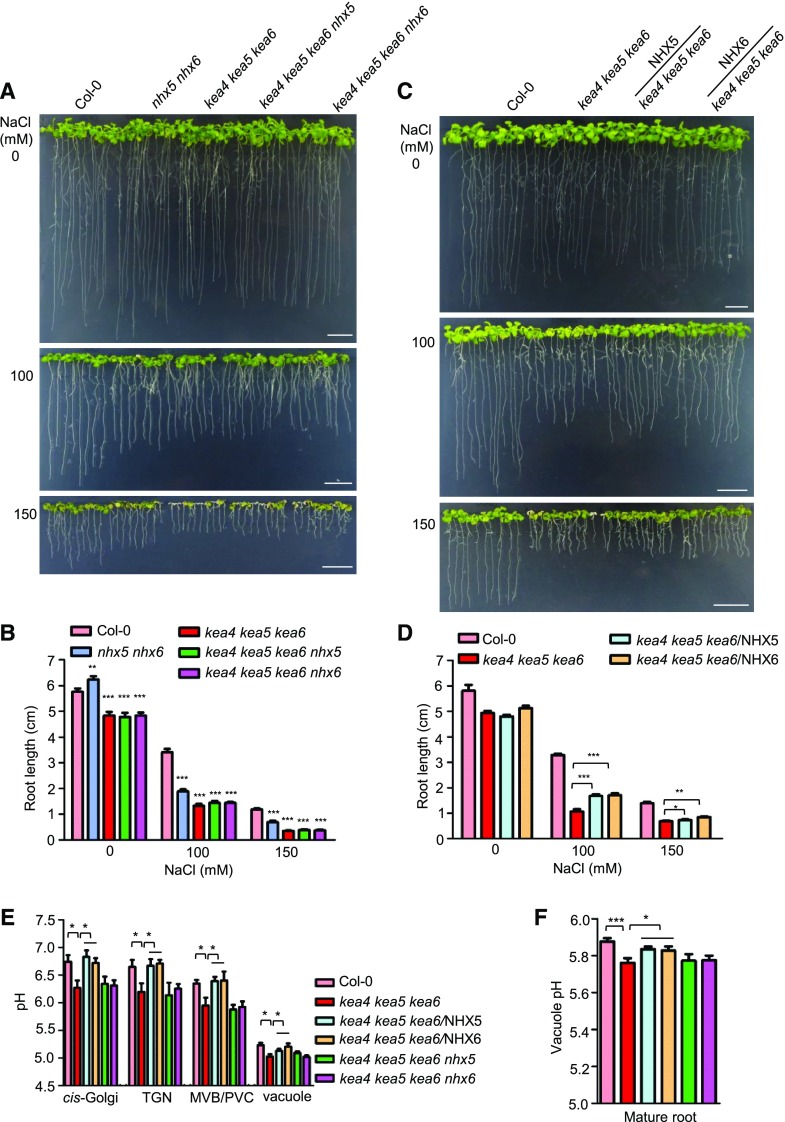
Since KEA4, KEA5, KEA6, NHX5, and NHX6 are localized to Golgi, TGN, and PVC/MVB, we first addressed the question of whether these two types of transporters function coordinately in maintaining pH homeostasis in endosomal compartments and to convey salt tolerance. We produced the quadruple mutants kea4 kea5 kea6 nhx5 and kea4 kea5 kea6 nhx6 and the quintuple mutant kea4 kea5 kea6 nhx5 nhx6 by genetic crossing. The quintuple mutant displayed severe defects in development and produced no seeds (Supplemental Fig. S9), suggesting complementary and critical roles of the endosomal KEA and NHX antiporters in growth and development. Therefore, the quadruple mutants were used to perform further analyses. As shown in Figure 9, A and B, the quadruple mutants grew like the triple kea4 kea5 kea6 mutant on medium with or without NaCl stress. This similar growth phenotype between the quadruple mutants and the triple mutants is likely caused by the functional redundancy between NHX5 and NHX6 (Bassil et al., 2011a). We also generated the suppressor lines kea4 kea5 kea6/NHX5 and kea4 kea5 kea6/NHX6 by expressing the NHX5 and NHX6 genes in the triple mutant kea4 kea5 kea6. There was no obvious difference between the overexpression lines and triple mutants without salt stress (Fig. 9, C and D). However, under salt stress, the overexpression lines grew better than the triple mutant: the root length of the kea4 kea5 kea6/NHX5 and kea4 kea5 kea6/NHX6 lines was increased by 58% and 60% at 100 mm NaCl and by 9% and 19% at 150 mm NaCl, respectively, compared with the triple mutant (Fig. 9, C and D). In addition, at 150 mm NaCl, the kea4 kea5 kea6 mutant displayed growth arrest and strong leaf paleness that were suppressed by the overexpression of NHX5 or NHX6 (Fig. 9C). These results indicate that the endosomal antiporters KEA4, KEA5, KEA6, NHX5, and NHX6 act coordinately in plant salt tolerance.
Figure 9.
Interactions between the endosomal KEA and NHX antiporters. A, The quadruple mutants kea4 kea5 kea6 nhx5 and kea4 kea5 kea6 nhx6 were grown on 1/2 MS medium for 4 d, and then seedlings were transferred to 1/2 MS medium containing 0, 100, or 150 mm NaCl. Photographs were taken at 10 d under salt stress. Bars = 1 cm. B, Root length for seedlings from A (mean ± se; n = 8 seedlings). C, The overexpression lines of kea4 kea5 kea6/NHX5 and kea4 kea5 kea6/NHX6 were grown on 1/2 MS medium for 4 d, and then seedlings were transferred to 1/2 MS medium containing 0, 100, or 150 mm NaCl. Photographs were taken at 10 d under salt stress. Bars = 1 cm. D, Root length for seedlings from C (mean ± se; n = 8 seedlings). E, The luminal pH of the Golgi, TGN, PVC/MVB, and vacuole in the quadruple mutants and overexpression lines was determined using organelle-specific pHluorin-based pH sensors (mean ± se; n ≥ 20 protoplasts). The method was the same as in Figure 8. F, Vacuolar pH of the mature root measured by BCECF-AM (mean ± se; n = 10 seedlings). The method was the same as in Figure 8. In B and D to F, statistics by Student’s t test are shown: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
We also determined the pH of the endosomal compartments of the quadruple mutants kea4 kea5 kea6 nhx5 and kea4 kea5 kea6 nhx6 and the overexpression lines kea4 kea5 kea6/NHX5 and kea4 kea5 kea6/NHX6 using the pHluorin-based pH sensors and the pH indicator BCECF as described in Figure 8. Compared with the triple kea4 kea5 kea6 mutant, the pH in the Golgi, TGN, PVC/MVB, and vacuole was not affected in the quadruple mutants kea4 kea5 kea6 nhx5 and kea4 kea5 kea6 nhx6 (Fig. 9E). However, overexpression of NHX5 or NHX6 in the triple mutant increased the pH in the Golgi, TGN, PVC/MVB, and vacuole compared with the triple mutant kea4 kea5 kea6 (Fig. 9E). Measurements of vacuolar pH with BCECF produced similar results. No significant difference in vacuole pH was observed between the triple mutant kea4 kea5 kea6 (pH 5.76) and the quadruple mutants kea4 kea5 kea6 nhx5 (pH 5.77) and kea4 kea5 kea6 (pH 5.77; Fig. 9F). The overexpression lines kea4 kea5 kea6/NHX5 (pH 5.83) and kea4 kea5 kea6/NHX6 (pH 5.82) had increased pH compared with the kea4 kea5 kea6 mutant, recovering a vacuolar pH similar to the wild type (pH 5.87; Fig. 9F). These results suggest that KEA4, KEA5, KEA6, NHX5, and NHX6 work together to maintain the endosomal pH homeostasis.
Role of the Endomembrane Compartment in Salt Accumulation
Although NHX1 and NHX2 are ultimately trafficked to the tonoplast, AtNHX1 was shown to complement the protein-sorting phenotype of the single-copy gene mutant nhx1 of the yeast Saccharomyces cerevisiae while in transit along the endomembrane system (Hernández et al., 2009). Hence, altering the expression of NHX1 and NHX2 in the kea4 kea5 kea6 triple mutant background could facilitate the assessment of functional interactions between KEA and NHX antiporters along the entire endomembrane system and in vacuolar salt sequestration. If the delivery of salts via the endosomal system to the vacuole was compromised in the kea4 kea5 kea6 mutant plants coincidentally with reduced vacuolar salt sequestration by the removal of NHX1 and NHX2, a more severe growth impairment should be expected. Conversely, enhancing vacuolar salt sequestration by the overexpression of NHX1 and NHX2 could recover the growth of the kea4 kea5 kea6 mutant in salt.
We first tested whether KEA proteins could complement the deletion of the single-copy nhx1 gene of S. cerevisiae. The nhx1 mutation renders yeast cells sensitive to the cationic drug hygromycin B, which is suppressed by Arabidopsis NHX1, NHX2, and NHX5 family members (Yokoi et al., 2002). Neither the full-length KEA proteins nor truncated ∆KEA4 and ∆KEA6 restored resistance to hygromycin, but ∆KEA5 partly suppressed the nhx1 phenotype (Supplemental Fig. S10A). The yeast NHX1 protein is localized primarily to the PVC (Nass and Rao, 1998; Hernández et al., 2009). Inspection of the subcellular localization of GFP-fused KEA proteins showed that all full-length and N-terminal truncated KEA proteins except ∆KEA5 were retained in the perinuclear ER. Only ∆KEA5 could progress successfully to the PVC, visualized as one to two late endosomal compartments per cell adjacent to the central vacuole (Supplemental Fig. S10B). These results indicate that KEA proteins can functionally substitute for the K/H antiport activity of NHXs as long as the protein locates to the proper subcellular compartment.
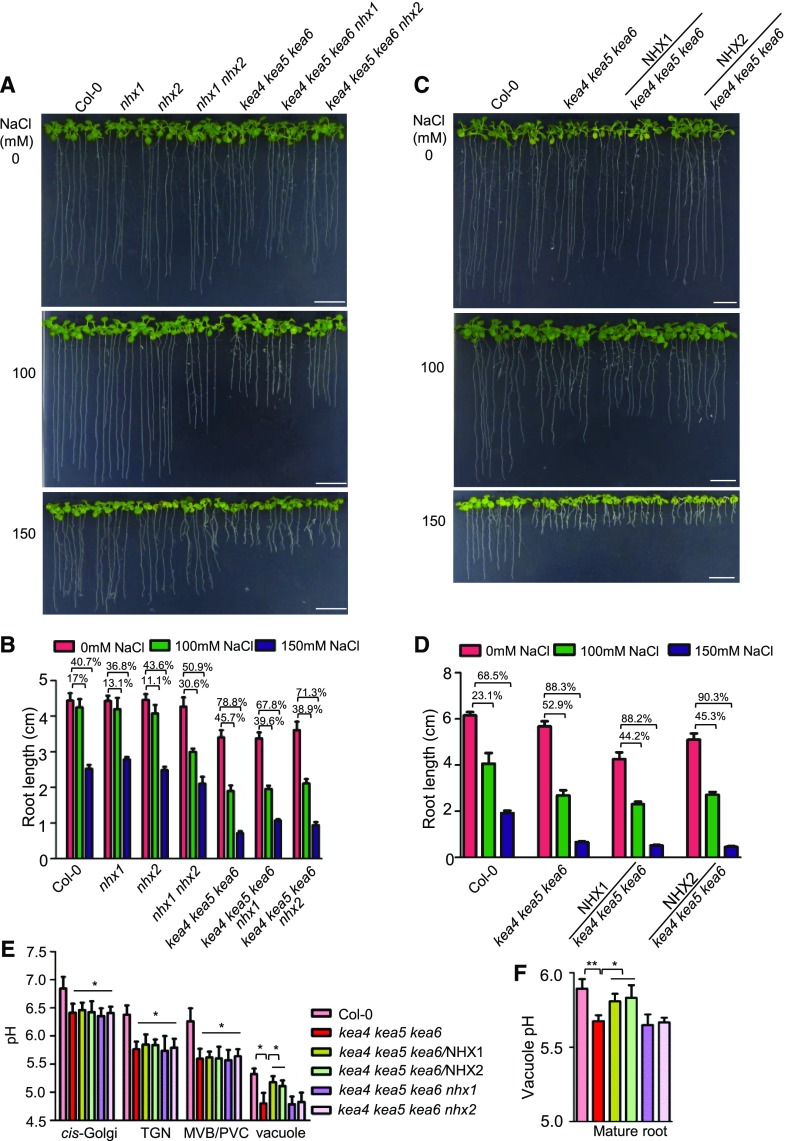
Next, we generated two quadruple mutants, kea4 kea5 kea6 nhx1 and kea4 kea5 kea6 nhx2, by genetic crossing. As shown in Figure 10, A and B, the growth of the quadruple mutants was similar to that of the triple mutants on medium without salt. Unexpectedly, the quadruple mutants grew better than the triple mutants under salt stress. We then expressed the NHX1 and NHX2 genes in the triple mutant kea4 kea5 kea6 to generate the overexpression lines kea4 kea5 kea6/NHX1 and kea4 kea5 kea6/NHX2. As shown in Figure 10, C and D, overexpression of NHX1 or NHX2 did not increase salt tolerance significantly. These results suggest that the endomembrane compartments and the vacuole function independently in enabling salt tolerance and that the endosomal compartments do not pass their salt into the vacuole.
Figure 10.
Interactions between the endosomal and vacuolar antiporters. A, The quadruple mutants. The quadruple mutants kea4 kea5 kea6 nhx1 and kea4 kea5 kea6 nhx2 were grown on 1/2 MS medium for 4 d, then the seedlings were transferred to 1/2 MS medium containing 0, 100, and 150 mm NaCl. Photographs were taken at 10 d under salt stress. Bars = 1 cm. B, Inhibition of root growth for seedlings from A (mean ± se; n = 8 seedlings). C, Overexpression lines. The overexpression lines kea4 kea5 kea6/NHX1 and kea4 kea5 kea6/NHX2 were grown on 1/2 MS medium for 4 d, then the seedlings were transferred to 1/2 MS medium containing 0, 100, and 150 mm NaCl. Photographs were taken at 10 d under salt stress. Bars = 1 cm. D, Inhibition of root growth for seedlings from C (mean ± se; n = 8 seedlings). E, pH measurement. pH of the Golgi, TGN, PVC/MVB, and vacuole in quadruple mutants and overexpression lines was determined (mean ± se; n ≥ 20 protoplasts). The method was the same as in Figure 8. F, Vacuolar pH of the mature root measured by BCECF-AM (mean ± se; n = 10 seedlings). The method was the same as in Figure 8. In E and F, statistics by Student’s t test are shown: *, P < 0.05 and **, P < 0.01.
We also determined the pH of the endosomal compartments of the quadruple mutants kea4 kea5 kea6 nhx1 and kea4 kea5 kea6 nhx2 and of the overexpression lines kea4 kea5 kea6/NHX1 and kea4 kea5 kea6/NHX2 using the pHluorin-based pH sensors and the fluorescein-based ratiometric pH indicator BCECF as described in Figure 8. As shown in Figure 10E, the kea4 kea5 kea6 mutant had a significantly reduced pH in the endomembrane compartments compared with the wild type, which is consistent with the independent measurements given in Figure 8. Compared with the triple kea4 kea5 kea6 mutant, the pH in the Golgi, TGN, PVC/MVB, and vacuole was not affected in the quadruple mutants kea4 kea5 kea6 nhx1 and kea4 kea5 kea6 nhx2 (Fig. 10E). Overexpression of NHX1 or NHX2 did not change the pH in Golgi, TGN, and PVC/MVB compared with the triple mutant kea4 kea5 kea6 (Fig. 10E). However, the overexpression lines kea4 kea5 kea6/NHX1 and kea4 kea5 kea6/NHX2 had increased pH in the vacuole (pH 5.1 and pH 5, respectively) compared with the kea4 kea5 kea6 mutant (pH 4.8), thus restoring a vacuolar pH similar to the wild type (pH 5.3). BCECF measurements of the vacuolar pH produced similar results to the pHluorin method. No significant difference was observed between the triple mutant kea4 kea5 kea6 (pH 5.67) and the quadruple mutants kea4 kea5 kea6 nhx1 (pH 5.65) and kea4 kea5 kea6 nhx2 (pH 5.66; Fig. 10F), whereas the overexpression lines kea4 kea5 kea6/NHX1 (pH 5.81) and kea4 kea5 kea6/NHX2 (pH 5.83) increased the luminal pH relative to the kea4 kea5 kea6 mutant, recovering the vacuolar pH to wild-type levels (pH 5.89; Fig. 10F). These results suggest that the NHX1 and NHX2 antiporters do not affect the pH of the endosomal compartments while in transit but increase vacuolar pH at their destination.
DISCUSSION
Arabidopsis KEA4, KEA5, and KEA6 Are Endosomal K+/H+ Antiporters That Mediate Ion and pH Homeostasis in Endomembrane Compartments
We have studied the gene expression, subcellular localization, and function of the KEA4, KEA5, and KEA6 proteins from Arabidopsis and found that, in stark contrast to other members of the KEA family that are imported to the chloroplast, the KEA4, KEA5, and KEA6 group is localized to the Golgi, TGN, and PVC/MVB (Fig. 7; Supplemental Fig. S8), indicating a function in the endomembrane compartments. The kea4 kea5 kea6 mutants had small rosettes and short seedlings, pointing to an important role of KEA4, KEA5, and KEA6 in plant growth and development (Fig. 1). We further found that kea4 kea5 kea6 mutants were sensitive to low K+ (Fig. 4), high Na+ (Figs. 5 and 6; Supplemental Fig. S6), and high Li+ (Supplemental Fig. S7) and that they accumulated more Na+ and less K+ under salt stress (Fig. 5, E and F), suggesting also a critical role in K+ nutrition and in salt tolerance. Our previous study showed that the Arabidopsis KEA genes conferred resistance to high K+ but not to Na+ and Li+ stress in a yeast expression system (Zheng et al., 2013), and here we have confirmed that proteins of the KEA4, KEA5, and KEA6 group mediated specific K+ transport but not Na+ transport in bacteria (Fig. 3). We also showed that the truncated ∆KEA5 protein reaches the PVC, where it successfully complemented the deficiency of the K+/H+ exchanger ScNHX1. Importantly, we found that the kea4 kea5 kea6 mutant had a significantly reduced pH in the Golgi, TGN, PVC, and vacuole (Figs. 8 and 9) and that the expression of the endosomal K+/H+ exchangers NHX5 and NHX6 restored the pH of endosomal compartments and salt tolerance. These data suggest that the effect of the kea4 kea5 kea6 mutations in the salt tolerance of Arabidopsis is a consequence of disturbed cellular K+ homeostasis and the malfunction of the endomembrane system due to altered luminal pH.
The KEA4, KEA5, and KEA6 proteins are predicted to be K+/H+ antiporters based on phylogenetic analysis (Mäser et al., 2001), and our data corroborated K+-specific transport. The more acidic luminal pH in the endomembrane system of the kea4 kea5 kea6 mutant (Fig. 8) revealed in this study strongly suggests a function of KEA4, KEA5, and KEA6 in luminal H+ efflux in exchange for cytosolic K+. The endosomal cation/proton exchangers NHX5 and NHX6 also are H+ shunts that help to alkalinize the Golgi and TGN compartments (Bassil et al., 2011a), while the tonoplast NHX1 and NHX2 proteins have been shown to catalyze K+/H+ exchange in vivo and to contribute the vacuolar alkalinization by extruding protons (Barragán et al., 2012; Andrés et al., 2014). Thus, the complementation of the kea4 kea5 kea6 mutant by NHX5 and NHX6 suggests that these H+ exchangers act coordinately in maintaining endosomal pH (Fig. 9). However, the lack of complementation of the kea4 kea5 kea6 mutant by NHX1 and NHX2 while in transit through the endomembrane system suggests that KEA4, KEA5, KEA6, NHX5, and NHX6 proteins fulfill specific roles in the operation of the endomembrane system that cannot be substituted by other K+/H+ exchangers (Fig. 10). The additive nature of kea4 kea5 kea6 and nhx5 nhx6 mutations also indicate that endosomal KEA and NHX proteins play partly overlapping or additive roles. Although RFP-tagged KEA4, KEA5, and KEA6 proteins were not visualized in the tonoplast, the vacuolar lumen of the kea4 kea5 kea6 mutant was abnormally acidic (Fig. 10), perhaps as a result of the continuous fusion of acidotic endosomal vesicles. However, overexpression of NHX1 or NHX2 restored near wild-type pH values in the vacuolar compartment. Overall, our results demonstrate that KEA4, KEA5, and KEA6 are endosomal ion transporters that mediate ion and pH homeostasis in the plant endomembrane compartments.
KEA and NHX Antiporters Function to Mediate Salt Tolerance
It has been well established that the vacuolar and plasma membrane Na+,K+/H+ antiporters of the NHX family play important roles in plant salt tolerance (Bassil and Blumwald, 2014). The vacuolar (NHX1–NHX4) and plasma membrane (NHX7/SALT OVERLY SENSITIVE1) Na+,K+/H+ exchangers act together to maintain low levels of Na+ in the cytoplasm (Apse et al., 1999; Shi et al., 2000; Zhang and Blumwald, 2001; Zhang et al., 2001; Qiu et al., 2002, 2003). Recently, the regulation of cellular Na+ homeostasis by NHX5 and NHX6, the endosomal Na+,K+/H+ antiporters in Arabidopsis, also was reported (Bassil et al., 2011a; Wang et al., 2015; Qiu, 2016a). The growth of nhx5 nhx6 seedlings was sensitive to salt stress, but the underlying mechanism could not be resolved (Bassil et al., 2011a). In this study, we demonstrate that KEA4, KEA5, and KEA6, which likely act as endosomal K+/H+ antiporters, also are essential to plant salt tolerance (Figs. 5 and 6; Supplemental Fig. S6). Since both the KEA4, KEA5, and KEA6 and NHX5 and NHX6 groups are endosomal antiporters, we examined genetically whether these two types of transporters act coordinately in facilitating pH homeostasis in endosomal compartments. We found that the quintuple mutant kea4 kea5 kea6 nhx5 nhx6 was retarded significantly in growth and bore no seeds (Supplemental Fig. S9), suggesting additive and essential roles of the endosomal KEA and NHX antiporters in plant development. In addition, we found that overexpression of NHX5 or NHX6 enhanced the salt tolerance (Fig. 9, C and D) and recovered the pH of endosomal compartments in kea4 kea5 kea6 mutants to wild-type levels (Fig. 9, E and F). Together, these results suggest that KEA4, KEA5, KEA6, NHX5, and NHX6 work coordinately to maintain the salt and pH homeostasis in the endosomal compartments. It is intriguing that plant endosomes demand the coordinated function of a large array of K+/H+ exchangers, including at least KEA4, KEA5, KEA6, NHX5, NHX6, and several cation/H+ exchangers (Sze and Chanroj, 2018). These transporters may be regulated differently and have distinct substrate affinities, pH dependence, and electrogenicity, and their coordinated activity could ensure that a steady-state pH at each endosomal compartment is maintained by balancing the rates of H+ pumping and H+ leakage (Sze and Chanroj, 2018). In addition to pH homeostasis, K+/H+ antiporters load K+ into endomembrane vesicles to support cation-dependent activities.
The Endomembrane Compartment as a Target of Salt Toxicity
Since the endomembrane compartments are the key players in vesicle trafficking along the endocytic and secretory pathways, the increased salt sensitivity of the endosomal antiporter mutants may reflect the impairment in protein sorting to various cell membranes, thus affecting both regulatory and effector proteins important in the salt stress response (Krebs et al., 2010; Schumacher, 2014; Baral et al., 2015b; Sze and Chanroj, 2018). The alternative mechanism, that the endomembrane compartments are sodium-storing organelles that accumulate salt (Schumacher, 2014; Baral et al., 2015b), remains to be illustrated. Moreover, if this were the case, then the question becomes the fate of the salt incorporated into the endomembrane compartments. Will the salt pass on to the vacuole or will it stay in the endosomes? Endocytosis is greatly stimulated under salinity stress (Golani et al., 2013; Baral et al., 2015a, 2015b), and proper functioning of the subsequent endosomal trafficking is indispensable for salt tolerance (Ebine et al., 2011). However, although the uptake of membrane-impermeant compounds by fluid endocytosis and delivery to vacuoles has been reported (Hamaji et al., 2009; Etxeberria et al., 2012), the hypothetical shuttling of apoplastic salts to the vacuolar lumen via endocytic vesicles has not been substantiated experimentally. To tackle this question, we investigated the interactions between the endomembrane compartments and the vacuole by the analysis of genetic interactions between KEA4, KEA5, KEA6, NHX1, and NHX2 proteins. Our results showed that overexpression of the vacuolar antiporters NHX1 and NHX2 could not recover the growth of the kea4 kea5 kea6 triple mutants (Fig. 10C), indicating that any enhancement in Na+ accumulation in the vacuole did not improve tolerance to salt stress. In addition, the growth of the quadruple mutants kea4 kea5 kea6 nhx1 and kea4 kea5 kea6 nhx2 was improved under salt stress instead of worsening (Fig. 10A), suggesting that the endomembrane compartments and the vacuole are independent in salt accumulation and that the endomembrane compartments may not pass their salt into the vacuole. This relative independence between the endosomal compartments and the vacuole was further confirmed by the pH analysis. The pH assay indicated that neither the quadruple mutants nor the overexpression lines kea4 kea5 kea6/NHX1 and kea4 kea5 kea6/NHX2 showed any changes in pH in their endosomal compartments relative to the triple kea4 kea5 kea6 mutant (Fig. 10E). Moreover, the expression of NHX1 mutant proteins with enhanced K+ selectivity over Na+ imparted salt tolerance to the yeast ∆nhx1 mutant, demonstrating that the endomembrane system is a target of Na+ toxicity rather than an ion storage compartment (Hernández et al., 2009). These results, together with the observation that the kea4 kea5 kea6 mutant plants had greater Na+ and lower K+ contents than the wild type, indicate that the salt accumulated in the endomembrane compartment would make this cellular system vulnerable to Na+ toxicity, as suggested previously (Hernández et al., 2009). In wild-type Arabidopsis, the KEA4, KEA5, and KEA6 proteins, by favoring the transport of K+ over Na+ (Fig. 3), would alleviate the uptake of Na+ and prevent salt-induced toxicity.
The fact that the growth of the quadruple mutants kea4 kea5 kea6 nhx1 and kea4 kea5 kea6 nhx2 was improved under salt stress may reflect the importance of cytosolic K+ in preventing Na+ toxicity in the endosomal system, since NHX1 and NHX2 have been reported to mediate H+-linked K+ transport into the vacuole in plants and the nhx1 nhx2 mutant of Arabidopsis had a greater concentration of cytosolic K+ due to impaired vacuolar import (Barragán et al., 2012). Therefore, knocking out the activity of NHX1 or NHX2 would restrict K+ movement into the vacuole and, thus, could increase the levels of K+ in the cytoplasm, making it more available to uptake into the endomembrane compartments by transporters other than KEA4, KEA5, and KEA6, for instance NHX5 and NHX6 or endosomal members of the large cation/H+ exchanger family (Bassil et al., 2011a; Chanroj et al., 2011, 2012). Collectively, these results indicate that the endomembrane compartments may be salt-accumulating organelles prone to suffer Na+ toxicity and that maintaining K+ homeostasis is critical for their function.
Overall, our results suggest that the endosomal proteins KEA4, KEA5, and KEA6 are K+/H+ exchangers critical to plant development and salt tolerance. Further studies are needed to understand the precise and specific physiological functions of the various types of endosomal K+ transporters as well as the molecular mechanism governing salt entry into the endomembrane compartments in plants.
MATERIALS AND METHODS
Escherichia coli Strains, Plasmids, and Growth Test
For functional expression in E. coli, cDNAs encoding N-terminal deletions of KEA4, KEA5, and KEA6 were amplified by PCR using the following primers: BAD-KEA4F and BAD-KEA4R, BAD-KEA5F and BAD-KEA5R, and BAD-KEA6F and BAD-KEA6R (Supplemental Table S1) and then subcloned into the bacterial plasmid pBAD24, an arabinose-inducible expression vector (Guzman et al., 1995). E. coli strains EP432 (melBLid, ΔnhaA1::kan, ΔnhaB1::cat, ΔlacZY, thr1; Pinner et al., 1993) and MJF276 (F− ΔkdpABC5 thi rha lacZ trkD1 kefB157 kefC::Tn10; Ferguson et al., 1993) were used as the host for the expression of Arabidopsis (Arabidopsis thaliana) KEA genes. For routine growth, bacterial cells were grown in LBK medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 100 mm KCl). For complementation tests of NaCl and KCl tolerance, 4 µL of saturated overnight cultures of EP432 cells in LBK medium transformed with the indicated plasmids was inoculated in 2 mL of LBK medium buffered with 50 mm 1,3 bis-[tris(hydroxymethyl)-methylamino]propane (BTP) at pH 7.5 and supplemented with 0.1 mm arabinose and the indicated KCl and NaCl concentrations. Plates were incubated for 48 h at 25°C. The viability of MJF276 cells, lacking the Kef transporters, to treatment with 0.1 mm NEM was determined as described previously (Ferguson et al., 1997) with the exceptions that K0.2 minimal medium contained glycerol (0.2%, w/t) instead of Glc to avoid the repression of the PBAD promoter and that 0.1 mm arabinose was added before NEM treatment. Recovery of cells was conducted on K10 minimal medium plates supplemented with Glc (0.2%, w/t).
Expression, Complementation, and Subcellular Localization in Yeast Cells
Yeast (Saccharomyces cerevisiae) strain WX1 (∆nhx1::TRP1; Quintero et al., 2000) was used to test the function and the subcellular localization of wild-type and truncated KEA proteins. First, a Gal-inducible yeast expression vector for generating recombinant GFP-tagged proteins was constructed by subcloning a PCR fragment containing the open reading frame of the enhanced GFP (oligonucleotides GFP_XbaIF and GFP_XhoIR; Supplemental Table S1) between the XhoI and XbaI sites of the polylinker of pYES2 (Invitrogen). This new plasmid vector was named pYES2GFP. The cDNAs encoding the full sequence or the N-terminal deletions of KEA4, KEA5, and KEA6 were amplified by PCR using primers YESKEA4F and YESKEA4R, YESKEA5F and YESKEA5R, YESKEA6F and YESKEA6R, ntKEA4F and YESKE4R, ntKEA5F and YESKEA5R, and ntKEA6F and YESKEA6R (Supplemental Table S1). Amplicons were then subcloned into the yeast expression vector pYES2GFP to achieve C-terminal GFP fusions. Yeast transformation was performed using a standard lithium acetate-polyethylene glycol method. Transformed cells were selected and propagated in standard YNB (yeast nitrogen base) minimal medium without uracil. To test growth in the presence of hygromycin B, strains were cultured overnight in YPGal medium (1% yeast extract, 2% peptone, and 2% Gal) and cells were diluted decimally in distilled water. Five-microliter aliquots were spotted onto YPGal plates supplemented with various concentrations of hygromycin B and grown for 3 to 4 d at 28°C. A Leica DM6000 fluorescence microscope was used to analyze the subcellular localization of GFP-fused KEA proteins.
Plant Materials and Growth Conditions
Arabidopsis Col-0, mutants, and transgenic lines were used in this study. In the growth chamber, plants were grown on compost (Pindstrup Substrate) and subirrigated with tap water. Greenhouse conditions were as follows: 16-h-light/8-h-dark cycles, light intensity of 100 μmol m−2 s−1 photosynthetically active radiation, temperature of 22°C, and relative humidity of 50% ± 10%.
For plate-grown plants, Arabidopsis seeds were surface sterilized with 20% (v/v) bleach. After cold treatment at 4°C for 3 d in the dark, the seeds were germinated on plates with 1/2 MS medium containing 1% agar, pH 5.8.
For Na+ and Li+ treatments, 4-d-old seedlings grown on 1/2 MS medium were transferred to 1/2 MS medium supplemented with various concentrations of NaCl or LiCl. Data were recorded 10 d after the transfer.
For growth at low potassium, seeds were plated to a modified MS medium containing various concentrations of KCl (Pandey et al., 2007). The modified MS medium contains K+-free one-twentieth-strength major salts and 1× minor salts (Pandey et al., 2007). The K+-free medium was prepared by replacing MS salts with the modified MS salts as described by Pandey et al. (2007). Varying levels of K+ in the medium were achieved by adding appropriate amounts of KCl to the K+-free medium. For the solidification of MS medium, 1% ultra-pure agarose was used (Pandey et al., 2007).
For flowering time measurements, the day of the inflorescence opening from the first flower per plant was scored (Lazaro et al., 2012).
Salt treatments for the plants grown in soil were performed as described (Qiu et al., 2002; Apse et al., 2003). Briefly, 2-week-old plants grown in soil (60-mL pots) were watered with 100 mL of either distilled water (the control group) or 200 mm NaCl every 3 d to thoroughly wet the soil in the pot and leach 10 to 20 mL per pot. The treatment lasted for 4 weeks.
All plant growth experiments in this study were repeated three times.
Generation of the kea4 kea5 kea6 Triple Mutant and Complementation Assay
T-DNA insertion lines were ordered from the Arabidopsis Biological Resource Center (ABRC). SALK lines used in this work were kea4-1 (SALK_012529C), kea4-3 (SALK_129985), kea5-1 (SALK_140807C), kea5-2 (SALK_082201), kea6-1 (SALK_141501C), and kea6-3 (SALK_137412). Insertion mutant information was obtained from the SIGnAL Web site (http://signal.salk.edu) and confirmed experimentally. Positions of T-DNA insertion sites are shown in Supplemental Figure S1A. Mutant kea4-1 and kea4-3 plants have the T-DNA insertions at nucleotides +815 and +404 relative to the start codon. The T-DNA insertion lines kea5-1 and kea5-2 contain the insertions at nucleotides +515 and +3,309 relative to the start codon, whereas mutants kea6-1 and kea6-3 carry the insertions at nucleotides +1,539 and +3,794. Homozygous mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci.
The double and triple mutant lines were generated from the single mutants by genetic crossing, including kea4-1 kea5-1, kea4-1 kea6-1, kea5-1 kea6-1, kea4-1 kea5-1 kea6-1, kea4-3 kea5-2, kea4-3 kea6-3, kea5-2 kea6-3, and kea4-3 kea5-2 kea6-3. The homozygous mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci.
For complementation assays, the genomic DNA was extracted from rosette leaves using the Plant Genomic DNA Extraction Kit (TaKaRa). The KEA4, KEA5, and KEA6 genes (without stop codons) were amplified by PCR from the genomic DNA using the following primers: GW-KEA4F and GW-KEA4R, GW-KEA5F and GW-KEA5R, and GW-KEA6F and GW-KEA6R (Supplemental Table S2).
The PCR products were cloned into the vector pDONR/Zeo (Invitrogen). The entry vectors (pDONR-gKEA4, pDONR-gKEA5, and pDONR-gKEA6) were recombined into the expression vector pUBC-RFP using Gateway technology (Invitrogen) to produce pUBC-gKEA4-RFP, pUBC-gKEA5-RFP, and pUBC-gKEA6-RFP, which were fused to the N termini of RFP, driven by the UBIQUITIN10 (UBQ10) promoter. These plasmids were transformed into Agrobacterium tumefaciens strain GV3101, and the resulting bacterial clones were used to transform the kea4-1 kea5-1 kea6-1 mutant for complementation assays by the floral dip procedure (Clough and Bent, 1998). The expression constructs were introduced into Arabidopsis Col-0 plants to generate the overexpression plants. Transgenic plants were selected in soil by Basta (0.002%) after 1 week. The homozygous lines of T3 progeny were selected for experiments.
RT-qPCR Analysis of the T-DNA Insertion Lines
Total RNA was extracted using the Plant RNA Extraction Kit (TaKaRa) from the seedlings of Col-0, kea4-1, kea5-1, kea6-1, kea4-3, kea5-2, kea6-3, kea4-1 kea5-1 kea6-1, and kea4-3 kea5-2 kea6-3 after germination for 7 d. One microgram of total RNA of each sample was used to synthesize cDNA using the M-MLV kit (TaKaRa). RT-qPCR was carried out to test the expression level of each allelic gene. The primers used for RT-qPCR were as follows: KEA4-F1 and KEA4-R1, KEA5-F2 and KEA5-R2, KEA5-F3 and KEA5-R3, KEA6-F4 and KEA6-R4, and KEA6-F5 and KEA6-R5. ACTIN2 was amplified as a control by primers Actin2F and Actin2R. The primers are listed in Supplemental Table S3.
RT-qPCR Analysis of Gene Expression
Col-0 seedlings were grown on 1/2 MS plates for 7 d or in soil for 6 weeks. Plant organs or tissues, including leaves, roots, stems, flowers, and siliques, were collected to isolate total RNA using the Plant RNA Extraction Kit (TaKaRa). The first-strand cDNA was synthesized from total RNA (1 µg) using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa) and was used as a template for qPCR amplification. qPCR amplification was performed with the CFX96 system (Bio-Rad) using SYBR Premix Ex Taq (TaKaRa). The Arabidopsis ACTIN7 gene was used as an internal control, and differences in product levels among the tested samples during the linear amplification phase were used to calculate differential gene expression (Mei et al., 2012). The gene-specific primers were as follows: qKEA4F and qKEA4R, qKEA5F and qKEA5R, and qKEA6F and qKEA6R. ACTIN7 was amplified as a control by primers Actin7F and Actin7R. The primers are listed in Supplemental Table S4.
GUS Staining Assay
Genomic DNA was isolated from Arabidopsis Col-0 plants using the Plant Genomic DNA Extraction Kit (TaKaRa). To clone the KEA4 promoter, 2,040 bp upstream of the open reading frame was amplified by PCR with the primers GW-ProKEA4F and GW-ProKEA4R. To clone the KEA5 promoter, 1,545 bp upstream of the open reading frame was amplified by PCR with the primers GW-ProKEA5F and GW-ProKEA5R. For the KEA6 promoter, a 1,500-bp promoter region was amplified by PCR with the primers GW-ProKEA6F and GW-ProKEA6R. The primers are listed in Supplemental Table S5. The resulting fragments were inserted into the vector pDONR/Zeo (Invitrogen). The entry vectors (pDONR-ProKEA4, pDONR-ProKEA5, and pDONR-ProKEA6) were recombined into the expression vector pBIB-GUS using Gateway technology (Invitrogen) to produce pBIB-ProKEA4-GUS, pBIB-ProKEA5-GUS, and pBIB-ProKEA6-GUS. The plasmids were transformed into A. tumefaciens strain GV3101 and then introduced into Col-0 by the floral dip procedure (Clough and Bent, 1998). The transgenic plants were selected in soil by Basta (0.002%) after 1 week. Three homozygous lines of T3 progeny were selected for the GUS staining assay. Primers of the cloned KEA4, KEA5, and KEA6 promoters were used to verify the presence of transgenes in the selected lines. For the histochemical GUS assay, whole young seedlings grown on 1/2 MS plates and various organs of mature plants were used. The plant materials were immersed in GUS staining solution (50 mm sodium phosphate buffer, pH 7, 1 mm EDTA, 0.1% Triton X-100, 2 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 5 mm ferricyanide, and 5 mm ferrocyanide) and incubated overnight at 37°C. Then, the materials were washed in 75% ethanol to clear chlorophyll. Photographs were taken using a Leica stereoscopic zoom microscope (DFC420C).
Subcellular Localization Assay of KEA4, KEA5, and KEA6 in Stably Transformed Arabidopsis Seedlings
For the stable transformation assays with Arabidopsis, the plasmids pUBC-gKEA4-RFP, pUBC-gKEA5-RFP, and pUBC-gKEA6-RFP (RFP tags were fused at the C termini of KEA4, KEA5, and KEA6) were transformed into A. tumefaciens GV3101 and introduced into Col-0 by the floral dip procedure (Clough and Bent, 1998). The transgenic plants were screened by Basta spray, and the Basta-positive seedlings were reconfirmed by fluorescence analysis and PCR amplification.
To generate the double reporter lines coexpressing SYP32, SYP43, or VAMP727 with pUBC-gKEA4-RFP, pUBC-gKEA5-RFP, and pUBC-gKEA6-RFP, the homozygous parents were crossed with SYP32-YFP, GFP-SYP43 (Ebine et al., 2008), or GFP-VAMP727 (Ebine et al., 2008) as pollen donors. The GFP, YFP, and RFP signals were visualized in the roots of F1 seedlings by a Leica TCS SP5 laser scanning confocal microscope system. The GFP excitation wavelength was 488 nm and emission was 500 to 530 nm, the YFP excitation wavelength was 514 nm and emission was 520 to 550 nm, and the RFP excitation wavelength was 561 nm and emission was 595 to 620 nm. Sequential scanning was used for double labeling to avoid cross talk between channels. The SYP32-YFP line (CS781656) was ordered from the ABRC. Colocalization analysis was performed using the FIJI plugin coloc2 (http://fiji.sc/Coloc_2) as described (Bassil et al., 2011a; Reguera et al., 2015).
Subcellular Localization Assay of KEA4, KEA5, and KEA6 in the Epidermal Cells of Nicotiana benthamiana Leaves
Total RNA was extracted from rosette leaves using the Plant RNA Extraction Kit (TaKaRa). The first-strand cDNA was synthesized from total RNA (1 µg) using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa) and was used as a template for PCR amplification. The KEA4, KEA5, and KEA6 genes (without stop codons) were amplified by PCR from the cDNA using the following primers (Supplemental Table S2): GW-KEA4F and GW-KEA4R, GW-KEA5F and GW-KEA5R, and GW-KEA6F and GW-KEA6R.
The PCR products were cloned into the vector pDONR/Zeo (Invitrogen). The entry vectors (pDONR-KEA4, pDONR-KEA5, and pDONR-KEA6) were recombined into the expression vector pBIB-RFP using Gateway technology (Invitrogen) to produce pBIB-KEA4-RFP, pBIB-KEA5-RFP, and pBIB-KEA6-RFP, which were fused to the N termini of RFP, driven by the 35S promoter. These plasmids were transformed into A. tumefaciens strain GV3101. Fully expanded 3-week-old N. benthamiana leaves were infiltrated with the A. tumefaciens containing pBIB-KEAs-RFP and Golgi marker Man49-GFP, TGN marker SYP61-GFP, or PVC/MVB marker ARA6-GFP, which was diluted to an OD600 of 0.6 to 0.8 with the solution (10 mm MES, pH 5.6, 10 mm MgCl2, and 150 μm acetosyringone). After 2 to 4 d, the fluorescence signals of the infiltrated leaves were analyzed with the Leica TCS SP5 confocal microscope. The GFP and excitation wavelength was 488 nm and emission was 500 to 530 nm, and the RFP excitation wavelength was 561 nm and emission was 595 to 620 nm. Sequential scanning was used for double labeling to avoid cross talk between channels. Man49-GFP (CD3-963) was purchased from the ABRC.
Determination of Na+ and K+ Contents
Seedlings of Col-0, kea4-1 kea5-1 kea6-1, kea4-1 kea5-1 kea6-1/KEA4, kea4-1 kea5-1 kea6-1/KEA5, and kea4-1 kea5-1 kea6-1/KEA6 lines were grown on 1/2 MS with or without 100 mm NaCl for 10 d. The seedlings then were collected, washed briefly five times with deionized water, and dried at 80°C for 48 h and weighed. Dried samples were digested with HNO3, and the Na+ concentration was determined with an atomic absorption spectrophotometer (Shi et al., 2002).
Generation of the Quadruple Mutants and Overexpression of NHX1 or NHX2 in the kea4 kea5 kea6 Triple Mutant
The T-DNA insertion line nhx1-2 (SALK_065623) was purchased from the ABRC. The T-DNA insertion site in line nhx1-2 occurred at nucleotide +1,246 relative to the start codon, as described previously (Barragán et al., 2012). The nhx1-2 nhx2-1 double mutant was generated in a previous study (Barragán et al., 2012). The nhx2-1 allele (SALK_036114) has a T-DNA insertion site at nucleotide +1,246 relative to the start codon (Barragán et al., 2012). The quadruple mutant kea4-1 kea5-1 kea6-1 nhx1-2 was generated by crossing kea4-1 kea5-1 kea6-1 with the single mutant nhx1-2 as the pollen donor. The second quadruple mutant line kea4-1 kea5-1 kea6-1 nhx2-1 was generated by crossing kea4-1 kea5-1 kea6-1 with the double mutant nhx1-2 nhx2-1 using the latter as the pollen donor. The homozygous quadruple mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci.
To prepare the overexpression lines, genomic DNA was extracted from rosette leaves using the Plant Genomic DNA Extraction Kit (TaKaRa). The NHX1 and NHX2 genes (without stop codons) were amplified by PCR from the genomic DNA using the following primers: GW-NHX1F and GW-NHX1R and GW-NHX2F and GW-NHX2R. The primers are listed in Supplemental Table S2. The PCR products were cloned into the vector pDONR/Zeo (Invitrogen). The entry vectors (pDONR-gNHX1 and pDONR-gNHX2) were recombined into the expression vector pUBC-RFP using Gateway technology (Invitrogen) to produce pUBC-gNHX1-RFP and pUBC-gNHX2-RFP, which were fused to the N termini of RFP, driven by the UBQ10 promoter. These plasmids were transformed into A. tumefaciens strain GV3101, and the resulting bacterial clones were used to transform the kea4-1 kea5-1 kea6-1 mutant to generate the overexpression lines by the floral dip procedure (Clough and Bent, 1998). Transgenic plants were selected in soil by Basta (0.002%) after 1 week. The homozygous lines of T3 progeny were selected for experiments.
Generation of the Quadruple Mutants kea4 kea5 kea6 nhx5 and kea4 kea5 kea6 nhx6, the Quintuple Mutant kea4 kea5 kea6 nhx5 nhx6, and Overexpression of NHX5 or NHX6 in the kea4 kea5 kea6 Mutant
The T-DNA insertion lines nhx5-1 (Wisc-DsLox345-348M8) and nhx6-1 (SALK_113129C) were purchased from the ABRC. The T-DNA insertion site in lines nhx5-1 and nhx6-1 occurred at nucleotides +1,504 and +3,809 relative to the start codon, respectively (Wang et al., 2015). The homozygous mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci (Wang et al., 2015). The quadruple mutants kea4-1 kea5-1 kea6-1 nhx5-1 and kea4-1 kea5-1 kea6-1 nhx6-1 were generated by crossing kea4-1 kea5-1 kea6-1 with nhx5-1 or nhx6-1. The nhx5-1 nhx6-1 double mutant was generated in a previous study (Wang et al., 2015). The quintuple mutant kea4-1 kea5-1 kea6-1 nhx5-1 nhx6-1 was generated by crossing the triple mutant kea4-1 kea5-1 kea6-1 with the double mutant nhx5-1 nhx6-1. The homozygous quadruple and quintuple mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci.
To prepare the overexpression lines, the plasmids pUBC-gNHX5-RFP and pUBC-gNHX6-RFP, fused to the N termini of RFP and driven by the UBQ10 promoter, were transformed into A. tumefaciens strain GV3101 (Fan et al., 2018). The resulting bacterial clones were used to transform kea4-1 kea5-1 kea6-1 to generate the overexpression lines by the floral dip procedure (Clough and Bent, 1998). Transgenic plants were selected in soil by Basta (0.002%) 1 week after sowing. The homozygous lines of T3 progeny were selected for experiments.
pH Measurement with pHluorin
To measure the luminal pH of the Golgi, TGN, PVC, and vacuole, the pH probes Man1-PRpHluorin, PRpHluorin-BP80 (Y612A), PRpHluorin-AtVSR2, and aleurain-PRpHluorin, respectively, were transiently expressed in the Arabidopsis protoplasts (Yoo et al., 2007; Shen et al., 2013). The protoplasts were prepared from 3-week-old seedlings of Col-0, kea4-1 kea5-1 kea6-1, kea4-1 kea5-1 kea6-1/KEA4, kea4-1 kea5-1 kea6-1/KEA5, kea4-1 kea5-1 kea6-1/KEA6, kea4 kea5 kea6 nhx1, kea4 kea5 kea6 nhx2, kea4 kea5 kea6/NHX1, and kea4 kea5 kea6/NHX2. The signals of the probes at an emission wavelength of 500 to 530 nm were recorded with dual-excitation wavelengths at 405 and 488 nm, respectively, and obtained a ratio of 405:488 nm to calculate the pH using the calibration curve. In vivo calibration was achieved from the same protoplasts expressing the PRpHluorins for pH measurement. To take the fluorescence image, protoplasts were incubated for 5 min in WI protoplast buffer (0.5 m mannitol and 20 mm KCl; Yoo et al., 2007) with 25 μm nigericin, 60 mm KCl, and 10 mm MES/HEPES Bis-Tris-propane, adjusted to various pH values ranging from 5 to 8 for each calibration point (Martinière et al., 2013; Reguera et al., 2015). Fluorescence images were acquired using the TCS SP5 laser scanning confocal microscope system equipped with a 20× objective by the sequential line scanning mode. The pH profile of the protoplast was indicated by pseudocolored images.
Vacuolar pH Measurement with BCECF-AM
The pH-sensitive fluorescent dye BCECF-AM was used to measure the vacuolar pH in root cells (Bassil et al., 2011b; Wang et al., 2015). Four-day-old seedlings grown on vertical plates were collected and incubated in liquid medium containing one-tenth-strength MS medium, 0.5% Suc, 10 mm MES (pH 5.8), 10 μm BCECF-AM, and 0.02% pluronic F-127 (Molecular Probes) for 1 h at 22°C in darkness. The seedlings were washed once for 10 min before microscopy. Dye fluorescence images were obtained using a Leica confocal SP8 microscope. The fluorophore was excited at 458 and 488 nm, and single emission between 530 and 550 nm was detected for all images. Mature root cells were collected for the images. After background correction, the integrated pixel intensity was measured for both the 458- and 488-nm excited images. The ratio values were used to calculate the pH based on the calibration curve using ImageJ software. For in situ pH calibration, 4-d-old seedlings were incubated for 15 min in pH equilibration buffer containing 50 mm MES-BTP (pH 5–6.5) or 50 mm HEPES-BTP (pH 7–7.5) and 50 mm ammonium acetate (Yoshida, 1994; Swanson and Jones, 1996). The average ratio values were determined from 10 individual seedlings.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: KEA4 (At2g19600), KEA5 (At5g51710), KEA6 (At5g11800), NHX1 (At5g27150), NHX2 (At3g05030), NHX5 (At1g54370), and NHX6 (At1g79610).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. T-DNA insertion mutants of the KEA4, KEA5, and KEA6 genes.
Supplemental Figure S2. Growth phenotypes at different stages in soil.
Supplemental Figure S3. The triple mutant kea4 kea5 kea6 flowers early.
Supplemental Figure S4. RT-qPCR assay of the organ-specific expression of KEA4, KEA5, and KEA6 in Arabidopsis.
Supplemental Figure S5. Sequence alignment of KEA4, KEA5, and KEA6 to bacterial cation antiporters of the CPA2 family.
Supplemental Figure S6. Seed germination of the kea4 kea5 kea6 mutant is sensitive to salt stress.
Supplemental Figure S7. KEA4, KEA5, and KEA6 are critical for Li+ homeostasis.
Supplemental Figure S8. Transient coexpression of KEA4, KEA5, and KEA6 with the endosomal markers in epidermal cells of the N. benthamiana leaf.
Supplemental Figure S9. The quintuple mutant kea4 kea5 kea6 nhx5 nhx6 is defective in growth and does not generate seeds.
Supplemental Figure S10. Complementation of the S. cerevisiae nhx1 mutant by N-terminal truncated KEA4, KEA5, and KEA6.
Supplemental Table S1. Primers for expression in E. coli and yeast.
Supplemental Table S2. Primers for cloning genes.
Supplemental Table S3. Primers for RT-PCR.
Supplemental Table S4. Primers for RT-qPCR.
Supplemental Table S5. Primers for GUS staining.
Supplemental Movie S1. Time-lapse movie indicating the trafficking of the KEA4-positive bodies in root cells.
Supplemental Movie S2. Time-lapse movie indicating the trafficking of the KEA5-positive bodies in root cells.
Supplemental Movie S3. Time-lapse movie indicating the trafficking of the KEA6-positive bodies in root cells.
Acknowledgments
We thank Dr. Takashi Ueda for GFP-SYP43/syp43 and GFP-VAMP727/vamp727 lines, Dr. Qingqiu Gong for SYP61-GFP and ARA6-GFP vectors, and Dr. Samantha Miller for the bacterial strain MJF276.
Footnotes
This work was supported by the National Natural Science Foundation of China (31571464, 31371438, and 31070222 to Q.-S.Q.), the National Basic Research Program of China (973 project, 2013CB429904 to Q.-S.Q.), and the Research Fund for the Doctoral Program of Higher Education of China (20130211110001 to Q.-S.Q.). F.J.Q. and J.M.P. were supported by grants from the Spanish Ministry MINECO cofinanced by the European Regional Development Fund (BIO2015-70946-R and BFU2015-64671-R) and by the Rural Development Administration, Republic of Korea, SSAC (PJ01318205).
References
- Andrés Z, Pérez-Hormaeche J, Leidi EO, Schlücking K, Steinhorst L, McLachlan DH, Schumacher K, Hetherington AM, Kudla J, Cubero B, et al. (2014) Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc Natl Acad Sci USA 111: E1806–E1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36: 229–239 [DOI] [PubMed] [Google Scholar]
- Aranda-Sicilia MN, Cagnac O, Chanroj S, Sze H, Rodríguez-Rosales MP, Venema K (2012) Arabidopsis KEA2, a homolog of bacterial KefC, encodes a K+/H+ antiporter with a chloroplast transit peptide. Biochim Biophys Acta 1818: 2362–2371 [DOI] [PubMed] [Google Scholar]
- Aranda-Sicilia MN, Aboukila A, Armbruster U, Cagnac O, Schumann T, Kunz HH, Jahns P, Rodríguez-Rosales MP, Sze H, Venema K (2016) Envelope K+/H+ antiporters AtKEA1 and AtKEA2 function in plastid development. Plant Physiol 172: 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Carrillo LR, Venema K, Pavlovic L, Schmidtmann E, Kornfeld A, Jahns P, Berry JA, Kramer DM, Jonikas MC (2014) Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat Commun 5: 5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Avni A (2009) EHD2 inhibits ligand-induced endocytosis and signaling of the leucine-rich repeat receptor-like protein LeEix2. Plant J 59: 600–611 [DOI] [PubMed] [Google Scholar]
- Baral A, Irani NG, Fujimoto M, Nakano A, Mayor S, Mathew MK (2015a) Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 27: 1297–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral A, Shruthi KS, Mathew MK (2015b) Vesicular trafficking and salinity responses in plants. IUBMB Life 67: 677–686 [DOI] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Blumwald E (2014) The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr Opin Plant Biol 22: 1–6 [DOI] [PubMed] [Google Scholar]
- Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E (2011a) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23: 224–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Tajima H, Liang YC, Ohto MA, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E (2011b) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23: 3482–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, Grinstein S, Orlowski J (2010) Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11: 50–61 [DOI] [PubMed] [Google Scholar]
- Chanroj S, Lu Y, Padmanaban S, Nanatani K, Uozumi N, Rao R, Sze H (2011) Plant-specific cation/H+ exchanger 17 and its homologs are endomembrane K+ transporters with roles in protein sorting. J Biol Chem 286: 33931–33941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanroj S, Wang G, Venema K, Zhang MW, Delwiche CF, Sze H (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front Plant Sci 3: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Contento AL, Bassham DC (2012) Structure and function of endosomes in plant cells. J Cell Sci 125: 3511–3518 [DOI] [PubMed] [Google Scholar]
- Demaurex N. (2002) pH homeostasis of cellular organelles. News Physiol Sci 17: 1–5 [DOI] [PubMed] [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. (2008) A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Etxeberria E, Pozueta-Romero J, Gonzalez P (2012) In and out of the plant storage vacuole. Plant Sci 190: 52–61 [DOI] [PubMed] [Google Scholar]
- Fan L, Zhao L, Hu W, Li W, Novák O, Strnad M, Simon S, Friml J, Shen J, Jiang L, et al. (2018) Na+,K+/H+ antiporters regulate the pH of endoplasmic reticulum and auxin-mediated development. Plant Cell Environ 41: 850–864 [DOI] [PubMed] [Google Scholar]
- Ferguson GP, Munro AW, Douglas RM, McLaggan D, Booth IR (1993) Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol Microbiol 9: 1297–1303 [DOI] [PubMed] [Google Scholar]
- Ferguson GP, Nikolaev Y, McLaggan D, Maclean M, Booth IR (1997) Survival during exposure to the electrophilic reagent N-ethylmaleimide in Escherichia coli: Role of KefB and KefC potassium channels. J Bacteriol 179: 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golani Y, Kaye Y, Gilhar O, Ercetin M, Gillaspy G, Levine A (2013) Inositol polyphosphate phosphatidylinositol 5-phosphatase9 (At5ptase9) controls plant salt tolerance by regulating endocytosis. Mol Plant 6: 1781–1794 [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji K, Nagira M, Yoshida K, Ohnishi M, Oda Y, Uemura T, Goh T, Sato MH, Morita MT, Tasaka M, et al. (2009) Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol 50: 2023–2033 [DOI] [PubMed] [Google Scholar]
- Hernández A, Jiang X, Cubero B, Nieto PM, Bressan RA, Hasegawa PM, Pardo JM (2009) Mutants of the Arabidopsis thaliana cation/H+ antiporter AtNHX1 conferring increased salt tolerance in yeast: The endosome/prevacuolar compartment is a target for salt toxicity. J Biol Chem 284: 14276–14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. (2004) Membrane trafficking in plants. Annu Rev Cell Dev Biol 20: 481–504 [DOI] [PubMed] [Google Scholar]
- Kang HG, Oh CS, Sato M, Katagiri F, Glazebrook J, Takahashi H, Kachroo P, Martin GB, Klessig DF (2010) Endosome-associated CRT1 functions early in resistance gene-mediated defense signaling in Arabidopsis and tobacco. Plant Cell 22: 918–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, Al-Rasheid KA, Marten I, Stierhof YD, Hedrich R, Schumacher K (2010) Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz HH, Gierth M, Herdean A, Satoh-Cruz M, Kramer DM, Spetea C, Schroeder JI (2014) Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc Natl Acad Sci USA 111: 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61: 495–506 [DOI] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF (2004) A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J Neurosci 24: 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshansky V, Futai M (2008) The V-type H+-ATPase in vesicular trafficking: Targeting, regulation and function. Curr Opin Cell Biol 20: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Bassil E, Jublanc E, Alcon C, Reguera M, Sentenac H, Blumwald E, Paris N (2013) In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell 25: 4028–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin T, Bassil E, Zhang S, Blumwald E (2014) Vacuolar Na+/H+ NHX-type antiporters are required for cellular K+ homeostasis, microtubule organization and directional root growth. Plants (Basel) 3: 409–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Jia WJ, Chu YJ, Xue HW (2012) Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res 22: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, Rao R (1998) Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast: Implications for vacuole biogenesis. J Biol Chem 273: 21054–21060 [DOI] [PubMed] [Google Scholar]
- Orlowski J, Grinstein S (2007) Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr Opin Cell Biol 19: 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Spitzer C (2008) Endosomal functions in plants. Traffic 9: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, Luan S (2007) CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res 17: 411–421 [DOI] [PubMed] [Google Scholar]
- Paroutis P, Touret N, Grinstein S (2004) The pH of the secretory pathway: Measurement, determinants, and regulation. Physiology (Bethesda) 19: 207–215 [DOI] [PubMed] [Google Scholar]
- Pinner E, Kotler Y, Padan E, Schuldiner S (1993) Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem 268: 1729–1734 [PubMed] [Google Scholar]
- Qiu QS. (2016a) Plant endosomal NHX antiporters: Activity and function. Plant Signal Behav 11: e1147643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS. (2016b) AtNHX5 and AtNHX6: Roles in protein transport. Plant Signal Behav 11: e1184810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Barkla BJ, Vera-Estrella R, Zhu JK, Schumaker KS (2003) Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol 132: 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Blatt MR, Pardo JM (2000) Functional conservation between yeast and plant endosomal Na(+)/H(+) antiporters. FEBS Lett 471: 224–228 [DOI] [PubMed] [Google Scholar]
- Radchenko MV, Waditee R, Oshimi S, Fukuhara M, Takabe T, Nakamura T (2006) Cloning, functional expression and primary characterization of Vibrio parahaemolyticus K+/H+ antiporter genes in Escherichia coli. Mol Microbiol 59: 651–663 [DOI] [PubMed] [Google Scholar]
- Reguera M, Bassil E, Tajima H, Wimmer M, Chanoca A, Otegui MS, Paris N, Blumwald E (2015) pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27: 1200–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Voss U, Jürgens G (2009) Post-Golgi traffic in plants. Traffic 10: 819–828 [DOI] [PubMed] [Google Scholar]
- Rose JK, Lee SJ (2010) Straying off the highway: Trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol 153: 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K. (2014) pH in the plant endomembrane system: An import and export business. Curr Opin Plant Biol 22: 71–76 [DOI] [PubMed] [Google Scholar]
- Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, Jiang L (2013) Organelle pH in the Arabidopsis endomembrane system. Mol Plant 6: 1419–1437 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AB, Kunz HH, Yang E, Schroeder JI (2016) Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc Natl Acad Sci USA 113: E5242–E5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson SJ, Jones RL (1996) Gibberellic Acid Induces Vacuolar Acidification in Barley Aleurone. Plant Cell 8: 2211–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Chanroj S (2018) Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol 177: 875–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wu X, Liu Y, Qiu QS (2015) AtNHX5 and AtNHX6 control cellular K+ and pH homeostasis in Arabidopsis: Three conserved acidic residues are essential for K+ transport. PLoS ONE 10: e0144716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ebine K, Ueda T, Qiu QS (2016) AtNHX5 and AtNHX6 are required for the subcellular localization of the SNARE complex that mediates the trafficking of seed storage proteins in Arabidopsis. PLoS ONE 11: e0151658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30: 529–539 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida S. (1994) Low Temperature-Induced Cytoplasmic Acidosis in Cultured Mung Bean (Vigna radiata [L.] Wilczek) Cells. Plant Physiol 104: 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Pan T, Fan L, Qiu QS (2013) A novel AtKEA gene family, homolog of bacterial K+/H+ antiporters, plays potential roles in K+ homeostasis and osmotic adjustment in Arabidopsis. PLoS ONE 8: e81463. [DOI] [PMC free article] [PubMed] [Google Scholar]