Abstract
Background
There is growing interest in the use of rapid blood culture identification (BCID) in antimicrobial stewardship programs (ASPs). Although many studies have looked at its clinical and economic utility, its comparative utility in gram-positive and gram-negative blood stream infections (BSIs) has not been as well characterized.
Methods
The study was a quasi-experimental retrospective study at the Mayo Clinic in Phoenix, Arizona. All adult patients with positive blood cultures before BCID implementation (June 2015 to December 2015) and after BCID implementation (June 2016 to December 2016) were included. The outcomes of interest included time to first appropriate antibiotic escalation, time to first appropriate antibiotic de-escalation, time to organism identification, length of stay, infectious diseases consultation, discharge disposition, and in-hospital mortality.
Results
In total, 203 patients were included in this study. There was a significant difference in the time to organism identification between the pre- and post-BCID cohorts (27.1 hours vs 3.3 hours, P < .0001). BCID did not significantly reduce the time to first appropriate antimicrobial escalation or de-escalation for either gram-positive BSIs (GP-BSIs) or gram-negative BSIs (GN-BSIs). Providers were more likely to escalate antimicrobial therapy in GP-BSIs after gram stain and more likely to de-escalate therapy in GN-BSIs after susceptibilities. Although there were no significant differences in changes in antimicrobial therapy for organism identification by BCID vs traditional methods, more than one-quarter of providers (28.1%) made changes after organism identification. There were no differences in hospital length of stay or in-hospital mortality comparing pre- vs post-BCID.
Conclusions
Although BCID significantly reduced the time to identification for both GP-BSIs and GN-BSIs, BCID did not reduce the time to first appropriate antimicrobial escalation and de-escalation.
Keywords: antimicrobial stewardship, gram-negative bacteremia, gram-positive bacteremia, rapid blood culture identification
Blood stream infections (BSIs) are life-threatening events that require effective treatment for optimal outcomes. Often, patients are on multiple antimicrobial drugs until the offending pathogen is identified. Thus, shortening the time to identification (ID) and antimicrobial susceptibility testing (AST) is essential to reducing unnecessary exposure to antimicrobial drugs [1]. Rapid blood culture identification (BCID) panels provide an opportunity to improve use of antimicrobial drugs and improve patient outcomes. These BCID panels are also a new tool for effective implementation of antimicrobial stewardship programs (ASPs) with active surveillance and proactive intervention by an infectious diseases specialist or pharmacist.
Observational studies have shown that rapid organism identification in BSI is associated with a decrease in mortality, length of stay (LOS), and cost [2–10]. These benefits are largely derived from appropriate antimicrobial escalation, timely antimicrobial de-escalation, and utilization of narrow-spectrum antimicrobials resulting in shorter lengths of stay, less treatment of contaminant blood cultures, and reduced cost of antibiotics. A large single-center prospective study confirmed the utility of rapid polymerase chain reaction–based BCID panels in reducing unnecessary antibiotic use, particularly in conjunction with ASP [11].
Currently, many BCID panels utilize gene testing to determine antimicrobial resistance in certain gram-positive and gram-negative organisms, such as the mecA, vanA, and vanB genes for methicillin and vancomycin resistance in staphylococci and enterococci, and the blaKPC gene for carbapenem resistance in Enterobacteriaceae [12]. The purpose of laboratory testing is to provide actionable information in a rapid manner to improve patient care. The goal of the present study was to evaluate the clinical utility of BCID panels in gram-positive vs gram-negative BSI, with special attention to the timing and composition of antimicrobial therapy.
METHODS
Study Design
This study was a quasi-experimental retrospective study at a medium-sized (268–inpatient bed) academic tertiary referral hospital with a high volume of solid and bone marrow transplant patients. All adult (>18 years of age) patients with positive blood cultures before BCID implementation (June 2015 to December 2015) and after BCID implementation (June 2016 to December 2016) were included. Exclusion criteria included nonbacterial BSI, contaminants, mixed gram-positive/gram-negative BSI, expiration or hospice enrollment before hospitalization, history of a previous positive blood culture with the same organism (ie, species) within 90 days, and any patients who were not admitted to our institution or had positive blood cultures obtained as an outpatient.
ASP interventions before availability of BCID included prospective review and feedback regarding restricted and targeted antimicrobials, intravenous to oral switch, and dose optimization of certain antimicrobials, which were performed during business hours. When the laboratory deployed the BCID test, there was concurrent implementation of an intervention related to the BCID result. BCID results were e-mailed to members of the ASP group to verify that the patient was on effective therapy and to make recommendations if there were opportunities to de-escalate therapy. Positive BCID results were generally reviewed on the same day or the next business day if after hours or on weekends. Escalation in antimicrobial therapy was defined as the addition of an additional antibiotic or substitution of an antibiotic with broader coverage. De-escalation was defined as the removal of an antibiotic or substitution of an antibiotic with narrower coverage. Changes that were neither escalation nor de-escalation were substitutions of antibiotics to cover completely different organisms. Notably, the BCID result e-mail and review by the ASP group were implemented for routine care and not specifically for this study. The study was approved by the Mayo Clinic Institutional Review Board.
Blood Culture Methods
The routine identification is an overnight subculture, followed by identification by mass spectrometry (Bruker Daltonics, Billerica, MA) and phenotypic susceptibilities by broth dilution using the Phoenix instrument (BD, Franklin Lakes, NJ). The lab does not have rapid phenotypic susceptibility methods in place. For rapid blood culture ID, our institution utilized the FilmArray BCID (BioFire Diagnostics, LLC, Salt Lake City, UT), which was performed on all positive blood cultures beginning on June 1, 2016. The BCID can identify Staphylococcus spp., Staphylococcus aureus, Streptococcus spp., Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus pneumoniae, Enterococcus spp., Listeria monocytogenes, Enterobacteriaceae, Escherichia coli, Enterobacter cloacae complex, Klebsiella oxytoca, Klebsiella pneumoniae, Serratia spp., Proteus spp., Acinetobacter baumannii, Haemophilus influenzae, Neisseria meningitidis, Pseudomonas aeruginosa, Candida albicans, Candida glabrata, Candida krusei, Candida parapsilosis, and Candida tropicalis and 4 antibiotic resistance genes, mecA, vanA, and vanB (vanA/B), and blaKPC. Blood culture gram stain results were immediately phoned to the patient’s nurse, both before and after BCID implementation. The time between gram stain and BCID result was about 2 hours. The BCID results were entered into the medical record, but there was no active notification of the patient’s caregiver team. In addition, the BCID results were e-mailed to the ASP team, which reviewed the result in <8 hours during working hours, <16 hours for after hours weekdays, and Monday morning for results reported over the weekend.
Outcome
The outcomes of interest included (1) time to first appropriate antibiotic escalation (initiation of broader-spectrum antibiotics), (2) time to first appropriate antibiotic de-escalation (alteration to narrow-spectrum antibiotics), (3) time to organism identification (the time of organism identification either by conventional testing or BCID from the time of culture positivity), (4) LOS, (5) infectious diseases consultation, and (6) therapy adjustments following each stage of BSI investigation (gram stain, ID, AST).
Statistical Analysis
Continuous data are presented as medians with interquartile ranges unless otherwise specified. Categorical data are presented as frequencies and percentages. Data were assessed using graphical and descriptive analysis to evaluate the distributions and assess for outliers. The Wilcoxon rank-sum test was used for continuous variables. The Fisher exact test or chi-square test was used for categorical variables. Results were considered statistically significant at a (2-tailed) P value of <.05. All statistical analyses were completed using SAS Studio 3.7 (SAS Institute Inc., Cary, NC).
RESULTS
Patient Characteristics and Comorbidities
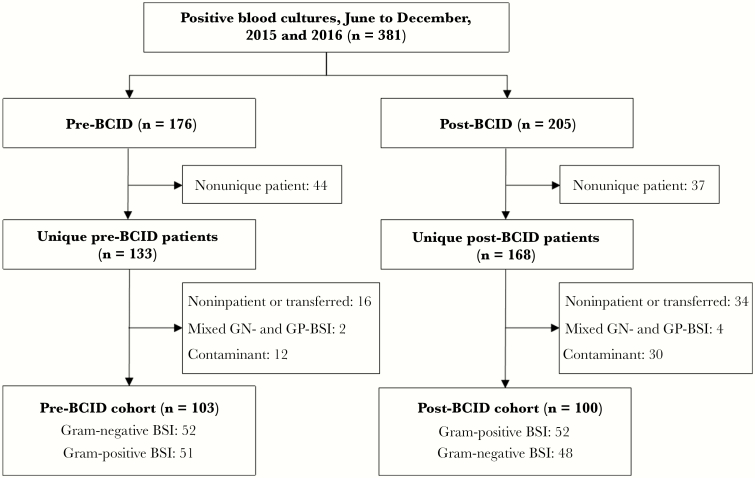
There were 381 positive blood cultures identified in the 2 aforementioned time periods, pre- and post-BCID (118 and 137 positive blood cultures, respectively). After exclusion of duplicates, contaminants, and noninpatient encounters, 203 patients (mean age, 65.0 ± 15.2 years; 35.0% female) were included. The study CONSORT diagram showing the derivation of the study cohort is shown in Figure 1. For most demographic and medical comorbidity variables, there were no significant differences between gram-negative and gram-positive BSIs. The data are summarized in Table 1.
Figure 1.
Study CONSORT diagram. CONSORT diagram for the derivation of the final cohort. Abbreviations: BCID, rapid blood culture identification panel; BSI, blood stream infection; GN-BSI, gram-negative blood stream infection; GP-BSI, gram-positive blood stream infection.
Table 1.
Patient Baseline and BSI Characteristics
| Gram-Negative | Gram-Positive | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pre-BCID (n = 52) | Post-BCID (n = 52) | P Valuea | Total (n = 104) | Pre-BCID (n = 51) | Post-BCID (n = 48) | P Valueb | Total (n = 99) | P Valuec | |
| Patient characteristics | |||||||||
| Age, y | 68.1 (60.8–77.9) | 67.4 (54.4–75.0) | .2762d | 67.7 (58.4–76.7) | 60.5 (48.2–72.1) | 66 (55.4–77.6) | .1226d | 64.4 (54.4–75.7) | .2039d |
| Female sex, No. (%) | 18 (34.6) | 25 (48.1) | .1634e | 43 (41.3) | 12 (23.5) | 16 (33.3) | .2790e | 28 (28.3) | .0511e |
| Race, No. (%) | .9024f | .6483f | .4132f | ||||||
| American Indian/Alaskan Native | 1 (1.9) | 1 (1.9) | 2 (1.9) | 2 (3.9) | 1 (2.1) | 3 (3.0) | |||
| Asian | 1 (1.9) | 2 (3.8) | 3 (2.9) | 1 (2.0) | 0 (0.0) | 1 (1.0) | |||
| Black/African/African American | 1 (1.9) | 1 (1.9) | 2 (1.9) | 4 (7.8) | 2 (4.2) | 6 (6.1) | |||
| Native Hawaiian/Pacific Island | 0 (0.0) | 1 (1.9) | 1 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
| Other | 2 (3.8) | 0 (0.0) | 2 (1.9) | 1 (2.0) | 3 (6.3) | 4 (4.0) | |||
| Unknown | 0 (0.0) | 1 (1.9) | 1 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
| White | 47 (90.4) | 46 (88.5) | 93 (89.4) | 43 (84.3) | 42 (87.5) | 85 (85.9) | |||
| CCI original | 7 (6–11) | 5 (3–8) | .0038d | 6.5 (4–10) | 6 (3–8) | 7 (4–9) | .1701d | 6 (3–8) | .4306d |
| CCI updated | 6 (4–10) | 4 (2- 6.5) | .0019d | 5 (3–9) | 4 (2–7) | 5.5 (3–7) | .2660d | 4 (3–7) | .3922d |
| Uncomplicated DM, No. (%) | 7 (13.5) | 6 (11.5) | .7668e | 13 (12.5) | 5 (9.8) | 6 (12.5) | .6697e | 11 (11.1) | .7593e |
| Complicated DM, No. (%) | 10 (19.2) | 13 (25.0) | .4784e | 23 (22.1) | 12 (23.5) | 17 (35.4) | .1940e | 29 (29.3) | .2416e |
| Mild liver disease, No. (%) | 22 (42.3) | 15 (28.8) | .1516e | 37 (35.6) | 13 (25.5) | 8 (16.7) | .2832e | 21 (21.2) | .0235e |
| Severe liver disease, No. (%) | 12 (23.1) | 5 (9.6) | .0634e | 17 (16.3) | 5 (9.8) | 13 (27.1) | .0259e | 18 (18.2) | .7293e |
| Malignancy, No. (%) | 34 (65.4) | 24 (46.2) | .0483e | 58 (55.8) | 23 (45.1) | 24 (50.0) | .6255e | 47 (47.5) | .2372e |
| Metastatic malignancy, No. (%) | 14 (26.9) | 8 (15.4) | .1497e | 22 (21.2) | 6 (11.8) | 8 (16.7) | .4842e | 14 (14.1) | .1910e |
| HIV/AIDS, No. (%) | 0 (0) | 0 (0) | - | 0 (0) | 0 (0) | 0 (0) | - | 0 (0) | - |
| CKD stage 3+, No. (%) | 31 (59.6) | 30 (57.7) | .8422e | 61 (58.7) | 27 (52.9) | 27 (56.3) | .7411e | 54 (54.5) | .5549e |
| Congestive heart failure, No. (%) | 21 (40.4) | 18 (34.6) | .5434e | 39 (37.5) | 23 (45.1) | 19 (39.6) | .5790e | 42 (42.4) | .4739e |
| Coronary artery disease, No. (%) | 17 (32.7) | 10 (19.2) | .1174e | 27 (26.0) | 14 (27.5) | 16 (33.3) | .5245e | 30 (30.3) | .4914e |
| COPD, No. (%) | 16 (30.8) | 16 (30.8) | 1.0000e | 32 (30.8) | 17 (33.3) | 14 (29.2) | .6550e | 31 (31.3) | .9333e |
| PVD, No. (%) | 28 (53.8) | 20 (38.5) | .1156e | 48 (46.2) | 21 (41.2) | 22 (45.8) | .6404e | 43 (43.4) | .6969e |
| CVA/TIA, No. (%) | 17 (32.7) | 9 (17.3) | .0700e | 26 (25.0) | 11 (21.6) | 12 (25.0) | .6862e | 23 (23.2) | .7686e |
| Dementia, No. (%) | 1 (1.9) | 1 (1.9) | 1.0000f | 2 (1.9) | 4 (7.8) | 5 (10.4) | .7359f | 9 (9.1) | .0241e |
| Hemiplegia, No. (%) | 4 (7.7) | 1 (1.9) | .3627f | 5 (4.8) | 4 (7.8) | 2 (4.2) | .6786f | 6 (6.1) | .6935e |
| Peptic ulcer disease, No. (%) | 5 (9.6) | 5 (9.6) | 1.0000e | 10 (9.6) | 7 (13.7) | 5 (10.4) | .6142e | 12 (12.1) | .5659e |
| Connective tissue disease, No. (%) | 4 (7.7) | 5 (9.6) | 1.0000f | 9 (8.7) | 4 (7.8) | 1 (2.1) | .3632f | 5 (5.1) | .3112e |
| BSI source, No. (%) | |||||||||
| CVC | 1 (1.9) | 2 (3.8) | 1.0000f | 3 (2.9) | 18 (35.3) | 8 (16.7) | .0353e | 26 (26.3) | <.0001e |
| Urinary | 21 (40.4) | 33 (63.5) | .0185e | 54 (51.9) | 2 (3.9) | 2 (4.2) | 1.0000f | 4 (4.0) | <.0001e |
| Respiratory | 2 (3.8) | 4 (7.7) | .6781f | 6 (5.8) | 4 (7.8) | 1 (2.1) | .3632f | 5 (5.1) | .8211e |
| Intra-abdominal | 16 (30.8) | 7 (13.5) | .0335e | 23 (22.1) | 3 (5.9) | 8 (16.7) | .0879e | 11 (11.1) | .0358e |
| Skin | 3 (5.8) | 1 (1.9) | .6176f | 4 (3.8) | 0 (0.0) | 11 (22.9) | .0003e | 11 (11.1) | .0479e |
| Surgical site | 1 (1.9) | 0 (0.0) | 1.0000f | 1 (1.0) | 1 (2.0) | 1 (2.1) | 1.0000f | 2 (2.0) | .6140f |
| Other | 1 (1.9) | 1 (1.9) | 1.0000f | 2 (1.9) | 10 (19.6) | 7 (14.6) | .5077e | 17 (17.2) | .0002e |
| Unknown | 7 (13.5) | 5 (9.6) | .5393e | 12 (11.5) | 13 (25.5) | 10 (20.8) | .5835e | 23 (23.2) | .0275e |
| Concurrent infections, No. (%) | |||||||||
| CVC | 1 (1.9) | 1 (1.9) | 1.0000f | 2 (1.9) | 1 (2.0) | 0 (0.0) | 1.0000f | 1 (1.0) | 1.0000f |
| Urinary | 0 (0.0) | 3 (5.8) | .2427f | 3 (2.9) | 7 (13.7) | 7 (14.6) | .9026e | 14 (14.1) | .0038e |
| Respiratory | 5 (9.6) | 2 (3.8) | .4367f | 7 (6.7) | 9 (17.6) | 5 (10.4) | .3021e | 14 (14.1) | .0831e |
| Intra-abdominal | 6 (11.5) | 2 (3.8) | .2695f | 8 (7.7) | 5 (9.8) | 1 (2.1) | .2056f | 6 (6.1) | .6465e |
| Skin | 1 (1.9) | 0 (0.0) | 1.0000f | 1 (1.0) | 1 (2.0) | 5 (10.4) | .1050f | 6 (6.1) | .0601f |
| Surgical site | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Other | 4 (7.7) | 1 (1.9) | .3627f | 5 (4.8) | 6 (11.8) | 7 (14.6) | .6781e | 13 (13.1) | .0370e |
| Required ICU, No. (%) | 18 (34.6) | 12 (23.1) | .1941e | 30 (28.8) | 17 (33.3) | 9 (18.8) | .0994e | 26 (26.3) | .6806e |
| ID consult within 72 h, No. (%) | 15 (28.8) | 11 (21.2) | .3650e | 26 (25.0) | 30 (58.8) | 30 (62.5) | .7083e | 60 (60.6) | <.0001e |
Data are presented as median (interquartile range) unless otherwise specified.
Abbreviations: CCI, Charlson Comorbidity Index; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; CVA/TIA, cerebrovascular accident/transient ischemic attack; CVC, central venous catheter; DM, diabetes mellitus; ICU, intensive care unit; ID, infectious diseases; PVD, peripheral vascular disease.
aTest between pre-BCID and post-BCID for gram-negative.
bTest between pre-BCID and post-BCID for gram-positive.
cTest between gram-negative and gram-positive.
dWilcoxon test.
eChi-square test.
fFisher exact test.
Microbiology Characteristics
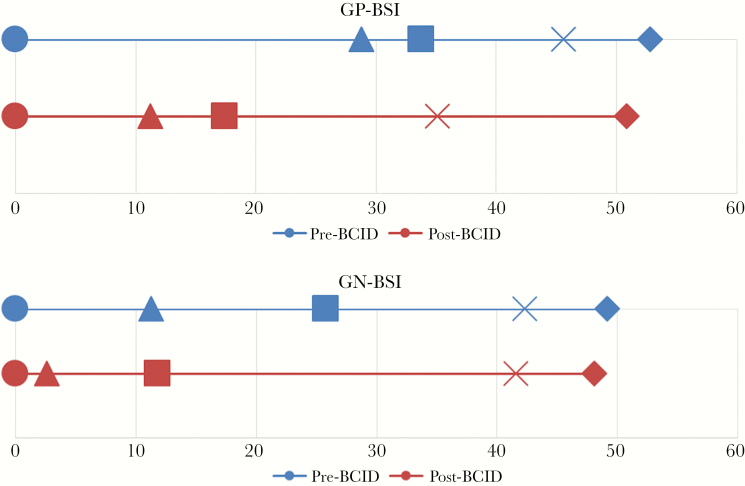
Overall, 48.4% were gram-positive BSIs, and 51.6% were gram-negative BSIs. The most frequently encountered organisms were S. aureus, S. epidermidis, E. coli, and K. pneumoniae. The distribution is shown in Table 2. The overall median time from collection to culture positivity (interquartile range [IQR]) was 12.0 (10–16) hours, the median time from positivity to identification was 21.7 (3.4–29.7) hours, and the median time from positivity to AST was 49.5 (43.5–55.3) hours. The median time to culture positivity (P = .092) and median time to susceptibility (P = .061) were not significantly different between all 4 cohorts. As expected, there was a significant difference in the time to organism identification between the pre- and post-BCID cohorts (27.1 hours vs 3.3 hours, P < .0001). Gram-negative blood stream infections (GN-BSIs) had shorter median times to identification (14.5 hours vs 25.5 hours, P < .0001) and susceptibilities (48.3 hours vs 52.4 hours, P = .0085) when compared with gram-positive blood stream infections (GP-BSI). The data are summarized in Table 3 and Figure 2.
Table 2.
Microbiology of BSIs Pre- and Post-BCID
| Pre-BCID (n = 107) |
Post-BCID (n = 106) |
|
|---|---|---|
| Blood culture pathogens | ||
| Gram-positive organisms (n = 103) | 50 (48.5) | 53 (51.5) |
| Bacillus cereus | 1 | 0 |
| Clostridium perfringens | 1 | 1 |
| Eggerthella lenta | 1 | 0 |
| Enterococcus faecalis (not VRE) | 7 | 6 |
| Enterococcus faecium (not VRE) | 0 | 5 |
| Enterococcus faecium (VRE) | 2 | 0 |
| Parvimonas micra | 1 | 0 |
| Staphylococcus aureus (MRSA) | 6 | 6 |
| Staphylococcus aureus (MSSA) | 13 | 11 |
| Staphylococcus epidermidis | 10 | 4 |
| Staphylococcus haemolyticus | 1 | 0 |
| Staphylococcus simulans | 0 | 1 |
| Staphylococcus spp. (coagulase-negative) | 0 | 2 |
| Streptococcus agalactiae | 1 | 2 |
| Streptococcus anginosus spp. | 1 | 2 |
| Streptococcus equinus | 0 | 1 |
| Streptococcus gallolyticus | 1 | 0 |
| Streptococcus group G | 0 | 1 |
| Streptococcus mitis spp. | 1 | 4 |
| Streptococcus parasanguinis | 0 | 1 |
| Streptococcus pneumoniae | 2 | 0 |
| Streptococcus pyogenes | 1 | 3 |
| Streptococcus sanguinis | 0 | 1 |
| Viridans streptococcus spp. | 0 | 2 |
| Gram-negative organisms (n = 110) | 57 (51.8) | 53 (48.2) |
| Acinetobacter baumannii | 2 | 1 |
| Bacteroides fragilis | 1 | 0 |
| Campylobacter lari/subantarcticus | 1 | 0 |
| Citrobacter freundii complex | 1 | 0 |
| Citrobacter koseri | 0 | 1 |
| Enterobacter cloacae complex | 2 | 5 |
| Escherichia coli | 22 | 20 |
| Escherichia coli (MDR) | 5 | 0 |
| Klebsiella oxytoca | 0 | 2 |
| Raoultella ornithinolytica | 1 | 0 |
| Klebsiella pneumoniae | 14 | 15 |
| Proteus mirabilis | 1 | 1 |
| Pseudomonas aeruginosa | 4 | 8 |
| Stenotrophomonas maltophilia | 2 | 0 |
| Veillonella spp. | 1 | 0 |
Abbreviations: BCID, rapid blood culture identification; BSI, blood stream infection; MDR, multidrug-resistant; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; VRE, vancomycin-resistent Enterococcus.
Table 3.
Antimicrobial Stewardship and Clinical Outcomes by Gram Stain
| Gram-Negative | Gram-Positive | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pre-BCID (n = 52) | Post-BCID (n = 52) | P Valuea | Total (n = 104) | Pre-BCID (n = 51) | Post-BCID (n = 48) | P Valueb | Total (n = 99) | P Valuec | |
| T0, h | 11 (10–12.5) | 12 (10–15.5) | .1851d | 11.8 (10–14) | 13 (10–18) | 13 (11–16) | .8249 | 13 (11–17) | .0323d |
| TTID, h | 25.8 (18.9–30.3) | 2.6 (2.3–3.1) | <.0001d | 14.5 (2.6–26.7) | 28.8 (23.1–45.7) | 17.4 (3.7–31.8) | .0002 | 25.5 (13.3–36.5) | <.0001d |
| TTS, h | 49.2 (42.6–53.1) | 48.1 (41.4–52.1) | .6134d | 48.3 (42.1–52.5) | 52.8 (46.3–66.9) | 50.9 (44.8–79.5) | .7720 | 52.4 (44.8–74.1) | .0085d |
| Time to first appropriate de-escalation, h | 42.3 (33.0–50.0) | 41.6 (34.1–50.3) | .6866d | 42.1 (33.0–50.2) | 45.6 (31.9–55.6) | 35.1 (20.2–45.8) | .0557 | 38.1 (29.2–51.3) | .4932d |
| Time to first appropriate escalation, h | 11.3 (11.3–11.3) | 11.8 (11.3–36.0) | 1.0000d | 11.6 (11.3–36.0) | 33.8 (5.3–82.0) | 11.2 (5.2–59.5) | .7015 | 20.2 (5.3–59.5) | .6501d |
| Antibiotic change, T0-TTID, No. (%) | 4 (7.7) | 1 (1.9) | .3627f | 5 (4.8) | 8 (15.7) | 10 (20.8) | .5069e | 18 (18.2) | .0027e |
| Antibiotic change, TTID-TTS, No. (%) | 11 (21.2) | 14 (26.9) | .4912e | 25 (24.0) | 15 (29.4) | 17 (35.4) | .5232e | 32 (32.3) | .1892e |
| Antibiotic change, post-TTS, No. (%) | 30 (57.7) | 30 (57.7) | 1.0000e | 60 (57.7) | 18 (35.3) | 17 (35.4) | .9898e | 35 (35.4) | .0014e |
| Any de-escalation, No. (%) | 38 (73.1) | 33 (63.5) | .2921e | 71 (68.3) | 32 (62.7) | 32 (66.7) | .6833e | 64 (64.6) | .5846e |
| Any escalation, No. (%) | 1 (1.9) | 5 (9.6) | .2050f | 6 (5.8) | 7 (13.7) | 7 (14.6) | .9026e | 14 (14.1) | .0454e |
| No antibiotic change, No. (%) | 12 (23.1) | 12 (23.1) | 1.0000e | 24 (23.1) | 14 (27.5) | 11 (22.9) | .6038e | 25 (25.3) | .7173e |
| Length of stay, d | 4 (3–8) | 4 (3–6) | .1116d | 4 (3–7) | 8 (5–17) | 6 (4.8–12.5) | .1441 | 7 (5–15) | <.0001d |
| Discharge to home, No. (%) | 39 (75.0) | 43 (82.7) | .3368e | 82 (78.8) | 34 (66.7) | 32 (66.7) | 1.0000e | 66 (66.7) | .0510e |
| Discharge to facility, No. (%) | 11 (21.2) | 7 (13.5) | .2998e | 18 (17.3) | 15 (29.4) | 13 (27.1) | .7971e | 28 (28.3) | .0619e |
| In-hospital death, No. (%) | 2 (3.8) | 3 (5.8) | 1.0000f | 5 (4.8) | 2 (3.9) | 3 (6.3) | .6716f | 5 (5.1) | 1.0000f |
Data are presented as median (interquartile range) unless otherwise specified.
Abbreviations: BCID, rapid blood culture identification panel; T0, time to positive gram stain; TTS, time to susceptibilities; TTID, time to identification.
aTest between pre-BCID and post-BCID for gram-negative.
bTest between pre-BCID and post-BCID for gram-positive.
cTest between gram-negative and gram-positive.
dWilcoxon test.
eChi-square test.
fFisher exact test.
Figure 2.
Timeline of key microbiological and clinical events from time to positivity and gram stain. The timeline of key events includes time to positive gram stain (circle), time to first escalation (triangle), time to identification (square), time to first de-escalation (x), and time to susceptibilities (diamond). Times are represented in hours. There were no significant differences in any of the time points for each key event for pre– vs post–rapid blood culture identification panel by gram stain. Abbreviations: BCID, rapid blood culture identification panel; GN-BSI, gram-negative blood stream infection; GP-BSI, gram-positive blood stream infection.
Antimicrobial Stewardship Outcomes
The most frequently used antibiotics for empiric therapies include vancomycin (62.3%), piperacillin-tazobactam (41.5%), levofloxacin (24.0%), cefepime (12.3%), and ceftriaxone (9.0%). Changes in antimicrobial therapy were assessed in 3 distinct time periods: (1) change before ID (time to gram stain to time to identification), (2) change after ID but before AST (time to identification to time to susceptibilities), and (3) change after AST (time to susceptibilities to hospital discharge). For both GN-BSIs and GP-BSIs, the median time to organism ID was significantly shorter post-BCID (2.6 hours vs 25.8 hours, P < .0001, and 17.4 hours vs 28.8 hours, P = .0002, respectively). However, BCID implementation did not lead to significant changes in antimicrobial therapy during any of the three assessed time periods. Moreover, there were no significant differences in the times to first appropriate escalation and de-escalation. However, when comparing GN-BSIs vs GP-BSIs as a whole, there were significant differences in changes before ID (4.8% vs 18.2%, P = .0027) and after AST (57.7% vs 35.4%, P = .0014). Specifically, there were significantly more escalations made after gram stain for GP-BSIs and more de-escalations made after AST for GN-BSIs. In GP-BSIs, 2 de-escalations were associated with a result of mecA-negative, and the rest were associated with a GN etiology. Overall, there were no significant differences in length of stay, patient disposition on discharge, or in-hospital mortality. The data are summarized in Table 3.
DISCUSSION
In this study, we sought to evaluate the clinical utility of BCID in antimicrobial stewardship. The main findings in our study were that (1) BCID significantly reduced the time to organism identification for both gram-positive and gram-negative BSIs as expected, (2) BCID did not significantly reduce the time to first appropriate antimicrobial escalation or de-escalation for either GP-BSIs or GN-BSIs, (3) providers were more likely to escalate antimicrobial therapy in GP-BSIs after gram stain and more likely to de-escalate therapy in GN-BSIs after susceptibilities, (4) there were no differences in LOS or in-hospital mortality comparing pre- vs post-BCID, and lastly (5) although there were no significant differences in changes in antimicrobial therapy after organism identification by BCID or gram stain status, more than one-quarter of providers (28.1%) still made changes after organism identification.
We present 1 of the first studies to analyze the effect of BCID on GN-BSI and GP-BSI separately on antimicrobial decision-making and associated outcomes. We found that there were no statistically significant differences in the time to first appropriate escalation and de-escalation among GN-BSIs and GP-BSIs after BCID implementation. However, there was a strong trend among GP-BSIs for a reduced time to de-escalation, which is important in the era of increasingly resistant organisms. In addition, there was a trend among GP-BSIs toward a reduced LOS, which is valuable not only for the patient but for the hospital and society overall with reduced use of health care services. For GN-BSIs, our study demonstrated an overall lack of statistically significant impact of BCID on decision-making for antimicrobial therapy and a lack of impact on patient outcomes. Broad-spectrum antibiotics were commonly used for empiric therapy, explaining little need for escalation. BCID had limited information regarding GN resistance mutations, which may explain why providers were reluctant to de-escalate antibiotics before AST despite ASP intervention.
Interestingly, this study has also provided additional quantitative insight on clinical practice in the treatment of BSIs. For example, in response to gram stain results, providers were more likely to escalate antibiotics in gram-positive BSIs. They were also more likely to wait to de-escalate antibiotics in gram-negative BSIs until after susceptibility results.
Based on the current evidence presented in this study, there were no significant differences in the time to first appropriate antimicrobial change. However, many providers (more than one-quarter) still changed antimicrobial therapy after organism identification, indicating that the identification of the organism likely plays a role in tailoring antimicrobial therapies. As expected, a greater proportion of providers were more likely to change antimicrobial therapy after susceptibility results.
Further study should be performed on the utility of BCID by gram stain morphology in larger samples. Since the time to susceptibilities appears to be highly important in antimicrobial stewardship, further research and development of accurate and rapid prediction of susceptibilities may provide the most impactful information sooner. One example of this type of technology, the Accelerate Pheno test, was recently cleared by the Food and Drug Administration [13].
Limitations
There were limitations to this study. First, this was a retrospective study with a modest sample size. Although we attempted to mitigate this with the method of cohort selection, we cannot fully exclude the possibility of bias. Likewise, the sample size does not allow for subgroup analysis. Second, the caregiver team was not immediately notified of the BCID result, which likely delayed their awareness of the result. Third, management of BSIs was largely left to the discretion of individual providers, with the ASP group providing recommendations. Fourth, our tertiary referral center population, generally older and with multiple significant medical comorbidities including active malignancies and solid organ and bone marrow transplantation, may not be representative of the general population. Lastly, we were unable to account for informal consultations with the hospital infectious diseases consultation service.
CONCLUSIONS
As expected, BCID significantly reduced the time to identification for both GP-BSIs and GN-BSIs. Although BCID did not significantly reduce the time to first appropriate antimicrobial escalation and de-escalation, there was a strong trend toward a clinical impact for GP-BSI but not for GN-BSI. It is possible that the implementation of both BCID and a highly active ASP may improve clinical outcomes in both GP-BSIs and GN-BSIs. Further investigation of the clinical utility of BCID for specific organisms and development of more rapid methods of susceptibility prediction are warranted.
Acknowledgments
The authors wish to thank the dedicated staff of the Mayo Clinic Arizona Microbiology Laboratory.
Financial support. This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lutgring JD, Bittencourt C, McElvania TeKippe E, et al. . Evaluation of the accelerate pheno system: results from two academic medical centers. J Clin Microbiol. 2018; 56 pii: e01672–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Perez KK, Olsen RJ, Musick WL, et al. . Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med 2013; 137:1247–54. [DOI] [PubMed] [Google Scholar]
- 3. Perez KK, Olsen RJ, Musick WL, et al. . Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant gram-negative bacteremia. J Infect 2014; 69:216–25. [DOI] [PubMed] [Google Scholar]
- 4. Huang AM, Newton D, Kunapuli A, et al. . Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 2013; 57:1237–45. [DOI] [PubMed] [Google Scholar]
- 5. Forrest GN, Roghmann MC, Toombs LS, et al. . Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother 2008; 52:3558–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen DT, Yeh E, Perry S, et al. . Real-time PCR testing for mecA reduces vancomycin usage and length of hospitalization for patients infected with methicillin-sensitive staphylococci. J Clin Microbiol 2010; 48:785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clerc O, Prod’hom G, Vogne C, et al. . Impact of matrix-assisted laser desorption ionization time-of-flight mass spectrometry on the clinical management of patients with gram-negative bacteremia: a prospective observational study. Clin Infect Dis 2013; 56:1101–7. [DOI] [PubMed] [Google Scholar]
- 8. Galar A, Leiva J, Espinosa M, et al. . Clinical and economic evaluation of the impact of rapid microbiological diagnostic testing. J Infect 2012; 65:302–9. [DOI] [PubMed] [Google Scholar]
- 9. Veesenmeyer AF, Olson JA, Hersh AL, et al. . A retrospective study of the impact of rapid diagnostic testing on time to pathogen identification and antibiotic use for children with positive blood cultures. Infect Dis Ther 2016; 5:555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pliakos EE, Andreatos N, Shehadeh F, et al. . The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev. 2018 May 30;31. pii:e00095-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banerjee R, Teng CB, Cunningham SA, et al. . Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shanmugam P, Meenakshisundaram J, Jayaraman P. blaKPC gene detection in clinical isolates of carbapenem resistant Enterobacteriaceae in a tertiary care hospital. J Clin Diagn Res 2013; 7:2736–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pancholi P, Carroll KC, Buchan BW, et al. . Multicenter evaluation of the accelerate phenotest BC kit for rapid identification and phenotypic antimicrobial susceptibility testing using morphokinetic cellular analysis. J Clin Microbiol. 2018; 56 pii: e01329–17. [DOI] [PMC free article] [PubMed] [Google Scholar]