Abstract
The use of copper complexes for redox and oxidative-based mechanisms in therapeutic strategies is an important field of multidisciplinary research. Here, a novel Cu(II) complex [Cu(TPMA)(Phen)](ClO4)2 (Cu-TPMA-Phen, where TPMA = tris-(2-pyridylmethyl)amine and Phen = 1,10-phenanthroline) was studied using both the free and encapsulated forms. A hollow pH-sensitive drug-delivery system was synthesized, characterized, and used to encapsulate and release the copper complex, thus allowing for the comparison with the free drug. The human neuroblastoma-derived cell line NB100 was treated with 5 μM Cu-PMA-Phen for 24 h, pointing to the consequences on mono- and polyunsaturated fatty acids (MUFA and PUFA) present in the membrane lipidome, coupled with cell viability and death pathways (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium viability assay, flow cytometry, microscopy, caspase activation). In parallel, the Cu-TPMA-Phen reactivity with the fatty acid moieties of phospholipids was studied using the liposome model to work in a biomimetic environment. The main results concerned: (i) the membrane lipidome in treated cells, involving remodeling with a specific increase of saturated fatty acids (SFAs) and a decrease of MUFA, but not PUFA; (ii) cytotoxic events and lipidome changes did not occur for the encapsulated Cu-TPMA-Phen, showing the influence of such nanocarriers on drug activity; and (iii) the liposome behavior confirmed that MUFA and PUFA fatty acid moieties in membranes are not affected by oxidative and isomerization reactions, proving the different reactivities of thiyl radicals generated from amphiphilic and hydrophilic thiols and Cu-TPMA-Phen. This study gives preliminary but important elements of copper(II) complex reactivity in cellular and biomimetic models, pointing mainly to the effects on membrane reactivity and remodeling based on the balance between SFA and MUFA in cell membranes that are subjects of strong interest for chemotherapeutic activities as well as connected to nutritional strategies.
1. Introduction
Among antitumoral active metallodrugs, copper(II) complexes are particularly interesting due to the fact that they can undergo redox activity upon in vivo reduction to Cu(I) and, together with DNA binding properties, cause genome damage by mediating DNA strand cleavage.1−7 The increased requirement of copper in cancer cells for redox metabolism and its catalytical properties to generate reactive oxygen species (ROS) are the biological and chemical basis, respectively, for the anticancer activity of these drugs, which act as artificial metallonucleases (AMNs) for the sequence-specific disruption of gene function.1,8,9
One of the first discovered antitumoral drugs of this kind was bleomycin (BLM). Its capabilities to both chelate redox-active metal centers (i.e. Fe(III) or Cu(II)) and subsequently bind to DNA endow metallobleomycin with AMN activity, whereby strand cleavage is mediated in the presence of O2 and a reductant by a metal-centered BLM oxidant.10 This drug has been recently investigated by some of us highlighting that: (i) the bleomycin–iron complex can interact not only with DNA but also with the membrane lipids due to the ligand lipophilic characteristics and (ii) the presence of thiols, as biologically relevant reducing agent, leads to the generation of thiyl radicals under aerobic conditions.11,12 These two events were found to induce a profound membrane remodeling of fatty acids, with saturated fatty acid (SFA) content increased at the expense of mono- and polyunsaturated fatty acids (MUFA and PUFA). Together with the process of lipid peroxidation induced by ROS production, the cis–trans interconversion of the geometry of unsaturated fatty acids in membranes occurred catalyzed by thiyl radicals generated endogenously under condition of stress.13,14 The biomimetic model of liposomes, made of SFA-, MUFA-, and PUFA-containing phospholipids, was also reported to model the behavior of lipid isomerization, as well as the PUFA consumption, under biologically related free-radical and oxidative conditions. The reactivity information helped for the mechanistic interpretation of experiments carried out in cell cultures.11
Lipid metabolism is of crucial importance for cancer cells due to their active proliferation, and a large amount of lipids made of SFA, MUFA, and PUFA for the synthesis of new membranes is needed. The biosynthesis of SFA, such as palmitic acid (16:0) (produced by fatty acid synthase) or stearic acid (18:0), and their subsequent desaturation to MUFA (palmitoleic acid (Δ-9 16:1) and oleic acid (Δ-9 18:1)) are now highlighted for their influence on the biophysical nature of the tumor cell membrane and on cancer cell signaling in proliferation and survival.15−19 With SFA, MUFA, and PUFA being present as dietary intakes, the fatty acid balance can also influence drug effects and interactions. Under these aspects, lipidomic studies provide important information for new discoveries with practical consequences in cancer treatments.20 In such aspects of fatty acid metabolism, the influence of copper complexes is not known.
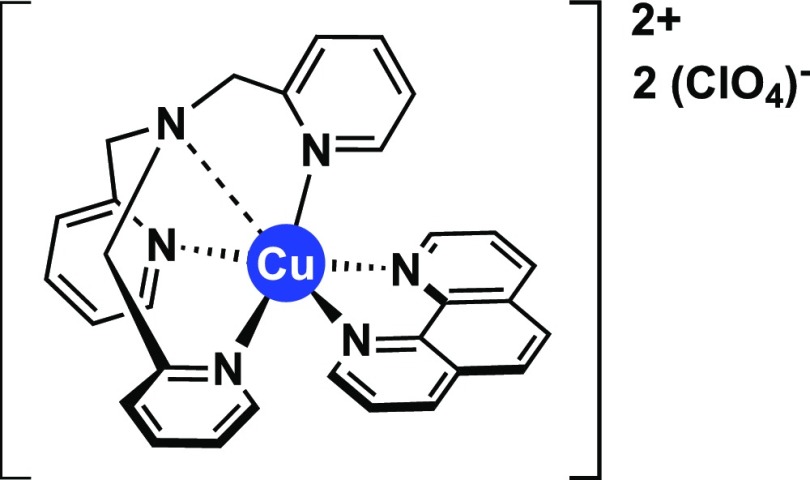
On the basis of these premises, the biomimetic and biological effects of the synthetic chemical nuclease [Cu(TPMA)(Phen)](ClO4)2 (Cu-TPMA-Phen, where TPMA = tris-(2-pyridylmethyl)amine and Phen = 1,10-phenanthroline) were examined (Figure 1). We have recently reported the synthesis and potent DNA cleavage properties of Cu-TPMA-Phen,21 which was rationally designed to combine the catalytic stabilizing effect of TPMA22−24 with the DNA oxidation properties of copper(II) phenanthroline.24−27 As the Cu-TPMA-Phen molecule belongs to a series of metal-containing reagents that induce chemical DNA scission, it may well fall into the group of AMNs with cytotoxic properties against human cancer cells.1,2,28,29
Figure 1.
Molecular structure of Cu-TPMA-Phen.
The cell model used in our biological and cytotoxicity evaluation of Cu-TPMA-Phen was the human neuroblastoma-derived NB100 cell line. Cell death pathway (apoptosis vs necrosis) triggered by Cu-TPMA-Phen was investigated. In parallel to cytotoxicity experiments, membrane fatty acids were analyzed by gas chromatography (GC), to obtain (i) the membrane profiles related to the drug administration and (ii) an insight into the fatty acid pathways that are influenced by the drug.
As means of comparison with the in vitro effects, a biomimetic model of liposomes treated with the copper complex was also used, as previously described for bleomycin.11 The radical and oxidative processes were followed up using a variety of reaction conditions, thereby providing a molecular basis to observe these processes.
Finally, the use of this complex was carried out in nanoscale drug-delivery systems, capable of releasing drugs in both retarded and protected ways. In recent research, drug encapsulation systems were successful at releasing the therapy directly into cancer cells, providing a valuable strategy to overcome the lack of selectivity in conventional chemotherapeutics.30,31 The properties of these systems are crucial for cancer targeting32−34 as much as the biological environment characteristics, which influence the accumulation of the delivery system and the drug effects.35 The capability of responding to a number of specific stimuli, related to particular tumor characteristics, such as acidic pH, different temperature, or reductive environment, is an important advancement in this field and can be utilized to achieve a specific drug delivery.36,37 Indeed, one of the major features of tumor tissues is the acidic extracellular pH that is due to lactate secretion from anaerobic glycolysis. To date, many polymeric pH-responsive delivery systems such as nanoparticles38,39 and micelles40,41 have been developed to take advantage of the pH values related to pathological conditions. These systems present physical properties such as swelling/deswelling, particle disruption and aggregation, and protonation/deprotonation of the main functional groups that change in response to different environmental conditions. By choosing the most appropriate components for their synthesis, properties can be fine-tuned to achieve the release of the loaded drug only under the pH condition of interest.37 Here, a hollow pH-sensitive drug-delivery system was synthesized, characterized, and used to encapsulate and release Cu-TPMA-Phen. Cell experiments were carried out comparing free and encapsulated Cu-TPMA-Phen formulations.
The interesting potentialities of copper complexes are delineated below by a multidisciplinary approach using chemical, biological, and pharmacological knowledge and the know-how to extend the boundary of this fast-growing research field.
2. Results and Discussion
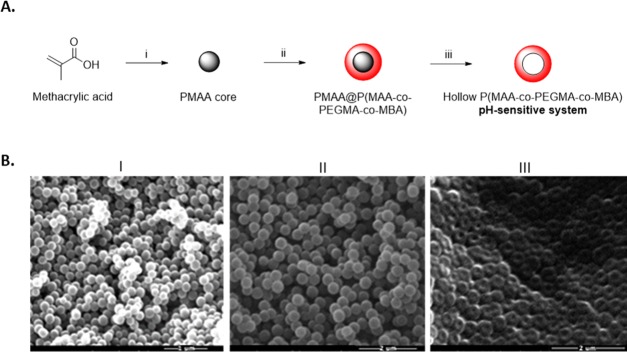
2.1. Synthesis of Hollow P(MAA-co-PEGMA-co-MBA) Nanocontainers (NCs)
The hollow pH-sensitive NCs were synthesized by a three-step process (Figure 2A). First, the poly(methacrylic acid) (PMAA) cores were obtained with the distillation–precipitation polymerization method. These cores were sacrificial templates synthesized with no cross-linking agent, to be easily removed at the end of the synthetic pathway.42,43 The second step was the synthesis of the pH-sensitive shell with the distillation–precipitation polymerization method, giving a core–shell structure.44,45 Finally, the cores were removed to obtain the hollow pH-sensitive NCs. The NCs owe the pH sensitivity to methacrylic acid (MAA), used as the main monomer for the synthesis of the shell: its polymer presents carboxylic groups that can be protonated or deprotonated depending on the pH of the medium (pKa ca. 4.5) and thus differently interact with the surrounding environment.46 Other components were used in the synthetic process for the stability of the final system. N,N′-Methylenebisacrylamide (MBA) was used as a cross-linking agent, to maintain the structure of the hollow NCs in water; poly(ethylene glycol)methyl ether methacrylate (PEGMA) was used as a hydrophilic, nontoxic component, which is known to show resistance against nonspecific protein adsorption and prolong in vivo circulation time of drug-delivery systems.47
Figure 2.
(A) Scheme of the synthesis of the pH-sensitive NCs: (i) distillation–precipitation polymerization method using MAA, 2,2′-azobisisobutyronitrile (AIBN), and N2 bubbling in ACN; (ii) distillation–precipitation polymerization method using MAA, PEGMA, MBA, and N2 bubbling in acetonitrile (ACN); and (iii) core removal in EtOH/H2O dist. (B) Scanning electron microscopy (SEM) pictures taken after each synthesis step: (I) PMAA cores; (II) PMAA@P(MAA-co-PEGMA-co-MBA); and (III) hollow P(MAA-co-PEGMA-co-MBA) NCs.
SEM was used to investigate the size, shape, and the degree of polydispersity of the samples. Figure 2B depicts the nanoparticles as spherical and monodisperse. In particular, the diameters of the cores and PMAA@P(MAA-co-PEGMA-co-MBA) were 190 ± 15 and 350 ± 15 nm, respectively, and the size of the hollow P(MAA-co-PEGMA-co-MBA) was approximately 350 ± 15 nm. The successful coating of the PMAA cores was confirmed by the increase in size between the NPs in I and II (from roughly 190 to 350 nm), whereas the central cavity of the NCs in III proved the success of the core removal procedure. We noted that the hollow NCs in Figure 2III appeared flattened; therefore, the measurements of their diameter were taken as an approximation. Finally, the synthesis of the pH-sensitive shell and the removal of the cores were also confirmed with Fourier transform infrared (FT-IR) (Figure S1 in the Supporting Information).
2.2. Loading and Release Behavior
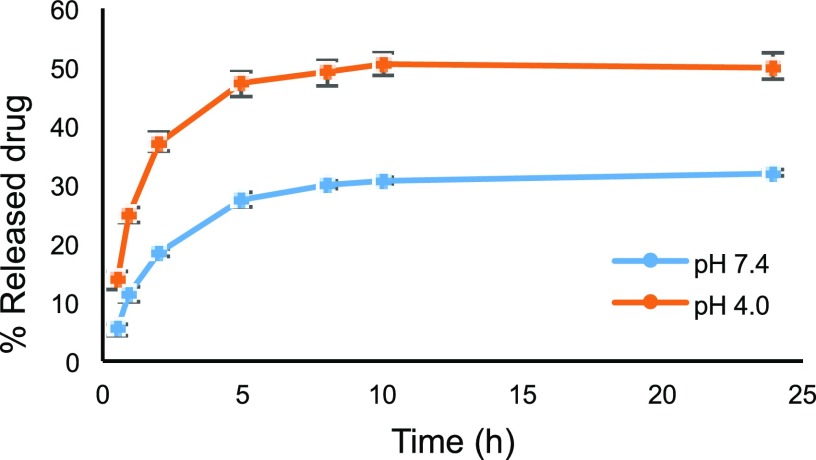
The loading process depends on the capability of the drug to interact with the hollow NCs, specifically via electrostatic interactions and hydrogen-bonding interactions involving the pendant functional groups of the NCs and the solubilized drug.48,49 Cu-TPMA-Phen is a water-insoluble molecule, and, to run the encapsulation experiments, it was necessary to find the adequate aqueous conditions that could both properly suspend the NCs and completely solubilize the drug. Cu-TPMA-Phen was therefore solubilized in the smallest possible volume of ACN before mixing it with the NCs suspension in buffer. To solubilize 2 mg of Cu-TPMA-Phen needed for the experiment, 50 μL of ACN was used and this solution was then added to a previously prepared suspension of 1 mg of NCs in 950 μL of phosphate-buffered saline (PBS). The final encapsulation conditions were: 2 mg of Cu-TPMA-Phen and 1 mg of NCs in 1 mL of a mixture of PBS/ACN (19:1). The encapsulation process resulted in encapsulation efficiency (EE%) and loading capacity (LC%) to be, respectively, 36.4 ± 5.2 and 42.0 ± 3.3. This corresponds to the encapsulation of 0.724 mg (0.988 μmol) of Cu-TPMA-Phen/mg of NCs. The main interaction involved in the encapsulation of Cu-TPMA-Phen is probably the electrostatic interaction between the negatively charged carboxylate anions of PMAA and the 2+ positive charge, which the complex assumes upon dissolution of the perchlorate counterions. In addition, considering the electron paramagnetic resonance (EPR) spectra and X-ray analysis previously reported,21 a distal pyridine nitrogen donor atom of TMPA was identified within the coordination complex and may therefore interact with the NCs through hydrogen bonding. In terms of loading, the large central cavity of the NCs plays a fundamental role in accommodating the drug,48 which explains the overall impressive results of the encapsulation process. The Cu-TPMA-Phen release profile from the hollow NCs was studied in both acidic and slightly basic environments to prove the pH-sensitive drug-delivery system. To release and detect the Cu-TPMA-Phen with no interference owing to its solubility, two mixtures of buffer and ACN were used. In particular, the release in acidic pH was studied in a mixture of 0.1 M citrate buffer of pH 4 and ACN (19:1, v/v), whereas the slightly basic release medium was a mixture of PBS and ACN (19:1, v/v). After 24 h, the amount of released drug was, respectively, 50 and 32%, as shown in Figure 3. To investigate whether the amount of ACN used for preparing the media affects the drug-release profile, the same experiment was carried out by increasing the amount of ACN in the mixtures up to 9:1. The drug-release profiles obtained in these conditions were identical. The carboxylic groups of PMAA are the key for the interpretation of these results. Indeed, they are mostly protonated at pH 4 (pKa ∼ 4.5); therefore, they cannot interact with the positively charged Cu-TPMA-Phen complex, causing the release. In addition, it is known that at pH 4, the hydrogen-bonding interactions are weaker than in neutral conditions,44 facilitating the release.
Figure 3.
Cu-TPMA-Phen release profile of the pH-sensitive NCs.
2.3. EPR Study of Released Cu-TPMA-Phen
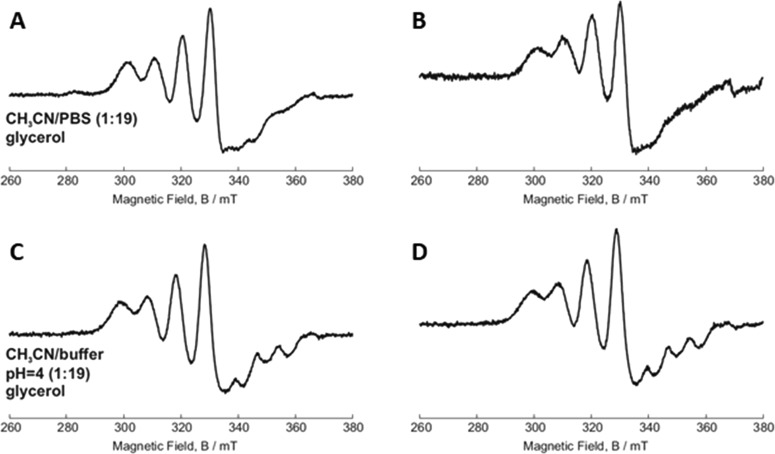
The EPR technique was used to investigate the structure of the free and released compounds and highlight possible modifications occurring during the encapsulation/release processes.21 Once Cu-TPMA-Phen is released from the hollow NCs, it is necessary to verify whether its starting conformation is kept, to exert the biological activity. Figure 4A,C shows the spectra of free Cu-TPMA-Phen dissolved in the release media in ACN/PBS (1:19) and ACN/0.1 M citrate buffer at pH 4 (1:19), respectively. In these conditions, Cu-TPMA-Phen shows a clear trigonal bipyramidal conformation, with five atoms coordinating the Cu(II) center. This structure is due to interactions between water and the distal nitrogen atom outlined above. As such, when this nitrogen donor does not coordinate the Cu(II) center, its lone pair is available to form a hydrogen bond with a vicinal water molecule. In this condition, the N is ligated outside the first coordination sphere, which results in a five-coordination structure and a clear trigonal bipyramidal spectrum.50−52Figure 4B,D shows the EPR spectra of released Cu-TPMA-Phen in ACN/PBS (1:19) and ACN/0.1 M citrate buffer pH 4 (1:19), respectively. The spectra are almost identical to the free complex, which is a strong indication that the encapsulation and release processes do not affect the structure of the complex.
Figure 4.
Continuous-wave (cw) EPR of free and released Cu-TPMA-Phen: (A) free Cu-TPMA-Phen in ACN/PBS (1:19); (B) released Cu-TPMA-Phen in ACN/PBS (1:19); (C) free Cu-TPMA-Phen in ACN/0.1 M citrate buffer of pH 4 (1:19); and (D) released Cu-TPMA-Phen in ACN/0.1 M citrate buffer pH 4 (1:19).
2.4. Intracellular Distribution of Cu-TPMA-Phen-Loaded Nanocontainers
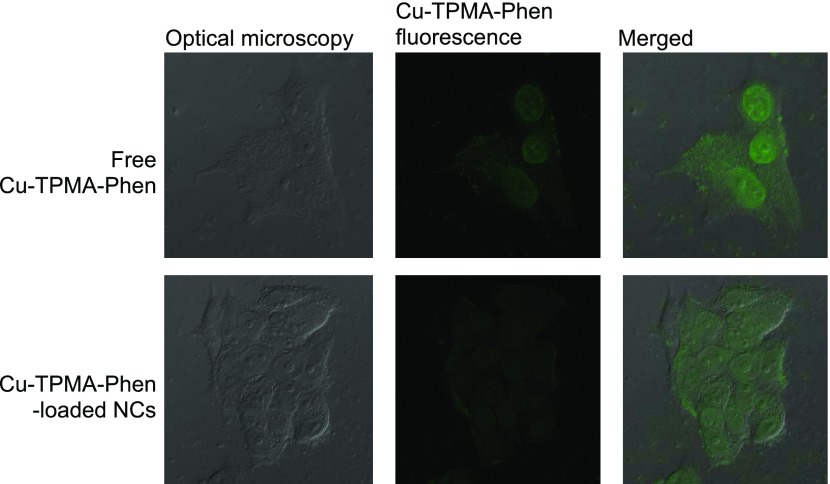
Confocal laser scanning microscopy (CLSM) was used to investigate the drug release and the effect of the encapsulation on the intra- and subcellular localization of Cu-TPMA-Phen by taking advantage of its intrinsic fluorescence signal, which was successfully applied in related studies involving daunorubicin.53 As shown in Figure 5, after 2 h treatment, both free Cu-TPMA-Phen and Cu-TPMA-Phen released from the pH-sensitive NCs were localized mostly in the nuclei of MCF7 cancer cells. Considering the intensities, the signal belonging to the encapsulated Cu-TPMA-Phen is less intense than that belonging to free drug. These observations suggest that the pH-sensitive NCs do not affect the intracellular localization of the drug, and NC is a drug delivery system that transports and delivers the drug without affecting its activity.54 On the other hand, the difference in intensity was expected: after 2 h in acidic environment (such as endosomes–lysosomes, by virtue of which the NCs are most likely internalized), the NCs release was roughly 35% of the encapsulated drug.
Figure 5.
Confocal laser scanning confocal microscopy images of MCF7 cells for 2 h treated with: (a–c) free Cu-TPMA-Phen (concentration 10 μM) and (d–f) Cu-TPMA-Phen-loaded NCs (drug concentration 10 μM).
2.5. Cytotoxicity of Cu-TPMA-Phen, Free or Encapsulated in pH-Sensitive Nanocontainers
Cell viability assays were performed to assess the toxicity effect of Cu-TPMA-Phen, when the latter is encapsulated in pH-sensitive nanocarriers. The encapsulation of this complex in nanocontainers was carried out as described above. The resulting viability curves were compared to those obtained after treatment with free Cu-TPMA-Phen at the same concentration range, 0.1–30 μM, as shown in Figure 6. Incubation with 3, 10, and 30 μM Cu-TPMA-Phen for 24 h results in significantly different viabilities between cells treated with free and encapsulated complexes (p < 0.0001, 3 and 10 μM and p < 0.01, 30 μM) (Figure S4 in the Supporting Information for the analogous experiments at 48 h). However, it should be considered that these differences do not indicate a lower activity of the NCs because the maximum release is at pH 4.0, which is a condition easily obtainable in the lysosomal compartment of the tumoral cells, but quite far from cell culture conditions.37,55 These differences in cytotoxicity can be attributed to the less amount of bioavailable drug. In fact, as shown in Figure 3, only one-third of Cu-TPMA-Phen is released from nanocontainers at the physiologic pH 7.4, which is similar to cell culture conditions.
Figure 6.
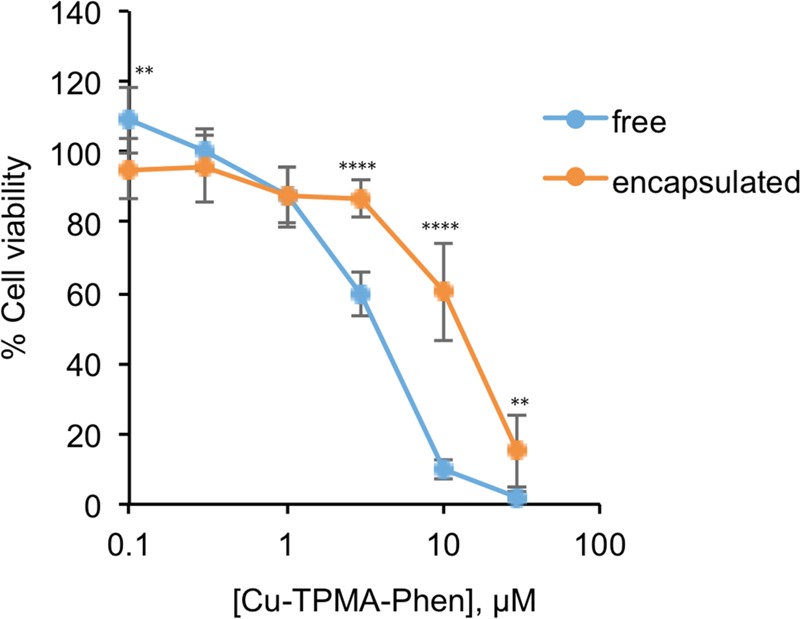
Dose-dependent response of NB100 cells treated with free or encapsulated Cu-TPMA-Phen for 24 h. The results are presented as mean ± standard deviation (SD) of three independent experiments performed in triplicate, representing the percentage of control values obtained from cultures grown in the absence of the complex. Statistical analysis was performed with unpaired t test. ** p < 0.01, **** p < 0.0001.
2.6. Effects of Cu-TPMA-Phen on Cell Viability
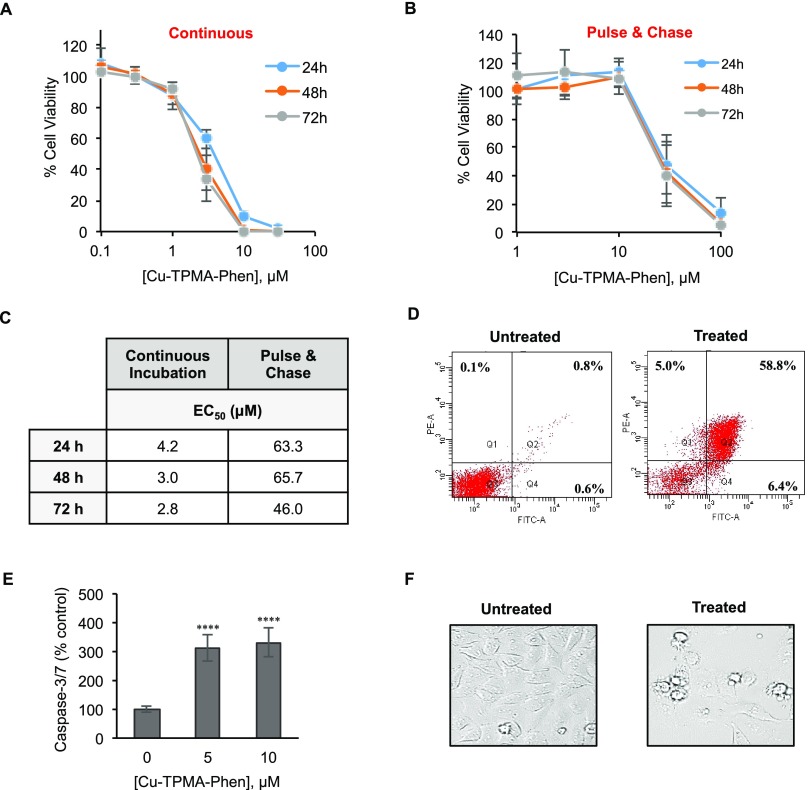
Neuroblastoma cells (NB100) were exposed to different concentrations of Cu-TPMA-Phen (0.1–30 μM) for 24, 48, and 72 h. Cell toxicity of free Cu-TPMA-Phen was determined using an 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell viability assay.56,57 Dose-dependent curves were derived (Figure 7A), and the half-maximal effective concentration (EC50) values were calculated. The EC50 was 4.2 μM (R2 = 0.97) after 24 h of continuous incubation with the complex. Cell viability was also evaluated in a pulse and chase experiment, in which NB100 cells were treated for 2 h with various concentrations of Cu-TPMA-Phen (1–100 μM) and then washed and incubated with complete medium for 24, 48, and 72 h (Figure 7B). The concentrations effective to reduce cell viability of 50% are reported in Figure 7C. It should be noted that 2 h of exposure to the complex can be enough to ensure a strong cytotoxicity. In fact, the comparison of EC50 calculated for continuous and pulse and chase experiments shows only one log difference.
Figure 7.
Cytotoxic effect of Cu-TPMA-Phen on NB100 cells. (A) Cell viability curves for NB100 cells continuously exposed to different doses of Cu-TPMA-Phen for 24, 48, and 72 h. (B) Cell viability pulse and chase curves of NB100 cells exposed at various doses of Cu-TPMA-Phen for 2 h and then incubated in complete medium for 24, 48, and 72 h. Cell viability was measured by MTS assay and expressed as a percentage of untreated cells. The results in (A) and (B) are presented as mean ± SD of three independent experiments performed in triplicate. (C) Half-maximal effective concentration (EC50) values were calculated by nonlinear regression with standard slope analysis. (D) The cell death pathway triggered by 5 μM Cu-TPMA-Phen was evaluated on NB100 cells after 24 h of treatment by Annexin V/propidium iodide (PI) staining and flow cytometry analysis. Fluorescein isothiocyanate (FITC)-A channel is used for the detection of Annexin V–enhanced green fluorescent protein (EGFP) fluorescence. Phycoerythrin (PE)-A channel is used for the detection of PI fluorescence. Representative plots of Annexin V/PI staining are shown; apoptotic (Q4, Annexin V+/PI−), necrotic (Q1, Annexin V–/PI+), and late-stage apoptotic cells (Q2, Annexin V+/PI+). (E) Caspase 3/7 activation in NB100 cells exposed to 5 and 10 μM Cu-TPMA-Phen for 24 h. The expression of activated caspases is reported as a percentage of untreated cell values. Mean ± SD of three independent experiments, each in triplicate, are given. Statistical significance was determined by unpaired t test (**** p < 0.0001). (F) Morphological analysis of NB100 cells treated with 5 μM Cu-TPMA-Phen for 24 h, using phase contrast microscopy (400×).
To further study the cell death pathway, cytofluorimetric analysis of Annexin V/PI double staining of NB100 cells treated with Cu-TPMA-Phen was carried out. This analysis indicated that NB100 cells treated for 24 h with 5 μM Cu-TPMA-Phen underwent apoptotic cell death. Treated and untreated cells were stained with Annexin V–FITC and PI to differentiate apoptosis versus necrosis. After treatment with 5 μM Cu-TPMA-Phen for 24 h, 59% of the copper complex, two-thirds of cell population, was in apoptosis and 5% of cells underwent necrotic death (Figure 7D). The low amount of necrotic cells measured in our experiments can represent an advantage for a possible therapeutic use of this complex. In fact, necrosis, contrary to apoptosis, causes inflammation that can be responsible for unwanted toxicity toward surrounding normal tissue. In parallel, to confirm the apoptotic cell death pathway, the caspase 3/7 activity was assessed in NB100 cells treated with 5 and 10 μM Cu-TPMA-Phen for 24 h in comparison with untreated (control) cells (Figure 7E). At both concentrations, caspases 3/7 were strongly activated in Cu-TPMA-Phen-treated cells, reaching values higher than 300% that of control cells. Finally, cell morphology was analyzed by phase contrast microscopy on NB100 cells incubated with 5 μM Cu-TPMA-Phen for 24 h. Treated cells showed typical apoptotic morphological features (Figure 7F).
2.7. Effects of Cu-TPMA-Phen on Membrane Lipidome
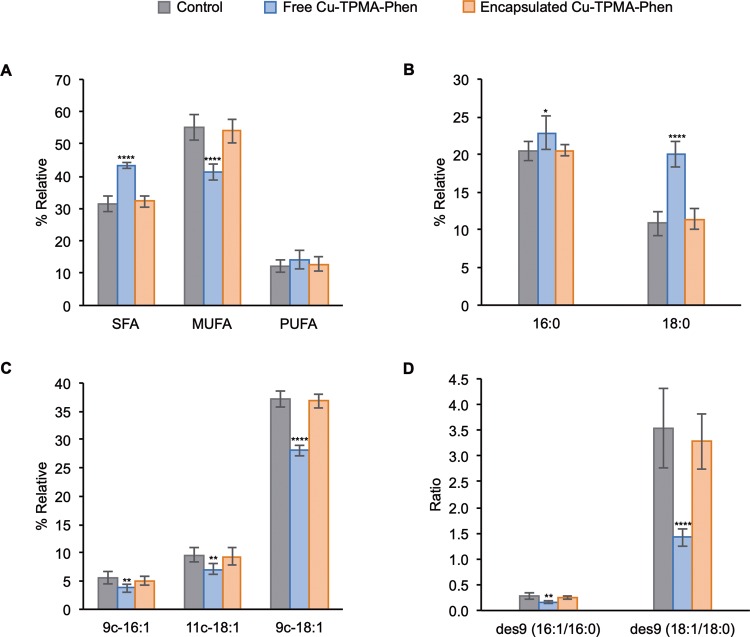
Using the cytotoxicity parameters identified above, NB100 cells were treated with 5 μM Cu-TPMA-Phen for 24 h (N = 6) and underwent fatty acid-based membrane lipidomic analyses. Membrane fatty acids were isolated, derivatized, and analyzed by gas chromatography (see Table S1 for details). Membrane fatty acid-based lipidomics analysis on NB100 after 24 h treatment revealed a significant increase of saturated fatty acids (SFAs) (p < 0.0001) accompanied by a parallel decrease of their monounsaturated (MUFA) counterparts (p < 0.0001) (Figure 8A). The family of polyunsaturated fatty acids (PUFAs) did not show significant alterations between treated and untreated cells. In particular, the main members of SFA family, palmitic and stearic acids, are significantly increased (Figure 8B), whereas the members of MUFA family, palmitoleic (9c-16:1), vaccenic (11c-18:1), and oleic (9c-18:1) acids, showed significantly decreased levels in NB100 cells exposed to Cu-TPMA-Phen (Figure 8C). The enzymatic activity of stearoyl-coA desaturase, which catalyzes the conversion of saturated fatty acids to monounsaturated fatty acids, calculated by the product-to-precursor fatty acid ratio, was estimated to be 2-fold decreased (p < 0.0001) (Figure 8D).58,59 Membrane lipidomics analysis was also performed on NB100 cells treated with 5 μM of pH-sensitive nanocarriers encapsulated Cu-TPMA-Phen. In this case, the impact on membrane lipidome presents no significant difference between untreated and treated cells (Figure 8A–D). The membrane lipidomic experiments were carried out also in the breast cancer-derived MCF7 cell line. The aim of this was to ascertain that the above-described membrane remodeling is not specific for the neuroblastoma cell line NB100, but can be extended to other cancer models. MCF7 cells were exposed to 10 μM Cu-TPMA-Phen, followed by membrane fatty acid analysis. Interestingly, Cu-TPMA-Phen shows a similar effect on cell membrane for both cell lines, although MCF7 and NB100 are cells of different origins, carcinoma and neuroblastoma, respectively (Figure S3 in the Supporting Information).
Figure 8.
Fatty acid-based membrane lipidomics on NB100 cells treated with 5 μM free or encapsulated Cu-TPMA-Phen for 24 h. (A) Relative distribution of fatty acid families; SFA: saturated fatty acid, MUFA: monounsaturated fatty acid, PUFA: polyunsaturated fatty acid. (B) Palmitic (16:0) and stearic (18:0) acids trends in treated cells. (C) Palmitoleic (9c-16:1), vaccenic (11c-18:1), and oleic (9c-18:1) patterns in treated cells. (D) Estimation of Δ-9 desaturase (des9) activity by the ratios palmitoleic/palmitic and oleic/stearic. Values represent mean ± SD (n = 6). Statistical significance was calculated with unpaired t test against the control groups. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
The results obtained from the viability and apoptosis experiments together with the results obtained from membrane fatty acid-based lipidomics suggest an interesting behavior arising from oxidative conditions typically associated with copper complex exposure.1,60 In fact, cell membranes exposed for 24 h to the free Cu-TPMA-Phen do not show diminution of the PUFA residues of phospholipids, thus suggesting that no interaction of these oxidizing molecules occurs. Instead, the increase of SFA was associated with MUFA diminution, which is not the most oxidizable lipid. Therefore, such decreases may be attributed to a “metabolic” rather than a “chemical” oxidative effect of Cu-TPMA-Phen. The increase of SFA can change membrane properties toward less permeability and fluidity.61 The changes of membrane properties can trigger several signals that lead to apoptosis, as demonstrated by supplementation of palmitic acid to NB100 cells.43
The inhibition of desaturase is known to induce cancer cell death and is indeed inspiring new pharmacological strategies in anticancer therapy.62 In the case of Cu-TPMA-Phen complex, more work is needed to understand the molecular basis of such membrane remodeling, its possible involvement in desaturase inhibition, and the association of this remodeling with the apoptotic fate here observed.
In this work, we observed the absence of effects of copper complex when encapsulated in pH-sensitive nanocarriers, which do not release sufficient drug doses for membrane remodeling, but suggest further studies on the control of oxidative effects by drug delivery in the sense of enhanced tumor targeting.63
2.8. Role of Cu-TPMA-Phen in the Geometrical Isomerization of 1-Palmitoyl 2-oleoyl Phosphatidylcholine (POPC) in Large Unilamellar Vesicles Obtained with the Extrusion Technique (LUVETs)
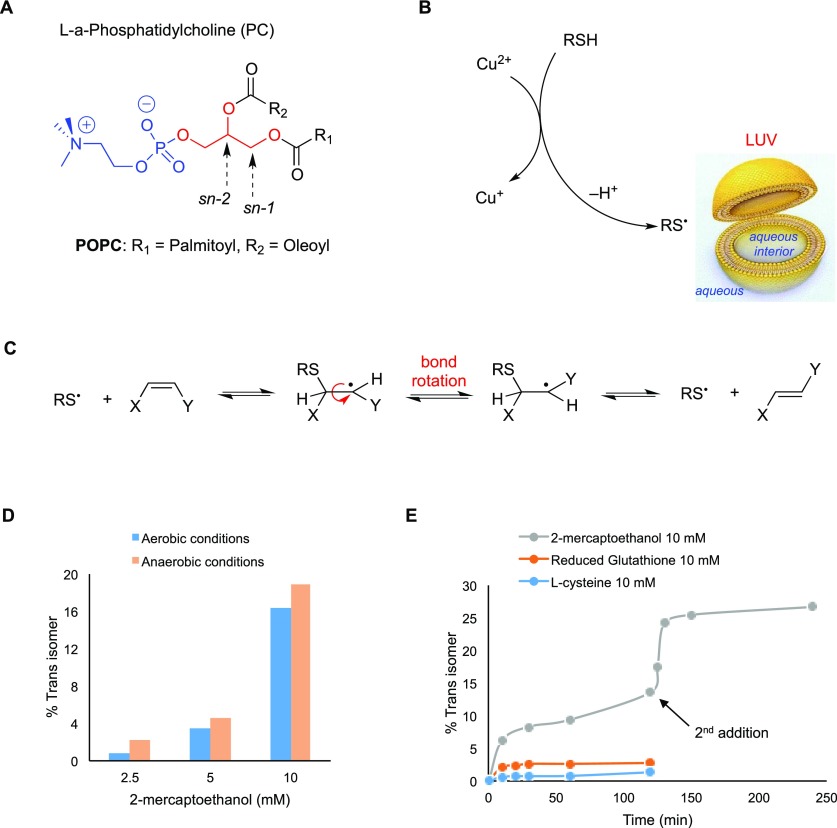
It has been recently established that the well-known bleomycin–iron complex does not generate oxidative damage only to target DNA, but it also induces a profound membrane remodeling at the level of the fatty acid constituents.11,12 The reaction of metal complexes with thiols, which are active biomolecules, can lead to the generation of thiyl radicals that either catalyze a cis–trans isomerization of double bonds and/or initiate a lipid peroxidation.64 The biomimetic model of liposomes—made of phospholipids—was designed and used to follow the fatty acid fate.11 In this section, we applied this biomimetic model to study the reaction of Cu-TPMA-Phen with thiols in vesicle suspensions made of l-α-phosphatidylcholine derivatives (Figure 9).
Figure 9.
(A) Molecular structures of l-α-phosphatidylcholine; in the sn-1 and sn-2 positions of the glycerol moiety, the two fatty acid residues are attached, whereas in the sn-3 position, the polar head group is connected. (B) A large unilamellar vesicle (LUV) and the generation of thiyl radicals by reaction of thiol with Cu-TPMA-Phen. (C) Reaction mechanism for the cis–trans isomerization catalyzed by thiyl radicals. (D) Trans isomer (elaidate) formation in POPC vesicles treated with 0.15 mM Cu-TPMA-Phen and different concentrations of 2-mercaptoethanol under aerobic or anaerobic conditions. (E) Trans isomer (elaidate) formation in POPC vesicles treated with 10 mM concentration of different thiols, 0.15 mM Cu-TPMA-Phen, and incubated at 37 °C under aerobic conditions. In the case of 2-mercaptoethanol, after 120 min, another 10 mM thiol was added.
As far as the membrane biomimetic model is concerned, large unilamellar vesicles were obtained by the extrusion technique (LUVETs).65 The vesicles were suspended in an aqueous medium containing Cu-TPMA-Phen and 2-mercaptoethanol or biothiol, like l-cysteine (CySH) and glutathione (GSH), applying anaerobic or aerobic conditions. The fatty acid content of the vesicles was SFA and MUFA in the form of oleic acid, when POPC was the phospholipid used for vesicle formation. In the liposomes composed of soybean lecithin, different percentages of SFA, MUFA, and PUFA fatty acid residues were evaluated.66
By mixing Cu-TPMA-Phen with 2-mercaptoethanol in aqueous solution, the formation of an adduct is instantaneous, as evidenced by UV–vis spectral features. In fact, the absorption band of Cu(II) complex with a maximum at 626 nm decreased, and at the same time, a new absorbance at 412 nm due to Cu(I) complex arose with the formation of the corresponding disulfide (Figures 9B and S4 in the Supporting Information), in analogy with the reaction of ascorbic acid. It is important to underline that, in the LUVET suspensions made of POPC, when the thiol solution was added at once to the solution in the presence of Cu-TPMA-Phen, the trans isomer of the oleic moiety (i.e., 9-trans-18:1 as a result of the cis–trans isomerization) was observed only in traces, even after prolonged incubation (4 h at 37 °C) (Figure 9C). However, adding the thiol dropwise (by a syringe pump, 0.5 mM/min), a substantial increase of the trans isomer was detected.
Different ratios of Cu-TPMA-Phen and 2-mercaptoethanol were then used as reagents in LUVET suspensions. Figure 9D shows the outcome of the reaction between 0.15 mM Cu-TPMA-Phen and 2.5, 5, and 10 mM of thiol under aerobic or anaerobic conditions, where as much as 19% of trans isomer formation was reached. The yield from the initial fatty acid composition to the final mixture was quantitatively determined by GC analysis. The progressive increase in the amount of Cu-TPMA-Phen up to 1 mM showed a parallel decrease in trans isomer formation (Figure S5 in the Supporting Information).
Next, we considered the effect of biothiols, such as l-cysteine and glutathione, in the same system. These two compounds differ from 2-mercaptoethanol since they have a hydrophilic nature. Figure 9E shows the time profiles of 2-mercaptoethanol, CySH, and GSH in LUVETs under identical conditions. Comparison of the three different thiols suggests that the isomerization rate follows the lipophilicity order of the three compounds (i.e., 2-mercaptoethanol > GSH > CySH) and indicates that the CyS• radical is unable to migrate into the lipid compartment.67,68Figure 9E also shows that further addition of 2-mercaptoethanol after 2 h with the remaining Cu(II) produced more thiyl radicals and, consequently, more of the trans isomers. After replacing POPC with soybean lecithin (SFA 18%, MUFA 13%, and PUFA 69%) in the biomimetic model described above, it was observed that (i) up to 120 min, the trans isomers of MUFA and PUFA are below 0.6 and 4%, respectively, independently from the presence or absence of oxygen and (ii) there is a consumption of 4 and 6% of PUFA moieties in aerobic and anaerobic conditions, respectively. These results concerning the oxidative and cis–trans lipid isomerization processes, obtained in the biomimetic model of POPC vesicles, are informative of the molecular reactivity of the complex, thereby explaining at least, in part, the absence of trans isomers.
3. Conclusions
An integrated approach was used to examine the biological reactivity of a novel artificial nuclease [Cu(TPMA)(Phen)](ClO4)2 using free and encapsulated drug forms within liposome and cellular models. A nanoscale hollow pH-sensitive drug-delivery system was successfully used to encapsulate and release, without changing the copper complex structure as identified by EPR experiments. Membrane fatty acid reactivity and the fatty acid remodeling were observed in these models, respectively, analyzing SFA, MUFA, and PUFA moieties. The lipidome analysis of NB100 cells indicated the involvement of SFA and MUFA only in the free drug supplementation, whereas no changes were observed when the drug release was controlled by the nanocontainer delivery system. The parallelism between cellular and liposome models was expedient to determine that MUFA and PUFA fatty acid moieties are not affected by oxidative and isomerization reactions in the presence of this copper(II) complex. More work is needed to understand the molecular basis of the observed membrane remodeling, its possible involvement in desaturase inhibition, and the association of this remodeling with the cell death pathways. Our results can contribute to the understanding of the behavior of metal complexes, in particular indicating the interactions of metallome with membrane lipidomics.
4. Materials and Methods
4.1. Materials
Methacrylic acid (MAA, 99%) was obtained from Acros Organics and used after purification by distillation under vacuum. Poly(ethylene glycol)methyl ether methacrylate (PEGMA; Mn = 475) was obtained from Sigma-Aldrich and used without further purification. 2,2′-Azobisisobutyronitrile (AIBN, 98%) and N,N′-methylenebisacrylamide (MBA, 96%) were purchased from Acros Organics and used as received. Acetonitrile (ACN) was used as received from Sigma-Aldrich. Cu-TPMA-Phen was prepared according to published procedure.21
Caspase activity was evaluated using the luminescent kit Caspase-Glo 3/7 assay (Promega Corporation, Fitchburg, WI). Morphological membrane changes were detected using Annexin V–EGFP/PI detection kit (BioVision, Mt. View, CA). Viability was measured using the colorimetric CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega). The CellTiter 96 Aqueous One Solution Reagent contains the tetrazolium compound 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) and an electron-coupling reagent (1-methoxy phenazine methosulfate). 1-Palmitoyl 2-oleoyl phosphatidylcholine (POPC) and soybean lecithin were commercially available from Larodan (Sweden). Other reagents used were from Merck (Darmstadt, Germany), Carlo Erba (Milano, Italy), and Sigma-Aldrich. Roswell Park Memorial Institute (RPMI) 1640, fetal bovine serum (FBS), l-glutamine, antibiotics, trypan blue, n-hexane, chloroform, and methanol were purchased from Sigma-Aldrich (St. Louis, MO). Flasks and plates were from Falcon, BD Biosciences, NJ. Trypsin–ethylenediaminetetraacetic acid (EDTA) was from BioWhittaker Europe, Verviers, Belgium. CellTiter 96 Aqueous One Solution Cell Proliferation Assay and Caspase Luminescent Assay were from Promega Corporation, Madison, WI. Viability was evaluated by measuring absorbance at 492 nm by a microtiter plate reader Multiskan EX (Thermo LabSystems, Basingstoke, U.K.). Phase contrast microscopy was carried out with a Wilovert Standard PH 20 (HUND, Wetzlar, Germany) and a digital camera from Motic Microscopes, China.
4.2. Synthesis and Characterization of Nanocontainers
The synthesis of the PMAA cores was achieved via distillation–precipitation polymerization. In particular, MAA (2.1 g; 24.4 mmol) was dissolved in 200 mL of ACN and stirred at 75 °C for 30 min under nitrogen atmosphere. AIBN (0.3 g; 1.8 mmol) was then added, and the flask content turned into a milky suspension. Afterward, distillation was started at a temperature of 95–100 °C. Once 20 mL of distilled ACN was collected, the reaction was stopped. The final product was obtained after three cycles of centrifugation and resuspension in ACN (5 min × 8000 rpm). A second distillation–precipitation polymerization process was used for the synthesis of the core–shell system. The PMAA cores (0.15 g) were suspended in 200 mL of ACN and stirred at 75 °C for 30 min under nitrogen atmosphere. MAA (0.53 g; 6.1 mmol) was added, and, after 10 min, PEGMA (0.16 g; 0.3 mmol; 5 mol % of MAA) and MBA (0.15 g; 0.97 mmol; 16 mol % of MAA) were added as well. After 30 min, AIBN (0.09 g; 0.5 mmol; 8 mol % of MAA) was then added. After 10 min, the temperature was raised to 95–100 °C to start the distillation. Once 30 mL of distilled ACN was collected, the reaction was stopped. The final product was obtained after three cycles of centrifugation and resuspension in ACN (5 min × 5000 rpm). Regarding the core removal procedure, 250 mg of PMAA@P(MAA-co-PEGMA-co-MBA) was suspended in 200 mL of a mixture of 1:1 (v/v) EtOH/H2O and stirred at room temperature overnight. The product was purified by three cycles of centrifugation and resuspension (5 min × 5000 rpm). Scanning electron microscopy (SEM) pictures were taken using an FEI Inspect nanoscope with tungsten filament operating at 25 kV. FT-IR spectra were recorded with a PerkinElmer Precisely Spectrum 100 spectrometer. The hydrodynamic diameter and the ζ potential of the NCs in distilled water were measured with dynamic light scattering (DLS) (Malvern Instruments Series, nano-ZS with multipurpose titrator). The concentration of the sample was 0.1 mg/mL, and the given results were the average value of 10 measurements, with 20 s integration time. Sonication was performed with an ultrasonic bath (Elmasonic S 30H).
4.3. Drug Loading
Typically, 1 mg of NCs was suspended in 950 μL of phosphate-buffered saline (PBS). Then, 2 mg of Cu-TPMA-Phen previously solubilized in 50 μL of ACN was added. The final suspension, containing 1 mg of NCs and 2 mg of Cu-TPMA-Phen in 1 mL of mixture of 19:1 (v/v) PBS/ACN, was kept under gentle magnetic stirring (ca. 300 rpm) for 24 h. At the end of the process, the nonencapsulated portion of Cu-TPMA-Phen was removed with five cycles of centrifugations and resuspensions in a fresh mixture PBS/ACN (5 min × 11 000 rpm). The amount of encapsulated Cu-TPMA-Phen was determined by UV spectroscopy (Helios Thermo Electron Corporation) and calculated by the difference of Cu-TPMA-Phen concentration between the original Cu-TPMA-Phen solution and the supernatants containing the nonencapsulated drug. The calculations were based on a calibration curve of Cu-TPMA-Phen obtained in the same solvent mixture (λmax 262 nm). Encapsulation efficiency (EE%) and loading capacity (LC%) were the parameters used to evaluate the process (eqs 1 and 2)
| 1 |
| 2 |
4.4. In Vitro Release Study
The drug release profile of Cu-TPMA-Phen-loaded NCs was studied with the dialysis bag method (dialysis tube molecular weight cutoff 140 kDa). The release experiments were carried out in acidic conditions (mixture of citrate buffer pH 4.0, 0.1 M and ACN—19:1) and in slightly basic conditions (mixture of PBS and ACN—19:1) to prove the pH sensitivity of the system. Generally, 1 mg of loaded NCs was suspended in distilled water, split into two dialysis bags, and incubated in 25 mL of each release medium. At different time points (30 min, 1 h, 2 h, 5 h, 8 h, 10 h, and 24 h), 1 mL was collected from each solution and the concentration of the samples was measured using UV–vis spectroscopy. The calculations were made upon a calibration curve of Cu-TPMA-Phen recorded in each buffer (λmax 262 nm).
4.5. EPR Study of Drug Released
Continuous-wave (cw) EPR measurements at X-band were performed on a Bruker ESP 380E spectrometer equipped with an EN 4118X-MD4 Bruker resonator. Experimental conditions: microwave (mw) frequency, 9.715 GHz; mw power incident to the cavity, 20 μW; modulation frequency, 100 kHz; modulation amplitude, 0.1 mT; temperature, 70 K. Measurements at cryogenic temperatures were performed using a helium cryostat from Oxford Inc. The microwave frequency was measured using an HP 5350B microwave frequency counter, and the temperature was stabilized using an Oxford ITC4 temperature controller. The cw EPR spectra were recorded in frozen solutions at 70 K, and the samples were in a concentration range between 1 and 5 mM. Glycerol was added as a glassing agent, whose ratio with the solutions was 1:4. The standard release procedure described above was modified to obtain the required concentration (1–5 mM) of the released complex in the media. In particular, 0.5 mg of loaded NCs was suspended in distilled water, put into a dialysis bag, and incubated in 0.3 mL of each release medium. After 24 h, the resulting media were collected, glycerol was added, and the resulting samples were then analyzed.
4.6. Confocal Laser Scanning Microscopy
Confocal laser scanning microscopy (CLSM) was used to evaluate the uptake of the pH-sensitive NCs and the subsequent release and localization of Cu-TPMA-Phen. MCF7 cells were inoculated on 22 mm coverslips placed into six-well culture plates (5 × 106 cells/well) and grown for 24 h in 1.5 mL of complete growth medium. Free Cu-TPMA-Phen (10 μM) or Cu-TPMA-Phen-loaded NCs (drug concentration 10 μM) were then added. After 2 h incubation, the medium was removed and the coverslips were washed twice with PBS, 1 mL of 4% paraformaldehyde for 8 min in PBS, and PBS again, before placing them onto microscope slides. The instrument used for the experiment was a Leica TCS SP8 MP, an inverted confocal microscope with Acousto-Optical Beam Splitter; for the excitation and multiband spectral detector Argon—excitation at 458, 476, 488, 496, and 514 nm DPSS 561—excitation at 561 nm (RED). Multiphoton IR laser Mai Tai DeepSee obtained from Spectral Physics, with excitation at 780 nm, was used.
4.7. Cell Cultures
The activity of Cu-TPMA-Phen was assayed on NB100 cells that were derived from a human primary neuroblastoma.56,57 Cells were cultured as a monolayer at 37 °C in a humidified atmosphere at 5% CO2 in complete medium (RPMI 1640 supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 units/mL penicillin, and 0.1 mg/mL streptomycin). Cultures were maintained in the log phase of growth with a viability of >95% and checked for the absence of Mycoplasma infection. The viability was checked before each experiment by trypan blue (BioWhittaker, Verviers, Belgium) dye exclusion. Before any treatment, cells were incubated for 24 h. Flasks and plates were from Falcon (Franklin Lakes, NJ). All of the other cell culture reagents were from Sigma-Aldrich. RPMI 1640 was purchased from Sigma-Aldrich. Trypsin–EDTA, l-glutamine, penicillin–streptomycin solution, and heat-inactivated fetal bovine serum (FBS) were obtained from Biochrom KG.
4.8. Cell Viability Assay
Cell viability was evaluated using the colorimetric CellTiter 96 Aqueous One Solution Cell Proliferation Assay. Cells (2 × 104/well) were seeded in 96-well microtiter plates in 100 μL of complete medium. After 24 h, the cells were incubated in the absence or presence of Cu-TPMA-Phen at various concentrations in complete medium. After the indicated times, 20 μL/well of kit solution was added. After 1–2 h of incubation at 37 °C, the absorbance at 492 nm was measured by a microtiter plate reader Multiskan EX (Thermo LabSystems, Helsinki, Finland). In continuous incubation experiments, the cells were exposed to Cu-TPMA-Phen for 24, 48, and 72 h at concentration ranging from 0.1 to 30 μM. In pulse and chase experiments, the cells were treated with Cu-TPMA-Phen for 2 h at concentration ranging from 0.1 to 100 μM and then incubated in complete medium for total time of 24 and 48 h. Half-maximal effective concentration (EC50) was determined by standard slope analysis without normalization.
4.9. Evaluation of Apoptosis
The cell death pathway (apoptotic vs necrotic) was assessed using a flow cytometry Annexin V/PI detection kit and by a luminescent reagent detecting caspase activity.56 For flow cytometry experiments, cells (2 × 105/3 mL) were seeded in 25 cm2 flasks and, after 24 h incubation with 5 μM Cu-TPMA-Phen, the cellsPhen were treated with Annexin V–EGFP and PI and analyzed by flow cytometry.56 The apoptotic (Annexin V+/PI−), necrotic (Annexin V–/PI+), and late-stage apoptotic cells (Annexin V+/PI+) were counted by the instrument and reported on scatter plots. The caspase-3/7 activity was assessed by the luminescent Caspase-Glo 3/7 Assay.56 Briefly, cells (2 × 103/well) were seeded in 96-well microtiter plates in 100 μL of complete medium. After 24 h, the cells were treated with 5 μM Cu-TPMA-Phen. After further 24 h incubation, 100 μL/well of caspase kit reagent was added. After 20 min, the luminescence was measured by a Fluoroskan Ascent FL (Thermo LabSystems), and the values were normalized to cell viability. The morphological features of the treated cells were analyzed through phase contrast microscopy, directly in a 96-well plate, using an inverted microscope Nikon Eclipse TS100 (Nikon, Melville, NY).
4.10. Phospholipid Extraction and Fatty Acid Analysis
To analyze the effect of Cu-TPMA-Phen treatment on membrane fatty acids, 0.8 × 106 cells were seeded in 25 cm2 flasks in 5 mL of complete medium. After 24 h of incubation, medium supplemented with 5 μM Cu-TPMA-Phen was added. The cells were harvested and washed twice with ice-cold PBS. Cell pellet was resuspended in 1 mL of Milli-Q H2O and centrifuged at 14 000 rpm for 15 min at 4 °C. Membrane pellet was dissolved in 2:1 chloroform/methanol, followed by the Folch extraction method. Lipid extract was examined by thin-layer chromatography (n-hexane/diethyl ether/acetic acid 70/30/1) to determine the purity of the phospholipid fraction. The phospholipid extract was then treated with 0.5 M KOH/MeOH for 10 min at room temperature under stirring for the derivatization of fatty acid residues of the phospholipids into their corresponding fatty acid methyl esters (FAMEs). After transesterification, FAMEs were extracted with n-hexane; n-hexane phase was dehydrated with anhydrous Na2SO4, evaporated, and analyzed by gas chromatography (Agilent 6850, Milan) equipped with a 60 m × 0.25 mm × 0.25 μm (50% cyanopropyl)-methylpolysiloxane column (DB23, Agilent) and a flame ionization detector, with an injector temperature of 230 °C and a split injection of 50:1. Oven temperature started from 165 °C, held for 3 min, followed by an increase of 1 °C/min up to 195 °C, held for 40 min, followed by a second increase of 10 °C/min up to 240 °C, and held for 10 min. A constant pressure mode (29 psi) with helium as carrier gas was used. Methyl esters were identified by comparison with the retention times of commercially available standards or trans fatty acid references, obtained as described elsewhere. The list of the examined FAME (corresponding to chromatographic peak areas >97%) in membrane PL is reported in Table S1 as % relative percentages ± standard deviation (SD).
4.11. LUVET Preparation
Phosphatidylcholine (PC) chloroform solution (53 mg dissolved in 3 mL) was evaporated to a thin film in a test tube under a stream of argon and then kept under high vacuum for 30 min at room temperature. To obtain a final concentration of 70 mM phospholipid content, 1 mL of tridistilled water was added. As a result, multilamellar vesicles were formed by vortex stirring for 7 min under an argon atmosphere. Large unilamellar vesicles (LUVs) with a mean diameter of 156–158 nm were prepared by extrusion technique using LiposoFast and a 200 nm polycarbonate membrane filter as described previously. The size of the liposomes was measured using dynamic light scattering (DLS) methodology. The LUVET stock suspensions were transferred into a vial and stored at 4 °C for a maximum of 2 weeks.
4.12. Isomerization of PC in LUVET
The total volume for every reaction was 1 mL of LUVET stock (phospholipid concentration of 1 mM). More specifically, an aliquot of 14.5 μL fatty acid content from the stock solution was added to tridistilled water in the reaction vessel. To the liposome suspension, the copper complex was transferred (0.15 mM) and the reaction remained under stirring for 2 min. From stock solutions in tridistilled water and final concentration in the reaction 10 mM, the thiol, 2-mercaptoethanol, l-cysteine (CySH), or reduced glutathione (GSH), was added to the reaction dropwise (0.5 mM/min) using a syringe pump. Each reaction vessel was incubated at 37 °C, and, to follow the formation of trans fatty acid residues, the samples were analyzed at different times. The workup of the vesicles was made with 2:1 chloroform/methanol, extracting and collecting the organic phases dried over anhydrous sodium sulfate and evaporating the solvent under vacuum at room temperature. The phospholipids extracts were treated with 0.5 M KOH/MeOH, in a transesterification type of reaction for 10 min at ambient temperature. The reaction was quenched with the addition of tridistilled water, followed by an extraction with n-hexane. The organic layer containing the corresponding fatty acid methyl esters (FAMEs) was analyzed by GC for the determination of the cis–trans isomer content. For the experiments under anaerobic conditions, all of the solutions were degassed with argon for 15 min and the addition of all reagents took place under a stream of argon. The anaerobic conditions were maintained during the incubation period by creating pressure of argon inside the reaction vial.
4.13. Statistical Analysis
Fatty acid values represent the mean ± SD. Statistical analysis was conducted using GraphPad Prism 7.02 software for Windows, GraphPad Software, La Jolla, CA. The data were analyzed with unpaired t test. Statistical significance was based on 95% confidence intervals (p ≤ 0.05).
Glossary
Abbreviations
- ACN
acetonitrile
- AIBN
2,2′-azobisisobutyronitrile
- AMN
artificial metallonuclease
- BLM
bleomycin
- CLSM
confocal laser scanning microscopy
- CySH
l-cysteine
- DLS
dynamic light scattering
- EC50
half-minimal effective concentration
- EDTA
ethylenediaminetetraacetic acid
- EE%
encapsulation efficiency %
- EPR
electron paramagnetic resonance
- FAME
fatty acid methyl esters
- FBS
heat-inactivated fetal bovine serum
- FT-IR
Fourier transform infrared spectroscopy
- GSH
glutathione
- LC%
loading capacity %
- LUV
large unilamellar vesicles
- LUVET
large unilamellar vesicles obtained with the extrusion technique
- MAA
methacrylic acid
- MBA
N,N′-methylenebisacrylamide
- MeOH
methanol
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- MUFA
monounsaturated fatty acids
- NC
nanocontainer
- PBS
phosphate-buffered saline
- PEGMA
poly(ethylene glycol)methyl ether methacrylate
- Phen
1,10-phenantroline
- PMAA
poly-methacrylic acid
- POPC
1-palmitoyl 2-oleoyl phosphatidylcholine
- PUFA
polyunsaturated fatty acids
- ROS
reactive oxygen species
- SEM
scanning electron microscopy
- SFA
saturated fatty acids
- TPMA
tris-(2-pyridylmethyl)amine
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b02526.
FT-IR spectra of pH-sensitive NCs; DLS analysis of pH-sensitive NCs; MTS assay on NB100 cells treated with free and encapsulated Cu-TPMA-Phen; relative percentages and indices of membrane fatty acids methyl esters (FAMEs) of NB100 cells treated with 5 μM free or encapsulated Cu-TPMA-Phen; fatty acid-based membrane lipidomics on MCF7 cells treated with 10 μM free Cu-TPMA-Phen for 24 h; reduction of Cu(II) in Cu-TPMA-Phen to Cu(I); and trans isomer formation in POPC vesicles treated with 10 mM 2-mercaptoethanol and different concentrations of Cu-TPMA-Phen under aerobic or anaerobic conditions (PDF)
Author Present Address
∇ Inorganic Chemistry Laboratory, Chemistry Department, National and Kapodistrian University of Athens, Panepistimiopolis, Zografou, 15771 Athens, Greece (E.K.E.).
Author Contributions
# G.T. and M.L. contributed equally to this work.
Author Contributions
C.C. formulated the idea and prepared the initial draft; C.C. and C.F. designed the methodology and created the models; G.T., M.L., G.M.(1), M.B., and N.Z.F. performed experiments; A.M., E.K.E., G.M.(2), L.P., A.B., and A.K. supervised the data acquisition; C.C., G.T., M.L., and G.M.(1) planned the activities, organized the data, and prepared all graphics. All authors participated in the data discussion and ameliorated the draft. C.F., A.B., A.K., and C.C. acquired financial support.
C.C., C.F., and A.K. acknowledge funding from the Marie Skłodowska-Curie Innovative Training Network (ITN) ClickGene (H2020-MSCA-ITN-2014-642023). G.T., M.L., G.M.(1), and N.Z.F. are Ph.D. students within the ClickGene Network (http://www.clickgene.eu).
The authors declare no competing financial interest.
Supplementary Material
References
- Slator C.; Molphy Z.; McKee V.; Long C.; Brown T.; Kellett A. Di-copper metallodrugs promote NCI-60 chemotherapy via singlet oxygen and superoxide production with tandem TA/TA and AT/AT oligonucleotide discrimination. Nucleic Acids Res. 2018, 46, 2733–2750. 10.1093/nar/gky105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slator C.; Barron N.; Howe O.; Kellett A. [Cu(o-phthalate)(phenanthroline)] exhibits unique superoxide-mediated NCI-60 chemotherapeutic action through genomic DNA damage and mitochondrial dysfunction. ACS Chem. Biol. 2016, 11, 159–171. 10.1021/acschembio.5b00513. [DOI] [PubMed] [Google Scholar]
- Molphy Z.; Prisecaru A.; Slator C.; Barron N.; McCann M.; Colleran J.; Chandran D.; Gathergood N.; Kellett A. Copper phenanthrene oxidative chemical nucleases. Inorg. Chem. 2014, 53, 5392–5404. 10.1021/ic500914j. [DOI] [PubMed] [Google Scholar]
- Molphy Z.; Slator C.; Chatgilialoglu C.; Kellett A. DNA oxidation profiles of copper phenanthrene chemical nucleases. Front. Chem. 2015, 3, 28 10.3389/fchem.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellett A.; Howe O.; O’Connor M.; McCann M.; Creaven B. S.; McClean S.; Foltyn-Arfa Kia A.; Casey A.; Devereux M. Radical-induced DNA damage by cytotoxic square-planar copper (II) complexes incorporating o-phthalate and 1,10-phenanthroline or 2, 2'-dipyridyl. Free Radic. Biol. Med. 2012, 53, 564–576. 10.1016/j.freeradbiomed.2012.05.034. [DOI] [PubMed] [Google Scholar]
- Marzano C.; Pellei M.; Tisato F.; Santini C. Copper complexes as anticancer agents. Anti-Cancer Agents Med. Chem. 2009, 9, 185–211. 10.2174/187152009787313837. [DOI] [PubMed] [Google Scholar]
- Santini C.; Pellei M.; Gandin V.; Porchia M.; Tisato F.; Marzano C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. 10.1021/cr400135x. [DOI] [PubMed] [Google Scholar]
- Fantoni N.; Lauria T.; Kellett A. Genome Engineering with Synthetic Copper Nucleases. Synlett 2015, 26, 2623–2626. 10.1055/s-0035-1560709. [DOI] [Google Scholar]
- Pitié M.; Pratviel G. Activation of DNA carbon–hydrogen bonds by metal complexes. Chem. Rev. 2010, 110, 1018–1059. 10.1021/cr900247m. [DOI] [PubMed] [Google Scholar]
- Chen J.; Stubbe J. Bleomycins: towards better therapeutics. Nat. Rev. Cancer 2005, 5, 102–112. 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- Cort A.; Ozben T.; Sansone A.; Barata-Vallejo S.; Chatgilialoglu C.; Ferreri C. Bleomycin-induced trans lipid formation in cell membranes and in liposome models. Org. Biomol. Chem. 2015, 13, 1100–1105. 10.1039/C4OB01924E. [DOI] [PubMed] [Google Scholar]
- Cort A.; Ozben T.; Melchiorre M.; Chatgilialoglu C.; Ferreri C.; Sansone A. Effects of bleomycin and antioxidants on the fatty acid profile of testicular cancer cell membranes. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 434–441. 10.1016/j.bbamem.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Ferreri C.; Chatgilialoglu C. Geometrical trans lipid isomers: a new target for lipidomics. ChemBioChem 2005, 6, 1722–1734. 10.1002/cbic.200500109. [DOI] [PubMed] [Google Scholar]
- Chatgilialoglu C.; Ferreri C.; Melchiorre M.; Sansone A.; Torreggiani A. Lipid geometrical isomerism: from chemistry to biology and diagnostics. Chem. Rev. 2014, 114, 255–284. 10.1021/cr4002287. [DOI] [PubMed] [Google Scholar]
- van Meer G.; et al. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopperton K. E.; Duncan R. E.; Bazinet R. P.; Archer M. C. Fatty acid synthase plays a role in cancer metabolism beyond providing fatty acids for phospholipid synthesis or sustaining elevations in glycolytic activity. Exp. Cell Res. 2014, 320, 302–310. 10.1016/j.yexcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Zalba S.; ten Hagen T. L. M. Cell membrane modulation as adjuvant in cancer therapy. Cancer Treat. Rev. 2017, 52, 48–57. 10.1016/j.ctrv.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst J. J.; Cross J. R.; Fan J.; de Stanchina E.; Mathew R.; White E. P.; Thompson C. B.; Rabinowitz J. D. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8882–8887. 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves A. C.; Ribeiro D.; Nunes C.; Reis S. Biophysics in cancer: the relevance of drug-membrane interaction studies. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 2231–2244. 10.1016/j.bbamem.2016.06.025. [DOI] [PubMed] [Google Scholar]
- Beloribi-Djefaflia S.; Vasseur S.; Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantoni N.; Molphy Z.; Slator C.; Menounou G.; Toniolo G.; Mitrikas G.; McKee V.; Chatgilialoglu C.; Kellett A.. Polypyridyl-based copper phenanthrene complexes: a new type of stabilized artificial chemical nuclease. Chem. - Eur. J. 2018, 24, 10.1002/chem.201804084. [DOI] [PubMed] [Google Scholar]
- Li L.; Karlin K. D.; Rokita S. E. Changing selectivity of DNA oxidation from deoxyribose to guanine by ligand design and a new binuclear copper complex. J. Am. Chem. Soc. 2005, 127, 520–521. 10.1021/ja044209e. [DOI] [PubMed] [Google Scholar]
- Humphreys K. J.; Karlin K. D.; Rokita S. E. Efficient and specific strand scission of DNA by a dinuclear copper complex: comparative reactivity of complexes with linked tris(2-pyridylmethyl)amine moieties. J. Am. Chem. Soc. 2002, 124, 6009–6019. 10.1021/ja020039z. [DOI] [PubMed] [Google Scholar]
- Thyagarajan S.; Murthy N. N.; Sarjeant A. A. N.; Karlin K. D.; Rokita S. E. Selective DNA strand scission with binuclear copper complexes: implications for an active Cu2-O2 species. J. Am. Chem. Soc. 2006, 128, 7003–7008. 10.1021/ja061014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larragy R.; Fitzgerald J.; Prisecaru A.; McKee V.; Leonard P.; Kellett A. Protein engineering with artificial chemical nucleases. Chem. Commun. 2015, 51, 12908–12911. 10.1039/C5CC04615G. [DOI] [PubMed] [Google Scholar]
- Prisecaru A.; Devereux M.; Barron N.; McCann M.; Colleran J.; Casey A.; McKee V.; Kellett A. Potent oxidative DNA cleavage by the di-copper cytotoxin: [Cu2(μ-terephthalate)(1,10-phen)4]2+. Chem. Commun. 2012, 48, 6906–6908. 10.1039/c2cc31023f. [DOI] [PubMed] [Google Scholar]
- Sigman D. S.; Mazumder A.; Perrin D. M. Chemical nucleases. Chem. Rev. 1993, 93, 2295–2316. 10.1021/cr00022a011. [DOI] [Google Scholar]
- Serment-Guerrero J.; Cano-Sanchez P.; Reyes-Perez E.; Velazquez-Garcia F.; Bravo-Gomez M. E.; Ruiz-Azuara L. Genotoxicity of the copper antineoplastic coordination complexes casiopeinas. Toxicol. In Vitro 2011, 25, 1376–1384. 10.1016/j.tiv.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Pratviel G.; Bernadou J.; Meunier B. Carbon—hydrogen bonds of DNA sugar units as targets for chemical nucleases and drugs. Angew. Chem., Int. Ed. 1995, 34, 746–769. 10.1002/anie.199507461. [DOI] [Google Scholar]
- Fleige E.; Quadir M. A.; Haag R. Stimuli-responsive polymeric nanocarriers for the controlled transport of active compounds: concepts and applications. Adv. Drug Delivery Rev. 2012, 64, 866–884. 10.1016/j.addr.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Cheng R.; Meng F.; Deng C.; Klok H. A.; Zhong Z. Dual and multi-stimuli responsive polymeric nanoparticles for programmed site-specific drug delivery. Biomaterials 2013, 34, 3647–3657. 10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- Deshayes S.; Maurizot V.; Clochard M. C.; Baudin C.; Berthelot T.; Esnouf S.; Lairez D.; Moenner M.; Déléris G. “Click” conjugation of peptide on the surface of polymeric nanoparticles for targeting tumor angiogenesis. Pharm. Res. 2011, 28, 1631–1642. 10.1007/s11095-011-0398-5. [DOI] [PubMed] [Google Scholar]
- Steichen S. D.; Caldorera-Moore M.; Peppas N. A. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur. J. Pharm. Sci. 2013, 48, 416–427. 10.1016/j.ejps.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon-Peppas L.; Blanchette J. O. Nanoparticle and targeted systems for cancer therapy. Adv. Drug Delivery Rev. 2012, 64, 206–212. 10.1016/j.addr.2012.09.033. [DOI] [PubMed] [Google Scholar]
- Fang J.; Nakamura H.; Maeda H. The EPR effect: unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Delivery Rev. 2011, 63, 136–151. 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Metaxa A. F.; Efthimiadou E. K.; Boukos N.; Fragogeorgi E. A.; Loudos G.; Kordas G. Microspheres based on – folic acid modified – hydroxypropyl cellulose and synthetic multi-responsive bio-copolymer for targeted cancer therapy: controlled release of daunorubicin, in vitro and in vivo studies. J. Colloid Interface Sci. 2014, 435, 171–181. 10.1016/j.jcis.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Ganta S.; Devalapally H.; Shahiwala A.; Amiji M. A review of stimuli-responsive nanocarriers for drug and gene delivery. J. Controlled Release 2008, 126, 187–204. 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Tapeinos C.; Efthimiadou E. K.; Boukos N.; Charitidis C. A.; Koklioti M.; Kordas G. Microspheres as therapeutic delivery agents: synthesis and biological evaluation of pH responsiveness. J. Mater. Chem. B 2013, 194–203. 10.1039/C2TB00013J. [DOI] [PubMed] [Google Scholar]
- Yang X.; Chen L.; Huang B.; Bai F.; Yang X. Synthesis of pH-sensitive hollow polymer microspheres and their application as drug carriers. Polymer 2009, 50, 3556–3563. 10.1016/j.polymer.2009.06.027. [DOI] [Google Scholar]
- Agut W.; Brûlet A.; Schatz C.; Taton D.; Lecommandoux S. pH and temperature pesponsive polymeric micelles and polymersomes by self-assembly of poly[2-(dimethylamino)ethyl methacrylate]-b-poly(glutamic acid) double hydrophilic block copolymers. Langmuir 2010, 26, 10546–10554. 10.1021/la1005693. [DOI] [PubMed] [Google Scholar]
- Gallon E.; Matini T.; Sasso L.; Mantovani G.; Armiñan de Benito A.; Sanchis J.; Caliceti P.; Alexander C.; Vicent M. J.; Salmaso S. Triblock copolymer nanovesicles for pH-responsive targeted delivery and controlled release of siRNA to cancer cells. Biomacromolecules 2015, 16, 1924–1937. 10.1021/acs.biomac.5b00286. [DOI] [PubMed] [Google Scholar]
- Bai F.; Huang B.; Yang X.; Huang W. Synthesis of monodisperse poly(methacrylic acid) microspheres by distillation – precipitation polymerization. Eur. Polym. J. 2007, 43, 3923–3932. 10.1016/j.eurpolymj.2007.06.024. [DOI] [Google Scholar]
- Chatzipavlidis A.; Bilalis P.; Efthimiadou E. K.; Boukos N.; Kordas G. C. Sacrificial template-directed fabrication of superparamagnetic polymer microcontainers for pH-activated controlled release of daunorubicin. Langmuir 2011, 27, 8478–8485. 10.1021/la201240h. [DOI] [PubMed] [Google Scholar]
- Yang X.; Chen L.; Huang B.; Bai F.; Yang X. Synthesis of pH-sensitive hollow polymer microspheres and their application as drug carriers. Polymer 2009, 50, 3556–3563. 10.1016/j.polymer.2009.06.027. [DOI] [Google Scholar]
- Efthimiadou E. K.; Tziveleka L. A.; Bilalis P.; Kordas G. Novel PLA modification of organic microcontainers based on ring opening polymerization: synthesis, characterization, biocompatibility and drug loading/release properties. Int. J. Pharm. 2012, 428, 134–142. 10.1016/j.ijpharm.2012.02.030. [DOI] [PubMed] [Google Scholar]
- Qiu Y.; Park K. Environment-sensitive hydrogels for drug delivery. Adv. Drug Delivery Rev. 2012, 64, 49–60. 10.1016/j.addr.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Bilalis P.; Boukos N.; Kordas G. C. Novel PEGylated pH-sensitive polymeric hollow microspheres. Mater. Lett. 2012, 67, 180–183. 10.1016/j.matlet.2011.09.062. [DOI] [Google Scholar]
- Kang X.-J.; Dai Y.-L.; Ma P.-A.; Yang D.-M.; Li C.-X.; Hou Z.-Y.; Cheng Z.-Y.; Lin J. Poly(acrylic acid)-modified Fe3O4 microspheres for magnetic-targeted and pH-triggered anticancer drug delivery. Chem. – Eur. J. 2012, 18, 15676–15682. 10.1002/chem.201202433. [DOI] [PubMed] [Google Scholar]
- Metaxa A. F.; Efthimiadou E. K.; Kordas G. Cellulose-based drug carriers for cancer therapy: cytotoxic evaluation in cancer and healthy cells. Mater. Lett. 2014, 132, 432–435. 10.1016/j.matlet.2014.06.134. [DOI] [Google Scholar]
- Jiang F.; Karlin K. D.; Peisach J. An electron spin echo envelope modulation (ESEEM) study of electron-nuclear hyperfine and nuclear quadrupole interactions of dZ2 ground state copper(II) complexes with substituted imidazoles. Inorg. Chem. 1993, 32, 2576–2582. 10.1021/ic00063a060. [DOI] [Google Scholar]
- Kokoszka G.; Karlin K. D.; Padula F.; Baranowski J.; Goldstein C. EPR of copper(II) complexes with tripodal ligands: dynamical properties. Inorg. Chem. 1984, 23, 4378–4380. 10.1021/ic00193a054. [DOI] [Google Scholar]
- Hathaway B. J.; Billing D. E. The electronic properties and stereochemistry of mono-nuclear complexes of the copper(II) ion. Coord. Chem. Rev. 1970, 5, 143–207. 10.1016/S0010-8545(00)80135-6. [DOI] [Google Scholar]
- Toniolo G.; Efthimiadou E. K.; Kordas G.; Chatgilialoglu C. Development of multi-layered and multi-sensitive polymeric nanocontainers for cancer therapy: in vitro evaluation. Sci. Rep. 2018, 8, 14704 10.1038/s41598-018-32890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tziveleka L. A.; Bilalis P.; Chatzipavlidis A.; Boukos N.; Kordas G. Development of multiple stimuli responsive magnetic polymer nanocontainers as efficient drug delivery systems. Macromol. Biosci. 2014, 14, 131–141. 10.1002/mabi.201300212. [DOI] [PubMed] [Google Scholar]
- Bishop C. J.; Kozielski K. L.; Green J. J. Exploring the role of polymer structure on intracellular nucleic acid delivery via polymeric nanoparticles. J. Controlled Release 2015, 219, 488–499. 10.1016/j.jconrel.2015.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito L.; Bortolotti M.; Pedrazzi M.; Mercatelli D.; Battelli M. G.; Bolognesi A. Apoptosis and necroptosis induced by stenodactylin in neuroblastoma cells can be completely prevented through caspase inhibition plus catalase or necrostatin-1. Phytomedicine 2016, 23, 32–41. 10.1016/j.phymed.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Bolognesi A.; Chatgilialoglu A.; Polito L.; Ferreri C. Membrane lipidome reorganization correlates with the fate of neuroblastoma cells supplemented with fatty acids. PLoS One 2013, 8, e55537 10.1371/journal.pone.0055537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton C. M.; Ntambi J. M. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E28–E37. 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren P.; Sierra-Johnson J.; Gertow K.; Rosell M.; Vessby B.; de Faire U.; Hamsten A.; Hellenius M.-L.; Fisher R. M. Fatty acid desaturases in human adipose tissue: relationships between gene expression, desaturation indexes and insulin resistance. Diabetologia 2008, 51, 328–335. 10.1007/s00125-007-0876-9. [DOI] [PubMed] [Google Scholar]
- Slator C.; Molphy Z.; McKee V.; Kellett A. Triggering autophagic cell death with a di-manganese(II) developmental therapeutic. Redox Biol. 2017, 12, 150–161. 10.1016/j.redox.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulucci G.; Cohen O.; Daniel B.; Sansone A.; Petropoulou P. I.; Filou S.; Spyridonidis A.; Pani G.; De Spirito M.; Chatgilialoglu C.; Ferreri C.; Kypreos K. E.; Sasson S. Fatty acid-related modulations of membrane fluidity in cells: detection and implications. Free Radic. Res. 2016, 50, S40–S50. 10.1080/10715762.2016.1231403. [DOI] [PubMed] [Google Scholar]
- Scaglia N.; Chisholm J. W.; Igal R. A. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS One 2009, 4, e6812 10.1371/journal.pone.0006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y.; Hao L.; Hu W.; Ran Y.; Bai Y.; Zhang L. Novel multifunctional pH-sensitive nanoparticles loaded into microbubbles as drug delivery vehicles for enhanced tumor targeting. Sci. Rep. 2016, 6, 29321 10.1038/srep29321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatgilialoglu C.; Ferreri C.; Guerra M.; Samadi A.; Bowry V. W. The reaction of thiyl radical with methyl linoleate: completing the Picture. J. Am. Chem. Soc. 2017, 139, 4704–4714. 10.1021/jacs.6b11320. [DOI] [PubMed] [Google Scholar]
- Ferreri C.; Costantino C.; Perrotta L.; Landi L.; Mulazzani Q. G.; Chatgilialoglu C. Cis–trans isomerization of polyunsaturated fatty acid residues in phospholipids catalyzed by thiyl radicals. J. Am. Chem. Soc. 2001, 123, 4459–4468. 10.1021/ja0040969. [DOI] [PubMed] [Google Scholar]
- Godoy C. A.; Valiente M.; Pons R.; Montalvo G. Effect of fatty acids on self-assembly of soybean lecithin systems. Colloids Surf., B 2015, 131, 21–28. 10.1016/j.colsurfb.2015.03.065. [DOI] [PubMed] [Google Scholar]
- Ferreri C.; Costantino C.; Chatgilialoglu C.; Ferreri C.; Landi L.; Mulazzani Q. G. The thiyl radical-mediated isomerization of cis-monounsaturated fatty acid residues in phospholipids: a novel path of membrane damage?. Chem. Commun. 1999, 407–408. 10.1039/a809674k. [DOI] [Google Scholar]
- Chatgilialoglu C.; Ferreri C.; Ballestri M.; Mulazzani Q. G.; Landi L. Cis–trans isomerization of monounsaturated fatty acid residues in phospholipids by thiyl radicals. J. Am. Chem. Soc. 2000, 122, 4593–4601. 10.1021/ja994169s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.