bri1-301 is a temperature-sensitive misfolded brassinosteroid receptor that requires kinase activity to promote growth and is rapidly degraded in the endoplasmic reticulum and on the cell surface.
Abstract
BRASSINOSTEROID-INSENSITIVE1 (BRI1) is a leucine-rich-repeat receptor-like kinase that functions as the cell surface receptor for brassinosteroids (BRs). Previous studies showed that BRI1 requires its kinase activity to transduce the extracellular BR signal into the nucleus. Among the many reported mutant bri1 alleles, bri1-301 is unique, as its glycine-989-to-isoleucine mutation completely inhibits its kinase activity in vitro but only gives rise to a weak dwarf phenotype compared with strong or null bri1 alleles, raising the question of whether kinase activity is essential for the biological function of BRI1. Here, we show that the Arabidopsis (Arabidopsis thaliana) bri1-301 mutant receptor exhibits weak BR-triggered phosphorylation in vivo and absolutely requires its kinase activity for the limited growth that occurs in the bri1-301 mutant. We also show that bri1-301 is a temperature-sensitive misfolded protein that is rapidly degraded in the endoplasmic reticulum and at the plasma membrane by yet unknown mechanisms. A temperature increase from 22°C to 29°C reduced the protein stability and biochemical activity of bri1-301, likely due to temperature-enhanced protein misfolding. The bri1-301 protein could be used as a model to study the degradation machinery for misfolded membrane proteins with cytosolic structural lesions and the plasma membrane-associated protein quality-control mechanism.
Brassinosteroids (BRs) are important plant growth hormones that regulate a wide range of plant developmental and physiological processes, such as pollen development, seed germination, vascular differentiation, hypocotyl/petiole elongation, senescence, stomata development, and flowering time (Clouse, 2011). Genetic, biochemical, and structural biology studies have shown that these plant steroids are perceived at the cell surface by BRASSINOSTEROID-INSENSITIVE1 (BRI1), a leucine-rich-repeat receptor-like kinase (LRR-RLK; Li and Chory, 1997; He et al., 2000; Wang et al., 2001; Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). Upon BR binding, BRI1 heterodimerizes with and transphosphorylates BRI1-ASSOCIATED RECEPTOR KINASE (BAK1), a similar but smaller LRR-RLK (Li et al., 2002; Nam and Li, 2002), to initiate a protein phosphorylation-mediated signaling cascade, which controls the activities of several key transcription factors regulating the expression of thousands of BR-responsive genes important for plant growth (for review, see Belkhadir and Jaillais, 2015). Loss-of-function mutations in BR-biosynthesis enzymes or BRI1 result in characteristic BR-deficient/insensitive phenotypes that include short hypocotyls in the dark, dwarf stature in the light, altered vascular development, prolonged vegetative phase, and reduced male fertility (Clouse, 1996; Li et al., 1996; Szekeres et al., 1996; Li and Chory, 1997).
BRI1 is the only BR signaling component identified by loss-of-function mutations in multiple forward genetic screens (Clouse, 1996; Li and Chory, 1997; Noguchi et al., 1999), and over 30 mutant bri1 alleles have been reported so far (Friedrichsen et al., 2000; Wang et al., 2001, 2005; Xu et al., 2008; Kang et al., 2010; Shang et al., 2011; She et al., 2011; Santiago et al., 2013; Bojar et al., 2014; Sun et al., 2017), most of which carry missense mutations in the BR-binding extracellular domain and the cytoplasmic kinase domain and exhibit dwarf phenotypes of varying strength (Table 1). Previous biochemical and structural studies have demonstrated or suggested that some bri1 mutations affect BRI1 binding with its ligand or its coreceptor BAK1 (He et al., 2000; Wang et al., 2001; Santiago et al., 2013; Bojar et al., 2014), some prevent BRI1 intracellular trafficking (Jin et al., 2007; Hong et al., 2008), while others inhibit BRI1’s kinase activity (Friedrichsen et al., 2000; Wang et al., 2005; Kang et al., 2010; Sun et al., 2017). Among these characterized bri1 alleles, bri1-301 is very interesting, as this mutant was generated by ethylmethanesulfonate mutagenesis, which often causes G-to-A and C-to-T transitions (Greene et al., 2003), but carries a GG-AT two-nucleotide change in the BRI1 gene, resulting in the Gly-989-Ile (G989I) missense mutation in the kinase domain (Xu et al., 2008). Morphologically, it is one of the weakest known bri1 mutants reported so far. However, two previous studies show that bri1-301 has an inactive kinase when assayed by in vitro phosphorylation or in yeast cells that coexpressed the full-length proteins of BRI1/bri1-301 and the wild-type BAK1 (Xu et al., 2008; Kang et al., 2010). It is interesting that a recently reported bri1 allele, known as bri1-702, carrying a mutation of Pro-1050-Ser, exhibited significantly reduced but detectable in vitro autophosphorylation activity but caused a growth phenotype stronger than bri1-301 (Sun et al., 2017). That study also included a transgenic experiment showing that the GFP-tagged BAK1 was constitutively phosphorylated in bri1-301 but not in a T-DNA insertional null bri1 mutant background (Sun et al., 2017), suggesting that bri1-301 might be an active kinase in Arabidopsis (Arabidopsis thaliana). Because the detected BAK1 phosphorylation might not be catalyzed by bri1-301 but a yet unknown bri1-301-dependent mechanism, it remains to be determined if bri1-301 requires its kinase activity for the observed weak growth phenotype. It is also worth mentioning that another recently reported mutant BR receptor, bri1-707 (with the same Gly-989 residue mutated to Glu), exhibits slightly reduced in vitro autophosphorylation activity but does not cause any detectable growth alteration (Sun et al., 2017).
Table 1. Summary of previously reported bri1 alleles.
Ecotypes are as follows: Col-0, Columbia-0; Ws-2, Wassilewskija-2.
| bri1 Allele | Mutation Site | Dwarfism | Ecotype | Biochemical Effect | Reference |
|---|---|---|---|---|---|
| bri1-1 | G2725A | Strong | Col-0 | A909T, extremely weak in vivo BL-stimulated BAK1 phosphorylation | Clouse et al. (1996); Wang et al. (2008) |
| bri1-2 (cbb2) | Unknown | Strong | C24 | Unknown | Kauschmann et al. (1996) |
| bri1-3 | 4-bp deletion at 2,745 | Strong | Ws-2 | Premature termination | Noguchi et al. (1999) |
| bri1-4 | 10-bp deletion at 459 | Strong | Ws-2 | Premature termination | Noguchi et al. (1999) |
| bri1-5 | G206A | Weak | Ws-2 | C69Y, ER retention | Noguchi et al. (1999) |
| bri1-5R1 | G260A | Weak | bri1-5 Ws-2 | Partially suppresses the bri1-5 mutation | Belkhadir et al. (2010) |
| bri1-6/bri1-119 | G1931A | Weak | Enkheim-2 | G644D, unknown | Noguchi et al. (1999); Friedrichsen et al. (2000) |
| bri1-7 | G1838A | Weak | Ws-2 | G613S, unknown | Noguchi et al. (1999) |
| bri1-8/bri1-108-102 | G2948A | Intermediate | Ws-2/Col-0 | R983N, no detectable in vitro kinase activity | Noguchi et al. (1999) |
| bri1-9 | C1985T | Weak | Ws-2/Col-0 | S662F, ER retention | Noguchi et al. (1999); Jin et al. (2007) |
| bri1-101 | G3232A | Strong | Col-0 | E1078K, extremely weak in vitro kinase activity | Li and Chory (1997); Friedrichsen et al. (2000) |
| bri1-102 | C2249T | Strong | Col-0 | T750I | Friedrichsen et al. (2000) |
| bri1-103/104 | G3091A | Strong | Col-0 | A1031T | Li and Chory (1997); Friedrichsen et al. (2000) |
| bri1-105-107 | C3175T | Strong | Col-0 | Q1059Stop | Li and Chory (1997); Friedrichsen et al. (2000) |
| bri1-113 | G1832A | Strong | Col-0 | G611E | Li and Chory (1997) |
| bri1-114/116 | C1747T | Strong | Col-0 | Q583Stop | Friedrichsen et al. (2000) |
| bri1-115 | G3143A | Strong | Col-0 | G1048D | Li and Chory (1997) |
| bri1-117/118 | G3415A | Strong | Col-0 | D1139N | Friedrichsen et al. (2000) |
| bri1-120 | T1196C | Weak | Landsberg erecta | S399F in the 13th LRR | Shang et al. (2011) |
| bri1-201 | G1831A | Strong | Ws-2 | G611R | Domagalska et al. (2007) |
| bri1-202 | C2854T | Strong | Ws-2 | R952W | Domagalska et al. (2007) |
| bri1-301 | GG2965/6AT | Very weak | Col-0 | No detectable kinase activity in vitro or in yeast cells | Xu et al. (2008); Kang et al. (2010); this study |
| bri1-401 | G2714A/G3582A | Strong | Ws | G895R/G1194E | Tanaka et al. (2005) |
| bri1-701 | T-DNA insertion at 1,245 | Strong | Col-0 | Knockout of BRI1 | Gou et al. (2012) |
| bri1-702 | C3148T | Weak | Col-0 | P1050S, reduced in vitro kinase activity | Sun et al. (2017) |
| bri1-703 | G3166A | Strong | Col-0 | E1056K, no detectable in vitro kinase activity | Sun et al. (2017) |
| bri1-704 | G3079A | Strong | Col-0 | D1027N, no detectable in vitro kinase activity | Sun et al. (2017) |
| bri1-705 | C2156T | Subtle | Col-0 | P719L, likely affects the interaction of BL, BRI1, and BAK1 | Sun et al. (2017) |
| bri1-706 | C758T | Subtle | Col-0 | S253F | Sun et al. (2017) |
| bri1-708 | C2947G | Strong | Col-0 | R983G, no detectable in vitro kinase activity | Sun et al. (2017) |
| bri1-709 | G2543A | Strong | Col-0 | W848Stop, premature termination | Sun et al. (2017) |
| bri1-710 | G1858A | Subtle | Col-0 | G620R | Sun et al. (2017) |
| bri1-711 | G2236A | Subtle | Col-0 | G746S | Sun et al. (2017) |
Because bri1-301 is widely used in many studies investigating BR signaling and its cross talk with many other plant signaling processes (Li and Nam, 2002; Nam and Li, 2002; Kim et al., 2007; Wang et al., 2009, 2016; Kang et al., 2010; Schwessinger et al., 2011; Albrecht et al., 2012; Shi et al., 2015; Unterholzner et al., 2015; Ha et al., 2016; Hao et al., 2016), we decided to investigate the mechanism of the bri1-301 mutation. During our study, we also discovered that bri1-301 exhibited a severe growth defect when grown in a 29°C growth chamber. Our biochemical and genetic experiments revealed that bri1-301 is a temperature-sensitive misfolded BR receptor that is unstable in the endoplasmic reticulum (ER) and at the plasma membrane (PM) and requires its kinase activity for the observed weak growth phenotype. Our study suggests that bri1-301 may serve as a model protein to study the PM-associated quality-control mechanism in Arabidopsis.
RESULTS
The Weak Growth Phenotype of bri1-301 Is Caused by the bri1-301 Mutation
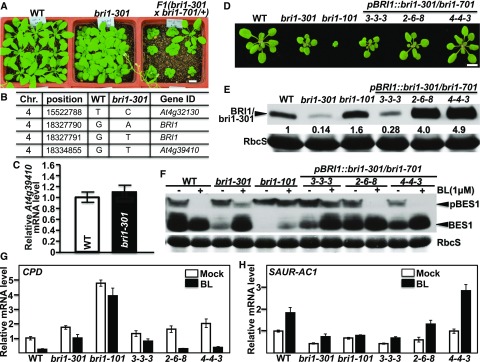
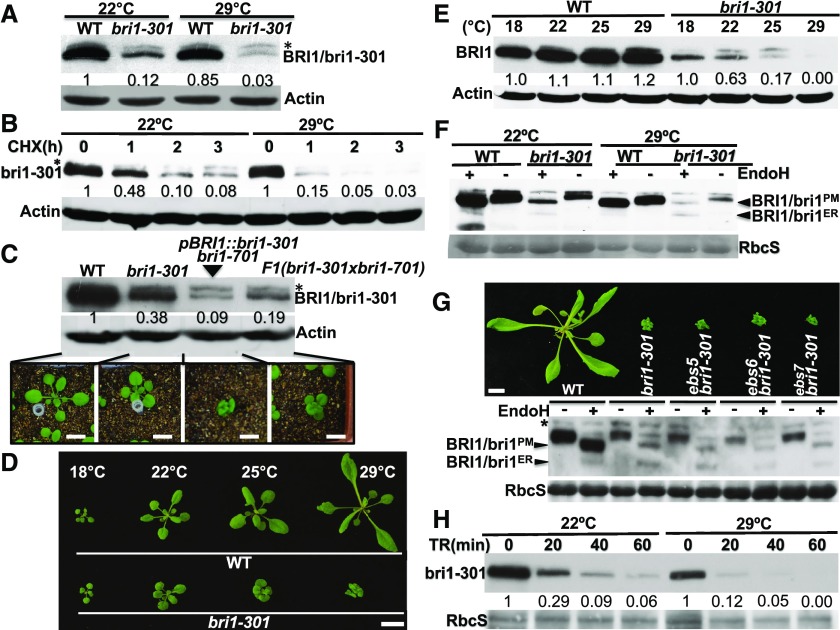
Because no severe dwarf phenotype segregated out in our previous genetic studies with the bri1-301 mutant (Li and Nam, 2002; Nam and Li, 2002; Wang et al., 2009; Kang et al., 2010), we initially hypothesized that the weak growth phenotype of the bri1-301 mutant might be caused by a linked unknown mutation that suppresses the dwarf phenotype of the "kinase-dead" BR receptor. To test this hypothesis, we crossed bri1-301 with the heterozygous T-DNA insertional mutant bri1-701/+ and discovered that the bri1-301/bri1-701 heterozygous mutants were smaller than bri1-301 (Fig. 1A), suggesting that the enhanced dwarfism could be caused by eliminating a recessive suppressor mutation or by reducing bri1-301 abundance. Genome sequencing of the bri1-301 mutant revealed three nucleotide changes in the bottom 3,100-kb region of chromosome 4 (15,522,789–18,660,050; Fig. 1B), including the two-nucleotide change (GG-AT at positions of 18,327,790–18,327,791) in the BRI1 gene (At4g39400; Xu et al., 2008) and a single-nucleotide change (G-T at position 18,334,855) in the immediate downstream gene (At4g39410) of BRI1. Because the third nucleotide change occurs in the annotated promoter region of At4g39410 that encodes an Arabidopsis WRKY transcription factor, AtWRKY13, and thus might affect At4g39410 transcript abundance, we performed reverse transcription quantitative PCR (RT-qPCR) and found that the G-T nucleotide change had little effect on At4g39410 transcript abundance (Fig. 1C), suggesting that the weak growth phenotype of bri1-301 is unlikely to be caused by the G-T mutation but rather is the result of the two-nucleotide change in the BRI1 gene.
Figure 1.
The G989I mutation of BRI1 is responsible for the weak growth phenotype of bri1-301. A, Photographs of 26-d-old soil-grown plants in a 22°C growth chamber. B, Nucleotide changes between bri1-301 and its wild-type control (WT) within the bottom 3,000-kb region of chromosome 4. C, RT-qPCR analysis of the relative transcript abundance of At4g39410 between 10-d-old seedlings of the wild type and bri1-301 grown in a 22°C growth room. D, Photographs of 18-d-old soil-grown seedlings in a 22°C growth room. E, Immunoblot analysis of BRI1/bri1-301 abundance of the Arabidopsis seedlings shown in D. The numbers shown below the anti-BRI1 strip are the relative values of anti-BRI1 signals after normalization with the signal intensity of the corresponding Coomassie Blue-stained RbcS bands. The signal intensity of each band was quantified by ImageJ. F, Immunoblot analysis of BES1 phosphorylation status. Eight-day-old seedlings grown on one-half-strength Murashige and Skoog (1/2 MS) medium were carefully transferred into liquid 1/2 MS medium supplemented with or without 1 μm brassinolide (BL) and incubated for 2 h. Their total protein extracts were separated by 10% SDS-PAGE and analyzed by Coomassie Blue staining or immunoblotting with an anti-BES1 antibody. G and H, RT-qPCR analyses of the relative transcript abundance of CPD (G) and SAUR-AC1 (H) using total RNAs extracted from 8-d-old seedlings treated with or without 1 μm BL for 2 h. The transcript abundance of CPD or SAUR-AC1 in nontreated wild-type seedlings was set as 1. In C, G, and H, for each sample, the RT-qPCR assays were repeated four times, and the error bars denote ±sd. Bars in A and D = 1 cm.
Further support for our conclusion came from our transgenic experiment. We generated a pBRI1::bri1-301 construct (driven by the BRI1 promoter), transformed it into heterozygous bri1-701 plants, and analyzed the phenotypes of the resulting transgenic lines that were homozygous for the bri1-701 mutation. As shown in Figure 1D, the pBRI1::bri1-301 construct complemented the null bri1-701 mutation, and the growth phenotypes observed with the pBRI1::bri1-301 bri1-701 transgenic lines matched well with bri1-301 protein abundance, with the wild type-like lines accumulating higher amounts of bri1-301 (Fig. 1E). It is important to note that a line exhibiting bri1-301-like morphology contained a similar amount of bri1-301 protein as the bri1-301 mutant (Fig. 1, D and E). Importantly, this line also exhibited similar BR sensitivity to the bri1-301 mutant revealed by the BR-induced BRI1-EMS-SUPPRESSOR1 (BES1) dephosphorylation assay (Mora-García et al., 2004) and by RT-qPCR analysis of three BR-responsive genes, CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD; Mathur et al., 1998), DWARF4 (DWF4; Bancoş et al., 2002), and SMALL-AUXIN-UP-RNA-ARABIDOPSIS COLUMBIA1 (SAUR-AC1; Nakamura et al., 2003; Fig. 1, F–H; Supplemental Fig. S1A). Consistent with their growth morphology, the two wild-type-like pBRI1::bri1-301 bri1-701 transgenic lines displayed wild-type-like BR sensitivity, as shown in Figure 1, F to H, and Supplemental Figure S1A. Taken together, these results showed that the weak growth phenotype of bri1-301 is due to the G989I substitution in BRI1 rather by the G-T mutation in the At4g39410 promoter.
The bri1-301 Protein Requires Its Kinase Activity for Its Weak Growth Phenotype
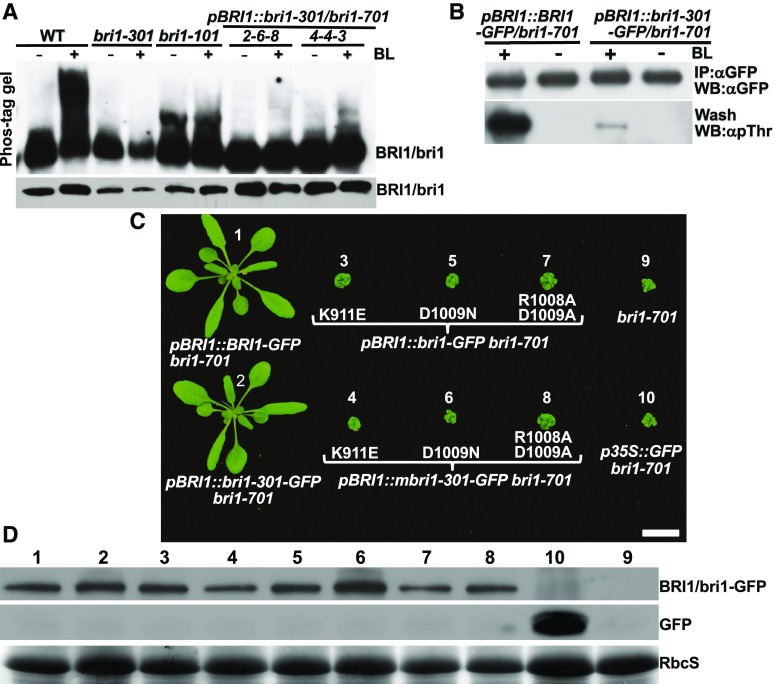
Our second hypothesis to explain the discrepancy between the nondetectable kinase activity of bri1-301 in vitro or in yeast cells and its weak growth phenotype is that bri1-301 is a rather active kinase in vivo. To test this hypothesis, we performed two different experiments capable of detecting in vivo phosphorylated proteins. First, we performed Phos-tag immunoblotting with total proteins extracted from seedlings of the wild type, bri1-301, and bri1-101 treated with or without 1 μm BL (the most active member of the BR family). Due to the presence of Phos-tag, a selective phosphate-binding tag molecule, in the SDS-polyacrylamide gel, phosphorylated proteins move slower than their corresponding nonphosphorylated forms (Kinoshita et al., 2009). As shown in Figure 2A, a 4-h treatment with 1 μm BL caused the appearance of highly abundant slower-moving BRI1 bands in the wild-type sample, while the same treatment resulted in no detectable slower-moving bands in the bri1-301 sample. To address the concern that our failure to detect slower-moving bri1-301 bands could be due simply to the lower abundance of total bri1-301 proteins (Fig. 2A), we included two wild-type-looking pBRI1::bri1-301 bri1-701 transgenic lines with similar or higher bri1-301 abundance than the wild-type BRI1 abundance (Fig. 2, A and B). As shown in Figure 2A, the 4-h BL treatment did show slower-moving bri1-301 bands in the two transgenic lines; however, the abundance of these slower-moving bands was much lower than that of the corresponding wild-type BRI1 bands, suggesting that bri1-301 exhibits a very low level of BR-induced phosphorylation. This conclusion was further supported by our second immunoblot experiment with a pBRI1::BRI1-GFP bri1-701 transgenic line and a wild-type-like pBRI1::bri1-301-GFP bri1-701 transgenic line (Fig. 2B) using an anti-pThr antibody capable of detecting phosphorylated Thr residues in BRI1 and its coreceptor BAK1 (Wang et al., 2005).
Figure 2.
bri1-301 is weakly phosphorylated in vivo and requires its kinase activity for the weak growth phenotype. A, Phos-tag assay of BL-triggered BRI1/bri1 phosphorylation (top strip; for details, see “Materials and Methods”) and immunoblot analysis of BRI1/bri1-301 abundance (bottom strip). WT, Wild type. B, Immunoblot analysis of BL-induced BRI1/bri1-301 phosphorylation by an anti-pThr antibody with 18-d-old seedlings treated with or without 1 μm BL. IP, immunoprecipitation; WB, western blot. C, Photographs of 26-d-old soil-grown seedlings of transgenic bri1-701 lines. Bar = 1 cm. D, Immunoblot analysis of the transgenically expressed BRI1/bri1-301-GFP fusion proteins in the lines shown in C. The numbers shown above the blots correspond to the numbered lines shown in C. The bottom strip shows the Coomassie Blue-stained RbcS bands for the loading control.
The detected bri1-301 phosphorylation could be caused by its autophosphorylation activity or transphosphorylation by a BRI1-associated kinase, such as BAK1 that functions as a BRI1 coreceptor (Li et al., 2002; Nam and Li, 2002). To directly test if bri1-301 requires its kinase activity for the observed weak growth phenotype, we mutated three residues in the kinase domain of BRI1 known to be essential for its kinase activity: Lys-911 mutated to Glu (K911E), which is known to form the salt bridge with Glu-927 essential to maintain an active conformation of BRI1 (Bojar et al., 2014); Arg-1008 mutated to Asn (R1008N) or Ala (R1008A); and Asp-1009 mutated to Ala (D1009A) in combination with the R1008A mutation of the catalytic His-Arg-Asp loop motif, known to be highly conserved throughout the protein kinase family (Hanks et al., 1988). We individually transformed the mutated pBRI1::BRI1-GFP or pBRI1::bri1-301-GFP transgene into the heterozygous bri1-701 plants and screened T1 plants homozygous for the bri1-701 mutation. We discovered that every homozygous bri1-701 transgenic plant was morphologically identical to the parental bri1-701 mutant (Fig. 2C), indicating that the kinase activity is absolutely required for the weak growth phenotype of the mutant bri1-301 receptor.
bri1-301 Is an Unstable Protein in the ER and on the PM
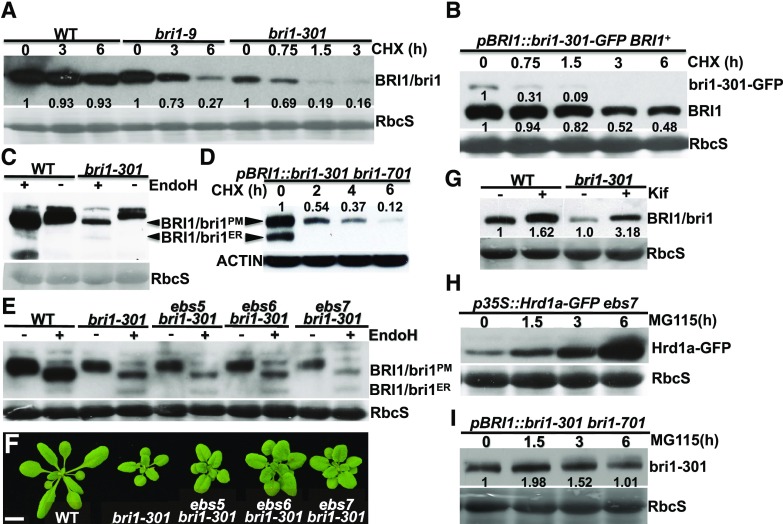
During our experiment assaying the BR-induced BRI1/bri1-301 phosphorylation, we discovered that the protein abundance of bri1-301 was much lower than that of wild-type BRI1 (Figs. 1E and 2A), suggesting that bri1-301 is an unstable protein. To determine if the low bri1-301 abundance is caused by increased degradation or decreased biosynthesis of bri1-301, we performed a cycloheximide (CHX)-chasing experiment by treating Arabidopsis seedlings of bri1-9, bri1-301, and their wild-type control with CHX, a widely used protein biosynthesis inhibitor. As shown in Figure 3A, while the wild-type BRI1 protein was rather stable, bri1-301 was rapidly degraded in the treated bri1-301 seedlings. Interestingly, it seemed that bri1-301 disappeared more rapidly than the ER-retained bri1-9, which is degraded by an endoplasmic reticulum-associated degradation (ERAD) mechanism (Hong et al., 2009). A similar study was performed with a transgenic line that expresses a GFP-tagged bri1-301 (driven by the BRI1 promoter) in a wild-type background. As shown in Figure 3B, the GFP-tagged bri1-301 degraded much more rapidly than the endogenous wild-type BRI1. Taken together, our experiments demonstrated that bri1-301 is rapidly degraded in Arabidopsis plants.
Figure 3.
bri1-301 is an unstable protein that is degraded in the ER and at the PM. A, Immunoblot analysis of BRI1/bri1 stability after treatment with CHX. B, Immunoblot analysis of the protein stability of the endogenous BRI1 and the transgenically expressed bri1-301-GFP fusion proteins. C and D, Endo-H analysis of BRI1/bri1-301 and transgenically expressed bri1-301 in the bri1-701 background. E, Immunoblot analysis of BRI1/bri1-301 abundance with total proteins treated with or without Endo-H. F, Photographs of 18-d-old soil-grown plants in a 22°C growth room. Bar = 1 cm. G, Immunoblot analysis of the impact of Kif treatment on BRI1/bri1301 abundance. H and I, Immunoblot analysis of the effect of MG115 on the stability of Hrd1a-GFP (H; used as a positive control) and bri1-301 (I). In A to C, E, and G to I, Coomassie Blue staining of duplicate gels was used as the loading control, while in D, immunoblotting of a duplicate blot with anti-ACTIN antibody served as the loading control. In A, B, D, and G to I, the numbers on the gel images or in the space between gel images are relative values of the anti-BRI1 signals (against the nontreated sample of the same genotype) after normalization with the signal intensity of the corresponding RbcS or ACTIN band. ImageJ was used to quantify the signal intensity of each band. WT, wild type.
Our earlier endoglycosidase H (Endo-H) assay revealed that bri1-301 is not retained in the ER but is localized mainly on the PM (Hong et al., 2008), as Endo-H is an endoglycosidase capable of cleaving high-mannose (Man)-type Asn-linked glycans (N-glycans) of ER-retained glycoproteins but not the Golgi-processed complex-type N-glycans (Robbins et al., 1984). However, Figure 3C shows a low but detectable level of the Endo-H-sensitive form of bri1-301 on the immunoblot, suggesting that the G989I mutation leads to the presence of a very small pool of bri1-301 proteins in the ER. Consistently, a similar Endo-H assay of bri1-301 from a wild-type-looking pBRI1:bri1-301 bri1-701 line revealed the presence of a high level of an ER-retained form of bri1-301, which was degraded much faster than the PM-localized bri1-301 proteins (Fig. 3D). We hypothesized that bri1-301 is a misfolded protein with a cytosolic structural lesion that is not efficiently retained in the ER. In addition, its ER-retained form is rapidly degraded and remains unstable even when it reaches the PM.
To investigate if the low abundance of the ER-retained bri1-301 pool is due to rapid degradation by the ERAD pathway known for degrading bri1-5 and bri1-9, two ER-retained mutant BR receptors (Jin et al., 2007; Hong et al., 2008), we crossed bri1-301 with three previously reported Arabidopsis ERAD mutants: EMS-mutagenized bri1 suppressor5 (ebs5), ebs6, and ebs7 (Su et al., 2011, 2012; Liu et al., 2015). EBS5, an ER transmembrane protein, works together with the ER luminal lectin EBS6, which binds to α1,2-Man-trimmed N-glycans to recruit a terminally misfolded glycoprotein to the ER membrane-anchored ubiquitin ligase HMG-CoA-reductase degradation protein1 (Hrd1), while EBS7 regulates the protein stability of Hrd1 (Liu and Li, 2014; Liu et al., 2015). As shown in Figure 3, E and F, none of the three ERAD mutations was able to suppress the bri1-301 growth phenotype and to stabilize the ER-retained form of bri1-301, indicating that degradation of the ER-retained bri1-301 does not involve the ERAD machinery that degrades bri1-5 and bri1-9. Surprisingly, treatment of the bri1-301 mutant with kifunensine (Kif), a widely used inhibitor of α1,2-mannosidases and ERAD of glycoproteins (Elbein et al., 1990), increased the protein abundance of bri1-301, suggesting the involvement of N-glycosylation in bri1-301 degradation (Fig. 3G). Because Kif inhibits not only the ER-mediated α1,2-Man trimming of terminally misfolded glycoproteins but also the Golgi-mediated α1,2-Man trimming of correctly folded glycoproteins that are destined to the PM or other cellular organelles, it is possible that Kif treatment might affect the activity of yet unknown glycoproteins to degrade bri1-301. It is worth noting that treatment with MG115, which could block the degradation of the Arabidopsis ER-localized E3 ligase Hrd1a that becomes unstable in an ebs7 mutant background (Liu et al., 2015; Fig. 3H), showed very limited impact on bri1-301 abundance (Fig. 3I), suggesting that the cytosolic proteasome might not make a major contribution to degrading the ER-retained or PM-localized bri1-301.
The bri1-301 Mutant Is Highly Sensitive to Higher Temperatures
During our study, we noticed that bri1-301 grown in a 22°C growth chamber was a weak dwarf but became an extreme dwarf when grown in a 29°C growth chamber (Fig. 4A), suggesting that bri1-301 is a temperature-sensitive mutant. Indeed, when grown in a 25°C growth chamber, the rosette size of bri1-301 was between that of the 22°C-grown and 29°C-grown bri1-301 mutants (Fig. 4B). We also tested the temperature sensitivity of other bri1 mutants. As shown in Figure 4C, while the temperature increase of 22°C to 29°C stimulated the growth of the wild type and weak bri1 mutants such as bri1-5, bri1-702, and bri1-707, which was likely mediated by high-temperature-induced auxin biosynthesis (Gray et al., 1998; Koini et al., 2009), it had little impact on stronger bri1 mutants, including bri1-9, bri1-101, bri1-701, bri1-703, bri1-704, and bri1-708. It is interesting that the temperature-triggered severe dwarfism was not observed in the dark (Fig. 4D). A quantitative assay, which measures the sensitivity of dark-grown seedlings to a BR biosynthesis inhibitor, brassinazole (BRZ; Asami et al., 2000; Nagata et al., 2000), showed that the temperature shift had little effect on the BRZ sensitivity of the weak receptor mutant, although the 22°C to 29°C temperature shift did enhance the BRZ resistance of the dark-grown wild-type seedlings (Fig. 4E).
Figure 4.
bri1-301 is a thermosensitive dwarf mutant. A, Photographs of 26-d-old soil-grown plants in a 22°C growth chamber (left pot) and in a 29°C growth chamber (right pot). B, Photographs of 20-d-old soil-grown plants at three different temperatures. C, Photographs of 18-d-old (top) and 23-d-old (bottom) soil-grown wild-type (WT) and various bri1 mutant plants in 22°C and 29°C growth chambers. D, Photographs of 5-d-old 22°C- and 29°C-grown etiolated seedlings. E, Quantitative analysis of seedling sensitivity to BRZ. Five-day-old dark-grown seedlings were carefully removed from petri dishes and photographed, and their hypocotyl lengths were measured by ImageJ. The data points are relative hypocotyl lengths of seedlings grown on BRZ-containing medium in comparison with seedlings grown on normal 1/2 MS medium at 22°C or 29°C. For each data point, the average length was obtained through three replicates of 20 seedlings each. The error bars indicate ±sd. Bars in A to D = 1 cm.
The Thermosensitivity of bri1-301 Is Caused by the G989I Mutation of BRI1
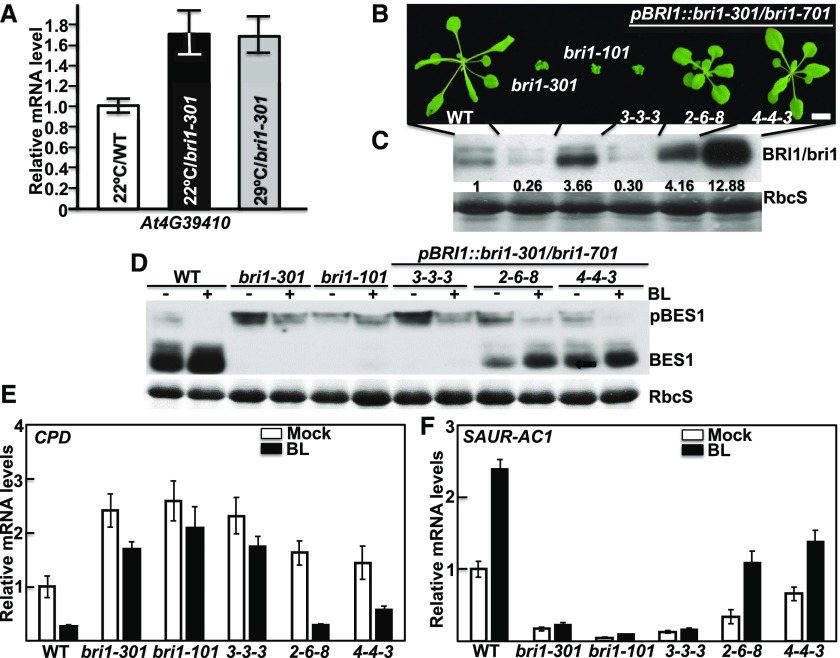
There are two possible explanations for the observed 29°C-triggered growth inhibition of bri1-301. It might be due to an unknown mutation that confers warm temperature-induced growth inhibition to bri1-301, or it may be caused by a warm temperature-triggered reduction in the activity and/or protein abundance of bri1-301. To investigate the first possibility, we crossed bri1-301 with the wild type and bri1-701, analyzed their F2 offspring (more than 3,000 F2 individual seedlings for each cross), and discovered a close linkage of the thermosensitive phenotype with the BRI1 locus. As mentioned before, there are only three detected nucleotide changes in the bottom 3,100-kb region of chromosome 4: two in the BRI1 gene and one in the promoter region of At4g39410 located immediately downstream of the BRI1 locus (Fig. 1B). Therefore, we performed RT-qPCR analysis of the At4g39410 gene with the 22°C- and 29°C-grown bri1-301 mutants but detected no obvious difference in its transcript abundance between temperatures (Fig. 5A), suggesting that the temperature-triggered severe dwarfism of bri1-301 is likely due to the G989I mutation of bri1-301 but not the nucleotide change in the At4g39410 promoter. Further support came from our analysis of the growth phenotypes of the above-mentioned pBRI1::bri1-301 bri1-701 transgenic lines grown in the 29°C growth chamber. As shown in Figure 5B, the bri1-301-like pBRI1::bri1-301 bri1-701 line (3-3-3), which contains a similar amount of bri1-301 to the bri1-301 single mutant (Fig. 5C), was an extreme dwarf resembling the 29°C-grown bri1-301 (Fig. 5B). Interestingly, the two other pBRI1::bri1-301 bri1-701 transgenic lines (2-6-8 and 4-4-3), which accumulated higher levels of bri1-301 mRNA and bri1-301 protein than the wild type (Fig. 5C; Supplemental Fig. S2), were larger than the 22°C-grown bri1-301 mutant and morphologically similar to the 22°C-grown wild-type plants (Fig. 5B). Consistent with the morphological phenotypes, the BR-induced BES1 dephosphorylation assay and RT-qPCR experiments of three BR-responsive genes showed that the 29°C-grown wild-type-looking pBRI1::bri1-301 bri1-701 lines still responded to BL, whereas the 29°C-grown seedlings of bri1-301 and the bri1-301-like pBRI1::bri1-301 bri1-701 line became insensitive to the exogenously applied BL (Fig. 5, D–F; Supplemental Fig. S1B). Together, these results demonstrated that the 29°C-triggered growth inhibition is caused by the bri1-301 mutation itself. In addition, our experiments revealed that bri1-301 remains active in promoting plant growth even at 29°C, when its protein abundance is high enough to compensate for its reduced activity at a higher growth temperature (Fig. 5, B and C).
Figure 5.
The G989I mutation is responsible for warm temperature-triggered severe dwarfism. A, RT-qPCR analysis of At4g39410 transcript abundance. B, Photographs of 20-d-old soil-grown plants in a 29°C growth chamber. Bar = 1 cm. C, Immunoblot analysis of BRI1/bri1-301 protein abundance in the plants shown in B. The numbers shown on the anti-BRI1 strip are the values of anti-BRI1 signal intensity relative to the wild-type (WT) BRI1 after normalization with the signal intensity of the corresponding Coomassie Blue-stained RbcS bands. The signal intensity of each band was quantified by ImageJ. D, Immunoblot analysis of BL-triggered changes in BES1 phosphorylation status. E and F, RT-qPCR analysis of the relative transcript abundance of CPD and SAUR-AC1 in 29°C-grown seedlings treated with or without 1 μm BL for 2 h. In A, E, and F, the error bars represent ±sd, and each bar is the average of four replicate RT-qPCR results.
The Warmer Temperature-Enhanced Protein Misfolding Is Likely Responsible for the Severe Dwarfism of 29°C-Grown bri1-301
Given the fact that the 29°C-induced severe dwarf phenotype of bri1-301 could be suppressed by increased production of the mutant BR receptor (Fig. 5, B and C), we suspected that the warm temperature-triggered dwarfism enhancement might be caused by a reduction in the activity and/or abundance of bri1-301. We first compared the protein abundance of the mutant BR receptor between 22°C- and 29°C-grown bri1-301 seedlings. As shown in Figure 6A, the temperature increase of 22°C to 29°C had a marginal impact on the protein level of the wild-type BRI1 but caused an ∼4-fold reduction in bri1-301 abundance. A subsequent CHX-chasing experiment showed that bri1-301 was degraded slightly faster in the 29°C-grown bri1-301 seedlings than in the 22°C-grown seedlings (Fig. 6B). Additional support for the suspected causal relationship between reduced bri1-301 abundance and severe dwarfism came from our immunoblot experiment with 22°C-grown seedlings of bri1-301, a pBRI1::bri1-301 bri1-701 transgenic line, the F1 offspring of the bri1-301 × bri1-701 cross, and their wild-type control plus our analysis of BRI1/bri1-301 abundance of wild-type and bri1-301 seedlings grown at 18°C, 22°C, 25°C, and 29°C. Figure 6C reveals an association between BRI1 abundance and the severity of dwarfism among 22°C-grown seedlings of various genotypes, while Figure 6, D and E, show that bri1-301 abundance decreased with the increased severity of dwarfism when the growth temperature increased.
Figure 6.
The 22°C to 29°C temperature increase reduces bri1-301 stability. A, Immunoblot analysis of BRI1/bri1-301 protein abundance. B, Immunoblot analysis of bri1-301 protein stability after the CHX treatment. C, Immunoblot analysis of BRI1/bri1-301 protein abundance in 18-d-old soil-grown seedlings (shown below the gel strips) of the wild type (WT), bri1-301, a pBRI1::bri1-301 bri1-701 line, and an F1 plant of the bri1-301 × bri1-701 cross. D, Photographs of 18-d-old seedlings grown at different temperatures. E, Immunoblot analysis of BRI1/bri1-301 protein abundance in the seedlings shown in D. F, Endo-H analysis of BRI1/bri1-301 proteins with total proteins extracted from wild-type and bri1-301 seedlings grown in 22°C and 29°C growth chambers. G, Photographs (top) and immunoblot analysis (bottom) of 20-d-old seedlings grown in soil at 29°C. H, Immunoblot analysis of trypsin sensitivity of the bri1-301 protein from the 22°C- and 29°C-grown bri1-301 mutant. TR, trypsin. In A to C, E, and H, the numbers shown in the space between two gel strips are BRI1/bri1 abundance (relative to that of the wild-type BRI1 in lane 1 in A and C, to that of nontreated samples in B and H, or to that of 18°C-grown seedlings of the same genotype in E) after normalization with the signals of corresponding ACTIN (A–C and E) or RbcS (H) bands. Bars in C, D, and G = 1 cm.
Based on our findings of the temperature-dependent reduction in the protein abundance of bri1-301 and its increased degradation rate at 29°C, we hypothesized that bri1-301 is a temperature-sensitive misfolded BR receptor that becomes even more misfolded at higher temperatures. If so, we would expect that more bri1-301 proteins would be retained in the ER. To test this hypothesis, we performed an Endo-H experiment, which indeed showed an increased ratio of ER-retained bri1-301 (Endo-H-sensitive form) to the PM-localized bri1-301 (Endo-H-resistant form) despite a remarkable reduction in the total amount of bri1-301 by the temperature increase (Fig. 6F). Similar to what was observed in 22°C-grown seedlings, degradation of the ER-retained bri1-301 in 29°C-grown seedlings did not involve EBS5, EBS6, or EBS7 (Fig. 6G). Consistently, the morphology of 29°C-grown ebs5 bri1-301, ebs6 bri1-301, or ebs7 bri1-301 was indistinguishable from that of the 29°C-grown bri1-301 single mutant (Fig. 6G). Additional support for a more misfolded state of bri1-301 in 29°C-grown bri1-301 mutants came from a trypsin sensitivity assay, which is widely used to assess if a protein is misfolded compared with its correctly folded conformer (https://bio-protocol.org/e1953). As shown in Figure 6H, the bri1-301 protein extracted from the 29°C-grown seedlings was more sensitive to the trypsin digestion than that extracted from the 22°C-grown seedlings. Together, these results strongly suggested that bri1-301 becomes more misfolded at 29°C, leading to its increased ER retention and further reduced protein stability on the PM.
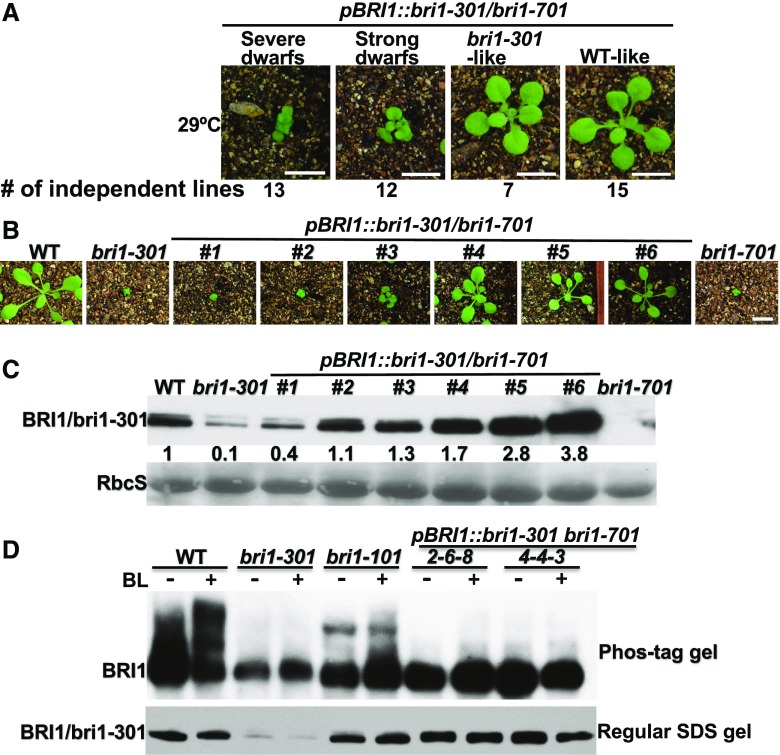
The temperature-enhanced misfolding and increased degradation of bri1-301 suggested that the activity of the mutant receptor also could be reduced by higher growth temperatures. Consistent with this assessment, morphological and immunoblot analyses of additional 29°C-grown pBRI1::bri1-301 bri1-701 transgenic lines, which were generated by an independent transformation experiment (Fig. 7A), showed that two transgenic lines (#1 and #2, with ∼4- and 11-fold higher bri1-301 abundance than that of the 29°C-grown bri1-301 mutant, respectively) were still severe dwarfs (Fig. 7, B and C). If the temperature increase had only affected bri1-301 stability, a 29°C-grown pBRI1::bri1-301 bri1-701 transgenic line accumulating greater than 4 times more bri1-301 than the 29°C-grown bri1-301 would be morphologically similar to the 22°C-grown bri1-301, as the bri1-301 abundance of 22°C-grown bri1-301 was ∼4-fold higher than that of the 29°C-grown bri1-301 (Fig. 6A). Further support for a temperature-triggered reduction of bri1-301 activity came from a Phos-tag experiment that measured the BR-induced BRI1/bri1-301 phosphorylation level in 29°C-grown seedlings. As shown in Figure 7D, despite the similar abundance of transgenically expressed bri1-301 in the bri1-701 mutant background to that of wild-type BRI1 in the wild-type control, the BL treatment resulted in no detectable BR-triggered bri1-301 phosphorylation in the two pBRI1::bri1-301 bri1-701 transgenic lines. This is quite different from what was observed with the same two transgenic lines grown in a 22°C growth chamber (Fig. 2A). Together, these two experiments strongly suggested that the temperature-enhanced bri1-301 misfolding not only increases the degradation rate of the mutant BR receptor but also reduces its biochemical activity.
Figure 7.
The 22°C to 29°C temperature increase also inhibits bri1-301 activity. A, Phenotypic grouping of transgenic pBRI1::bri1-301 bri1-701 lines grown in a 29°C growth chamber. The numbers indicate the numbers of transgenic lines (out of the same transformation experiment) exhibiting similar growth morphology to the plant shown above. B, Photographs of 18-d-old soil-grown plants in the 29°C growth chamber. C, Immunoblot analysis of BRI1/bri1-301 protein abundance of the plants shown in B. The numbers shown between the two gel strips are the relative signal intensity of the bri1-301 bands after normalization with the signals of the corresponding Coomassie Blue-stained RbcS bands. The signal intensity of each band was measured by ImageJ. D, Immunoblot analysis of BRI1/bri1-301 phosphorylation status in 29°C-grown plants of the two characterized wild-type-like pBRI1::bri1-301 bri1-701 lines shown in Figure 1D. The top strip is the anti-BRI1 immunoblot from a Phos-tag gel, while the bottom strip is the anti-BRI1 immunoblot from a regular SDS-PAGE of the same set of protein samples. WT, wild type.
DISCUSSION
The Mutant bri1-301 Receptor Requires Its Kinase Activity to Promote Plant Growth
Among various reported alleles of the Arabidopsis BR receptor, bri1-301 is very interesting because it lacks a detectable auto/transphosphorylation activity in vitro or in yeast cells but only causes a weak growth phenotype (Xu et al., 2008; Kang et al., 2010; Sun et al., 2017), which led us to question whether the kinase activity is required for BRI1’s physiological activity. In this study, we eliminated the possibility of a linked suppressor mutation being responsible for the apparent inconsistency between an inactive kinase and the weak growth phenotype and demonstrated by a transgenic approach that the weak growth phenotype of bri1-301 is caused by the G989I mutation in the BRI1 protein. Our analysis of in vivo bri1-301 phosphorylation with an anti-pThr antibody and Phos-tag gel electrophoresis revealed that bri1-301 was very weakly phosphorylated in response to BR treatment, which is quite different from what was reported recently for the strong BR-independent BAK1 phosphorylation in the bri1-301 mutant background (Sun et al., 2017). Although we could not tell if the detected bri1-301 phosphorylation was caused by autophosphorylation of bri1-301 or transphosphorylation by a BAK1 or other yet unknown BRI1-interating kinases, our transgenic experiments using several kinase-dead mutant pBRI1::bri1-301 constructs that were transformed into the null bri1-701 mutant clearly demonstrated that bri1-301 absolutely requires its kinase activity for the weak growth phenotype, as all of the transgenic lines expressing these mutant pBRI1::bri1-301 transgenes were morphologically indistinguishable from bri1-701 (Fig. 2C).
The G989I Mutation Might Cause a Structural Defect of the BRI1 Kinase Domain That Is Partially Stabilized by Chaperones/Cochaperones in Planta
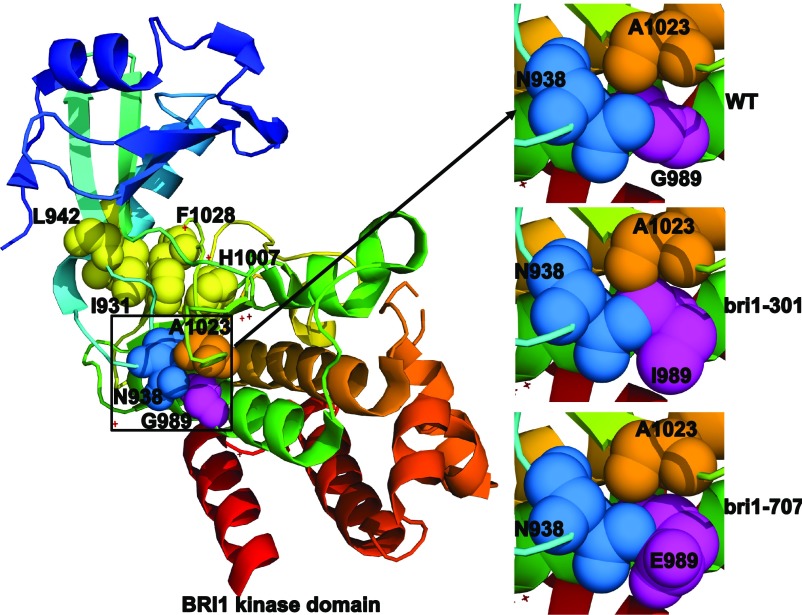
The Gly-989 residue mutated in bri1-301 is located in the conserved kinase subdomain VIa (Li and Chory, 1997). While this residue is not conserved among the plant LRR-RLKs, it is absolutely conserved between BRI1 and its plant homologs (Sun et al., 2017). Analysis of the BRI1 kinase’s crystal structure revealed that this residue is solvent exposed in the middle of the αE helix and is tightly packed against Asn-938 and Ala-1023 residues (Bojar et al., 2014; Fig. 8). Asn-938 occupies a strategic position in the αC-β4 loop that facilitates the interaction of Leu-942 of the β4 strand with Ile-931 of the αC helix. Ala-1023 is the first residue of the β8 strand that flanks the highly conserved Asp-Phe-Gly motif of the activation loop, with the Asp-1027 residue interacting with the ATP-bound Mg2+ ion and Phe-1028 making hydrophobic interactions with Ile-931 of the αC helix and His-1007 of the His-Arg-Asp motif of the catalytic loop (Fig. 8). Leu-942, Ile-931, Phe-1028, and His-1007 form the so-called regulatory spine (R-spine) that is a hallmark of activated protein kinases (Taylor and Kornev, 2011). We suspected that the substitution of a small Gly-989 residue by a large hydrophobic Ile residue might affect the spatial positioning of the R-spine residues, thus preventing the assembly of the R-spine and greatly inhibiting BRI1’s kinase activity. It is interesting that the same Gly-989 residue is changed to Glu in a newly discovered bri1 allele, bri1-707, that has no observable growth phenotype (Sun et al., 2017). Structural analysis by PyMol (http://www.pymol.org) revealed that the G989E substitution only slightly affects its packing with Asn-938 but not with Ala-1023 (Fig. 8), explaining its slightly reduced in vitro kinase activity and its slight impact on plant growth (Sun et al., 2017). This revelation is consistent with the recent finding that Gly-989 is often replaced by Glu and Asp in other Arabidopsis LRR-RLKs (Sun et al., 2017). It is quite possible that certain chaperones and/or cochaperones, such as HEAT SHOCK PROTEIN70 (HSP70)/90s, could buffer the structural defect of bri1-301, similar to what was demonstrated previously for the Arabidopsis HSP90s (Queitsch et al., 2002; Sangster et al., 2008). Consistent with our hypothesis, a loss-of-function mutation in Arabidopsis TWISTED DWARF1, a 42-kD FK506-binding protein (FKBP42) known to interact with HSP90 (Kamphausen et al., 2002), was shown recently to exhibit a strong synergistic interaction with bri1-301 (Zhao et al., 2016). These chaperones and cochaperones might work together to allow the mutant bri1-301 protein to exhibit a low kinase activity in planta, which is necessary and sufficient to confer the weak growth phenotype. Previous mathematical modeling coupled with a root growth stimulation assay suggested that the receptor activity of bri1-301 is only ∼1% to 3% that of the wild-type BRI1 (van Esse et al., 2012). It remains to be tested if a 97% to 99% reduction in the BRI1 protein abundance could recapitulate the bri1-301 phenotype.
Figure 8.
The G989I mutation likely causes a structural change in the BRI1 kinase domain. Shown on the left is the colored ribbon model of the crystal structure of the BRI1 kinase domain (Protein Data Bank no. 5LPY). The four hydrophobic amino acids that make up the conserved regulatory spine are shown with yellow spheres, Gly-989 is shown with purple spheres, Asn-938 is represented by pale blue spheres, and Ala-1023 is indicated by orange spheres. The relative positioning of the three tightly packed residues in wild-type BRI1 (WT), bri1-301, and bri1-707 is shown on the right. The mutagenesis of Gly-898 to Ile-989 (in bri1-301) or Glu-989 (in bri1-707) was performed with PyMol (http://www.pymol.org).
The Rapid Degradation of the ER-Retained Form of bri1-301 Is Not Mediated by the ERAD Machinery That Degrades bri1-5and bri1-9
In this study, we discovered the presence of a small pool of the ER-retained form of bri1-301 at 22°C (Fig. 3, C and D), which was greatly increased when the mutant was grown at 29°C (Fig. 6F). A CHX-chasing experiment with a bri1-301-overexpressing bri1-701 (pBRI1::bri1-301 bri1-701) transgenic line indicated that the ER-retained form of bri1-301 is degraded very rapidly (Fig. 3D). These results suggested that Arabidopsis contains an inefficient quality-control system that recognizes and retains a transmembrane protein carrying a cytosolic structural defect yet houses a very efficient degradation system to remove such an ERAD-C client (an ERAD client carrying a structural defect in its cytosolic domain). Consistent with what has been learned in yeast cells (Carvalho et al., 2006), the degradation of such an ERAD-C substrate does not involve components of the highly conserved ERAD machinery that degrades ERAD-L and ERAD-M substrates carrying structural abnormalities in their luminal domain and transmembrane segment, respectively. Loss-of-function mutations of EBS5, EBS6, and EBS7 did not suppress the bri1-301 dwarfism or increase the accumulation of the ER-retained form of bri1-301 at 22°C or 29°C (Figs. 3, E and F, and 6G). In yeast and mammalian cells, the removal of ERAD-C clients involves a different ERAD system that builds around a different ER membrane-anchored E3 ligase known as Degradation of alpha factor2-10 (Doa10; yeast) or Membrane associated RING-CH-type finger6 (MARCH6; mammalian cells; Hassink et al., 2005; Carvalho et al., 2006). Interestingly, the Arabidopsis genome encodes two homologs of the Doa10/MARCH6 protein, including SUPPRESSOR OF DRY2 DEFECTS1 (SUD1; also known as ECERIFERUM9) that was shown recently to be involved in cuticular wax biosynthesis (Lü et al., 2012; Doblas et al., 2013; Zhao et al., 2014). Further investigation is needed to fully understand the Arabidopsis quality-control mechanism that recognizes and degrades an ERAD-C substrate. A simple genetic cross with the T-DNA insertional mutants of SUD1 and its homolog will reveal if rapid removal of the ER-retained bri1-301 proteins involves SUD1 and/or its close homolog, while proteomic studies of a transgenically expressed bri1-301-GFP fusion protein could identify proteins involved in recognizing and retaining bri1-301 in the ER.
bri1-301 Could Be a Model Protein to Study the PM-Associated Protein Quality-Control Mechanism in a Plant Model Organism
Likely due to the presence of an inefficient ER recognition and retention system for an ERAD-C client, a large amount of bri1-301 proteins successfully reach the PM. However, the G989I mutation that makes the ER-retained bri1-301 protein extremely unstable also leads to accelerated degradation of the PM-localized bri1-301 (Fig. 3D), suggesting the existence of a PM-associated quality-control (PMQC) system that detects the structural lesion of bri1-301 and targets the mutant BR receptor for degradation. The PMQC system is an integral part of the cellular proteostasis network that constantly monitors the folding status of all cellular proteins and removes their incorrectly folded and structurally damaged conformers at all subcellular locations (Babst, 2014). Recent studies in yeast and mammalian cells have revealed at least two PMQC mechanisms for removing the structurally unstable proteins from the cell surface via ubiquitination, endocytosis, and lysosomal degradation (Okiyoneda et al., 2011; Apaja and Lukacs, 2014). These studies have implicated two E3 ubiquitin ligases in ubiquitinating the structurally defective PM proteins, including the mammalian Carboxy terminus of Hsc70-interacting protein (CHIP), a U-box domain-containing E3 ligase (Edkins, 2015), and the yeast HECT-type E3 ligase Reverses Spt-phenotype5 (Zhao et al., 2013). The Arabidopsis genome encodes a potential CHIP homolog known as AtCHIP1, which was implicated in stress tolerance, plant immunity, and chloroplast function (Yan et al., 2003; Luo et al., 2006; Shen et al., 2007a, 2007b; Wei et al., 2015; Copeland et al., 2016), and at least seven HECT-type E3 ligases (Downes et al., 2003; Marín, 2013). Compared with what has been learned about the PMQC system in yeast and mammalian cells, almost nothing is known about a similar system in plants. Despite its faster degradation rate than the wild-type BRI1, bri1-301 might share the same degradation pathway with wild-type BRI1, which was suggested recently to involve K63-linked polyubiquitination, endocytosis, and vacuolar degradation for its removal from the PM in a ligand-independent manner (Martins et al., 2015). The bri1-301 mutant could be used as a convenient system for genetic dissection of a plant PMQC mechanism.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotypes Columbia-0 and Wassilewskija-2 were used as the wild-type controls for genetic, transgenic, and phenotypic analyses. The Arabidopsis mutants that were used in this study were all described previously, including bri1-5 (Noguchi et al., 1999), bri1-9 (Jin et al., 2007), bri1-101 (Li and Chory, 1997), bri1-301 (Xu et al., 2008), bri1-701 (Gou et al., 2012), bri1-702, bri1-703, bri1-704, bri1-707, and bri1-708 (Sun et al., 2017), ebs5 (Su et al., 2011), ebs6 (Su et al., 2012), and ebs7 (Liu et al., 2015). Seed sterilization and seedling growth were performed following previously described protocols (Li et al., 2001).
Generation of Plasmid Constructs and Transgenic Plants
The pBRI1::BRI1-GFP construct was described previously (Friedrichsen et al., 2000) and was used to generate the pBRI1::BRI1 plasmid by deleting the GFP-coding sequence using the ClonExpress II One Step cloning kit (Vazyme) and the BRI1::BRI1 primer set (Supplemental Table S1). Both plasmids were used as templates to generate pBRI1::bri1-301-GFP, pBRI1::bri1-301, and other mutant constructs via a PCR-driven overlapping extension mutagenesis approach (Heckman and Pease, 2007) with mutagenesis primers and flanking primers listed in Supplemental Table S1. These newly created plasmids were fully sequenced to ensure no PCR-introduced error and used subsequently to transform the null bri1-701 mutant and its wild-type control via the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Every analyzed transgenic plant was genotypically verified by PCR, and the expression of bri1-301/BRI1 or their GFP fusions was analyzed by immunoblotting.
RNA Analysis
Total RNAs were isolated from 8-d-old Arabidopsis seedlings grown on 1/2 MS medium with the RNeasy plant mini kit (Qiagen) and were subsequently converted into first-strand cDNAs using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s recommended protocols. To analyze the transcript abundance of the genes of interest, 0.5 μL of the first-strand cDNA templates was used for RT-qPCR amplification on a Bio-Rad CFX96 Touch system with the primer sets shown in Supplemental Table S1 and iTaq Universal SYBR Green Supermix (Bio-Rad). The ACTIN transcript was amplified using the ACTIN primer set (Supplemental Table S1) for an internal reference. For each transcript, the RT-qPCR assay was repeated four times.
Protein Analysis
Total proteins were extracted from whole seedlings grown on 1/2 MS agar medium or rosette leaves of soil-grown plants. Briefly, 30 mg of plant tissue for each sample was quickly frozen in liquid nitrogen, cryogenically ground into fine powder with steel beads in a mixer mill (MM400; Retsch), and dissolved in 150 μL of 2× SDS loading buffer (100 mm Tris-HCl, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, 0.02% [w/v] Bromophenol Blue, and 100 mm DTT). The mixed crude extracts were heated at 95°C for 10 min and centrifuged immediately at 10,000g for 10 min. The cleared supernatants were used immediately for gel electrophoresis or treated with or without 1,000 units of Endo-Hf in 1× G5 buffer (New England Biolabs) for 1 h at 37°C. The treated or nontreated total proteins were separated by SDS-PAGE, transferred onto a polyvinylidene difluoride membrane (Bio-Rad), and analyzed by Coomassie Blue staining (for loading controls) or immunoblotting with antibodies generated against BRI1 (Mora-García et al., 2004), GFP (632381; Clontech), BES1 (Mora-García et al., 2004), or ACTIN (CW0264; Beijing CWBio). The quantification of relative BRI1/bri1 abundance was performed using ImageJ (https://imagej.nih.gov/ij/) with scanned images of Coomassie Blue-stained gels or immunoblots after normalizing anti-BRI1 signals with the signal intensity of the corresponding RbcS or anti-ACTIN bands.
To perform coimmunoprecipitation experiments, seedlings were collected, immediately ground into fine powder in liquid nitrogen, and extracted with the immunoprecipitation buffer (2–3 mL g−1 plant tissue; 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.2% [v/v] Triton X-100, 1 mm phenylmethylsulfonyl fluoride [Sigma-Aldrich], 1× cOmplete protease inhibitor cocktail [Roche], and 1× phosphatase inhibitor cocktail [BioTool Chemicals] that was added only for the immunoprecipitation experiments to detect protein phosphorylation). The dissolved crude extracts were centrifuged twice at 20,000g for 15 min each at 4°C. The resulting supernatants were collected and incubated with prewashed anti-GFP monoclonal antibody-agarose beads (D153-8; MBL) at 4°C for 4 h. The agarose beads were subsequently washed four times with the immunoprecipitation buffer and four additional times with the washing buffer (the immunoprecipitation buffer excluding Triton X-100). The proteins remaining on the beads were removed with 2× SDS loading buffer and 5 min of boiling at 95°C. After 10 min of centrifugation, the supernatants were subject to SDS-PAGE and analyzed by Coomassie Blue staining or immunoblotting with appropriate antibodies.
To analyze the phosphorylation status of BRI1 and bri1-301, total protein extracts were loaded onto Zn2+-Phos-tag SDS-PAGE gels containing 50 μm Phos-tag Acrylamide (Wako Laboratory Chemicals). The detailed procedure was performed according to a manufacturer’s recommended protocol (http://www.wako-chem.co.jp/english/labchem/product/life/Phos-tag/pdf/AAL107_v12.pdf).
Treatment with Chemicals
To analyze BRZ sensitivity, sterilized Arabidopsis seeds were planted directly on 1/2 MS agar medium containing varying concentrations of BRZ (TCI Chemicals). After 2 h of light exposure, the petri dishes were wrapped with light-proof aluminum foil and kept in total darkness for 5 d in a 22°C or 29°C growth chamber for seed germination and seedling growth. Seedlings were removed carefully from the petri dishes, photographed immediately, and their hypocotyl lengths were quantified by ImageJ (https://imagej.nih.gov/ij/). To investigate the effect of BL on the BES1 phosphorylation status and transcript abundance of selected BR-responsive genes, 8-d-old light-grown seedlings were removed carefully from petri dishes, immediately transferred into liquid 1/2 MS medium supplemented with or without 1 μm BL (Wako Chemicals), incubated for 2 h, and subsequently harvested and placed in liquid nitrogen for the extraction of total proteins and RNAs or storage in a −80°C freezer. To analyze the protein stability of BRI1 and its mutant variants, 2-week-old seedlings grown on 1/2 MS agar medium were transferred carefully into liquid 1/2 MS medium supplemented with or without 180 μm CHX (Sigma-Aldrich), 50 μm Kif (Toronto Research Chemicals), or 80 μm MG115 (Sigma-Aldrich), incubated for different times, and subsequently collected into liquid nitrogen for total protein extraction.
Endo-H and Trypsin Sensitivity Assays
The Endo-H assay was performed according to a previously described procedure (Hong et al., 2008). To carry out the trypsin sensitivity assay, 10-d-old seedlings grown on 1/2 MS agar medium were collected, immediately ground into a fine powder in liquid nitrogen, dissolved in 100 μL of phosphate-buffered saline solution (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4, pH 7.4) containing 0.4% (v/v) Triton X-100, and centrifuged at 10,000g for 5 min at 4°C to remove cellular debris and insoluble materials. The resulting supernatant was mixed with trypsin (Sigma-Aldrich) at 5 μg mL−1 and incubated on ice. Aliquots of 30 μL of the reaction mixtures were removed at different time points, immediately mixed with 20 μL of 2× SDS loading buffer for denaturing by 15 min of boiling, and subsequently frozen in liquid nitrogen. The frozen samples were reheated, separated by SDS-PAGE, and subsequently analyzed by immunoblotting.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: BRI1, AAC49810.1; CPD, NM120651; DWF4, AF044216; SAUR-AC1, S70188.1; and WRKY13, NM120101.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR analysis of DWF4 transcript abundance.
Supplemental Figure S2. RT-qPCR analysis of BRI1/bri1-301 transcript abundance.
Supplemental Table S1. Oligonucleotides used in this study.
Acknowledgments
We thank Dr. Yanhai Yin for supplying the anti-BES1 antibody and Dr. Jia Li for seeds of Arabidopsis bri1-701-704, bri1-707, and bri1-708 mutants and for discussing his similar study with us. We also thank additional members of the Li laboratory for stimulating discussion throughout this study.
Footnotes
This work was partially supported by a grant from the Chinese Academy of Sciences (2012CSP004 to J.L.), grants from the Natural Science Foundation of China (NSFC31600996 to L.L. and NSFC31730019 to J.L.), and the Shanghai Center for Plant Stress Biology.
Articles can be viewed without a subscription.
References
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apaja PM, Lukacs GL (2014) Protein homeostasis at the plasma membrane. Physiology (Bethesda) 29: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. (2014) Quality control at the plasma membrane: One mechanism does not fit all. J Cell Biol 205: 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancoş S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206: 522–540 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Durbak A, Wierzba M, Schmitz RJ, Aguirre A, Michel R, Rowe S, Fujioka S, Tax FE (2010) Intragenic suppression of a trafficking-defective brassinosteroid receptor mutant in Arabidopsis. Genetics 185: 1283–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojar D, Martinez J, Santiago J, Rybin V, Bayliss R, Hothorn M (2014) Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J 78: 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA (2006) Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroids. The Arabidopsis Book 9: e0151, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland C, Ao K, Huang Y, Tong M, Li X (2016) The evolutionarily conserved E3 ubiquitin ligase AtCHIP contributes to plant immunity. Front Plant Sci 7: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Amorim-Silva V, Posé D, Rosado A, Esteban A, Arró M, Azevedo H, Bombarely A, Borsani O, Valpuesta V, et al. (2013) The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in Arabidopsis. Plant Cell 25: 728–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ (2007) Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35: 729–742 [DOI] [PubMed] [Google Scholar]
- Edkins AL. (2015) CHIP: A co-chaperone for degradation by the proteasome. Subcell Biochem 78: 219–242 [DOI] [PubMed] [Google Scholar]
- Elbein AD, Tropea JE, Mitchell M, Kaushal GP (1990) Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J Biol Chem 265: 15599–15605 [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y, Shang Y, Nam KH (2016) Brassinosteroids modulate ABA-induced stomatal closure in Arabidopsis. J Exp Bot 67: 6297–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Hao Y, Wang H, Qiao S, Leng L, Wang X (2016) Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc Natl Acad Sci USA 113: 10418–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, Coleman CS, Bartee E, Früh K, Chau V, et al. (2005) TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J 388: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J (2000) Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363 [DOI] [PubMed] [Google Scholar]
- Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932 [DOI] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Fitchette AC, Xia Y, Monk AM, Faye L, Li J (2009) Mutations of an α1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell 21: 3792–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam KH, Li J (2007) Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphausen T, Fanghänel J, Neumann D, Schulz B, Rahfeld JU (2002) Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J 32: 263–276 [DOI] [PubMed] [Google Scholar]
- Kang B, Wang H, Nam KH, Li J, Li J (2010) Activation-tagged suppressors of a weak brassinosteroid receptor mutant. Mol Plant 3: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T (1996) Genetic evidence for an essential role of brassinosteroids in plant development. Plant J 9: 701–713 [Google Scholar]
- Kim TW, Lee SM, Joo SH, Yun HS, Lee Y, Kaufman PB, Kirakosyan A, Kim SH, Nam KH, Lee JS, et al. (2007) Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ 30: 679–689 [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T (2009) Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc 4: 1513–1521 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li J (2014) Endoplasmic reticulum-mediated protein quality control in Arabidopsis. Front Plant Sci 5: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Wang D, Su W, Liu L, Wang M, Li J (2015) EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc Natl Acad Sci USA 112: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, Bangarusamy DK, Wang G, Rowland O, Juenger T, et al. (2012) Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, Zhang H (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46: 649–657 [DOI] [PubMed] [Google Scholar]
- Marín I. (2013) Evolution of plant HECT ubiquitin ligases. PLoS ONE 8: e68536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaître B, Jaillais Y, Chory J, Geldner N, et al. (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al. (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Min YK, Nakano T, Asami T, Yoshida S (2000) Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinazole, induces some characteristics of light-grown plants. Planta 211: 781–790 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Shimada Y, Goda H, Fujiwara MT, Asami T, Yoshida S (2003) AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis. FEBS Lett 553: 28–32 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Apaja PM, Lukacs GL (2011) Protein quality control at the plasma membrane. Curr Opin Cell Biol 23: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Robbins PW, Trimble RB, Wirth DF, Hering C, Maley F, Maley GF, Das R, Gibson BW, Royal N, Biemann K (1984) Primary structure of the Streptomyces enzyme endo-beta-N-acetylglucosaminidase H. J Biol Chem 259: 7577–7583 [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Lee HN, Watanabe E, Schellenberg K, Morneau K, Wang H, Undurraga S, Queitsch C, Lindquist S (2008) HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 2969–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Lee MM, Li J, Nam KH (2011) Characterization of cp3 reveals a new bri1 allele, bri1-120, and the importance of the LRR domain of BRI1 mediating BR signaling. BMC Plant Biol 11: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G, Adam Z, Zhang H (2007a) The E3 ligase AtCHIP ubiquitylates FtsH1, a component of the chloroplast FtsH protease, and affects protein degradation in chloroplasts. Plant J 52: 309–321 [DOI] [PubMed] [Google Scholar]
- Shen G, Yan J, Pasapula V, Luo J, He C, Clarke AK, Zhang H (2007b) The chloroplast protease subunit ClpP4 is a substrate of the E3 ligase AtCHIP and plays an important role in chloroplast function. Plant J 49: 228–237 [DOI] [PubMed] [Google Scholar]
- Shi C, Qi C, Ren H, Huang A, Hei S, She X (2015) Ethylene mediates brassinosteroid-induced stomatal closure via Gα protein-activated hydrogen peroxide and nitric oxide production in Arabidopsis. Plant J 82: 280–301 [DOI] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J (2011) Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc Natl Acad Sci USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J (2012) The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol Plant 5: 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Yan K, Han JT, Tao L, Lv MH, Shi T, He YX, Wierzba M, Tax FE, Li J (2017) Scanning for new BRI1 mutations via TILLING analysis. Plant Physiol 174: 1881–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP (2011) Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci 36: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG, Mayer KF, Sieberer T, Poppenberger B (2015) Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 27: 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse GW, van Mourik S, Stigter H, ten Hove CA, Molenaar J, de Vries SC (2012) A mathematical model for BRASSINOSTEROID INSENSITIVE1-mediated signaling in root growth and hypocotyl elongation. Plant Physiol 160: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Liu M, Yuan M, Oses-Prieto JA, Cai X, Sun Y, Burlingame AL, Wang ZY, Tang W (2016) The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B′ subunits. Mol Plant 9: 148–157 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wei J, Qiu X, Chen L, Hu W, Hu R, Chen J, Sun L, Li L, Zhang H, Lv Z, et al. (2015) The E3 ligase AtCHIP positively regulates Clp proteolytic subunit homeostasis. J Exp Bot 66: 5809–5820 [DOI] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H (2003) AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol 132: 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lv M, Feng Z, Campbell T, Liscum E, Li J (2016) TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol Plant 9: 582–592 [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhang H, Cui P, Ding F, Wang G, Li R, Jenks MA, Lü S, Xiong L (2014) The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in Arabidopsis. Plant Physiol 165: 1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Macgurn JA, Liu M, Emr S (2013) The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. eLife 2: e00459. [DOI] [PMC free article] [PubMed] [Google Scholar]