Abstract
Up to half of all patients do not respond to pharmacological treatment as intended. A substantial fraction of these inter-individual differences is due to heritable factors and a growing number of associations between genetic variations and drug response phenotypes have been identified. Importantly, the rapid progress in Next Generation Sequencing technologies in recent years unveiled the true complexity of the genetic landscape in pharmacogenes with tens of thousands of rare genetic variants. As each individual was found to harbor numerous such rare variants they are anticipated to be important contributors to the genetically encoded inter-individual variability in drug effects. The fundamental challenge however is their functional interpretation due to the sheer scale of the problem that renders systematic experimental characterization of these variants currently unfeasible. Here, we review concepts and important progress in the development of computational prediction methods that allow to evaluate the effect of amino acid sequence alterations in drug metabolizing enzymes and transporters. In addition, we discuss recent advances in the interpretation of functional effects of non-coding variants, such as variations in splice sites, regulatory regions and miRNA binding sites. We anticipate that these methodologies will provide a useful toolkit to facilitate the integration of the vast extent of rare genetic variability into drug response predictions in a precision medicine framework.
Keywords: precision medicine, personalized medicine, variant effect prediction, ADME, NGS, rare variant analysis, noncoding variation, pharmacogenomics
Introduction
Inter-individual differences in drug response are clinically important phenomena that result in reduced efficacy or adverse reactions in 25–50% of all patients and genetic factors have been estimated to account for around 20–30% of these (Spear et al., 2001; Sim et al., 2013). Fueled by technological advances in Next-Generation Sequencing (NGS) technologies, the application of comprehensive sequencing approaches is on the rise for various applications, including studies of biodiversity, population genetics and biomedical research (Levy and Myers, 2016). Furthermore, plummeting costs to < 1,000 USD per human genome and increasing worldwide sequencing capacities that we estimate to exceed 100 petabases per year (1015 bases corresponding to the size of around 100,000 human genomes) open tremendous possibilities for NGS to revolutionize precision medicine.
Strikingly, these massive NGS data sets revealed that individuals harbored on average more than 3.7 million single nucleotide variants (SNVs) and more than 350,000 insertions and deletions across different populations, emphasizing the substantial variability of the human genome (The 1000 Genomes Project Consortium, 2012). Particularly genes involved in drug absorption, distribution, metabolism and excretion (ADME) proved to be highly diverse and genetically complex (Fujikura et al., 2015; Bush et al., 2016; Kozyra et al., 2017). Across 208 ADME genes more than 69,000 SNVs have been described, 98.5% of these being rare with minor allele frequencies (MAF) < 1% (Ingelman-Sundberg et al., 2018). The overall pharmacogenetic variability was highly population specific, particularly for isolated populations, such as Ashkenazi Jews (Ahn and Park, 2017; Kozyra et al., 2017; Zhou and Lauschke, 2018). Given this enormous pharmacogenetic variability, one of the key frontiers of contemporary pharmacogenomics is the translation of these comprehensive genomic data into clinically actionable treatment recommendations (Lauschke and Ingelman-Sundberg, 2016a, 2018).
Heterologous expression in cell lines followed by quantitative determination of gene product functionality using appropriate end points is considered as the gold standard strategy to characterize the functional impact of pharmacogenetic variants. Furthermore, epidemiological association studies can provide additional indications about the consequences of genetic variants on drug metabolism related phenotypes in vivo. However, for the functional interpretation of rare variants these approaches suffer from multiple shortcomings:
These methods are generally low throughput and are not compatible with the interrogation of tens of thousands of variants.
Experimental characterizations are time consuming, expensive and require specially trained technical staff, which renders them unsuitable for the rapid functional interpretation of the pharmacogenotype of an individual patient at the point of care.
Epidemiological analyses require a sufficient number of patients who carry the allele, which drastically limits their feasibility for rare genetic variant studies.
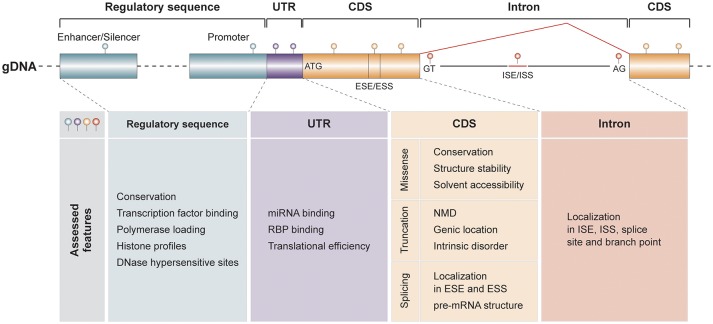
Thus, in the absence of viable experimental strategies, computational prediction methodologies are routinely used to predict the functional impact of genetic variants. Most of these algorithms focus on predicting the functional consequences of variants that result in amino acid substitutions. However, recently much progress has also been made regarding the interpretation of non-coding variants that affect splice sites, promoters, enhancers or miRNA binding sites (Figure 1).
Figure 1.
Overview of features that can be assessed by current computational prediction methods. Different parameters and features are assessed for genetic variants depending on whether they are localized in putatively regulatory sequences, untranslated regions (UTR) of the gene, its coding sequences (CDS) or within introns. ESE/ESS, exonic splicing enhancer/silencer; ISE/ISS, intronic splicing enhancer/silencer; NMD, nonsense-mediated decay; RBP, RNA binding protein.
Prediction algorithms are generally trained on pathogenic variant sets and most tools base their conclusions, at least in part, on the evolutionary conservation of the respective sequence. Importantly however, pharmacogenes are hallmarked by low evolutionary conservation and are generally not associated with human disease. These peculiarities result is specific problems for the interpretation of pharmacogenetic variants. Here, we provide an updated overview of computational approaches for the functional interpretation of genetic variants, specifically focusing on their suitability for pharmacogenetic predictions. We describe the underlying statistical frameworks and discuss their different bases for decision-making. Furthermore, we highlight important progress particularly in the interpretation of non-coding genetic variability. We conclude that computational tools are essential for the functional interpretation of an individual's pharmacogenotype and that their further improvement constitutes one of the most important frontiers for the clinical implementation of NGS-based genotyping.
Interpretation of Variants Resulting in Amino Acid Exchanges
Genetic variants that result in amino acid substitution, henceforth termed missense variants, can impact the functionality of the respective protein by various mechanisms, including alterations in active sites, structural destabilization due to protein misfolding, perturbations in solvent accessibility or modification of post-translational processing. Each individual harbors 10,000–12,000 missense variants, many of which are rare (The 1000 Genomes Project Consortium, 2015). These rare variants have been suggested as important modulators of complex disease risk (Kryukov et al., 2007) and inter-individual differences in drug response (Kozyra et al., 2017). Among all variant classes, missense variants are the most extensively studied and a plethora of computational methods is available for their functional interpretation. Conceptually, these algorithms predict the functional impact of missense variants based on sequence information, primarily evolutionary conservation of the respective residues, and/or structural information of the corresponding gene product. In the following, we highlight recent progress, provide an overview of available tools and discuss their utility for pharmacogenetic predictions. For methodological details we refer the interested reader to excellent recent reviews (Ng and Henikoff, 2006; Peterson et al., 2013; Tang and Thomas, 2016).
Predictions Based on Sequence Information
Evolutionary conservation scores are calculated by analyzing the evolutionary variation dynamics of DNA or amino acid sequences among homologs with the hypothesis that the extent of conservation is a strong predictor of the importance of the respective sequence for structure and function of the corresponding gene product. Thus, positions with a high evolutionary rate are thought to be dispensable, whereas slowly evolving, i.e., conserved sequences indicate a selective pressure against variation in these regions and thus deleterious effects if mutated.
Evolutionary conservation as a metric to distinguish deleterious from neutral variants is considered by most computational prediction algorithms. The majority of approaches that focus on the functional interpretation of missense variants utilize amino acid sequence alignment, whereas others utilize nucleotide sequence alignments or a combination of both methods (Table 1). While alignment of amino acid sequence proved to be effective for the analysis of missense variants, genomic sequence alignments provide additional versatility and allow to extend functional interpretations to variant classes that do not alter the amino acid sequence, such as synonymous and regulatory variants. Notably, commonly used conservation-based functionality predictors do not consider sequence interdependencies. Explicit integration of residue dependency information obtained from multiple sequence alignments was however recently shown to improve predictive performance (Hopf et al., 2017), emphasizing the added value of complementing conservation based functionality predictions with variant interaction data.
Table 1.
Methods to predict the functional effect of missense variants based on sequence information.
| Algorithm | Model | Basis of decision | Model training or evaluation | References |
|---|---|---|---|---|
| SIFT | Direct | Prediction of functionality based on sequence conservation metrics that make use of Dirichlet priors | Variants from protein specific studies (LacI, HIV-1 Protease and Bacteriophage T4 Lysozyme) | Ng and Henikoff, 2001 |
| PANTHER | HMM | Sequence conservation analysis using HMM | Variants from HGMD and dbSNP as deleterious and functionally neutral variants, respectively | Thomas et al., 2003 |
| MAPP | Direct | Quantification of the physicochemical characteristics at each position of the amino acid sequence based on observed evolutionary variation | Protein specific studies (LacI, HIV-1 Protease, HIV reverse transcriptase and Bacteriophage T4 Lysozyme) | Stone and Sidow, 2005 |
| PhastCons | HMM | Identification of conserved elements using a two-state phylogenetic HMM | Calibration on genomes from four model species (human, D. melanogaster, C. elegans, and S. cerevisiae) | Siepel et al., 2005 |
| SNPs3D | SVM | Variant effect prediction based on amino acid sequence conservation metrics and folded state stability of protein structure | Variants from HGMD and dbSNP as deleterious and functionally neutral variants, respectively | Yue et al., 2006 |
| PhD-SNP | SVM | Prediction of variant pathogenicity based on sequence profiles | Variants from HumVar and HumVarProf datasets | Capriotti et al., 2006 |
| SiPhy | HMM | Sequence conservation analysis using HMM | ENCODE Phase I regions | Garber et al., 2009 |
| LRT | Direct | Evolutionary conservation model across 32 vertebrates | Variants in three sequenced human genomes | Chun and Fay, 2009 |
| SNPs&GO | SVM | Variant effect prediction based on sequence information, evolutionary conservation and defined gene ontology score | Variants from SwissProt | Calabrese et al., 2009 |
| B-SIFT | Direct | Sequence conservation metrics that calculate the difference between wild-type and mutant allele | Variants from SwissProt database and protein specific study (Dnase I) | Lee et al., 2009 |
| PolyPhen-2 | NB | Considering sequence conservation, Structure parameters such as hydrophobic propensity and B factor | Variants fromn HumDiv and HumVar from UniProt Database | Adzhubei et al., 2010 |
| MutationTaster | NB | Prediction of mutation pathogenicity based on evolutionary conservation, splice-site changes, loss of protein features and changes that affect expression levels | Variants from OMIM database, HGMD and the literature as pathogenic set and neutral variants from dbSNP as controls | Schwarz et al., 2014 |
| MutationAssessor | Direct | Evolutionary conservation patterns within protein families and across species using combinatorial entropy | Variants from UniProt database (HumSaVar) | Reva et al., 2011 |
| Condel | Direct | Integration of five algorithms (SIFT, PolyPhen-2 MAPP, MutationAssessor, and Log R Pfam E-value) into single output score | Variants from HumVar, HumDiv, Cosmic database, IARC TP53 database | González-Pérez and López-Bigas, 2011 |
| PROVEAN | Direct | Alignment-based score that can also assess in-frame insertions, deletions, and multiple amino acid substitutions | Missense variants and indels, replacements from UniProt database | Choi et al., 2012 |
| FATHMM | HMM | Identification of pathogenic variants based on sequence conservation, protein domain-based information and species-specific pathogenicity weights. Also suitable for prediction of non-coding variations. | Variants from the HGMD and Uniprot databases | Shihab et al., 2013, 2015; Rogers et al., 2018 |
| VEST | RF | Prioritization of variants underlying Mendelian diseases | Rare variants from HGMD database as pathogenic set and variants from ESP | Carter et al., 2013 |
| Evolutionary Action | Direct | Prediction of variant effects on evolutionary fitness using a formal genotype-phenotype perturbation equation | Variants from 1000 Genomes Project | Katsonis and Lichtarge, 2014 |
| MetaSVM | SVM | Ensemble score integrating nine functionality predictors (SIFT, PolyPhen-2, GERP++, MutationTaster, MutationAssessor, FATHMM, LRT, SiPhy and PhyloP) | Variants causing Mendelian diseases as pathogenic set and variants that are not associated with any phenotypes as controls, all from Uniprot database | Dong et al., 2015 |
| MetaLR | RM | Same as MetaSVM but using logistic regression instead of SVM. | Dong et al., 2015 | |
| SuSPect | SVM | Sequence conservation metrics, structure features and additional network information | Variants from Humsavar database | Yates et al., 2014 |
| PredictSNP | EL | Ensemble score integrating six functionality predictors (MAPP, PhD-SNP, PolyPhen-1, PolyPhen-2, SIFT and SNAP) | Variants mainly from SwissProt, HGMD, dbSNP and Humsavar database | Bendl et al., 2014 |
| SNAP2 | NN | Prediction of amino acid variations based on amino acid properties, predicted binding residues, predicted disordered and low-complexity regions, proximity to N- and C-terminus, statistical contact potentials, co-evolving positions, secondary structure and solvent accessibility | Variants from PMD, Swiss-Prot, OMIM, HumVar and protein specific data sets (LacI) | Hecht et al., 2015 |
| REVEL | RF | Ensemble method tailored specifically for the prediction of rare genetic variant effects integrating MutPred, FATHMM, VEST, PolyPhen, SIFT, PROVEAN, MutationAssessor, MutationTaster, LRT, GERP, SiPhy, phyloP, and phastCons | Variants from HGMD as pathogenic set and neutral variants from ESP as controls | Ioannidis et al., 2016 |
| ConSurf | Empirical Bayesian method and maximum likelihood estimation | Mapping of evolutionarily conserved residues on protein surfaces by estimating the evolutionary rates of each nucleic acid and amino acid sequence position using multiple sequence alignments. Also offers RNA secondary structure predictions. | Protein with at least five known 3D structure homologs and precise annotation of their functional sites (with different nature) | Ashkenazy et al., 2016 |
| VIPUR | RM | Combination of sequence- and structure-based features to identify and functionally interpret deleterious variants | Variants from HumDiv and UniProt with clear evidence of protein disruption | Baugh et al., 2016 |
| Envision | GTB | Decision tree ensemble-based tool using a stochastic gradient boosting learning algorithm | Variants from nine large-scale experimental mutagenesis datasets in eight proteins | Gray et al., 2018 |
| EVmutation | Direct | Unsupervised method exploiting sequence conservation by incorporating interaction information between all pairs of residues in protein | 34 data sets from 21 proteins and a tRNA gene extracted from 27 publications | Hopf et al., 2017 |
| PredSAV | GTB | Identification of pathogenic variants based on sequence, structure, residue-contact networks as well as structural neighborhood features | Human variants from Uniprot and OMIM as pathogenic set and Ensemble variants as neutral controls | Pan et al., 2017 |
| SNPMuSiC | NN | Structure stability based, implement PoPMuSiC and HoTMuSiC on the basis of 13 statistical potentials (distence potentials, solvent accessibility potentials and torsion potentials) and 2 biophysical characteristics (solvent accessibility of mutated residue and difference in volume) | Variants from dbSNP, SwissVar and HumSaVar datasets | Ancien et al., 2018 |
| DEOGEN2 | RF | Integration of 11 scores and metrices into one meta-score, considering evolutionary features, folding predictions, domain information as well as gene features to identify deleterious variants | Training and test on variants from the UniProt Humsavar16 dataset | Raimondi et al., 2017 |
| ADME prediction framework | Direct | Integration of prediction scores from five orthogonal algorithms (LRT, MutationAssessor, PROVEAN, VEST3 and CADD) using parameters optimized for pharmacogenes | Training and validation specifically on experimentally characterized pharmacogenetic data sets from 43 ADME genes | Zhou et al., 2018 |
HMM, hidden Markov model; SVM, support vector machine; NB, naïve Bayes classifier; EL, ensemble learning; RF, random forest; RM, regression model; NN, neural networks; GTB, gradient tree boosting; HGMD, Human Gene Mutation Database; OMIM, Online Mendelian Inheritance in Man; ESP, Exome Sequencing Project; PMD, Protein Mutant Database.
On the basis of multiple sequence alignments, algorithms derive their functionality predictions either based on direct theoretical models, or by various machine-learning approaches. The former methods predict the functional impact of variants based on phenomenological scores derived from theoretical models that are known a priori. In contrast, machine learning methods search for patterns in multi-dimensional training data sets consisting of labeled deleterious and benign variations, which will then be used as the basis to generate predictions on new unlabeled data. Machine learning approaches include support vector machines, random forests, artificial neural networks, naive Bayes approaches, gradient tree boosting and regression models. With increasing wealth of large-scale data sets to learn from, machine learning methods become increasingly popular as versatile tools to generate predictive models in many areas of biomedicine (Camacho et al., 2018).
Commonly used algorithms are generally designed to flag deleterious variants, which are mostly assumed to result in a reduced gene product function, and their performance of gain-of-function variants is substantially worse (Flanagan et al., 2010). Notably, the algorithm B-SIFT, a modified version of the widely used SIFT tool (Ng and Henikoff, 2001), was developed to overcome this limitation (Lee et al., 2009). Conceptually, B-SIFT identifies increased functionality variants based on protein sequence alignments by scoring whether a given mutation results in a change commonly present in protein homologs and the tool successfully identified experimentally validated gain-of-function variants in cancer.
While computational missense variant predictors are generally reported to achieve high predictive accuracies with areas under the receiver operating characteristic curve (AUCROC) that often pivot around 0.9, drastic drops in performance to AUCROC of 0.5–0.75 have been reported on independent, functionally determined human variant datasets (Mahmood et al., 2017). These findings were corroborated by a recent cross-comparison of 23 methods based on three independent pathogenicity datasets in which the authors found that REVEL and VEST3 performed overall best, whereas the most commonly used methods SIFT and PolyPhen-2 performed only medially (Li et al., 2018). Furthermore, no functional consequences could be detected using various in vitro or in vivo tools for 40% of variants predicted to be deleterious by common functionality prediction tools (Miosge et al., 2015). Thus, while current tools have proven powerful in clinical diagnostics to prioritize potentially causative mutations in genetic diseases for further analyses (Boycott et al., 2013), their predictive power is not yet sufficient to predict functional variant effects without substantial subsequent validations.
Importantly, the quality of prediction models critically relies on accurate training data sets. For instance, models are commonly generated using training sets of pathogenic variants as positive controls and polymorphisms identified to be common in large-scale sequencing projects as negative, i.e., functionally neutral variants. For pharmacogenetic predictions such a strategy is associated with multiple problems: Firstly, training on disease-associated data sets will, in the best case, result in prediction models that accurately predict the pathogenicity of variants. However, only very few ADME genes are directly associated with disease, suggesting that pathogenicity is not the right endpoint to inform about variant effects in the pharmacogenetic arena. Secondly, while evolutionary conservation constitutes a useful metric to predict functional consequences in genes under purifying selection, evolutionary conservation in pharmacogenes is generally much lower (Fujikura, 2016), indicating that conservation cannot reliably inform about functional impacts of variations in pharmacogenes. Finally, the choice of common polymorphisms as neutral training sets is problematic. Genetic variants that occur with high frequencies are not necessarily functionally neutral, particularly in pharmacogenetic loci, as evidenced by a multitude of high-frequency loss of function variants in CYP genes, such as CYP3A5*3 (MAF = 95% in Europeans), CYP2C19*2 (MAF = 34% in South Asians) and CYP2D6*4 (MAF = 16% in Latinos) (Zhou et al., 2017).
The indicated problems incentivized us to develop a prediction framework tailored specifically toward pharmacogenetic functionality assessment (Zhou et al., 2018). Specifically, the model was devised using a two-step procedure: Firstly, functionality classification threshold of 18 commonly used functional prediction algorithms were optimized by leveraging a dataset of 337 experimentally characterized pharmacogenetic variants using 5-fold cross validations. In a second step, we integrated the best performing orthogonal algorithms following a strategy that had been shown to further improve predictive accuracy (Martelotto et al., 2014). The resulting method achieved 93% for both sensitivity and specificity for both loss-of-function and functionally neutral variants. Moreover, the returned score can provide quantitative estimates of the effect of the variant in question on gene function, thus facilitating the functional and personalized interpretation of an individual's NGS-based pharmacogenome.
Recent progress in large-scale experimental mutagenesis screens provides a promising approach to further expand the development of powerful training resources for missense variant effect predictors. While such a strategy has already been used to develop a prediction method based on 10 proteins from different species with disparate structures (Gray et al., 2018), we propose that deep mutational scanning data from ADME proteins is likely to substantially refine the resulting model for pharmacogenetic predictions. For such an endeavor, we recommend to use multiple substrates for each protein, as correlations between prediction and experiments improved with more comprehensive interrogation of protein function (Gallion et al., 2017). Combined with ADME-optimized prediction models, we envision that such an approach can further enhance the predictive accuracy of in silico methods and yield sufficiently accurate tools to allow for the clinical implementation of computational pharmacogenetic predictions.
Utilization of Structural Data
While evolutionary conservation scores can provide useful metrics to assess the pathogenicity of missense variants, they have limitations when applied to the less conserved genes, such as most ADME genes, which prompted the search for additional orthogonal in silico methods. To this end, the analysis of predicted or experimental structural data provides an appealing concept, as the correct folding of polypeptide chains into three-dimensional tertiary structures is of paramount importance for their biological functions. Structure-based approaches either directly use known crystal or NMR structures, preferably at high resolution < 2–3 Å (Wlodawer et al., 2008) or, should such data not be available, leverage knowledge of the experimental 3D structures of homologous sequences (Table 2).
Table 2.
Methods to predict the functional effect of missense variants based primarily on structural features.
| Algorithm | Model | Basis of decision | Model training or evaluation | References |
|---|---|---|---|---|
| SDM | Direct | Predicts variant effects on thermal protein stability using conformationally constrained environment-specific substitution tables derived from 2,054 protein family sequence and structure alignments from the TOCCATA database | Validated on 2,690 SNVs from 132 different protein structures. | Topham et al., 1997; Pandurangan et al., 2017 |
| I-Mutant | SVM | Protein structure or sequence-based prediction of point mutation effects on protein stability | Training and testing on thermodynamic experimental data of free energy changes of protein stability upon mutation from the ProTherm database | Capriotti et al., 2005 |
| HOPE | Direct | Analyzes the structural and functional effects of point mutations based on available crystal structures, homology modeling and sequence information. | Evaluated using case studies. | Venselaar et al., 2010 |
| mCSM | RM | Translation of distance patterns between atoms into graph-based signatures providing data that is complementary to potential energy based approaches | Prediction of protein stability changes, protein-protein and protein-nucleic acid interactions and pathogenicity based on an array of preexisting experimental data sets | Pires et al., 2014b |
| DUET | SVM | SVM predictor that integrates mCSM and SDM in a consensus prediction | Benchmarking again mCSM and SDM alone on p53 data set. | Pires et al., 2014a |
| STRUM | GTB | Predicts variant effects on protein stability based on 3D models constructed by iterative threading assembly refinement simulations | Evaluated on 3,421 experimentally determined mutations distributed across 150 proteins. | Quan et al., 2016 |
| ELASPIC | GTB | Predicts effects of mutations on protein folding and protein–protein interactions using homology modeling of domains and domain–domain interactions | Performance analysis via case study using EP300 mutations found in COSMIC | Witvliet et al., 2016 |
| SAAFEC | RM | Prediction of effects of amino acid changes on folding free energy using a Molecular Mechanics Poisson-Boltzmann approach | Training and testing on thermodynamic experimental data of free energy changes of protein stability upon mutation from the ProTherm database | Getov et al., 2016 |
SVM, support vector machine; RM, regression model; GTB, gradient tree boosting.
The effect of variants is predicted by how the folding free energy difference between the unfolded and folded states (ΔG°) is modified upon point mutations (ΔΔG°) with negative and positive values of ΔΔG° indicating destabilizing and stabilizing mutations, respectively. In recent years a large number of mechanistically diverse approaches have been presented, with machine learning-based strategies being most prevalent. SDM constitutes a statistical potential energy function that can estimate variant effects on protein stability (Topham et al., 1997). This approach pioneered the knowledge-based prediction of mutation effects on protein stability and has also been successfully used in combination with machine learning techniques (Pires et al., 2014a). An updated version of the tool, SDM2 (Pandurangan et al., 2017), with a 5-fold increase in underlying structural information as well as extensions for interaction modeling can be accessed through a free, publically available web server interface. Similarly, the algorithm HOPE (Venselaar et al., 2010) can calculate structural and functional effects of amino acid exchanges based on homology modeling. It should be however noted that most of the current tools are strongly biased toward the detection of destabilizing effects (Pucci et al., 2018).
Approximately 70% of the human proteome can be structurally modeled by homology (Somody et al., 2017). Yet, the number of resolved 3D structures for genes involved in drug ADME remains relatively low, at least in part due to the membrane bound nature of many of these proteins. Furthermore, as many metabolic enzymes, such as cytochrome p450s (CYPs) exhibit marked active-site flexibility, which often results in ligand-induced conformational changes, prediction of variant effects based on direct structural data is difficult for these proteins and substrate-specific effects have to be considered. Thus, while the prediction of amino acid exchanges on substrate metabolism remain difficult, folding stability of variant proteins of interest can be estimated using existing computational tools based on sequence homology modeling (Kulshreshtha et al., 2016).
Evaluation of Truncation Variants
Drug metabolizing enzymes and transporters have been found to harbor a multitude of truncation variants, such as micro-insertions and micro-deletions (indels) causing frameshifts, stop-gain and start-lost variants. Some of these variants are clinically relevant and occur with high frequencies in specific populations, including the stop-gain variant CYP2C19*3 in East Asians and the frameshift variants CYP2D6*3 and CYP2D6*6 in Europeans (Zhou et al., 2017). As most pharmacogenes have only minor endogenous functions, they are under low evolutionary pressure and, consequently, such loss-of-function variants are often not selected against (Lauschke et al., 2017). Moreover, it has been speculated that pharmacogenetic loss-of-function alleles can even be selected for in modern humans, possibly due to reduced bioactivation of dietary toxicants (Fujikura, 2016). Truncation variants are commonly assumed to have deleterious effects and only few studies have been presented that provide approaches to quantitatively assess the functional consequences of such mutations (Cline and Karchin, 2011).
Early bioinformatic tools, such as LOFTEE, prioritize truncation variants based on a set of empirical rules, including whether the variant of interest occurs in the last 5% of transcript or whether the truncating allele is the ancestral state (MacArthur et al., 2012). Other approaches, such as Likelihood-ratio scoring (Zia and Moses, 2011), SIFT Indel (Hu and Ng, 2012) and NutVar (Rausell et al., 2014), primarily utilize the evolutionary conservation of amino acid residues. However, predictive performance of these tools for loss-of-function mutations is limited when trained on only missense mutations. Moreover, these methods are trained on genes that have high-quality annotations, which poses problems for the functional interpretation of truncation variants in genes for which such annotations are not readily available.
To overcome these shortcomings, CADD was developed by integrating many diverse functional genomics annotations into a single score for each variant, which allows to estimate the impact of all classes of genetic variation, including truncating variants (Kircher et al., 2014). Newer approaches, such as DDIG-in (Folkman et al., 2015) and VEST-Indel (Douville et al., 2016) supplement conservation-based features with information about sequence and structural properties at nucleotide and protein levels as well as intrinsic disorder predictions from the region affected by stop gain and frameshift variants. Notably, the recently developed tool ALoFT (Annotation of Loss-of-Function Transcripts) can categorize the pathogenic importance of putative loss-of-function mutations by integrating variant information with redundancy and haplosufficiency data of the corresponding gene (Balasubramanian et al., 2017). However, aforementioned methods are primarily focused on distinguishing benign and disease-causing mutations. Thus, future studies are needed to evaluate whether this emphasis on the pathogenicity of variants might affect the performance of these methods regarding the functionality prediction of truncating variants in genes not associated with disease, such as many ADME genes.
In addition to impacts on functional and structural properties of proteins, truncating variants can affect nonsense-mediated mRNA decay (NMD). NMD is a conserved translation-dependent mechanism that is responsible for recognizing and eliminating aberrant mRNA transcripts to prevent the production of truncated peptides, thereby playing a critical role in preventing the accumulation of misfolded protein and subsequent initiation of the unfolded protein response (UPR) (Kervestin and Jacobson, 2012; Schoenberg and Maquat, 2012). Recently, Hsu et al. presented NMD Classifier, a tool for the systematic classification of NMD events, which was reported to correctly identify 99.3% of the NMD-causing transcript structural changes (Hsu et al., 2017). The incorporation of this information alongside functional estimates is expected to not only increase discriminative power but also to suggest the nature of the functional impact of a given variant. Interestingly, there is evidence that NMD efficiency varies between individuals and that these differences correlate with response to NMD inhibitors in cystic fibrosis patients (Linde et al., 2007; Kerem et al., 2008). While this phenomenon has to the best of our knowledge not been explicitly tested in the context of pharmacogenomics, inter-individual differences in NMD magnitude could, at least in part, explain the large differences in drug response between patients with loss-of-function genotypes (Jukić et al., 2018) and thus have important implications for therapy.
In summary, much progress has been made regarding the functional interpretation of variants causing truncations of the corresponding gene product and current computational tools are able to incorporate a variety of features into their predictions, including evolutionary conservation, sequence and structural information as well as putative effects on NMD. However, it remains to be demonstrated whether these available tools will also be suitable for the prediction of effects of truncation variants in poorly conserved pharmacogenetic loci.
Prediction of Aberrant Splicing Events
Splicing of pre-mRNA is a critical step during mRNA maturation in which introns are excised and exons are ligated. This process necessitates the presence of 5′ and 3′ splicing signals and branch point sequence and is further regulated by exonic and intronic splicing enhancer/silencer (ESE/ESS and ISE/ISS, respectively) (Lee and Rio, 2015; Shi, 2017). Mutations in these regions can disrupt the splicing process and result in aberrantly processed transcripts, which can trigger NMD or result in the production of dysfunctional proteins. The functional importance of genetic variants in splice sites is emphasized by estimates that around 15% of human pathogenic mutations cause dysregulation of splicing (Baralle et al., 2009).
Variants located in canonical splice sites are considered having the largest effect on splicing events. Therefore, a multitude of computational algorithms were developed to handle the prediction of 5′ and 3′ splice site, such as NNSplice (Reese et al., 1997), MaxEntScan (Yeo and Burge, 2004), GeneSplicer (Pertea et al., 2001), and SplicePort (Dogan et al., 2007; Table 3). Moreover, variants outside splice sites can have substantial effects on splicing (Soukarieh et al., 2016) and a variety of computational methods have been developed to predict the effect of such regulatory sequences. Examples are sequence the conservation-based algorithm Skippy (Woolfe et al., 2010) and the machine learning tools MutPred Splice (Mort et al., 2014), scSNVEL (Jian et al., 2014b), SPANR (Xiong et al., 2015), and CryptSplice (Lee et al., 2017). Further tools are available for the identification of branch point sequences (Corvelo et al., 2010; Zhang et al., 2017). Lastly, the secondary structure of pre-mRNAs can interfere with splice-site recognition, modulate spliceosome binding or can facilitate splicing efficiency by bringing splice donors and acceptors into close proximity (Warf and Berglund, 2010). Consequently, genetic variants that alter pre-mRNA structure were found to promote alternative splicing (Wan et al., 2014), incentivizing the incorporation of structural information provided by tools, such as TurboFold (Harmanci et al., 2011) or CentroidFold (Sato et al., 2009), into variant effect predictions. For a more detailed description of structural RNA analyses we refer the interested reader to excellent recent reviews (Jian et al., 2014a; Lorenz et al., 2016; Ohno et al., 2018).
Table 3.
Tools for the prediction of variant effects on splicing, transcript levels or translation.
| Algorithm | Application | Basis of decision | Model training or evaluation | References |
|---|---|---|---|---|
| NMD Classifier | NMD | Prediction of NMD for a given transcript based on comparison to most similar coding transcript | Simulation-based evaluation based on screening artificial transcript structure-altering events | Hsu et al., 2017 |
| NNSplice | Splicing (splice sites) | Sequence splice site analysis using HMM | Distinguish splice site sequences from sequences in the neighborhood of real splice sites | Reese et al., 1997 |
| MaxEntScan | Splicing (splice sites) | Splice site analysis by modeling short sequence motifs using the maximum entropy principle with constraints estimated from available data. | 1,821 transcripts unambiguously aligned across the entire coding region, spanning a total of 12,715 introns | Yeo and Burge, 2004 |
| GeneSplicer | Splicing (splice sites) | Splice site prediction using maximal dependence decomposition with the addition of markov model to capture dependencies among neighboring bases | Annotated genes from the Exon-Intron Database | Pertea et al., 2001 |
| SplicePort | Splicing (splice sites) | Splice site prediction using C-modified least squares learning based on positional and compositional sequence features | Training on 4,000 pre-mRNA human RefSeq sequences and test on B2Hum data set | Dogan et al., 2007 |
| Skippy | Splicing (regulatory sequences) | Prediction of variants causing exon skipping, exon inclusion or ectopic splice site activation based on sequence information, proximity to splice junctions and evolutionary constraint of the peri-variant region | Multiple exonic splicing regulatory elements datasets as positive data and HapMap variants as splicing-neutral variants | Woolfe et al., 2010 |
| MutPred Splice | Splicing (regulatory sequences) | Prediction of auxiliary splice sequences using multiple variant-, flanking exon- and gene-based features | Splicing variants from HGMD as pathogenic set and non-splicing variants from both HGMD and 1000G as neutral controls | Mort et al., 2014 |
| scSNVEL | Splicing (splice sites) | Ensemble prediction using 8 algorithms using random forest learning | Splice variants from HGMD, SpliceDisease database and DBASS as pathogenic set and variants not implicated in splicing from both HGMD and 1000G as controls | Jian et al., 2014b |
| SPANR | Splicing (splice sites and splice regulatory sequences) | Integrating 1,393 sequence features from each exon and its neighboring introns and exons to identify splice sites as well as intronic and exonic splice regulators | 10,689 exons that displayed evidence of alternative splicing | Xiong et al., 2015 |
| CryptSplice | Splicing (splice sites) | Prediction of cryptic splice-site activation using an SVM model | Sequences from the annotated NN269 and HS3D splice datasets with positive sequence in splice sites and control sequence outside splice sites | Lee et al., 2017 |
| Corvelo et al. | Splicing (branch points) | Analysis of splice site sequence conservation and position bias using SVM | A set of 8,156 conserved putative branch point sequences from 7 mammalian species | Corvelo et al., 2010 |
| BPP | Splicing (branch points) | Identification of branch point motifs by integrating information on the branch point sequence and the polypyrimidine tract | Intron sequences longer than 300 nucleotides | Zhang et al., 2017 |
| TurboFold | Splicing (pre-mRNA structure) | Probabilistic method that integrates comparative sequence analyses with thermodynamic folding models | Thorough benchmarking against three methods that estimate base pairing probabilities and eight tools for structural predictions based on known RNA structures | Harmanci et al., 2011 |
| CentroidFold | Splicing (pre-mRNA structure) | RNA secondary structure prediction using the γ-centroid estimator | Validation based on 151 RNA experimentally determined RNA structures | Sato et al., 2009 |
| mrSNP | miRNA binding | miRNA binding energy calculations for reference and variant containing sequence and report of binding difference | Evaluation based on variants that map to miRNA targets predicted by TargetScan | Deveci et al., 2014 |
| PinPor | RBP binding | Bayesian network approach that incorporates information about sequence features, stabilization of RNA secondary structure and evolutionary conservation | Inframe indels from HGMD as pathogenic and common indels from 1000G as neutral controls | Zhang et al., 2014 |
HGMD, Human Gene Mutation Database; 1000G = 1000 Genomes Project; DBASS, Database for Aberrant Splice Sites; NMD, nonsense-mediated decay; HMM, hidden Markov model; RBP, RNA binding protein.
In ADME genes, dysregulation of splicing has long been recognized as a cause for inter-individual variability drug metabolism (Hanioka et al., 1990) and toxicity (Raida et al., 2001) and the liver was found to be is among the tissues with highest levels of alternative splicing activity (Yeo et al., 2004). As splicing is highly tissue specific, these data indicate that algorithms for the prediction of variant splice effects in pharmacogenetics should ideally be trained on positive control sets for which aberrant splicing is confirmed in the tissue of interest, i.e., primarily liver. To this end, the GTEx project (GTEx Consortium, 2017) provides a rich resource that has already been successfully utilized for the identification of tissue-specific splice events in pharmacogenes (Chhibber et al., 2017).
In summary, the toolkit of available computational algorithms for the prediction of variant effects on splicing has rapidly grown and by now allows not only to evaluate direct impact on splice sites, but also to assess mutations in regulatory splice enhancers and silencers, as well as branch points. For the application of these methods for pharmacogenomics there is a need to benchmark available tools on splice variants in ADME genes. Moreover, we anticipate that the utilization of tissue-specific expression data will further refine splice site predictions.
Functional Impact of Variants in Untranslated Regions
miRNAs play important roles in the regulation of mRNA stability and translation. miRNA-mRNA interaction occurs through conserved miRNA binding sites in the 3′-UTRs and at least 10% of all SNPs are located in 3′-UTRs and might affect complementary miRNA-mRNA pairing (Xiao et al., 2009). Furthermore, miRNAs have been shown to be important modulators of ADME gene expression profiles (Rieger et al., 2013). Therefore, functional interpretation of genetic variations within miRNA target sites constitutes an important factor for the prediction of the fate of corresponding transcript. Thus, to evaluate the potential relevance of genetic polymorphisms in UTRs various databases, such as the polymiRTS Database 3.0 (Bhattacharya et al., 2014) or MirSNP (Liu et al., 2012), provide useful resources that contains a collection of experimentally confirmed SNPs and indels not only in miRNA target sites but also in miRNA seed regions responsible for mRNA binding. Furthermore, a variety of other SNP effect prediction servers are publically available (Fehlmann et al., 2017).
In case no experimental data is available, various computational tools can be used to predict possible disruption of the miRNA-mRNA pairing for a given variant (Table 3). MicroSNiPer (Barenboim et al., 2010) and ImiRP (Ryan et al., 2016) identify and predict such disruptions by comparing the mutant 3′-UTR sequences with major variant databases. Similarly, mrSNP can predict the effect of any variant identified in NGS-based projects on miRNA-target transcript interaction (Deveci et al., 2014). However, it is important to note that miRNA target predictions seem to have a high false-positive rate (Pinzón et al., 2017), suggesting that these problems might be lingering for studies utilizing miRNA-target databases without stringent experimental validations. Besides predicting the effect of genetic variants in putative miRNA target sites, multiple online tools are available for inverse approaches, analyzing variants in miRNAs or pre-miRNAs for possible deleterious effects. For more comprehensive collection of miRNA related variant interpretation tools the reader is referred to the recent reviews and online resources (Akhtar et al., 2016; Moszynska et al., 2017).
In addition, recent approaches expanded the methodological portfolio beyond miRNA binding site prediction to include effects of UTR variants on binding of RNA-binding proteins (RBPs), translational efficacy and ribosomal loading. Effects of indels on RBP binding can be evaluated using PinPor, which has been demonstrated to have some success in distinguishing disease-causing and neutral indels (Zhang et al., 2014). Furthermore, Sample et al. presented the preprint of a deep learning approach based on experimental polysome profiling to predict the impact of UTR sequence on translation (Sample et al., 2018). These developments nicely indicate the diversification of parameters that can incorporated into variant effect predictions, thus further refining biological interpretation of NGS data sets.
Analysis of Regulatory Variants
Non-coding regions account for more than 99% of the human genome and, consequently, their consideration substantially expands the analysis space of computational predictions. Variants in non-coding regions can affect regulatory elements, such as promoters, enhancers, silencers, and insulators, which, in turn, may alter their affinity to transcription factor or remodel the local chromatin structure (Zhang and Lupski, 2015; Deplancke et al., 2016). Accurate prediction of the functional consequences of such variants constitutes one of the major challenges in human genetics.
To interpret noncoding variants, a variety of different strategies have been presented. The first approaches, such as SiPhy (Garber et al., 2009), PhyloP (Pollard et al., 2010), PhastCons (Siepel et al., 2005), GERP++ (Davydov et al., 2010), or SCONE (Asthana et al., 2007), were based on evolutionary constraint using sequence alignments. However, the observation that no enhanced constraints were identified in regulatory elements at the level of DNA sequence despite conserved transcription factor binding led to the realization that conservation of regulatory regions can only be a weak indicator of the functional effects of SNVs in regulatory regions (Schmidt et al., 2010; Arbiza et al., 2013). Consequently, conservation metrics were complemented with additional functional genomics features, such as the sequence and genic context, transcription factor binding profiles (Johnson et al., 2007), histone modification data (Zhang et al., 2010) and DNase I hypersensitive sites (Boyle et al., 2008) in an attempt to improve prediction quality. Based on these rich data sets, a variety of ensemble classifiers were developed using various machine learning approaches that aim to distinguish neutral from pathogenic variants, including GWAVA (Ritchie et al., 2014), CADD (Kircher et al., 2014), FATHMM (Shihab et al., 2013, 2015; Rogers et al., 2018), DANN (Quang et al., 2015), DIVAN (Chen et al., 2016), and Genomiser (Smedley et al., 2016) (Table 4).
Table 4.
Algorithms for the functional interpretation of regulatory variants.
| Algorithm | Model | Application | Model training | Features | References |
|---|---|---|---|---|---|
| FATHMM | HMM | Pathogenic variants | HGMD regulatory variants as pathogenic set and common 1000G variants as controls | Evolutionary conservation data (PhastCons and PhyloP), chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data, genome segmentation, frequency data (1000G and ESP) and information about genic and sequence context | Shihab et al., 2013, 2015; Rogers et al., 2018 |
| GWAVA | RF | Pathogenic variants | HGMD regulatory variants as pathogenic set and common 1000G variants as controls | Evolutionary conservation data (GERP), chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data, genome segmentation, frequency data (1000G) and information about genic and sequence context | Ritchie et al., 2014 |
| CADD | SVM | Deleterious variants | Sites with MAF < 5% where for which the human genome differed from the inferred human-chimp ancestral genome and equal number of simulated variants | Evolutionary conservation data (GERP++, PhastCons and PhyloP), chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data, genome segmentation, frequency data (1000G and ESP) and information about genic and sequence context | Kircher et al., 2014 |
| DANN | NN | Deleterious variants | Same as CADD but using deep neural networks instead of linear SVM. | Quang et al., 2015 | |
| DeepSEA | NN | Variants that affect gene expression | HGMD regulatory variants, eQTLs and NHGRI GWAS phenotype-associated SNPs | Evolutionary conservation data (GERP++, PhastCons and PhyloP), chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data | Zhou and Troyanskaya, 2015 |
| gkm-SVM | SVM | Variants that affect gene expression | Tissue-specific enhancer sequences marked by H3K4me1 from length-, GC content- and repeat-matched random control | Definition of tissue-specific regulatory dictionary based on chromatin accessibility (DNase-HSS) and H3K4me1 ChIP-Seq data | Lee et al., 2015 |
| fitCons | INSIGHT | Prediction of cis-regulatory elements | Unsupervised classifier that clusters genomic regions on the basis of functional genomic data and then estimates a probability of fitness consequences for each group from associated patterns of genetic polymorphism and divergence. | Evolutionary conservation data (GERP, PhastCons and PhyloP), chromatin accessibility (DNase-HSS), TF binding and histone modification ChIP-Seq data, genome segmentation and RNA-Seq data | Gulko et al., 2015 |
| GenoCanyon | US | Identification of functional regions | Unsupervised classifier based on the estimated proportion of functional regions in the human genome. | Evolutionary conservation data (GERP and PhyloP), chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data | Lu et al., 2015 |
| DIVAN | EL | Disease-specific risk variants | Disease-specific regulatory NHGRI GWAS SNPs and common 1000G variants or benign GWAS SNPs as controls | Chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data | Chen et al., 2016 |
| Genomiser | RF | Mendelian disease | Sites with MAF < 5% where for which the human genome differed from the inferred human-chimp ancestral genome as functionally neutral variation and 453 positive variants based on literature review | Evolutionary conservation data (GERP++, PhastCons and PhyloP), chromatin accessibility (DNase-HSS), TF binding and histone modification ChIP-Seq data, frequency data (1000G and ESP) and information about enhancer context from FANTOM5 | Smedley et al., 2016 |
| Eigen | US | Effect of variants on gene expression and disease risk | Unsupervised classifier based on the blockwise conditional independence between annotations given the functional impact of the variant. | Evolutionary conservation data (GERP, PhastCons and PhyloP), chromatin accessibility (DNase-HSS and FAIRE-Seq), TF binding and histone modification ChIP-Seq data and frequency data (1000G) | Ionita-Laza et al., 2016 |
RF, random forest; SVM, support vector machine; HMM, hidden Markov model; EL, ensemble learning; NN, neural networks; INSIGHT, Inference of Natural Selection from Interspersed Genomically Coherent Elements Gronau et al., 2011; US, unsupervised; HGMD, Human Gene Mutation Database; 1000G, 1000 Genomes Project; ESP, Exome Sequencing Project; TF, transcription factor; HSS, hypersensitive site; FAIRE, Formaldehyde-Assisted Isolation of Regulatory Elements Giresi et al., 2007; NHGRI, National Human Genome Research Institute.
In contrast, other methods, such as gkm-SVM (Lee et al., 2015) and DeepSEA (Zhou and Troyanskaya, 2015) have been developed to predict regulatory elements based on primary sequence alone. Trained on publically available cell type-specific chromatin data provided by ENCODE (The ENCODE Project Consortium, 2012) and the Roadmap Epigenomics Project (Roadmap Epigenomics Consortium et al., 2015) as well as transcription factor binding patterns accessible via JASPAR (Khan et al., 2018), these algorithms predict to what extent a genetic variant will cause changes to the local chromatin profiles and how these effects translate into functional consequences. The resulting data demonstrate that inferring consequences from functional genomics data is highly cell type and context specific and relies on biologically appropriate training sets. These convincing findings incentivize the generation of functional genomics data from carefully phenotyped human tissues involved in drug ADME to derive tissue-specific regulatory lexica and we envision that training machine learning approaches on these data sets will substantially increase the power of regulatory pharmacogenetic prediction classifiers.
As with coding variants, the use of potentially biased training sets and multi-dimensional circularity between training and test data constitutes an inherent problem for current variant prediction tools (Grimm et al., 2015). For instance, a variety of algorithms consider common variants from the 1000 Genomes project as functionally neutral control sets for model training. However, while these variants are likely to be depleted of pathogenic variants in haploinsufficient genes, many common variants entail functional consequences in their respective gene product, particularly if the gene is rapidly evolving, such as many CYP genes. Similar problems arise when the model is trained using phenotype associated GWAS polymorphisms as functional variant sets, as only 5.5% of GWAS index SNPs are estimated to be causal whereas the remainder is only in linkage disequilibrium with the true functional variant in the locus (Farh et al., 2015).
To overcome these problems, unsupervised approaches have been developed that do not rely on the labeling of training data, thereby reducing the dependence on preexisting variant classifications and existing models of mutation. These unsupervised models, such as GenoCanyon (Lu et al., 2015) and Eigen (Ionita-Laza et al., 2016), represent powerful tools for the genome-wide interpretation of variants. However, as they are calibrated on genome-wide data, it remains to be determined whether gene class-specific peculiarities, such as low evolutionary conservation in ADME genes, might affect the predictive accuracy of these approaches for pharmacogenetic applications.
Conclusions
Technical progress in NGS technology has resulted in its routine application in medical genetics and clinical diagnostics. In contrast, clinical implementation of NGS-based pharmacogenomics is largely lagging behind (Lauschke and Ingelman-Sundberg, 2016b; Ji et al., 2018). Most importantly, in order to utilize the major advantage of NGS-based genotyping, which is the discovery of the entire panorama of the individual's genetic portfolio, tools have to be in place, which allow to translate these variability data into functional consequences and clinical recommendations. Whereas, the identification of rare putatively deleterious mutations in congenital diseases is aided by clear phenotypic alterations of the affected patient and the possibility to perform comparative genomic analyses of unaffected family members, pharmacogenomic phenotypes are generally more difficult to detect as they only present in a given context, such as exposure to specific medications. In the absence of drug response associations or experimental characterizations that support the functional interpretation of rare variants, there is thus an urgent need for reliable computational prediction tools to fill this space.
Importantly, recent developments in computational variant effect prediction methods promise to narrow the gap to meet the exacting demands on genomics applications in the clinics. Machine learning constitutes an important tool kit to fully harness the power of large data sets provided by NGS. However, these approaches rely on accurate labeling of input variants, i.e., training data need to be correctly classified into deleterious and functionally neutral variants. Thus, we advocate for approaches that leverage smaller data sets of variants for which comprehensive experimental or functional genomic data is available instead of training algorithms on large but functionally poorly annotated data, such as treating all common polymorphisms identified in the 1000 Genomes Project as functionally neutral. In addition, we endorse previous appeals for the sharing of codes and data sets, which will enable comparative benchmarking of newly developed tools and algorithms and will accelerate research progress within the area of computational pharmacogenomics and beyond (Kalinin et al., 2018).
The functional consequences of missense variants have been most extensively studied. Respective methods base their predictions on evolutionary conservation and structural information of the polypeptide encoded by the respective gene. Importantly, while evolutionary conservation is a suitable measure to inform about the deleteriousness of a variant, i.e., its effect on organismal fitness, it is not suitable for the prediction of variant effects in genes under low selective pressure, such as most pharmacogenes. Recognition of these conceptual problems resulted in the development of computational predictors trained specifically on ADME missense variants (Zhou et al., 2018). We envision that these approaches will become more powerful with increasing functionally annotated pharmacogenetic variant data.
Furthermore, multiple strategies have been developed to analyze the functional impact of variants in non-coding regions of the genome, which are increasingly recognized as a substantial contributor to inter-individual variability. An increasing number of algorithms is by now available that base their predictions on a multitude of different parameters, including effects on miRNA binding or translational efficiency, modulation of splicing and impacts on transcriptional events by disruption of transcription factor binding sites or polymerase loading (Figure 1). While these developments provide a methodological arsenal to comprehensively characterize all different classes of genetic variants, these methods are generally trained on pathogenic variant sets and have not been benchmarked on independent data sets. Thus, their predictive power for pharmacogenetic assessments remains to be evaluated.
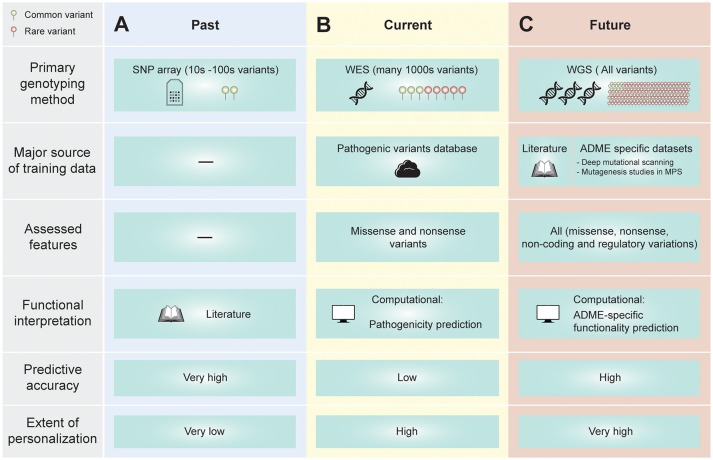
The prediction of drug metabolism phenotypes based on the genotype of the individual has made tremendous progress over the last decades (Figure 2). Conventional approaches use data from few candidate variants for which substantial in vitro or in vivo characterization data was available to predict drug response. While this strategy has been successful in incorporating common pharmacogenetic variability into clinical decision-making, they fail to address functional effects of the vast extent of rare genetic variants. To also include rare variants, pilot programs were initiated in which WES was used to comprehensively interrogate the genetic landscape of pharmacogenomic loci (Bielinski et al., 2014). However, analyses were restricted to pharmacogenetic missense variants and the effects of SNVs with unknown functional relevance were interpreted using computational models trained on pathogenic data sets with negative impacts on the accuracy of phenotype predictions, as discussed above. Thus, while these strategies constitute an important step toward the further personalization of genotype-guided treatment decisions their predictive accuracy is rather low.
Figure 2.
The past, present and future of pharmacogenetic phenotype predictions. (A) Conventionally, pharmacogenetic predictions were based on the interrogation of few common candidate SNPs, whose functional effects were predicted based on extensive literature evidence, resulting in high predictive accuracy but only few considered variations. (B) With increasing prevalence of whole exome sequencing (WES), a multitude of pharmacogenetic variants with unknown functional relevance are identified. These variants can be interpreted using computational methods. However, current algorithms are generally trained to detect the pathogenicity rather than the functionality of queried variants, resulting in overall relatively low predictive accuracy. Furthermore, only effects of missense and nonsense variants are evaluated. (C) In the near future, whole genome sequencing (WGS) will become the predominant genotyping methodology, revealing not only coding variants but also variants in regulatory regions and introns. To facilitate interpretation of this data, we envision that pharmacogenetic predictors will be directly trained on functionally annotated ADME data sets. Emerging technologies, such as deep mutational scanning for the systematic interrogation of missense variants or mutagenesis screens in microphysiological systems (MPS) for the characterization of variants in regulatory regions, provide powerful tools to generate these data, boosting the predictive performance of data hungry machine learning tools. These advances allow to go beyond the interpretation of missense and nonsense variants and to include also non-coding and regulatory variations into pharmacogenetic assessments.
We expect that technological, methodological and analytical progress will contribute to a further refinement of NGS-guided drug treatment in the near future. Firstly, technological advances will result in an increasing dissemination of WGS, which facilitates the incorporation of the entire profile of an individual's genetic variability, including regulatory variants, into pharmacogenetic predictions. Secondly, we envision that novel high-throughput methodologies for functional characterizations, such as deep mutational scanning, will provide powerful approaches to generate large functionally annotated pharmacogenetic variant data sets. In addition, recent advances in the development of microphysiological systems (MPS) that allow to model key target tissues associated with drug metabolism or safety provide (Ewart et al., 2018) provide promising tools to generate tissue-specific and human-relevant data sets for studies of gene-drug interactions (Ingelman-Sundberg and Lauschke, 2018). Using this integrated wealth of functional pharmacogenetic data to train machine learning models aspires to provide high-accuracy predictions based on the entire genetic variability landscape of the respective patient.
Importantly, leveraging this information as guidance for clinical decision-making promises to increase treatment efficacy and reduce the risks of adverse events in carriers of pharmacogenetic variants whose effects have not been experimentally evaluated. Current market analysis estimates suggests that implementation of artificial intelligence into the clinical decision support toolbox might increase average life expectancy in the Western World by 0.2–1.3 years and reduce total health care expenditures by 5–9%, corresponding to 2 trillion to 10 trillion USD globally per year (Bughin et al., 2017). However, in order to realize these exciting prospects, there is a need for prospective, randomized controlled trials that evaluate patient outcomes and cost-effectiveness of such preemptive advice across genes, drugs and health care systems.
In summary, computational prediction methods are essential for the implementation of NGS into clinical decision-making. While much progress has been made and a plethora of conceptually diverse tools is already available, there is a need to develop specialized methods that are optimized for the prediction of variant functionality rather than pathogenicity and are calibrated specifically on pharmacogenetic data. We envision that technological, methodological and analytical advances will soon allow to comprehensively predict variant effects with sufficient accuracy to justify the design of trials in which the clinical value of NGS-guided treatment decisions can be tested in a prospective setting.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
VL is co-founder and owner of HepaPredict AB. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The work in the authors' laboratory is supported by the Swedish Research Council [grant agreement numbers: 2016-01153 and 2016-01154], by the Strategic Research Programme in Diabetes at Karolinska Institutet, by the European Union's Horizon 2020 research and innovation program U-PGx [grant agreement No. 668353], and by the Lennart Philipson and Harald och Greta Jeansson Foundations.
References
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., et al. (2010). A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn E., Park T. (2017). Analysis of population-specific pharmacogenomic variants using next-generation sequencing data. Sci. Rep. 7:8416. 10.1038/s41598-017-08468-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M. M., Micolucci L., Islam M. S., Olivieri F., Procopio A. D. (2016). Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 44, 24–44. 10.1093/nar/gkv1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancien F., Pucci F., Godfroid M., Rooman M. (2018). Prediction and interpretation of deleterious coding variants in terms of protein structural stability. Sci. Rep. 8:4480. 10.1038/s41598-018-22531-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbiza L., Gronau I., Aksoy B. A., Hubisz M. J., Gulko B., Keinan A., et al. (2013). Genome-wide inference of natural selection on human transcription factor binding sites. Nat. Genet. 45, 723–729. 10.1038/ng.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S., Roytberg M., Stamatoyannopoulos J., Sunyaev S. (2007). Analysis of sequence conservation at nucleotide resolution. PLoS Comput. Biol. 3:e254. 10.1371/journal.pcbi.0030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Fu Y., Pawashe M., McGillivray P., Jin M., Liu J., et al. (2017). Using ALoFT to determine the impact of putative loss-of-function variants in protein-coding genes. Nat. Commun. 8:382. 10.1038/s41467-017-00443-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle D., Lucassen A., Buratti E. (2009). Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep. 10, 810–816. 10.1038/embor.2009.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenboim M., Zoltick B. J., Guo Y., Weinberger D. R. (2010). MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum. Mutat. 31, 1223–1232. 10.1002/humu.21349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh E. H., Simmons-Edler R., Müller C. L., Alford R. F., Volfovsky N., Lash A. E., et al. (2016). Robust classification of protein variation using structural modelling and large-scale data integration. Nucleic Acids Res. 44, 2501–2513. 10.1093/nar/gkw120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E. D., Zendulka J., et al. (2014). PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 10:e1003440. 10.1371/journal.pcbi.1003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Ziebarth J. D., Cui Y. (2014). PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 42, D86–D91. 10.1093/nar/gkt1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinski S. J., Olson J. E., Pathak J., Weinshilboum R. M., Wang L., Lyke K. J., et al. (2014). Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin. Proc. 89, 25–33. 10.1016/j.mayocp.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott K. M., Vanstone M. R., Bulman D. E., MacKenzie A. E. (2013). Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 14, 681–691. 10.1038/nrg3555 [DOI] [PubMed] [Google Scholar]
- Boyle A. P., Davis S., Shulha H. P., Meltzer P., Margulies E. H., Weng Z., et al. (2008). High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell 132, 311–322. 10.1016/j.cell.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bughin J., Hazan E., Ramaswamy S., Chui S., Allas T., Dahlström P., et al. (2017). Artificial Intelligence the Next Digital Frontier? McKinsey and Company Global Institute. [Google Scholar]
- Bush W. S., Crosslin D. R., Owusu-Obeng A., Wallace J., Almoguera B., Basford M. A., et al. (2016). Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin. Pharmacol. Therapeut. 100, 160–169. 10.1002/cpt.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese R., Capriotti E., Fariselli P., Martelli P. L., Casadio R. (2009). Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 30, 1237–1244. 10.1002/humu.21047 [DOI] [PubMed] [Google Scholar]
- Camacho D. M., Collins K. M., Powers R. K., Costello J. C., Collins J. J. (2018). Next-generation machine learning for biological networks. Cell 173, 1581–1592. 10.1016/j.cell.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Casadio R. (2006). Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22, 2729–2734. 10.1093/bioinformatics/btl423 [DOI] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. (2005). I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 33, W306–W310. 10.1093/nar/gki375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H., Douville C., Stenson P. D., Cooper D. N., Karchin R. (2013). Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics 14(Suppl. 3), S3. 10.1186/1471-2164-14-S3-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Jin P., Qin Z. S. (2016). DIVAN: accurate identification of non-coding disease-specific risk variants using multi-omics profiles. Genome Biol. 17:252. 10.1186/s13059-016-1112-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber A., French C. E., Yee S. W., Gamazon E. R., Theusch E., Qin X., et al. (2017). Transcriptomic variation of pharmacogenes in multiple human tissues and lymphoblastoid cell lines. Pharmacogenomics J. 17, 137–145. 10.1038/tpj.2015.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G. E., Murphy S., Miller J. R., Chan A. P. (2012). Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7:e46688. 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S., Fay J. C. (2009). Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S., Karchin R. (2011). Using bioinformatics to predict the functional impact of SNVs. Bioinformatics 27, 441–448. 10.1093/bioinformatics/btq695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvelo A., Hallegger M., Smith C. W., Eyras E. (2010). Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 6:e1001016. 10.1371/journal.pcbi.1001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov E. V., Goode D. L., Sirota M., Cooper G. M., Sidow A., Batzoglou S. (2010). Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6:e1001025–e1001013. 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Alpern D., Gardeux V. (2016). The genetics of transcription factor DNA binding variation. Cell 166, 538–554. 10.1016/j.cell.2016.07.012 [DOI] [PubMed] [Google Scholar]
- Deveci M., Catalyürek U. V., Toland A. E. (2014). mrSNP: software to detect SNP effects on microRNA binding. BMC Bioinformatics 15:73. 10.1186/1471-2105-15-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan R. I., Getoor L., Wilbur W. J., Mount S. M. (2007). SplicePort–an interactive splice-site analysis tool. Nucleic Acids Res. 35, W285–W291. 10.1093/nar/gkm407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., et al. (2015). Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137. 10.1093/hmg/ddu733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville C., Masica D. L., Stenson P. D., Cooper D. N., Gygax D. M., Kim R., et al. (2016). Assessing the pathogenicity of insertion and deletion variants with the variant effect scoring tool (VEST-Indel). Hum. Mutat. 37, 28–35. 10.1002/humu.22911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart L., Dehne E.-M., Fabre K., Gibbs S., Hickman J., Hornberg E., et al. (2018). Application of microphysiological systems to enhance safety assessment in drug discovery. Annu. Rev. Pharmacol. Toxicol. 58, 65–82. 10.1146/annurev-pharmtox-010617-052722 [DOI] [PubMed] [Google Scholar]
- Farh K. K., Marson A., Zhu J., Kleinewietfeld M., Housley W. J., Beik S., et al. (2015). Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343. 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlmann T., Sahay S., Keller A., Backes C. (2017). A review of databases predicting the effects of SNPs in miRNA genes or miRNA-binding sites. Brief. Bioinformatics. 10.1093/bib/bbx155. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Flanagan S. E., Patch A.-M., Ellard S. (2010). Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet. Test. Mol. Biomarkers 14, 533–537. 10.1089/gtmb.2010.0036 [DOI] [PubMed] [Google Scholar]
- Folkman L., Yang Y., Li Z., Stantic B., Sattar A., Mort M., et al. (2015). DDIG-in: detecting disease-causing genetic variations due to frameshifting indels and nonsense mutations employing sequence and structural properties at nucleotide and protein levels. Bioinformatics 31, 1599–1606. 10.1093/bioinformatics/btu862 [DOI] [PubMed] [Google Scholar]
- Fujikura K. (2016). Premature termination codons in modern human genomes. Sci. Rep. 6:22468. 10.1038/srep22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura K., Ingelman-Sundberg M., Lauschke V. M. (2015). Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet. Genomics 25, 584–594. 10.1097/FPC.0000000000000172 [DOI] [PubMed] [Google Scholar]
- Gallion J., Koire A., Katsonis P., Schoenegge A.-M., Bouvier M., Lichtarge O. (2017). Predicting phenotype from genotype: improving accuracy through more robust experimental and computational modeling. Hum. Mutat. 38, 569–580. 10.1002/humu.23193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M., Guttman M., Clamp M., Zody M. C., Friedman N., Xie X. (2009). Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–i62. 10.1093/bioinformatics/btp190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getov I., Petukh M., Alexov E. (2016). SAAFEC: predicting the effect of single point mutations on protein folding free energy using a knowledge-modified MM/PBSA approach. Int. J. Mol. Sci. 17, 512–514. 10.3390/ijms17040512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi P. G., Kim J., McDaniell R. M., Iyer V. R., Lieb J. D. (2007). FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885. 10.1101/gr.5533506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pérez A., López-Bigas N. (2011). Improving the assessment of the outcome of Nonsynonymous SNVs with a consensus deleteriousness score, condel. Am. J. Hum. Genet. 88, 440–449. 10.1016/j.ajhg.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray V. E., Hause R. J., Luebeck J., Shendure J., Fowler D. M. (2018). Quantitative missense variant effect prediction using large-scale mutagenesis data. Cell Syst. 6, 116–124.e3. 10.1016/j.cels.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. G., Azencott C.-A., Aicheler F., Gieraths U., MacArthur D. G., Samocha K. E., et al. (2015). The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 36, 513–523. 10.1002/humu.22768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronau I., Arbiza L., Mohammed J., Siepel A. (2011). Inference of natural selection from interspersed genomic elements based on polymorphism and divergence. Mol. Biol. Evol. 30, 1159–1171. 10.1093/molbev/mst019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2017). Genetic effects on gene expression across human tissues. Nature 550, 204-213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulko B., Hubisz M. J., Gronau I., Siepel A. (2015). A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 47, 276–283. 10.1038/ng.3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanioka N., Kimura S., Meyer U. A., Gonzalez F. J. (1990). The Human Cyp2d locus associated with a common genetic-defect in drug oxidation-a G1934-]a base change in intron-3 of a mutant Cyp2d6 allele results in an Aberrant-3' Splice Recognition site. Am. J. Hum. Genet. 47, 994–1001. [PMC free article] [PubMed] [Google Scholar]
- Harmanci A. O., Sharma G., Mathews D. H. (2011). TurboFold: iterative probabilistic estimation of secondary structures for multiple RNA sequences. BMC Bioinformatics 12:108. 10.1186/1471-2105-12-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M., Bromberg Y., Rost B. (2015). Better prediction of functional effects for sequence variants. BMC Genomics 16(Suppl 8):S1. 10.1186/1471-2164-16-S8-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T. A., Ingraham J. B., Poelwijk F. J., Schärfe C. P., Springer M., Sander C., et al. (2017). Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135. 10.1038/nbt.3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.-K., Lin H.-Y., Chen F.-C. (2017). NMD Classifier: A reliable and systematic classification tool for nonsense-mediated decay events. PLoS ONE 12:e0174798. 10.1371/journal.pone.0174798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Ng P. C. (2012). Predicting the effects of frameshifting indels. Genome Biol. 13, R9. 10.1186/gb-2012-13-2-r9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M., Lauschke V. M. (2018). Human liver spheroids in chemically defined conditions for studies of gene–drug, drug–drug and disease–drug interactions. Pharmacogenomics 19, 1133–1138. 10.2217/pgs-2018-0096 [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M., Mkrtchian S., Zhou Y., Lauschke V. M. (2018). Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genomics 12:26. 10.1186/s40246-018-0157-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis N. M., Rothstein J. H., Pejaver V., Middha S., McDonnell S. K., Baheti S., et al. (2016). REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885. 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I., McCallum K., Xu B., Buxbaum J. D. (2016). A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Rev. Clin. Oncol. 48, 214–220. 10.1038/ng.3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Si Y., McMillin G. A., Lyon E. (2018). Clinical pharmacogenomics testing in the era of next generation sequencing: challenges and opportunities for precision medicine. Expert Rev. Mol. Diagn. 18, 411–421. 10.1080/14737159.2018.1461561 [DOI] [PubMed] [Google Scholar]
- Jian X., Boerwinkle E., Liu X. (2014a). In silico tools for splicing defect prediction: a survey from the viewpoint of end users. Genetics in Medicine 16, 497–503. 10.1038/gim.2013.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X., Boerwinkle E., Liu X. (2014b). In silico prediction of splice-altering single nucleotide, variants in the human genome. Nucleic Acids Res. 42, 13534–13544. 10.1093/nar/gku1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502. 10.1126/science.1141319 [DOI] [PubMed] [Google Scholar]
- Jukić M. M., Haslemo T., Molden E., Ingelman-Sundberg M. (2018). Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a retrospective study based on 2,087 patients. Am. J. Psychiatry 175, 463–470. 10.1176/appi.ajp.2017.17050550 [DOI] [PubMed] [Google Scholar]
- Kalinin A. A., Higgins G. A., Reamaroon N., Soroushmehr S., Allyn-Feuer A., Dinov I. D., et al. (2018). Deep learning in pharmacogenomics: from gene regulation to patient stratification. Pharmacogenomics 19, 629–650. 10.2217/pgs-2018-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P., Lichtarge O. (2014). A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res. 24, 2050–2058. 10.1101/gr.176214.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem E., Hirawat S., Armoni S., Yaakov Y., Shoseyov D., Cohen M., et al. (2008). Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet 372, 719–727. 10.1016/S0140-6736(08)61168-X [DOI] [PubMed] [Google Scholar]
- Kervestin S., Jacobson A. (2012). NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13, 700–712. 10.1038/nrm.3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J. A. R., et al. (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266. 10.1093/nar/gkx1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O'Roak B. J., Cooper G. M., Shendure J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Rev. Clin. Oncol. 46, 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyra M., Ingelman-Sundberg M., Lauschke V. M. (2017). Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet. Med. 19, 20–29. 10.1038/gim.2016.33 [DOI] [PubMed] [Google Scholar]
- Kryukov G. V., Pennacchio L. A., Sunyaev S. R. (2007). Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 80, 727–739. 10.1086/513473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha S., Chaudhary V., Goswami G. K., Mathur N. (2016). Computational approaches for predicting mutant protein stability. J. Comput. Aided Mol. Des. 30, 401–412. 10.1007/s10822-016-9914-3 [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Ingelman-Sundberg M. (2016a). Precision medicine and rare genetic variants. Trends Pharmacol. Sci. 37, 85–86. 10.1016/j.tips.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Ingelman-Sundberg M. (2016b). Requirements for comprehensive pharmacogenetic genotyping platforms. Pharmacogenomics 17, 917–924. 10.2217/pgs-2016-0023 [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Ingelman-Sundberg M. (2018). How to consider rare genetic variants in personalized drug therapy. Clin. Pharmacol. Therapeut. 19, 20 10.1002/cpt.976 [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Milani L., Ingelman-Sundberg M. (2017). Pharmacogenomic biomarkers for improved drug therapy-recent progress and future developments. AAPS J. 20, 4. 10.1208/s12248-017-0161-x [DOI] [PubMed] [Google Scholar]
- Lee D., Gorkin D. U., Baker M., Strober B. J., Asoni A. L., McCallion A. S., et al. (2015). A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 47, 955–961. 10.1038/ng.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Roos P., Sharma N., Atalar M., Evans T. A., Pellicore M. J., et al. (2017). Systematic computational identification of variants that activate exonic and intronic cryptic splice sites. Am. J. Hum. Genet. 100, 751–765. 10.1016/j.ajhg.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Zhang Y., Mukhyala K., Lazarus R. A., Zhang Z. (2009). Bi-directional SIFT predicts a subset of activating mutations. PLoS ONE 4:e8311. 10.1371/journal.pone.0008311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rio D. C. (2015). Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 84, 291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. E., Myers R. M. (2016). Advancements in next-generation sequencing. Annu. Rev. Genomics Hum. Genet. 17, 95–115. 10.1146/annurev-genom-083115-022413 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao T., Zhang Y., Zhang K., Shi L., Chen Y., et al. (2018). Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 46, 7793–7804. 10.1093/nar/gky678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L., Boelz S., Nissim-Rafinia M., Oren Y. S., Wilschanski M., Yaacov Y., et al. (2007). Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Invest. 117, 683–692. 10.1172/JCI28523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhang F., Li T., Lu M., Wang L., Yue W., et al. (2012). MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics 13:661. 10.1186/1471-2164-13-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R., Wolfinger M. T., Tanzer A., Hofacker I. L. (2016). Predicting RNA secondary structures from sequence and probing data. Methods 103, 86–98. 10.1016/j.ymeth.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Lu Q., Hu Y., Sun J., Cheng Y., Cheung K.-H., Zhao H. (2015). A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci. Rep. 5:10576. 10.1038/srep10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., et al. (2012). A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828. 10.1126/science.1215040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood K., Jung C.-H., Philip G., Georgeson P., Chung J., Pope B. J., et al. (2017). Variant effect prediction tools assessed using independent, functional assay-based datasets: implications for discovery and diagnostics. Hum. Genomics 11:10. 10.1186/s40246-017-0104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelotto L. G., Ng C. K., De Filippo M. R., Zhang Y., Piscuoglio S., Lim R. S., et al. (2014). Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 15:484. 10.1186/s13059-014-0484-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miosge L. A., Field M. A., Sontani Y., Cho V., Johnson S., Palkova A., et al. (2015). Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. U.S.A. 112, E5189–E5198. 10.1073/pnas.1511585112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M., Sterne-Weiler T., Li B., Ball E. V., Cooper D. N., Radivojac P., et al. (2014). MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol. 15:R19. 10.1186/gb-2014-15-1-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moszynska A., Gebert M., Collawn J. F., Bartoszewski R. (2017). SNPs in microRNA target sites and their potential role in human disease. Open Biol. 7:170019. 10.1098/rsob.170019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S. (2001). Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S. (2006). Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7, 61–80. 10.1146/annurev.genom.7.080505.115630 [DOI] [PubMed] [Google Scholar]
- Ohno K., Takeda J.-I., Masuda A. (2018). Rules and tools to predict the splicing effects of exonic and intronic mutations. Wiley Interdiscip. Rev. 9:e1451. 10.1002/wrna.1451 [DOI] [PubMed] [Google Scholar]
- Pan Y., Liu D., Deng L. (2017). Accurate prediction of functional effects for variants by combining gradient tree boosting with optimal neighborhood properties. PLoS ONE 12:e0179314. 10.1371/journal.pone.0179314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandurangan A. P., Ochoa-Montaño B., Ascher D. B., Blundell T. L. (2017). SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 45, W229–W235. 10.1093/nar/gkx439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Lin X., Salzberg S. L. (2001). GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 29, 1185–1190. 10.1093/nar/29.5.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A., Doughty E., Kann M. G. (2013). Towards precision medicine: advances in computational approaches for the analysis of human variants. J. Mol. Biol. 425, 4047–4063. 10.1016/j.jmb.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzón N., Li B., Martinez L., Sergeeva A., Presumey J., Apparailly F., et al. (2017). microRNA target prediction programs predict many false positives. Genome Res. 27, 234–245. 10.1101/gr.205146.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E., Ascher D. B., Blundell T. L. (2014a). DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 42, W314–W319. 10.1093/nar/gku411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E., Ascher D. B., Blundell T. L. (2014b). mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30, 335–342. 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A. (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F., Bernaerts K., Kwasigroch J. M., Rooman M. (2018). Quantification of biases in predictions of protein stability changes upon mutations. Bioinformatics 13:3031 10.1093/bioinformatics/bty348 [DOI] [PubMed] [Google Scholar]
- Quan L., Lv Q., Zhang Y. (2016). STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 32, 2936–2946. 10.1093/bioinformatics/btw361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D., Chen Y., Xie X. (2015). DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31, 761–763. 10.1093/bioinformatics/btu703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raida M., Schwabe W., Häusler P., Van Kuilenburg A. B., Van Gennip A. H., Behnke D., et al. (2001). Prevalence of a common point mutation in the Dihydropyrimidine dehydrogenase (DPD) gene within the 5'-splice donor site of intron 14 in patients with severe 5-fluorouracil (5-FU)-related toxicity compared with controls. Clin. Cancer Res. 7, 2832–2839. [PubMed] [Google Scholar]
- Raimondi D., Tanyalcin I., Ferté J., Gazzo A., Orlando G., Lenaerts T., et al. (2017). DEOGEN2: prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Res. 45, W201–W206. 10.1093/nar/gkx390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausell A., Mohammadi P., McLaren P. J., Bartha I., Xenarios I., Fellay J., et al. (2014). Analysis of stop-gain and frameshift variants in human innate immunity genes. PLoS Comput. Biol. 10:e1003757. 10.1371/journal.pcbi.1003757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M. G., Eeckman F. H., Kulp D., Haussler D. (1997). Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323. 10.1089/cmb.1997.4.311 [DOI] [PubMed] [Google Scholar]
- Reva B., Antipin Y., Sander C. (2011). Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39:e118. 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger J. K., Klein K., Winter S., Zanger U. M. (2013). Expression variability of absorption, distribution, metabolism, excretion-related micrornas in human liver: influence of nongenetic factors and association with gene expression. Drug Metab. Dispos. 41, 1752–1762. 10.1124/dmd.113.052126 [DOI] [PubMed] [Google Scholar]
- Ritchie G. R., Dunham I., Zeggini E., Flicek P. (2014). Functional annotation of noncoding sequence variants. Nat. Methods 11, 294–296. 10.1038/nmeth.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A. (2015). P, Integrative analysis of 111 reference human epigenomes. Nature 518, 317-330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. F., Shihab H. A., Mort M., Cooper D. N., Gaunt T. R., Campbell C. (2018). FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34, 511–513. 10.1093/bioinformatics/btx536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan B. C., Werner T. S., Howard P. L., Chow R. L. (2016). ImiRP: a computational approach to microRNA target site mutation. BMC Bioinformatics 17:190. 10.1186/s12859-016-1057-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample P. J., Wang B., Reid D. W., Presnyak V., McFadyen I., Morris D. R., et al. (2018). Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. bioRxiv. 10.1101/310375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Hamada M., Asai K., Mituyama T. (2009). CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37, W277–W280. 10.1093/nar/gkp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., Marshall A., et al. (2010). Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040. 10.1126/science.1186176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg D. R., Maquat L. E. (2012). Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 13, 246–259. 10.1038/nrg3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J. M., Cooper D. N., Schuelke M., Seelow D. (2014). MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362. 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- Shi Y. (2017). Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670. 10.1038/nrm.2017.86 [DOI] [PubMed] [Google Scholar]
- Shihab H. A., Gough J., Cooper D. N., Stenson P. D., Barker G. L., Edwards K. J., et al. (2013). Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34, 57–65. 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab H. A., Rogers M. F., Gough J., Mort M., Cooper D. N., Day I. N., et al. (2015). An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31, 1536–1543. 10.1093/bioinformatics/btv009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S. C., Kacevska M., Ingelman-Sundberg M. (2013). Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J. 13, 1–11. 10.1038/tpj.2012.45 [DOI] [PubMed] [Google Scholar]
- Smedley D., Schubach M., Jacobsen J. O. B., Köhler S., Zemojtel T., Spielmann M., et al. (2016). A whole-genome analysis framework for effective identification of pathogenic regulatory variants in mendelian disease. Am. J. Hum. Genet. 99, 595–606. 10.1016/j.ajhg.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somody J. C., MacKinnon S. S., Windemuth A. (2017). Structural coverage of the proteome for pharmaceutical applications. Drug Discov. Today 22, 1792–1799. 10.1016/j.drudis.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Soukarieh O., Gaildrat P., Hamieh M., Drouet A., Baert-Desurmont S., Frébourg T., et al. (2016). Exonic splicing mutations are more prevalent than currently estimated and can be predicted by using in silico tools. PLoS Genet. 12:e1005756 10.1371/journal.pgen.1005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear B. B., Heath-Chiozzi M., Huff J. (2001). Clinical application of pharmacogenetics. Trends Mol. Med. 7, 201–204. 10.1016/S1471-4914(01)01986-4 [DOI] [PubMed] [Google Scholar]
- Stone E. A., Sidow A. (2005). Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 15, 978–986. 10.1101/gr.3804205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Thomas P. D. (2016). Tools for predicting the functional impact of nonsynonymous genetic variation. Genetics 203, 635–647. 10.1534/genetics.116.190033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68-74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., et al. (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham C. M., Srinivasan N., Blundell T. L. (1997). Prediction of the stability of protein mutants based on structural environment-dependent amino acid substitution and propensity tables. Protein Eng. 10, 7–21. 10.1093/protein/10.1.7 [DOI] [PubMed] [Google Scholar]
- Venselaar H., Te Beek T. A., Kuipers R. K., Hekkelman M. L., Vriend G. (2010). Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics 11:548. 10.1186/1471-2105-11-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Qu K., Zhang Q. C., Flynn R. A., Manor O., Ouyang Z., et al. (2014). Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709. 10.1038/nature12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M. B., Berglund J. A. (2010). Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 35, 169–178. 10.1016/j.tibs.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvliet D. K., Strokach A., Giraldo-Forero A. F., Teyra J., Colak R., Kim P. M. (2016). ELASPIC web-server: proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity. Bioinformatics 32, 1589–1591. 10.1093/bioinformatics/btw031 [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Minor W., Dauter Z., Jaskolski M. (2008). Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 275, 1–21. 10.1111/j.1742-4658.2007.06178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Mullikin J. C., Elnitski L. (2010). Genomic features defining exonic variants that modulate splicing. Genome Biol. 11:R20. 10.1186/gb-2010-11-2-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Wigneshweraraj S. R., Weinzierl R., Wang Y.-P., Buck M. (2009). Construction and functional analyses of a comprehensive sigma54 site-directed mutant library using alanine-cysteine mutagenesis. Nucleic Acids Res. 37, 4482–4497. 10.1093/nar/gkp419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H. Y., Alipanahi B., Lee L. J., Bretschneider H., Merico D. R., Yuen R. K., et al. (2015). RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 347:1254806 10.1126/science.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates C. M., Filippis I., Kelley L. A., Sternberg M. J. (2014). SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J. Mol. Biol. 426, 2692–2701. 10.1016/j.jmb.2014.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Burge C. B. (2004). Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394. 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
- Yeo G., Holste D., Kreiman G., Burge C. B. (2004). Variation in alternative splicing across human tissues. Genome Biol. 5, R74–R15. 10.1186/gb-2004-5-10-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P., Melamud E., Moult J. (2006). SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics 7:166. 10.1186/1471-2105-7-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Lupski J. R. (2015). Non-coding genetic variants in human disease. Hum. Mol. Genet. 24, R102–R110. 10.1093/hmg/ddv259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fan X., Wang Y., Sun M.-A., Shao J., Guo D. (2017). BPP: a sequence-based algorithm for branch point prediction. Bioinformatics 33, 3166–3172. 10.1093/bioinformatics/btx401 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lin H., Zhao H., Hao Y., Mort M., Cooper D. N., et al. (2014). Impact of human pathogenic micro-insertions and micro-deletions on post-transcriptional regulation. Hum. Mol. Genet. 23, 3024–3034. 10.1093/hmg/ddu019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lv J., Liu H., Zhu J., Su J., Wu Q., et al. (2010). HHMD: the human histone modification database. Nucleic Acids Res. 38, D149–D154. 10.1093/nar/gkp968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Troyanskaya O. G. (2015). Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934. 10.1038/nmeth.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ingelman-Sundberg M., Lauschke V. M. (2017). Worldwide distribution of cytochrome P450 Alleles: a meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Therapeut. 102, 688–700. 10.1002/cpt.690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lauschke V. M. (2018). Comprehensive overview of the pharmacogenetic diversity in Ashkenazi Jews. J. Med. Genet. 55, 617–627. 10.1136/jmedgenet-2018-105429 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Mkrtchian S., Kumondai M., Hiratsuka M., Lauschke V. M. (2018). An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J. 28:1 10.1038/s41397-018-0044-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zia A., Moses A. M. (2011). Ranking insertion, deletion and nonsense mutations based on their effect on genetic information. BMC Bioinformatics 12:299. 10.1186/1471-2105-12-299 [DOI] [PMC free article] [PubMed] [Google Scholar]