Depletion of organellar thymidine kinase affects plastid genome replication and repair, leading to the accumulation of truncated genomes and the apparent mobilization of new replication origins.
Abstract
Thymidine kinase (TK) is a key enzyme of the salvage pathway that recycles thymidine nucleosides to produce deoxythymidine triphosphate. Here, we identified the single TK of maize (Zea mays), denoted CPTK1, as necessary in the replication of the plastidial genome (cpDNA), demonstrating the essential function of the salvage pathway during chloroplast biogenesis. CPTK1 localized to both plastids and mitochondria, and its absence resulted in an albino phenotype, reduced cpDNA copy number and a severe deficiency in plastidial ribosomes. Mitochondria were not affected, indicating they are less reliant on the salvage pathway. Arabidopsis (Arabidopsis thaliana) TKs, TK1A and TK1B, apparently resulted from a gene duplication after the divergence of monocots and dicots. Similar but less-severe effects were observed for Arabidopsis tk1a tk1b double mutants in comparison to those in maize cptk1. TK1B was important for cpDNA replication and repair in conditions of replicative stress but had little impact on the mitochondrial phenotype. In the maize cptk1 mutant, the DNA from the small single-copy region of the plastidial genome was reduced to a greater extent than other regions, suggesting preferential abortion of replication in this region. This was accompanied by the accumulation of truncated genomes that resulted, at least in part, from unfaithful microhomology-mediated repair. These and other results suggest that the loss of normal cpDNA replication elicits the mobilization of new replication origins around the rpoB (beta subunit of plastid-encoded RNA polymerase) transcription unit and imply that increased transcription at rpoB is associated with the initiation of cpDNA replication.
The faithful replication of nuclear and organellar genomes depends on the regulation of dNTP pools, which can be synthesized either de novo, by a pathway whose limiting step is catalyzed by ribonucleotide reductase, or by a salvage pathway (Kafer et al., 2004; Zrenner et al., 2006). The salvage pathway recycles nucleotides by converting deoxynucleosides into the corresponding nucleotides by the action of nucleotide kinases. The salvage pathway is more energetically efficient than de novo synthesis and may be the preferred pathway to meet high demands for DNA synthesis during rapid cell proliferation or for DNA repair (Moffatt et al., 2002). It is therefore not surprising that the de novo and salvage pathways are coordinated during seed germination in plants. Two distinct phases have been proposed: “salvage synthesis” at the inception of germination, and “de novo synthesis” at later stages of germination (Stasolla et al., 2003).
Thymidine kinase (TK) is a crucial enzyme in the pyrimidine salvage pathway. TK contributes to the deoxythymidine triphosphate (dTTP) pool by phosphorylation of deoxythymidine (dT) into dT monophosphate (dTMP), which is rapidly converted by mono- and diphosphate kinases to dTTP (Zrenner et al., 2006). In metazoan, there are two types of TKs, TK1 and TK2, which are unrelated in sequence and are found in the cytoplasm and mitochondria, respectively (Konrad et al., 2014). The Arabidopsis (Arabidopsis thaliana) genome encodes two paralogs of animal TK1, named either TK1A and TK1B (Clausen et al., 2012; Pedroza-García et al., 2015) or TK1 and TK2 (Xu et al., 2015). To avoid confusion with TK2 from animals, we use the TK1A/TK1B nomenclature. TK1A is cytosolic, while TK1B is found in mitochondria (Xu et al., 2015). While mutants for each gene are not affected in their development, the tk1A tk1B double mutants are albino and do not pass the seedling stage, suggesting that TK1 depletion in Arabidopsis affects chloroplast biogenesis (Clausen et al., 2012; Xu et al., 2015). Like animal TK1 that only phosphorylates dT and deoxyuridine, both TK1A and TK1B from Arabidopsis have narrow substrate specificities and can mainly phosphorylate dT, deoxyuridine, and deoxycytidine (dC), with much less affinity for dC (Clausen et al., 2012; Xu et al., 2015). A gene coding for a protein with similarities to human (Homo sapiens) TK2 exists in Arabidopsis, but this TK2-like protein is a multisubstrate deoxyribonucloside kinase (dNK) that can phosphorylate dA, dG and dC, but not thymidine (Clausen et al., 2012). Thus, the TK1-like enzymes are the sole proteins with TK activity in plants.
The importance of dTMP pools on faithful genome replication is patent in animals, where absence of de novo dTMP synthesis because of folate deficiency results in genome instability and deoxyuridine misincorporation into nuclear DNA (Blount et al., 1997). The mutation of the animal mitochondrial TK2 is associated with mitochondrial DNA (mtDNA)-depletion and severe mitochondrial dysfunction in humans and mice (Mus musculus; Zhou et al., 2008).
In plants, changes in nucleotide pools have major consequences on plant development, with a pronounced effect on chloroplast biogenesis. For example, the mutation of ribonucleotide reductase genes results in abnormal leaf and flower morphology, reduced root growth, and variegation (Garton et al., 2007). These phenotypes correlate with a deficiency in chloroplast division and a reduced copy number of the plastidial genome (cpDNA). Similarly, inactivation of an uracil phosphoribosyltransferase gene (UPP) results in chlorotic phenotypes because of deficient chloroplast biogenesis (Mainguet et al., 2009). UPP codes for the major uracil phosphoribosyltransferase activity of Arabidopsis, which is required for the salvage of pyrimidines by catalyzing the formation of uridine monophosphate. UPP is targeted to plastids, showing that uracil salvage takes place predominantly in chloroplasts.
While it is well established and unsurprising that nucleotide pools affect plant development and chloroplast biogenesis, the direct effects on the replication of the organellar genomes have not been addressed. Here, we describe a mutant deficient in the single TK of maize (Zea mays), chloroplast TK1 (CPTK1). The cptk1 mutant displays an albino seedling phenotype and a deficiency in plastidial translation. We show that CPTK1 is targeted to both chloroplasts and mitochondria and that its loss results in a severe reduction in cpDNA copy number, the magnitude of which varies among different regions of the genome. This variation appears to be due to the accumulation of truncated subgenomes and the recruitment of alternative origins of replication. Characterization of the Arabidopsis orthologs confirms the importance of the organellar TKs on cpDNA replication early in plant development and reveals an additional role in cpDNA repair.
RESULTS
A Chloroplast-Localized Salvage Pathway for Nucleotide Synthesis Is Required for Chloroplast Differentiation and Normal Accumulation of cpDNA
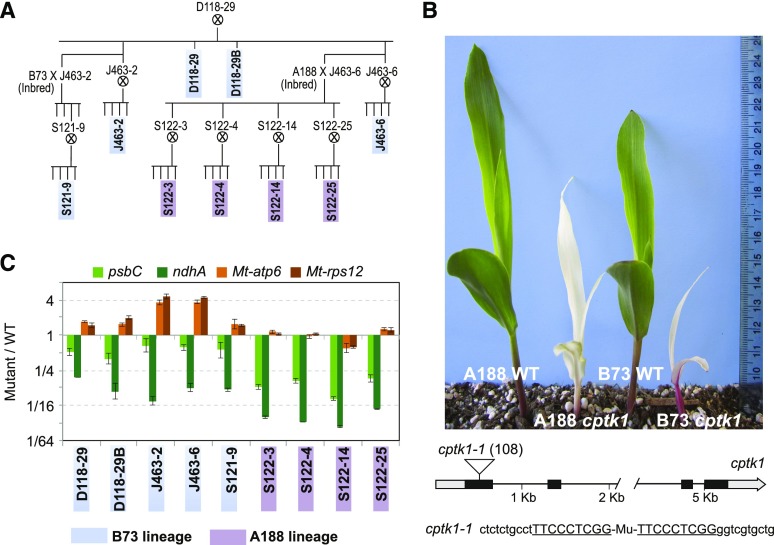
To identify genes involved in maintenance of cpDNA, we screened approximately 100 albino mutants selected from a large collection of nonphotosynthetic maize mutants (Belcher et al., 2015) for reduced cpDNA copy number (see Supplemental Fig. S3 of Udy et al. [2012]). We used quantitative PCR (qPCR) to quantify DNA from the PSII reaction center protein C (psbC) and NADH dehydrogenase subunit1 (ndhA) genes, which map to the large and small single-copy regions (LSC and SSC) of the chloroplast genome, respectively. Two mutant plants derived from closely related ears had a reduced amount of DNA from both genes. Interestingly, the reduction of ndhA DNA was more severe than that of psbC DNA (Fig. 1). Albino progeny of heterozygous siblings of these plants displayed an even more severe reduction in DNA from both loci (Fig. 1B). By contrast, mtDNA, which we evaluated by quantification of mitochondrial ATP synthase subunit 6 (atp6) and ribosomal protein S12 (rps12) sequences, was unchanged or slightly elevated in the affected plants. Increased mtDNA copy number was observed for many of the other albino mutants in the initial screen (see Supplemental Fig. S3 of Udy et al. [2012]), leading us to suspect that this is a secondary effect of chloroplast dysfunction.
Figure 1.
Albino phenotype and reduced cpDNA copy number in cptk1 plants. A, Pedigree showing the genealogical relationship between the assayed plants. B, Albino phenotype of homozygous cptk1 plants in two different inbred backgrounds. The diagram shows the cptk1 gene (GRMZM2G048821 in B73 genome v3) and the positions of the Mu insertion. The target site duplication is underlined. The nucleotide position with respect to the start codon is shown in parentheses. C, Quantification of mitochondrial and plastid DNA in cptk1 mutants. Each seedling is identified by the number of its family, whose relationships are diagrammed in A. Segments of the plastidial psbC and ndhA and mitochondrial atp6 and rps12 genes were assayed by qPCR. Results are the mean ± sd of technical triplicates and are represented as a ratio with respect to the wild-type (WT) in a log2 scale.
These mutants arose in maize lines with active Mutator (Mu) transposons. To identify the causal mutation, Mu insertions that cosegregate with the mutant phenotype were identified using a deep-sequencing approach (Williams-Carrier et al., 2010). An insertion in the first exon of gene GRMZM2G048821 was found in each of the two mutants identified in the original qPCR screen and in four other mutants from various branches of the same pedigree (Fig. 1A). This insertion was either heterozygous or absent in siblings of normal phenotype (Supplemental Fig. S1). The gene harboring this insertion, GRMZM2G048821, is a predicted ortholog of the Arabidopsis type 1 TKs TK1A and TK1B (see http://cas-pogs.uoregon.edu/#/pog/13546). Because a different maize gene had already been assigned the name tk1, we named this maize gene cptk1. We observed a significant decrease (P = 0.011, unpaired Student’s t test) in TK activity in the cptk1 mutant when total leaf extracts were assayed for TK activity (Supplemental Fig. S2), supporting the view that CPTK1 is an active TK enzyme.
A phylogenetic tree (Supplemental Fig. S3A) shows that type 1 TK is encoded by a single gene in nonflowering plants and in Poaceae (e.g. maize cptk1), while a second gene was acquired during the evolution of eudicots, giving the co-orthologs TK1A and TK1B found in Arabidopsis. Maize cptk1 and Arabidopsis TK1A have the same number and positions of introns, indicating that they derive from the same gene ancestor, while Arabidopsis TK1B lacks introns (Supplemental Fig. S3C). This suggests that TK1B originated by reverse-transcriptase mediated duplication of a TK1A-like ancestor after the divergence of monocots and dicots. Subcellular targeting predictions suggest that TK1 from nonflowering plants and monocots, and TK1B from eudicots are targeted to organelles, with a majority of the programs giving a chloroplastic prediction (Supplemental Fig. S3B). On the contrary, none of the TK1A sequences from eudicots are predicted to have organellar targeting sequences, suggesting that after the gene duplication event that created TK1B, the ancestral TK1A gene lost its N-terminal targeting sequence and assumed nonorganellar functions.
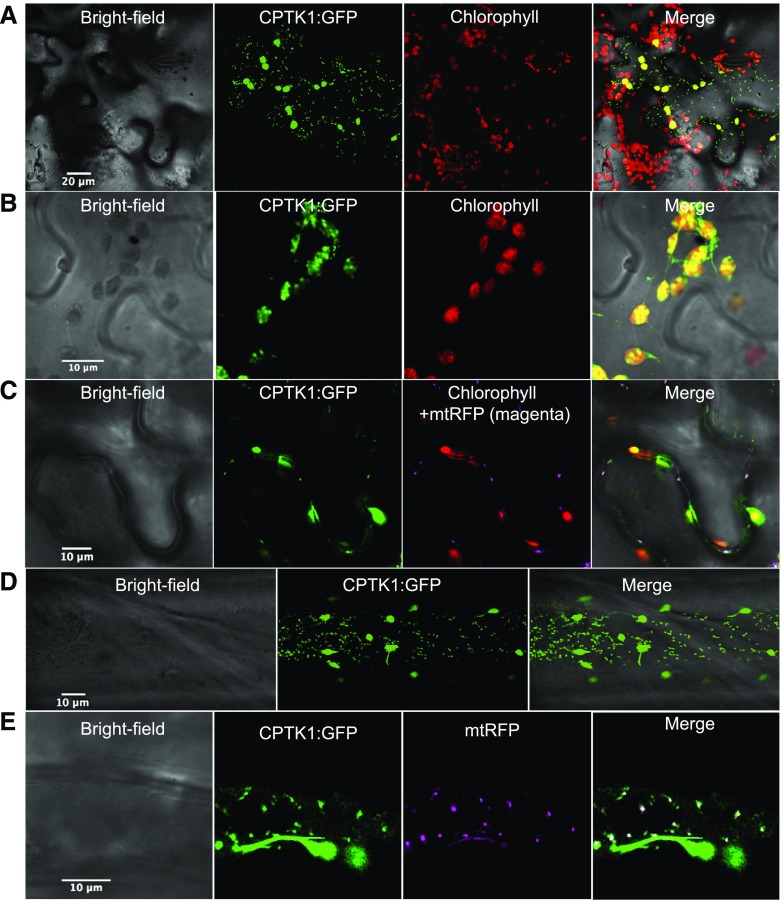
The cptk1 mutant phenotype and targeting predictions suggested that CPTK1 acts inside chloroplasts. Plastid-localization of a pyrimidine salvage pathway is in agreement with the presence of a nucleobase transporter in the plastid envelope, which provides plastids with the precursors for pyrimidine salvage (Witz et al., 2012). Indeed, the initial steps of pyrimidine and purine-nucleoside cleavage depend on nucleoside hydrolase1, which is located in the cytosol (Jung et al., 2009, 2011). The main salvage of uracil was also reported to take place in plastids (Mainguet et al., 2009). We tested the subcellular targeting of CPTK1 by fusing its coding sequence to eGFP and transiently expressing the fusion protein in Nicotiana benthamiana leaf epidermal cells and in onion (Allium cepa) epidermal cells (Fig. 2). CPTK1-GFP localized to both chloroplasts and mitochondria in N. benthamiana, as indicated by colocalization with chlorophyll autofluorescence and with mitochondrial-targeted DsRed, respectively (Fig. 2, A and C). The mitochondrial localization was only visible at enhanced exposure conditions where the plastidial signal was already saturated. At conditions optimal for plastid visualization the GFP signal was seen in bright foci that could be nucleoids and in stromules (Fig. 2B). Dual localization to mitochondria and plastids was also observed in onion epidermal cells (Fig. 2, D and E), where plastids were easily identified by distinctive stromules and mitochondria by their smaller size and characteristic cytoplasmic puncta.
Figure 2.
Organellar targeting of CPTK1. A, Dual targeting of CPTK1 fused to GFP into plastids and mitochondria in Nicotiana benthamiana leaves. B, Enlarged image of chloroplasts showing foci inside chloroplasts that might be nucleoids. C, Colocalization with cox4:dsRED (mtRFP) targeted to mitochondria. D and E, As in A and C, but in onion epidermal cells. A, scale bar 20 µm; B to E, scale bars 10 µm. WT, wild type.
In Arabidopsis, TK1A is reported to be a cytosolic protein, while TK1B is described as solely targeted to mitochondria (Xu et al., 2015). However, most TK1B protein sequences from eudicots are predicted to be targeted to chloroplasts, and peptides corresponding to TK1B have been identified in a proteome of Arabidopsis chloroplasts (Sun et al., 2009). To resolve this issue, we tested the localization of Arabidopsis TK1B:GFP in our experimental conditions. When imaged under standard conditions, we observed GFP only inside chloroplasts, often as puncta resembling chloroplast nucleoids (Supplemental Fig. S4). Thus, like maize CPTK1, Arabidopsis TK1B might also be targeted to chloroplasts.
Homozygous maize cptk1 mutants display an albino phenotype (Fig. 1B) and die after several weeks of growth, as is typical of nonphotosynthetic maize mutants. No chlorophyll fluorescence was observed in cptk1 leaves, and transmission electron microscopy (TEM) detected only undeveloped and/or degenerated plastids (Supplemental Fig. S5). Importantly, mitochondria seemed morphologically normal, both at the basal and apical sections of the leaf, consistent with the normal (or increased) levels of mtDNA. Thus, although CPTK1 is targeted to both organelles, the salvage pathway that depends on CPTK1 is important primarily for chloroplast biogenesis.
Impairment of cpDNA Replication in Maize cptk1 Plants
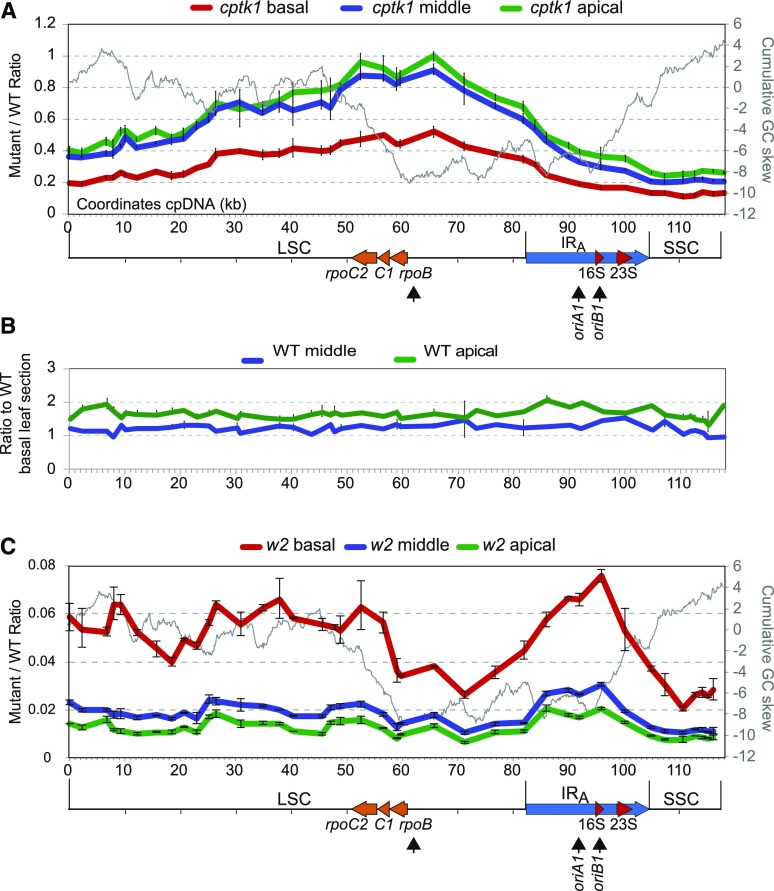
As noted above, qPCR data indicated position-dependent differences in the magnitude of the cpDNA deficiency in cptk1 mutants. These results were reminiscent of those seen previously in maize mutants deficient in chloroplast DNA polymerase (Udy et al., 2012) and suggested that the loss of CPTK1 causes a defect in cpDNA replication. To explore this possibility, the relative stoichiometry of cpDNA regions across the genome was scanned by qPCR using a set of primer pairs spaced several kb apart. DNA was examined from apical, middle, and basal sections of the leaf, which in wild-type plants harbor mature, young, and immature chloroplasts, respectively (Supplemental Fig. S6; Leech et al., 1973). The magnitude of cpDNA deficiency in cptk1 mutants varied along the genome (Fig. 3A). The deficiency was most severe in the SSC region and mildest near the beta subunit of RNA polymerase (rpoB) transcription unit, where the copy number was five times higher with respect to the wild type than was the SSC. A bidirectional gradient of increasingly severe DNA loss emanated from the rpoB gene region. The magnitude of the cpDNA deficiency was much less in the basal leaf section than in the middle and apical sections, but the topography of the copy number gradient was similar in all three leaf sections. This suggests that the replication defect responsible for the establishment of the copy number gradient occurred early in chloroplast biogenesis and was propagated through subsequent rounds of replication. We observed similar cpDNA copy number profiles in different cptk1 mutant individuals (Supplemental Fig. S7). By comparison, the stoichiometry of sequences along the cpDNA was similar among the different leaf sections in wild-type plants (Fig. 3B).
Figure 3.
Effect of cptk1 mutation on DNA abundance across the plastidial genome. A, Scanning of the cpDNA of cptk1 for changes in relative copy number of the different genome regions. Sequences along the cpDNA were quantified on the different leaf sectors defined in Supplemental Figure S6. Coordinates are those of the maize cpDNA sequence (accession NC_001666) with the LSC in reverse orientation, which seems to be the isomeric orientation of the genome consistent with the copy number gradient. The GC skew cumulative plot is superimposed as a gray line. The representation of the genome is shown below, with the position of the rRNA genes and of the rpoB, rpoC1, and rpoC2 genes marked. The position of the previously proposed origins of replication oriA and oriB are indicated, as well as the possible alternative replication origin active in cptk1 (arrow beneath rpoB). B, Comparison of position-dependent change in DNA copy number in middle and apical leaf sections as compared to the basal section in wild-type (WT) plants. Results showed no apparent stoichiometry changes, with variations across the genome in the range expected for technical errors. C, As in A for the w2-mum2 mutant deficient in the plastidial DNA polymerase (Udy et al., 2012). Values are the mean ± sd of technical triplicates.
We speculated that this copy number gradient along the genome results from premature arrests or pauses during cpDNA replication due to insufficient deoxynucleotide (dNTP) concentrations. According to this hypothesis, the position at which cpDNA copy number is maximal would correspond to the region of replication initiation (see, e.g. Nordman et al., 2014; Gowrishankar, 2015). However, the cpDNA region with the highest copy number in cptk1 mutants does not coincide with replication origins that have been described for maize and other plant species (Chiu and Sears, 1992; Lu et al., 1996; Kunnimalaiyaan and Nielsen, 1997; Oldenburg and Bendich, 2004, 2016; Day and Madesis, 2007). We explored the possibility that there is a relationship between the copy number gradient and GC skew (Fig. 3A), which is an asymmetry in G and C content between the two DNA strands. In bacteria, replication origins and termination regions typically map to minima and maxima in cumulative GC skew plots, respectively (Grigoriev, 1998; Gowrishankar, 2015). As described previously (Oldenburg and Bendich, 2016), the maize cpDNA GC skew plots have multiple minima and maxima, as expected for genomes that have multiple replication origins (Xia, 2012). The previously described origins of replication A and B (oriA and oriB) sequences are found at slight inflection points in the plot. However, a more prominent inflection point maps just upstream of the rpoB gene (Fig. 3A). It is interesting that the maximum in DNA copy number in cptk1 mutants coincides with this minimum in GC skew, suggesting that replication might initiate in this region in cptk1 mutants.
To elucidate the relationship between the cpDNA copy number gradient and DNA replication dynamics in cptk1 mutants, we performed a similar analysis on the maize white2 (w2) mutant w2-mum2 which is deficient in the plastidial DNA polymerase and exhibits a dramatic decrease in cpDNA copy number (Udy et al., 2012). Like cptk1, the w2-mum2 mutant is albino and does not grow past the seedling stage. However, the mechanisms that lead to reduced cpDNA copy number in cptk1 and w2-mum2 are fundamentally different, because in w2-mum2 it is the polymerase that is in limiting amounts and, therefore, replication arrests would not be caused by imbalanced dNTP concentrations. As reported previously (Udy et al., 2012), the DNA from the SSC region in w2-mum2 was more severely reduced than that from the LSC region in the basal leaf section. Interestingly, however, this gradient had a different topography than that in cptk1 (Fig. 3C). In particular, the gradient peak centered in the inverted repeat (IR) region in w2-mum2 near the oriB sequence proposed as an origin of replication. A change in copy number was also found near rpoB in the basal leaf section. Therefore, an additional or alternative replication origin in that region might also be used early in development under conditions of compromised cpDNA replication in genotypes other than cptk1.
Defects in cpDNA Replication Are Associated with Dramatic Changes in Plastidial RNA Levels and Loss of Photosynthetic Complexes in cptk1 Plants
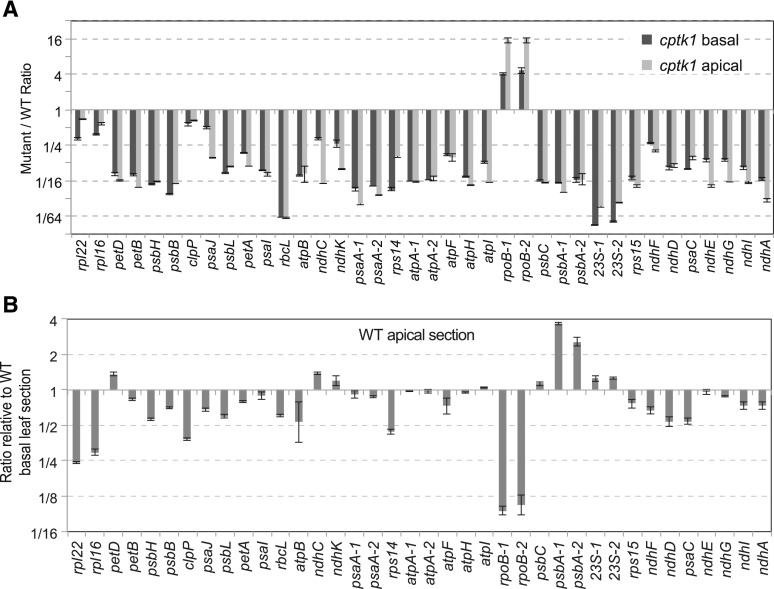
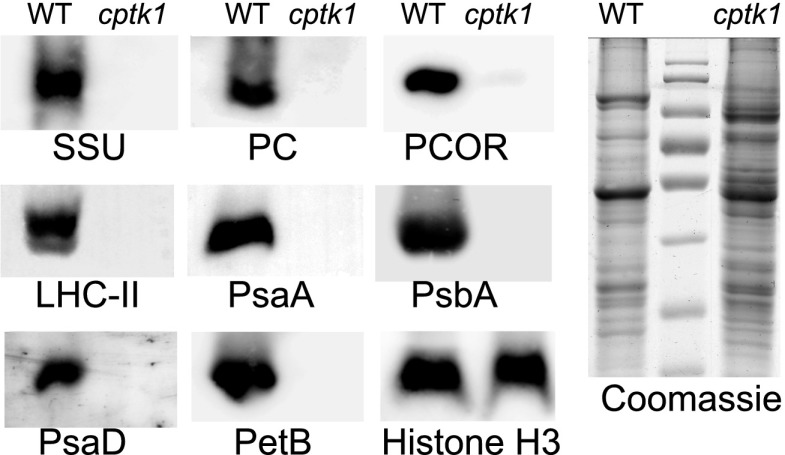
To assess how the reduction in cpDNA in cptk1 mutants affects the abundance of chloroplast-encoded RNAs and proteins, we tested the accumulation of representative chloroplast transcripts and proteins by RT-qPCR and immunoblot analysis, respectively. The RT-qPCR results showed a dramatic reduction in most cpDNA transcripts (Fig. 4A), and similar reductions between basal and apical sections of the leaf. There was no apparent relationship between the change in transcript level and the change in gene copy number. There was a major reduction in the abundance of the 23S rRNA in the basal section of the leaf (∼100-fold), which is expected to cause a severe defect in plastid translation. In general, the abundance of transcripts synthesized primarily by the plastid-encoded RNA polymerase were reduced, and those synthesized primarily by the nucleus-encoded polymerase (NEP) were increased. This change in transcript populations is typical of albino mutants that are deficient in plastidial ribosomes (e.g. Udy et al., 2012). Immunoblot analysis of cptk1 total protein extracts showed severe reductions in the levels of a variety of components of the photosynthetic apparatus, which was expected given the albino phenotype and lack of chloroplast rRNA (Fig. 5).
Figure 4.
Accumulation of plastidial transcripts in maize cptk1. A, Representative plastidial transcripts were quantified by RT-qPCR and normalized against a set of nuclear housekeeping genes. For several genes (psaA, atpA, rpoB, psbA, and rRNA 23S) two different transcript regions were quantified. Results from the different mutant leaf sectors were compared against the corresponding leaf sectors of wild type (WT). The genes are sorted according to their physical location in the cpDNA. B, Quantification of transcript abundance in the apical section as compared to the basal section in wild-type leaves. Results are the mean ± sd of technical triplicates and are represented in a log2 scale.
Figure 5.
Immunoblot analysis of components of various proteins related to chloroplast functions in cptk1 mutants. Total leaf proteins were extracted from the basal leaf section. SSU, small subunit of Rubisco; PC, plastocyanin; PCOR, protochlorophyllide oxidoreductase; PsaA, PSI reaction center subunit A; PsaD, PSI subunit D; PsbA, PSII protein D1; LHCII, 26-kD protein from light-harvesting complex II; AtpB, β-subunit of ATPase; H3, nuclear histone H3 used as loading control; WT, wild type. The Coomassie staining of a replicate gel is shown to the right. The center lane contains size standards.
Abortion of cpDNA Replication in cptk1 Mutants Results in the Accumulation of Aberrant Genomes Generated by Microhomology-Mediated Recombination
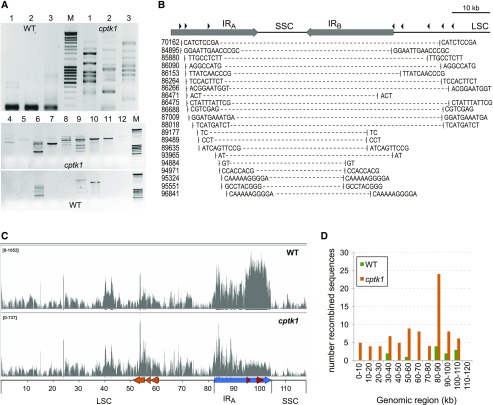
Stalled DNA replication can ultimately lead to dissociation of the replication complex, resulting in ssDNA gaps that can be processed into double-strand breaks (Aguilera and Gómez-González, 2008). If broken genomes are not repaired, replication cannot be restarted and DNA is lost. The observed reduction in certain cpDNA regions in cptk1 mutants could reflect genomic deletions that result from inaccurate repair or restart of replication at ectopic genomic sites. These deleted genomes might then be propagated in older tissues, where the need for the salvage pathway is apparently less critical for genome replication. This could explain why the same copy number gradient across the chloroplast genome was maintained from the base to tip of the leaf of cptk1 plants, even as the overall cpDNA copy number increased. To test this hypothesis, we used PCR to amplify predicted truncated genomes deleted for part of the IRs and the entire SSC region. Various primers flanking this region (Fig. 6) were tested for their ability to amplify DNA from the apical leaf section of wild-type and cptk1 plants. As expected based on the large distance between their binding sites in wild-type cpDNA, most primer pairs did not yield any product from DNA extracted from wild-type plants (Fig. 6A). By contrast, many products were amplified from cptk1 mutants. DNA fragments that only amplified from cptk1 DNA were cloned and sequenced. Most sequences resulted from the joining of two distant regions in the genome sharing small repeated sequences (2 to 14 nucleotides) at the junction borders (Fig. 6B). Such rearranged DNA sequences are characteristic products of error-prone microhomology-mediated repair (MHMR) pathways. It is very unlikely that these resulted from template shifting during PCR because analogous products were not produced from wild-type DNA.
Figure 6.
cpDNAs with large deletions accumulate in cptk1 mutants. A, Amplification of recombined cpDNA sequences by PCR using primers mapping far apart in the genome. B, Truncated cpDNA sequences specifically amplified from cptk1 resulting from microhomology-mediated repair processes. The cpDNA region analyzed is represented above, with the IRs (IRA and IRB), SSC, and part of the LSC regions. Scale bar (10 kb) is shown. Primers used for amplification in different combinations are indicated by the black arrowheads. The regions deleted in each sequence are represented, with the small repeats found at the junction borders. Coordinates on the genome are indicated on the left. (C) Coverage of reads mapped to the cpDNA following deep-sequencing of total DNA from wild-type (WT) and cptk1 leaves. The range is indicated on the upper left of the plots. D, Reads identified as corresponding to rearranged cpDNA molecules and their distribution among different genomic regions.
To confirm and extend these observations, we analyzed DNA from cptk1 and wild type by deep sequencing. We sequenced total DNA from the apical region of a seedling leaf, obtaining 47,137 and 27,378 150-nt reads mapping to the cpDNA of wild type and cptk1, respectively, out of a total of ∼1.5 million reads in each case. The data confirmed the reduced abundance of cpDNA in cptk1 and the particularly severe deficiency in the rDNA region (Fig. 6C). Stringent filtering for reads potentially mapping to two different single-copy regions followed by individual sequence alignments identified reads derived from rearranged cpDNA genomes, similar to the ones revealed by PCR. Several showed no repeated bases or just one at the junction border, suggesting they were produced by nonhomologous end joining. Very few such junctions were found in sequences derived from wild-type plants as compared to cptk1 (Supplemental Fig. S8). Importantly, the majority of these sequences in cptk1 mapped just upstream of the genomic region that is most strongly reduced in abundance, near the rRNA transcription unit (Fig. 6D). Thus, our data support the conclusion that collapse of cpDNA replication early in leaf development in cptk1 mutants leads to broken chloroplast genomes that are then repaired by MHMR or nonhomologous end-joining pathways. These truncated genomes are deficient in various essential genes and are therefore incapable of supporting chloroplast biogenesis.
TKs Are Also Required for Efficient cpDNA Replication and Chloroplast Biogenesis in Arabidopsis
To compare the functions of TK in maize and Arabidopsis, we also examined mutants of the two TK genes of Arabidopsis. We obtained several T-DNA mutant alleles for each gene, which we crossed in different combinations (Supplemental Fig. S9). We confirmed phenotypes described previously for the double mutants (Clausen et al., 2012; Xu et al., 2015): double mutants segregating from all reciprocal combinations that we tested (tk1A-2 tk1B-2, tk1A-2 tk1B-3, tk1B-2 tk1A-3, and tk1B-2 tk1A-2) had albino cotyledons and did not grow past the cotyledon stage, whereas TK1A/tk1A tk1B plants had pale virescent cotyledons and leaves but grew to maturity and set seeds. In contrast with maize cptk1, fluorescent microscopy showed that the albino leaves in Arabidopsis tk1A tk1B mutants included a few plastids showing chlorophyll fluorescence; TEM confirmed these to be chloroplasts, although much fewer in number, smaller in size, and with a less-developed membrane system than in wild type. Importantly, like in maize cptk1, mitochondria seemed morphologically normal (Supplemental Fig. S10).
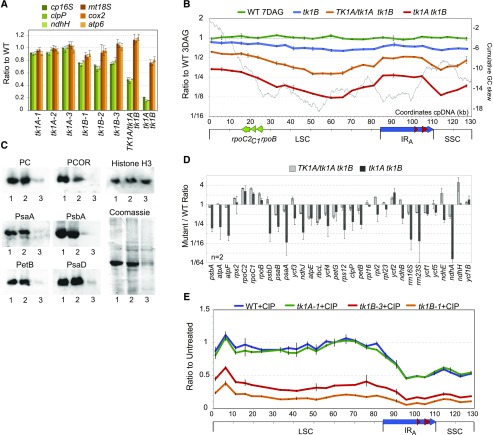
Quantification of mtDNA in small seedlings of the single Arabidopsis mutants showed little or no effect on the abundance of mtDNA (Fig. 7A). However, mtDNA was slightly reduced in the double mutant tk1A tk1B. Like in maize cptk1, the loss of TKs in Arabidopsis impacted the replication of the cpDNA, with up to 7-fold reductions in cpDNA copy number in the tk1A tk1B double mutant. We also assayed the stoichiometry of cpDNA sequences along the genome in tk1A tk1B by qPCR, using a set of 27 primer pairs spaced roughly 5 kb apart. A gradient in copy number along the genome was more pronounced in the double mutant but was also visible in TK1A/tk1A tk1B (Fig. 7B). This gradient had a maximum in the IR region, consistent with the idea that replication origins map close to the rRNA genes, as described for other eudicots (Chiu and Sears, 1992; Lu et al., 1996; Kunnimalaiyaan and Nielsen, 1997; Oldenburg and Bendich, 2004, 2016; Day and Madesis, 2007). A minimum in the GC skew plot of the Arabidopsis cpDNA was also found upstream of the rpoB operon, but contrarily to maize cptk1, there was no evidence that replication initiated from that locus in the Arabidopsis mutants. Because tk1A tk1B double mutants were delayed in growth compared to wild type, they were compared to slightly younger wild-type seedlings (7 d after germination [DAG] for tk1A tk1B versus 3 DAG for wild type). As a control, we verified that there was no difference in the data obtained for 7 DAG and 3 DAG seedlings in the wild type (Fig. 7B).
Figure 7.
Effect of Arabidopsis tk1 mutants on cpDNA abundance and expression. A, Copy number quantification of plastidial (16S rRNA, clpP, and ndhH) and mitochondrial (18S rRNA, cox2, and atp6) genes in the indicated single and double mutants. B, Scanning of Arabidopsis cpDNA for changes in relative abundance of different genome regions, as described in Figure 3 for maize. C, Accumulation of plastidial transcripts in Arabidopsis tk1 double mutants. The relative abundances of representative transcripts of the Arabidopsis plastidial genome were quantified by RT-qPCR and normalized against a set of nuclear housekeeping genes. The genes are sorted according to their physical location in the cpDNA. Results are represented in a log2 scale. Error bars are standard deviations from two biological replicates. D, Immunodetection of proteins from total seedling extracts with antibodies recognizing plastid-encoded and nucleus-encoded chloroplast proteins. 1, wild type; 2, TK1A/tk1A tk1B; 3, tk1A tk1B. E, Scanning of the cpDNA showing changes in relative copy numbers of the different genome regions between untreated and CIP-treated (0.75 µm) plants. Values in A, B, C, and E are the mean ± sd of technical triplicates.
At the transcriptome level, RT-qPCR results for the tk1A tk1B double mutants were analogous to those for the maize cptk1 mutant: in particular, there was an overall reduction of rRNAs and mRNAs encoding photosynthesis subunits and milder effects or some increase in RNAs transcribed by NEP (Fig. 7C). Similar to maize, in Arabidopsis there was no relationship between changes in transcript accumulation and the reduction in cpDNA copy numbers. A slight decrease in plastidial transcripts was also observed in TKA1/tk1A tk1B but much less than in the double homozygote mutant. At the protein level, a strong reduction of plastidial proteins was observed in the albino homozygous tk1A tk1B double mutant, although all tested proteins were detected (Fig. 7D), unlike in maize cptk1. These results are consistent with the microscopy data showing a few chlorophyll-containing chloroplasts in Arabidopsis tk1A tk1B. In agreement with the mild effect of the TKA1/tk1A tk1B mutation on the abundance of plastidial transcripts, we could not detect any reduction in chloroplast proteins in this mutant.
To test the effect of replication stress on the physiological manifestations of TK depletion, we treated the plants with ciprofloxacin (CIP), an inhibitor of the DNA gyrase that is targeted only to organelles in plants (Wall et al., 2004). CIP has a major effect on plastid development and recombination (Evans-Roberts et al., 2016) and its effect is exacerbated in mutants deficient in repair by recombination (Maréchal et al., 2009; Janicka et al., 2012). In wild-type and tk1A plants, the major effect of CIP was on the replication of the IR and SSC regions, with a clear 2-fold decrease in replication efficiency in the region of the rRNA genes; copy numbers of genes in the LSC region showed little if any change (Fig. 7E). However, in tk1B plants there was a major effect of CIP on overall cpDNA copy number, but still much reduced replication of the IRs and SSC regions. These results confirm that TK1B has important roles not only in plastidial genome replication but also in cpDNA repair.
DISCUSSION
The importance of the salvage pathway in the replication of organellar genomes in plants is poorly understood. The only prior reports addressing this issue involve an analysis of a mutant lacking ribonucleotide reductase. This mutation has a major effect on chloroplast biogenesis, with a more pronounced effect on the replication of the cpDNA than on the nuclear genome (Garton et al., 2007) and also affects the programmed degradation of the cpDNA during pollen development (Tang et al., 2012). Here, we show that in maize, the salvage pathway that depends on CPTK1 is essential for efficient cpDNA replication and chloroplast biogenesis. In Arabidopsis, the partial redundancy of the two TK1-type TKs has been demonstrated previously, and the albino phenotype of the double mutant had implied their importance in the development of plastids. We further show here that the main effect of the tk1A tk1B double mutation is on the replication of the plastidial (as opposed to the mitochondrial) genome.
The evolutionary origin of the different plant TK1 genes is complex. The conservation of intron positions between the single TK of Poaceae (CPTK1 in maize) and TK1A of eudicots shows that they are derived from the same ancestor. This ancestral protein was likely targeted to organelles, and it was probably after the duplication that gave rise to TK1B that TK1A lost its organellar targeting sequence. The mitochondrial and/or plastidial location of plant organellar TK1 remains controversial. Our data show unambiguously that maize CPTK1 localizes to both chloroplasts and mitochondria. By contrast, a sole mitochondrial location was described for Arabidopsis TK1B (Xu et al., 2015). However, targeting predictions and our transient expression results suggest that TK1B is also targeted to plastids. At present, we cannot explain these apparently contradictory results. Furthermore, the major effects of TK1 deficiency are in the chloroplast in Arabidopsis, as in maize. No meaningful change in mtDNA copy number was detected in maize cptk1 mutants, and only a slight decrease in mtDNA was observed in Arabidopsis tk1A tk1B double mutants. Moreover, mitochondrial morphology appeared normal in TEM images in mutants from both species. This implies that mtDNA replication can be supported by dTMP provided by de novo synthesis. By contrast, the major decrease observed in cpDNA copy number in maize and Arabidopsis reveals that replication of the cpDNA requires the salvage pathway. This requirement is likely due to the high demand for DNA synthesis inside developing chloroplasts: plastid DNA is rapidly amplified after germination and constitutes a large proportion of the cellular DNA in developed leaf tissue.
The loss of TK1 has remarkable effects on the dynamics of cpDNA replication, both in maize and Arabidopsis. The reduction in copy number is differential along the genome, and in maize cptk1 there is a distinct gradient in copy numbers, with a minimum at the SSC region and a maximum in the region upstream of rpoB in the LSC region. Such differential DNA replication presumably results from low dTTP concentrations that cause slowing, pausing, or abortion of DNA polymerase progression. Maximum sequence copy number should then map to the locus where replication is reinitiated. Surprisingly, in maize cptk1, that locus differs from the proposed replication origins mapped in several flowering plants in the IRs of the cpDNA (IRA and IRB), close to rRNA gene sequences (Chiu and Sears, 1992; Lu et al., 1996; Kunnimalaiyaan and Nielsen, 1997; Oldenburg and Bendich, 2004; Day and Madesis, 2007). These putative promoter regions often coincide with the extremities of linear cpDNA molecules that were mapped both in maize and tobacco (Nicotiana tabacum; Oldenburg and Bendich, 2004, 2016; Scharff and Koop, 2006). However, targeted inactivation of these replication origins in transplastomic tobacco gave viable plants (Mühlbauer et al., 2002; Scharff and Koop, 2007), implying that there are additional origins of replication and/or alternative mechanisms of plastid DNA replication that can be recruited. The existence of alternative mechanisms of cpDNA replication was originally revealed by electron microscope observation of maize and pea (Pisum sativum) cpDNA, which showed both Cairns and rolling-circle intermediates of replication (Kolodner and Tewari, 1975). Our observations suggest that in cptk1 plants, there is indeed an alternative replication origin that is preferentially used. A possible scenario is that early in leaf development in cptk1, because of replication arrests due to low nucleotide concentrations, there is reinitiation of replication at ectopic locations, resulting in the accumulation of truncated genomes, most of which are deleted for the canonical replication origins. The fact that the copy number gradient present in the basal section is seen also at the leaf tip suggests that cpDNA replication is less dependent on the salvage pathway later in leaf development, such that the truncated genomes generated early on are propagated using an alternative replication origin.
Interestingly, there is a relationship between cpDNA regions that are rapidly transcribed and the regions where replication apparently initiates. It is tempting to speculate that high transcription activity is necessary to locally melt the DNA molecule and recruit the chloroplast replisome and/or to synthesize RNA primers. In bacteria, it has been shown that cells lacking the origin of replication oriC or chromosomal replication initiator protein (DnaA) can survive by means of a pathway called constitutive stable DNA replication, which involves transcription-associated R-loops that act as primers for DNA synthesis and establishment of new replication forks (Kogoma, 1997; Gowrishankar, 2015). R-loops similarly initiate the replication of ColE1 plasmids (Fukuoh et al., 1997) and of the human mitochondrial genome (Shadel and Clayton, 1997). In chloroplasts of wild-type plants and in developed tissues it could be that plastid-encoded RNA polymerase-mediated transcription of the rRNA genes provides synthesis of primers for replication. In cptk1 it could be the high activity of the NEP at the rpoB operon that promotes replication initiation under conditions that trigger retrograde activation of rpo transcription.
In Arabidopsis, the loss of TKs also has a dramatic effect on cpDNA replication. However, the gradient in copy number is less pronounced than in maize, and it peaks at the expected region of replication initiation in the IR. This suggests that pausing in replication is less deleterious than in maize and that the replication complex is able to restart without loss of genomic information. Maintenance of the organellar genomes of Arabidopsis relies on two DNA polymerases that are redundant for replication functions, while one is preferentially required for repair (Parent et al., 2011). They have been shown to have remarkable capability to bypass apurinic/apyrimidinic DNA lesions, an activity that could be important in preventing replication fork collapse when the polymerase meets such unrepaired lesions (Baruch-Torres and Brieba, 2017). In maize, however, the product of the w2 gene seems to be the sole enzyme responsible for cpDNA replication (Udy et al., 2012). It is possible that either one or both organellar DNA polymerases of Arabidopsis are less sensitive to low nucleotide concentrations than is the plastidial DNA polymerase of maize. As a consequence, there is still some gene expression and translation in the few remaining plastids of Arabidopsis tk1A tk1B.
The process by which truncated cpDNA molecules accumulate in cptk1 seems to be linked to repair pathways that rely on micro sequence homologies to reprime replication, known as MHMR or microhomology-mediated break-induced replication (Hastings et al., 2009). MHMR has been shown to be a predominant pathway of repair in plant chloroplasts. In Arabidopsis, it was shown that specific cleavage of the cpDNA with a plastid targeted I-CreII endonuclease leads to repair of the specific double-strand break by MHMR (Kwon et al., 2010). Plastid and mitochondrial Arabidopsis genomes rearranged by MHMR were also revealed in mutants that affect organellar genome maintenance (Maréchal et al., 2009; Parent et al., 2011; Janicka et al., 2012). A genome-wide analysis of such type of events further suggested that short-range U-turn-like inversions could be the major outcome of replication-dependent repair processes in Arabidopsis plastids (Zampini et al., 2015). Because of the two alternative isomeric configurations of the cpDNA generated by recombination between the IR regions, our results cannot distinguish between the possibilities that the rearranged sequences in cptk1 result from a long-range deletion involving sequence microhomologies in direct orientation across the SSC and the second IR copy, as represented in Figure 6, or from short range U-turn inversion of replication involving sequences inversely oriented in the genome. But whatever the process, the outcome would be the same, which is the loss of the nonreplicated cpDNA sequences.
The effects of replication stress on cpDNA also elucidate aspects of cpDNA replication. Reduced dNTPs in tk1 mutants, reduced DNA polymerase in w2, or gyrase inhibition in CIP-treated Arabidopsis all resulted in reduction of the rRNA gene sequences and of the entire SSC region. The likely explanation is that replication of the cpDNA proceeds bidirectionally, with one replication fork progressing from the IR region into the LSC and another into the SSC. The preferential loss of SSC sequences suggests that these are more prone to pausing and/or collapse in replication stress conditions. It is possible that the highly transcribed rRNA genes trigger transcription-replication conflicts in conditions of suboptimal fork progression, but this remains speculative. These are important questions for future work.
MATERIALS AND METHODS
Plant Material
The cptk1 mutation arose in maize (Zea mays) lines harboring active Mu transposons. Heterozygotes were crossed to inbred lines, and the F1 progeny were self-pollinated to recover ears segregating homozygous mutants. DNA was isolated from mutant individuals from each of four F2 ears and analyzed by Mu-Illumina sequencing to map all of the Mu insertions in each plant (Williams-Carrier et al., 2010). An insertion in GRMZM2G048821 (B73 RefGen_v3) was found in all six of the mutant individuals shown in the pedigree in Figure 1. Subsequent gene-specific PCR showed this insertion to be absent from closely related +/+ ears and that phenotypically normal plants on ears segregating cptk1 were either +/+ or +/− (Supplemental Fig. S1).
For phenotypic analyses, maize plants were grown in a growth chamber under 16 h light/8 h dark photoperiod. Tissue was harvested from the second leaf when the third leaf was beginning to emerge from the whorl. The second leaf was divided into basal, middle, and apical sections (see Supplemental Fig. S6). Material from three individuals was pooled and used for DNA, RNA, and protein extractions, as previously described (Udy et al., 2012).
RNA was extracted using TRI Reagent (Molecular Research Centre) and samples were depleted from contaminating genomic DNA by treatment with RQ1 RNase-free DNase (Promega). For RT-qPCR experiments, 3 µg of RNA were reverse-transcribed with Superscript III Reverse Transcriptase (Life Technologies) according to the manufacturer’s protocol using random hexamers. For immunoblots, total leaf proteins were extracted in the presence of 2% (w/v) sodium dodecyl sulfate and 4 m urea and precipitated with methanol-chloroform before sodium dodecyl sulfate-polyacrylamide gel electrophoresis fractionation and transfer to Immobilon-P membranes. Chemiluminescence was detected and quantified with a Fusion-FX7 camera system (Vilber Lourmat).
Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutant lines, all in the Col-0 background, were obtained from the Nottingham Arabidopsis Stock Centre (tk1A-1 [SALK_097767], tk1A-2 [GK-401F10], tk1A-3 [SALK_094632], tk1B-1 [SALK_074256], tk1B-2 [GK-143D03], tk1B-3 [GK-457H01]). Plant genotypes were determined by PCR using gene and T-DNA specific primers. Seeds were stratified for 3 days at 4°C and plants were grown on soil or on MS255 medium (Duchefa) supplemented with 1% (w/v) Suc at 22°C. Seeds were cultivated in vitro, and young seedlings were selected according to their characteristic phenotypes of pale green and albino cotyledons, respectively. RNA, DNA, and proteins were extracted from the same samples. For the replicative stress assay, surface-sterilized seeds were spread on plates supplemented with different concentrations of CIP (Sigma-Aldrich). DNA was extracted using the cetyltrimethylammonium bromide method (Murray and Thompson, 1980).
Phylogenetic and Bioinformatics Analysis
Bacterial and plant sequences were identified in the databases by BLASTP and TBLASTN with maize and Arabidopsis sequences as queries. Alignments were constructed with ClustalW implemented in the Macvector package using the GonneT matrix. Phylogenetic trees were built with PhyML v3.1 online software (www.phylogeny.fr) using the neighbor-joining method implemented in the BioNJ program. Graphical representations were performed with TreeDyn (v198.3). Subcellular targeting predictions were obtained with TargetP, Predotar, and MultiLoc2 (http://urgi.versailles.inra.fr/predotar/predotar.html, http://www.cbs.dtu.dk/services/TargetP, http://abi.inf.uni-tuebingen.de/Services/MultiLoc2). GC skew plots were calculated with GCSkew (http://genskew.csb.univie.ac.at/).
Sequencing
Total DNA of the upper leaf sections of wild-type and cptk1 plants was quantified with a QuBit Fluorometer (Life Technologies). Libraries were prepared with the Nextera XT Library Prep Kit according to manufacturer’s recommendations (Illumina) using 1 ng of each DNA sample. Final libraries were quantified with the QuBit Fluorometer (Life Technologies), checked on a Bioanalyzer 2100 (Agilent) and sequenced on a Illumina Miseq system (2 × 150 paired end reads).
Intracellular Localization
Sequences encoding the maize CPTK1 and Arabidopsis TK1B were cloned into the pUCAP-GFP vector, upstream and in frame with eGFP. Young leaves of Nicotiana benthamiana and onion (Allium cepa) epidermal cells were transfected by bombardment with a Biolistic PDS-1000/He System (Bio-Rad). The pCKmRFP plasmid (Vermel et al., 2002), which encodes mitochondrial-targeted RFP, was cotransfected. After 24 h, the fluorescence of GFP and chlorophyll was observed at 505 to 540 nm and beyond 650 nm, respectively, after excitation at 488 nm on a Zeiss LSM700 confocal microscope. The DsRED fluorescence was excited at 555 nm and observed at 560 to 615 nm.
Measure of TK Activity
TK enzymatic activity was determined mainly as described in Pedroza-García et al. (2015). Soluble proteins from the second leaf of wild-type and cptk1 individual plants were extracted in extraction buffer (70 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 25 mm KCl, 5 mm EDTA, 5% gycerol, 15 mm β-mercaptoethanol, 0.1% Triton X-100, complete protease inhibitor cocktail [Roche]). After grinding, the mix was centrifuged at 15,000g for 10 min, and the soluble protein concentration quantified by Bradford (Bio-Rad protein assay reagent). For the activity test, a reaction mix of 0.5 mL containing 100 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 10 mm NaF, 10 mm ATP, 2 mm β-mercaptoethanol, 150 μg of soluble protein extract, and 2 µm [methyl-3H]-thymidine was incubated at 37°C for 90 min. At the end of the reaction, 5 volumes of precipitation solution (100 mm LaCl3; 5 mm triethanolamine) were added and the precipitate was collected by centrifugation at 2,000g for 10 min. After three washing steps with precipitation solution, the precipitate was dissolved in 200 μL of 50 mm HCl, added to 3 mL scintillation liquid, and radioactivity quantified on a Beckman LS 6500 scintillation counter. The background of nonspecific radioactivity was determined for the wild-type extract without incubation.
qPCR Analysis
qPCR experiments were performed in a LightCycler480 (Roche) in a total volume of 6 µL containing 0.5 mm of each specific primer and 3 µL of SYBR Green I Master Mix (Roche Applied Science). The thermocycling program was as follows: a 7-min denaturing step at 95°C followed by 40 cycles of 10 s at 95°C, 15 s at 60°C, and 15 s at 72°C. The second derivative maximum method was used to determine Cp values, and PCR efficiencies were determined from DNA serial dilution curves or using LinRegPCR software (http://LinRegPCR.nl). Three technical replicates were performed for each experiment. Results of qPCR and RT-qPCR analysis of maize and Arabidopsis sequences were standardized as previously described (Udy et al., 2012; Wallet et al., 2015). Quantification of cpDNA copy numbers used a set of primer pairs located along the organellar genomes whose sequences and coordinates are given in Supplemental Table S1.
TEM
Leaf tissue samples were fixed overnight in 3% glutaraldehyde, then treated for 2 h with 10% (w/v) picric acid, 2 h with 2% uranyl acetate, and stained with 0.1% (v/v) osmium tetroxide in 150 mm phosphate buffer, pH 7.2. Samples were dehydrated through an ethanol series and infiltrated with EPON812 medium-grade resin (Polysciences). Polymerization was done for 48 h at 60°C. Ultrathin sections (70 mm) were cut with an Ultracut E microtome (Reichert) and collected on grids coated with formvar (Electron Microscopy Sciences). Samples were visualized with a Hitachi H-600 electron microscope operating at 75 kV.
Acknowledgments
We thank two anonymous reviewers for comments that significantly improved the article, Dr. Kristina Kühn (Humboldt Universität zu Berlin) for helpful discussions, Dr. Etienne Delannoy for antibodies and Abdelmalek Alioua for technical assistance with qPCR.
Accession Numbers
Maize cptk1, GRMZM2G048821; Arabidopsis TK1A, At3g07800; Arabidopsis TK1B, At5g23070; Maize cpDNA, NC_001666; Arabidopsis cpDNA, NC_000932.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Linkage between the Mu insertion in cptk1 and the albino phenotype
Supplemental Figure S2. TK activity is reduced in cptk1
Supplemental Figure S3. Evolution and organellar targeting of plant TK1 proteins
Supplemental Figure S4. Plastidial targeting of Arabidopsis TK1B fused to eGFP
Supplemental Figure S5. Morphology of organelles in maize cptk1 leaves
Supplemental Figure S6. Leaf sections of the second leaf of the maize whorl utilized for analysis
Supplemental Figure S7. Scanning of relative copy numbers of the different cpDNA regions in two independent cptk1 plants
Supplemental Figure S8. Alignments to the cpDNA of sequences corresponding to rearranged genomes identified from DNAseq data
Supplemental Figure S9. Phenotypes of Arabidopsis TK1 mutants
Supplemental Figure S10. Morphology of organelles in Arabidopsis tk1A tk1B double mutants
Supplemental Table S1. Primers
Footnotes
This work was supported by grant IOS-1339130 to A.B. from the US National Science Foundation. J.M.G benefits from funding from the French government as part of the program "Investments for the Future", under the framework of the Labex consortium MitoCross [ANR-11-LABX- 0057_MITOCROSS].
Articles can be viewed without a subscription.
References
- Aguilera A, Gómez-González B (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Baruch-Torres N, Brieba LG (2017) Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res 45: 10751–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher S, Williams-Carrier R, Stiffler N, Barkan A (2015) Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim Biophys Acta 1847: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 94: 3290–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Sears BB (1992) Electron microscopic localization of replication origins in Oenothera chloroplast DNA. Mol Gen Genet 232: 33–39 [DOI] [PubMed] [Google Scholar]
- Clausen AR, Girandon L, Ali A, Knecht W, Rozpedowska E, Sandrini MP, Andreasson E, Munch-Petersen B, Piškur J (2012) Two thymidine kinases and one multisubstrate deoxyribonucleoside kinase salvage DNA precursors in Arabidopsis thaliana. FEBS J 279: 3889–3897 [DOI] [PubMed] [Google Scholar]
- Day A, Madesis P (2007) DNA replication, recombination, and repair in plastids. Top Curr Genet 19: 65–119 [Google Scholar]
- Evans-Roberts KM, Mitchenall LA, Wall MK, Leroux J, Mylne JS, Maxwell A (2016) DNA gyrase is the target for the quinolone drug ciprofloxacin in Arabidopsis thaliana. J Biol Chem 291: 3136–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H (1997) ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J 16: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ (2007) crinkled leaves 8—a mutation in the large subunit of ribonucleotide reductase--leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. Plant J 50: 118–127 [DOI] [PubMed] [Google Scholar]
- Gowrishankar J. (2015) End of the beginning: elongation and termination features of alternative modes of chromosomal replication initiation in bacteria. PLoS Genet 11: e1004909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res 26: 2286–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicka S, Kühn K, Le Ret M, Bonnard G, Imbault P, Augustyniak H, Gualberto JM (2012) A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J 72: 423–435 [DOI] [PubMed] [Google Scholar]
- Jung B, Flörchinger M, Kunz HH, Traub M, Wartenberg R, Jeblick W, Neuhaus HE, Möhlmann T (2009) Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 21: 876–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B, Hoffmann C, Möhlmann T (2011) Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of purines. Plant J 65: 703–711 [DOI] [PubMed] [Google Scholar]
- Kafer C, Zhou L, Santoso D, Guirgis A, Weers B, Park S, Thornburg R (2004) Regulation of pyrimidine metabolism in plants. Front Biosci 9: 1611–1625 [DOI] [PubMed] [Google Scholar]
- Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61: 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Tewari KK (1975) Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature 256: 708–711 [DOI] [PubMed] [Google Scholar]
- Konrad A, Lai J, Mutahir Z, Piškur J, Liberles DA (2014) The phylogenetic distribution and evolution of enzymes within the thymidine kinase 2-like gene family in metazoa. J Mol Evol 78: 202–216 [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL (1997) Fine mapping of replication origins (ori A and ori B) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res 25: 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Huq E, Herrin DL (2010) Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci USA 107: 13954–13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech RM, Rumsby MG, Thomson WW (1973) Plastid differentiation, acyl lipid, and Fatty Acid changes in developing green maize leaves. Plant Physiol 52: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Kunnimalaiyaan M, Nielsen BL (1996) Characterization of replication origins flanking the 23S rRNA gene in tobacco chloroplast DNA. Plant Mol Biol 32: 693–706 [DOI] [PubMed] [Google Scholar]
- Mainguet SE, Gakière B, Majira A, Pelletier S, Bringel F, Guérard F, Caboche M, Berthomé R, Renou JP (2009) Uracil salvage is necessary for early Arabidopsis development. Plant J 60: 280–291 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C (2002) Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol 128: 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer SK, Lössl A, Tzekova L, Zou Z, Koop HU (2002) Functional analysis of plastid DNA replication origins in tobacco by targeted inactivation. Plant J 32: 175–184 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman JT, Kozhevnikova EN, Verrijzer CP, Pindyurin AV, Andreyeva EN, Shloma VV, Zhimulev IF, Orr-Weaver TL (2014) DNA copy-number control through inhibition of replication fork progression. Cell Reports 9: 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ (2004) Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol 335: 953–970 [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ (2016) The linear plastid chromosomes of maize: terminal sequences, structures, and implications for DNA replication. Curr Genet 62: 431–442 [DOI] [PubMed] [Google Scholar]
- Parent JS, Lepage E, Brisson N (2011) Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol 156: 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza-García JA, Nájera-Martínez M, de la Paz Sanchez M, Plasencia J (2015) Arabidopsis thaliana thymidine kinase 1a is ubiquitously expressed during development and contributes to confer tolerance to genotoxic stress. Plant Mol Biol 87: 303–315 [DOI] [PubMed] [Google Scholar]
- Scharff LB, Koop HU (2006) Linear molecules of tobacco ptDNA end at known replication origins and additional loci. Plant Mol Biol 62: 611–621 [DOI] [PubMed] [Google Scholar]
- Scharff LB, Koop HU (2007) Targeted inactivation of the tobacco plastome origins of replication A and B. Plant J 50: 782–794 [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66: 409–435 [DOI] [PubMed] [Google Scholar]
- Stasolla C, Katahira R, Thorpe TA, Ashihara H (2003) Purine and pyrimidine nucleotide metabolism in higher plants. J Plant Physiol 160: 1271–1295 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LY, Matsushima R, Sakamoto W (2012) Mutations defective in ribonucleotide reductase activity interfere with pollen plastid DNA degradation mediated by DPD1 exonuclease. Plant J 70: 637–649 [DOI] [PubMed] [Google Scholar]
- Udy DB, Belcher S, Williams-Carrier R, Gualberto JM, Barkan A (2012) Effects of reduced chloroplast gene copy number on chloroplast gene expression in maize. Plant Physiol 160: 1420–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermel M, Guermann B, Delage L, Grienenberger JM, Maréchal-Drouard L, Gualberto JM (2002) A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc Natl Acad Sci USA 99: 5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MK, Mitchenall LA, Maxwell A (2004) Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc Natl Acad Sci USA 101: 7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallet C, Le Ret M, Bergdoll M, Bichara M, Dietrich A, Gualberto JM (2015) The RECG1 DNA translocase is a key factor in recombination surveillance, repair, and segregation of the mitochondrial DNA in Arabidopsis. Plant Cell 27: 2907–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde RA, Coalter R, Barkan A (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Witz S, Jung B, Fürst S, Möhlmann T (2012) De novo pyrimidine nucleotide synthesis mainly occurs outside of plastids, but a previously undiscovered nucleobase importer provides substrates for the essential salvage pathway in Arabidopsis. Plant Cell 24: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. (2012) DNA replication and strand asymmetry in prokaryotic and mitochondrial genomes. Curr Genomics 13: 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang L, Yang DL, Li Q, He Z (2015) Thymidine kinases share a conserved function for nucleotide salvage and play an essential role in Arabidopsis thaliana growth and development. New Phytol 208: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Zampini É, Lepage É, Tremblay-Belzile S, Truche S, Brisson N (2015) Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res 25: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Solaroli N, Bjerke M, Stewart JB, Rozell B, Johansson M, Karlsson A (2008) Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum Mol Genet 17: 2329–2335 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57: 805–836 [DOI] [PubMed] [Google Scholar]