Abstract
The crystal structure of a construct consisting of the FERM and SH2-like domains of the human Janus kinase 1 (JAK1) bound to a fragment of the intracellular domain of the interferon λ receptor 1 (IFNLR1) has been determined at the nominal resolution of 2.1 Å. In this structure the receptor peptide forms an 85-Å long extended chain, in which both the previously identified box1 and box2 regions bind simultaneously to the FERM and SH2-like domains of JAK1. Both domains of JAK1 are generally well ordered, with regions not seen in the crystal structure limited to loops located away from the receptor-binding regions. The structure provides much more complete and accurate picture of the interactions between JAK1 and IFNLR1 than those given in earlier reports, illuminating the molecular basis of the JAK–cytokine receptor association. A glutamate residue adjacent to the box2 region in IFNLR1 mimics the mode of binding of a phosphotyrosine in classical SH2 domains. It was shown here that a deletion of residues within the box1 region of the receptor abolishes stable interactions with JAK1, although it was previously shown that box2 alone is sufficient to stabilize a similar complex of the interferon-α receptor and TYK2.
Keywords: JAK/STAT signaling, cytokine receptors, Janus kinases, protein-protein interactions, structure comparisons
Introduction
The JAK/STAT signaling pathways play very important roles in a variety of biological processes, leading to a long-standing interest in understanding the mechanistic details of their activity. Defects in the cytokine/interferon-JAK/STAT pathways may lead to immunodeficiency, inflammatory diseases, autoimmune disorders, hematological defects, myoproliferative diseases, a variety of leukemias and lymphomas, as well as to inability to fight viral infections [1–3]. The critical role of the molecules involved in these pathways is well-illustrated by various diseases and disorders, often debilitating, resulting from defects in any of the participating molecules that include, among others, cytokines (interferons), cytokine (interferon) receptors, Janus kinases (JAKs), and STATs [4]. Mechanisms of creating biological responses initiated by binding of cytokines and interferons to their receptors have been investigated in structural terms for more than 25 years, starting from the pioneering work on the structure of the complex of the human growth hormone with the extracellular part of its receptor [5]. In greatly simplified terms, a signal is initiated by binding of the ligand to the extracellular parts of one or more transmembrane-spanning receptors. As a result, the receptor molecules oligomerize (in most cases forming homo- or heterodimers), or change the conformation of the pre-existing oligomers [6]. While the initial interaction involves the receptor ectodomains only, effects of dimerization (or oligomerization) span across the cellular membrane to regions housing the intracellular domains of the receptors, leading to cross-phosphorylation of the associated Janus kinases (JAKs). Multidomain JAK molecules interact with receptors through their FERM and SH2-like domains, triggering a series of phosphorylation events resulting in the activation of their kinase domains. The latter phosphorylate STAT molecules that subsequently disengage from the membrane-associated complex and transduce the signal to the nucleus. It is clear that the interface between the cytoplasmic domain of a receptor and the specific JAK molecule plays a role of an adaptor, critical for conveying the extracellular signal to the interior of the cell. A structural description of this interface is important for understanding these mechanistic aspects of JAK/STAT signaling.
The human genome encodes four JAK molecules, JAK1–3 and TYK2, more than 20 cytokine and interferon receptors, and even more cytokines and interferons. This imbalance in molecular variation of the interacting components that define a signaling pathway leads to promiscuity, which, in vivo, is controlled by different regulation of individual proteins [7]. However, deeper understanding of how the JAK/STAT signaling pathways achieve specificities necessary to control and coordinate complex biological processes in which they participate requires further, more detailed and extensive studies. In contrast to a large number of structures reported for cytokines and interferons [8–10], their complexes with the ectodomains of cytokine/interferon receptors [5, 11–14], and of the kinase [15] and pseudokinase [16] domains of JAKs, structural data for the regions corresponding to the JAK-receptor interface are very limited. The crystal structure of TYK2(FERM-SH2) fragment with a short peptide derived from the IFN-α receptor 1 (IFNAR1) was the first published report addressing this topic [17]. Very recently, when this manuscript was under preparation, a second report appeared, describing complexes of JAK1(FERM-SH2) region with two peptides, the first one corresponding to a region of IFN-λ receptor 1 (IFNLR1), and the second to a chimera consisting of a short section of IFNLR1 fused to a fragment of IL10RA [18]. Additionally, the structure of the FERM-SH2 module of human JAK2, not associated with a receptor, was recently published [19].
Interferons λ1, λ2, λ3, and λ4 (also known as IL-28 and IL-29) signal by forming heterodimeric complexes with their primary receptor IFNLR1 and a shared IL-10R2 chain (IL10R2), the latter also serving as the second subunit of the IL-10, IL-22, and IL-26 receptor complexes. This class of signaling molecules exhibits antiviral activity and involvement in cancer progression. As examples of the latter property, interferons λ were shown to reduce proliferation of various cancer cell lines including HCT116 cells [20], glioblastoma LN319 cells [21], or aneuroendocrine BON1 tumor cells [22]. We have previously determined the crystal structure of a complex between IFN-λ1 and the extracellular domain of the IFNLR1 [23]. Here we describe an investigation of the structural basis of the intracellular signaling processes that are initiated by an interaction of the intracellular parts of the receptor chain with a member of the JAK family.
It has been previously shown that the receptor IFNLR1 associates with the kinase JAK1 [24]. A majority of the intracellular domains of cytokine/interferon receptors interact with the JAK-family kinases primarily through two regions, named box1 and box2. These regions were originally defined on the basis of single-site mutations and truncation of the receptors and by analysis of conservation of their sequences. Murakami et al. [25] studied signal transduction induced by IL-6 and pointed out the existence of the two specific JAK1 binding sites in the first 61 residues of the intracellular fragment of the shared receptor gp130. For many receptors the signature of box1 includes a motif PxxPxP preceded by two hydrophobic residues, although the exact boundaries of box1 vary in different publications [26–28]. Moreover, sequence conservation of the box1 region is fairly low in different receptors. Box2 is located 10–50 residues downstream from box1 and its sequence conservation is even lower, with the exception of a frequently present glutamate residue flanked by a number of hydrophobic amino acids [25, 26]. Whereas until recently only indirect mutation data were used to delineate the extent of these boxes, structural basis of the interactions involving box2 was provided by the structure of the IFNAR1/TYK2 complex [17]. In turn, box1 interactions were recently described for IFNLR1/JAK1 and IL10RA/JAK1 [18].
The regions of the JAK kinases interacting with the intracellular parts of the receptors include the FERM and SH2-like domains. The latter domain has been originally identified in JAKs by sequence analysis and homology searches [29, 30]. The function of the SH2-like domains in JAKs was, however, unclear, as the lack of the canonical β5B arginine in some of them (i.e. TYK2) put into question their ability to bind phosphorylated tyrosine, the classical role of SH2 domains [31]. Furthermore, mutation of β5B arginine to lysine in JAK1 does not have an adverse effect on its function [31]. Parallel in vitro studies have shown that the SH2-like domain is important for intimate association between JAKs and receptors [25, 32]. A structural similarity to classical SH2 domains, details of the interactions with cytokine receptors, and a potential (or its lack) for a pTyr-binding by SH2-like domains in JAKs could only be illuminated by structural studies of these proteins. Recently, although several structures of JAK fragments containing SH2-like domains have been published [17–19], some questions regarding the structural details of the SH2-like domains and their role in interactions with the receptors still remain. For instance, TYK2, described in the complex with IFNAR1 [17], lacks the β5B Arg that is present in most of JAK kinases and almost all classical SH2 domains. In the case of JAK2, the published structure represents only a receptor-free FERM-SH2-like construct and the interactions with the receptor cannot be directly evaluated [19]. In the most recently published structures of JAK1 complexes, a large section of the receptor and several regions of the SH2-like domain are presumably disordered [18].
This report provides the first description of structural details for the simultaneous binding of both the box1 and box2 regions of an intracellular fragment of a type II cytokine receptor to the FERM and SH2-like domains of a JAK kinase.
Results and discussion
Interactions between fragments of JAK1 and IFNLR1
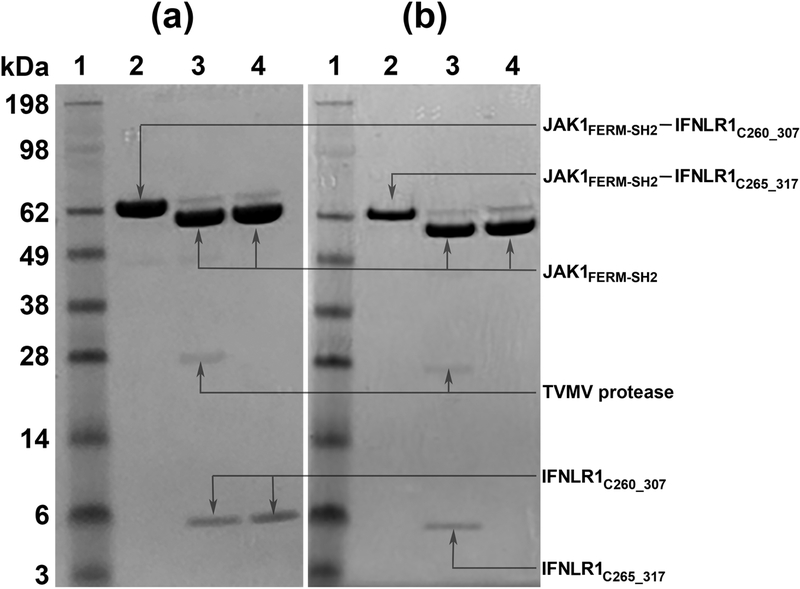
Ten IFNLR1/JAK1 constructs containing receptor peptides of different length were expressed and purified in order to identify the boundaries of the intracellular region of IFNLR1 sufficient to form a stable complex with the fragment of JAK1 comprising the FERM and SH2-like domains. The primary goal of these experiments was to design the most suitable and shortest construct for crystallographic experiments. The amino acid sequences of the receptor peptides are shown in Table 1. The longest peptide, extending furthest in both directions, includes the fragment of IFNLR1 spanning ArgR260 to ArgR317. Whereas both JAK1 and the receptor fragments were expressed as single-chain fusion proteins, they could be cleaved with TVMV protease at a specific sequence engineered in the middle of the linker region. After completion of the cleavage process, each reaction mixture was applied on a gel filtration column and the dominant single peak was subjected to the SDS-Page analysis. Gel images for the complexes involving two representative receptor peptides are shown in Fig. 1. The first example illustrates the complex-forming construct C260_307, IFNLR1(R260Arg-R307Arg), and the second corresponds to the non-interacting construct C265_317, IFNLR1(R265Arg-R317Arg). The results obtained for all ten receptor peptides are summarized in Table 1.
Table 1.
Definitions of the IFNLR1 fragments fused to the FERM and SH2-like domains of JAK1 in various expression constructs, and their interactions with the FERM/SH2 fragments of JAK1.
| Construct | Amino acid sequence of the IFNLR1 fragment | Binds to JAK1* |
|---|---|---|
| C260_292 | RAKMPRALDFSGHTHPVATFQPSRPESVNDLFL | Yes |
| C260_297 | RAKMPRALDFSGHTHPVATFQPSRPESVNDLFLCPQKE | Yes |
| C260_307 | RAKMPRALDFSGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPR | Yes |
| C260_312 | RAKMPRALDFSGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPRVRAPA | Yes |
| C260_317 | RAKMPRALDFSGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPRVRAPATQQTR | Yes |
| C265_307 | .....RALDFSGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPR | No |
| C265_312 | .....RALDFSGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPRVRAPA | No |
| C265_317 | .....RALDFSGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPRVRAPATQQTR | No |
| C270_307 | ..........SGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPR | No |
| C270_312 | ..........SGHTHPVATFQPSRPESVNDLFLCPQKELTRGVRPTPRVRAPA | No |
This column indicates whether a receptor construct co-elutes with JAK1(FERM-SH2) during gel-filtration chromatography after cleavage with TVMV protease. This effect is monitored by an analysis of chromatographic fractions with the SDS-page stained with Coomassie.
Figure 1.
Analysis of the interactions between the receptor peptide and JAK1. In each panel lane 1 represents the molecular weight markers (SeaBlue II, Life Technologies); lane 2, uncleaved JAK1/IFNLR1 constructs; lane 3, the reaction mixtures after cleavage with TVMV protease; lane 4, components of a major single peak from the gel filtration chromatography on the mixtures applied to lane 3. The gel shown in panel A is for the construct JAK1/C260_307, whereas the gel for the construct JAK1/C265_317 is shown in panel B. The protein components associated with the visible bands are labeled. Based on these results, it is evident that C260_307 forms a complex with JAK1 (panel a) whereas no stable interactions exist between JAK1 and C265_317 (panel b).
Structural analysis of the IFNLR1/JAK1 complex, presented in sections that follow, helped in assigning the region ProR264–PheR269 as box1 and a fragment including several residues flanking GluR285 as box2. As seen in Table 1, the receptor peptides beginning with ArgR265 or SerR270 lack a part or the entire box1. Because none of these peptides binds stably to JAK1, we concluded that box1 is required for a stable, specific interaction between JAK1 and IFNLR1. Since all receptor peptides used in our studies include a sequence corresponding to box2, we postulate that box2 alone is insufficient for interaction with JAK1. Whereas our data cannot provide the answer whether the box1 region of IFNLR1 is by itself sufficient for stable binding to JAK1, this question is partly answered by Ferrao et al. [18]. Using results of pull-down experiments, they showed that the receptor section missing the entire box2 region forms only weak complexes with JAK1. Based on the results from the binding studies, we selected the IFNLR1/JAK1 construct in which the receptor is represented by the peptide C260_307 for further structural analysis.
Processing and analysis of the diffraction data
Crystals of the IFNLR1/JAK1 complex were grown as shown in Methods and diffraction data were collected from more than 15 individual crystals. Whereas complete datasets extending to the resolution of 2.8 Å could be obtained in most cases, diffraction at a higher resolution, up to 2.1 Å, was observed for only a few crystals. Even for these crystals, the completeness of data extending beyond 2.7 Å was diminishing with resolution (see Tables 2,3). Additionally, crystals suffered extensive radiation damage, presenting a challenge to collect complete datasets even at medium-to-low resolution. Visual inspection of X-ray images indicated an anisotropic distribution of the diffraction pattern, which was subsequently confirmed when the X-ray datasets were analyzed using the Diffraction Anisotropy Server (https://services.mbi.ucla.edu/anisoscale/). It was found that for all crystals the resolution of diffraction data was consistently lower along the b*-direction compared to two other directions, a* and c* (asterisks denotes that directions refer to the reciprocal space). As illustrated in Table 3, resolution in a* and c* directions approaches 2.1–2.2 Å whereas the resolution in the b* direction extends to just 2.7–3.3 Å, depending on the crystal. Subsequent analysis of the crystal packing explained the physical/molecular basis of the anisotropy observed for diffraction data, since protein molecules were found to form flat sheets perpendicular to the b cell axis. Weak interactions between these sheets allowed for their easy movement relative to each other. Furthermore, we observed that crystals were very thin in the direction of the b cell axis. In addition to the anisotropy, completeness and redundancy of the data collected from a single crystal was further decreased due to radiation damage. The latter problem could, however, be mitigated by merging data from several crystals. The validity of this approach was confirmed by the statistics shown in Table 3. This merged dataset was used during building and refinement of the IFNLR1/JAK1 structure. Although the nominal resolution of the final structure is 2.1 Å, the complete diffraction information extends only to about 2.6–2.7 Å. However, the additional 11,731 reflections measured within the resolution shell 2.7 – 2.1 Å account for 38% of total experimental data and their contribution significantly improved the quality of both the electron density and the final model, with both being much better than expected for a typical medium-resolution structure. A similar improvement was reported before for another system [33].
Table 2.
Statistics for diffraction data collection and processing.
| Diffraction source | Beamline 22-ID, SER-CAT, APS, ANL, IL, USA |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Detector | Rayonix 300HS high speed CCD |
| Crystal-to-detector distance (mm) | 270 |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 360& |
| Exposure time per image (s) | 1.0 |
| Space group | C2 |
| a, b, c (Å) | 165.70, 53.8, 87.39 |
| α, β, γ (°) | 90, 114.72, 90 |
| Mosaicity (°) | 0.25 – 0.90 |
| Resolution range (Å) | 99.00 – 2.10 (2.10 – 2.18)* |
| Total No. of reflections | 384,056 |
| No. of unique reflections | 40,748 |
| Completeness (%) | 98.6 (89.7) |
| Multiplicity | 9.4 (8.0) |
| <I/σ(I)> | 27.4 (1.94) |
| Rmerge† | 0.077 (0.634) |
| Rp.i.m‡ | 0.025 (0.218) |
| CC1/2 | 0.98 (0.89) |
| Selected statistics after elliptical truncation due to anisotropy | |
| (for the resolution range 99.0 – 2.10 Å) | |
| No. of unique reflections | 31,136 |
| <I/σ(I)>, highest resolution shell | 6.97 |
| Completeness (%) | 75.5 (25.9) |
| (for the resolution range 99.0 – 2.70 Å) | |
| No. of unique reflections | 19,405 |
| <I/σ(I)>, highest resolution shell | 24.8 |
| Completeness (%) | 99.4 |
Rotation ranges for individual crystals varied between 120 and 180°. Value reported in this Table is the result of merging the images collected from five crystals.
Values in parentheses are for the highest resolution shell.
Rmerge = Σ(|(I - <I>)| / Σ(I).
Estimated Rp.i.m. = Rmerge[N/(N-1)]1/2, where N is the multiplicity of data.
Table 3.
Statistics for five X-ray data sets collected from single crystals of Jak1–IFN-λR1. All data were processed with HKL3000 using the common resolution ranges, 50–2.1Å and the same overall protocol.
| Set | Unit cell | #refl. tot. Red (HiRes) Compl. (HiRes) |
I/σ(I) (HiRes) Rscale (HiRes) Rpim (HiRes) |
Data for resolution 2.55 Å (data for resolution 2.73 Å) |
Data from ANSO server (UCLA) https://services.mbi.ucla.edu/anisoscale/ |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compl [%] | I/σ(I) | Rscale | #refl. tot | Resolution along direction | ||||||
| a* | b* | c* | ||||||||
| m5p10 | a = 165.70 b = 53.82 c = 87.40 β = 114.72 |
28,810 3.2 (2.6) 82.7 (41.2) |
18.1 (1.72) 0.058 (0.468) 0.036 (0.324) |
90.9 (98.9) |
4.78 (8.41) |
0.189 (0.154) |
28,379 | 2.1 | 2.7 | 2.1 |
| m5p11 | a = 165.53 b = 53.23 c = 87.31 β = 114.66 |
24,638 3.3 (2.2) 75.3 (18.7) |
15.2 (1.30) 0.068 (0.481) 0.041 (0.339) |
74.9 (87.3) |
3.57 (5.07) |
0.245 (0.194) |
24,157 | 2.1 | 3.3 | 2.1 |
| n2p4 | a = 166.25 b = 54.26 c = 87.43 β = 114.72 |
24,732 2.9 (1.6) 84.8 (52.6) |
13.2 (0.77) 0.078 (0.873) 0.050 (0.724) |
92.3 (97.7) |
3.60 (5.23) |
0.390 (0.286) |
24,732 | 2.2 | 2.8 | 2.1 |
| n2p6 | a = 166.58 b = 54.30 c = 87.72 β = 114.77 |
25,896 3.4 (2.6) 80.0 (43.3) |
14.7 (1.33) 0.076 (0.656) 0.048 (0.451) |
83.8 (97.1) |
3.95 (5.05) |
0.296 (0.224) |
25,650 | 2.1 | 3.1 | 2.1 |
| n2p8 | a = 166.27 b = 53.66 c = 87.40 β = 114.70 |
27,981 3.2 (2.4) 85.9 (54.1) |
18.7 (2.31) 0.054 (0.394) 0.035 (0.296) |
95.2 (99.7) |
5.29 (7.82) |
0.234 (0.177) |
27,378 | 2.1 | 2.8 | 2.1 |
| merged | a = 165.70 b = 53.80 c = 87.39 β = 114.72 |
40,748 9.4 (8.0) 98.6 (89.7) |
27.4 (1.94) 0.077 (0.634) 0.025 (0.218) |
100 (100) |
10.2 (13.4) |
0.229 (0.165) |
31,136 | 2.1 | 2.8 | 2.1 |
The structure of the JAK1 component in the IFNLR1/JAK1 complex
The construct IFNLR1/JAK1, used in this investigation, spans residues 31–577 of the human JAK1, followed by a linker that includes six SG repeats interspersed with the TVMV protease cleavage site. The first residue that could be traced in the electron density map is Gly35, and the last visible residue is Glu563. Several areas of the chain are not ordered sufficiently well to be included in the final model. The missing segments correspond to residues 132–145, 211–215, 297–307, and 330–361 of the FERM domain, as well as residues 485–491 of the SH2 domain. The disordered regions represent loops that are not stabilized by either intramolecular or crystal contacts, and all are distant from the putative epitope for binding the IFNLR1 peptide.
The ordered parts of the FERM domain (residues 35–439) and the SH2-like domain (referred to simply as SH2 throughout this paper, residues 440–563) make intimate contacts with each other (Fig. 2). The FERM domain itself is composed of three subdomains, highlighted in Fig. 2.
Figure 2.
A cartoon showing the chain tracing and the molecular surface of the FERM/SH2 fragment of the human JAK1. The molecular surface of the FERM domain is green, with the chain of the N-terminal ubiquitin-like domain (F1) shown in dark orange, acyl-coenzyme A binding-like domain (F2) in green, and the plextrin-homology-like domain (F3) in blue. The molecular surface of the SH2-like domain is pink and the chain tracing orange. The amino acid sequence of the JAK1(FERM/SH2) construct used in this study is shown below the cartoon. Residues from specific domains are colored according to the same scheme as in the image above. Fragments of the sequence of the protein that are not included in the crystal structure are shown in lighter font and underlined.
A superposition of 417 Cα atoms from our model of JAK1 on the equivalent coordinates of TYK2 (PDB ID 4po6; [17]) yields rmsd of 1.35 Å. The recently published models of the corresponding regions of JAK1 [18] are less complete than our structure, especially for parts of the SH2 domain. A comparison of our structure with 5ixd yields rmsd of 0.82 Å for 402 Cα pairs, and 0.79 Å when compared with 5ixi, where 436 Cα pairs were included. Unlike the structure described here, the two models presented by Ferrao et al. contain a strand that consists of residues 297–304, not visible in our structure, which extends the large β sheet in a symmetry-related molecule through antiparallel contacts with residues 289–294. Whereas the latter stretch of residues is present and structurally similar in all three structures, the packing of molecules in the crystals of our complex does not allow extension of the above-mentioned β sheet. In addition, the turn between strands 289–295 and 297–304 is modeled somewhat differently in 5ixd and in 5ixi, resulting in a difference of 4.5 Å between positions of the Cα atoms of Asn297 in both structures. This region is quite different from that observed in our structure, in which crystal packing would not be compatible with either conformation presented by Ferrao et al. In particular, the tip of this loop would create severe clashes with the turn formed by residues 434–438 of a symmetry-related molecule. The latter region, in turn, is nearly identical in the three structures of JAK1 as well as in the structure of TYK2. On the other hand, the conformation of residues 283–295 in TYK2 (equivalent to 294 and following in JAK1) is completely different from the conformations presented in any models of JAK1. Residues 297–304 are involved in extensive crystal contacts with helices 227–231 and 235–243 in 5ixd, and create an extended β sheet with their own symmetry mate in 5ixi. Therefore, it is very likely that this segment of JAK1 is highly flexible and the specific conformations observed in the two structures of the receptor complexes of JAK1 are serendipitously stabilized by crystal contacts.
The recently published structure of the unliganded FERM/SH2 construct of JAK2 [19] is based on 3 Å data and contains eight molecules in the asymmetric unit. For the comparisons with the other structures we utilized the most complete molecule A. Superposition of our JAK1 model on molecule A of JAK2 yields the rmsd of 1.55 Å for 411 Cα pairs, somewhat higher than for TYK2. This larger difference may, however, be due to both the lower resolution of the JAK2 structure and many minor differences in the conformations of specific regions. A comparison does not indicate gross topological differences, i.e. changes in relative positions of whole domains, between the complexed and unliganded states of JAK molecule. Several of the disordered loops that are not visible in the structures of the IFNLR1/JAK1 complex were fully traced in the structure of JAK2. The equivalent of the missing loop 132–145 in JAK1 is a much shorter loop 137–140 in JAK2, stabilized by an interaction with a symmetry-related molecule. Similarly, a very well-defined helical turn 201–204 in JAK2, held in place by extensive interactions with residues 73–76 of the adjacent molecule B, corresponds to missing residues 211–215 in JAK1. The regions corresponding to the disordered loops 297–307 and 330–361 in JAK1 are also missing in the structure of JAK2. Finally, the loop formed by residues 485–491 of the SH2 domain and not modeled in JAK1 is much shorter in JAK2 and its conformation is also stabilized by intermolecular interactions.
Interactions between JAK1 and the IFNLR1 peptide
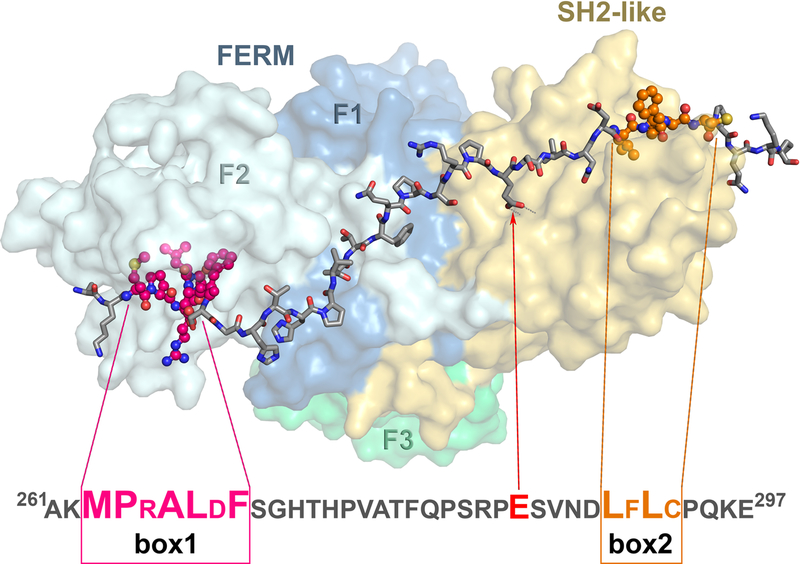
In our construct, the fragment of JAK1 consisting of the FERM and SH2 domains and a 17-residue-long linker is followed by a 48-residue-long section (residues R260-R307) of the intracellular domain of IFNLR1, thereafter referred to as “the receptor peptide”, with its sequence numbers preceded by letter “R”. With the exception of ArgR260 and the C-terminal segment starting at LeuR298, a majority of the residues of the receptor peptide could be unambiguously traced in the electron density map, although the electron density corresponding to Gly271-His274 was discontinuous when contoured at 1σ level. Enhanced flexibility of this stretch of the receptor peptide can be linked to very few interactions that it forms with JAK1. Despite less well-defined electron density, this fragment could be modeled with an acceptable geometry, resulting in the model of a contiguous receptor peptide extending from AlaR261 through GluR297 (Fig. 3), thus containing both box1 and box2 residues [25]. The last modeled residue in the receptor peptide projects towards a large open channel in the crystal, therefore the residues that follow are most likely disordered. The length of the visible part of the receptor peptide is over 85 Å and the surface area buried by its interaction with JAK1 is 3390 Å2.
Figure 3.
A cartoon representation of the IFNLR1/JAK1 complex. The JAK1 component, consisting of the FERM domain (colored in three shades of blue and green, indicating different subdomains which are labeled) and the SH2-like domain (colored yellow), is shown in the molecular surface representation. The IFNLR1 receptor fragment is shown in stick representation with the regions of box1 and box2 highlighted in dark pink and orange, respectively. Additionally, GluR285 that interacts with β5B Arg from the SH2 domain is indicated by a red arrow. The sequence for a fragment of IFNLR1 visible in the crystal structure is shown below the cartoon with box1, box2, and GluR285 highlighted. Residues involved in close interactions with JAK1 are indicated by enlarged font,
The N-terminal part of the receptor peptide forms a distorted 310 helical turn involving ProR264-LeuR267 and is located inside a wide open channel, with sides formed by a helix-turn-helix motif in the FERM domain (residues 243–273), and the bottom lined by residues 184–195, contributed by a longer helix of the FERM domain. The electron density is defined well (Fig. 4a). The side chain of LeuR267 is wedged inside a hydrophobic pocket formed primarily by the side chains of Val194, Phe247, and Phe251. ProR264 and the side chain of MetR263 also point towards that pocket, whereas the side chain of ArgR265 faces a solvent channel and is not involved in direct interactions with JAK1. The side chain of PheR269 occupies another hydrophobic pocket consisting of Phe251, Val261, Leu266, and the hydrophobic part of the side chain of Lys269. The conformation of the receptor peptide is also stabilized by an intramolecular hydrogen bond between the carbonyl oxygen of AspR268 and the NZ of LysR269. With the exception of HisR274, which forms a putative weak hydrogen bond with the carboxylate of Asp121, the section the receptor peptide consisting of residues SerR270-ValR276 does not interact with JAK1. Since this part of the receptor peptide is stabilized by only a single weak interaction, the electron density for this stretch of residues is comparatively unclear.
Figure 4.
The coordinates and the corresponding difference electron density maps contoured at the levels 1.3σ (2Fo-Fc, gray), 3.0σ (Fo-Fc, green), and −3.0σ (Fo-Fc, red) for the regions of the interactions between IFNLR1 (Cα atoms colored gray) and JAK1 (Cα atoms colored green). (A) A stereo image of the box1 region in IFNLR1 interacting with the neighboring residues belonging to the FERM domain of JAK1. (B) Interactions in the area corresponding to box2.
The residues that follow in the chain of the receptor make hydrogen-bonded interactions with the residues of JAK1. The carbonyl oxygen of AlaR277 interacts with the amide nitrogen of Leu150 through a water molecule, while direct hydrogen bonds are present between N of PheR279 and O of Leu150, as well as O of PheR279 and N of Ala152. The aromatic ring of PheR279 is located in a hydrophobic pocket created by the side chains of Leu73, Thr147, and Pro148. The electron density for the stretch between ProR281 and ProR284 is very well-defined for the prolines and the main chain of the residues between them, but it is missing for the side chains of SerR282 and ArgR283 – none of these residues interacts with JAK1.
The side chain of GluR285 makes a salt bridge with the guanidinium group of Arg466 (Fig. 4b). The latter residue is very well ordered, due to restraining hydrogen bonds with OG1 of Thr478 and OD1 of Asn497, as well O of Ile444, mediated by a water molecule. The main chain of the peptide is further stabilized by hydrogen bonds involving N of AsnR288 and O of Asn497, OD1 of AsnR288 and N of Asn497, as well as a water-mediated interaction between O of AsnR288 and N of Gln499. The side chain of ValR287 is stabilized by hydrophobic interactions with the non-polar section of the Gln499 side chain. The carbonyl oxygen of AspR289 is hydrogen bonded to OG1 of Thr533, while the next several residues of the receptor peptide form antiparallel β-strand interactions with the main chain of the strand in the SH2 domain (N of PheR291 with O of Arg532; O of PheR291 with N of Arg532; N of CysR293 with O of Ile530; O of CysR293 with N of Ile530; and N of Gln295 with O of Lys528). This part of the chain is additionally stabilized through crystal contacts with its counterpart in a symmetry-related molecule that involve both hydrogen bonds and hydrophobic interactions. The only stabilizing interaction for the C-terminal part of the receptor peptide involves a putative water-mediated contact between NZ of LysR296 and N of LeuR290 of a symmetry-related molecule and does not interact with the JAK1 molecule.
A comparison of the structure and interactions of the SH2-like domain of JAK1 to its classical counterparts
In the model discussed here the structure of the JAK1-SH2 domain is complete, with the exception of seven residues within the tip of the CD loop. As seen in Fig. 5, the overall topology of the JAK1-SH2 domain is the same as of p56lck, a classical SRC-like SH2 domain, in which two α-helices (αA and αB) sandwich the central three-stranded antiparallel sheet (βBCD). Among the main differences between the JAK1-SH2 and p56lck domains is the length of the BG loop, which in JAK1 forms a significantly longer β-hairpin. Also, the CD loop in JAK1-SH2 is longer than in p56lck, although it is partially disordered in our structure. Details of the interactions between the JAK1-SH2 domain and the IFNLR1 are shown in Supplementary Fig. S1.
Figure 5.
The SH2 domain of human JAK1 (blue) superimposed on p56lck [43], a classical SRC-like SH2 domain (transparent orange). The JAK1-SH2 domain is shown in a complex with the fragment (SerR282-GlnR295) of a cognate receptor, IFN-λR1 (green), while the p56lck is shown in the complex with a specific pTyr-containing peptide (orange). The secondary structure elements and the intervening loops, named accordingly to the SH2-specific convention, are marked with black labels. Locations of two terminal residues and those flanking a disordered CD-loop in JAK1-SH2 are indicated with arrows and blue labels. Two sites, the primary binding site of pTyr and the secondary (specificity) site in classical SH2 domains that overlaps with the Box2-binding site in JAKs, are marked with boxes and magnified in side panels. (a) Interactions of GluR285, the residue mimicking pTyr in complexes of classical SH2 domains, with JAK1-SH2 residues. Polar interactions are indicated with cyan dashed lines. The receptor residues (dark green) are shown in ball-and-stick representation. (b) An equivalent site in p56lck (light gray, in stick representation) showing an extensive network of interactions contributed by the pTyr residue from the ligand (dark gray, ball-and-stick representation). A number of SH2-ligand interactions within both sites, suggests much tighter binding in the case of the lckp56-pTyr complex. (c) Interactions between Box2 section of IFNλR1 (Leu290-Phe-Leu-Cys293, shown in dark green as ball-and-sticks) and a hydrophobic pocket of JAK1-SH2 (light green, sticks). Polar contacts are indicated with dashed lines. (d) The equivalent part of the p56lck complex, with the ligand residues labeled both with specific names as well as positions relative to pTyr (in parentheses).
In general, positioning of the receptor molecule against the JAK1-SH2 domain is similar to that of a pTyr-containing peptide bound to p56lck. Both ligand molecules are assuming extended conformations and are oriented perpendicularly to the plane of the central sheet, associating with the respective SH2 domains via interactions with two sites located on the opposite faces of the βBCD sheet. Similar observations were noted earlier for the complex formed between TYK2 with IFNAR1 [17]. The interaction between IFNLR1 and JAK1-SH2 is quite extensive and buries ~676 Å2 of the molecular surface of JAK1. Several significant differences are found between specific interactions of the IFNLR1 with JAK1-SH2 and the phosphorylated ligands bound to classical SH2 domains. The primary binding site in the latter domain evolved to accommodate pTyr, and the phosphorylated phenyl group contributes multiple polar interactions (see Fig. 5b). The role of pTyr in IFNLR1 is played by GluR285, which forms a salt bridge with β5B Arg. Due to the differences in the size of side chains, the position of GluR285 in the JAK1-receptor complex is shifted by two residues relative to the position of pTyr in p56lck. Additionally, the side chain of pTyr cannot be presented to the β5B site in JAK1 in the same way as in p56lck because of potential collision with Leu476 (see Fig. 5a). A comparison of the interactions within the β5B sites of JAK1-SH2 and p56lck (Figs. 5a,b) suggests that in JAK1 a contribution of this site to the binding strength of the ligand is much less than in typical SH2 domains. This conclusion agrees with an earlier report showing that mutation of β5B Arg to Lys does not significantly affect the interaction of JAK1 with IFNLR1 [31]. Our structural data indicate that a lysine residue occupying this position would most likely retain the ability to interact with the side chain of GluR285. It is also tempting to suggest that the presence of β5B Arg in JAK1-SH2 is by itself not sufficient to assure strong binding of pTyr-containing ligands. Other differences distributed throughout the site, such as the presence of Leu476, could interfere with pTyr binding. The observed mode of interactions between β5B Arg (JAK1) and GluR285 (IFNLR1) was not described in the previously studied complexes of SH2 domains since, for example, the network of interactions described in the TYK2/IFNAR1 complex [17] differed (see Supplementary Fig. S2a). The lack of structural conservation of the interactions within the β5B site across different JAKs suggests a lesser role of this site in binding to the receptor.
As noted previously [17], the SH2 domains in JAKs very likely evolved in order to support constitutive binding to receptors, as opposed to temporary interactions with pTyr seen for typical SH2 domains. Further support for this idea can be found through analysis of the site located on the opposite side of the βBCD sheet. Typically, this secondary binding site provides additional ligand specificity of SH2 domains. The shape of this specificity site is defined by several hydrophobic residues to assure a good fit for a side chain of the Y+3 residue of a phosphorylated ligand, such as Ile207 in the case of p56lck, shown in Fig. 5d. Whereas this interaction adds stability to binding of a phosphorylated ligand by a classical SH2 domain, its contribution is secondary compared to the β5B site. In the case of JAKs, the region of a receptor that interacts with this secondary site largely overlaps a hydrophobic section of the box2 motif (LeuR290-PheR291-LeuR292-CysR293). The main interface in JAK1-SH2 is formed by the βx strand from the long BG loop and the box2 section of IFNLR1 that accommodates β-conformation, extending to three strands the anti-parallel sheet (see Fig. 5c). The network of interactions within this interface is much more extensive than in the case of a typical SH2 domain. In addition to hydrophobic interactions contributed by the side chains LeuR290 and LeuR292 from the box2 motif, the receptor participates in six hydrogen bonds stabilizing the binding (also see Supplementary Fig. S2b). It is apparent that a contribution of interactions within this site to the strength of binding of the receptor by the JAK1 molecule exceeds the contribution provided by the β5B site. This seems to be also true for TYK2 and likely for other JAKs, see Supplementary Fig. S2c), but not for typical SH2 domains.
The structural analysis presented here shows that the SH2 domain provides major contribution to the association of JAK1 and IFNLR1 molecules and explains why the canonical β5B Arg is dispensable for this binding. As postulated previously [31], extensive interactions between JAK1-SH2 and the receptor provide a rational explanation for the central role of the entire SH2 domain in this process.
Comparisons of the binding sites of receptor peptides to Janus kinases
Two regions of the intracellular parts of cytokine/interleukin receptors have been identified as playing important roles in binding the members of the Janus family of kinases. These regions, characterized by only very limited sequence conservation, have been named box1 and box2 [25]. The construct used previously for structural studies of the complex between TYK2 and the intracellular part of the IFNAR1 does not include box1 residues, since its starting point is at residue IleR478, about 20 residues past the end of the putative transmembrane helix. Nevertheless, this fragment of IFNAR1 was sufficient to create a stable complex with the FERM and SH2 domains of TYK2. Whereas both boxes were included in the peptides used to determine the recently published structures of the IFNLR1/JAK1 and IL10RA/JAK1 complexes [18], only box1 region was involved in biologically relevant interactions. Residues belonging to box2 were disordered in the former complex and were involved in serendipitous intermolecular contacts in the latter one (see below). Thus the structure of the IFNLR1/JAK1 complex described here is the first one in which 1:1 binding of the receptor peptide to the FERM/SH2 construct of a Janus kinase can be visualized based on crystallographic data (Figs. 3,6).
Figure 6.
A comparison of the mode of binding of the peptides representing the intracellular domains of interferon receptors to JAK-family kinases. The chain tracing of the secondary structure of the FERM/SH2 domains of JAK1 is shown in wheat. Residues R261-R297 of the intracellular part of the IFNLR1 are shown in stick representation, with carbon atoms colored green. The carbon atoms in the corresponding peptide from the PDB file 5ixd (residues R250-R270) are colored orange, whereas the carbon atoms of the IFNAR1 peptide (residues R489-R507 in 4po6) are colored magenta. The latter peptide is shown after superposition of the coordinates of TYK2 (not shown) on JAK1.
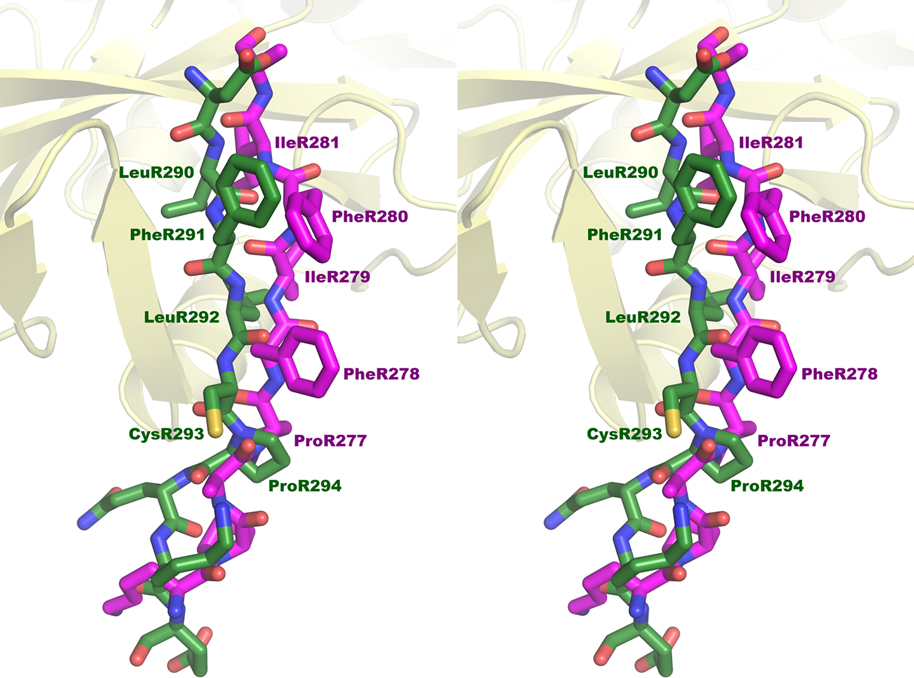
The residues that belong to the poorly conserved box1 general motif PxxPxP (residues ProR264 through PheR269 in IFNLR1), which is found in most intracellular domains of the cytokine and interferon receptors that bind JAK1, were present in both the structure described here and in the two structures determined by Ferrao et al. [18]. The extent of the box1-containing fragment is different in these two investigations. Whereas in both constructs described by Ferrao et al. the receptor peptide begins with LysR250 [18], in the construct used in our studies the starting residue is ArgR260, and the first residue modeled in the electron density is AlaR261 (Fig. 7a). Another difference between the two approaches is the mode of presentation of the receptor peptide to the JAK1 component. Whereas in our construct a receptor peptide is linked to the JAK1 fragment by a flexible linker (containing several SG repeats, with the TVMV cleavage site in the middle), in the complexes described by Ferrao et al. the JAK1 and IFNLR1 (or IL10RA) components were expressed independently prior to formation of the complexes [18]. The N-terminal sections of the receptor peptides in the latter structures were intimately involved in crystal contacts which could have influenced their conformation. The linker residues present in our structure that could have potentially mimicked the first 10 residues of the box1-containing region in 5ixd are not visible. This observation suggests that residues R250-R260 are dispensable for creating a stable complex. This conclusion is in agreement with the results of Ferrao et al. [18], who noted that mutation of TrpR257 to Ala did not affect the affinity of the interaction in a significant way, even though the side chain of this residue in the model 5ixd was involved in quite extensive contacts with JAK1, including a hydrogen bond with Glu186.
Figure 7.
A comparison of the mode of binding of IFNLR1 and IL10RA to JAK1. (a) The chain tracing showing the secondary structure of JAK1 (in wheat) with the Cα trace of the IFNLR1 peptide (green), as well as the IL10RA peptide (orange) bound through box1, and a symmetry-related molecule of this peptide bound in an opposite direction to box2 epitope. (b) A detailed superposition of the box1 residues of IFNLR1 (green) and IL10RA (orange) interacting with JAK1. (c) Superposition of the residues in IFNLR1 (green) and in IL10RA (magenta) interacting with the box2 epitope of JAK1. In case of IFNLR1 these residues represent the box2 region of the receptor, whereas residues in IL10RA are contributed by a symmetry-related chain running in an opposite direction, an artifact that was noted in the original publication [18].
The conformation of the residues from the box1-defining motif, here PxxLxF, is very similar in all three structures, although minor differences (which could have been due to the limited resolution of diffraction data used in all three structural studies) can be seen. The structure of the PxxLxF motif of IFNLR1 together with interacting residues from JAK1, determined in this study is shown in Fig. 4a, and a superposition with the corresponding region of the IL10RA/JAK1 complex in Fig. 7b. The carbonyl oxygen of LysR262 (LysR265 in IL10RA) makes hydrogen bonds with Asn187 of JAK1 in 5ixd and 5ixi (with OD1 in the latter structure, most likely due to its incorrect assignment), whereas the corresponding distance in our structure exceeds 4 Å, indicating that no hydrogen bond is present. The guanidinium moiety of ArgR265 makes a salt bridge with JAK1 Asp184 in 5ixd but not in our structure, while the side chain of the equivalent SerR268 in 5ixi makes a long hydrogen bond with the carboxylate of Glu188. Both AlaR266 in IFNLR1 and the equivalent ValR269 in IL10RA face a hydrophobic pocket. Both LeuR267 and PheR269 (both conserved in IL10RA) are involved in virtually identical interactions with in the JAK1 pockets, whereas AspR268 makes a clear intramolecular hydrogen bond with the amide nitrogen of SerR270, with no equivalent bond present in the 5ixd model (in which SerR270 is the last visible residue, with differently modeled peptide bond). This hydrogen bond also has no equivalent in 5ixi, where the equivalent residue is LeuR271; nevertheless, the conformation of the peptide of LysR273 is very close to SerR270 in our structure.
All three structures that include the conserved box1 motif corroborate its importance to stabilizing the binding between JAK1 and the intracellular parts of the cytokine/interferon receptors. As shown in our binding experiments, constructs of IFNLR1 starting with ArgR265 or SerR270, thus lacking part of whole of box1 motif, did not bind to JAK1 (Table 1), whereas the construct starting at ArgR260 bound strongly enough to be useful in crystallization experiments. The first ten residues preceding ArgR260 (the sequence predicted to start immediately following the transmembrane helix in IFNLR1) do not seem to be critical for the affinity of the interaction between the two partners. All these results are in agreement with the results of Ferrao et al. [18] and with other available mutation data.
Residues R276-R297 form an extended chain that interacts at its N terminal region with residues 149–151 of the FERM domain of IFNLR1, but a majority of its interactions involve the SH2 domain. This stretch of residues contains the putative box2, but the sequence bears very little conservation among various cytokine and interferon receptors. A single residue, highly conserved across different cytokine receptors is GluR285 (equivalent to GluR497 in IFNAR1). The structure of the interface between box2 from IFNLR1 and JAK1, determined in this study, together with primary stabilizing interactions between the two fragments, is shown in Fig. 4b. In our structure both carboxylate oxygens of GluR285 form an ion pair with the NH1 and NH2 nitrogens of the guanidinium group of Arg466. The side chain of Arg466 is well ordered and its conformation is stabilized by several additional hydrogen bonds (NE-OG1 of Thr478; NH2-OD1 of Asn497; and a water-mediated hydrogen-bonded interaction of NH1 with the carbonyl oxygen of Ile444 – Fig. 4b). Thus Arg466 in the SH2 domain of JAK1 seems to be especially well posed for stabilizing the interactions with the conserved glutamate of the receptor. Interestingly, the conformation of the side chain of Arg426 in JAK2 [19], corresponding to Arg466 in JAK1, is virtually identical and well-supported by the electron density in both structures. Inspection of the JAK2 Fo-Fc electron density map indicates a possible presence of a water molecule which was not modeled, held in place by hydrogen bonds provided by NH1 of Arg426 to the carbonyl oxygens of Ile404 and Cys427. The other stabilizing interactions for this crucial residue were also not modeled in JAK2, but may nevertheless be present. Thr438 in JAK2 corresponds to Thr478 in JAK1 and its side chain could potentially be rotated to form the same hydrogen bond with the NE atom of the guanidinium group. Whereas the other residue that stabilizes Arg466 in JAK1, Asn497, is replaced by His451 in JAK2, it is possible that by turning the imidazole ring by 180°, the presence of a stabilizing N-N hydrogen bond could be postulated. The model of the SH2 domain of JAK1 is partially disordered in 5ixd (for example, Thr478 is not present) and thus the rotamer of the side chain of Arg466 is not the same, but it is almost identical in the better-ordered structure of the complex with the hybrid receptor peptide (5ixi), including the presence of the stabilizing water molecule. It may be concluded that Arg466 is strongly held in place in both JAK1 and JAK2, ready to form a structurally-conserved salt bridge with the glutamate present in the cytoplasmic domains of most receptors to which these kinases bind.
A part of the box1 epitope on JAK1 binds several residues from a symmetry-related peptide in the crystal structure of the IL10RA/JAK1 complex (Fig. 7c). However, the chain runs in a direction opposite to the one observed in the IFNLR1/JAK1 complex described here. Since the side chains of several identical or similar residues are oriented similarly in both structures, we may conclude that the binding observed in the former structure may have contributed to creation of the crystal lattice by serendipitously mimicking the binding of part of the box2 residues, but it was not contributed by the actual box2 residues in IL10RA.
Although GluR497 was also described as making extensive polar interactions with the side chain hydroxyl oxygens of Ser476 and Thr477 of the SH2 domain of IFNAR1 [17], such interactions would most likely not be as strong and specific as the ion pair found in the structure presented here. It is of interest, though, that although the interactions that involve box2 are sufficient for the formation of a stable complex between TYK2 and IFNAR1 [17], they are not sufficient to stabilize the complex between JAK1 and IFNLR1 (Table 1 and [18]), raising a question of the importance of the contribution of the interactions of GluR285 to its stability.
Materials and methods
Expression and purification of the JAK1–IFN-λR1 fusion protein (JAK1-IFNLR1)
The cDNAs encoding human JAK1 (Cat. No. MHS1010–202694799) and human IFNLR1 (Cat. No. MHS6278–213246004) were purchased from Open Biosystems, GE Dharmacon. All oligonucleotide primers used in this study were purchased from Integrated DNA Technologies. All DNA sequence analyses were performed by Macrogen Inc. The sequence encoding the FERM and SH2 domains of the human JAK1 (a.a. 31 – 577) was PCR amplified using primers J-fwd (5’-CCCCCCATGGCTGAGAACCTGTACTTCCAGGGCGCCCCTGAGCCAGGGGTG-3’) and J-rev (5’-TCCACTACCAGATCCGCTACCACTACCTGATCCAGACCGATCGAAACTCAGCTGGCTC-3’). The sequence encoding a fragment of the IFNLR1 (a.a. 270–307) was PCR amplified using primers I-fwd (5’-TCTGGATCAGGTAGTGGTAGCGGATCTGGTAGTGGATCCGGTCACACCCACCCC-3’) and I-rev (5’-GGGGCTCGAGCTATTATCTGGGGGTGGGACGGAC-3’). The PCR reactions were performed according to a standard protocol using the PfuUltra II Hotstart PCR Master Mix (Cat. No. 600850 Agilent Technologies). Primers J-rev and I-fwd, in addition to regions overlapping sequences of JAK1 and IFNLR1, respectively, also contained complementary sequences (36 bases) encoding a twelve residue long linker, (SG)6. In turn, primers J-fwd and I-rev were flanked by the NcoI and XhoI restriction sites, respectively, facilitating ligation of the final product with an expression vector. Furthermore, a sequence encoding the tobacco etch virus (TEV) protease recognition sequence, ENLYFQG, was inserted downstream of the NcoI site. The PCR assembly of the amplified fragments, in the presence of primers J-fwd and I-rev, created the construct encoding the FERM and SH2 domains of human JAK1 followed by a flexible linker and a 38 a.a. fragment of human IFNLR1. The product, amplified, gel-purified, and nicked with restriction enzymes (Cat. No. R3193S and R0146S, New England BioLabs Inc.) was cloned into the pET-32a(+) expression plasmid (Cat. No. 69015, EMD Millipore). The ORF in the expression vector encoded the thioredoxin-tag, His6-tag, a TEV protease cleavage site, and the fused JAK1 and IFNLR1 fragments, connected by a flexible linker.
The expression vector was transformed into Rosetta 2 (DE3) competent cells (Cat. No. 71400, EMD Millipore), and pilot expression experiments were conducted at both 310 K (Pilot37) and 289 K (Pilot16). In both cases cells were cultured in a Luria-Bertani (LB) medium, supplemented with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol at 310 K until they reached a mid-log phase (A600=0.5). At this stage, the temperature of the Pilot16 culture was lowered to 289 K and expression in both cultures was induced by the addition of isopropyl β-D-thiogalactopyranoside, IPTG, (Cat. No. AB00841, American Bioanalytical) to a final concentration of 0.5 mM. Growth of the Pilot37 continued for additional 4 hours with intermittent extraction of samples for analysis, whereas the culture Pilot16 was cultivated overnight. The large-scale expression was conducted according to the protocol established for the culture Pilot16. The cells were harvested by centrifugation and stored at 193 K for further processing. About 25 g of cells were thawed and suspended in 200 mL of buffer containing 50 mM Tris, pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM TCEP, 25 mM imidazole, 0.5 mM PMSF and EDTA-Free protease inhibitor (Cat.No. 04693132001, Roche Diagnostics). All subsequent steps were performed at the temperature of 278 K.
After lysing cells in a microfluidizer and addition of CHAPS to the final concentration of 0.25% (w/w), the suspension was incubated for 1 hour. The lysate was then cleared by centrifugation at 15,000 rpm for 1 hour followed by filtration through 0.45 μm membrane. In the first purification step, protein was recovered with the His60 Ni Superflow resin (Cat. No. 635660, TaKaRa) by rocking the suspension for 1 hour. After pouring the suspension on a gravity column and washing with the lysis buffer, protein was eluted with a solution of 250 mM imidazole in the lysis buffer. Combined fractions containing Trx-Jak1-IFNLR1 were then concentrated and applied on the HiLoad Superdex 200 16/60 PG column (Prod. No. 28989335, GE Healthcare Sci.) equilibrated with the buffer containing 50 mM Tris, pH 8.0, 0.3 M NaCl, 10% glycerol, and 1 mM TCEP. Combined fractions of Trx-Jak1-IFNλR1 were subjected to an overnight cleavage with the His6-TEV protease at 1 mg/ml. The next morning, the reaction mixture was applied on the 5 ml HisTrap FF column (Prod. No. 17-5286-01, GE Healthcare Sci.) equilibrated with the buffer containing 50 mM Tris, pH 8.0, 0.3 M NaCl, 10% glycerol, and 25 mM imidazole. The eluent fractions containing JAK1-IFNLR1 were collected, concentrated, and subjected to the final purification step using the HiLoad Superdex 200 16/60 PG column equilibrated with the buffer containing 25 mM Tris, 150 mM NaCl, and 1 mM TCEP. Purified protein was concentrated to 12 mg/ml and either used immediately for crystallization experiments or flash-frozen in liquid nitrogen and stored 193 K. All purification steps were monitored by SDS-page electrophoresis and the final sample was analyzed by mass spectrometry.
As discussed in the Results section, the recombinant preparation obtained from this construct could not be crystallized, due to the lack of interactions between the JAK1 and IFNLR1 fragments. In particular, the boundaries of the receptor fragment required further delineation. To facilitate systematic analysis of the interactions between both proteins prior the crystallization attempts, we introduced the tobacco vein mottling virus (TVMV) protease recognition sequence, TVRFQ, into the linker region. This modification was achieved by the QuikChange site-directed mutagenesis (Agilent Technologies) with the aid of two primers, 5’-TCTGGATCAGGTAGTGGTACCGTGCGTTTCCAGAGCGGATCTGGTAGTGGA-3’ and 5’-TCCACTACCAGATCCGCTCTGGAAACGCACGGTACCACTACCTGATCCAGA-3’. As a results of this change the linker region was extended from twelve amino acids (SGSGSGSGSGSGSG) to 17 (SGSGSGTVRFQSGSGSG). This construct served as a template for subsequent modifications leading to variants encoding fragments of IFN-λR1 that varied by length. Using the QuikChange site directed mutagenesis and different sets of oligonucleotide primers, we prepared 10 constructs differing by the length of a receptor fragment and described in Table 1. All proteins were expressed and purified according to the scheme described above. The construct C260_307 was successfully crystallized and subjected to detail structural analysis.
Binding of IFNLR1 fragments to JAK1
For binding experiments, fusions of Trx-His6-JAK1(FERM-SH2) with sections of IFNLR1 of different length were cloned, expressed, and purified as described above, leading to the constructs described by a general formula JAK1-(SG)3-TVMV-(SG)3-[IFNLR1]x. The sequences of the receptor peptides, [IFNLR1]x, are shown in Table 1. Each of the constructs was subjected to cleavage with TVMV protease, breaking a covalent linkage between JAK1 and IFNLR1 fragments. The cleavage reaction was conducted for 8 h at 4 °C in a buffer containing 50 mM Tris, 10% glycerol, 200 mM NaCl, pH 8.0, in the presence of 200 μg of TVMV protease. Completeness of the cleavage was monitored by SDS-Page electrophoresis. The reaction mixture was applied on the gel filtration HiLoad Superdex 200 16/60 PG column and eluted with a buffer containing 50 mM Tris, pH 8.0, 200 mM NaCl, 1 mM TCEP. The presence of the interaction between both fragments was deduced from the elution profile.
Crystallization
Preliminary crystallization trials were performed at 293 K using the Phenix crystallization robot (Art Robbins Instruments) and a wide range of commercial crystallization screens. The sitting droplets were prepared by mixing the buffered protein solutions (7 – 13 mg/ml in 50 mM HEPES pH 7.5, and 150 mM NaCl) with the crystal screen solutions. On average, the volume of each droplet was about 0.25 μl with the volume ratios of protein to reservoir solutions varying between 2:1 and 1:2. Crystals appeared under several different conditions, usually containing medium molecular weight polyethylene glycol (PEG) as a precipitant, medium concentration (between 0.1 and 0.2 M) of salt, and pH ranging from 6.5 to 8.5, depending on the buffer. In all cases the crystals were shaped as thin plates. Optimized crystals with approximate dimensions 0.1×0.5×0.01 mm3 were grown in the hanging droplets formed by mixing 1.5 μl of protein solution (10 mg/ml) with the equal volume of precipitant solution composed of 18% (w/v) PEG and 0.2 M ammonium formate. Individual crystals were harvested using Litholoops (Molecular Dimensions), briefly flashed in the precipitant solution, enriched with 27% (v/v) of glycerol, and finally frozen in a liquid nitrogen (100 K) for subsequent diffraction studies.
Collection of X-ray data
X-ray diffraction data were collected at the Advanced Photon Source in Argonne National Laboratory, (Argonne, IL, USA). All diffraction experiments were conducted at 100 K. The experimental images processed with subsequent scaling of reflection intensities using the programs HKL2000 and HKL3000 (HKL Research Inc.) [34, 35]. Details of the collection of experimental data and statistics from subsequent processing are presented in Tables 2 and 3.
Structure solution and refinement
Using the structure of the TYK2 molecule from the IFNAR1/TYK2 PDB deposit 4po6 as the search model [17], a solution of the IFNLR1/JAK1 complex could be easily identified by the molecular replacement (MR) method with the program PHASER [36]. An early dataset collected at the resolution extending to only 2.75 Å was used in MR. The solution was characterized by the values of Z-score and LLG equal to 15 and 503, respectively. Using another dataset, the solution was then rebuilt, including conversion to the correct amino acid sequence, with the aid of the programs PHENIX [37]and MR Rosetta [38], and refined at the resolution 2.5 Å using the program Refmac5 [39]. At this stage the R-value and Rfree were 0.21 and 0.30, respectively, and the stereochemistry of the model was mostly correct. Inspection of the electron density maps clearly indicated the location of the IFNLR1 receptor fragment on the surface of the JAK1 molecule. However, due to the limited experimental information and a significant bias of the MR-based model, further improvement of the IFNLR1/JAK1 structure was very slow and gained impetus only after better X-ray data became available (see above and Table 3). In the next step the structure was subjected again to automatic rebuilding using the programs MR Rosetta [38] and ARP/wARP [40]. Complementary information, aimed at minimizing the initial bias, was obtained from the composite omit maps calculated with the program Phenix [37]. Combined information from the models generated by either of the two automated approaches, with the omit maps and subsequent manual corrections aided by the program Coot [41], allowed us to build a model of the IFNLR1/JAK1 structure that could be refined using the program Refmac5 to the final values of R and Rfree of 0.169 and 0.230, respectively, with acceptable stereochemistry. Details of the refinement statistics are shown in Table 4. Figures showing the structural models and electron density were prepared with the program PyMol [42].
Table 4.
Refinement statistics for the structure of the IFNLR1/ JAK1 complex
| Resolution range (Å) | 79.38 – 2.10 (2.15 – 2.10)# |
| Completeness (%) | 75.5 (20.6) |
| σ cutoff | none |
| No. of reflections, working set | 30,056 (598) |
| No. of reflections, test set | 1080 (26) |
| Final Rcryst | 0.169 (0.190) |
| Final Rfree | 0.230 (0.248) |
| No. of non-H atoms | |
| Protein | 4036 |
| Water | 185 |
| Total | 4221 |
| R.m.s. deviations | |
| Bonds (Å) | 0.018 |
| Angles (°) | 1.852 |
| Average B-factors (Å2) | |
| Protein (JAK1, IFNLR1) | 50.7 (49.3, 68.3) |
| Water | 45.8 |
| Total | 50.5 |
| Ramachandran plot§ | |
| Favored regions (%) | 96.1 |
| Additionally allowed (%) | 3.9 |
| Number of complexes in a.u. | 1 |
| PDB code | 5l04 |
Statistics were calculated with the Web-based version of MolProbity (http://molprobity.biochem.duke.edu/ [44])
Outermost shell
Supplementary Material
Acknowledgements
This project was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31–109-Eng-38.
Footnotes
Accession numbers
The coordinates and structure factors of the IFNLR1/JAK1 complex have been deposited in the Protein Data Bank (accession code 5l04)
References
- [1].O’Sullivan LA, Liongue C, Lewis RS, Stephenson SE, Ward AC. Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol Immunol. 2007;44:2497–506. [DOI] [PubMed] [Google Scholar]
- [2].Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14:504–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schwartz DM, Bonelli M, Gadina M, O’Shea JJ. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nat Rev Rheumatol. 2016;12:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: Crystal structure of the complex. Science. 1992;255:306–12. [DOI] [PubMed] [Google Scholar]
- [6].Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344:1249783. [DOI] [PubMed] [Google Scholar]
- [7].Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN. The molecular regulation of Janus kinase (JAK) activation. Biochem J. 2014;462:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Walsh ST. Structural insights into the common gamma-chain family of cytokines and receptors from the interleukin-7 pathway. Immunol Rev. 2012;250:303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Broughton SE, Dhagat U, Hercus TR, Nero TL, Grimbaldeston MA, Bonder CS, et al. The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunol Rev. 2012;250:277–302. [DOI] [PubMed] [Google Scholar]
- [10].Zdanov A Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev. 2010;21:325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zdanov A. Structure and function of IL-10 and IL-10 receptor. In: Marincola F, editor. Interleukin-10: Landes Bioscience; 2006. p. 1–10. [Google Scholar]
- [12].Chow D, He X, Snow AL, Rose-John S, Garcia KC. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–5. [DOI] [PubMed] [Google Scholar]
- [13].Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Spangler JB, Moraga I, Mendoza JL, Garcia KC. Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol. 2015;33:139–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Boggon TJ, Li Y, Manley PW, Eck MJ. Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog. Blood. 2005;106:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014;21:443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ferrao R, Wallweber HJ, Ho H, Tam C, Franke Y, Quinn J, et al. The Structural Basis for Class II Cytokine Receptor Recognition by JAK1. Structure. 2016;24:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McNally R, Toms AV, Eck MJ. Crystal Structure of the FERM-SH2 Module of Human Jak2. PLoS One. 2016;11:e0156218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, et al. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol. 2005;289:G960–G8. [DOI] [PubMed] [Google Scholar]
- [21].Meager A, Visvalingam K, Dilger P, Bryan D, Wadhwa M. Biological activity of interleukins-28 and −29: comparison with type I interferons. Cytokine. 2005;31:109–18. [DOI] [PubMed] [Google Scholar]
- [22].Zitzmann K, Brand S, Baehs S, Goke B, Meinecke J, Spottl G, et al. Novel interferon-lambdas induce antiproliferative effects in neuroendocrine tumor cells. Biochem Biophys Res Commun. 2006;344:1334–41. [DOI] [PubMed] [Google Scholar]
- [23].Miknis ZJ, Magracheva E, Li W, Zdanov A, Kotenko SV, Wlodawer A. Crystal structure of human interferon-lambda1 in complex with its high-affinity receptor interferon-lR1. J Mol Biol. 2010;404:650–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem. 2004;279:32269–74. [DOI] [PubMed] [Google Scholar]
- [25].Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, et al. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci U S A. 1991;88:11349–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Usacheva A, Sandoval R, Domanski P, Kotenko SV, Nelms K, Goldsmith MA, et al. Contribution of the Box 1 and Box 2 motifs of cytokine receptors to Jak1 association and activation. J Biol Chem. 2002;277:48220–6. [DOI] [PubMed] [Google Scholar]
- [27].Haan C, Heinrich PC, Behrmann I. Structural requirements of the interleukin-6 signal transducer gp130 for its interaction with Janus kinase 1: the receptor is crucial for kinase activation. Biochem J. 2002;361:105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Floss DM, Klocker T, Schroder J, Lamertz L, Mrotzek S, Strobl B, et al. Defining the functional binding sites of Interleukin 12 receptor beta1 and Interleukin 23 receptor to Janus kinases. Mol Biol Cell. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zurcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kampa D, Burnside J. Computational and functional analysis of the putative SH2 domain in Janus Kinases. Biochem Biophys Res Commun. 2000;278:175–82. [DOI] [PubMed] [Google Scholar]
- [31].Radtke S, Haan S, Jorissen A, Hermanns HM, Diefenbach S, Smyczek T, et al. The Jak1 SH2 domain does not fulfill a classical SH2 function in Jak/STAT signaling but plays a structural role for receptor interaction and up-regulation of receptor surface expression. J Biol Chem. 2005;280:25760–8. [DOI] [PubMed] [Google Scholar]
- [32].Richter MR, Dumenil G, Uze G, Fellous M, Pellegrini S. Specific Contribution of Tyk2 JH Regions to the Binding and the Expression of the Interferon α/β Receptor Component IFNAR1 J Biol Chem. 1998;273:24723–9. [DOI] [PubMed] [Google Scholar]
- [33].Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. [DOI] [PubMed] [Google Scholar]
- [35].Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr. 2006;D62:859–66. [DOI] [PubMed] [Google Scholar]
- [36].McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. 2007;D63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].DiMaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, et al. Improved molecular replacement by density and energy guided protein structure optimization. Nature. 2011;473:540–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. 2011;D67:355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Struct Biol. 1999;6:458–63. [DOI] [PubMed] [Google Scholar]
- [41].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–32. [DOI] [PubMed] [Google Scholar]
- [42].DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- [43].Eck MJ, Shoelson SE, Harrison SC. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature. 1993;362:87–91. [DOI] [PubMed] [Google Scholar]
- [44].Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.