The microtubule associated protein, CLASP, impacts polymer dynamics leading to a slowdown in microtubule array pattern transitions without altering the ability to form specific array patterns.
Abstract
Acentrosomal plant microtubule arrays form patterns at the cell cortex that influence cellular morphogenesis by templating the deposition of cell wall materials, but the molecular basis by which the microtubules form the cortical array patterns remains largely unknown. Loss of the Arabidopsis (Arabidopsis thaliana) microtubule-associated protein, CYTOPLASMIC LINKER ASSOCIATED PROTEIN (AtCLASP), results in cellular growth anisotropy defects in hypocotyl cells. We examined the microtubule array patterning in atclasp-1 null mutants and discovered a significant defect in the timing of transitions between array patterns but no substantive defect in the array patterns per se. Detailed analysis and computational modeling of the microtubule dynamics in two atclasp-1 fluorescent tubulin marker lines revealed marker-dependent effects on depolymerization and catastrophe frequency predicted to alter the steady-state microtubule population. Quantitative in vivo analysis of the underlying microtubule array architecture showed that AtCLASP is required to maintain the number of growing microtubule plus ends during transitions between array patterns. We propose that AtCLASP plays a critical role in cellular morphogenesis through actions on new microtubules that facilitate array transitions.
Microtubules are highly conserved polymers in eukaryotic cells that form relatively rigid tubes over length scales capable of directing intracellular traffic. In vascular plants, microtubules form a 2D network attached to the cytoplasmic side of the plasma membrane (Newcomb, 1969; Hardham and Gunning, 1978; Ehrhardt and Shaw, 2006). This acentriolar, interphase array plays a key role in defining plant cell morphology by patterning the deposition of cell wall materials that influence how the cell expands under turgor pressure (Green, 1962; Baskin, 2001; Paredez et al., 2006; Lloyd, 2011). While much is known about the properties of individual microtubules, how plant cells organize the cortical array into functional patterns for the deposition of wall materials is poorly understood (Cyr, 1994; Wasteneys, 2000; Ehrhardt and Shaw, 2006; Hashimoto and Kato, 2006; Lloyd and Chan, 2008; Shaw, 2013).
The majority of cortical microtubules nucleate from γ-tubulin complexes at the plasma membrane associated with preexisting microtubules (Shaw et al., 2003; Murata and Hasebe, 2007; Chan et al., 2009; Nakamura et al., 2010). Although individual polymers appear motile, the microtubules are physically anchored at the membrane and move via treadmilling; the more dynamic plus ends bias the dynamic instability to gain polymers and the minus ends slowly depolymerize (Shaw et al., 2003; Shaw and Lucas, 2011). The plasma membrane association leads to direct physical interactions of cortical microtubules, leading to crossing over, induced catastrophes, severing, and bundling (Dixit and Cyr, 2004; Wightman and Turner, 2007; Nakamura et al., 2010; Lucas et al., 2011). The microtubule polymerization dynamics and the outcomes of the physical interactions are influenced by microtubule-associated proteins (MAPs) and cellular cues (Ambrose et al., 2011; Sambade et al., 2012; Lindeboom et al., 2013; Vineyard et al., 2013). How MAPs and local cellular activities work together to create specific array patterns is the subject of significant interest (Wasteneys, 2000; Ehrhardt and Shaw, 2006; Baulin et al., 2007; Lucas and Shaw, 2008; Sedbrook and Kaloriti, 2008; Tindemans et al., 2010; Deinum et al., 2011; Lloyd, 2011).
The CYTOPLASMIC LINKER ASSOCIATED PROTEIN (CLASP) is a well-studied MAP found across plant, fungal, and animal systems (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005; Al-Bassam and Chang, 2011; Ruan and Wasteneys, 2014). In mitotic animal cells, CLASP has been described chiefly as a tip-tracking protein that promotes the addition of tubulin to kinetochore microtubules (Maiato et al., 2005) and stabilizes plus ends (Mimori-Kiyosue et al., 2005). CLASP interacts with membranes during interphase with differing levels of microtubule lattice and tip localization under the control of kinase activities (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005; Efimov et al., 2007). CLASP associates with microtubules directly through a TOG domain and via other plus end-binding proteins, including END BINDING1 (EB1) and CYTOPLASMIC LINKER PROTEINS (e.g. CLIP170; Patel et al., 2012). Extensive biochemical analyses of CLASP activity have shown widely differing effects on microtubules, with the majority indicating an ability to alter catastrophe and/or rescue (Akhmanova et al., 2001; Al-Bassam et al., 2010; Al-Bassam and Chang, 2011; Moriwaki and Goshima, 2016).
The model plant Arabidopsis (Arabidopsis thaliana) has a single CLASP gene (Ambrose et al., 2007; Kirik et al., 2007). AtCLASP shares two functional domains with human (Homo sapiens) CLASP, a microtubule-binding TOG domain (23% identity, 42% similarity) and a C-terminal CLIP domain (28% identity, 52% similarity), although there is no known orthologous CLIP gene in Arabidopsis (Ambrose et al., 2007). AtCLASP is expressed throughout the plant, and a native promoter-driven YFP-AtCLASP localizes irregularly along the microtubule lattice, plus end, and to membrane patches, similar to animal systems (Ambrose et al., 2007, 2011; Ambrose and Wasteneys, 2008). Overexpression leads to high levels of microtubule decoration, highly curved microtubules, and polymerization defects, indicating that AtCLASP binding is sensitive to expression levels and can lead to changes in the microtubule steady state (Kirik et al., 2007). The atclasp-1 mutant has a T-DNA insertion in exon 13 (Ambrose et al., 2007; Kirik et al., 2007) and appears to be a null allele based on expression studies (Ambrose et al., 2007; Kirik et al., 2007). The mutant is short in stature with reduced anisotropic axial cell growth and diminished cell division in the elongation zones, consistent with other mutants having microtubule defects (Ambrose et al., 2007; Kirik et al., 2007).
Initial cellular studies of atclasp-1 did not identify pronounced cortical microtubule array organization defects in axially growing root or hypocotyl cells (Ambrose et al., 2007; Kirik et al., 2007). Subsequent work showed that cortical arrays in small leaf epidermal pavement cells appeared hyperparallel in atclasp-1 (Ambrose and Wasteneys, 2008). This defect was attributed initially to AtCLASP’s potential role in attaching microtubules to the cell cortex, although this interpretation was later revised in modeling studies (Ambrose and Wasteneys, 2008; Allard et al., 2010a, 2010b). Additional work found AtCLASP enrichment at the peripheral edge of the cell face in young leaf epidermis and root cells (Ambrose et al., 2011). AtCLASP was shown to facilitate polymerization over pronounced cell edges in smaller cells, where the absence of AtCLASP at specific cell edges reduced microtubules at that edge through an increase in catastrophe frequency (Ambrose et al., 2011). More recent work suggests that AtCLASP may indirectly regulate cortical microtubule array organization by influencing the apical-basal polarization of PINOID2 proteins (auxin gradient mediators; Ambrose et al., 2013; Ruan and Wasteneys, 2014). AtCLASP also has been genetically linked to the role of SABRE, a protein found among many eukaryotic phyla having a role in orchestrating cell division planes (Pietra et al., 2013).
In this cell biological study, we examined the microtubule array patterning in atclasp-1 mutants that show a specific defect in cell growth anisotropy. Using the axially growing hypocotyl cells, we wanted to determine how the molecular action of AtCLASP, proposed from biochemical studies of animal and yeast CLASP proteins, affected array patterning and microtubule properties in vivo. Using new imaging tools and a set of microtubule markers, we identified a primary function for AtCLASP in transitioning the acentrosomal microtubule arrays between patterns. Extensive in vivo analysis indicated that AtCLASP modulates microtubule plus end dynamics that are critical for new microtubules in new orientations to affect the array pattern transitions.
RESULTS
The atclasp-1 Growth Anisotropy Phenotype Is Conditioned by the Microtubule Marker
We ultimately wanted to determine the mechanisms and extent to which individual microtubule array patterns contribute to the anisotropic extension of the plant cell wall. The atclasp-1 loss-of-function mutant exhibits growth defects at the cell and organ levels, including more isotropic hypocotyl cells and stunted hypocotyl elongation (Ambrose et al., 2007; Kirik et al., 2007). To quantitatively evaluate microtubule patterning and architecture, we crossed the atclasp-1 tDNA mutant (Alonso et al., 2003; Kirik et al., 2007) into our well-characterized transgenic lines expressing an Arabidopsis BETA-1-TUBULIN (GFP-TUB1) or an EB1 (EB1-GFP) microtubule marker (Mathur et al., 2003). We additionally acquired ALPHA-5-TUBULIN (YFP-TUA5) lines for comparisons with prior atclasp-1 studies (Lindeboom et al., 2013; Nakamura et al., 2018). The tDNA position in the AT2G20190 locus and the microtubule marker identity were verified in whole-genome sequencing of the mutant lines at ∼10× coverage.
The magnitude of the atclasp-1 growth phenotype was assessed by measuring the hypocotyl lengths of 6-d-old seedlings grown under a 24-h light regime at 22°C (Vineyard et al., 2013). We observed significant differences in wild-type hypocotyl length between reference and transgenic lines in all cases (Fig. 1A; P < 0.01, Student’s t test in all comparisons). Compared with the wild-type Columbia (Col-0) reference line (0.912 ± 0.118 mm; n = 70), the GFP-TUB1 line was 8% shorter (0.842 ± 125 mm; n = 50), the YFP-TUA5 line was 28% shorter (0.654 ± 0.083 mm; n = 50), and the EB1-GFP line was 37% taller (1.245 ± 0.248 mm; n = 40). Comparing mutant atclasp-1 hypocotyl lengths with the parent lines in each case, the Col-0 reference was 17% shorter (0.755 ± 0.136 mm; n = 43), the GFP-TUB1 line was 8% shorter (0.771 ± 0.127 mm; n = 49), the YFP-TUA5 line was 12% longer (0.731 ± 0.123 mm; n = 48), and the EB1-GFP line was 34% shorter (0.825 ± 0.158 mm; n = 48). We found no significant difference between the three atclasp-1 mutant lines and the atclasp-1 Col-0 reference hypocotyl length (P > 0.03). Thus, the plant background associated with the microtubule marker altered the apparent magnitude of the atclasp-1 hypocotyl growth defect and, in the case of YFP-TUA5, appeared to reverse the organ-level growth phenotype.
Figure 1.
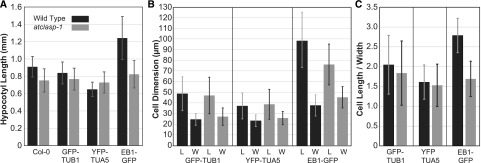
The cell expansion phenotype of the atclasp-1 mutant is altered differently by different microtubule marker transgenes. A, Average hypocotyl lengths ± sd for 6-d-old, light-grown, wild-type and atclasp-1 seedlings in the Col-0, GFP-TUB1, YFP-TUA5, and EB1-GFP backgrounds. Differences between the wild type and atclasp-1 are significant in all backgrounds (P < 0.01 [n > 40 in all cases], Student’s t test used for all growth assays), and differences between all transgenic wild-type backgrounds and the Col-0 parent line are significant in all cases (P < 0.01). B, Mean cell length (L) and width (W) ± sd for fluorescent tubulin-expressing lines in the wild-type and atclasp-1 backgrounds (n = 14 plants, n > 530 cells for tubulin lines; n = 6 plants, n > 230 cells for EB1-GFP lines). Cell lengths between the wild type and the mutant in both fluorescent tubulin backgrounds are not statistically different (P > 0.05 in both cases), while the atclasp-1 cells are wider on average compared with the wild type in both backgrounds (P < 0.001). Cell lengths and widths are significantly different (P < 0.001) between the wild type and the mutant in the EB1-GFP background. C, Cellular anisotropy expressed as cell length divided by cell width ± sd for fluorescent tubulin-expressing lines.
Noting the differences in hypocotyl length, we compared the fluorescent marker lines for defects in cellular growth anisotropy. We measured cell length and width for all cells from two adjacent hypocotyl cell files (Fig. 1B). The cell lengths in the GFP-TUB1 wild-type (48.9 ± 16 µm; n = 14 plants/537 cells) and mutant (47.2 ± 17.1 µm; n = 14/544) lines were not significantly different (P > 0.05), but the mutant trended toward shorter lengths. The cell lengths in the YFP-TUA5 wild-type (37.4 ± 12.2 µm; n = 14/536) and mutant (38.9 ± 14 µm; n = 14/553) lines also were not significantly different (P > 0.05), but the mutant trended toward longer lengths, in agreement with the organ-level measurement. The EB1-GFP cells were significantly longer (P > 0.001) in the wild type (99.2 ± 27.4 µm; n = 6/233) compared with the mutant (75.8 ± 19.2 µm; n = 6/253). Cell widths (Fig. 1B) were significantly different both for the GFP-TUB1 wild type (24.8 ± 5.5 µm) compared with the mutant (27.3 ± 8.1 µm) and for the YFP-TUA5 wild type (23.5 ± 5.4 µm) compared with the mutant (26.0 ± 6.2 µm), with the mutant being wider in both cases (P < 0.01). The EB1-GFP wild-type width (37.2 ± 9.4 µm) was significantly different (P < 0.01) compared with the mutant width (45.5 ± 9.8 µm). Comparing the cell length-to-width ratios (i.e. anisotropy) for all lines (Fig. 1C), cells in the atclasp-1 mutants consistently showed a significant reduction in cell shape anisotropy (P < 0.001).
While both mutant backgrounds showed less growth anisotropy, we hypothesized that the taller hypocotyls observed in the YFP-TUA5 atclasp-1 mutant were due to more total growth in the YFP-TUA5 mutant background. Using the product of cell length and width to estimate cell size (e.g. cell face area), the GFP-TUB1 atclasp-1 mutant was 6.3% larger than the wild type, and the YFP-TUA5 mutant was 15% larger. These data indicate that atclasp-1 leads to the same defect in cell shape in both backgrounds (i.e. less anisotropic) but additionally show that the absence of AtCLASP in the YFP-TUA5 background, grown under these conditions, results in more total cell growth than the wild-type parent, making the hypocotyl longer in the mutant.
Microtubule Array Pattern Is Specified Independently of AtCLASP
Since cortical microtubules have been shown to direct the orientation of cellulose deposition, we hypothesized that the cortical microtubule arrays in the atclasp-1 cells would show defects in array pattern related to the cellular growth anisotropy phenotype. To test this central hypothesis, we imaged light-grown, 6-d-old wild-type and atclasp-1 hypocotyls from apex to base (Fig. 2, A and B) and visually classified array patterns (Fig. 2, C–F) from two adjacent cell files in both GFP-TUB1- and YFP-TUA5-expressing lines. The microtubule arrays showed a characteristic distribution (Liu et al., 2013; Vineyard et al., 2013; Atkinson et al., 2014) of transverse (0°–15°), oblique (16°–75°), and longitudinal (76°–90°) coaligned patterns, with a separate class having no visibly dominant microtubule coalignment, termed basket arrays (Vineyard et al., 2013).
Figure 2.
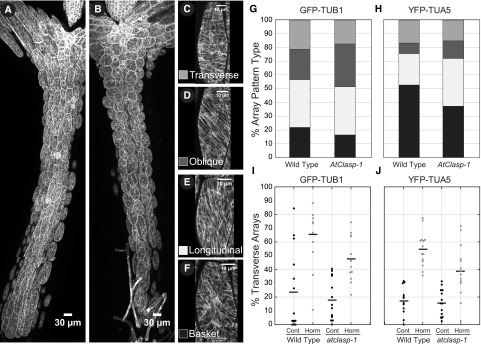
atclasp-1 has only small effects on the distribution of microtubule array pattern types in light-grown hypocotyl cells. A and B, Maximum projection of tiled confocal microscopy images from wild-type (A) and atclasp-1 mutant (B) 6-d-old light-grown seedlings expressing a GFP-TUB1 microtubule marker. Bars = 30 µm. C to F, Examples of cells showing the four dominant microtubule array patterns. Bars = 10 µm. G and H, Stacked bar graphs showing the accumulated percentage of cells with each array pattern from 14 untreated hypocotyls (n > 530 cells for each line) in GFP-TUB1 and YFP-TUA5 lines, with the color key given in C to F. I and J, Dot plots showing the percentage of transverse patterned cells per hypocotyl induced by mock treatment or combined IAA and GA4 treatment after 2 h in GFP-TUB1 and YFP-TUA5 backgrounds. Bars represent mean values.
The accumulated percentages of each array pattern indicated that wild-type (n = 14 plants, 532 cells) and atclasp-1 (n = 14 plants, 514 cells) lines in the GFP-TUB1 background exhibited similar distributions of longitudinal (32% versus 35%, respectively), oblique (23% versus 33%, respectively), transverse (24% versus 18%, respectively), and noncoaligned (21% versus 14%, respectively) array patterns (Fig. 2G). The GFP-TUB1 atclasp-1 mutant showed a weak (less than 10%) shift toward more coalignment, predominantly into oblique patterns. The YFP-TUA5 background (Fig. 2H) exhibited a stronger relative shift toward more coaligned patterns, with wild-type (n = 14 plants, 532 cells) and atclasp-1 (n = 14 plants, 529 cells) lines exhibiting longitudinal (23% versus 34%, respectively), oblique (7% versus 13%, respectively), transverse (16% versus 16%, respectively), and noncoaligned (53% versus 37%, respectively) arrays. We found no evidence for novel array patterns in either background. Contrary to our hypothesis, we found that AtCLASP is not required to create specific microtubule array patterns normally observed in 6-d-old light-grown hypocotyl cells but may influence the percentage of cells exhibiting each pattern class.
Auxin Induces Transverse Microtubule Patterns in atclasp-1 Mutants
We wanted to determine if AtCLASP was required to make the correct pattern at the correct time. For this experiment, we used plant growth-inducing hormones that signal the cortical arrays to form transverse patterns correlated with rapid axial growth (Vineyard et al., 2013). We performed end-point assays comparing the percentage of transverse array patterns observed after a 2-h treatment with both 0.5 µm indole 3-acetic-acid (IAA) and 5 µm GA4 or mock solvent using both GFP-TUB1 and YFP-TUA5 lines. Data from two adjacent hypocotyl cell files were visually classified for array pattern. Hormone induction resulted in a significant increase in transverse array patterns at 2 h for both transgenic backgrounds (Fig. 2, I and J). Hormone induction in the GFP-TUB1 parent lines increased transverse pattern frequency from 22% to 65% (n = 14 hypocotyls, 537 cells and n = 11 hypocotyls, 389 cells; P < 0.001), and the YFP-TUA5 lines showed an increase from 16% to 55% (n = 14 hypocotyls, 532 cells and n = 14 hypocotyls, 515 cells; P < 0.001). Hormone addition to the atclasp-1 mutant in the GFP-TUB1 lines increased the fraction of transverse patterns from 17% to 48% (n = 14 hypocotyls, 536 cells and n = 13 hypocotyls, 493 cells; P < 0.001), while the same treatment in the YFP-TUA5 atclasp-1 lines increased the fraction from 16% to 39% transverse patterns (n = 14 hypocotyls, 529 cells and n = 14 hypocotyls, 563 cells; P < 0.001). These experiments revealed that, in both transgenic marker backgrounds, the atclasp-1 mutant formed the correct pattern in response to exogenously applied IAA/GA4 but failed to produce the same fraction of transverse arrays per plant as the wild-type parent in the 2-h period.
AtCLASP Affects the Frequency of Array Pattern Transitions
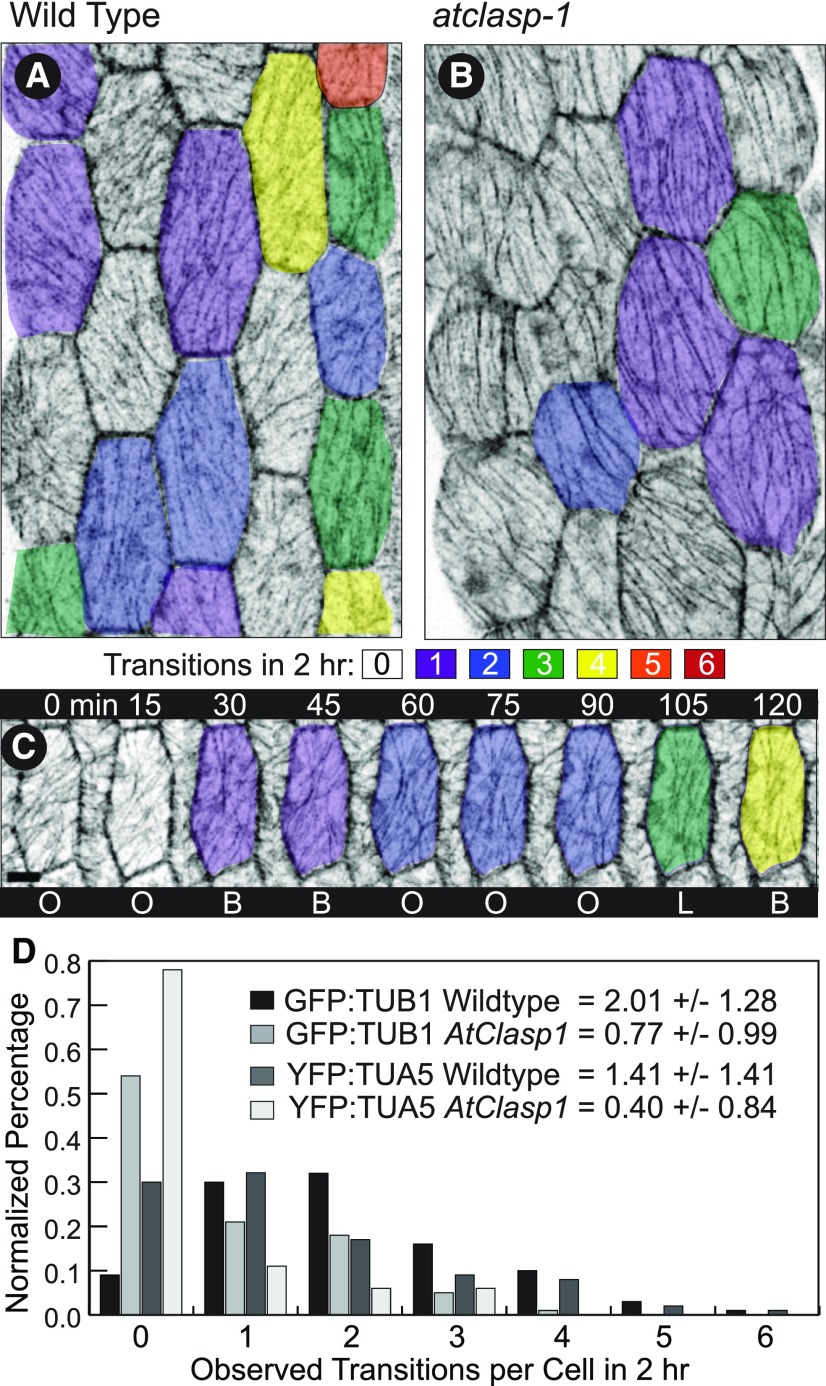
We hypothesized from our fixed time point observations that AtCLASP was not required to create a specific pattern but could be involved in transitioning the array to new patterns. To address this hypothesis, we made time-course observations of untreated GFP-TUB1 and YFP-TUA5 seedlings and measured the frequency of pattern transition in the wild type and atclasp-1 mutants (Fig. 3). We imaged the middle portion of 6-d-old, light-grown hypocotyls (Fig. 3, A and B) every 15 min for 2 h and visually assigned patterns at each time point from a maximum projection of the 3D image stack (Fig. 3C). The pattern classifications for each cell then were collated over the 2-h time course, and transitions between array patterns were counted (Fig. 3, A and B). The average number of transitions in 2 h for the GFP-TUB1 wild type was 2.01 ± 1.28 (n = 101 cells) while the atclasp-1 cells averaged 0.77 ± 0.99 (n = 112 cells), a greater than 2-fold difference (Fig. 3D). The distribution of array transitions per cell indicated that only 9% of the wild-type cells failed to make a transition in 2 h whereas 54% of the atclasp-1 arrays did not transition (Fig. 3D). Repeating the experiment with the YFP-TUA5 lines resulted in an average of 1.41 ± 1.38 (n = 156 cells) transitions per 2 h in the wild type and 0.4 ± 0.84 (n = 179 cells) transitions in the atclasp-1 mutant (Fig. 3D). Approximately 30% of the wild-type cells failed to transition in the 2-h time course whereas nearly 78% failed in the mutant. In conclusion, the loss of AtCLASP lowered the frequency of microtubule array pattern transitions to less than half of the wild-type value in both transgenic backgrounds. The transition frequency defect correlates with the cell growth anisotropy defect, where fewer array transitions corresponded with less growth anisotropy in both transgenic backgrounds.
Figure 3.
Microtubule arrays transition between patterns less frequently in atclasp-1 cells. A and B, Hypocotyl cells from projected confocal image stacks displayed with inverted contrast showing the number of transitions between pattern types over a 2-h period for the wild type (A) and atclasp-1 (B). The color code is shown in the key, and images were taken at 15-min intervals. C, A single wild-type cell imaged at 15-min intervals with color coding showing four transitions over 120 min. D, Interleaved histograms of pattern transition frequencies for the wild type and atclasp-1 mutants in both GFP-TUB1 and YFP-TUA5 lines with the corresponding mean number of observed transitions ± sd per 120 min. B, basket; L, longitudinal; O, oblique.
atclasp-1 Mutants Show Background-Specific Defects in Microtubule Dynamics
The CLASP protein family has been shown in both in vivo and in vitro experiments to alter microtubule dynamic properties in a variety of ways (Grallert et al., 2006; Bratman and Chang, 2007; Ambrose and Wasteneys, 2008; Al-Bassam et al., 2010; Ambrose et al., 2011). We hypothesized that the observed defect in array transition frequency could result from defects in the dynamic properties of the polymers. Therefore, we measured dynamic instability parameters for both GFP-TUB1 and YFP-TUA5 lines using time-lapse data from confocal microscopy. Frame-to-frame length changes for unbundled microtubules imaged at 2- or 4-s intervals were measured from kymographs using MATLAB scripts (Shaw et al., 2003).
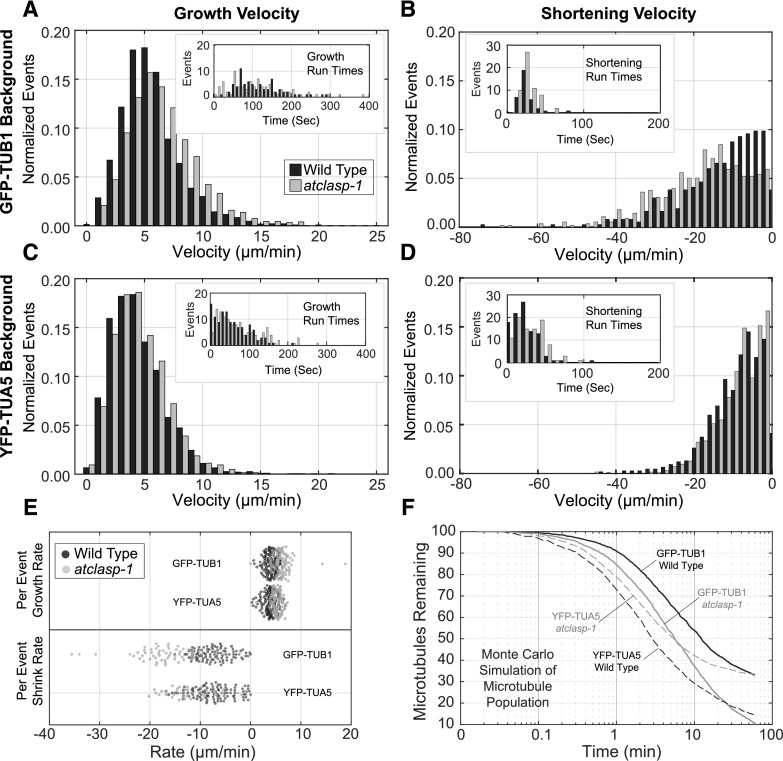
Microtubule plus end growth rates for the GFP-TUB1 lines were ∼15% faster in the atclasp-1 mutant when compared with the wild type (Table 1; Fig. 4A; P < 0.001), and the average shortening rates were 35% faster than in the wild type (Fig. 4B; P < 0.001). Growth rates for the YFP-TUA5 atclasp-1 mutant and wild type were statistically indistinguishable (Fig. 4C; P = 0.11), where shortening in the mutant was 10% slower than in the wild type, rather than faster (Fig. 4D; P < 0.001). Both polymerization and depolymerization were substantially slower in the YFP-TUA5 lines (−22% growth, −30% shortening for the wild type; P < 0.001) when compared with the GFP-TUB1 background. For comparisons, the velocities of microtubules labeled with EB1-GFP were calculated from tracked data, with the wild type and mutant showing virtually the same velocity (Table 1) and appearing 10% faster than the GFP-TUB1 lines and 28% faster than the YFP-TUA4 lines.
Table 1. Microtubule dynamics measurements.
| Plant | Growth | Runs/Events | Shortening | Runs/Events | Catastrophes per Minute | Events/Seconds | Rescues per Minute | Events/Seconds |
|---|---|---|---|---|---|---|---|---|
| µm min−1 | n | µm min−1 | n | n | n | |||
| GFP-TUB1 lines | ||||||||
| Wild type | 5.41 ± 2.6 | 76/3,425 | 13.4 ± 10.7 | 37/365 | 0.163 | 24/8,864 | 0.640 | 9/844 |
| atclasp-1 | 6.29 ± 3.1 | 85/3,589 | 18.3 ± 11.8 | 58/555 | 0.231 | 36/9,362 | 0.672 | 16/1,428 |
| YFP-TUA5 lines | ||||||||
| Wild type | 4.25 ± 2.3 | 123/3,371 | 9.4 ± 7.1 | 100/1,069 | 0.572 | 68/7,138 | 1.473 | 55/2,240 |
| atclasp-1 | 4.34 ± 2.3 | 122/4,308 | 8.4 ± 6.2 | 93/1,257 | 0.475 | 71/8,966 | 1.397 | 60/2,573 |
| EB1-GFP lines | ||||||||
| Wild type | 5.9 ± 1.5 | 6,515 | – | – | – | – | – | – |
| atclasp-1 | 5.8 ± 1.4 | 4,868 | – | – | – | – | – | – |
Figure 4.
Dynamic properties of the cortical microtubules in the wild-type and atclasp-1 mutant lines in GFP-TUB1 and YFP-TUA5 backgrounds. A to D, Interleaved histograms of growth and shortening velocities from wild-type and atclasp-1 cells (normalized for total counts) in the GFP-TUB1 (A and B) and YFP-TUA5 (C and D) backgrounds. Insets show distributions of observed growth run times (A and C) or shortening run times (B and D) used for the calculation of catastrophe and rescue frequencies, respectively. Tabular data are presented in Table 1. E, Distribution of growth and shortening velocities taken for each growth or shortening run for the wild type and mutant in GFP-TUB1 and YFP-TUA5 backgrounds. F, Monte Carlo simulation showing the loss of microtubules from an initial population of 100 (average of 25 trials for each curve) using mean microtubule dynamics parameters from wild-type and atclasp-1 plants in the GFP-TUB1 and YFP-TUA5 backgrounds. Time in log scale for a 60-min simulation was taken at 1-s intervals.
Microtubule plus end catastrophe and rescue frequencies were determined by dividing the total observed events by the total time accumulated in growth or shortening (Table 1). The GFP-TUB1 wild type exhibited 20% to 25% fewer catastrophe events per time than the atclasp-1 mutant but showed approximately the same rescue frequency. Therefore, microtubules in the mutant averaged 30% shorter growth run times (260 versus 369 s) before catastrophe, whereas both the wild type and the mutant rescued after ∼90 s of depolymerization. The YFP-TUA5 wild type had ∼15% more catastrophe events per unit of time than the atclasp-1 mutant, with only a marginal (∼5%) difference in the relative number of rescue events. The average growth run time was 20% longer in the mutant (126 versus 105 s) and approximately equal (42 s) for average shortening run times. The distribution of recorded run times, paired for each wild-type and mutant data set (Fig. 4, A–D, insets), were statistically insignificantly different from each other (P > 0.01) in all cases. They also showed mean distribution times substantially shorter than the calculated respective catastrophe or rescue run lengths, indicating equivalently sampled data sets in each case where the majority of runs did not show a catastrophe or rescue event.
The distribution of frame-to-frame shortening velocities for the GFP-TUB1 lines at the plus end showed a specific deficiency for slower shortening events (less than 15 µm min−1) in the atclasp-1 background, whereas the faster velocities (greater than 15 µm min−1) shared approximately the same distribution (Fig. 4B). To determine if the rate changes were specific to individual microtubules or mixed over the population of depolymerization events, we plotted the mean rate for each contiguous growth or shortening sequence independent of run length time (Fig. 4E). The distributions for shortening rates in the GFP-TUB1 lines clearly showed segregation of the wild-type and mutant events, indicating that the faster velocities in the mutant were not due to a subpopulation of microtubules. Nearly all depolymerization events in the YFP-TUA5 lines occurred at slower (less than 20 µm min−1) velocities when compared with the GFP-TUB1 lines, potentially obscuring the effect of atclasp-1 in the YFP-TUA5 background.
The aggregate minus end depolymerization rate, including pause and shortening, sets the rate of microtubule treadmilling in this system. The minus end depolymerization rate for the GFP-TUB1 wild type (1.3 ± 1.4 µm min−1; n = 526) and atclasp-1 mutant (1.3 ± 1.1 µm min−1; n = 357) were statistically indistinguishable, with the wild type trending slightly longer for the percentage time spent in shortening (40% versus 30%). The YFP-TUA5 line showed substantially slower minus end depolymerization, with the wild type (0.47 ± 0.37 µm min−1; n = 809) shortening faster than the mutant (0.21 ± 0.24 µm min−1; n = 1,666) and trending toward less time in the shortening phase (46% versus 60%). Thus, the GFP-TUB1 lines treadmilled at ∼0.5 µm min−1, whereas the YFP-TUA5 lines were on the order of ∼0.1 to 0.3 µm min−1. These data indicate that microtubules in the YFP-TUA5 lines remain in their original track for a longer time period relative to polymers in the GFP-TUB1 lines, exclusive of microtubule lifetime.
To estimate what effect the dynamic instability parameters would have on the microtubule lifetime, we performed Monte Carlo simulations using the average measured parameter values and examined microtubule loss over time (Fig. 4F). Nucleating 100 microtubules with the same probability of minus end detachment for treadmilling, we plotted the mean number of microtubules remaining over time from 25 replicate simulations for each parameter set. In the absence of other effectors or subunit limitations, the simulations predicted decay curves with 50% loss achieved between 2 and 11 min and a percentage of the polymer population reaching lengths that make extinction unlikely on a normal time scale. The GFP-TUB1 lines showed relatively long-lived polymers, with the loss of AtCLASP expected to shorten the lifespan of microtubules where, for example, 54% of wild-type polymers were remaining at 10 min versus 37% for the mutant. The YFP-TUA5 lines showed a generally shorter predicted lifetime, with the atclasp-1 parameters leading to longer-lived, rather than shorter-lived, microtubules relative to the wild type (Fig. 4F).
In sum, we find differences between the in vivo dynamics measurements that are predicted to alter the steady-state population of cortical microtubules, partly consistent with biochemical studies. However, our data do not support the prediction that the general dynamicity (i.e. total subunit turnover) of the microtubules specifically controls or limits the frequency of array transition. Note that the shorter microtubule lifetimes predicted for the GFP-TUB1 atclasp-1 mutant correlate with fewer array transitions. But the YFP-TUA5 atclasp-1 lines show longer predicted lifetimes than the wild-type parent and still show the decreased array transition frequencies, indicating a more specific role for AtCLASP. The increased microtubule dynamicity in the YFP-TUA5 atclasp-1 line, however, does correlate with the unexpectedly longer hypocotyls (i.e. larger cells), suggesting a relationship between microtubule dynamics and cell size somewhat independent of cell shape.
AtCLASP Modulates the Number of Growing Microtubule Plus Ends
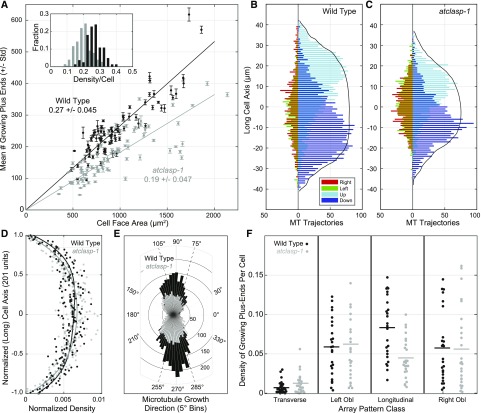
We hypothesized that AtCLASP generally changes the steady-state microtubule population and may further have a specific effect on microtubules related to array transitions. To test these predictions, we used an EB1-GFP transgenic line that labels growing microtubule plus ends to assess the number, position, and direction of growing microtubules on the outer periclinal cell face (Elliott and Shaw, 2018). To estimate the effect of AtCLASP on microtubule number, we counted the mean number of EB1-GFP foci, averaged over five sequential frames, as a function of cell face area in the wild type and atclasp-1 mutants (Fig. 5A). Wild-type EB1-GFP-expressing cells averaged 0.27 ± 0.045 per µm2 growing microtubule plus ends (n = 84 cells), whereas the atclasp-1 cells averaged 0.19 ± 0.047 per µm2 (n = 78 cells) on the outer periclinal cell face. The approximately linear scaling (Fig. 5A) indicated that the atclasp-1 mutant averaged 30% fewer growing plus ends, independent of cell face area. Histograms of the mean density per cell face showed nearly identical normal distributions, centered at different average values (Fig. 5A, inset). In agreement with our predictions from the dynamic instability analysis, AtCLASP alters the steady-state microtubule population and is required to maintain a substantial fraction of the growing microtubule plus ends.
Figure 5.
AtCLASP increases the density of longitudinally oriented microtubule plus ends across the cell face. A, Scatterplot relating the mean ± sd number of EB1-GFP foci per cell face (from five consecutive images) to cell face area for the wild type and atclasp-1 mutants. The inset histogram shows EB1-GFP comet densities per cell for wild-type and mutant lines. Trend lines represent mean density plotted from the origin (n > 80 cells). B and C, Interleaved histograms showing cumulative distributions of growing microtubule (MT) plus ends on the cell’s long axis by growth direction, taken in 90° quadrants (up, down, left, and right), for wild-type and atclasp-1 seedlings (n = 25/25 cells). Longitudinal directions are to the right of the ordinate and transverse directions to the left. Solid black lines indicate the scaled cumulative cell area as a function of the long axis. D, Spatial distribution of growing plus ends as a function of the normalized long axis (21 bins) scaled for total counts. Trend lines represent fourth-order polynomial fits to the wild-type and mutant data showing no obvious difference in the spatial distribution. E, Rose plot showing the number of microtubule trajectories per angle relative to the cell axis in the wild type and the mutant. F, Dot plots showing the density of EB1-GFP foci per cell for each of four defined microtubule orientations (i.e. transverse, 0°–15°/165°–180°; left oblique, 16°–74°; longitudinal, 75°–105°; and right oblique, 106°–164°). Data in B to F are from the same 25 cells for the wild type and the atclasp-1 mutant.
To address whether AtCLASP plays a more specific role in managing the array dynamics, we asked if the deficiency in growing plus ends was a general effect or related to a subpopulation of the microtubules on the cell face. Prior analyses have shown that nontransverse cortical array patterns have a distinct underlying architecture (Chan et al., 2007; Sambade et al., 2012; Elliott and Shaw, 2018). The microtubules on the upper half of the cell face polymerize apically and plus ends on the lower half polymerize basally to form a split bipolar array architecture (Elliott and Shaw, 2018). We imaged transgenic EB1-GFP lines over short (10 frames, 3- to 4-s intervals) time-lapse sequences and tracked the EB1-GFP foci (Elliott and Shaw, 2018). Interleaved histograms showing the cumulative distribution of plus end trajectories, taken in 90° quadrants (i.e. up, down, left, and right), showed a distinct separation of apically and basally directed microtubules for both the wild type (n = 25 cells, 6,515 trajectories) and atclasp-1 (n = 25 cells, 4,868 trajectories) as a function of the cell’s long axis (Fig. 5, B and C), indicating a preservation of the overall split bipolar architecture.
We then evaluated the spatial distribution of all growing plus ends on the cells’ long axes by normalizing each cell length and then normalizing the cumulative number of mapped plus ends in each background for comparison (Fig. 5D). Trend lines, representing fourth-order polynomial fits, indicated that the relative distribution of plus ends on the cell face was not substantively different, cumulatively, between the wild type and atclasp-1, even though the mutant lines had 30% fewer growing plus ends. We next asked if AtCLASP affected microtubules proportionately in all spatial orientations with reference to the cell’s growth axis. Polar histograms of all microtubule trajectories for wild-type and atclasp-1 lines (Fig. 5E) showed that growing microtubules in the wild type appeared to concentrate more in longitudinal directions relative to the atclasp-1 line. Converting direction (1°–360°) to orientation (1°–180°) and determining the density of growing plus ends in transverse (0°–15° and 165°–180°), left-oblique (106°–164°), longitudinal (75°–165°), or right-oblique (16°–74°) orientations from each cell (Fig. 5F), we found that the absence of AtCLASP had a specific and significant effect on the density of only the longitudinally oriented polymers (P < 0.001). Therefore, we conclude that AtCLASP alters the steady-state microtubule population in a specific capacity related to the maintenance of longitudinally oriented polymers.
AtCLASP Affects the Reorientation of the Split Bipolar Microtubule Arrays
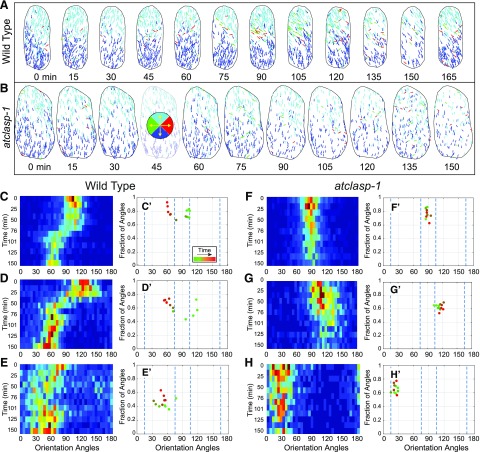
Based on our observations of microtubule pattern, dynamics, and array architecture, we hypothesized that AtCLASP is required to make or maintain new microtubules in new orientations for array transitions. Time-course studies using EB1-GFP in light-grown cells have depicted constant shifts in array orientation (Chan et al., 2007; Sambade et al., 2012; Elliott and Shaw, 2018), with a recent quantitative study showing continuous gradual transitions between longitudinal and oblique patterns (Elliott and Shaw, 2018). Using this technique, we asked how microtubule array orientation and coalignment changed over time in the atclasp-1 mutant, where ∼30% of the longitudinally oriented microtubule plus ends are missing (Fig. 6, A and B). Trajectory maps were made from time-lapse data (10 3D stacks every 3 s for 30 s) acquired every 15 min for 2 h. Microtubule growth trajectories showed array orientation changes for wild-type plants, where new microtubules appeared on the apical half of the cell face at each interval in one new orientation and new microtubules on the basal half appeared in the opposite (relative) orientation (Fig. 6A). The coordinated appearance of new microtubules on both halves of the cell face results in a gradual transition of the split bipolar array orientation in the wild type. The atclasp-1 trajectory maps showed maintenance of the split bipolar array architecture but no evidence for new microtubules appearing in new orientations to affect a change in array orientation (Fig. 6B).
Figure 6.
Microtubule arrays in atclasp-1 show comparatively slow changes in array orientation and coalignment over time. A and B, Time-course analysis of a single wild-type (A) and atclasp-1 mutant (B) cell showing EB1-GFP trajectories color coded by growth direction at ∼15-min intervals over a 150-min period. The key in B shows the color coding trajectory. C to H, Histograms of microtubule orientation (1°–180°) taken at ∼15-min intervals determined from EB1-GFP growth trajectories plotted as heat maps with time running from top to bottom for three wild-type (C–E) and three atclasp-1 (F–H) cells starting in the same pattern. C′ to H′, Plots of the relative degree of array coalignment over the dominant microtubule orientation angle, color coded green to red for time, for the wild type (C′–E′) and atclasp-1 (F′–H′) using a ±21° window.
For quantitative comparisons, plus end growth direction (1°–360°) was converted to orientation angle (1°–180°), and histograms for all orientations at each time point were collected as heat maps for wild-type (Fig. 6, C–E) and atclasp-1 (Fig. 6, F–H) cells, with time progressing from top to bottom. The relative degree of microtubule coalignment at each time point was quantified further by applying a sliding (±20°) window to each histogram to find the range of orientation angles containing the highest fraction of plus end trajectories (Elliott and Shaw, 2018). The central angle from that window, representing the dominant orientation angle for the array, was plotted against the fraction of all plus ends contained within the window (representing the degree of array coalignment) for wild-type (Fig. 6, C′–F′) and atclasp-1 mutant (Fig. 6, G′–J′) cells.
Data from three representative wild-type control cells (n = 5 time courses each) showed gradual shifts between longitudinal and oblique patterns (Fig. 6, C–E) over a progression of relative array coalignment values (Fig. 6, C′–E′), in agreement with prior work (Chan et al., 2007; Elliott and Shaw, 2018). Time-course data from three atclasp-1 cells (Fig. 6, F–H) with arrays starting in approximately the same orientation as the corresponding wild-type cells (i.e. longitudinal, right oblique, and left oblique, respectively) maintained a relatively constant degree of array coalignment (Fig. 6, F′–H′). Consistent with the pattern transition data for atclasp-1 in the fluorescent tubulin-expressing lines, these cells failed to show a gradual reorientation over time, remaining essentially fixed within a 10° angular range over a 2-h period (note in Fig. 6 that the cell in A is used in C and the cell in B is used in F). Moreover, the cortical microtubules maintained an average level of coalignment (Fig. 6, F′–H′) that did not indicate failed or aborted efforts to transition the pattern. These data show that, in light-grown atclasp-1 cells lacking 30% of the longitudinally oriented growing microtubule plus ends, there is a corresponding absence of microtubule plus ends appearing in a new orientation on the upper and lower halves of the cell face to affect the reorientation of the array.
AtCLASP Protects Microtubules from KATANIN-Dependent Severing at Crossovers
Our data suggest a model where the action of AtCLASP on the dynamic properties of the microtubules leads to a specific loss of new or nascent polymers required for transitioning the array to a new pattern. To further probe the potential functions of AtCLASP on this microtubule population, we examined the outcome of microtubule-microtubule interactions in light-grown cells undergoing gradual changes in array orientation. We hypothesized that the persistent generation of microtubules in a new orientation during array transitions would produce microtubule-microtubule interactions resulting in microtubule bundling or crossovers. Bundling typically reinforced the existing array pattern by adding new polymers to the existing order. We speculated that successful microtubule crossovers permit array reorientation. We examined the outcome of microtubule crossover events in light-grown seedlings as a function of position on the cell face. We further examined the outcome related to the microtubule-microtubule crossover angle and the angle of the polymer relative to the cell’s primary growth axis. We imaged wild-type and atclasp-1 cells in both the GFP-TUB1 and YFP-TUA5 backgrounds at 2-s intervals for 4 to 6 min and analyzed data with a MATLAB script to mark all crossover events that resolved through either severing or depolymerization. Crossover events were collected from an equivalent number of GFP-TUB1 cells (n = 15), having a nearly equivalent total cell face area for the wild type (19,680 um2) and atclasp-1 (18,673 um2) as well as equivalent numbers of YFP-TUA5 cells (n = 13) with more disparate wild-type (9,593 um2) and atclasp-1 (16,212 um2) cumulative areas.
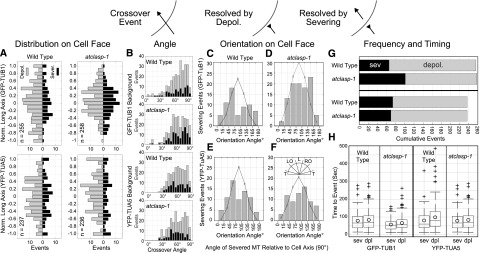
The spatial distribution of resolved crossovers, as a function of the cell’s long axis, showed a trend toward higher counts in the middle 50% of the cell face in all lines after normalizing for cell length (Fig. 7A). In the absence of AtCLASP, the GFP-TUB1 lines showed an obvious increase toward the middle of the cell, whereas the YFP-TUA5 lines did not. The distribution of angles between crossed microtubules (Fig. 7B) was not substantially different between the four lines, suggesting that any effects on microtubule bundling angle (Dixit and Cyr, 2004; Zhang et al., 2013) did not propagate to the level of crossover angle in the atclasp-1 lines. The crossover angle also had no obvious effect on whether the event resolved through severing or depolymerization in wild-type or atclasp-1 lines (Fig. 7B).
Figure 7.
The atclasp-1 mutant shows background-dependent defects in microtubule severing at crossover sites. The schematic indicates microtubule crossover as a black arrow resolved through depolymerization (reversed arrow) or through severing at the crossover site. A, Spatial distribution of crossover events normalized to the cell’s long axis showing events resolved by depolymerization (gray, left of ordinate) and severing (black, right of ordinate) in GFP-TUB1 and YFP-TUA5 backgrounds for wild-type and atclasp-1 cells. B, Angle of microtubule crossover for all backgrounds in stacked histograms showing depolymerization (gray) and severing (black) events. C to F, Histograms showing the cumulative number of severing events for microtubules over a range of microtubule orientation angles relative to the long axis of the cell. Gray lines represent expected distributions of orientation angles estimated from EB1-GFP wild-type and mutant lines (Fig. 5) and scaled to the number of severing events in each histogram. The key in F gives the spatial orientation of histogram bins. G, Stacked bar graphs showing the fraction of crossover sites that were resolved by either severing (black) or depolymerization (gray) for wild-type and atclasp-1 mutant cells in GFP-TUB1 (n = 256/258) and YFP-TUA5 (n = 237/236) backgrounds. The difference between the wild type and the mutant in the GFP-TUB1 background is significant at P < 0.001 and not significant in the YFP-TUA5 background at P = 0.05 (Student’s t test). H, Box-and-whisker plots showing the amount of time in each crossover recorded prior to a severing or depolymerization event. Boxes represent quartiles, whiskers indicate ranges, and circles represent arithmetic means.
Noting the specific effect of AtCLASP on longitudinally oriented microtubules (Fig. 5F), we examined the orientation of microtubules severed at crossovers with respect to the long axis of the cell (Fig. 7, C–F). The expected distributions of all microtubule orientation angles for wild-type and atclasp-1 lines were estimated from EB1-GFP trajectories (Fig. 5) and approximated normal distributions centered at longitudinal (90°), with the wild type (90.7° ± 34.1°, n = 6,515) less spread than atclasp-1 (89.3° ± 40.6°, n = 4,868). Comparing the orientation angles of the severed polymers with the expected distributions for all polymers and scaled to the number of severing events in each case (Fig. 7, C–F, line plots), we observed an obvious deficiency in severing events for longitudinally oriented polymers in both GFP-TUB1 (Fig. 7C) and YFP-TUA5 (Fig. 7E) wild-type lines. Loss of AtCLASP in the GFP-TUB1 background resulted in longitudinal polymers sharing a greater proportion of the severing events (Fig. 7D). Loss of AtCLASP in the YFP-TUA5 line also led to a higher relative number of severing events occurring for longitudinally oriented polymers (Fig. 7F), but to a lesser extent than for the GFP-TUB1 lines. These data indicate that the distribution of severing events at crossover sites in wild-type lines is not random but becomes more randomized in the absence of AtCLASP.
The total number of resolved crossover events observed in the GFP-TUB1 wild-type and atclasp-1 lines, using equivalent cell face areas and imaging times, was nearly identical. However, severing occurred for 26% of the events in the wild type (n = 65/255; 15 cells) compared with 40% in the atclasp-1 mutant (n = 101/258; 15 cells), indicating a significantly higher fraction of severing events in the mutant (Fig. 7G; P < 0.001, Z-score test). Analysis of the YFP-TuA1 lines indicated no significant difference (P > 0.05) in how the crossover events were resolved (Fig. 7G), with the wild type severing in 31% of cases (74/237; n = 13 cells) and atclasp-1 severing in 29% of cases (69/236; n = 13 cells). We interpret these data to indicate that polymer severing at crossover sites can be affected by AtCLASP but likely only plays an auxiliary role in facilitating the array transitions.
To determine if the increased number of severing events in the GFP-TUB1 atclasp-1 lines arose because of higher severing activity or because of more opportunity for severing (i.e. time spent in the crossover), we recorded the time leading up to each severing or depolymerization event (Fig. 7H). Data include crossovers present at the start of the image series together with crossovers that initiated within the experiment. The mean time to severing in the wild type (68.9 ± 57.5 s) was significantly longer than that in the atclasp-1 mutant (47 ± 32.6 s; P < 0.01). Moreover, the mean time to depolymerization for the atclasp-1 mutant (56.3 ± 41.8 s) was significantly shorter than that for the wild type (73.7 ± 55.5 s; P < 0.01). These results show that, for an equivalent number of crossover events in the GFP-TUB1 background, the microtubules in the wild type remained crossed over for a longer average time than in the mutant, having more opportunity for severing, not less. Therefore, we conclude that severing activity is higher in GFP-TUB1-expressing cells lacking the AtCLASP protein. The crossover durations in the YFP-TUA5 lines for severing in the wild type (70.6 ± 57.8 s) and the mutant (75.5 ± 71.2 s) were not statistically different (P > 0.05), and the times to depolymerization for the wild type (88.7 ± 76 s) and the mutant (101.3 ± 84.6 s) also were not significantly different (P > 0.05).
To determine if the increased severing frequency in the atclasp-1 mutant was dependent upon KATANIN p60 (McNally et al., 2000; Bouquin et al., 2003; Zhang et al., 2013), we created an atclasp-1 katanin p60 double mutant in the GFP-TUB1 background. We observed 87 crossover events resolved through depolymerization and found no evidence for severing at crossovers (n = 10 cells, 10,428 µm2 area), consistent with prior data for the katanin single mutant (Zhang et al., 2013). The mean time to depolymerization was 105 ± 83.9 s, which was longer than the related times for the wild type and the atclasp-1 single mutant. Thus, the higher severing activity at crossover sites observed in the GFP-TUB1 atclasp-1 mutant requires KATANIN activity.
DISCUSSION
AtCLASP Facilitates Cortical Microtubule Pattern Transitions
The loss of AtCLASP results in more isotropic plant cell shapes (Ambrose et al., 2007; Kirik et al., 2007) similar to treatments of wild-type seedlings with low doses of microtubule-specific drugs, such as oryzalin or taxol (Baskin et al., 1994). In this regard, the atclasp-1 hypocotyl growth phenotype generally follows the predictions made from the microtubule-microfibril coalignment hypothesis (Green, 1962; Ledbetter and Porter, 1963). Under this hypothesis, the interphase microtubules form patterns at the cell cortex that template the deposition of cellulose into the cell wall, creating anisotropic wall materials when the microtubules are strongly coaligned (Green, 1962; Cyr, 1994). For plant tissues, microtubule coalignment that is transverse to the plant axis would allow conjoined cells to expand axially along their lateral walls, producing axial extension of organs such as the hypocotyl and root.
Central to the microtubule-microfibril hypothesis is the idea that the microtubule pattern persists at the cell cortex for an appropriate amount of time to orient microfibril deposition in a functional capacity. Cellulose deposition rates, based on measurements of cellulose synthase movement, are on the order of 3 to 4 µm over a 10-min interval (Paredez et al., 2006). Time-course studies have shown that microtubules in light-grown cells rarely form static array patterns at the cell cortex (Chan et al., 2007; Elliott and Shaw, 2018), changing orientation in a gradual (Elliott and Shaw, 2018) or sometimes dramatic (Chan et al., 2007; Sambade et al., 2012; Vineyard et al., 2013) fashion over a 10-min time span. Therefore, we reason that the ability of the microtubules to influence the net orientation of cellulose deposition should depend upon both the array pattern, per se, and the time over which the pattern is resident at the cell cortex.
We found that the dominant macroscopic defect for the cortical microtubule arrays in the atclasp-1 mutant was the reduced frequency of array transitions between patterns. The absence of AtCLASP in both the GFP-TUB1 and YFP-TUA5 fluorescent tubulin marker lines resulted in less than half the number of pattern transitions per time interval when compared with the wild-type parents. The longitudinal and oblique patterns in the EB1-GFP atclasp-1 mutant shifted the array orientation only a few degrees over a 2-h time course compared with 40° to 60° for the wild type. We found no evidence for novel array patterns or for arrays that transitioned to new patterns at the same velocity as the wild type, but less frequently. Our evidence, consistent across three different microtubule marker lines, indicates that AtCLASP acts generally to increase the rate at which the cortical microtubule arrays change pattern on the outer hypocotyl cell face. We argue that observing the same array patterning phenotype in all three backgrounds strongly supports the case for the defect arising through the loss of AtCLASP and not because of the tubulin probe or differences in the Col-0 parent lines.
Surprisingly, given the evident defect in transition frequency, the atclasp-1 mutants formed all of the cortical array patterns normally observed in wild-type, light-grown hypocotyl cells and in nearly the same relative numbers for each respective background (Vineyard et al., 2013). In agreement with prior work (Ambrose and Wasteneys, 2008; Ambrose et al., 2011), we found a measurably higher percentage of arrays with coaligned patterns that we interpret as a defect in making new polymers in new orientations. The nontransverse array patterns in the mutant exhibited the correct underlying split bipolar array architecture (Elliott and Shaw, 2018) and transitioned to the correct transverse pattern when treated with IAA/GA4, albeit to a lesser extent than in the wild type, in the 2-h allotted time. Our findings, therefore, place AtCLASP into a category of proteins that facilitate array transitions, rather than a protein that explicitly specifies a microtubule array pattern. Moreover, our study indicates that the inability to transition the array patterns on an appropriate time scale, rather than an inability to form the actual patterns, correlates with a specific defect in cellular growth anisotropy. These data extend our conceptual framework for the microtubule-microfibril coalignment hypothesis by showing the importance of timing with regard to the residency time and the transitioning of array patterns.
AtCLASP Modulates Microtubule Disassembly
A defining feature of the CLASP gene family is the tubulin-binding TOG domain (Al-Bassam et al., 2010; Al-Bassam and Chang, 2011). In vitro studies with a CLASP protein from fission yeast showed a concentration-dependent ability to increase rescue frequency, decrease catastrophe frequency, and slow the microtubule depolymerization rate (Bratman and Chang, 2007; Al-Bassam et al., 2010). Other studies, in combination with other MAPs, have indicated that CLASP reconstitutes the pausing of microtubules observed in vivo (Moriwaki and Goshima, 2016). We examined how the loss of AtCLASP affected in vivo microtubule dynamics in two plant backgrounds that have substantially different microtubule dynamics properties. A comparison of how AtCLASP loss alters the base dynamic properties in these lines provides a broader interpretation of how AtCLASP is able to modulate the microtubule plus ends.
The GFP-TUB1 lines exhibited faster microtubule growth and shortening rates with less frequent catastrophe and rescue when compared with the YFP-TUA5 lines. Differences could be due to the tubulin isoform, fluorescent protein, linker, expression levels, or many other effects that go beyond the scope of this study. We assume that both transgenes alter microtubule dynamics to some extent but argue that the GFP-TUB1 lines likely report dynamics closer to native values, based on our comparison of hypocotyl growth phenotypes with the reference Col-0 plants.
atclasp-1 in the GFP-TUB1 background exhibited changes in shortening velocity and catastrophe frequency consistent with in vitro CLASP studies (Al-Bassam et al., 2010), but we observed no significant change in rescue frequency under these conditions. The faster (35%) microtubule shortening rate was due to the loss of slower depolymerization events (less than 15 µm min−1), indicating a specific defect rather than a general retardation of shortening. AtCLASP was observed previously to have microtubule tip-biased localization with discontinuous labeling along the microtubule lattice (Akhmanova et al., 2001; Ambrose et al., 2007; Kirik et al., 2007). Therefore, we propose that AtCLASP most likely directly retards depolymerization just after catastrophe and in regions along the polymer with higher AtCLASP binding (Kirik et al., 2007). A stabilizing role for AtCLASP on the polymer lattice would additionally explain the higher catastrophe frequency found in the GFP-TUB1 atclasp-1 mutant lines. We expected that faster depolymerization would make rescue events less frequent but did not find evidence in support of this hypothesis. Faster subunit loss in the shortening phases, we propose, could account for this observation if the increased concentration of free tubulin subunits from shortening improves the odds of rescue and leads to the ∼10% increase in microtubule growth rate.
The wild-type YFP-TUA5 lines showed substantially slower rates of polymer growth and shortening than the GFP-TUB1 lines, with markedly higher levels of catastrophe and rescue. We further report a slower rate of minus end depolymerization. We infer from these in vivo data that the YFP-TUA5 transgene slows polymer disassembly and assembly. In accordance with theory, slow polymer growth should result in a higher catastrophe frequency, as we observed, owing to a shorter GTP cap. Increasing catastrophe on its own, however, should liberate more free tubulin, leading to faster growth rates, which is the opposite of what we observed. Similarly, slower polymer disassembly could increase the odds of rescue, but higher rescue frequency alone would not be expected to slow the rate of polymer disassembly. The absence of AtCLASP in this YFP-TUA5 background had only small effects on dynamics and actually increased the predicted lifetime of the polymers. We reason here that the loss of a stabilizing factor (i.e. AtCLASP) in a plant line with intrinsically slower kinetic properties actually could increase the overall dynamics of the polymer system. We did not observe the same loss of slower depolymerization events in the YFP-TUA5 mutant that we observed in the atclasp-1 GFP-TUB1 line. We propose that the inherently slower disassembly in the YFP-TUA5 line masks or partly obviates the stabilizing action of AtCLASP observed in the GFP-TUB1 lines, where the absence of shortening events greater than 15 µm min−1 may preclude the observation of slower depolymerization in the YFP-TUA5 atclasp-1 background.
Monte Carlo simulations using the measured dynamics parameters to examine the persistence of treadmilling microtubules in a population led to a set of predictions about the GFP-TUB1 and YFP-TUA5 lines. The more general prediction was that the GFP-TUB1 lines would have more persistent microtubules than the YFP-TUA5 lines. This outcome broadly correlates with the finding of longer hypocotyls and more frequent array transitions in the GFP-TUB1 lines when compared with the YFP-TUA5 lines. The more specific prediction was that the atclasp-1 mutant in the YFP-TUA5 background would have more persistent microtubules than the wild-type parent. This outcome correlates with our observation of slightly longer hypocotyls in the atclasp-1 YFP-TUA5 mutant when compared with the wild type, owing to more total growth in the mutant lines. Based on these observations, we propose that cellular growth anisotropy is driven more directly by the AtCLASP-dependent frequency of array pattern transitions, which is related to the tenets of the microtubule-microfibril coalignment hypothesis. The total cell size, however, showed a dependency on the dynamic properties of the microtubules, where at present we have no theory to guide our expectations.
AtCLASP Changes the Steady-State Microtubule Population
How the split bipolar array gradually changes orientation or rotates between longitudinal and oblique patterns has not been elucidated (Elliott and Shaw, 2018). Since cortical microtubules do not slide on the cell cortex (Shaw et al., 2003), changing array orientation requires the concerted nucleation or redirection of treadmilling microtubules in a new orientation. To preserve the split bipolarity, microtubules on the upper and lower cell face halves must be created in opposite directions with reference to the cell face (e.g. leftward for the upper half and rightward for the lower half of the cell face for a counterclockwise transition; Chan et al., 2007; Sambade et al., 2012; Elliott and Shaw, 2018). Our study shows that AtCLASP is required for this process.
A limited number of mechanisms have been described that could contribute to the observed transitions. Microtubules could nucleate from existing polymers at an angle (i.e. branched nucleation; Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010; Kirik et al., 2012) or sweep in from the cell’s lateral sides (Chan et al., 2007; Sambade et al., 2012; Vineyard et al., 2013). Microtubule plus ends could be dislodged from the cell cortex and reoriented upon reassociation (Ambrose and Wasteneys, 2008) or selectively lost in all other orientations (Ambrose et al., 2011). More recently, transitions to a longitudinal pattern in response to light transiently amplified the microtubules in a perpendicular orientation through KATANIN-mediated severing at crossover sites with rescue of the newly created plus end (Lindeboom et al., 2013). In all cases, a new mechanism would need to be proposed to account for creating or maintaining microtubules in only one direction on each half of the cell face (Murata and Hasebe, 2007; Nakamura et al., 2010).
Our observations of the atclasp-1 mutant provide significant insight into how these mechanisms relate to pattern transitions. We found that transitions are not a direct consequence of the mechanisms creating the split bipolar architecture. The atclasp-1 mutants retained the split bipolarity with a high degree of array coalignment and did not show episodic losses of microtubule coalignment to indicate failed attempts at reorientation. Moreover, atclasp-1 mutants did not redirect microtubules in random directions. We infer that AtCLASP is not decreasing the bundling acceptance angle (i.e. bundling instead of allowing microtubules to cross over) to increase transition frequency, based on the distribution of crossover angles for the microtubules. While an increase in catastrophe frequency was observed in the GFP-TUB1 atclasp-1 mutant, we could not ascribe the increase to induced catastrophes at sites of microtubule intersection or at cell edges. Furthermore, we ruled out mechanisms related to the culling of microtubules from old orientations because of the high degree of array coalignment and the fact that 30% of the growing plus ends were missing in the mutant, suggesting loss of polymers, not retention.
AtCLASP exerts a stronger effect on longitudinally oriented microtubules (Ambrose et al., 2011; Elliott and Shaw, 2018), suggesting a specific or more restricted role for AtCLASP on one class of microtubules. The specificity for longitudinal polymers is in agreement with prior work examining the cell edge-dependent effects of AtCLASP (Ambrose et al., 2011), although we found no real change to the distribution of coaligned array patterns and no spatial bias for plus end distribution on the cell face or at cell edges. A special role for AtCLASP in array transitions is suggested from experiments showing that the YFP-TUA5 atclasp-1 line was strongly retarded for transition frequency but showed a relative increase, not decrease, in predicted microtubule lifetime over the YFP-TUA5 wild type. Thus, the general effect of AtCLASP on microtubule dynamics does not fully explain the spatial biases observed for the microtubule phenotypes.
We reasoned that a common element to all mechanisms leading to the reorientation of the cortical arrays is an increased number of interactions between microtubules in the new orientation and those in the old pattern. Modeling studies have shown that interactions leading to bundling should preserve the old array orientation, limiting the ability to shift pattern (Dixit and Cyr, 2004; Baulin et al., 2007; Ehrhardt, 2008; Allard et al., 2010a; Eren et al., 2010). Therefore, we examined the position, orientation, and extent of interactions leading to microtubule crossovers. In both GFP-TUB1 and YFP-TUA5 backgrounds, the spatial distributions of the resolved crossovers were not altered in atclasp-1 mutants. However, longitudinal polymers experienced disproportionately fewer severing events in the wild type than we expected, given the overall distribution of microtubule orientation (Elliott and Shaw, 2018). Loss of AtCLASP led to a more uniform distribution of severing events across all orientations, which was observed more prominently in the GFP-TUB1 background. The effect was partly explained by the higher probability of KATANIN-dependent severing at crossover positions in the GFP-TUB1 background. The absence of a clear severing defect in the YFP-TUA5 atclasp-1 mutant under these growth conditions indicates that severing frequency alone is not the critical factor in controlling steady-state array transitions in this background.
In sum, we find that the in vivo role of AtCLASP includes a stabilizing role for the microtubule polymers in the interphase plant cortical array, acting to slow depolymerization, suppress catastrophe, and protect crossovers from KATANIN-dependent severing. The general actions of AtCLASP in this treadmilling system permit enough new microtubules to persist in new orientations to affect both the gradual steady-state transitions for the split bipolar arrays and the more transformative hormone-induced transitions into a transverse coalignment related to axial growth. We argue that the AtCLASP-dependent ability to make these array pattern transitions in a timely manner, rather than the ability to create the array pattern per se, leads to the prescribed defect in cellular growth anisotropy. While extending the microtubule-microfibril coalignment hypothesis neatly accounts for the observed cellular anisotropy defect, we find evidence that the dynamic properties of the microtubules, separable from creating microtubule pattern, may influence total cell size.
MATERIALS AND METHODS
Plant Materials and Seedling Culture
Arabidopsis (Arabidopsis thaliana) seeds were sterilized in 83% (v/v) ethanol and 1.5% (v/v) hydrogen peroxide and plated on 1% (w/v) agar plates containing 0.5× Murashige and Skoog (MS) Basal Salt Mixture (Sigma-Aldrich). Plates with seed were refrigerated in the dark at 4°C for 1 d and then germinated and grown under constant illumination at 22°C. Wild-type Arabidopsis plants expressing a 35S-GFP-TUB1 transgene (Elliott and Shaw, 2018) or 35S-GFP-EB1 (Mathur et al., 2003) were crossed to atclasp-1 lines originally supplied from the Arabidopsis stock center (Kirik et al., 2007), and F2 lines with the growth phenotype and fluorescence were propagated for stable F3 fluorescent lines for experiments. YFP-TUA5 lines were a generous gift from David Ehrhardt (Stanford University). Double atclasp-1 katanin p60 mutants were derived by crossing the GFP-TUB1 atclasp-1 line to a katanin p60 mutant in the same GFP-TUB1 background followed by selection for phenotype and genotyping. Whole-genome sequencing was performed using libraries made from GFP-TUB1 atclasp-1, YFP-TUA5 atclasp-1, and EB1-GFP atclasp-1 lines for Illumina sequencing to ∼10× coverage. Postassembly identification of the tDNA insertion site and the fluorescent protein junction were used to verify the mutation and fluorescent reporter in each case.
Hypocotyl length was determined from calibrated stereomicroscope images using the segmented-line tool in FIJI. For cell length and width, confocal image stacks were taken with a 40× objective lens using two to three fields of view and stitched together in Photoshop before import into a MATLAB script. Cells were analyzed by selecting a position on the apex, base, and left and right sides of each cell and calculating the distance from the pixel to micrometer calibration. Effective width was from the widest cross section of the cell.
Microtubule Dynamic Instability Studies
Six-day-old light-grown seedlings were transferred to liquid 0.5× MS medium and equilibrated to the liquid at 22°C for 20 to 40 min in the light. Wild-type and atclasp-1 seedlings were mounted on the same glass slides with 0.5× MS liquid medium, where two layers of double-sided tape were used on either side of the slide to adhere a coverslip to form an imaging chamber. Confocal images were generated using a 100× oil-immersion objective on a Yokogawa CSU-10 spinning disk confocal on an inverted Nikon microscope, a 60× water-immersion objective on a Nikon A1 laser scanning confocal, and a 100× silicon oil objective on an Olympus OSR with a W1 spinning disk. Images were taken at 2- or 4-s intervals for 4 to 12 min in all cases. Images were converted from manufacturer-specific files to tiff files using FIJI and imported into MATLAB (MathWorks) with temporal and spatial information entered by hand. A Graphical User Interface (GUI) developed in MATLAB was used to import image series and to trace over single microtubules by hand to generate kymographs. The microtubule ends then were tracked on the kymograph, and the positions were computationally related back to the image series for hand editing as described previously (Shaw and Lucas, 2011). Frame-to-frame distances for microtubule tip movement were recorded, converted to micrometers (from stage micrometer) per minute, and exported to a second GUI for analysis. The contribution of dynamics information from different imaging sources was tested as a source of error both by examining the data from each microscope independently and by randomly bootstrapping the microtubules imported into the analysis at increasingly lower fractions of the total data set. All figure generation was performed with custom scripts in MATLAB and imported as vector-based graphics into Illustrator (Adobe Systems).
Microtubule Severing at Crossover Site Studies
Six-day-old light-grown seedlings were grown and prepared on glass slides as described for the microtubule dynamic instability studies. Confocal images were generated using a 100× oil-immersion objective on a Yokogawa CSU-10 spinning disk confocal on an inverted Nikon microscope and a 60× or 100× silicon oil objective on an Olympus OSR with a W1 spinning disk. Images were taken at 2- or 4-s intervals for 4 to 8 min in all cases. Images were converted to tiff files using FIJI and imported into MATLAB with temporal and spatial information entered by hand.
Microtubule Plus End Density
Six-day-old, light-grown EB1-GFP-expressing lines were transferred to liquid 0.5× MS medium and allowed to incubate in the light for 30 to 60 min. EB1-GFP lines then were mounted on glass slides with 0.5× MS liquid medium using vacuum grease to adhere the coverslips. Plants were allowed to equilibrate for 20 to 40 additional min in the light at 22°C. Confocal images were generated using a 60× water-immersion objective lens on a Nikon A1 inverted laser scanning confocal microscope. 3D image stacks consisting of five focal planes taken at 0.3- to 1.2-µm axial steps were captured at 8-s intervals for 40 s. Images for each five-frame time lapse were projected into a series of 2D images using FIJI and converted to tiff files. MATLAB code was developed to import images into a GUI, create a hand-drawn cell perimeter, and assist in the identification of all comets in each image. The identification of EB1-GFP comets was performed initially using optimized difference of Gaussian filters followed by intensity thresholding and center of mass calculations. Putative spot identifications then were overlain onto each image as a transparent marker in the GUI for hand editing to add or delete spots. The area was calculated from the cell perimeter and converted to square micrometers using a conversion factor from the software and verified with a stage micrometer. The density of EB1-GFP spots was calculated by dividing the mean number of plus ends from the five time points over the cell area and plotted as mean ± sd over the cell face area.
Microtubule Pattern Transition Frequency
Seedlings expressing a fluorescent tubulin marker were grown and mounted according to dynamics studies and were imaged using a 40× oil-immersion objective on a Nikon A1 inverted confocal laser scanning microscope. Image stacks were taken of the central region of the hypocotyl at 15-min intervals for 2 h and projected to 2D maximum intensity images using FIJI. Images were printed using a laser printer, and each cell on each page was scored for array pattern. The temporal sequence of transitions for each cell then was recorded to a spreadsheet for the determination of transition frequency.
Hormone Induction Assays
Seedlings grown according to dynamics studies were incubated in the light for 2 h in 2 mL of 0.5× MS liquid medium with or without a final concentration of 0.5 µm IAA (Sigma-Aldrich) and 10 µm GA4 (Sigma-Aldrich), as established previously (Vineyard et al., 2013). Seedlings then were mounted onto glass slides with 0.5× MS liquid medium, and vacuum grease was used to adhere the coverslips. Plants were immediately imaged with a 40× oil-immersion objective on a Nikon A1 inverted confocal laser scanning microscope. Image stacks were projected to 2D and stitched to create a representation of the entire hypocotyl length using the Nikon Elements Scan Large Image stitching function. Image data were imported into a MATLAB-based GUI developed to record the positions and investigator-assigned array patterns of all cells from two adjacent cell files running the length of the hypocotyl.
Accession Number
The accession number for the Arabidopsis cytoplasmic linker protein CLASP is AT2G20190.
Acknowledgments
We thank David Ehrhardt for lines. We thank Dr. Viktor Kirik for helpful advice and Dr. Jim Powers and the Indiana University Light Microscopy Imaging Center for invaluable help with this project. Sequencing was performed through contract with the Indiana University Center for Genomics and Bioinformatics, with expert help from Doug Rusch and Chris Hemmirich.
Footnotes
This work was supported by the National Science Foundation (MCB:1615907 to S.L.S.) and by a James P. Holland Fellowship, a Carlos Miller Graduate Fellowship in Plant Biology, and a graduate fellowship from the Indiana University Graduate School in cooperation with the Templeton Foundation to D.T.
Senior author.
References
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, et al. (2001) Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104: 923–935 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F (2011) Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol 21: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F (2010) CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell 19: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard JF, Ambrose JC, Wasteneys GO, Cytrynbaum EN (2010a) A mechanochemical model explains interactions between cortical microtubules in plants. Biophys J 99: 1082–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard JF, Wasteneys GO, Cytrynbaum EN (2010b) Mechanisms of self-organization of cortical microtubules in plants revealed by computational simulations. Mol Biol Cell 21: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ambrose JC, Wasteneys GO (2008) CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol Biol Cell 19: 4730–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO (2011) A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat Commun 2: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Ruan Y, Gardiner J, Tamblyn LM, Catching A, Kirik V, Marc J, Overall R, Wasteneys GO (2013) CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev Cell 24: 649–659 [DOI] [PubMed] [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO (2007) The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S, Kirik A, Kirik V (2014) Microtubule array reorientation in response to hormones does not involve changes in microtubule nucleation modes at the periclinal cell surface. J Exp Bot 65: 5867–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2001) On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE (1994) Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol 35: 935–942 [PubMed] [Google Scholar]
- Baulin VA, Marques CM, Thalmann F (2007) Collision induced spatial organization of microtubules. Biophys Chem 128: 231–244 [DOI] [PubMed] [Google Scholar]
- Bouquin T, Mattsson O, Naested H, Foster R, Mundy J (2003) The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J Cell Sci 116: 791–801 [DOI] [PubMed] [Google Scholar]
- Bratman SV, Chang F (2007) Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell 13: 812–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C (2007) Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat Cell Biol 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chan J, Sambade A, Calder G, Lloyd C (2009) Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21: 2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr RJ. (1994) Microtubules in plant morphogenesis: Role of the cortical array. Annu Rev Cell Biol 10: 153–180 [DOI] [PubMed] [Google Scholar]
- Deinum EE, Tindemans SH, Mulder BM (2011) Taking directions: the role of microtubule-bound nucleation in the self-organization of the plant cortical array. Phys Biol 8: 056002. [DOI] [PubMed] [Google Scholar]
- Dixit R, Cyr R (2004) Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. (2007) Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell 12: 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW. (2008) Straighten up and fly right: Microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr Opin Cell Biol 20: 107–116 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Shaw SL (2006) Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Elliott A, Shaw SL (2018) Microtubule array patterns have a common underlying architecture in hypocotyl cells. Plant Physiol 176: 307–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren EC, Dixit R, Gautam N (2010) A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol Biol Cell 21: 2674–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM (2006) S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev 20: 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PB. (1962) Mechanism for plant cellular morphogenesis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Hardham AR, Gunning BE (1978) Structure of cortical microtubule arrays in plant cells. J Cell Biol 77: 14–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Kato T (2006) Cortical control of plant microtubules. Curr Opin Plant Biol 9: 5–11 [DOI] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V (2012) TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Herrmann U, Parupalli C, Sedbrook JC, Ehrhardt DW, Hülskamp M (2007) CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J Cell Sci 120: 4416–4425 [DOI] [PubMed] [Google Scholar]
- Ledbetter MC, Porter KR (1963) A “microtubule” in plant cell fine structure. J Cell Biol 19: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom JJ, Nakamura M, Hibbel A, Shundyak K, Gutierrez R, Ketelaar T, Emons AM, Mulder BM, Kirik V, Ehrhardt DW (2013) A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 342: 1245533. [DOI] [PubMed] [Google Scholar]
- Liu X, Qin T, Ma Q, Sun J, Liu Z, Yuan M, Mao T (2013) Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell 25: 1740–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C. (2011) Dynamic microtubules and the texture of plant cell walls. Int Rev Cell Mol Biol 287: 287–329 [DOI] [PubMed] [Google Scholar]
- Lloyd C, Chan J (2008) The parallel lives of microtubules and cellulose microfibrils. Curr Opin Plant Biol 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Lucas J, Shaw SL (2008) Cortical microtubule arrays in the Arabidopsis seedling. Curr Opin Plant Biol 11: 94–98 [DOI] [PubMed] [Google Scholar]
- Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL (2011) Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell 23: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL (2005) Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol 7: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Srinivas BP, Hülskamp M (2003) A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr Biol 13: 1991–1997 [DOI] [PubMed] [Google Scholar]
- McNally KP, Bazirgan OA, McNally FJ (2000) Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J Cell Sci 113: 1623–1633 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. (2005) CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol 168: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki T, Goshima G (2016) Five factors can reconstitute all three phases of microtubule polymerization dynamics. J Cell Biol 215: 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Hasebe M (2007) Microtubule-dependent microtubule nucleation in plant cells. J Plant Res 120: 73–78 [DOI] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M (2005) Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ehrhardt DW, Hashimoto T (2010) Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat Cell Biol 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Lindeboom JJ, Saltini M, Mulder BM, Ehrhardt DW (2018) SPR2 protects minus ends to promote severing and reorientation of plant cortical microtubule arrays. J Cell Biol 217: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb EH. (1969) Plant microtubules. Annu Rev Plant Physiol 20: 253–288 [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Patel K, Nogales E, Heald R (2012) Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton (Hoboken) 69: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra S, Gustavsson A, Kiefer C, Kalmbach L, Hörstedt P, Ikeda Y, Stepanova AN, Alonso JM, Grebe M (2013) Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat Commun 4: 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Wasteneys GO (2014) CLASP: a microtubule-based integrator of the hormone-mediated transitions from cell division to elongation. Curr Opin Plant Biol 22: 149–158 [DOI] [PubMed] [Google Scholar]
- Sambade A, Pratap A, Buschmann H, Morris RJ, Lloyd C (2012) The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell 24: 192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Kaloriti D (2008) Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci 13: 303–310 [DOI] [PubMed] [Google Scholar]
- Shaw SL. (2013) Reorganization of the plant cortical microtubule array. Curr Opin Plant Biol 16: 693–697 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Lucas J (2011) Intrabundle microtubule dynamics in the Arabidopsis cortical array. Cytoskeleton (Hoboken) 68: 56–67 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW (2003) Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Tindemans SH, Hawkins RJ, Mulder BM (2010) Survival of the aligned: Ordering of the plant cortical microtubule array. Phys Rev Lett 104: 058103. [DOI] [PubMed] [Google Scholar]
- Vineyard L, Elliott A, Dhingra S, Lucas JR, Shaw SL (2013) Progressive transverse microtubule array organization in hormone-induced Arabidopsis hypocotyl cells. Plant Cell 25: 662–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO. (2000) The cytoskeleton and growth polarity. Curr Opin Plant Biol 3: 503–511 [DOI] [PubMed] [Google Scholar]
- Wightman R, Turner SR (2007) Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J 52: 742–751 [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM (2005) Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol 169: 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Fishel E, Bertroche T, Dixit R (2013) Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr Biol 23: 2191–2195 [DOI] [PubMed] [Google Scholar]